Summary
Biomass yield, salt tolerance and drought tolerance are important targets for alfalfa (Medicago sativa L.) improvement. Medicago truncatula has been developed into a model plant for alfalfa and other legumes. By screening a Tnt1 retrotransposon‐tagged M. truncatula mutant population, we identified three mutants with enhanced branching. Branch development determines shoot architecture which affects important plant functions such as light acquisition, resource use and ultimately impacts biomass production. Molecular analyses revealed that the mutations were caused by Tnt1 insertions in the SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE 8 (SPL8) gene. The M. truncatula spl8 mutants had increased biomass yield, while overexpression of SPL8 in M. truncatula suppressed branching and reduced biomass yield. Scanning electron microscopy (SEM) analysis showed that SPL8 inhibited branching by directly suppressing axillary bud formation. Based on the M. truncatula SPL8 sequence, alfalfa SPL8 (MsSPL8) was cloned and transgenic alfalfa plants were produced. MsSPL8 down‐regulated or up‐regulated alfalfa plants exhibited similar phenotypes to the M. truncatula mutants or overexpression lines, respectively. Specifically, the MsSPL8 down‐regulated alfalfa plants showed up to 43% increase in biomass yield in the first harvest. The impact was even more prominent in the second harvest, with up to 86% increase in biomass production compared to the control. Furthermore, down‐regulation of MsSPL8 led to enhanced salt and drought tolerance in transgenic alfalfa. Results from this research offer a valuable approach to simultaneously improve biomass production and abiotic stress tolerance in legumes.
Keywords: Medicago truncatula, alfalfa, Medicago sativa, forage legume, branching, biomass yield, salt tolerance, drought tolerance
Introduction
Alfalfa (Medicago sativa L.), known as the ‘Queen of Forages’ due to its remarkable adaptability, high biomass yield, exceptional nutritive value and notable capacity for biological nitrogen fixation, is one of the most important and widely cultivated forage crops around the world (Annicchiarico et al., 2015; Aung et al., 2015; Biazzi et al., 2017; Li et al., 2008; Russelle et al., 2007; Samac et al., 2006). In the last century, extensive efforts have been made to improve alfalfa biomass yield, but the achievements have been limited (Aung et al., 2015). The biomass yields of first/second harvests of recently released cultivars have not shown improvements compared to previous cultivars released 50 years ago (Volenec et al., 2002). Although biomass yield is a complex trait, use of biotechnology for alfalfa yield improvement has achieved success in recent years. For example, overexpression of microR156 in alfalfa increased biomass yield more than 10% (Aung et al., 2015). The efficiency of genetic engineering approaches for improving alfalfa biomass yield depends upon the identification of specific genes that control important agronomic traits (Aung et al., 2015; Volenec et al., 2002). Alfalfa is an obligate outcrossing and tetraploid species. Seeds from a plant are genetically different and heterogeneous, and genome sequence information is limited. Because of its genetic complexity, the identification of agronomic target genes in alfalfa is difficult (Aung et al., 2015).
A corresponding model system with simpler genetics has been developed to meet the challenges of modifying this complex agronomic crop. Medicago truncatula belongs to the Trifolieae tribe that includes major forage legumes such as alfalfa and clovers (Trifolium sp.). Plus it has an autogamous mode of reproduction and a short growth cycle, with the added benefit of a small, deeply sequenced, well‐annotated genome (Young et al., 2005, 2011). Because of these advantages, M. truncatula has been found to be an excellent model for legumes, especially for alfalfa. Various genetic and genomic resources have been developed in M. truncatula, including Tnt1 retrotransposon‐tagged mutants (>22 000 lines and 520 000 random insertions) and fast‐neutron mutants (>117 000 lines) (https://medicago-mutant.noble.org/mutant/index.php), ecotype collections (Cook, 1999), EST and genespace sequencing information (Young and Udvardi, 2009), and the Gene Expression Atlas (https://mtgea.noble.org/v2/index.php). All of these resources dramatically accelerate studies in M. truncatula and alfalfa. However, most previous research related primarily to bacterial and mycorrhizal symbioses, leaf development, disease resistance and seed development (Chai et al., 2016; Espinoza Ldel et al., 2012; Kang et al., 2016; Zhou et al., 2014). In contrast, studies on the regulation of aerial morphogenesis, especially of shoot architecture development, are very limited (Espinoza Ldel et al., 2012; Julier et al., 2007).
Branch development (branching) is a key determinant of shoot architecture which affects important plant functions like light acquisition, resource use and ultimately impacts biomass yield. The primary branch arising from the main shoot produces secondary, then tertiary and even higher order branches. Axillary buds are the sole originators of vegetative and floral branches (Domagalska and Leyser, 2011). Axillary buds arise in the leaf axil (the upper side of the region where the leaf joins to the stem) and exhibit two stages, initiation and outgrowth, ultimately forming the various branches (Bennett and Leyser, 2006). In the last three decades, outgrowth of axillary buds has been well characterized due to the identification of many mutations related to bud outgrowth (Costes et al., 2014; Domagalska and Leyser, 2011; Gomez‐Roldan et al., 2008). These intensive studies regarding the outgrowth of buds have revealed a global and complex regulation network of genetic, hormonal and environmental factors (Domagalska and Leyser, 2011; Guo et al., 2013; Kebrom et al., 2013; McSteen, 2009; Wang and Li, 2011). In contrast, the initiation of axillary buds appears to be exclusively genetically regulated without any implication of other contributing factors (Kebrom et al., 2013). Currently, only a few genes have been identified, including Lateral suppressor (Ls) and its orthologs [LATERAL SUPPRESSOR (LAS) and MONOCULM1 (MOC1)] (Cheng et al., 2006; Gallavotti et al., 2008; Li et al., 2003; Schumacher et al., 1999) and LAX PANICLE1 (LAX1) and its orthologs [BARREN STALK1 (BA1)] (Gallavotti et al., 2004; Komatsu et al., 2003; Yang et al., 2012).
SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE (SPL) family proteins are plant‐specific transcription factors, that share a highly conserved zinc ion‐containing DNA binding domain named the SBP‐box (Wang et al., 2009; Yamasaki et al., 2004). In Arabidopsis (Xie et al., 2006 ) and rice (Wang et al., 2009), respectively, 16 and 19 SPLs have been found. These SPLs are conserved across monocots and eudicots (Wang et al., 2009), but each individual member may function divergently in the regulation of various processes. For example, AtSPL3/4/5 redundantly regulates developmental ageing and phase transition in Arabidopsis (Jung et al., 2016; Yamaguchi et al., 2009); AtSPL9 controls the initiation of cauline leaf axillary meristems in Arabidopsis (Tian et al., 2014); AtSPL8/2/9/15 redundantly acts in pollen development and male fertility in Arabidopsis (Unte et al., 2003; Xing et al., 2010, 2013); OsSPL13 and OsSPL16 regulate grain size and shape in rice. OsSPL14, also known as IDEAL PLANT ARCHTECTURE1 (IPA1), has been found to promote panicle branching while suppressing basal branch formation in rice (Jiao et al., 2010; Miura et al., 2010), an ideal situation for grain yield increase but not necessarily for biomass production. Such tiller suppression is not due to genetic inhibition of the basal bud initiation or outgrowth, but due to a delay in the time that elapses between the formation of primordia (i.e. a prolonged plastochron) and to the associated regulatory effects of leaf development (Wang and Li, 2011).
In this study, we identified three mutants with enhanced branching from the M. truncatula Tnt1 mutant population. Molecular analysis revealed that these mutations were caused by Tnt1 insertions in different regions of MtSPL8. Overexpression of MtSPL8 in wild‐type M. truncatula resulted in a dramatic decrease in branch formation. Further analyses revealed that MtSPL8 controlled branching by directly inhibiting axillary bud initiation. Subsequently, alfalfa SPL8 gene (MsSPL8) was cloned, and the transgenic alfalfa lines with overexpression or down‐regulation of MsSPL8 exhibited similar phenotype to what was observed in M. truncatula transgenics or mutants. In particular, knockdown of MsSPL8 significantly increased biomass yield, promoted regrowth and enhanced salt and drought tolerance. Our results demonstrate simultaneous improvement of multiple important agronomic traits by genetic manipulation of a single SPL gene.
Results
An SPL gene regulates branching and shoot architecture
To identify mutants with shoot architecture alterations, over ten thousand independent lines of Tnt1 retrotransposon‐tagged M. truncatula populations were screened. Three mutant lines (NF7738, NF10281 and NF10498) were identified that exhibited enhanced branch development. The three mutants showed no obvious difference in very early development. Three weeks after sowing, these mutants started to develop more branches than control plants (ecotype R108) (Figure 1a). After 6–8 weeks, along with the initial formation of additional secondary branches the difference became more apparent as even greater numbers of secondary and tertiary branches were produced in the mutants (Figure 1b and c). Except for this phenotype, the three mutants showed no difference in other morphological traits, including seed germination, leaf pattern, leaf size, plant height, flower development, floral structure, or pod and seed shape and size.
Figure 1.
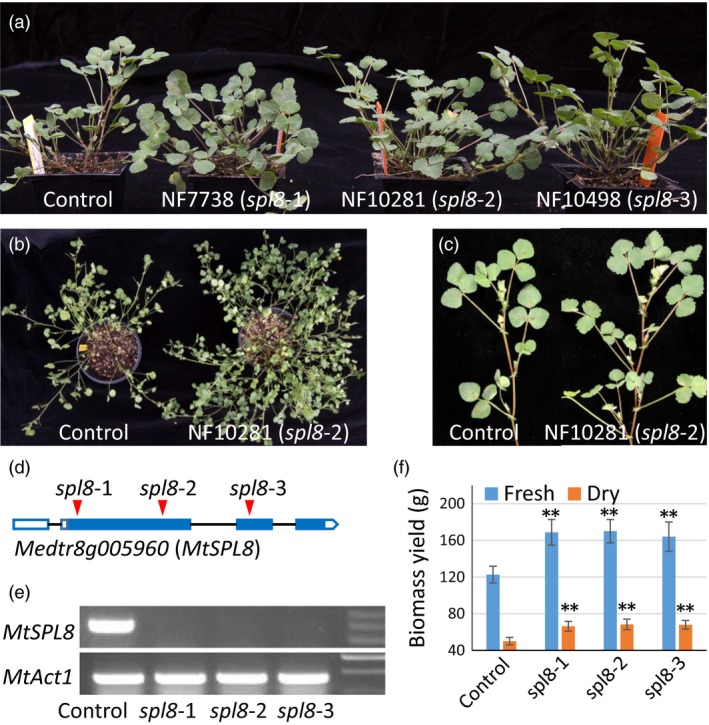
Phenotype and biomass yield of three Tnt1 mutants of SPL8 in M. truncatula. (a) Phenotype of 4‐week‐old mutant plants. (b) Eight‐week‐old plants of mutants and control. (c) Branch development in the control and spl8 mutants. (d) Tnt1 insertion sites in SPL8 of the three mutants. (e) Semiquantitative PCR indicates that MtSPL8 expression is abolished in spl8 mutants. (f) Fresh and dry biomass yields of mutant and control plants. Values represent mean ± S.D. of five biological replicates and are analyzed by Student's t‐test (**P < 0.01).
To determine which gene(s) underlie this phenotype of enhanced branching, we first investigated the three mutants for all possible defective genes. Based on a search of the Medicago truncatula Mutant Database (https://medicagomutant.noble.org/mutant/database.php), we found 35, 45 and 109 potential Tnt1 insertions in mutants NF7738, NF10281 and NF10498, respectively. By limiting the candidates to only the high confidence ones, the number of Tnt1 insertions decreased to 14, 17 and 29 in the three mutants. Further analysis showed that only two mutated genes (Medtr8g005960 and Medtr1g102390) were common to all three mutants. To verify which gene or genes caused the phenotype, each individual mutant was crossed with the wild type. The F1 plants did not show the phenotype; however, enhanced branching was observed in segregated F2 plants. Based on PCR verification (using primers across the Tnt1 insertion sites in the two genes), we found that all the F2 plants with phenotype contained the homozygous mutation only in Medtr8g005960. The three homozygous mutants had Tnt1 insertions in different exons of Medtr8g005960 (Figure 1d). Semiquantitative PCR (using selected primers from exons 2 and 3) showed that MtSPL8 expression was abolished in all mutants (Figure 1e). These results together suggest that only Medtr8g005960 corresponds to the enhanced branching phenotype. Sequence BLAST analysis suggested that Medtr8g005960 coded a SPL gene containing three exons (Figure 1d). In M. truncatula genome, a total of 11 SPL genes were found by BLAST analysis using the Medicago truncatula Mt4.0v1 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Mtruncatula) (Figure S1). Medtr8g005960 was named MtSPL8 because it is closely related to AtSPL8 in Arabidopsis (Figure S1). Correspondingly, the three mutants NF7738, NF10281 and NF10498 were named as spl8‐1, spl8‐2 and spl8‐3.
The expression pattern of MtSPL8 was analysed using the M. truncatula Gene Expression Atlas (https://mtgea.noble.org/v2/index.php). The expression of MtSPL8 exhibited distinct tissue specificity. It was predominantly expressed in the vegetative bud, flower and pod. In contrast, it showed very low expression in the other organs, with almost no expression in the root (Figure S2).
The spl8 mutants showed significantly more lateral branching and increased biomass yield
Repeated experiments demonstrated that the spl8 mutants began to form more branches three weeks after sowing, and the difference became more significant after 6 weeks or more (Figure 1c). To decipher details of this time‐critical factor, we investigated the development of primary, secondary and tertiary branches when the plants were 10 weeks old. At that time, both control and mutants showed nearly 50% flowering. Our data showed the three mutants had 24.5%–33.8% more primary branches than control (P < 0.05, Figure S3). The difference was even greater with respect to lateral branches (secondary + tertiary). Mutants had 50.2%–58.5% more secondary branches and 131.3%–179.7% more tertiary branches (Figure S3). These results suggest enhanced branching during development.
The spl8 mutants displayed more than 39.1% increase in fresh biomass yield (Figure 1f), and more than 32.3% increase in dry biomass (Figure 1f) due to the increased branching. The differences were highly significant (P < 0.01).
Overexpression of SPL8 suppressed branching
To further confirm its function, MtSPL8 was overexpressed in the wild type. Forty‐four independent antibiotic resistant plants were produced by Agrobacterium‐mediated transformation (Figure. S4). PCR analysis, using a forward primer designed from the CaMV35S promoter and a reverse primer designed from the 5′‐end of SPL8, showed bands of expected sizes in 42 transgenic events (Figure S4a). Based on SPL8 expression level (Figure S4b), 15 lines were selected and their T1 progenies were further analysed by PCR. The results confirmed transgenic nature of the T1 plants (Figure S4c). During their entire lifespan, these transgenic plants showed dramatically decreased primary branches and lateral branches (Figure 2a) and the phenotype remained stable in the T2 plants. Notably, there were five transgenic plants that showed very few branches and especially rare lateral branches (Figure 2a. SPL8OE‐14 and 18 are two representatives of these plants). Consistently, these five plants showed the highest expression levels of MtSPL8 (over 10‐fold more than control) among the transgenics (Figure 2b, Figure S4b). The overexpression plants were taller than the control and spl8 mutants (Figure S5a and b); however, their branch density (branch number/1 cm main stem) and total branch numbers were dramatically decreased (Figure 2c, Figure S5c). The differences were more significant in secondary and tertiary branching than for primary branches (P < 0.001 vs. P = 0.0265). Unsurprisingly, the biomass yield in the overexpression plants was also markedly decreased (Figure 2d and e).
Figure 2.
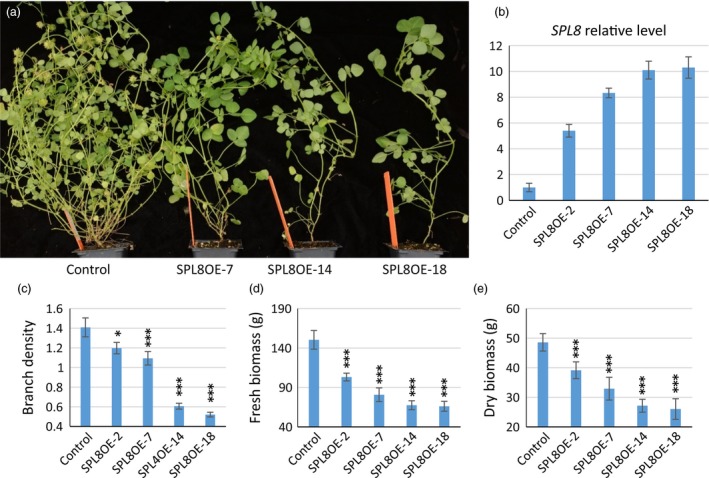
Effects of SPL8 overexpression in M. truncatula. (a) Mature plants of control and SPL8 overexpression transgenic lines (SPL8OE). (b) SPL8 relative expression levels in transgenic plants. (c) Branch density (primary branches produced in one centimeter of main stem) of overexpression lines and control. (d) Fresh biomass yield of overexpression lines and control. (e) Dry biomass yield of overexpression lines and control. Values represent means ± S.D. of three biological replicates and are analyzed by Student's t‐test (*P < 0.05, ***P < 0.001).
SPL8 directly regulates axillary bud initiation
From spl8 mutants to the overexpression plants, the results consistently indicated that SPL8 regulates branching (Figure S5). As branches are developed from axillary buds, the formation of axillary buds was investigated using scanning electron microscope (SEM). Shoot tips of 5‐week‐old plants were harvested and examined under SEM. This observation revealed that axillary buds were formed following the elongation of leaf primordia in the control. In most cases, a well‐developed axillary bud could easily be found at the axil of the oldest leaf primordia or the youngest juvenile leaf (Figure 3a). Compared to control, bud formation in spl8 mutants occurred much earlier, and the size of the bud appeared bigger relative to the control at the same position (Figure 3b). The results demonstrated that axillary bud initiation was significantly promoted in spl8 mutants. In contrast, no obvious axillary buds were found in the MtSPL8 overexpression plants (Figure 3c). To confirm this observation, we attempted to remove all juvenile leaves and other surrounding tissues, but no axillary bud was observed in the same position as that of the control (Figure 3c). Thus, overexpression of MtSPL8 markedly inhibited axillary bud initiation.
Figure 3.
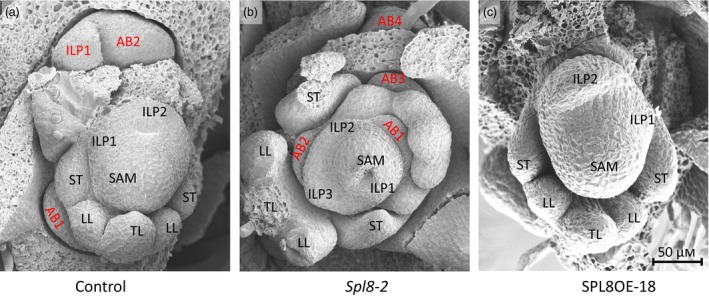
SAM development in spl8 mutant (spl8‐2), control and overexpression plant (SPL8OE‐18) in M. truncatula. (a) SAM structure in the control. (b) SAM structure in spl8. (c) SAM structure in SPL8OE with complete removal of all surrounding tissue and leaflets. AB, axillary bud; ILP, incipient leaf primordia; LL, lateral leaflet primordia; TL, terminal leaflet primordia; ST, stipule primordia.
Global gene expression profiling of SPL8 regulation
To understand the underlying mechanisms, shoot apical meristems (SAM) harvested from the five overexpression plants with highest MtSPL8 expression levels (Figure S4b) and the control were subjected to microarray analysis using Affymetrix GeneChip Medicago Genome Array. Microarray analysis revealed that 199 and 176 genes were significantly down‐ and upregulated in the transgenic plants (Tables S1 and S2). MapMan analysis revealed that stress response was the first most significantly regulated biological process. Twenty‐three of the 199 down‐regulated genes were associated with abiotic and biotic stress responses, such as defensin and defensin‐like genes, production of anthocyanin pigment 1 (PAP1), chalcone synthase (CHS) and dihydroflavonol‐4‐reductase (DFR) genes (Table S1). Endogenous signalling was identified as the second most significant process, which included 11 gibberellin (GA) signalling genes, such as GA2‐ox6 (encodes a major GA deactivator), GID1L2 (encodes a GA receptor) and GASRs (GA responsive genes). Transcriptional regulation was the third most significant process, which included many MADSs, MYBs and WRKYs. In addition, SCARECROW‐like 11, the LAS ortholog in Medicago, also showed a significant difference.
To confirm the microarray results, representative genes were chosen from these biological processes for further analysis using reverse transcription quantitative PCR (RT‐qPCR) in SAM and mature leaves. The results confirmed that all of these genes showed consistent and verifiable changes in the mutants, compared to control and overexpression plants in both SAM and leaves (Figure S6).
SPL8 is involved in the regulation of GA signalling
Many GA signalling genes were down‐regulated by SPL8 except for GA2ox6 which was up‐regulated, indicating a difference in bioactive GA accumulation. SAM and mature leaves harvested from spl8 mutants, control and overexpression plants were subjected to phytohormone quantification. The data confirmed that spl8 mutants did accumulate higher GA4 than control, while the overexpression plants consistently accumulated much less. The differences were more significant in SAM than in leaf tissues (Figure S7). However, GA1 was undetectable in both tissues. Both GA1 and GA4 are the bioactive GAs, but each has specific predominance in different species. This result also indicates that, in M. truncatula, the major bioactive GA is not GA1 but GA4. Compared to GA, both IAA and ABA showed no significant difference in either SAM or leaf (Figure S7).
Isolation of the SPL8 gene from alfalfa
Because many genes have high sequence similarity between M. truncatula and alfalfa, the full‐length mRNA sequence of SPL8 was isolated from alfalfa by 5′‐ and 3′‐RACE using primers derived from the M. truncatula SPL8. The resulting putative orthologous gene from alfalfa was designated as MsSPL8. Sequence analysis showed that there was 89.6% similarity between MtSPL8 and MsSPL8 (Figure S8). Analysis of deduced amino acid sequences revealed that the MsSPL8 protein contained 304 amino acids, showing 89.9% identity to MtSPL8, with identical sequences in their SBP domain (Figure S9). The MsSPL8 protein sequence was also highly similar to its putative ortholog in soybean, with only one amino acid difference in the SBP domain (Figure S9). These results suggest that SPL8 is highly conserved in legume species.
Modification of MsSPL8 expression significantly affects branching and biomass yield in alfalfa
To suppress its activity, a MsSPL8‐RNAi construct was introduced into alfalfa and 78 independent lines were produced by Agrobacterium‐mediated transformation. PCR analysis showed that 71 plants contained the target gene (Figure S10). RT‐qPCR analysis showed that the endogenous MsSPL8 level decreased to various levels, with over 50% reduction in 37 of the 71 transgenics and over 90% reduction in eight of the transgenics (Figure S11). These plants showed a steady increase in branching when compared with the control or transgenic plants without significant reduction in MsSPL8. Two lines (MsSPL8Ri‐57 and MsSPL8Ri‐10) with more than 50% and three lines (MsSPL8Ri‐37, MsSPL8Ri‐14 and MsSPL8Ri‐53) with more than 90% reduction in MsSPL8 level were selected for further analyses (Figure S12a and b).
To perform further characterization, four clonal plants were vegetatively propagated from each transgenic line using shoot cuttings. These propagated transgenic plants exhibited more branches, especially the plants with more than 90% reduction in MsSPL8 level (Figure 4a to c, Figure S12a and b). Consistently, MsSPL8 down‐regulation plants yielded more forage biomass than control, especially the lines with heavy down‐regulation of MsSPL8 (Figure 4d, Figure S12c and d). The fresh and dry biomass of the heavily down‐regulated lines (MsSPL8Ri‐37, MsSPL8Ri‐14, MsSPL8Ri‐53) increased 38.5%–45.2% and 38.4%–46.5%, respectively (Figure S12c and d).
Figure 4.
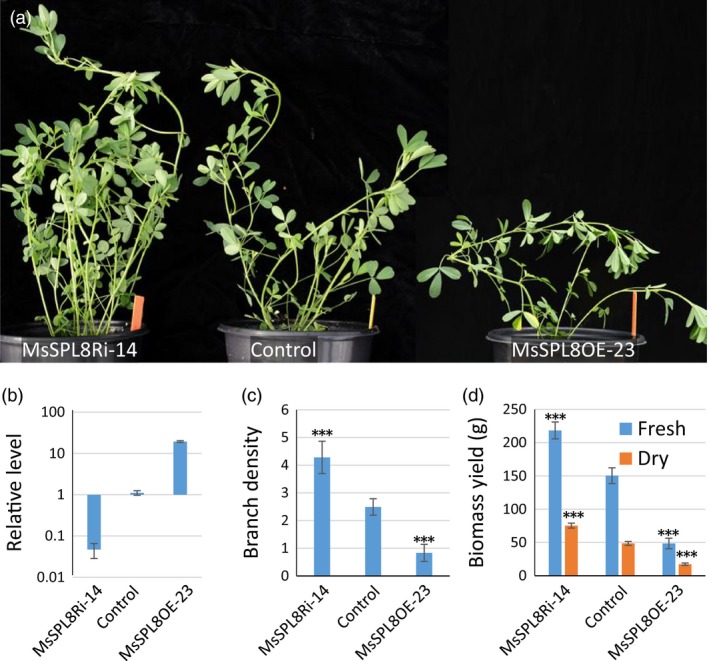
Genetic modification of MsSPL8 significantly altered shoot architecture and biomass yield in alfalfa (Medicago sativa). (a) Two‐month‐old plants of control, MsSPL8 down‐regulation (MsSPL8Ri‐14) and overexpression transgenics (MsSPL8OE‐23). (b) Relative expression levels of MsSPL8 in the MsSPL8Ri, control and MsSPL8OE plants. (c) Branch density (the total branch numbers produced in one centimeter of main stem) of the MsSPL8Ri, control and MsSPL8OE plants. (d) Fresh and dry biomass yields (gram) of the MsSPL8Ri, control and MsSPL8OE plants. Values represent means ± S.D. of six biological replicates and are analyzed by Student's t‐test (***P < 0.001).
In parallel, we also overexpressed MsSPL8 in alfalfa and produced 46 independent lines. PCR analysis showed that 44 of these were positive transgenic plants (Figure S13). These plants displayed the phenotype of decreased branches as seen in the MtSPL8 overexpression plants (Figure S14a, Figure 2). The case was more notable in four lines with more than 10‐fold increase in MsSPL8 level, almost no lateral branches were produced in these lines (MsSPL8OE‐23 is a representative example of the four lines, Figure S14a, Figure 4a). The extent of the decrease was negatively correlated with increased MsSPL8 levels (Figure S14b). Consequently, forage biomass yield was dramatically decreased (Figure S14c and d).
The differences became more notable when the MsSPL8 down‐regulation and overexpression plants were compared together (Figure 4). Side by side comparison strongly indicated that SPL8 regulated branch development and affected biomass yield (Figure 4c and d).
Down‐regulation of SPL8 markedly accelerated regrowth in alfalfa
Alfalfa plants are harvested multiple times per year. In this study, all harvests were performed when plants were at budding to early bloom stage. In three independent experiments, besides significantly improving branch formation, MsSPL8 down‐regulation plants showed no other morphological difference, including flowering time. This allowed synchronous harvest of the control and down‐regulation plants. Interestingly, after each harvest, more shoots developed quickly in MsSPL8 down‐regulation plants than in control plants (Figure 5a). This indicates that down‐regulation of MsSPL8 markedly accelerated regrowth. The increase in biomass yield in MsSPL8 down‐regulation plants was even more prominent in the second harvest (Figure 5b). With respect to the control, all three MsSPL8 down‐regulation plants showed 37.9%–43.2% increase in biomass yield in the first harvest while the increase was 65.7%–86.3% in the second harvest. The difference between the first and second harvests was significant (Figure 5c).
Figure 5.
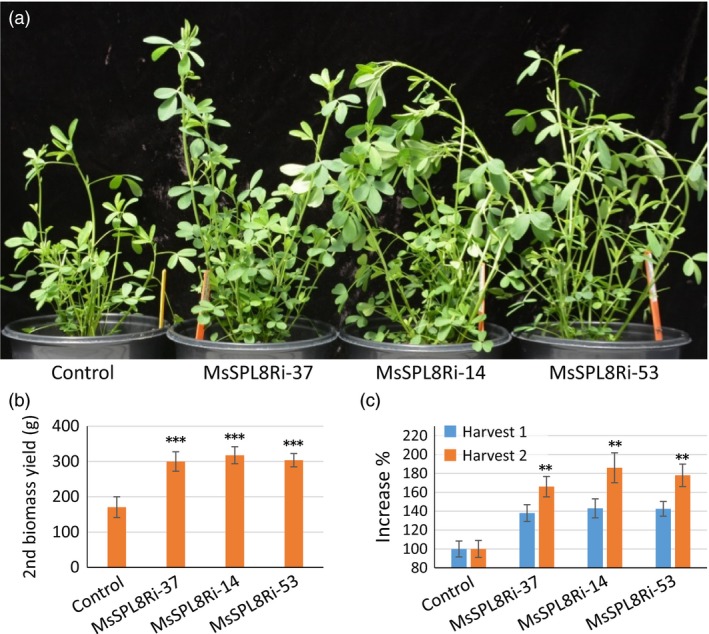
Down‐regulation of MsSPL8 markedly accelerated regrowth in alfalfa. (a) New shoots produced after cutting from the control and MsSPL8 down‐regulation transgenic plants (MsSPL8Ri). (b) Forage biomass (g) of the second harvest. (c) Percentage increase in biomass yield of the first and second harvests. Biomass of the second harvest is significantly higher than the first harvest in the transgenics. Values represent means ± S.D. of three biological replicates and are analyzed by Student's t‐test (**P < 0.01, ***P < 0.001).
The harvested biomass from three independent experiments was also analysed for forage quality. The results indicated that the MsSPL8 down‐regulation plants had no significant difference in protein content or lignin accumulation with respect to the control. Actually, two of the three lines even showed increased total digestible nutrients and relative feed value (Table S3).
Down‐regulation of SPL8 expression improved abiotic stress tolerance in alfalfa
Microarray analysis of M. truncatula revealed that many genes were significantly regulated by SPL8, including CHS, PAP1 and DFR, which are major anthocyanin genes and closely associated with stress response in plants (Cui et al., 2014; Gou et al., 2011b). To assess whether MsSPL8 affects these genes in a similar manner in alfalfa, we analysed their expression level in 12 independent alfalfa plants. RT‐qPCR data showed that all three genes were dramatically up‐regulated in the MsSPL8 down‐regulation plants and down‐regulated in the MsSPL8 overexpression plants (Figure S15a).
The effects of salt and drought stress on the transgenic plants were examined in the greenhouse. Under salt treatment (Figure 6a to c), MsSPL8 overexpression plants began turning yellow after just 1 week. Two weeks later, the control also began turning yellow and some MsSPL8 overexpression plants died (Figure 6b). After 3 weeks, more than 70% of overexpression plants and half of control plants were dying, by contrast, more than 90% of the MsSPL8 down‐regulation plants still survived and some even flowered (Figure 6c, Figure S15b).
Figure 6.
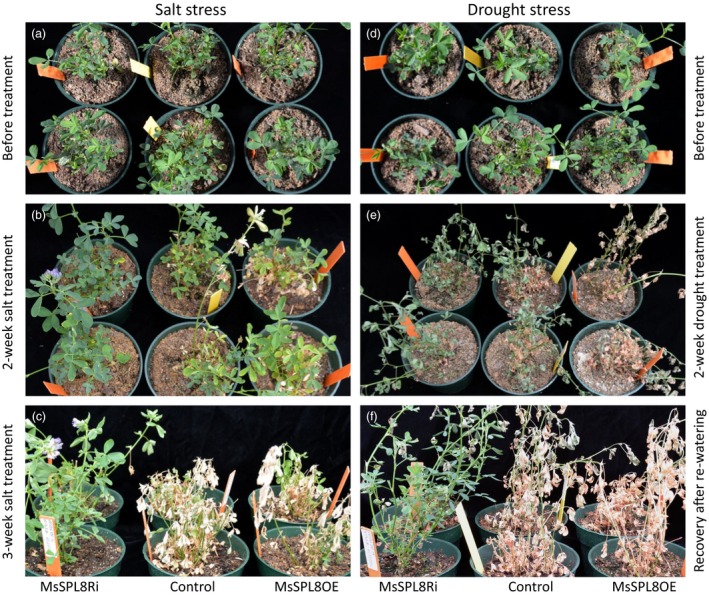
Effects of salt and drought stress on transgenic alfalfa plants down‐regulated with MsSPL8 (MsSPL8Ri) or up‐regulated with MsSPL8 (MsSPL8OE). (a) Plants before salt treatment. (b) Phenotype after 2‐week salt treatment. (c) Phenotype after 3‐week salt treatment. (d) Plants before drought treatment. (e) Phenotype after 2‐week drought treatment (no watering). (f) Six days of recovery after re‐watering from 2‐week drought treatment. Experiments are conducted with four biological replicates.
In the drought experiments (Figure 6d to f), 1 week after watering was stopped, all of the MsSPL8 overexpression and half of the control plants wilted. Two weeks later, over half of the overexpression plants were dead and most of the control became dehydrated and dead‐like, while the MsSPL8 down‐regulation plants just began to wilt (Figure 6e). Upon resumption of the normal watering scheme, the MsSPL8 down‐regulation plants recovered quickly and most of them survived. In contrast, more than one‐half to two‐thirds of the control and MsSPL8 overexpression plants were completely dead and did not recover with watering (Figure 6f, Figure S15b). These results showed that down‐regulation of SPL8 significantly enhanced stress tolerance in alfalfa; meanwhile, overexpression of SPL8 rendered plants more susceptible.
Discussion
Branch development and shoot architecture are critical for plants to compete for resources and to achieve maximum light capture and carbon generation, especially because modern agricultural practice employs high‐density stands to maximize yields (McSteen, 2009; McSteen and Leyser, 2005). In forage legumes such as M. truncatula and alfalfa, it is known that aerial morphogenesis including branch development and shoot architecture impacts biomass yield, production persistency, lodging tolerance and forage quality (Espinoza Ldel et al., 2012; Julier et al., 2007). The main challenge is how to control branch development and optimize plant architecture. A few mapping studies showed that branch development was positively correlated with forage biomass yield, and 24 quantitative trait loci (QTLs) were mapped for branch development in four recombinant inbred line (RIL) populations of M. truncatula (Espinoza Ldel et al., 2012; Julier et al., 2007). Except for these few resources, limited information is available on the genetic regulation of branch development and shoot architectures in legumes.
Sixteen SPL genes have been identified in Arabidopsis (Xie et al., 2006). Orthologs have also been identified in various species based on their highly conserved nature across monocots and eudicots (Wang et al., 2009). The SPL family also shows divergent functions in the regulation of various processes, throughout phase transition, microspore development, trichome development, grain shaping and anthocyanin biosynthesis (Gou et al., 2011b; Jung et al., 2016; Si et al., 2016; Wang et al., 2015; Yu et al., 2010). However, very little is known about their importance to axillary bud initiation and branch formation. In rice, OsSPL14 has been reported to decrease tiller number (basal branching) while promoting panicle branching (Jiao et al., 2010). However, OsSPL14 suppresses tillering neither through axillary bud initiation nor through bud outgrowth, but by prolonging the plastochron (Wang and Li, 2011). In the current study, three mutants with enhanced branching were identified through screening a M. truncatula Tnt1 mutant population. The enhanced branching phenotypes were caused by Tnt1 insertions in different exons of SPL8. Further investigation revealed that the mutated SPL8 enhanced branch development through the promotion of axillary bud formation. Consistently, overexpression of SPL8 suppressed axillary bud development and subsequently inhibited branching which ultimately resulted in an alteration of shoot architecture. The SPL8 ortholog was isolated from alfalfa. Modifying MsSPL8 expression in alfalfa also significantly affected branching and biomass yield. The increase in biomass yield in MsSPL8 down‐regulated alfalfa plants was 38%–43% in the first harvest, while the impact was more significant in the second harvest, with 66%–86% increase in biomass production. Alfalfa plants are typically grown in close proximity under field conditions, increased branching of the transgenics will lead to improved canopy architecture which allows better capture of sunlight. Furthermore, increased branching may allow reduced plant density and thus reduced seeding rate. It should be noted that enhanced regrowth after cutting is a particularly important trait in alfalfa. Most of the alfalfa grown in the USA is used for hay production, which requires multiple harvests per year. In some cases, alfalfa is also directly used for grazing by animals. Enhanced regrowth is very beneficial for either hay production or for grazing purpose.
Various studies indicate that LAS/Ls/MOC1 and LAX1/BA1/ROX represent the two pathways that are conserved in the regulation of bud initiation in dicots and monocots (Oikawa and Kyozuka, 2009; Tanaka et al., 2015). The details of the underlying mechanisms are still unknown. Our microarray and RT‐qPCR analyses suggest that SPL8 has no affect on the Medicago LAX1 ortholog but significantly regulates the LAS ortholog. AtSPL9 has been shown to directly suppress LAS in Arabidopsis (Tian et al., 2014). LAS/Ls orthologs encode the GRAS family nuclear proteins (including Scarecrow, SCR; Gibberellin insensitive, GAI; and repressor of gal‐3, RGA), which play a central role in the GA response and appear to be crosstalk points with other signals (Achard et al., 2006). A dramatic increase in GA levels was found in the ls mutant (Schumacher et al., 1999). Our data showed that GA levels were significantly higher in the spl8 mutants while much lower in the MtSPL8 overexpression plants, indicating that the GA signalling pathway is regulated by SPL8 from the upstream GA receptor GID1L2 to the downstream GA responsive genes GRASs. GA receptor perceives and binds endogenous GA, and the binding then induces the formation of GID1–GA–DELLA protein complex and finally triggers the subsequent signal transduction (Shimada et al., 2008). Furthermore, GA2‐ox6 is significantly up‐regulated by SPL8. GA2‐oxidase (GA2‐ox) is the major GA deactivation enzyme (Yamaguchi, 2008). Overexpression of GA2‐ox resulted in a dramatic decrease of bioactive GAs (Gou et al., 2010, 2011a). Taken together, these evidences consistently suggest that SPL8 regulation likely proceeds by inhibiting LAS/Ls/MOC1 pathway. SPL8 regulation appears to crosstalk with GA signalling, and GA2‐ox is possibly the key node of this crosstalk regulation. Further study of the interaction between SPL8, LAS and GA2‐ox may enable us to understand this mechanism in detail.
Alfalfa is the fourth most widely grown crop in the United States behind only corn, wheat and soya beans. Compared to the other crops, alfalfa has a relatively high level of drought tolerance (Zhang et al., 2005). Even so, drought tolerance is still a key challenge in improving alfalfa productivity (Arshad et al., 2017; Lei et al., 2017). Salinity is a major threat to alfalfa production (Arshad et al., 2017). Previous studies have gained some valuable information. For example, overexpression of WXP1, a gene related to wax accumulation, enhanced drought and dehydration tolerance in alfalfa, but the transgenic plants also showed moderately slow growth (Zhang et al., 2005). Overexpression of miR156 conferred salt tolerance in alfalfa, but plant height and flowering time were negatively affected in the transgenics (Arshad et al., 2017). In our study, the down‐regulation of MsSPL8 markedly enhanced drought and salt tolerance in alfalfa without any negative morphological or developmental changes. In future studies, these very promising results in controlled growth conditions will have to be tested and confirmed under field conditions.
In summary, our study revealed a new mechanism for regulating branch development and shoot architecture in the model legume M. truncatula, and we have successfully applied this knowledge to alfalfa improvement. This study demonstrated that down‐regulation of MsSPL8 significantly enhanced branching by promoting axillary bud formation and, consequently, increased forage biomass yield and promoted regrowth after cutting. Furthermore, down‐regulation of MsSPL8 also notably enhanced salt and drought tolerance in transgenic alfalfa. Results from this research offer a valuable approach to simultaneously improve biomass production and abiotic stress tolerance in plants. This study illustrates how knowledge gained from a model system can be used to genetically improve a commercial crop.
Experimental procedures
Plant materials and growth conditions
Medicago truncatula ecotype R108 was used as the wild type. Generation of the M. truncatula Tnt1 insertional mutant population was described previously by Tadege et al. (2008). Mutant and wild‐type seeds were scarified with concentrated sulphuric acid and treated at 4 °C for 5 days on filter paper. Small plantlets were transferred to Metro‐Mix 830 soil mix and grown in the greenhouse at 24/22 °C (day/night) temperature with 16 h light (390 μE/m2/s). An alfalfa (Medicago sativa) genotype, Regen SY‐4D, was used for Agrobacterium tumefaciens‐mediated transformation to produce transgenic plants (Fu et al., 2015). Both transgenic and wild‐type alfalfa plants were vegetatively propagated using shoot cuttings. All plants were grown in the greenhouse at 24/22 °C (day/night) temperature with 16 h light (390 μE/m2/s).
Screening of M. truncatula branching mutants and cloning of SPL8
The three mutant lines, NF7738, NF10281 and NF10498, were identified from a M. truncatula Tnt1‐insertion population based on enhanced branch development. Tnt1 flanking sequences of the mutants were found by searching the Medicago truncatula Mutant Database (https://medicago-mutant.noble.org/mutant/database.php). The putative Tnt1 flanking sequences were further verified by PCR amplification (using primers identified from the M. truncatula genome sequence that spanned across the Tnt1 insertion sites). The PCR products were purified and cloned into pGEM‐T Easy Vector (Promega) and sequenced using Sanger dideoxy sequencing. The flanking sequences were BLAST searched against the M. truncatula genome sequence at the NCBI database. The genomic sequence of SPL8 was obtained from the M. truncatula R108 database (http://www.medicagohapmap.org/tools/r108_blastform).
For overexpression, the coding sequences of SPL8 were obtained through RT‐PCR amplification using primers MtSPL8‐F and MtSPL8‐R (Table S4). The fragment was inserted into pENTR/D‐TOPO cloning vector (Invitrogen) and transferred into the pEarleyGate 100 gateway vector (driven by CaMV35S promoter) by attL/attR recombination reactions (Invitrogen). The verified constructs were used to transform M. truncatula ecotype R108 using leaf explants (Crane et al., 2006). PCR analysis of the regenerated plants was carried out using a forward primer selected from the CaMV35S promoter (35Spromoter‐F) and a reverse primer selected from the 5′‐end of MtSPL8 (MtSPL8‐R1) (Table S4).
Gene expression quantification
Reverse transcription quantitative PCR (RT‐qPCR) was performed to analyse transcript abundance of various genes. Total RNA was extracted from various tissues by TRI‐Reagent (Invitrogen) and subjected to reverse transcription with Superscript III Kit (Invitrogen). SYBR Green (Applied Biosystems, Foster City, CA) was used as the reporter dye. The primers used for RT‐qPCR are listed in Table S4. M. truncatula actin2 gene (TC107326) was used as an internal control. The normalized data were analysed using Student's t‐test.
Microarray analysis
Total RNA samples from three biological replicates of the selected MtSPL8 overexpression transgenic events and the wild‐type R108 were extracted from shoot apical meristems of 6‐week‐old plants using Spectrum™ Plant Total RNA Kit (Sigma‐Aldrich). 500 ng RNA was amplified and labelled using the GeneChip 3′ IVT Express Kit (Affymetrix, Santa Clara, CA) and hybridized to M. truncatula Affymetrix chips. Data normalization was conducted using the robust multi‐array average (RMA). Data analysis of differentially expressed probe sets on the chip was performed by associative analysis as described previously (Dozmorov and Centola, 2003). Hierarchical analysis was used to identify genes with a positive correlation between phenotype and gene expression.
Characterization of plant growth and development
Branch numbers were measured from three biological replicates of each tested line when the plants were 10 weeks old. Fresh biomass was measured when plants were in the budding to early bloom stage (up to 10% in bloom). The harvested biomass was dried in an oven at 45 °C for 96 h before measuring the dried biomass. The data were analysed using Student's t‐test.
Microscopy analysis and photography
Vegetative bud samples were harvested and immediately fixed in 3% glutaraldehyde (in 25 mm phosphate buffer (pH 7.0) overnight and dehydrated in graded ethanol series. The fixed and dried samples were observed using Hitachi TM‐3000 scanning electron microscope (SEM). Light microscopy was performed using a Nikon SMZ 1500 stereomicroscope.
Phytohormone quantification
Meristem tissues of vegetative buds and mature leaves were harvested from wild‐type R108, spl8 mutants and SPL8 overexpression transgenic lines when plants were 6 weeks old. Harvested samples were frozen in liquid N2 and ground immediately. Fifty mg of each sample was applied for hormone quantification with HPLC‐MS analysis as described by Pan et al. (2010).
Isolation of MsSPL8 and creation of MsSPL8 modified transgenic alfalfa
Purified mRNA from vegetative buds of the alfalfa genotype, Regen SY‐4D, was used for cDNA synthesis. The full‐length mRNA sequence of MsSPL8 was isolated from the alfalfa cDNA by 5′‐ and 3′‐RACE using primers produced from M. truncatula SPL8 (Table S4).
To knockdown MsSPL8 in alfalfa, a 443‐bp fragment of MsSPL8 was PCR‐amplified from alfalfa cDNA using primers MsSPL8Ri‐F and MsSPL8Ri‐R (Table S4). The fragment was inserted into pENTR/D‐TOPO cloning vector (Invitrogen) and transferred into the pANDA35HK vector (Li et al., 2010) by attL/attR recombination reactions (Invitrogen). For overexpression, the coding sequence of MsSPL8 was obtained through RT‐PCR amplification using primers MsSPL8cDNA‐F and MsSPL8cDNA‐R (Table S4) and cloned into the pEarleyGate 100 gateway vector driven by CaMV35S promoter. The verified constructs were transferred into Agrobacterium strain EHA105 using the freezing/heat‐shock method. Transgenic alfalfa plants were obtained by Agrobacterium‐mediated transformation as previously reported (Fu et al., 2015). PCR analysis of the regenerated plants was carried out using a forward primer selected from the CaMV35S promoter (35Spromoter‐F) and a reverse primer selected from the 5′‐end of MsSPL8 (MsSPL8‐R1).
Forage analysis of transgenic alfalfa lines
Transgenic and control alfalfa plants were grown in the soil with full nutrition. Two‐month‐old plants were harvested, and fresh biomass yield was measured immediately. The materials were dried in an oven at 45 °C for 96 h to measure dried biomass. The samples were then ground through a Thomas‐Wiley Laboratory Mill (Lehman Scientific) with a 1‐mm sieve. Near‐infrared reflectance spectroscopy (NIRS) was performed using a Foss NIRS 6500 monochromator with a scanning range of 1100–2500 nm (Foss NIR Systems). Each sample was scanned eight times, and the average spectra were used for calibration. Mathematical and statistical treatments of all spectra were performed with WinISI III calibration development software (Foss NIR Systems). The existing commercial NIRS prediction equations (07AHY50) developed by the NIRS Forage and Feed Testing Consortium were employed to calculate quality characteristics of alfalfa. The precision of NIRS has been assessed by regression analysis of the predicted values and actual determined values. All data were analysed using the SAS GLM procedure (SAS Institute). Statistical significance was determined by Student's t‐test.
Drought and salt treatments
Both transgenic and control alfalfa plants were propagated using shoot cuttings. Uniformed seedlings were transplanted to 4.5‐inch pots filled with Metro‐Mix 830 soil mix and grown in the greenhouse. Three weeks later, well‐established and similarly sized plants (with eight replicates for each experiment) were selected from each line for two kinds of treatments. For salt treatment, all pots were completely soaked in 1.5% NaCl solution for 3 h every 4 days for 3 weeks. For drought treatment, all plants were completely soaked in water for three hours, and then watering was withheld for 2 weeks before re‐watering. The experiments were repeated at least three times for all measurements.
Supporting information
Figure S1 Phylogenetic analysis of SPL orthologs in Arabidopsis, rice and M. truncatula.
Figure S2 Expression patterns of SPL8 in M. truncatula, produced from the M. truncatula Gene Expression Atlas (https://mtgea.noble.org/v2/index.php).
Figure S3 Comparison of branch development between control and the three Tnt1 mutants of SPL8 in M. truncatula.
Figure S4 PCR analysis of regenerated M. truncatula plants after Agrobacterium‐mediated transformation with the SPL8 overexpression vector.
Figure S5 Branching and shoot architecture were significantly altered in SPL8 knockout mutants and overexpression transgenics in M. truncatula.
Figure S6 Relative expression levels of candidate genes in spl8 mutants, control and SPL8 overexpression lines (SPL8OE) in M. truncatula.
Figure S7 Phytohormone accumulation in spl8 mutants, control and SPL8 overexpression lines (SPL8OE).
Figure S8 Sequence comparison of MsSPL8 (M. sativa SY4D) with SPL8 (M. truncatula R108).
Figure S9 Amino acid alignment of MsSPL8 and SPL8 proteins and their ortholog in soybean (Glycine max).
Figure S10 PCR analysis of regenerated alfalfa plants after Agrobacterium‐mediated transformation with the MsSPL8 RNAi vector.
Figure S11 Relative expression levels of MsSPL8 in the transgenic downregulation plants of alfalfa (MsSPL8Ri).
Figure S12 Transgenic alfalfa showing increased biomass phenotype with downregulation of MsSPL8.
Figure S13 PCR analysis of regenerated alfalfa plants after Agrobacterium‐mediated transformation with the MsSPL8 overexpression vector.
Figure S14 Transgenic alfalfa showing decreased biomass phenotype with overexpression of MsSPL8.
Figure S15 Effects of abiotic stresses on MsSPL8 transgenic alfalfa plants.
Table S1 Downregulated genes with abundance changed more than 2.5 folds in SPL8 overexpression plants relative to the control.
Table S2 Upregulated genes with abundance changed more than 2.5 folds in SPL8 overexpression plants relative to the control.
Table S3 Forage quality analysis of alfalfa plants.
Table S4 Primers used in this study.
Acknowledgements
We thank Bonnie Watson and David Huhman for their assistance in hormone quantification, and Dennis Walker for forage quality analysis. This research was supported by the BioEnergy Science Center and Noble Research Institute, LLC. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. The authors declare no conflict of interest.
References
- Achard, P. , Cheng, H. , De Grauwe, L. , Decat, J. , Schoutteten, H. , Moritz, T. , Van Der Straeten, D. et al (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science, 311, 91–94. [DOI] [PubMed] [Google Scholar]
- Annicchiarico, P. , Nazzicari, N. , Li, X. , Wei, Y. , Pecetti, L. and Brummer, E.C. (2015) Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genom. 16, 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad, M. , Gruber, M.Y. , Wall, K. and Hannoufa, A. (2017) An insight into microRNA156 role in salinity stress responses of alfalfa. Front. Plant Sci. 8, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung, B. , Gruber, M.Y. , Amyot, L. , Omari, K. , Bertrand, A. and Hannoufa, A. (2015) MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. [DOI] [PubMed] [Google Scholar]
- Bennett, T. and Leyser, O. (2006) Something on the side: axillary meristems and plant development. Plant Mol. Biol. 60, 843–854. [DOI] [PubMed] [Google Scholar]
- Biazzi, E. , Nazzicari, N. , Pecetti, L. , Brummer, E.C. , Palmonari, A. , Tava, A. and Annicchiarico, P. (2017) Genome‐wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLoS ONE, 12, e0169234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, M. , Zhou, C. , Molina, I. , Fu, C. , Nakashima, J. , Li, G. , Zhang, W. et al (2016) A Class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. USA, 113, 6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Dai, X. and Zhao, Y. (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D.R. (1999) Medicago truncatula–a model in the making!. Curr. Opin. Plant Biol. 2, 301–304. [DOI] [PubMed] [Google Scholar]
- Costes, E. , Crespel, L. , Denoyes, B. , Morel, P. , Demene, M.N. , Lauri, P.E. and Wenden, B. (2014) Bud structure, position and fate generate various branching patterns along shoots of closely related Rosaceae species: a review. Front Plant Sci. 5, 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane, C. , Wright, E. , Dixon, R.A. and Wang, Z.‐Y. (2006) Transgenic Medicago truncatula plants obtained from Agrobacterium tumefaciens‐transformed roots and Agrobacterium rhizogenes‐transformed hair roots. Planta, 223, 1344–1354. [DOI] [PubMed] [Google Scholar]
- Cui, L.G. , Shan, J.X. , Shi, M. , Gao, J.P. and Lin, H.X. (2014) The miR156‐SPL9‐DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. [DOI] [PubMed] [Google Scholar]
- Domagalska, M.A. and Leyser, O. (2011) Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell. Bio. 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Dozmorov, I. and Centola, M. (2003) An associative analysis of gene expression array data. Bioinformatics, 19, 204–211. [DOI] [PubMed] [Google Scholar]
- Espinoza Ldel, C. , Huguet, T. and Julier, B. (2012) Multi‐population QTL detection for aerial morphogenetic traits in the model legume Medicago truncatula. Theor. Appl. Genet. 124, 739–754. [DOI] [PubMed] [Google Scholar]
- Fu, C. , Hernandez, T. , Zhou, C. and Wang, Z.Y. (2015) Alfalfa (Medicago sativa L.). Methods Mol. Biol. 1223, 213–221. [DOI] [PubMed] [Google Scholar]
- Gallavotti, A. , Zhao, Q. , Kyozuka, J. , Meeley, R.B. , Ritter, M.K. , Doebley, J.F. , Pe, M.E. et al (2004) The role of barren stalk1 in the architecture of maize. Nature, 432, 630–635. [DOI] [PubMed] [Google Scholar]
- Gallavotti, A. , Barazesh, S. , Malcomber, S. , Hall, D. , Jackson, D. , Schmidt, R.J. and McSteen, P. (2008) Sparse inflorescence1 encodes a monocot‐specific YUCCA‐like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA, 105, 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Roldan, V. , Fermas, S. , Brewer, P.B. , Puech‐Pages, V. , Dun, E.A. , Pillot, J.P. , Letisse, F. et al (2008) Strigolactone inhibition of shoot branching. Nature, 455, 189–194. [DOI] [PubMed] [Google Scholar]
- Gou, J. , Strauss, S.H. , Tsai, C.J. , Fang, K. , Chen, Y. , Jiang, X. and Busov, V.B. (2010) Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell, 22, 623–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J. , Ma, C. , Kadmiel, M. , Gai, Y. , Strauss, S. , Jiang, X. and Busov, V. (2011a) Tissue‐specific expression of Populus C(19) GA 2‐oxidases differentially regulate above‐ and below‐ground biomass growth through control of bioactive GA concentrations. New Phytol. 192, 626–639. [DOI] [PubMed] [Google Scholar]
- Gou, J.Y. , Felippes, F.F. , Liu, C.J. , Weigel, D. , Wang, J. and Wang, J.W. (2011b) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156‐targeted SPL transcription factor. Plant Cell, 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S.Y. , Xu, Y.Y. , Liu, H.H. , Mao, Z.W. , Zhang, C. , Ma, Y. , Zhang, Q.R. et al (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 4, 1566 https://doi.org/10.1038/ncomms2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y.Q. , Wang, Y.H. , Xue, D.W. , Wang, J. , Yan, M.X. , Liu, G.F. , Dong, G.J. et al (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–U536. [DOI] [PubMed] [Google Scholar]
- Julier, B. , Huguet, T. , Chardon, F. , Ayadi, R. , Pierre, J.B. , Prosperi, J.M. , Barre, P. et al (2007) Identification of quantitative trait loci influencing aerial morphogenesis in the model legume Medicago truncatula. Theor. Appl. Genet. 114, 1391–1406. [DOI] [PubMed] [Google Scholar]
- Jung, J.H. , Lee, H.J. , Ryu, J.Y. and Park, C.M. (2016) SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT‐FD module in Arabidopsis flowering. Mol. Plant, 9, 1647–1659. [DOI] [PubMed] [Google Scholar]
- Kang, Y. , Li, M. , Sinharoy, S. and Verdier, J. (2016) A snapshot of functional genetic studies in Medicago truncatula . Front Plant Sci. 7, 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom, T.H. , Spielmeyer, W. and Finnegan, E.J. (2013) Grasses provide new insights into regulation of shoot branching. Trends Plant Sci. 18, 41–48. [DOI] [PubMed] [Google Scholar]
- Komatsu, K. , Maekawa, M. , Ujiie, S. , Satake, Y. , Furutani, I. , Okamoto, H. , Shimamoto, K. et al (2003) LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA, 100, 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Hannoufa, A. and Yu, P. (2017) The use of gene modification and advanced molecular structure analyses towards improving alfalfa forage. Int. J. Mol. Sci. 18, 298 https://doi.org/10.3390/ijms18020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X.Y. , Qian, Q. , Fu, Z.M. , Wang, Y.H. , Xiong, G.S. , Zeng, D.L. , Wang, X.Q. et al (2003) Control of tillering in rice. Nature, 422, 618–621. [DOI] [PubMed] [Google Scholar]
- Li, X. , Weng, J. and Chapple, C. (2008) Improvement of biomass through lignin modification. Plant J. 54, 569–581. [DOI] [PubMed] [Google Scholar]
- Li, J. , Todd, T.C. and Trick, H.N. (2010) Rapid in planta evaluation of root expressed transgenes in chimeric soybean plants. Plant Cell Rep. 29, 113–123. [DOI] [PubMed] [Google Scholar]
- McSteen, P. (2009) Hormonal regulation of branching in grasses. Plant Physiol. 149, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen, P. and Leyser, O. (2005) Shoot branching. Annu. Rev. Plant Biol. 56, 353–374. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Ikeda, M. , Matsubara, A. , Song, X.J. , Ito, M. , Asano, K. , Matsuoka, M. et al (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549 U102. [DOI] [PubMed] [Google Scholar]
- Oikawa, T. and Kyozuka, J. (2009) Two‐step regulation of LAX PANICLE1 protein accumulation in Axillary Meristem formation in rice. Plant Cell, 21, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Welti, R. and Wang, X. (2010) Quantitative analysis of major plant hormones in crude plant extracts by high‐performance liquid chromatography‐mass spectrometry. Nat. Protoc. 5, 986–992. [DOI] [PubMed] [Google Scholar]
- Russelle, M.P. , Lamb, J.F.S. , Turyk, N.B. , Shaw, B.H. and Pearson, B. (2007) Managing nitrogen contaminated soils: Benefits of N‐2‐fixing alfalfa. Agronomy J. 99, 738–746. [Google Scholar]
- Samac, D.A. , Jung, H.G. and Lamb, J.F.S. (2006) Development of alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts. Alcoholic Fuels, 112, 79–98. [Google Scholar]
- Schumacher, K. , Schmitt, T. , Rossberg, M. , Schmitz, G. and Theres, K. (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA, 96, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, A. , Ueguchi‐Tanaka, M. , Nakatsu, T. , Nakajima, M. , Naoe, Y. , Ohmiya, H. , Kato, H. et al (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature, 456, 520–523. [DOI] [PubMed] [Google Scholar]
- Si, L. , Chen, J. , Huang, X. , Gong, H. , Luo, J. , Hou, Q. , Zhou, T. et al (2016) OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. [DOI] [PubMed] [Google Scholar]
- Tadege, M. , Wen, J.Q. , He, J. , Tu, H.D. , Kwak, Y. , Eschstruth, A. , Cayrel, A. et al (2008) Large‐scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . Plant J. 54, 335–347. [DOI] [PubMed] [Google Scholar]
- Tanaka, W. , Ohmori, Y. , Ushijima, T. , Matsusaka, H. , Matsushita, T. , Kumamaru, T. , Kawano, S. et al (2015) Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell, 27, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C. , Zhang, X. , He, J. , Yu, H. , Wang, Y. , Shi, B. , Han, Y. et al (2014) An organ boundary‐enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 10, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unte, U.S. , Sorensen, A.M. , Pesaresi, P. , Gandikota, M. , Leister, D. , Saedler, H. and Huijser, P. (2003) SPL8, an SBP‐Box gene that affects pollen sac development in Arabidopsis. Plant Cell, 15, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volenec, J.J. , Cunningham, S.M. , Haagenson, D.M. , Berg, W.K. , Joern, B.C. and Wiersma, D.W. (2002) Physiological genetics of alfalfa improvement: past failures, future prospects. Field. Crop. Res. 75, 97–110. [Google Scholar]
- Wang, Y.H. and Li, J.Y. (2011) Branching in rice. Curr. Opin. Plant Biol. 14, 94–99. [DOI] [PubMed] [Google Scholar]
- Wang, J.W. , Czech, B. and Weigel, D. (2009) miR156‐regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang, S.K. , Li, S. , Liu, Q. , Wu, K. , Zhang, J.Q. , Wang, S.S. , Wang, Y. et al (2015) The OsSPL16‐GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. [DOI] [PubMed] [Google Scholar]
- Xie, K.B. , Wu, C.Q. and Xiong, L.Z. (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter‐binding‐like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S.P. , Salinas, M. , Hohmann, S. , Berndtgen, R. and Huijser, P. (2010) miR156‐targeted and nontargeted SBP‐box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell, 22, 3935–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, S. , Salinas, M. , Garcia‐Molina, A. , Hohmann, S. , Berndtgen, R. and Huijser, P. (2013) SPL8 and miR156‐targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 75, 566–577. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008) Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, A. , Wu, M.‐F. , Yang, L. , Wu, G. , Poethig, R.S. and Wagner, D. (2009) The microRNA‐regulated SBP‐Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell, 17, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, K. , Kigawa, T. , Inoue, M. , Tateno, M. , Yamasaki, T. , Yabuki, T. , Aoki, M. et al (2004) A novel zinc‐binding motif revealed by solution structures of DNA‐binding domains of Arabidopsis SBP‐family transcription factors. J. Mol. Biol. 337, 49–63. [DOI] [PubMed] [Google Scholar]
- Yang, F. , Wang, Q. , Schmitz, G. , Muller, D. and Theres, K. (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71, 61–70. [DOI] [PubMed] [Google Scholar]
- Young, N.D. and Udvardi, M. (2009) Translating Medicago truncatula genomics to crop legumes. Curr. Opin. Plant Biol. 12, 193–201. [DOI] [PubMed] [Google Scholar]
- Young, N.D. , Cannon, S.B. , Sato, S. , Kim, D. , Cook, D.R. , Town, C.D. , Roe, B.A. et al (2005) Sequencing the Genespaces of Medicago truncatula and Lotus japonicus . Plant Physiol. 137, 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, N.D. , Debellé, F. , Oldroyd, G.E.D. , Geurts, R. , Cannon, S.B. , Udvardi, M.K. , Benedito, V.A. et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature, 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, N. , Cai, W.J. , Wang, S. , Shan, C.M. , Wang, L.J. and Chen, X.Y. (2010) Temporal control of trichome distribution by microRNA156‐targeted SPL genes in Arabidopsis thaliana. Plant Cell, 22, 2322–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Y. , Broeckling, C.D. , Blancaflor, E.B. , Sledge, M.K. , Sumner, L.W. and Wang, Z.Y. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain‐containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707. [DOI] [PubMed] [Google Scholar]
- Zhou, C. , Han, L. , Li, G. , Chai, M. , Fu, C. , Cheng, X. , Wen, J. et al (2014) STM/BP‐Like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula . Plant Cell, 26, 1464–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic analysis of SPL orthologs in Arabidopsis, rice and M. truncatula.
Figure S2 Expression patterns of SPL8 in M. truncatula, produced from the M. truncatula Gene Expression Atlas (https://mtgea.noble.org/v2/index.php).
Figure S3 Comparison of branch development between control and the three Tnt1 mutants of SPL8 in M. truncatula.
Figure S4 PCR analysis of regenerated M. truncatula plants after Agrobacterium‐mediated transformation with the SPL8 overexpression vector.
Figure S5 Branching and shoot architecture were significantly altered in SPL8 knockout mutants and overexpression transgenics in M. truncatula.
Figure S6 Relative expression levels of candidate genes in spl8 mutants, control and SPL8 overexpression lines (SPL8OE) in M. truncatula.
Figure S7 Phytohormone accumulation in spl8 mutants, control and SPL8 overexpression lines (SPL8OE).
Figure S8 Sequence comparison of MsSPL8 (M. sativa SY4D) with SPL8 (M. truncatula R108).
Figure S9 Amino acid alignment of MsSPL8 and SPL8 proteins and their ortholog in soybean (Glycine max).
Figure S10 PCR analysis of regenerated alfalfa plants after Agrobacterium‐mediated transformation with the MsSPL8 RNAi vector.
Figure S11 Relative expression levels of MsSPL8 in the transgenic downregulation plants of alfalfa (MsSPL8Ri).
Figure S12 Transgenic alfalfa showing increased biomass phenotype with downregulation of MsSPL8.
Figure S13 PCR analysis of regenerated alfalfa plants after Agrobacterium‐mediated transformation with the MsSPL8 overexpression vector.
Figure S14 Transgenic alfalfa showing decreased biomass phenotype with overexpression of MsSPL8.
Figure S15 Effects of abiotic stresses on MsSPL8 transgenic alfalfa plants.
Table S1 Downregulated genes with abundance changed more than 2.5 folds in SPL8 overexpression plants relative to the control.
Table S2 Upregulated genes with abundance changed more than 2.5 folds in SPL8 overexpression plants relative to the control.
Table S3 Forage quality analysis of alfalfa plants.
Table S4 Primers used in this study.


