Abstract
Rab GTPases are key regulators of membrane trafficking, and many are activated by guanine nucleotide exchange factors bearing a differentially expressed in normal and neoplastic cells (DENN) domain. By activating the small GTPase Rab12, DENN domain–containing protein 3 (DENND3) functions in autophagy. Here, we identified a structural domain (which we name PHenn) containing a pleckstrin homology subdomain that binds actin and is required for DENND3 function in autophagy. We found that a hydrophobic patch on an extended β-turn of the PHenn domain mediates an intramolecular interaction with the DENN domain of DENND3. We also show that DENND3 binds actin through a surface of positively charged residues on the PHenn domain. Substitutions that blocked either DENN or actin binding compromised the role of DENND3 in autophagy. These results provide new mechanistic insight into the structural determinants regulating DENND3 in autophagy and lay the foundation for future investigations of the DENN protein family.
Keywords: guanine-nucleotide exchange factor (GEF), Rab, autophagy, structural biology, structure-function, actin, DENN domain, DENND3, GTPase, PH domain, PHenn domain, Rab12, intramolecular interaction
Introduction
Small GTPases, key regulators of cellular function, are found in five classes, Ras, Rho, Ran, Rab, and Arf (1, 2). Ras and Rho family members function in signal transduction, Ran mediates nucleocytoplasmic transport, and Arf and Rab proteins are regulators of intracellular vesicle trafficking (1, 2). All of these small GTPases cycle between inactive GDP-bound and active GTP-bound forms. In their GTP-bound form, GTPases interact with effectors that carry out their cellular functions. Guanine nucleotide exchange factors (GEFs)4 catalyze the exchange of GDP to GTP to activate small GTPases, and as such, GEF activity is often tightly regulated by physiological activities. For example, through conformational changes, the catalytic activity of GEFs can be released from autoinhibition, and through subcellular translocation, GEFs can be recruited to specific membranes to recruit and activate their substrates. Phosphorylation is another mechanism to regulate GEFs in response to specific physiological stimulation (1, 3).
With 26 members, many of them involved in human diseases, the differentially expressed in normal and neoplastic cells (DENN) domain–containing proteins are the largest family of Rab GEFs (4–8). We previously demonstrated that DENN domain-containing protein 3 (DENND3), a GEF for Rab12, is regulated through phosphorylation during macroautophagy (9, 10). Macroautophagy, hereafter referred to as autophagy, is a conserved process functioning in degradation of macromolecules. Upon specific stimulation, such as starvation, selective cellular materials are sequestered into a double membrane organelle, the autophagosome. Following fusion with lysosomes, the materials enclosed in the autophagosome are degraded, and the resulting products, such as amino acids, are released into the cytosol for reuse (11). DENND3 plays a vital role in this process. Upon starvation, the most upstream kinase in the autophagy process, Unc-51–like kinase (ULK), phosphorylates DENND3, activating its GEF activity toward Rab12. Active Rab12 facilitates the traffic of autophagosomes toward lysosomes (9). In addition to ULK-mediated phosphorylation, DENND3 is regulated by an intramolecular interaction that autoinhibits its GEF activity at the steady state (12, 13). Phosphorylation of Tyr-940 within DENND3 counters the intramolecular interaction and renders an open conformation of DENND3, resulting in enhanced GEF activity toward Rab12 (12, 13).
Tyr-940 resides in a region of DENND3 thought to be unstructured. Through generation of a series of protein constructs followed by expression studies, we identified a stable region surrounding Tyr-940 and solved its 3D structure at 1.85 Å resolution. Based on its structure and function, we propose the term PHenn, for PH-like domain binding to a DENN domain. We demonstrate that the PHenn domain functions as a novel actin-binding module and that the actin and DENN domain interactions are required for the role of DENND3 in autophagy.
Results and discussion
Structure of a PHenn domain of DENND3
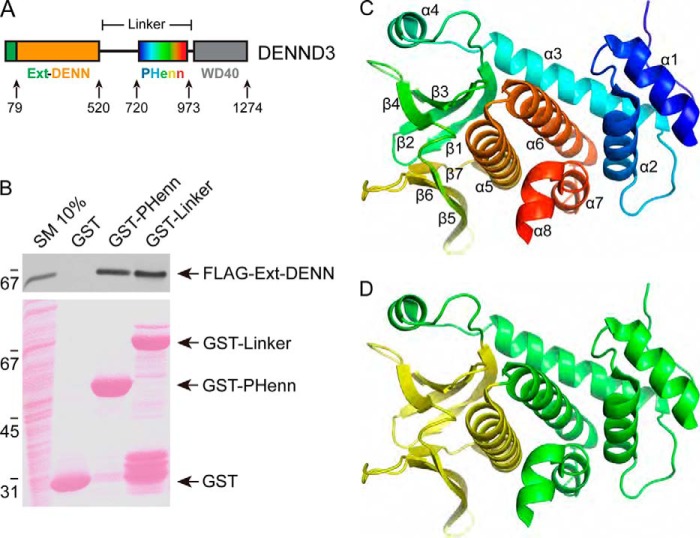
The N terminus of DENND3 has a DENN domain, which is preceded by an N-terminal 79-amino acid extension. We refer to the two regions collectively as the extended-DENN (Ext-DENN) domain. The C terminus contains a WD40 repeats domain (Fig. 1A). The region between these domains is termed the linker. We previously demonstrated an intramolecular interaction between Ext-DENN and the linker that autoinhibits DENND3 GEF activity at steady state (12). Importantly, the phosphorylation status of Tyr-940 near the C terminus of the linker regulates this intramolecular interaction. Using PSIPRED, a secondary structure prediction tool (14), we noticed that the region surrounding Tyr-940 contains clustered α-helices and β-sheets, a sign of a structured domain. We thus sought to solve the structure of the linker through crystallography. The purified full-length linker failed to produce protein amenable to crystal formation. We thus generated a series of constructs involving progressive deletion of the N-terminal side of the linker until we mapped a minimal region that retained binding to the Ext-DENN and was suitable for structural studies. The construct, encoding residues 720–973, bound to FLAG-Ext-DENN in pulldown experiments to the same extent as full-length linker (Fig. 1B). Crystals generated from the 720–973 construct diffracted to better than 2 Å resolution. To determine the phases, the protein was labeled with selenomethionine, and the structure was solved using the single-wavelength anomalous dispersion method and refined to 1.85 Å. Statistics of data collection and refinement are shown in Table S1. The structure, termed a PHenn domain, is present in two copies in the asymmetric unit. The molecules are very similar to each other with a root mean square deviation of atomic positions of 0.63 Å over 206 Cα atoms (Fig. S1A). We observed residues 722–946 (chain A) and 723–947 (chain B) in the electron density map. The N-terminal residues 720–721 and the C-terminal residues 948–973 are absent in the model due to disorder. The structure shows a large helical bundle capped by a sandwich of two β-sheets stacked perpendicularly to each other (Fig. 1C). The β-sheets in combination with the following helix form a characteristic pleckstrin homology (PH)-like fold (yellow in Fig. 1D). The PH-like fold is an integral part of the larger domain, as it is preceded by four α-helices and followed by another three α-helices, all of which make up the helical bundle (Fig. 1D).
Figure 1.
Structure of the PHenn domain of DENND3. A, schematic diagram of DENND3 with an N-terminal DENN domain with its preceding extension, a C-terminal WD40 domain, and the PHenn domain 720–973, which is the minimal fragment required for binding the Ext-DENN. B, HEK-293T cells were transfected with FLAG-Ext-DENN, and lysates were incubated with GST, GST-PHenn(720–973), or GST-linker amino acids 538–973 coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG antibody. An aliquot of the lysate (starting material; SM) equal to 10% of that added to the beads was analyzed in parallel. C, schematic representation of the PHenn domain of DENND3 in rainbow colors, from blue at the N terminus to red at the C terminus. Secondary structure elements are labeled. D, the PHenn domain is composed of a PH domain (yellow) surrounded by N- and C-terminal helices (green).
Previous studies demonstrated a key role for Tyr-940 in regulating the GEF activity of the DENN domain via an intramolecular interaction with the Ext-DENN region (12). Tyr-940 is positioned at the surface made by the junction of four helices on the opposite site of the PH subdomain (Fig. S1B). The side chain of Tyr-940 is solvent-accessible and is primed for contacts with the Ext-DENN (Fig. S1B). Moreover, when evolutionarily conserved regions of the PHenn domain were mapped onto the surface of the structure (gray, poorly conserved regions; green, highly conserved regions), we noticed that one of the most highly conserved surfaces is centered on Tyr-940 (Fig. S1C). This further highlights the physiological relevance of the intramolecular interaction.
A hydrophobic β-turn in the PHenn domain is involved in the intramolecular interaction
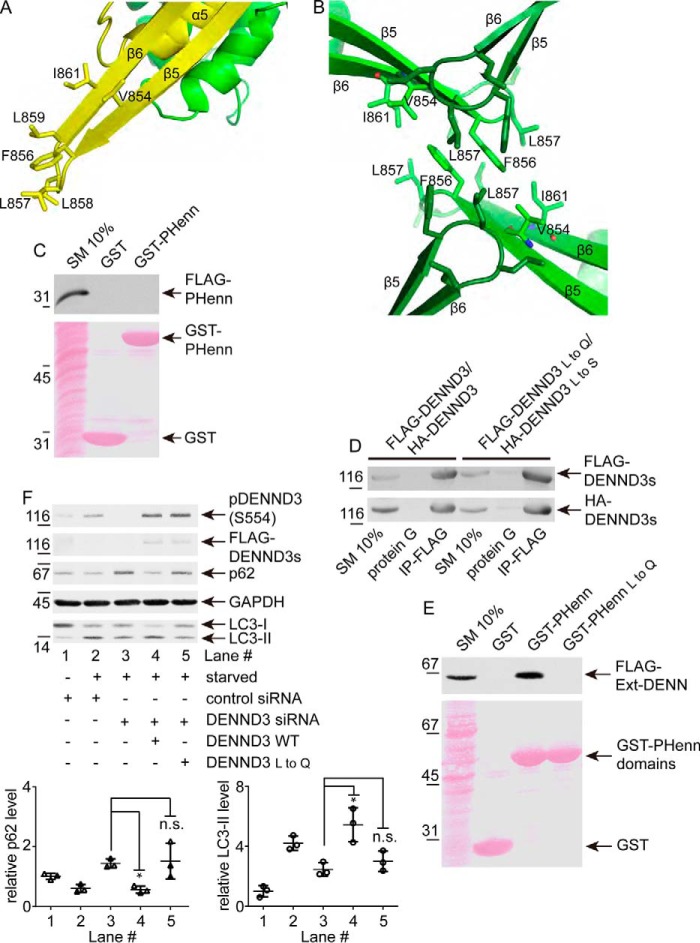
The most unusual structural feature of the PHenn domain is a cluster of hydrophobic residues (Phe-856, Leu-857, Leu-858, and Leu-859) that make up a hydrophobic β-turn that protrudes from the structure (Fig. 2A). On the outside surface of the β-sheet, the hydrophobic surface extends on either side by Val-854 and Ile-861 (Fig. 2A). Interestingly, in the crystal, the hydrophobic β-turn is stabilized by interactions between DENND3 molecules (Fig. 2B). We previously observed that DENND3 self-associates (12) and wondered whether the PHenn domain contributes to DENND3 oligomerization. We thus performed pulldown experiments using the GST-PHenn domain with cell lysates expressing FLAG-tagged PHenn domain but detected no interaction (Fig. 2C). To test a possible role of the hydrophobic β-turn in the context of the full-length protein, we mutated both Leu-857 and Leu-858 to either glutamine or serine (labeled L to Q or L to S in Fig. 2). The interaction of full-length FLAG-tagged and HA-tagged DENND3 proteins was not altered by the mutations (Fig. 2D). These results indicate that the hydrophobic β-turn is not involved in the oligomerization of DENND3. The interaction between the PHenn domains seen in the crystal is probably limited to the process of crystal formation.
Figure 2.
A hydrophobic β-turn in the PHenn domain is required for DENN domain binding but not DENND3 oligomerization. A, PHenn β-strands 5 and 6 form an extended hydrophobic surface. B, the β5-β6 loops are involved in hydrophobic contacts between adjacent PHenn domains in the protein crystals. C, the PHenn domain does not oligomerize. HEK-293T cells were transfected with FLAG-PHenn domain 716–973, and lysates were incubated with GST or GST-PHenn domain 716–973 coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG antibody. An aliquot of the lysate (starting material; SM) equal to 10% of that added to the beads was analyzed in parallel. D, mutations in the PHenn hydrophobic loop do not impair DENND3 oligomerization. Lysates from HEK-293T cells co-transfected with FLAG-DENND3 and HA-DENND3 or mutants FLAG-DENND3 L857Q/L858Q (L to Q) and HA-DENND3 L857S/L858S (L to S) were incubated with protein G beads alone or protein G beads coupled to anti-FLAG antibody (IP-FLAG). Proteins bound specifically to the beads were processed for Western blot with anti-FLAG and anti-HA antibodies. An aliquot of the cell lysate (starting material) equal to 10% of that added to the beads was analyzed in parallel. E, mutations in the hydrophobic loop impair DENN domain binding. HEK-293T cells were transfected with FLAG-Ext-DENN, and lysates were incubated with GST, GST-PHenn domain wildtype, or GST-PHenn domain with the mutation of L857Q/L858Q coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-FLAG antibody. An aliquot of the lysate (starting material) equal to 10% of that added to the beads was analyzed in parallel. F, mutations in the hydrophobic loop impair DENND3-dependent autophagy. Cells were treated with control siRNA or with siRNA for human DENND3. Additionally, cells treated with DENND3 siRNA were subsequently mock-transfected or transfected with wildtype mouse DENND3 or mouse DENND3 with L857Q/L858Q mutations. The cells were subsequently starved with EBSS, and then lysates were processed for Western blotting with anti-pSer-554 of DENND3, anti-FLAG, anti-p62, anti-LC3, or anti-GAPDH antibody. Relative p62 and LC3-II levels were determined from three repeats. Bars, mean ± S.D. (error bars). Statistical analysis employed one-way analysis of variance followed by Tukey's post hoc test. *, p < 0.05.
To test whether the hydrophobic β-turn is involved in the intramolecular interaction between PHenn domain and the Ext-DENN, we performed pulldown experiments using the GST-PHenn domain with cell lysates expressing FLAG-tagged Ext-DENN. As shown in Fig. 2E, the binding between Ext-DENN and the PHenn domain was abolished by the L857Q/L858Q double mutation in the PHenn domain, suggesting that the hydrophobic β-turn is critical for the intramolecular interaction.
Release of the intramolecular interaction, leading to up-regulation of GEF activity, would leave the hydrophobic cluster exposed. In some cases, clustered hydrophobic residues insert into lipid bilayers, such as what is seen in the N-BAR domain of endophilin, which anchors into membranes through hydrophobic residues in its N-terminal amphipathic helix (15, 16). We wondered whether the hydrophobic β-turn would insert into membranes upon release of the intramolecular interaction. We thus compared the membrane pool of wildtype DENND3 with that of DENND3 with L857Q/L858Q mutations using subcellular fractionation experiments. Postnuclear supernatants from lysates expressing the DENND3 proteins were centrifuged at 200,000 × g, and the membrane (pellet) and cytosolic (supernatant) fractions were analyzed. Whereas Na+K+-ATPase, an integral membrane protein was found in the pellet and GAPDH, a cytosolic protein, was found mainly in the supernatant, the L857Q/L858Q mutant was found mainly in the membrane fraction, the same as wildtype DENND3 (Fig. S2, A and B). Thus, the hydrophobic β-turn does not appear to be a determinant of membrane association of DENND3.
PH domains are well-known for their property to bind phosphatidylinositol (17). We thus tested for the association of the PHenn domain with an array of lipid species using the GST-PHenn fusion protein overlay assay with hydrophobic membrane strips spotted with various lipid species (Fig. S2C). As for GST, the GST-PHenn domain failed to bind any lipid species, whereas the GST-PH domain of PLC-δ1 bound to phosphatidylinositol 4,5-bisphosphate as expected (18) (Fig. S2C). This suggests that the PHenn domain of DENND3 does not possess lipid-binding activity.
To investigate whether the hydrophobic β-turn is involved in the role of DENND3 in autophagy, we performed knockdown/rescue experiments. As shown previously (9), upon starvation, reduced p62 levels and increased LC3 lipidation levels indicate the induction of autophagy (Fig. 2F, lanes 1 and 2). Knockdown of DENND3 compromises autophagy, as indicated by the reduced level of lipidated LC3 and the elevated level of p62 (Fig. 2F, lane 3). Importantly, the inhibited autophagy resulting from DENND3 knockdown is rescued with re-expression of wildtype DENND3 (Fig. 2F, lane 4), but not with the L857Q and L858Q mutant, which has compromised hydrophobicity at the β-turn of the PHenn domain (Fig. 2F, lane 5). This suggests that the hydrophobic β-turn is involved in the role of DENND3 in starvation-induced autophagy, supporting the hypothesis that, besides the intramolecular interaction, the β-turn interacts with an unknown binding partner when it is not occupied by the Ext-DENN region and that this interaction is required for the proper function of DENND3 in autophagy.
The PHenn domain of DENND3 is a novel actin-binding module
Structure-based alignment using the Dali server (19), which compares a query protein 3D structure against structures in the Protein Data Bank, revealed that the PHenn domain has structural similarity to the FERM domain from sorting nexin-17 (20) and focal adhesion kinase (21) (Fig. S3). FERM domains consist of three subdomains, a ubiquitin-like fold, an acyl-CoA–binding protein–like fold, and a PH domain fold (22). FERM domains are frequently found in actin-binding proteins, and it is reported that the isolated PH subdomain of FERM domains binds to filamentous actin (F-actin) (23). We thus tested whether the PHenn domain of DENND3 binds to F-actin. Purified GST-PHenn domain, GST alone, or purified actinin, a known F-actin–binding protein, was incubated with purified, preassembled F-actin and then subjected to ultracentrifugation, which is capable of pelleting F-actin but not monomeric actin. After centrifugation in the absence of F-actin, purified GST, GST-PHenn domain, and actinin remained in the supernatant (Fig. 3A, lanes 1, 3, and 13). In contrast, in the presence of F-actin, the DENND3 GST-PHenn domain sedimented (Fig. 3A, lane 10), as did actinin (Fig. 3A, lane 12), but GST remained in the supernatant (Fig. 3A, lane 8). Purified full-length DENND3 also co-sedimented with F-actin (Fig. 3B), and the GST-PHenn domain bound to endogenous actin from rat tissue lysates (Fig. 3C). Together, these data demonstrate that the PHenn domain is a novel actin-binding module.
Figure 3.
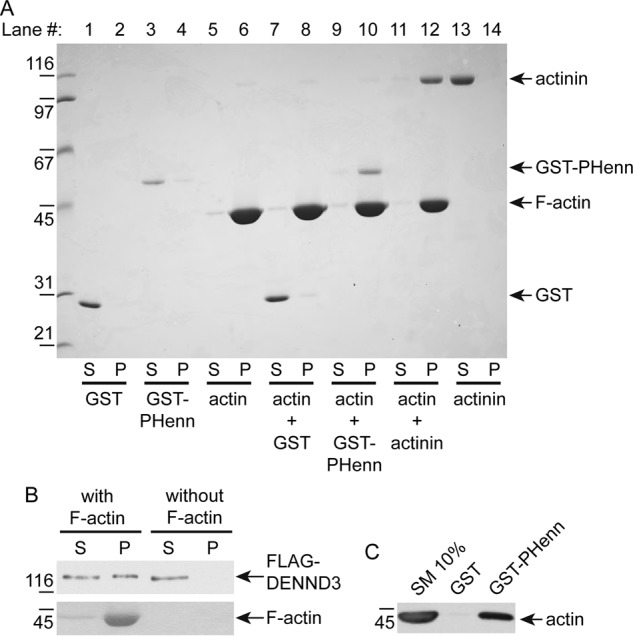
The PHenn domain of DENND3 is a novel actin-binding module. A, purified PHenn domain binds to F-actin. GST-PHenn domain 720–973 (codon-optimized construct), negative control GST, or positive control actinin was incubated with or without F-actin, followed by centrifugation capable of pelleting F-actin. Protein bound to F-actin was co-sedimented during the centrifugation. Supernatant (S) and pellet (P) were processed for SDS-PAGE, and gel was stained by Coomassie Blue. B, full-length DENND3 binds to F-actin. Purified FLAG-tagged DENND3 was cleared by centrifugation at 240,800 × g for 15 min at 4 °C and incubated with or without F-actin, followed by centrifugation at 148,300 × g for 15 min at 24 °C. Protein bound to F-actin co-sedimented during the centrifugation. Supernatant (S) and pellet (P) were processed for Western blotting with an anti-FLAG antibody. C, GST-PHenn binds actin in tissue lysates. Rat lung lysates were incubated with GST or GST-PHenn domain 720–973 (codon-optimized construct) coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-actin antibody. An aliquot of the lysate (starting material; SM) equal to 10% of that added to the beads was analyzed in parallel.
Actin binding is required for the function of DENND3 in autophagy
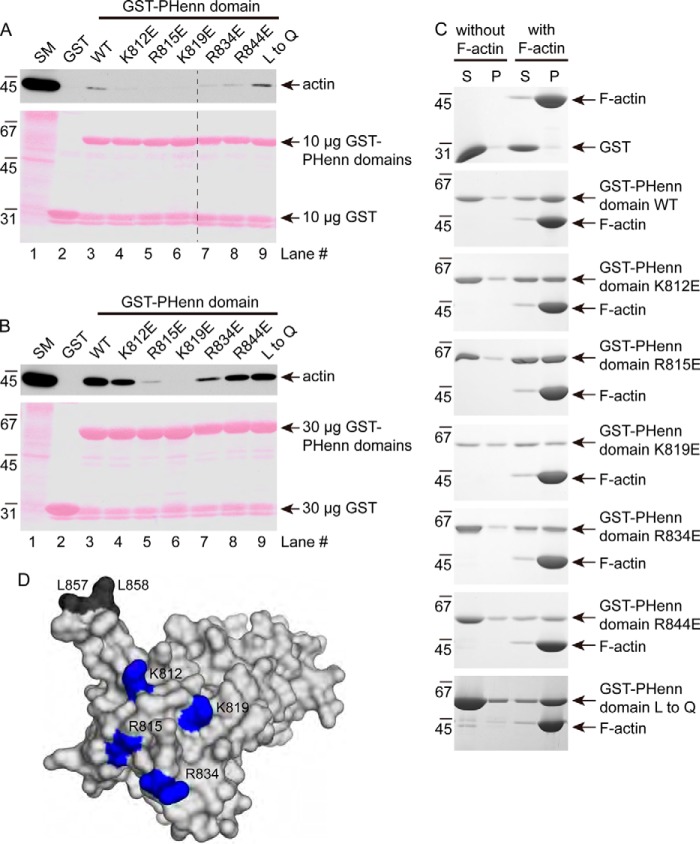
To investigate whether actin binding is required for the role of DENND3 in autophagy, we sought to map the interaction between actin and DENND3 through site-directed mutagenesis. Bruton's tyrosine kinase interacts with F-actin through basic amino acids close to the N terminus of its PH domain (24). We thus mutated surface-exposed Arg and Lys residues around the N-terminal region of the first β-sheet of the PHenn domain of DENND3. We screened for actin binding by pulldown experiments with GST fusion proteins. As shown in Fig. 4A, 10 μg of the GST-PHenn domain with mutations at Lys-812, Arg-815, Lys-819, Arg-834, or Arg-844 has compromised actin binding compared with wildtype protein. In contrast, mutation of both Leu-857 and Leu-858 to glutamine in the β-turn of the PHenn domain does not influence binding (Fig. 4A, compare lane 3 with lane 9). Even with increased amounts of the GST-PHenn domain up to 30 μg, R815E and K819E still exhibit clear defects in binding to actin (Fig. 4B). We then performed actin sedimentation experiments with these mutants (Fig. 4C). Whereas the majority of wildtype PHenn domain and of the L857Q/L858Q mutant co-sediments with F-actin, the mutants of the basic residues have less co-sedimentation. Among these mutants, R844E, which resides on a loop connecting the first and second β-sheets, has only a subtle reduction of actin binding (Fig. 4, A and C). The basic residues with obvious defects in actin binding cluster on a surface of the PHenn domain, rendering a binding platform (Fig. 4D). To test the role of actin binding in autophagy, we performed knockdown/rescue experiments. As shown previously, knockdown of DENND3 counters the change of p62 and LC3 induced by starvation, indicating that autophagy is inhibited (Fig. 5A, lane 3). The compromised autophagy resulting from DENND3 knockdown is rescued with re-expression of wildtype DENND3 (Fig. 5A, lane 4), but not with the Arg-815 or Lys-819 mutants (Fig. 5A, lanes 5 and 6). These data demonstrate that actin binding is required for the role of DENND3 in autophagy.
Figure 4.
Mapping for actin-binding sites of the PHenn domain. A, positively charged residues in PHenn are required for actin binding. Rat lung lysates were incubated with 10 μg of GST, GST-PHenn domain 716–973, or GST-PHenn domain with mutation at Lys-812, Arg-815, Lys-819, Arg-834, Arg-844, or Leu-857 and Leu-858 (L to Q)) that was coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-actin antibody. An aliquot of the lysate (starting material; SM) equal to 2.5% of that added to the beads was analyzed in parallel. B, rat lung lysates were incubated with 30 μg of GST or GST-PHenn domains. Experiments were carried out as in A. C, loss of basic residues impairs co-sedimentation with actin. Purified GST, GST-PHenn domain (residues 716–973) wildtype, or GST-PHenn domain with mutation at Lys-812, Arg-815, Lys-819, Arg-834, Arg-844, or Leu-857 and Leu-858 was incubated with or without F-actin, followed by centrifugation capable of pelleting F-actin. Supernatant (S) and pellet (P) were processed for SDS-PAGE, and the gel was stained by Coomassie Blue. D, a surface model of the PHenn domain highlighting residues Lys-812, Arg-815, Lys-819, and Arg-834 with blue and Leu-857 and Leu-858 with dark gray.
Figure 5.
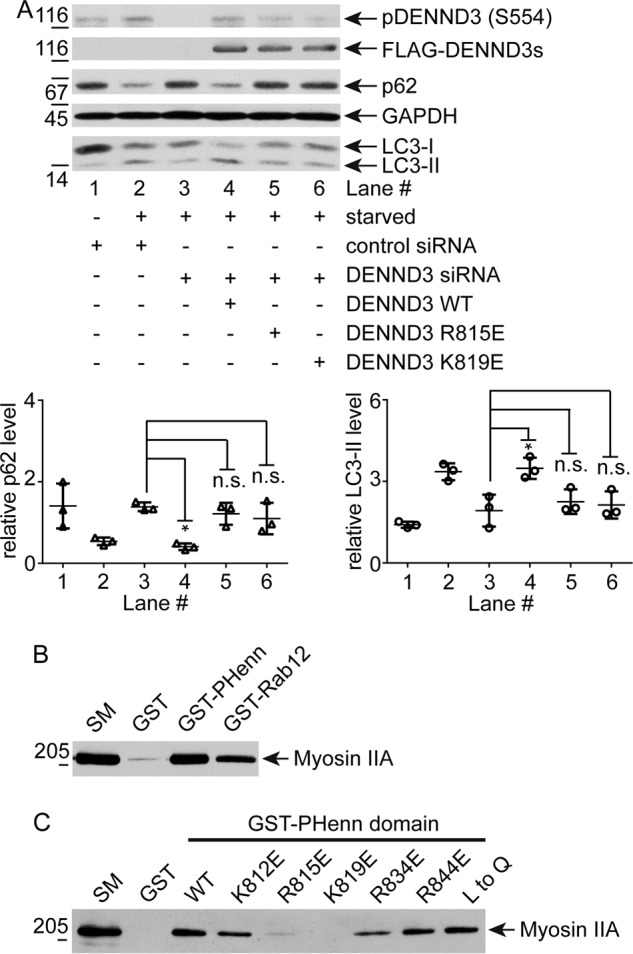
Actin binding is required for DENND3-mediated autophagy. A, defect in actin binding impairs DENND3-dependent autophagy. Cells were treated with control siRNA or with siRNA for human DENND3. Cells treated with DENND3 siRNA were subsequently mock-transfected or transfected with wildtype mouse DENND3 or mouse DENND3 with Arg-815/Lys-819 mutation. The cells were subsequently starved with EBSS, and then lysates were processed for Western blotting with anti-pSer-554 of DENND3, anti-FLAG, anti-p62, anti-LC3, or anti-GAPDH antibody. Relative p62 and LC3-II level were determined from three repeats. Bars, mean ± S.D. (error bars). Statistical analysis employed one-way analysis of variance followed by Tukey's post hoc test. *, p < 0.05; n.s., not significant. B, the DENND3 PHenn domain binds myosin. Rat lung lysates were incubated with GST, GST-PHenn domain 720–973 (codon-optimized construct), or GST-Rab12 coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-myosin IIA antibody. C, actin binding is required for the DENND3 PHenn domain to interact with myosin IIA. Rat lung lysates were incubated with 30 μg of GST, GST-PHenn domain, or GST-PHenn domain with mutation at Lys-812, Arg-815, Lys-819, Arg-834, Arg-844, or Leu-857 and Leu-858 (L to Q) that was coupled to glutathione-Sepharose beads. Proteins specifically bound to the beads were processed for Western blotting with anti-myosin IIA antibody.
Actin has essential roles in multiple aspects of autophagy, which includes providing tracks for myosin-based traffic (25). Previously, we had predicted that Rab12, the substrate of DENND3 that engages in autophagosome trafficking, associates with a motor protein to achieve its role in autophagy (9). Because we identified the PHenn domain of DENND3 as a novel actin-binding module, we tested whether Rab12 binds to actin motor protein myosin. Among the three non-muscle myosins, IIA, IIB, and IIC, that we tested (26), endogenous myosin IIA has robust binding with the GST-Rab12 fusion protein (Fig. 5B). In addition, we found that the GST-PHenn domain of DENND3 interacts with endogenous myosin IIA (Fig. 5B). Interestingly, PHenn domain mutants with defects in actin binding also have compromised interactions with myosin IIA (Fig. 5C), which suggests that actin may bridge the binding between DENND3 and myosin IIA. Upon starvation, ULK-mediated phosphorylation of DENND3 activates Rab12, which localizes at autophagosome and facilitates autophagosome trafficking (9). Considering that ULK-mediated activation of myosin II is also required for starvation-induced autophagy (27), autophagosome-associated Rab12 may traffic along actin through interacting with myosin IIA. In this way, this functional protein complex of myosin IIA, DENND3, and Rab12 lies downstream of ULK upon starvation. The close proximity within the protein complex may facilitate the immediate coupling of Rab12 with myosin IIA to transport autophagosome along actin to fuse with lysosome/endosome after Rab12 is recruited and activated by DENND3 during autophagy.
The PH domain is well-known as a phosphatidylinositol lipid-binding module (17). In fact, it has diverse functions in GEFs. For example, the PH domain of Ras-GRF binds to βγ subunits of heterotrimeric G proteins (28), and the PH domain of PDZ-RhoGEF binds to GTP-bound RhoA (29). There are often Dbl homology (DH)/PH domain tandems in GEFs for Rho family GTPases. The PH domain of Sos binds to the DH domain, a GEF domain, inhibiting the GEF activity of Sos toward Rac (30, 31). Here, for the first time, we find that a PH-like module binds to a DENN domain and thus autoinhibits the GEF activity, suggesting a common role of PH domains across different GEF families.
The intramolecular interaction within DENND3 may be conserved among other DENN proteins that possess PH domains, such as MTMR5 and MTMR13, whose mutation links to Charcot-Marie-Tooth disease (32). It will be interesting to test whether PH domains in these proteins also bind to actin. Our findings provide a paradigm for future investigations of the large family of DENN proteins.
Experimental procedures
Antibodies and reagents
FLAG and HA antibodies were from Sigma and BioLegend, respectively. Myosin IIA and LC3 antibodies were from Cell Signaling Technology. The rabbit polyclonal antibody against pSer-554 of DENND3 was custom-made under contract with Phosphosolutions. Na+K+-ATPase antibody was from Millipore. GST antibody was purchased from Sigma. Mouse DENND3 cDNA was from Imagenes. Amino acid region 716–973 of DENND3 was cloned into pGEX vector, creating the GST-PHenn domain. FLAG-DENND3, FLAG-Ext-DENN, GST-linker, etc. were generated as described previously (9, 12). Leu-857 and Leu-858 mutations of FLAG-DENND3 and the GST-PHenn domain were created by the QuikChange site-directed mutagenesis kit from Stratagene, the same as the mutation of Lys-812, Arg-815, Lys-819, Arg-834, or Arg-844 of the GST-PHenn domain.
Protein expression and purification for crystallography
Mouse DENND3 fragment 720–973 was codon-optimized for Escherichia coli expression (BioBasic) and inserted into pGEX-6P-1 vector. The plasmid was transformed into BL21 (DE3) and plated on LB-agar with ampicillin (100 mg/liters) for selection. A single colony was inoculated in 20 ml of LB medium and incubated in a 37 °C shaker overnight. The overnight culture was then inoculated into 1 liter of LB medium and grown at 37 °C. When A600 reached 0.8, the cell culture was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 18 h. For production of selenomethionine-labeled protein, the expression plasmid was transformed into the E. coli methionine auxotroph strain DL41 (DE3), and the protein was produced using LeMaster medium. After expression, the cell culture was pelleted at 7000 × g for 20 min and resuspended in PBS buffer (10 mm sodium phosphate, pH 7.4, 137 mm NaCl, 2.7 mm potassium chloride). Cells were lysed by sonication and centrifuged at 30,000 × g for 45 min. The supernatant was loaded onto glutathione-Sepharose resin (Qiagen, Valencia, CA), which was pre-equilibrated with PBS buffer. The protein/resin mixture was incubated at 4 °C for 30 min, followed by washes with PBS buffer. Subsequently, the GST-tagged protein was eluted with 20 mm glutathione, and the GST tag was cleaved with PreScission Protease, followed by purification using a size-exclusion Superdex 75 column (GE Healthcare), equilibrated with 20 mm MES, pH 6.5, 150 mm NaCl, 3 mm DTT.
Crystallization
DENND3(720–973) was concentrated to ∼10 mg/ml. Crystallization screens were performed in 24-well plates in a hanging-drop format using commercial Qiagen screens. Promising conditions were further explored by systematic modifications of the initial conditions within a narrow range. The best native crystals for DENND3(720–973) were obtained at 20 °C by equilibrating a 0.8-μl drop of protein at 10 mg/ml in 20 mm MES, pH 6.5, 150 mm NaCl, 3 mm DTT, with 0.8 μl of reservoir solution containing 0.24 m sodium malonate, pH 7.0, 20% PEG 3350, suspended over 1 ml of reservoir solution. The selenomethionine-labeled crystals for were obtained in similar condition. For cryoprotection, crystals were transferred into crystallization solution containing 25% (w/v) ethylene glycol. For data collection, crystals were picked up in a nylon loop and flash-frozen in an N2 cold stream (Oxford Cryosystem).
Structure determination and refinement
The selenium single-wavelength anomalous dispersion data set was collected using a single-wavelength (0.98 Å) regime on an ADSC Quantum-210 CCD detector (Area Detector Systems Corp.) at beamline A1 at the Cornell High-Energy Synchrotron Source (CHESS) (Table S1). Data processing and scaling were performed with HKL2000 (33). The anomalously scattering selenium substructure was determined using the program AUTOSOL in the PHENIX suite (34), which built ∼80% of the model. The model was further extended manually using the program Coot (35) and was improved by multiple cycles of refinement using the program PHENIX (34). Coordinates have been deposited in the RCSB Protein Data Bank with accession code 6B3Y.
Immunoprecipitation
Cells were collected in HEPES lysates buffer (20 mm HEPES, pH 7.4, 10 mm sodium fluoride, 0.5 mm sodium orthovanadate, 60 nm okadaic acid, 100 mm sodium chloride, 1% Triton X-100, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). Following 10 min on ice, lysates were spun at 238,700 × g for 15 min. The supernatant was incubated for ∼3 h at 4 °C with antibodies coupled to protein A or G Sepharose. Beads were subsequently washed three times with HEPES lysates buffer, and processed for SDS-PAGE. Samples were then analyzed through Western blotting.
Pulldown assays
Cell lysates prepared in the same way as described in immunoprecipitation were incubated for ∼3 h at 4 °C with GST or GST fusion proteins coupled to glutathione-Sepharose. The samples were subsequently washed and prepared for Western blotting as described under “Immunoprecipitation.”
Subcellular fractionation
HEK-293T cells were transfected overnight with FLAG-tagged wildtype DENND3 or L857Q/L858Q double mutant. Cells were then washed in PBS and collected in HEPES buffer (20 mm HEPES, pH 7.4, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). After a 10-min incubation on ice, lysates were passed through syringes with 25-gauge needles five times under positive pressure and then spun at 800 × g for 10 min. The supernatant was adjusted to 1.2 mg/ml and then subject to centrifugation at 200,000 × g (TLA-100 Beckman rotor) for 30 min at 4 °C. The resulting supernatant and pellet were processed for SDS-PAGE and Western blotting.
Lipid overlay assay
This assay was performed according to the manual of the PIP Strip membrane (Echelon Biosciences) with slight modifications. Briefly, the PIP Strip membrane was covered with blocking buffer (PBS, 0.1% (v/v) Tween 20, and 3% BSA) for 1 h at room temperature. The blocking buffer was changed for 1.8 μg/ml GST/GST-PHenn domain of DENND3 or 0.5 μg/ml GST-PH domain of PLC-δ1 (Echelon Biosciences) in blocking buffer for 1 h at room temperature and then anti-GST primary antibody, followed by secondary HRP antibody, and a wash with PBST buffer (PBS and 0.1% (v/v) Tween 20) between, processing as for Western blotting.
Actin sedimentation experiment
GST/GST-PHenn domains freshly purified or purified and then snap-frozen in liquid nitrogen and stocked at −80 °C (maximum one freeze thaw cycle) were used. The experiments were performed following the manual of the Actin Binding Protein Biochem Kit (Cytoskeleton) with modifications. Briefly, for preparing the F-actin, the lyophilized actin was resuspended to 1 mg/ml with General Actin Buffer (Cytoskeleton) and left on ice for 30 min. Actin Polymerization Buffer (Cytoskeleton) was then added. The polymerization reaction was kept at room temperature for 1 h. For preparing GST/GST-PHenn domains to be tested, 16 μl of purified GST/GST-PHenn domain wildtype or mutants (∼3 mg/ml) was diluted with the addition of 49 μl of General Actin Buffer and then subjected to centrifugation at 217,000 × g (TLA-100 Beckman rotor) for 1.5 h at 4 °C. 30 μl of the supernatant was gently mixed with 80 μl of F-actin or 80 μl of buffer of the F-actin, incubated at room temperature for 30 min, and then centrifuged at 148,300 × g (TLA-100 Beckman rotor) for 1 h at 24 °C. The resulting supernatant was immediately collected, followed by the addition of 27.5 μl of 5× Laemmli Sample Buffer. The resulting pellet was resuspended into 68.8 μl of double-distilled water. 2× Laemmli Sample Buffer was then added to the resuspension. After boiling the sample, 65-μl samples from the supernatant or the pellet were processed for SDS-PAGE, and the gel was stained by Coomassie Blue.
Knockdown and rescue experiments
Cells were treated with control non-targeting siRNA (AllStars Negative Control siRNA, Qiagen) or with a previously validated (9) siRNA targeting human DENND3 (5′- cgacggtttagttctgataaa-3′) for 3 days (Lipofectamine RNAiMAX, Thermo Fisher) before lysing the cells. Cells that were treated with DENND3 siRNA were subsequently mock-transfected or transfected with wildtype mouse DENND3 or mouse DENND3 with the L857Q/L858Q, R815E, or K819E mutation for 9 h (Lipofectamine 3000, Thermo Fisher) before the cells were starved with Earle's balanced salt solution (EBSS). Cells were then collected in HEPES lysis buffer (20 mm HEPES, pH 7.4, 10 mm sodium fluoride, 0.5 mm sodium orthovanadate, 60 nm okadaic acid, 100 mm sodium chloride, 1% Triton X-100, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.83 mm benzamidine, and 0.23 mm phenylmethylsulfonyl fluoride). Following 10 min on ice, lysates were spun at 238,700 × g for 15 min. The supernatant was processed for SDS-PAGE and then Western blotting.
Statistical evaluation
Experiments were repeated at least three times. Statistical analysis of the results was carried out by one-way analysis of variance, followed by Tukey's multiple-comparison test when appropriate. p < 0.05 was considered significant.
Author contributions
J. X., P. S. M., and K. G. conceptualization; J. X., G. K., P. S. M., and K. G. formal analysis; J. X. and G. K. investigation; J. X. and G .K. methodology; J. X. and G. K. writing-original draft; J. X., G. K., P. S. M., and K. G. writing-review and editing; P. S. M. and K. G. resources; P. S. M. and K. G. supervision; P. S. M. and K. G. funding acquisition; P. S. M. and K. G. project administration.
Supplementary Material
Acknowledgments
We thank Tin Pan, Maïwenn Beaugrand, Marie Ménade and Véronique Sauvé for fragment screening and identifying the crystallization conditions. Diffraction data were acquired at the Macromolecular Diffraction (MacCHESS) facility at the Cornell High Energy Synchrotron Source (CHESS). CHESS is supported by the National Science Foundation (NSF) and National Institutes of Health (NIH)/NIGMS, via NSF award DMR-0225180, and the MacCHESS resource is supported by NIH/NCRR award RR-01646.
This work was supported by a Foundation Grant from the Canadian Institutes of Health Research (to P. S. M.) and Canadian Institutes of Health Research Grant MOP-14219 (to K. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (code 6B3Y) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GEF
- guanine nucleotide exchange factor
- DENN
- differentially expressed in normal and neoplastic cells
- ULK
- Unc-51–like kinase
- PH
- pleckstrin homology
- DH
- Dbl homology
- EBSS
- Earle's balanced salt solution.
References
- 1. Cherfils J., and Zeghouf M. (2013) Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- 2. Wennerberg K., Rossman K. L., and Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- 3. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 4. Levine T. P., Daniels R. D., Gatta A. T., Wong L. H., and Hayes M. J. (2013) The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics 29, 499–503 10.1093/bioinformatics/bts725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang D., Iyer L. M., He F., and Aravind L. (2012) Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3, 283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marat A. L., Dokainish H., and McPherson P. S. (2011) DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 286, 13791–13800 10.1074/jbc.R110.217067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoshimura S., Gerondopoulos A., Linford A., Rigden D. J., and Barr F. A. (2010) Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191, 367–381 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allaire P. D., Marat A. L., Dall'Armi C., Di Paolo G., McPherson P. S., and Ritter B. (2010) The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol. Cell 37, 370–382 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu J., Fotouhi M., and McPherson P. S. (2015) Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 16, 709–718 10.15252/embr.201440006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu J., and McPherson P. S. (2015) DENND3: a signaling/trafficking interface in autophagy. Cell Cycle 14, 2717–2718 10.1080/15384101.2015.1071136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizushima N., Yoshimori T., and Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 12. Xu J., and McPherson P. S. (2017) Regulation of DENND3, the exchange factor for the small GTPase Rab12 through an intramolecular interaction. J. Biol. Chem. 292, 7274–7282 10.1074/jbc.M116.772434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caplan S. (2017) Into the linker's DENN: a tyrosine's control of autophagy. J. Biol. Chem. 292, 7283–7284 10.1074/jbc.H116.772434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 15. Gallop J. L., Jao C. C., Kent H. M., Butler P. J., Evans P. R., Langen R., and McMahon H. T. (2006) Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 25, 2898–2910 10.1038/sj.emboj.7601174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masuda M., Takeda S., Sone M., Ohki T., Mori H., Kamioka Y., and Mochizuki N. (2006) Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J. 25, 2889–2897 10.1038/sj.emboj.7601176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemmon M. A. (2007) Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 81–93 10.1042/BSS0740081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., and Schlessinger J. (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 92, 10472–10476 10.1073/pnas.92.23.10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stiegler A. L., Zhang R., Liu W., and Boggon T. J. (2014) Structural determinants for binding of sorting nexin 17 (SNX17) to the cytoplasmic adaptor protein Krev interaction trapped 1 (KRIT1). J. Biol. Chem. 289, 25362–25373 10.1074/jbc.M114.584011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ceccarelli D. F., Song H. K., Poy F., Schaller M. D., and Eck M. J. (2006) Crystal structure of the FERM domain of focal adhesion kinase. J. Biol. Chem. 281, 252–259 10.1074/jbc.M509188200 [DOI] [PubMed] [Google Scholar]
- 22. Hamada K., Shimizu T., Matsui T., Tsukita S., and Hakoshima T. (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 19, 4449–4462 10.1093/emboj/19.17.4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee H. S., Bellin R. M., Walker D. L., Patel B., Powers P., Liu H., Garcia-Alvarez B., de Pereda J. M., Liddington R. C., Volkmann N., Hanein D., Critchley D. R., and Robson R. M. (2004) Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 343, 771–784 10.1016/j.jmb.2004.08.069 [DOI] [PubMed] [Google Scholar]
- 24. Yao L., Janmey P., Frigeri L. G., Han W., Fujita J., Kawakami Y., Apgar J. R., and Kawakami T. (1999) Pleckstrin homology domains interact with filamentous actin. J. Biol. Chem. 274, 19752–19761 10.1074/jbc.274.28.19752 [DOI] [PubMed] [Google Scholar]
- 25. Kast D. J., and Dominguez R. (2017) The cytoskeleton-autophagy connection. Curr. Biol. 27, R318–R326 10.1016/j.cub.2017.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conti M. A., and Adelstein R. S. (2008) Nonmuscle myosin II moves in new directions. J. Cell Sci. 121, 11–18 10.1242/jcs.007112 [DOI] [PubMed] [Google Scholar]
- 27. Tang H. W., Wang Y. B., Wang S. L., Wu M. H., Lin S. Y., and Chen G. C. (2011) Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 30, 636–651 10.1038/emboj.2010.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Touhara K., Inglese J., Pitcher J. A., Shaw G., and Lefkowitz R. J. (1994) Binding of G protein βγ-subunits to pleckstrin homology domains. J. Biol. Chem. 269, 10217–10220 [PubMed] [Google Scholar]
- 29. Chen Z., Medina F., Liu M. Y., Thomas C., Sprang S. R., and Sternweis P. C. (2010) Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J. Biol. Chem. 285, 21070–21081 10.1074/jbc.M110.122549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soisson S. M., Nimnual A. S., Uy M., Bar-Sagi D., and Kuriyan J. (1998) Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell 95, 259–268 10.1016/S0092-8674(00)81756-0 [DOI] [PubMed] [Google Scholar]
- 31. Sondermann H., Soisson S. M., Boykevisch S., Yang S. S., Bar-Sagi D., and Kuriyan J. (2004) Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 10.1016/j.cell.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 32. Senderek J., Bergmann C., Weber S., Ketelsen U. P., Schorle H., Rudnik-Schöneborn S., Büttner R., Buchheim E., and Zerres K. (2003) Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum. Mol. Genet. 12, 349–356 10.1093/hmg/ddg030 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 34. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., et al. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 10.1016/j.ymeth.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.