AKT is emerging as a central player in tumorigenesis. In this issue of PNAS, Mayo and Donner (1) report on yet another function of AKT, involving regulation of the Mdm2/p53 pathway.
The first evidence pointing to a role of AKT in oncogenesis was provided by early studies of transforming viruses. A novel retrovirus, isolated from an AKR mouse T cell lymphoma (2), harbored transduced sequences of cellular origin (3). In 1991, our collaborative studies with Philip Tsichlis and Stephen Staal resulted in the cloning of the viral oncogene v-akt (4). The predicted oncoprotein contained viral Gag sequences fused to a kinase related to protein kinase C. The oncogenic potential of v-akt arises from the creation of a myristylation site at the amino terminus and consequent constitutive kinase activity (5). By different approaches, aimed at identifying novel protein kinases, two other groups independently cloned the identical cellular sequence at about the same time (6, 7). AKT is now known to define a family of closely related, highly conserved cellular homologues (reviewed in ref. 8). In human, these are designated AKT1, AKT2, and AKT3, located at chromosomes 14q32, 19q13, and 1q44, respectively (reviewed in ref. 9). The encoded proteins are serine/threonine kinases belonging to the protein kinase B (PKB) family, and the AKT1, AKT2, and AKT3 proteins are also known as PKBα, PKBβ, and PKBγ, respectively. Each AKT family member contains an amino-terminal pleckstrin homology (PH) domain, a short α-helical linker, and a carboxyl-terminal kinase domain (8). PH domains exist in diverse signaling molecules and permit anchorage of proteins to the cell membrane via phospholipid interactions (10).
The degree of functional redundancy between AKT1, AKT2, and AKT3 is currently unclear. Although each kinase responds similarly to various stimuli, their different tissue-specific expression patterns suggest distinct roles, e.g., compared to Akt1, Akt2 transcripts are especially abundant in highly insulin-responsive tissues such as brown fat (11). Moreover, Akt2 knockout mice exhibit impaired ability of insulin to lower blood glucose as a result of defects in the action of the hormone on liver and skeletal muscle (12). Expression of Akt1 and Akt3 does not compensate for loss of Akt2, thus establishing Akt2 as an essential gene for the maintenance of normal glucose homeostasis.
Mounting evidence suggests that AKT perturbations play an important role in human malignancy. In 1992, we reported the first recurrent involvement of an AKT gene in a human cancer, demonstrating amplification and overexpression of AKT2 in ovarian tumors and cell lines (13). Subsequent studies documented AKT2 amplification and/or mRNA overexpression in 10–20% of human ovarian and pancreatic cancers (14, 15) and activation of the AKT2 kinase in ≈40% of ovarian cancers (16). Overexpression of AKT2 can transform NIH 3T3 cells (17), and AKT2 antisense RNA inhibits the tumorigenic phenotype of cancer cells exhibiting amplified AKT2 (15). Amplification of AKT1 was observed in a human gastric cancer (3), and AKT1 kinase activity is often increased in prostate and breast cancers and is associated with a poor prognosis (18). To date, amplification of AKT3 has not been described. However, AKT3 mRNA is up-regulated in estrogen receptor-negative breast tumors, and increased AKT3 enzymatic activity was found in estrogen receptor-deficient breast cancer and androgen-insensitive prostate cancer cell lines (19), suggesting that AKT3 may contribute to the aggressiveness of steroid hormone-insensitive cancers.
There has been enormous interest in the mechanisms and cellular consequences of signal propagation from receptor tyrosine kinases to AKT (reviewed in refs. 8 and 20–28). The AKT kinases are major downstream targets of growth factor receptor tyrosine kinases that signal via phosphatidylinositol 3-kinase (PI3K).
AKT activation is a multistep process involving both membrane translocation and phosphorylation (29). The pleckstrin homology domain of AKT kinases has affinity for the 3′-phosphorylated phosphoinositides 3,4,5-trisphosphate (PI-3,4,5-P3) and PI-3,4,-P2 produced by PI3K, and they are activated specifically by the latter lipid. Phospholipid binding triggers the translocation of AKT kinases to the plasma membrane. Upon membrane localization, AKT molecules are phosphorylated at Thr-308/309 in the kinase activation loop and Ser-473/474 in the carboxyl-terminal tail. Thr-308/309 phosphorylation is necessary for AKT activation, and Ser-473/474 phosphorylation is only required for maximal activity. Phosphorylation on these residues is induced by growth factor stimulation and inhibited by the PI3K inhibitor, LY294002. Indeed, the kinase responsible for Thr-308/309 phosphorylation, PDK1 (for 3-phosphoinositide-dependent kinase) is activated by the PI3K lipid products PI-3,4,5-P3 and PI-3,4-P2. More controversial is the identity of PDK2, the kinase(s) responsible for Ser-473/474 phosphorylation (30). Interestingly, avian sarcoma virus 16 contains a potent transforming sequence derived from the cellular gene for the catalytic subunit of PI3K (31), and its human homologue, PIK3CA, was implicated as an oncogene in human ovarian cancer (32). Furthermore, the negative regulator of this pathway, the tumor suppressor PTEN, inhibits AKT activation by dephosphorylating PI-3,4,-P2/PI-3,4,5-P3 (reviewed in refs. 33 and 34).
Recent studies have revealed a burgeoning list of AKT substrates implicated in oncogenesis (reviewed in ref. 26). Among its pleiotropic effects, activated AKT is a well-established survival factor, exerting anti-apoptotic activity by preventing release of cytochrome c from mitochondria and inactivating forkhead transcription factors known to induce expression of pro-apoptotic factors such as Fas ligand. AKT phosphorylates and inactivates the pro-apoptotic factors BAD and pro-caspase-9. Moreover, AKT activates IκB kinase, a positive regulator of NF-κB, which results in transcription of anti-apoptotic genes. AKT kinases also phosphorylate and inactivate glycogen synthase kinase 3, thereby stimulating glycogen synthesis (35). AKT activation affects cell cycle progression, through regulation of cyclin D stability (36) and inhibition of p27Kip1 protein levels (37), and mRNA translation, via phosphorylation of 4E-BP1 and its dissociation from the mRNA cap binding protein elF4E (38). Furthermore, AKT mediates the activation of endothelial nitric oxide synthase, an important modulator of angiogenesis and vascular tone (39, 40). Germane to this, infection of the chicken wing web with RCAS retroviral vector expressing activated forms of mammalian Akt leads to the formation of hemangiosarcomas, malignant tumors of vascular cells (41). AKT activation also enhances telomerase activity via phosphorylation of the human telomerase reverse transcriptase subunit (42).
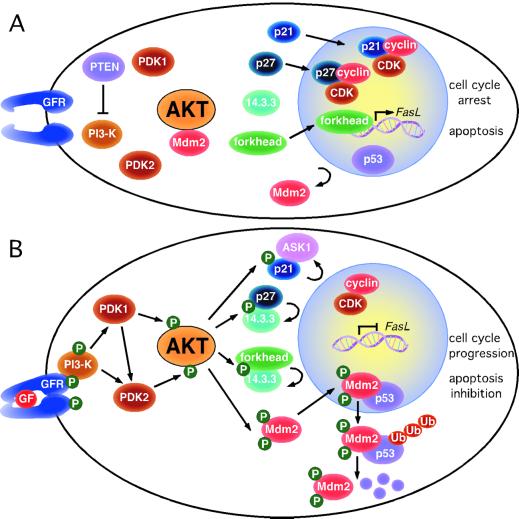
Until recently, it has been difficult to identify unifying themes in AKT substrates. Now, the study by Mayo and Donner (1) and other recent work (ref. 43 and G. Viglietto, personal communication) define an emerging mechanism driven by AKT phosphorylation, namely regulation of nucleo-cytoplasmic localization of critical substrates involved in cell cycling and apoptosis. Mayo and Donner show that phosphorylation by AKT is necessary for nuclear translocation of Mdm2. The oncoprotein Mdm2 and the tumor suppressor p53 are part of an autoregulatory feedback: Mdm2 transcription is induced by p53, and the Mdm2 protein, in turn, binds the p53 transactivation domain, inhibiting expression of p53-regulated genes involved in cell cycle arrest and apoptosis. In the absence of the p19/p14ARF tumor suppressor, the Mdm2-p53 complex shuttles from the nucleus to the cytoplasm where p53 is targeted for ubiquitin (Ub)/proteasome-mediated degradation (reviewed in ref. 44). Mayo and Donner demonstrate that in serum-starved cells, Mdm2 localizes in the cytoplasm in a complex with AKT. After growth factor stimulation, Mdm2 is phosphorylated by AKT, rapidly dissociates from the complex and enters the nucleus; this leads to reduction of both p53 levels and transactivation (Fig. 1). This study establishes a novel mitogen-regulated pathway linking PI3K/AKT and Mdm2/p53. However, this pathway only regulates nuclear entry of Mdm2, and additional components, e.g., relative levels of p19/p14ARF, are required for a full effect on p53-dependent cell cycle arrest/apoptosis.
Figure 1.
Phosphorylation by AKT regulates compartmentalization of multiple substrates involved in cell cycle progression and inhibition of apoptosis. (A) In serum-starved cells, the pro-apoptotic transcription factors of the forkhead family and cell cycle inhibitors p21 and p27 localize in the nucleus, whereas the oncoprotein Mdm2 is restrained in the cytoplasm. (B) After growth factor (GF) stimulation and phosphorylation by AKT, the subcellular localization of these AKT substrates is diametrically changed, contributing to cell cycle progression and inhibition of apoptosis. Cytoplasmic p21 can bind to the apoptosis signal-regulating kinase (ASK1), inhibiting apoptosis. In the absence of p19/p14ARF induction, the Mdm2-p53 complex shuttles into the cytoplasm where p53 is ubiquitinated (Ub) and targeted for degradation.
Whereas AKT acts in concert with the oncoprotein Mdm2, a recent study by Zhou et al. (43) indicates that AKT restrains the tumor suppressor p21WAF1. In breast cancer cells exhibiting AKT activation due to HER-2/neu overexpression, phosphorylation by AKT prevents nuclear localization of p21WAF1, separating this cell cycle inhibitor from its cyclin/cyclin-dependent kinase targets (Fig. 1). Thus, AKT activation antagonizes p21WAF1-mediated cell cycle arrest (43). Cytoplasmic p21 binds to the apoptosis-signal-regulating kinase (ASK1), inhibiting apoptosis. Similarly, recent work revealed that phosphorylation of p27Kip1 by AKT results in cytoplasmic retention of this cell cycle inhibitor and loss of its growth inhibition (G. Viglietto, personal communication). Cytoplasmic retention of AKT-phosphorylated p27Kip1 occurs, at least partly, by binding to the 14.3.3 scaffold protein (Fig. 1). Interestingly, binding to 14.3.3 had been previously reported for the forkhead family transcription factor FKHRL1 after AKT phosphorylation (45), which again is associated with cytoplasmic sequestration of the substrate (Fig. 1). In all these cases, regulation of substrate compartmentalization by AKT appears to be a consequence of phosphorylation near nuclear localization/nuclear export sequences, presumably affecting their net charge and/or conformation. In some cases, binding of the AKT-phosphorylated substrate to 14.3.3 may also affect subcellular localization.
As proposed by Hanahan and Weinberg (46), most tumor-related genetic/epigenetic changes are representative of a finite set of physiological alterations that collectively drive a cell toward malignancy. Based on the evidence outlined above and in Table 1, AKT signaling appears to play a prominent role in several processes considered hallmarks of cancer. Growth signal autonomy would not appear to be a direct effect of AKT signaling. However, overexpression of AKT may permit a tumor cell to become overly responsive to ambient levels of growth factors that normally would not provoke proliferation. Moreover, AKT activation can up-regulate insulin-like growth factor I receptor expression (47), and overexpression of growth factor receptors may facilitate oncogenic signaling (reviewed in ref. 46). AKT activation may contribute to tumor invasion/metastasis by stimulating secretion of matrix metalloproteinases (48).
Table 1.
Hallmarks of cancer and the multiple roles of AKT
| Cancer hallmarks (46) | Akt functions/substrates (in bold) |
|---|---|
| Acquired growth signal autonomy | Overexpression of AKT may mediate hyper-responsiveness to ambient levels of growth factors |
| Insensitivity to antigrowth signals | Promotes nuclear entry of Mdm2, thus inhibiting p53 pathway |
| Induces cytoplasmic localization of p21WAF1 and p27Kip1, promoting cell growth | |
| Stabilizes cyclin D1/D3 | |
| Inhibition of programmed cell death | Inactivates pro-apoptotic factors BAD and (pro)caspase-9 |
| Activates IKK, resulting in NF-κB transcription of anti-apoptotic genes | |
| Inactivates forkhead transcription factors, thereby inhibiting expression of Fas ligand | |
| Unlimited replicative potential | Enhances telomerase activity by phosphorylation of hTERT |
| Sustained angiogenesis | Activates eNOS, thus promoting angiogenesis |
| Tissue invasion and metastasis | Contributes to invasiveness by stimulating secretion of MMP |
IKK, IκB kinase; hTERT, human telomerase reverse transcriptase; eNOS, endothelial nitric oxide synthase; MMP, matrix metalloproteinase.
The involvement of AKT in diverse tumorigenic activities suggests that AKT activation alone might be sufficient to induce cancer. However, whereas overexpression of myristylated forms of Akt1, Akt2, and Akt3 are strongly oncogenic, wild-type forms of Akt are only poorly transforming (5, 41). Nevertheless, it is noteworthy that in human chronic myeloid leukemia a single chromosome change, leading to the creation of the BCR/ABL oncogenic tyrosine kinase, is considered sufficient to transform bone marrow cells, and activation of the PI3K/AKT pathway is essential for this process (49). Interestingly, in an experimental setting the oncogenic effects of Akt1, Akt2, and Akt3 were indistinguishable (41), suggesting that the downstream targets relevant to oncogenic transformation may be shared by the three AKT kinases. Whether the various AKT isoforms have some distinguishing substrates in human malignancy or have different, functionally pertinent binding affinities for other interacting proteins, such as the adaptor APPL (50), remains to be determined. Clearly, the expanding number of substrates implicated in various aspects of tumorigenesis highlights the central role of AKT kinases in many human cancers. For this reason, we anticipate that much attention will be given to the identification of inhibitors or modulators of the PI3K/AKT pathway, with the intention of developing novel therapeutic strategies directed at neoplasms exhibiting AKT activation.
Acknowledgments
We apologize for not citing original work of many colleagues because of space constraints. This work was supported by National Institutes of Health Grants CA77429 and CA06927.
Footnotes
See companion article on page 11598.
References
- 1.Mayo L D, Donner D B. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. . (First Published August 14, 2001; 10.1073/pnas.181181198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staal S P, Hartley J W, Rowe W P. Proc Natl Acad Sci USA. 1977;74:3065–3067. doi: 10.1073/pnas.74.7.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staal S P. Proc Natl Acad Sci USA. 1987;84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N N, Franke T F, Bellacosa A, Datta K, Gonzalez-Portal M E, Taguchi T, Testa J R, Tsichlis P N. Oncogene. 1993;8:1957–1963. [PubMed] [Google Scholar]
- 6.Coffer P J, Woodgett J R. Eur J Biochem. 1991;201:475–481. doi: 10.1111/j.1432-1033.1991.tb16305.x. [DOI] [PubMed] [Google Scholar]
- 7.Jones P F, Jakubowicz T, Pitossi F J, Maurer F, Hemmings B A. Proc Natl Acad Sci USA. 1991;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Testa J R, Bellacosa A. Leukemia Res. 1997;21:1027–1031. doi: 10.1016/s0145-2126(97)00093-3. [DOI] [PubMed] [Google Scholar]
- 9.Murthy S S, Tosolini A, Taguchi T, Testa J R. Cytogenet Cell Genet. 2000;88:38–40. doi: 10.1159/000015481. [DOI] [PubMed] [Google Scholar]
- 10.Haslam R J, Kolde H B, Hemmings B A. Nature (London) 1993;36:309–310. doi: 10.1038/363309b0. [DOI] [PubMed] [Google Scholar]
- 11.Altomare D A, Lyons G E, Mitsuuchi Y, Cheng J Q, Testa J R. Oncogene. 1998;16:2407–2411. doi: 10.1038/sj.onc.1201750. [DOI] [PubMed] [Google Scholar]
- 12.Cho H, Mu J, Kim J K, Thorvaldsen J L, Chu Q, Crenshaw E B, Kaestner K H, Bartolomei M S, Shulman G I, Birnbaum M J. Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J Q, Godwin A K, Bellacosa A, Taguchi T, Franke T F, Hamilton T C, Tsichlis P N, Testa J R. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellacosa A, de Feo D, Godwin A K, Bell D W, Cheng J Q, Altomare D A, Wan M, Dubeau L, Scambia G, Masciullo V, et al. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 15.Cheng J Q, Ruggeri B, Klein W M, Sonoda G, Altomare D A, Watson D K, Testa J R. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Z Q, Sun M, Feldman R I, Wang G, Ma X, Jiang C, Coppola D, Nicosia S V, Cheng J Q. Oncogene. 2000;19:2324–2330. doi: 10.1038/sj.onc.1203598. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J Q, Altomare D A, Klein M A, Lee W-C, Kruh G D, Lissy N A, Testa J R. Oncogene. 1997;14:2793–2801. doi: 10.1038/sj.onc.1201121. [DOI] [PubMed] [Google Scholar]
- 18.Sun M, Wang G, Paciga J E, Feldman R I, Yuan Z Q, Ma X L, Shelley S A, Jove R, Tsichlis P N, Nicosia S V, Cheng J Q. Am J Pathol. 2001;159:431–437. doi: 10.1016/s0002-9440(10)61714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakatani K, Thompson D A, Barthel A, Sakaue H, Liu W, Weigel R J, Roth R A. J Biol Chem. 1999;274:21528–21532. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- 20.Hemmings B A. Science. 1997;275:628–630. doi: 10.1126/science.275.5300.628. [DOI] [PubMed] [Google Scholar]
- 21.Franke T F, Kaplan D R, Cantley L C. Cell. 1997;88:435–437. doi: 10.1016/s0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- 22.Coffer P J, Jin J, Woodgett J R. Biochem J. 1998;335:1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alessi D R, Cohen P. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 24.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 25.Chan T O, Rittenhouse S E, Tsichlis P N. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 26.Datta S R, Brunet A, Greenberg M E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 27.Kops G J, Burgering B M. J Mol Med. 1999;77:656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 28.Kandel E S, Hay N. Exp Cell Res. 1999;253:210–229. doi: 10.1006/excr.1999.4690. [DOI] [PubMed] [Google Scholar]
- 29.Bellacosa A, Chan T O, Ahmed N N, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- 30.Chan, T. O. & Tsichlis, P. N. (January 23, 2001) Sciences STKE, http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/66/pe1. [DOI] [PubMed]
- 31.Chang H W, Aoki M, Fruman D, Auger K R, Bellacosa A, Tsichlis P N, Cantley L C, Roberts T M, Vogt P K. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 32.Shayesteh L, Lu Y, Kuo W L, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills G B, Gray J W. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 33.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Cristofano A, Pandolfi P P. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 35.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 36.Muise-Helmericks R C, Grimes H L, Bellacosa A, Malstrom S E, Tsichlis P N, Rosen N. J Biol Chem. 1998;273:29864–29872. doi: 10.1074/jbc.273.45.29864. [DOI] [PubMed] [Google Scholar]
- 37.Collado M, Medema R H, Garcia-Cao I, Dubuisson M L, Barradas M, Glassford J, Rivas C, Burgering B M, Serrano M, Lam E W. J Biol Chem. 2000;275:21960–21968. doi: 10.1074/jbc.M000759200. [DOI] [PubMed] [Google Scholar]
- 38.Sonenberg N, Gingras A C. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 39.Fulton D, Gratton J P, McCabe T J, Fontana J, Fujio Y, Walsh K, Franke T F, Papapetropoulos A, Sessa W C. Nature (London) 1999;399:597–601. doi: 10.1038/21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dimmeler S, Fleming I, Fisslthaler B, Hermann C, Busse R, Zeiher A M. Nature (London) 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- 41.Mende I, Malstrom S, Tsichlis P N, Vogt P K, Aoki M. Oncogene. 2001;20:4419–4423. doi: 10.1038/sj.onc.1204486. [DOI] [PubMed] [Google Scholar]
- 42.Kang S S, Kwon T, Kwon D Y, Do S I. J Biol Chem. 1999;274:13085–13090. doi: 10.1074/jbc.274.19.13085. [DOI] [PubMed] [Google Scholar]
- 43.Zhou B P, Liao Y, Xia W, Spohn B, Lee M H, Hung M C. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
- 44.Sherr C J, Weber J D. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 45.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg R A. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Tanno S, Tanno S, Mitsuuchi Y, Altomare D A, Xiao G H, Testa J R. Cancer Res. 2001;61:589–593. [PubMed] [Google Scholar]
- 48.Thant A A, Nawa A, Kikkawa F, Ichigotani Y, Zhang Y, Sein T T, Amin A R, Hamaguchi M. Clin Exp Metastasis. 2000;18:423–428. doi: 10.1023/a:1010921730952. [DOI] [PubMed] [Google Scholar]
- 49.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi J K, Trotta R, Wlodarski P, Perrotti D, Chan T O, et al. EMBO J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitsuuchi Y, Johnson S W, Tanno S, Golemis E A, Testa J R. Oncogene. 1999;18:4891–4898. doi: 10.1038/sj.onc.1203080. [DOI] [PubMed] [Google Scholar]