Abstract
Background
Treatment of Helicobacter pylori infection is often empiric; however, current guidelines for management of H. pylori infection advise against the use of standard Triple Therapy (clarithromycin, amoxicillin, and proton pump inhibitor) when clarithromycin resistance exceeds 20%. We developed and tested a new culture-free assay to detect clarithromycin resistance-conferring mutations to determine the prevalence of H. pylori clarithromycin resistance in patients from the United States Pacific Northwest.
Materials and Methods
Droplet digital PCR (ddPCR) was used to detect the H. pylori 23S rRNA gene, and resistance-conferring mutations, in archived, formalin-fixed, paraffin-embedded (FFPE) gastric tissue and to retrospectively determine the prevalence of clarithromycin-resistant H. pylori among 110 patients at an academic medical center in the Northwest United States between 2012–2014.
Results
Of 102 patients with the H. pylori 23S rRNA gene detected by the ddPCR assay, 45 (44%) had clarithromycin resistance mutations. Thirty-three of the 45 patients with clarithromycin resistance mutations had a mix of wild-type and resistance alleles. Prevalence of clarithromycin resistance mutations differed among racial groups and was highest among Asians, with mutations detected in 14 (67%) of the 21 patient samples.
Conclusions
The prevalence of clarithromycin resistance detected in this region exceeds 20%, indicating that standard Triple Therapy should not be the first line antibiotic treatment for H. pylori infection. Culture-free assays for detecting clarithromycin resistance mutations can be performed on archived tissue samples and will aid in informing tailored treatment for effective H. pylori eradication.
Keywords: Helicobacter pylori, clarithromycin resistance, culture-free assay, droplet digital PCR
INTRODUCTION
Helicobacter pylori infects approximately half the world’s population and one-third of the U.S. population 1 and can lead to peptic ulcer and gastric cancer. Clarithromycin is typically part of first-line treatment along with amoxicillin and a proton pump inhibitor (PPI) 2. Eradication of H. pylori infection is effective in resolving gastric and duodenal ulcers, 3 gastric MALT lymphomas, 4, 5 and in reducing progression of some pre-neoplastic lesions to gastric cancer 6, 7. However, rising rates of antibiotic resistance have negatively impacted the efficacy of H. pylori treatment 2.
H. pylori infection can be diagnosed by histological analysis of formalin-fixed gastric biopsy specimens collected during upper GI endoscopy. Formalin fixation precludes bacterial culture, and separate gastric biopsies for culture are rarely collected. Thus, antibiotic resistance testing is generally not performed before prescribing H. pylori treatment, leaving clinicians to rely on empiric treatment strategies, as local patterns of antibiotic resistance are often unknown. Current guidelines recommend against the use of standard Triple Therapy (clarithromycin, amoxicillin and PPI ) as first-line treatment when the local clarithromycin resistance rate exceeds 20% 8. If empiric first and second-line treatments fail to eradicate infection, patients may need to undergo repeat endoscopy, with biopsies collected for culture and antibiotic susceptibility testing, to guide further treatment 2.
Accordingly, a culture-free method to detect antibiotic resistance would be useful for determining resistance patterns in a population and for tailoring the treatment of individual patients. Recently, we reported the use of droplet digital PCR (ddPCR) to quantitatively detect the H. pylori 16S gene and distinguish between alleles of the cagA virulence gene from stool samples 9. ddPCR provides absolute quantitation and increased sensitivity for detecting rare targets by partitioning the PCR reaction into 10,000–100,000 droplets, and multiple fluorescent hydrolysis probes can differentiate alleles of a given gene 10.
To assess H. pylori antibiotic resistance, we developed a ddPCR assay to detect mutations that confer resistance to clarithromycin. Clarithromycin blocks bacterial protein synthesis by targeting the bacterial 23S ribosomal subunit, however, resistance can arise from point mutations in the 23S rRNA gene. The majority of clarithromycin-resistant H. pylori strains have one of three point mutations (A2142G, A2142C, and A2143G) in the 23S rRNA gene 11, 12. The ddPCR assay we developed uses multiple fluorescent probes to distinguish between the wild-type and the three common clarithromycin resistance 23S rRNA alleles, and quantifies the proportion of 23S rRNA gene copies in a sample with resistance mutations. This assay utilizes DNA extracted from archived, formalin-fixed paraffin-embedded (FFPE) gastric tissue samples, eliminating the need for H. pylori bacterial culture. The technique allows for retrospective examination of resistance rates in the population to inform empiric treatment, reported here for patients at University of Washington Medical Center in Seattle, Washington. Additionally, this culture-free assay can also be used for making tailored treatment decisions for individual patients either at the time of initial upper GI endoscopy or to avoid repeat endoscopy in the event of treatment failure.
MATERIALS AND METHODS
Study Populations and Specimen Collection
H. pylori positive gastric biopsy specimens were retrospectively identified from the University of Washington Medical Center using Current Procedural Terminology (CPT) billing codes. Chart review was performed to confirm diagnosis of H. pylori infection by histological analysis using immunofluorescence or Genta stain and identify patients with culture-based antibiotic susceptibility test results.
In total, 146 formalin-fixed, paraffin-embedded (FFPE) gastric biopsy specimens, collected between 2012 and 2014, were identified. Of these, 28 were determined to have an insufficient amount of remaining tissue to be used in the study. In addition, 8 specimens were only available as histology slides without FFPE tissue for use in the assay. From the remaining 110 gastric biopsy specimens, with histologically evident H. pylori diagnosis and unknown antibiotic susceptibility, two 10 μm curls from the archived FFPE gastric tissue were obtained for analysis. Additional review of a microbiology database identified six gastric biopsy specimens containing clarithromycin-resistant H. pylori, determined by the culture-based Epsilometer test (E-test). Two 10 μm curls were obtained from these six FFPE gastric tissues for testing the ddPCR assay.
Patient demographic data including age, sex, and race or nationality was extracted from the patient database. Patient race or nationality was grouped for analysis into one of six main categories: White, Black or African-American, Asian, Hispanic, Native American or Alaska Native, and Native Hawaiian or Other Pacific Islander. Study procedures were approved by the University of Washington Institutional Review Board.
Selection and Construction of Clarithromycin Resistant H. pylori
To obtain H. pylori strains having the three clarithromycin resistance-conferring mutations in the 23S rRNA gene (A2142G, A2142C, or A2143G) for testing the ddPCR assay, we either selected for clarithromycin resistance by passage of the wild-type strain G27 13 on selective media or constructed the strain by site-directed mutagenesis. For selection, H. pylori G27 was grown to mid-log in non-selective BB10 liquid media (Brucella broth (BD Biosciences) supplemented with 10% fetal bovine serum (GIBCO)) with shaking in a tri-gas incubator equilibrated to 10% CO2 and 10% O2. 2.6 × 1010 cfu was plated onto 10 horse blood agar plates containing 0.1 μg/ml clarithromycin and incubated in a tri-gas incubator for five days. Of 12 colonies obtained, one was successfully expanded. PCR and DNA sequencing with Hp23SF and Hp23SR (Supplementary Table 1) identified the A2143G mutation.
Strains having the A2142G and A2142C mutations in the 23S rRNA gene were engineered by first cloning a fragment of the 23S rRNA gene from H. pylori strain G27 (base pairs 786 through 907 of the G27 23S rRNA gene, amplified with primers Hp23SF and Hp23SR (Supplementary Table 1) into pCR2.1-TOPO plasmid (Invitrogen). The resulting plasmid was used for site-directed mutagenesis using primers A2142G and A2142C (Table1). The mutated plasmids were naturally transformed into G27 14. Mutant clones were selected by plating on horse blood agar plates containing 0.1 μg/ml clarithromycin followed by PCR and DNA sequencing for confirmation.
Table 1.
Demographic characteristics and H. pylori 23S rRNA genotype determined by the clarithromycin resistance ddPCR assay for 110 University of Washington Medical Center patients
| Patient Characteristic | No. (%) or mean (SD) n = 110 |
|
|---|---|---|
| Age (years) | 54 (SD=15) | |
| Sex | Male | 57 (52%) |
| Female | 53 (48%) | |
| Race | White | 53 (48%) |
| Black or African-American | 9 (8%) | |
| Asian | 23 (21%) | |
| Hispanic | 17 (15%) | |
| American Indian or Alaska Native | 3 (3%) | |
| Native Hawaiian or Other Pacific Islander | 5 (5%) | |
| 23S genotype | Wild-type allele only | 57 (52%) |
| Resistance allele only | 12 (11%) | |
| Mixed | 33 (30%) | |
| No 23S rRNA gene detected | 8 (7%) | |
Ethics statement
Mouse studies were performed under practices and procedures of Animal Biosafety Level 2, and carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and complies with all United States Department of Agriculture, Public Health Service, Washington State and local area animal welfare regulations. All activities were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee.
Processing of Mouse Stomachs H. pylori 23S rRNA gene polymorphism detection
Mouse stomachs were incubated in either liquid H. pylori culture, or sterile media for control. Fresh stomach tissues were harvested from uninfected C57BL/6 mice, opened along the lesser curvature, and divided in half. Approximately 107 CFU wild-type H. pylori, 23S rRNA mutant H pylori, or a mixture of wild-type and mutant bacteria in 100 μl BB10 (Brucella broth (BD Biosciences), supplemented with 10% fetal bovine serum (GIBCO), or 100 μl sterile BB10 were added to the luminal side of each half stomach and incubated for 30 minutes in a tri-gas incubator before fixation in 10% buffered formalin phosphate (Fisher). After fixation, the half stomachs were embedded in paraffin by FHCRC Experimental Histopathology Shared Resource and five 10 μm curls were cut from each block for DNA extraction.
Primer and Probe Design
The primer and probe sequences for the H. pylori 23S rRNA gene and human 18S rRNA gene ddPCR assays are listed in Supplementary Table 1. Probes were designed to distinguish between the H. pylori wild-type 23S rRNA allele and the three most common clarithromycin resistance-conferring mutations in the H. pylori 23S rRNA gene (A2143G, A2142G, and A2142C). The wild-type probe was labeled with hexachloro-fluorescein (HEX) and the three clarithromycin resistance mutation probes were labeled with 6-carboxyfluorescein (FAM). All four probes were included in a single reaction. For quantification of human DNA, a single HEX probe to the wild-type human 18S rRNA gene was utilized.
DNA Extraction and Droplet Digital PCR
DNA was extracted from curls of FFPE gastric tissue using the FFPE Tissue DNA Isolation Kit (MoBio) according to the manufacturer’s instructions. Two 10 μm curls were used for patient samples and five 10 μm curls for mouse samples. Droplet digital PCR was performed according to the manufacturer’s instructions with each 20 μL reaction containing 1x ddPCR Supermix for Probes (BioRad), 900 nM of each primer, 250 nM of each probe, and 10 μL DNA. For spike-in experiments 0.4 – 40 pg of H. pylori genomic DNA of the indicated ratio of genotype was diluted in 10 μL H2O. Droplets were generated using the QX200 Droplet Generator (BioRad), subject to thermal cycling, and then analyzed for fluorescent amplitude using the QX200 Droplet Reader (BioRad). Data was analyzed using the QuantaSoft software version 1.6.6 (BioRad) and thresholds were set manually by visually inspecting the amplitude plots for positive controls and placing the threshold between the cluster of positive droplets and the cluster of negative droplets.
For the clarithromycin resistance assay, thermal cycling conditions were 95°C for 10 minutes, 45 cycles of 94°C for 30 seconds and 58°C for 1 minute, and one cycle of 98°C for 10 minutes. The thresholds for positive droplets were set at 2000 for wild-type (HEX channel) and at 4000 for resistance mutations (FAM channel). To decrease non-specific detection of wild-type and resistance alleles, the concentration of wild-type and resistance alleles was set to zero if less than five positive droplets were detected in the respective channel.
For the 18S assay, thermal cycling conditions were 95°C for 10 minutes, 45 cycles of 94°C for 30 seconds and 59°C for 1 minute, and a final incubation at 98°C for 10 minutes. Data was analyzed with the positive threshold set to 2000.
Statistical Analysis
Age, sex, and race were compared between patients with clarithromycin resistance alleles and patients with only wild-type allele by Student’s t-test or chi-square test, as appropriate. Copy number of the 23S rRNA gene between racial groups was compared using the Wilcoxon rank-sum test. Statistical analyses were performed using SAS version 9.4 (SAS Institute, Inc.).
RESULTS
Droplet digital PCR is more sensitive than Sanger sequencing for detection of H. pylori 23S clarithromycin resistance alleles
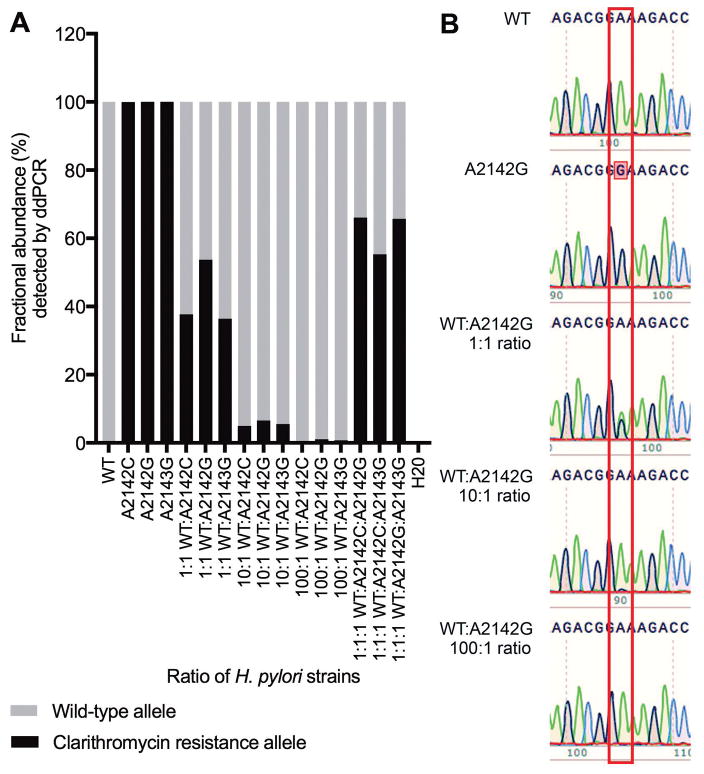
To assess the sensitivity of the H. pylori 23S rRNA clarithromycin resistance ddPCR assay to detect and quantify clarithromycin resistance alleles, we tested the assay on genomic DNA from a clarithromycin-sensitive wild-type strain and three isogenic clarithromycin-resistant strains spiked into water individually and in different ratios. The clarithromycin resistance ddPCR assay correctly distinguished between wild-type and resistance alleles. The assay also correctly quantified the ratios of wild-type and resistance alleles for all ratios that were tested, and it detected resistance mutations even when present in just 1% of the H. pylori DNA (Figure 1A). In comparison, Sanger sequencing of 23S rRNA gene PCR product, which is often used for genotypic testing of H. pylori clarithromycin resistance, was only able to detect a mixture of wild-type and resistance allele at a 1:1 ratio (Figure 1B).
Figure 1.
Detection of clarithromycin resistance alleles by ddPCR assay (A) and Sanger sequencing (B). DNA from H. pylori strains with different 23S rRNA gene alleles was spiked into water individually and in different ratios as indicated. A: Fractional abundance (%) of wild-type and clarithromycin resistance alleles detected by ddPCR assay. B: Chromatogram from Sanger sequencing of 23S rRNA gene PCR amplicon from water spiked with H. pylori DNA from wild-type and resistant strains (A2142G, A2142C, and A2143G) at the ratios indicated. A box is drawn around position 2142 of the H. pylori 23S rRNA gene: adenosine green peaks, guanosine black peaks. Data is representative of three independent experiments.
Detection and quantification of H. pylori 23S rRNA gene clarithromycin resistance alleles in FFPE gastric tissue
To test the ability of the H. pylori 23S rRNA gene clarithromycin resistance ddPCR assay to detect clarithromycin resistance alleles in DNA extracted from FFPE gastric tissue samples, we first assayed FFPE mouse gastric tissues incubated with H. pylori or sterile BB10 liquid media (control). A wild-type strain and three isogenic clarithromycin-resistant strains were added to the mouse half-stomachs individually and in different ratios. The clarithromycin resistance assay correctly distinguished between wild-type and mutant, and it detected approximately the correct ratios of wild-type and mutant bacteria for all the conditions tested (Figure 2).
Figure 2.
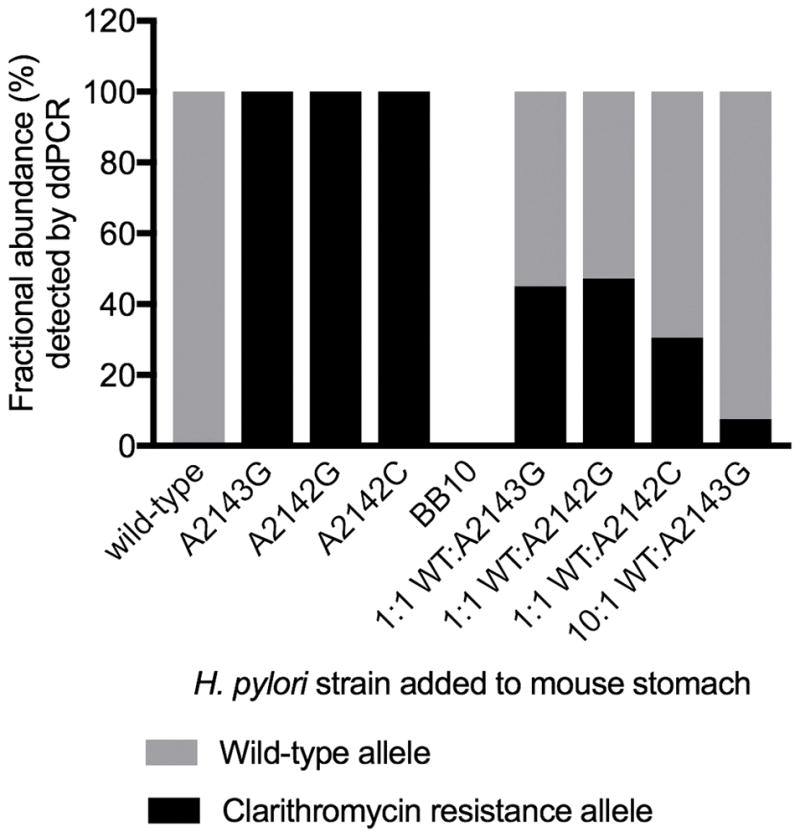
ddPCR assay detects expected frequency of wild-type and clarithromycin resistance alleles in formalin-fixed paraffin-embedded (FFPE) tissue. Fractional abundance (%) of wild-type and clarithromycin resistance alleles was detected by the ddPCR assay in FFPE mouse stomach tissue incubated with either H. pylori or sterile media (BB10). A wild-type H. pylori strain and isogenic mutants having one of three clarithromycin resistance-conferring mutations in the 23S rRNA gene (A2142G, A2142C, and A2143G) were incubated on the lumen side of the mouse stomach. Each strain was incubated individually and in different ratios as indicated. Data is from a single replicate experiment.
We further tested the clarithromycin resistance assay on archived FFPE gastric tissues from six patients at University of Washington Medical Center who had been previously determined to have clarithromycin-resistant H. pylori infection by culture-based susceptibility testing. The ddPCR assay detected clarithromycin resistance alleles in all six patient samples. Four of the patients had only clarithromycin resistance alleles detected. Two patients had a mix of clarithromycin sensitive and resistance alleles; one had 17% resistance alleles and the other had 99.9% resistance alleles.
Rate of clarithromycin resistance alleles detected in Seattle patient population exceeds clinical threshold for the use of clarithromycin in first-line therapy
To assess the prevalence of clarithromycin-resistant H. pylori infection among patients undergoing upper GI endoscopy at University of Washington Medical Center, we performed the clarithromycin resistance ddPCR assay on archived FFPE gastric tissue samples from 110 H. pylori-positive patients. The demographic characteristics of these patients are presented in Table 1. Of these 110 patients, the 23S rRNA gene was detected by ddPCR in 102 (93%) of the samples. To examine whether detection of the H. pylori 23S rRNA gene differed by amount of gastric tissue in the sample, the copy number of the human18S rRNA gene was compared for four samples with no H. pylori 23S rRNA gene detected and four samples each in the lowest, middle, and highest third of 23S rRNA gene copy number. Amount of gastric tissue in the sample as measured by copy number of 18S did not correlate with detection level of H. pylori 23S rRNA gene. Furthermore, three of the four samples that did not have the 23S rRNA gene detected had 18S copy numbers within the range of that measured for the 23S-positive samples (Figure 3). Of the 102 patients with the 23S rRNA gene detected, 57 (56%) had only wild-type 23S rRNA alleles detected, and 45 (44%) had clarithromycin resistance alleles detected. The 45 patients with clarithromycin resistance alleles included 12 with resistance alleles only and 33 with a mix of wild-type and resistance alleles (Table 1). Of those with a mix of wild-type and resistance alleles, the percent of 23S rRNA gene copies having a resistance mutation ranged from 0.02% to 99.7% with a median of 13%.
Figure 3.
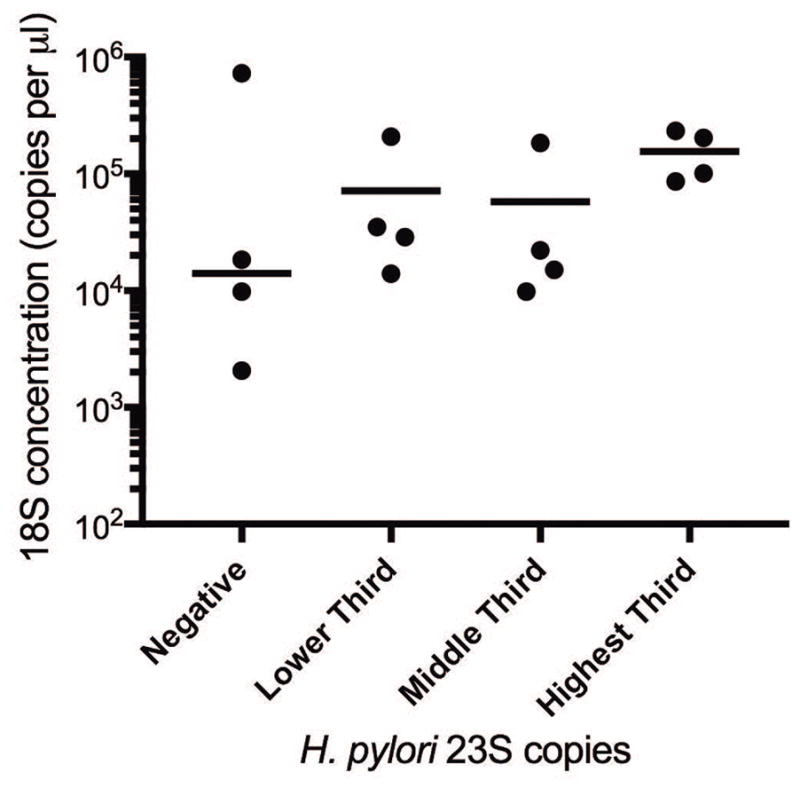
Detection of H. pylori 23S rRNA gene by ddPCR does not depend on amount of gastric tissue in formalin-fixed paraffin-embedded (FFPE) samples measured by human 18S rDNA gene. The human 18S rDNA gene concentration (copies per μL) was compared for four samples with no H. pylori 23S rRNA gene detected and four samples each in the lowest, middle, and highest third of 23S rRNA gene copy number. The median for each group is indicated by a horizontal line.
Prevalence of clarithromycin resistance alleles varied by race and was highest among Asian patients
Age and sex were not significantly different between patients with clarithromycin resistance alleles and patients with only wild-type alleles. While the prevalence of clarithromycin resistance was above 20% in all racial groups, a significantly higher proportion of Asian patients had clarithromycin resistance alleles compared to non-Asian patients. Of the 21 Asian patients, 14 (67%) had clarithromycin resistance alleles compared to 31 (38%) of the 81 non-Asian patients (chi-square, p = 0.02) (Table 2).
Table 2.
Comparison of demographic characteristics between 45 patients with clarithromycin resistance alleles and 57 patients with only the wild-type 23S rRNA allele detected by the ddPCR assay
| Patient Characteristic | No. (%) or mean (SD)
|
p | ||
|---|---|---|---|---|
| Resistance allele n = 45 |
Wild-type allele n = 57 |
|||
| Age | 52 (SD=15) | 55 (SD=16) | 0.4† | |
| Sex | Male | 22 (49%) | 30 (53%) | 0.7‡ |
| Female | 23 (51%) | 27 (47%) | ||
| Race | White | 15 (33%) | 32 (56%) | 0.02‡,§ |
| Black or African-American | 3 (7%) | 6 (11%) | ||
| Asian | 14 (31%) | 7 (12%) | ||
| Hispanic | 8 (18%) | 9 (16%) | ||
| American Indian or Alaska Native | 2 (4%) | 1 (2%) | ||
| Native Hawaiian or Other Pacific Islander | 3 (7%) | 2 (4%) | ||
p-value for Student’s t-test
p-value for chi-square test
p-value for comparison of Asian to non-Asian race
Since the limit of detection of clarithromycin resistance alleles depends on H. pylori 23S rRNA gene copy number, differences in H. pylori load among racial groups could contribute to differences in the proportion of patients with clarithromycin resistance alleles detected. However, the 23S rRNA gene copy number was not significantly different comparing Asians and non-Asians [median 23S rRNA gene copies per μL reaction (range), Asian: 66.8 (3.5 – 512); non-Asian: 44.9 (0.48 – 5407.6); Wilcoxon p-value = 0.6]. Furthermore, among patients with clarithromycin resistance alleles detected, the proportion of the H. pylori 23S rRNA gene copies having clarithromycin resistance mutations was not significantly different comparing the Asian patients and the non-Asian patients [median percent clarithromycin resistance alleles (range), Asian: 39.3 (0.41 – 100); non-Asian 69.1 (0.02 – 100); Wilcoxon p-value = 0.7] (Figure 4).
Figure 4.

Distribution and median values of H. pylori 23S rRNA copy number (Panel A) and percent of clarithromycin resistance alleles (Panel B) of 21 Asian and 81 non-Asian patients with H. pylori 23S rRNA gene detected by ddPCR. H. pylori 23S rRNA gene copy number and percent clarithromycin resistance alleles were compared using the Wilcoxon rank-sum test and p-values are indicated above the graph.
DISCUSSION
We have developed a ddPCR assay for detecting H. pylori clarithromycin resistance alleles in FFPE gastric tissue samples which can be used to retrospectively to examine the prevalence of clarithromycin resistance in a population, or to detect resistance in an individual patient in order to inform treatment. The prevalence of clarithromycin resistance among patients at a Northwestern U.S. hospital as measured by ddPCR analysis (44%) exceeds the proposed cutoff of 20%, indicating that standard Triple Therapy (clarithromycin, amoxicillin and PPI) should not be used as empiric first-line therapy in this patient population, unless patient-specific antibiotic susceptibility testing is first performed.
While the prevalence of clarithromycin resistance was above 20% in all racial groups, a higher prevalence was detected among Asian patients. Based on our analyses, this higher prevalence is not due to higher H. pylori loads as a possible cause of increased detection of clarithromycin resistance alleles. The higher prevalence of clarithromycin resistance alleles among Asian patients may be driven by representation in our study population of foreign-born patients from countries with a higher prevalence of clarithromycin resistance. Unfortunately, country of birth could not be determined in the retrospective review of medical records.
A recent study by Park et al. of four U.S. medical centers observed the overall prevalence of clarithromycin resistance alleles to be 32.3% 15. The prevalence of clarithromycin resistance alleles that we observed in the Seattle patient population (44%) is higher, however, the difference does not reach statistical significance (chi-square p = 0.07). The higher prevalence of clarithromycin resistance mutations in the Seattle patient population could be explained by differences in the assays used to detect clarithromycin resistance mutations. The Park et al. study used Sanger sequencing of the 23S rRNA gene that was PCR amplified from FFPE gastric tissue to categorize the samples as either sensitive or resistant 15. If there were a mixture of wild-type and resistance alleles, this would likely be missed by sequencing. The ddPCR assay used in the present study is very sensitive for detection of rare mutations in a mixed population, detecting clarithromycin resistance mutations that would have been misclassified as wild-type using the Sanger sequencing approach, leading to a higher prevalence of clarithromycin resistance estimated by this study.
Because culture-based antibiotic susceptibility testing of H. pylori is difficult and time-consuming, PCR-based methods to detect clarithromycin resistance-conferring mutations have been developed as an alternative. A real-time PCR assay detecting clarithromycin resistance mutations in gastric biopsies has been developed that can detect mixed infection of both wild-type and resistance alleles, but only if the resistance alleles make up at least 10% of the 23S rRNA gene copies. Failure to detect clarithromycin resistance mutations present in <10% of the H. pylori bacterial population resulted in 82% sensitivity compared to the gold standard of culture-based testing 16. The ddPCR assay is more sensitive for detection of resistance mutations even when present in a small proportion of the H. pylori population. In this study, mixed infection was detected in 33 (32%) of 102 patients, and 16 of these had mixed infections with clarithromycin resistance alleles making up less than 10% of the 23S rRNA gene copies. Unfortunately, culture-based susceptibility testing was not conducted for these patients; therefore, it is not possible to examine whether the ddPCR assay is more sensitive than the culture-based method for detecting the presence of resistance. A recent study by Oikawa et al. used pyrosequencing of gastric wash samples to detect presence of clarithromycin resistance alleles before and 1, 2, and 3 years after treatment and found that the percentage of alleles having resistance mutations increased during the follow-up period, suggesting that mixed infection prior to treatment resulted in eventual recrudescence in these patients 17. Future studies will need to be conducted to determine what proportion and bacterial load of clarithromycin-resistant H. pylori is necessary to cause treatment failure.
This study is limited by the lack of H. pylori culture analysis, antibiotic use history data, and follow-up data to confirm successful H. pylori eradication for the 110 patients. Although the ddPCR assay detected clarithromycin resistance mutations in all six patients for which culture-based susceptibility testing was conducted, future studies that compare the ddPCR assay to culture-based susceptibility testing for patients with clarithromycin-susceptible H. pylori and patients with clarithromycin-resistant H. pylori will be necessary to determine the sensitivity and specificity of the ddPCR assay. Patient data on previous antibiotic use will be useful for better understanding the influence of recent macrolide treatment on H. pylori clarithromycin resistance mutations.
Eradication of H. pylori is necessary for resolving associated diseases. When antibiotic susceptibility is unknown, choice of treatment regimen should be based on local patterns of antibiotic resistance. Methods to examine the prevalence of resistance using archived samples, such as the ddPCR assay presented in this study, are useful for determining whether standard Triple Therapy should be used in a given region. The high prevalence of clarithromycin resistance in the Seattle patient population indicates that alternative regimens should be used as first-line treatment for H. pylori infection, though clarithromycin treatment would likely be successful for the 56% of patients who had only wild-type 23S rRNA alleles. Development of methods to facilitate antibiotic resistance testing will be useful for informing tailored treatment of individual patients, as opposed to empiric treatment, particularly in regions with high clarithromycin resistance
Supplementary Material
Acknowledgments
The authors would like to thank Jutta Fero and Laura Martinez for their technical assistance.
STATEMENT OF INTERESTS
This study was supported in part by grant R01 AI054423 (NRS), K01 DK090103 (ST) and the Genomics Shared Resource of the NIH/NCI Cancer Center Support Grant P30 CA015704 from the NIH. The contents are solely the responsibility of the authors and do not represent the official views of the NIH.
Footnotes
Disclosures: none
References
- 1.Dooley CP, Cohen H, Fitzgibbons PL, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989;321:1562–6. doi: 10.1056/NEJM198912073212302. [DOI] [PubMed] [Google Scholar]
- 2.Gerrits MM, van Vliet AH, Kuipers EJ, et al. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 3.Ford AC, Gurusamy KS, Delaney B, et al. Eradication therapy for peptic ulcer disease in Helicobacter pylori-positive people. Cochrane Database Syst Rev. 2016;4:CD003840. doi: 10.1002/14651858.CD003840.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen LT, Lin JT, Tai JJ, et al. Long-term results of anti-Helicobacter pylori therapy in early-stage gastric high-grade transformed MALT lymphoma. J Natl Cancer Inst. 2005;97:1345–53. doi: 10.1093/jnci/dji277. [DOI] [PubMed] [Google Scholar]
- 5.Stathis A, Chini C, Bertoni F, et al. Long-term outcome following Helicobacter pylori eradication in a retrospective study of 105 patients with localized gastric marginal zone B-cell lymphoma of MALT type. Ann Oncol. 2009;20:1086–93. doi: 10.1093/annonc/mdn760. [DOI] [PubMed] [Google Scholar]
- 6.Wong BC, Lam SK, Wong WM, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291:187–94. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 7.Correa P, Fontham ET, Bravo JC, et al. Chemoprevention of gastric dysplasia: randomized trial of antioxidant supplements and anti-helicobacter pylori therapy. J Natl Cancer Inst. 2000;92:1881–8. doi: 10.1093/jnci/92.23.1881. [DOI] [PubMed] [Google Scholar]
- 8.Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646–64. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 9.Talarico S, Safaeian M, Gonzalez P, et al. Quantitative Detection and Genotyping of Helicobacter pylori from Stool using Droplet Digital PCR Reveals Variation in Bacterial Loads that Correlates with cagA Virulence Gene Carriage. Helicobacter. 2016;21:325–33. doi: 10.1111/hel.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone GG, Shortridge D, Flamm RK, et al. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–8. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 12.Versalovic J, Osato MS, Spakovsky K, et al. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–6. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 13.Baltrus DA, Amieva MR, Covacci A, et al. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447–8. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JY, Dunbar KB, Mitui M, et al. Helicobacter pylori Clarithromycin Resistance and Treatment Failure Are Common in the USA. Dig Dis Sci. 2016;61:2373–80. doi: 10.1007/s10620-016-4091-8. [DOI] [PubMed] [Google Scholar]
- 16.Schabereiter-Gurtner C, Hirschl AM, Dragosics B, et al. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol. 2004;42:4512–8. doi: 10.1128/JCM.42.10.4512-4518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oikawa R, Watanabe Y, Miyamoto S, et al. Enrichment of Helicobacter pylori mutant strains after eradication therapy analyzed by gastric wash-based quantitative pyrosequencing. Tumour Biol. 2017;39:1010428317734865. doi: 10.1177/1010428317734865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.