Abstract
We previously demonstrated the effects of azo dyes and their reduction metabolites on bacterial cell growth and cell viability. In this report, the effects of Orange II and Sudan III on gene expression profiling in Staphylococcus aureus ATCC BAA 1556 were analyzed using microarray and quantitative RT-PCR technology. Upon exposure to 6 μg/ml Orange II for 18 h, 21 genes were found to be differently expressed. Among them, 8 and 13 genes were up- and down-regulated, respectively. Most proteins encoded by these differentially expressed genes involve stress response caused by drug metabolism, oxidation, and alkaline shock indicating that S. aureus could adapt to Orange II exposure through a balance between up and down regulated gene expression. Whereas, after exposure to 6 μg/ml Sudan III for 18 h, 57 genes were differentially expressed. In which, 51 genes were up-regulated and 6 were down-regulated. Most proteins encoded by these differentially expressed genes involve in cell wall/membrane biogenesis and biosynthesis, nutrient uptake, transport and metabolite, and stress response, suggesting that Sudan III damages the bacterial cell wall or/and membrane due to binding of the dye. Further analysis indicated that all differentially expressed genes encoded membrane proteins were up-regulated and most of them serve as transporters. The result suggested that these genes might contribute to survival, persistence and growth in the presence of Sudan III. Only one gene msrA, which plays an important role in oxidative stress resistance, was found to be down-regulated after exposure to both Orange II and Sudan III. The present results suggested that both these two azo dyes can cause stress in S. aureus and the response of the bacterium to the stress is mainly related to characteristics of the azo dyes.
Keywords: Staphylococcus aureus, Orange II, Sudan III, Microarray, Gene expression
Introduction
Azo dyes consist of one or more azo groups (R1–N = N–R2), which are characterized by a diazotized amine coupled to an amine or a phenol [12, 79]. It is estimated that more than 2000 different azo dyes are ubiquitously used in various industrial sectors including the plastics, textile, paper, food, cosmetics, and pharmaceutical industries [11, 24, 58, 74]. Humans are exposed to these compounds through ingestion, inhalation, or skin contact. Thus the wide application of various azo dyes in foods, drugs and cosmetics, has raised a concern in regard to the impacts of colorants on human health and the environment. It has been demonstrated that some azo dyes are associated with bladder cancer in humans, splenic sarcomas, hepatocarcinomas, and nuclear anomalies in experimental animals, and chromosomal aberrations in mammalian cells [19, 37, 55]. In general, azo dyes are considered as xenobiotic compounds that are very recalcitrant with respect to biodegradation [74].
Nevertheless, more and more microorganisms have been found to be capable of degrading azo dyes under certain conditions [14, 24, 64, 73, 81]. The first step of azo dye degradation is reduction of the azo bond catalyzed by azoreductase, which has recently been reviewed [11]. Orange II (D&C Orange No. 4) is a water-soluble sulfonated dye, while Sudan III (D&C Red No. 17) is a water-insoluble diazo dye. Their chemical structures and properties are shown in Table 1. Both are approved for use in drug and cosmetic products as colorants [49]. There is evidence that water-soluble azo dyes can penetrate through the cell membrane and subsequently be reduced by azoreductase in the cytoplasm [23]. Whereas, for water-insoluble Sudan dyes, their degradation might be extracellular and/or membrane associated processes rather than an intracellular one [81]. It has been demonstrated that Escherichia coli is not able to efficiently degrade Sudan I, II, III, IV and Para Red due to lack of membrane azo reduction enzymes and membrane bound azo dyes are not efficiently transported into the bacterial cells [43, 81].
Table 1.
Chemical structures and properties of azo dyes
| Azo dye | Chemical structure | Molecular weight | Water Solubility | Melting point (°C) |
|---|---|---|---|---|
| Orange II |
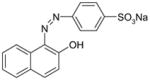
|
350.32 | Water-soluble (116 g/l) | 164 |
| Sudan III |
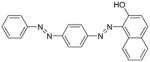
|
352.39 | Insoluble | 199 |
Human skin is an intricate habitat for a diverse population of microbiota including commensal and pathogenic bacteria contributing to both human health and disease [31]. Staphylococcus is one of the most prevalent aerobic genera on human skin. Our previous studies have shown that Staphylococcus aureus is capable of degrading both Orange II and Sudan III [57, 73]. In addition, we found that a tetrameric NADPH-dependent flavin azoreductase (Azo1) from S. aureus (ATCC 25923) metabolizes Methyl Red (MR), Orange II, Amaranth, Ponceau BS and Ponceau S [10]. No significant effect on cell growth and cell viability of the bacterium was found after exposure to 6 μg/ml Orange II and Sudan III in our previous study [57]. However, very little is known about the effects of the azo dyes on gene expression in S. aureus. Here we report genome-wide responses of S. aureus to azo dyes assessed by microarray and quantitative RT-PCR assays with differential gene expression profiling of the bacterium in the presence of water soluble Orange II and water insoluble Sudan III.
Materials and methods
Materials
Orange II (4-(2-hydroxy-1-naphthylazo) benzenesulfonic acid sodium salt), Sudan III (1-[4-(phenylazo)phenylazo]-2-naphthol), dimethyl sulfoxide (DMSO), isopropanol, ethanol, chloroform, and acid-washed glass beads (212–300 μm) were purchased from Sigma-Aldrich Co. Stock solutions of Orange II and Sudan III were freshly prepared by dissolving the chemicals in deionized water and DMSO, respectively.
Bacterial strain and culture conditions
S. aureus ATCC BAA 1556 was used for the experiments in this study. The strain was routinely cultured in Brain Heart Infusion (BHI) agar plate media. After 16–18 h of incubation at 37 °C, one colony was picked by a loop and inoculated into a 15-ml centrifuge tube containing 10 ml BHI medium. The culture was incubated in static conditions at 37 °C overnight for use as seed culture. The bacterial seed culture was inoculated into BHI medium with an inoculation ratio of 1 % (v/v), and then 40 ml aliquots of the medium were transferred to 50-ml Falcon centrifuge tubes. Stock solutions of Orange II or Sudan III were added to the BHI medium at final concentrations of 6 μg/ml (each in triplicate, all experiments were triplicate unless otherwise stated). The cultures without azo dyes but with an equal volume of deionized water or DMSO were inoculated with the bacterial strain as controls. The cultures were incubated at 37 °C without agitation. Decolorization of the dyes was monitored by spectrometry. For Orange II, 1 ml samples were collected from the cultures at various points in time, and centrifuged at 10,000×g for 3 min. Supernatants of the cultures were assayed by measuring the absorption in a Beckman Coulter DU 800 UV–visible spectrophotometer at 483 nm. For Sudan III, two volumes of absolute ethanol were added to 0.5 ml of the samples collected from the cultures. The mixtures were briefly vortexed, and then the samples were assayed at 500 nm following the procedure described above for Orange II. After 18 h incubation, the cultures were collected for total RNA isolation as described below.
Total RNA isolation and purification
The cultures were collected by centrifugation at 3200×g for 15 min. Cell pellets were washed with diethylpyrocarbonate-treated deionized water (DEPC-treated H2O, 0.1 %) twice. Each cell pellet was suspended in 1 ml DEPC-treated H2O and frozen at −70 °C for 20 min. Then acid-washed glass beads (450 mg) were added, and the cells were disrupted at top speed on a VORTEX-GENIE 2 (Scientific Industries, Inc.) at 4 °C for 30 min. The mixtures of disrupted cells and glass beads were transferred to 15-ml centrifuge tube. Then total RNA was extracted following the RNA-Bee™ (Tel-Test, Inc.) reagent procedure. Five ml RNA-Bee and 0.5 ml chloroform were added, mixed and incubated on ice for 15 min followed by centrifugation at 3,200×g for 30 min. The upper phase (aqueous layer) was transferred to a clean tube and equal volume of isopropanol was added, mixed gently and stored at −80 °C for 30 min or overnight at −20 °C. RNA precipitation was collected by centrifugation at 4 °C. The pellet was washed with 75 % ethanol (diluted by DEPC-treated H2O) twice by vortexing, dried by letting it stand on the bench for 5–10 min and dissolved in 100 μl DEPC-treated H2O. The total RNA was then purified according to the standard protocol provided with an RNeasy mini kit (QIAGEN). Contaminating genomic DNA in the RNA preparations was removed using DNA-free kit (Ambion). DNA removal was confirmed by PCR. The RNA quality and quantity were determined by agarose gel electrophoresis and by Nanodrop 1000 (Thermo Scientific), respectively. DNA-free RNA was stored at −80 °C.
Microarray
The microarray was constructed by MYcroarray Inc. using 3 catalog 3 × 15 K S. aureus subsp. aureus USA300 Microarrays. This strain name is referred to as S. aureus ATCC BAA 1556 in this study. The complete genome sequence of this strain is available at http://www.ncbi.nlm.nih.gov/nuccore/CP000255. Each 3 × 15 K microarray has three separate arrays composed of 16,320 spots, of which 12,860 spots (~45-mers) for S. aureus genes. The remaining 3,460 are control features (spike-in empty spots, positive controls, negative control probes, etc.). There are 5 identical replicates of each S. aureus probe such that a total of 2572 S. aureus genes are surveyed by each array (http://www.MYcroarray.com). Hybridization image on the slide was scanned using an Axon4000B scanner (Molecular Devices).
Statistical and bioinformatic analysis
The scanned hybridization images were quantified by GenePix Pro Software (version 6.1.0.4). Array data were normalized using scaling normalization to adjust the total or average intensity of each array to be approximately the same [82]. Microarray data analysis was conducted using a FDA microarray software, ArrayTrack [77]. Lists of differentially expressed genes were identified using a two group (Sample vs. Control) t test after excluding spots flagged as bad. The criteria of p value <0.05 and an absolute relative ratio ≥1.5-fold were applied. Additionally, Venn diagrams were used to examine the overlap of resulting lists of genes differentially expressed between the different sample groups.
The differentially expressed genes were sorted into functional categories based on clusters of orthologous groups (COGs) [45]. Subcellular localizations of proteins, encoded by differentially expressed genes, were predicted using PSORTb ver. 3.0 (http://psort.org) [85].
Validation of microarray data by quantitative real-time PCR (qRT-PCR)
Some genes (including one azoreductase gene-SAUSA300_0545 (azo1)) identified by the microarray were then randomly selected for confirmation by qRT-PCR. The special primers were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi), an enhanced web interface to Primer3. The sequences of primers used in this study are listed in Table 2. Total RNA (2.5–4.5 μg) was reverse transcribed into single-stranded cDNA using Transcriptor First-Strand cDNA Synthesis Kit (Roche Diagnostics) with both Anchored-oligo (dT) 18 primer and random hexamer primer, according to the manufacturer’s instructions. The reaction mixture in tube was incubated for 10 min at 25 °C, 30 min at 55 °C, 5 min at 85 °C and then placed the tube on the ice. The synthesized cDNA was stored at −20 °C and diluted 1/10 prior to use as template in qRT-PCR. qRT-PCR was performed using a LightCycler 480 system (Roche Diagnostics) and the LightCycler 480 SYBR Green I Master kit (Roche Diagnostics, 2×), using 2 μl of cDNA, 1 μl of each primer (10 μM) in a final volume of 20 μl. Reactions were done in triplicate wells for each sample. The amplification reactions consisted of denaturation (95 °C for 10 min), amplification repeated for 40 cycles (95 °C for 10 s, 52 °C for 20 s, 72 °C for 20 s), melting curve (95 °C for 5 s, 65 °C for 1 min with continuous fluorescence measurement at 97 °C) and cooling (40 °C for 10 s). The gene encoding DNA gyrase subunit A, gyrA, was used as the reference gene for normalization of cDNA amount. A single amplified PCRs product was confirmed by melting curve analysis.
Table 2.
Oligonucleotide primers used in qRT-PCR
| Locus tag or gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| SAUSA300_0006 (gyrA) | CAACCGTTGAAACAAATTGATCATC | CTGAGCGTAATGGTAATGTTGTATG |
| SAUSA300_1531 (phoH) | GGATGACAACATCGAAACTCTCT | GACGATATGAACCAATCTCAAGC |
| SAUSA300_0381 | GGGAACCCGCAGATGAC | TTTGCGATTCCCGTTTG |
| SAUSA300_1200 (glnR) | TGCGTGGCATCTACAATCAT | TAACGGATTTAACGCCAAGG |
| SAUSA300_1731 (pckA) | GTGATTCAACCTTCGTCTTCAAC | TGCATTACCTGATTCTGAACCTT |
| SAUSA300_0846 (argG) | GTGGCGCAGCATAAGGAT | GGGCATGGAGTCGTGAAG |
| SAUSA300_1808 | GTGGCGCAGCATAAGGAT | GGGCATGGAGTCGTGAAG |
| SAUSA300_0914 | ACTGCGATGTATGCACAAGC | CGAACCTACGCCAGAGAAAG |
| SAUSA300_1679 (acsA) | CAGTGCAACAAACGCCTTAAT | TGGACCATTTGAGGTTGAGTC |
| SAUSA300_1005 | GCACCCATTGTTGCACCT | CGCAAGCGATTAAATTTGC |
| SAUSA300_1897 | TTTGATTCGTATTGGCTTCTTGT | AGTATCATCCGAATATGGCAATG |
| SAUSA300_0619 | GGCGTCAGGTTTCGTTTTAC | CCTGGTGTTGCCCTATCATT |
| SAUSA300_0545 (azo1) | TGGCAGTGCACAAGTGAAT | CCGCCATTGCTTTCTCTTT |
Results and discussion
Global gene expression profiling of S. auresu in the presence of azo dyes
Our previous studies demonstrated that S. aureus USA strain FPR3757 is capable of degrading both water-soluble Orange II and water-insoluble Sudan III azo dyes [57]. In the present study, the degradation of the azo dyes was monitored by spectrometry. We observed that 45.3 and 38.6 % of Orange II and Sudan III were reduced when the cultures were incubated for 18 h, respectively, indicating the cells were metabolically active. At this stage, the cultures were collected for total RNA isolation. In order to investigate the effects of azo dyes on gene expression patterns in S. aureus, whole genome microarray analysis was performed. The complete genome sequence of S. aureus USA300 strain FPR3757 (namely ATCC BAA 1556) has been published [18]. Genes were considered to be differentially expressed based on a change at least 1.5-fold up- or down-regulated and p-value less than 0.05 in comparison with controls. The microarray data revealed that water-soluble Orange II (6 μg/ml) affected the expression of 21 genes in S. aureus at 18 h. While water-insoluble Sudan III (6 μg/ml) affected the expression of 57 genes under the same growth conditions. The genes affected by Orange II and Sudan III were about 0.82 and 2.22 % of the total genome, respectively. The effects of Sudan III on gene expression profile of S. aureus appeared to be more pronounced with 51 up- and 6 down-regulated genes, compared to 8 up- and 13 down-regulated genes after the Orange II exposure (Table 3). The Venn diagram (not shown) delineated the numbers of differentially expressed genes upon exposure to these two azo dyes. Among these differentially expressed genes, msrA (SAUSA300_1317, encoding methionine-S-sulfoxide reductase) was the only gene shared by exposure to both Orange II and Sudan III and it was down-regulated (Tables 4, 5). The data suggested that the effects of Orange II on S. aureus gene expression profile were different in compared with those of Sudan III, which is consistent with our previous observation that the mechanisms of the bacterial degradation of water-soluble and water-insoluble azo dyes might be distinct [81].
Table 3.
Summary of differentially expressed genes in S. aureus incubated with Orange II and Sudan III
| Azo dye (6 μg/ml) | Total number of genes affecteda
|
|||
|---|---|---|---|---|
| Up-regulated | Down-regulated | Total | % Genome | |
| Orange II | 8 | 13 | 21 | 0.82 |
| Sudan III | 51 | 6 | 57 | 2.22 |
The data were from triplicate incubations
A gene was identified as significant differentially expressed if p value <0.05 and the change ≥1.5-fold (up- or down-regulated) in comparison with control
The cultures were incubated for 18 h
Table 4.
Genes differentially expressed in the presence of Orange II
| Function and locus tag | Gene name | COG categorya | Description of gene product | p-value | Putative location | Fold change |
|---|---|---|---|---|---|---|
| Message storage and processing | ||||||
| SAUSA300_0785 | K | Acetyltransferase, GNAT family | 0.0414 | Cytoplasmic | −1.56 | |
| Cellular processes and signaling | ||||||
| SAUSA300_1531 | phoH | T | Phosphate starvation-induced protein, PhoH | 0.0211 | Cytoplasmic | 1.50 |
| SAUSA300_1989 | agrB | TK | Accessory gene regulatory protein B | 0.0495 | Cytoplasmic Membrane | −1.75 |
| SAUSA300_2468 | M | Acetyltransferase, GNAT family | 0.0291 | Unknown | −1.75 | |
| SAUSA300_1534 | O | Conserved hypothetical protein | 0.0328 | Cytoplasmic Membrane | −1.68 | |
| SAUSA300_1317 | msrA | O | Methionine-S-sulfoxide reductase | 0.0162 | Unknown | −1.71 |
| Metabolism | ||||||
| SAUSA300_0381 | C | Putative NAD(P)H-flavin oxidoreductase | 0.0279 | Cytoplasmic | 1.92 | |
| SAUSA300_0855 | mnhA | CP | Na(+)/H(+) antiporter subunit A | 0.037 | Cytoplasmic Membrane | −1.64 |
| SAUSA300_1585 | H | Conserved hypothetical protein | 0.0337 | Cytoplasmic | 1.52 | |
| SAUSA300_2225 | moaC | H | Molybdenum cofactor biosynthesis protein C | 0.0327 | Cytoplasmic | −1.53 |
| SAUSA300_1125 | acpP | IQ | Acyl carrier protein | 0.0439 | Cytoplasmic | −1.74 |
| Poorly characterized | ||||||
| SAUSA300_0748 | R | Conserved hypothetical protein | 0.0128 | Cytoplasmic | 1.59 | |
| SAUSA300_0604 | R | Hydrolase, alpha/beta hydrolase fold family | 0.0461 | Cytoplasmic | −2.07 | |
| SAUSA300_2590 | R | Conserved hypothetical protein | 0.0185 | Cytoplasmic | −1.75 | |
| SAUSA300_0113 | R | Immunoglobulin G binding protein A precursor | 0.049 | Cell wall | −1.62 | |
| SAUSA300_1249 | S | Conserved hypothetical protein | 0.039 | Cytoplasmic Membrane | −1.62 | |
| SAUSA300_2142 | asp23 | S | Alkaline shock protein 23 | 0.0322 | Unknown | 1.87 |
| SAUSA300_2257 | S | Conserved hypothetical protein | 0.0306 | Unknown | 1.71 | |
| Not in COGs | ||||||
| SAUSA300_1698 | – | Conserved hypothetical protein | 0.0211 | Unknown | 1.98 | |
| SAUSA300_1180 | – | Conserved hypothetical protein | 0.0174 | Cytoplasmic | −1.51 | |
| SAUSA300_1985 | sdrH | – | Serine-aspartate repeat family protein, SdrH | 0.0355 | Cell wall | 1.84 |
– Not in COGs
The COGs functional categories are shown as follows: message and storage and processing [includes transcription (K)], cellular processes and signaling [includes signal transduction mechanisms (T), cell wall/membrane/envelope biogenesis (M), post-translational modification, protein turnover, chaperones (O)). Metabolism (includes energy production and conversion (C), coenzyme transport and metabolism (H), lipid transport and metabolism (I), inorganic ion transport and metabolism (P), secondary metabolite biosynthesis, transport and catabolism (Q)]. Poorly characterized [includes general function prediction only (R), function unknown (S)]
Table 5.
Genes differentially expressed in the presence of Sudan III
| Function and locus tag | Gene name | COG categorya | Description of gene product | p value | Putative location | Fold change |
|---|---|---|---|---|---|---|
| Message storage and processing | ||||||
| SAUSA300_0334 | K | Transcriptional regulator, MarR family | 0.0402 | Unknown | 1.55 | |
| SAUSA300_1200 | glnR | K | Glutamine synthetase repressor | 0.0456 | Cytoplasmic | 1.73 |
| SAUSA300_1793 | L | Conserved hypothetical protein | 0.0379 | Cytoplasmic | 1.51 | |
| Cellular processes and signaling | ||||||
| SAUSA300_1855 | sgtB | M | Monofunctional glycosyltransferase | 0.0325 | Cytoplasmic membrane | 1.53 |
| SAUSA300_2314 | M | Conserved hypothetical protein | 0.0345 | Unknown | 1.51 | |
| SAUSA300_1512 | pbp3 | M | Penicillin-binding protein 3 | 0.0195 | Cytoplasmic membrane | 1.94 |
| SAUSA300_0394 | MG | FAD/NAD(P)-binding Rossmann fold Superfamily | 0.0185 | Cytoplasmic | −1.98 | |
| SAUSA300_0867 | spsA | U | Signal peptidase IA | 0.0117 | Cell wall | 1.65 |
| SAUSA300_1317 | msrA | O | Methionine-S-sulfoxide reductase | 0.0347 | Unknown | −1.55 |
| Metabolism | ||||||
| SAUSA300_0170 | C | Aldehyde dehydrogenase | 0.0329 | Cytoplasmic | 1.88 | |
| SAUSA300_1731 | pckA | C | Phosphoenolpyruvate carboxykinase (ATP) | 0.0392 | Cytoplasmic | 2.31 |
| SAUSA300_2329 | gltT | C | Proton/sodium-glutamate symport protein | 0.0412 | Cytoplasmic membrane | 1.67 |
| SAUSA300_2449 | G | Putative transporter | 0.0347 | Cytoplasmic membrane | 1.53 | |
| SAUSA300_1454 | zwf | G | Glucose-6-phosphate 1-dehydrogenase | 0.0309 | Cytoplasmic | 1.52 |
| SAUSA300_2105 | mtlF | G | PTS system, mannitol specific IIBC component | 0.0238 | Cytoplasmic membrane | 1.67 |
| SAUSA300_0337 | glpT | G | Glycerol-3-phosphate transporter | 0.0458 | Cytoplasmic membrane | 1.90 |
| SAUSA300_2385 | E | Putative membrane protein | 0.0175 | Cytoplasmic membrane | 2.20 | |
| SAUSA300_1231 | E | Gamma-aminobutyrate permease | 0.0241 | Cytoplasmic membrane | 1.56 | |
| SAUSA300_0864 | argG | E | Argininosuccinate synthase | 0.0075 | Cytoplasmic | 3.14 |
| SAUSA300_2538 | E | Amino acid permease family protein | 0.0103 | Cytoplasmic membrane | 1.91 | |
| SAUSA300_1808 | E | Amino acid ABC transporter, | 0.0332 | Cytoplasmic membrane | 2.45 | |
| SAUSA300_1916 | E | Aminotransferase | 0.0224 | Cytoplasmic | 1.51 | |
| SAUSA300_0914 | E | Sodium:alanine symporter family protein | 0.0058 | Cytoplasmic membrane | 2.33 | |
| SAUSA300_2006 | ilvD | EG | Dihydroxy-acid dehydratase | 0.0263 | Cytoplasmic | 2.06 |
| SAUSA300_1102 | gmk | F | Guanylate kinase | 0.0317 | Cytoplasmic | 2.23 |
| SAUSA300_1476 | accB | I | Acetyl-CoA carboxylase, biotin carboxyl carrier | 0.0277 | Cytoplasmic | 2.20 |
| SAUSA300_0229 | I | Putative acyl-CoA transferase FadX | 0.0003 | Unknown | 2.17 | |
| SAUSA300_1679 | acsA | I | Acetyl-coenzyme A synthetase | 0.0072 | Cytoplasmic | 2.18 |
| SAUSA300_1673 | I | 1-acyl-sn-glycerol-3-phosphate acyl-transferases | 0.0334 | Cytoplasmic | 1.51 | |
| SAUSA300_2176 | P | ABC transporter, ATP-binding protein | 0.0267 | Cytoplasmic membrane | 1.52 | |
| SAUSA300_1005 | P | Mn2+/Fe2+ transporter, NRAMP family | 0.0338 | Cytoplasmic membrane | 1.67 | |
| SAUSA300_1897 | P | Sodium-dependent transporter | 0.0399 | Cytoplasmic membrane | 2.16 | |
| SAUSA300_0619 | P | ABC transporter, permease protein | 0.0416 | Cytoplasmic membrane | 2.45 | |
| SAUSA300_1899 | Q | Conserved hypothetical protein | 0.0246 | Cytoplasmic | 1.52 | |
| Poorly characterized | ||||||
| SAUSA300_1700 | R | Polysaccharide biosynthesis protein | 0.0085 | Cytoplasmic membrane | 1.77 | |
| SAUSA300_2268 | R | Sodium/bile acid symporter family | 0.0351 | Cytoplasmic membrane | 1.61 | |
| SAUSA300_1229 | R | Hydrolase, haloacid dehalogenase-like family | 0.0439 | Cytoplasmic | 1.53 | |
| SAUSA300_1685 | R | Conserved hypothetical protein | 0.0459 | Cytoplasmic | −1.55 | |
| SAUSA300_1364 | engA | R | GTP-binding protein EngA | 0.0126 | Cytoplasmic membrane | 2.05 |
| SAUSA300_0013 | S | Putative membrane protein | 0 | Cytoplasmic membrane | 1.62 | |
| SAUSA300_0733 | S | degV family protein | 0.047 | Cytoplasmic | 1.67 | |
| SAUSA300_2418 | S | Conserved hypothetical protein | 0.0019 | Cytoplasmic | −1.53 | |
| SAUSA300_0330 | S | Putative transport protein SgaT | 0.0309 | Cytoplasmic membrane | 1.84 | |
| SAUSA300_2031 | S | Conserved hypothetical protein | 0.0086 | Cytoplasmic membrane | 1.59 | |
| SAUSA300_1284 | S | Conserved hypothetical protein | 0.0136 | Cytoplasmic | 1.59 | |
| Not in COGs | ||||||
| SAUSA300_pUSA010001 | – | Replication protein | 0.0346 | Cytoplasmic | 1.95 | |
| SAUSA300_pUSA030001 | repA | – | Replication initiator protein | 0.0459 | Cytoplasmic | 1.87 |
| SAUSA300_pUSA030007 | ermC | – | rRNA adenine N-6-methyltransferase | 0.0234 | Cytoplasmic | 2.02 |
| SAUSA300_0661 | – | Conserved hypothetical protein | 0.0107 | Cytoplasmic membrane | 1.67 | |
| SAUSA300_0642 | – | Conserved hypothetical protein | 0.0334 | Cytoplasmic membrane | 1.58 | |
| SAUSA300_0681 | – | Conserved hypothetical protein | 0.0182 | Unknown | 1.61 | |
| SAUSA300_1011 | – | Conserved hypothetical protein | 0.034 | Unknown | 1.64 | |
| SAUSA300_2355 | – | Putative lipoprotein | 0.0158 | Unknown | 2.09 | |
| SAUSA300_1680 | acuA | – | Acetoin utilization protein AcuA | 0.0027 | Cytoplasmic | 2.13 |
| SAUSA300_1461 | – | Conserved hypothetical protein | 0.0167 | Cytoplasmic membrane | 1.98 | |
| SAUSA300_1053 | – | Conserved hypothetical protein | 0.0061 | Unknown | −1.81 | |
| SAUSA300_1055 | efb | – | Fibrinogen-binding protein | 0.0211 | Extracellular | −1.58 |
– Not in COGs
The COGs functional categories are shown as follows: message and storage and processing [includes transcription (K), replication, recombination and repair (L)]. Cellular processes and signaling [includes cell wall/membrane/envelope biogenesis (M), intracellular trafficking, secretion and vesicular transport (U), post-translational modification, protein turnover, chaperones (O)]. Metabolism [includes energy production and conversion (C), carbohydrate transport and metabolism (G), amino acid transport and metabolism (E), nucleotide transport and metabolism (F), lipid transport and metabolism (I), inorganic ion transport and metabolism (P), secondary metabolites biosynthesis, transport and catabolism (Q)]. Poorly characterized (includes general function prediction only (R), function unknown (S))
Genes differentially expressed in the presence of Orange II
There were 21 differentially expressed genes in the presence of Orange II based on the statistical analysis. All 21 genes sorted by functional COG category are shown in Table 4. Out of these, approximate half of the differentially expressed genes (47.6 %) are in the poorly characterized category or not in COGs, and 1, 5 and 5 genes involve in message storage and processing (4.8 %), cellular processes and signaling (23.8 %) and metabolism (23. 8 %), respectively. Subcellular localizations of proteins encoded by 21 differentially expressed genes were predicted, of which 10 are cytoplasmic (47.6 %), 4 cytoplasmic membrane (19.1 %), 2 cell wall (9.5 %) and 5 unknown (23.8 %, Table 4).
In message storage and processing functional category, only one differentially expressed gene (SAUSA300_0785) encoding a cytoplasmic acetyltransferase in COG K (correspond to transcription) was down-regulated 1.56-fold. This acetyltransferase belongs to Gcn5-related N-acetyltransferase (GNAT) superfamily, which catalyzes the transfer of an acetyl group from acetyl coenzyme A (AcCoA) to an acceptor substrate and release both CoA and the acetylated product [20, 41]. They are important for pathogenic bacteria because they contribute to emergence of drug resistance by acetylating and rendering antibiotics inactive [20, 41]. Hence, the down-regulation of SAUSA300_0785 gene may result in drug-sensitive S. aureus cells.
Five genes were grouped into cellular processes and signaling functional category, of which, phoH (encoding phosphate starvation-induced protein located in cytoplasm, SAUSA300_1531) and agrB (encoding accessory gene regulatory protein B located in cytoplasmic membrane, SAUSA300_1989) classified into COG T (correspond to signal transduction mechanisms) were up- and down-regulated with 1.50- and 1.75-fold, respectively. The PhoH shares 63.2 % amino acid identity with PhoH (BSU25340) in Bacillus subtilis, which involves in phospholipid metabolism and RNA modification [35]. The S. aureus quorum-sensing system is encoded by the accessory gene regulator (agr) locus, which consists of 5 genes (hld, agrB, agrD, agrC, and agrA) [8]. Agr has been described previously to be associated with reduced vancomycin susceptibility as well as virulence in S. aureus [56, 63, 67]. Our finding implies that the vancomycin resistance and virulence in S. aureus may be decreased after Orange II exposure. SAUSA300_2468 encoding an acetyltransferase with unknown subcellular location in COG M (cell wall/membrane/envelope biogenesis) was detected to be 1.75-fold down-regulated. Another two genes, SAUSA300_1534 and msrA in COG O (post-translational modification, protein turnover, and chaperones) also showed to be down-regulated. msrA encodes a methionine-S-sulfoxide reductase, which is crucial for the protection of cellular damage from oxidative stress [6, 68, 72]. Moreover, for some bacterial pathogens, msr is a potential virulence determinant and the down-regulation of msrA could affect resistance to oxidative stress and virulence of S. aureus [21, 28, 68].
There were 5 differentially expressed genes involved in metabolism after Orange II exposure. Among them, 2 genes, SAUSA300_0381 and mnhA (SAUSA300_0855) fell into COG C (correspond to energy production and conversion). SAUSA300_0381 encoding a putative NAD(P) Hflavin oxidoreductase located in cytoplasm with 1.92-fold change in the present of Orange II. Whereas Na+/H+ antiporter subunit A encoded by mnhA was 1.64-fold down-regulated and located in cytoplasmic membrane. The putative NAD(P)H-flavin oxidoreductase (SAUSA300_0381) shares 99.6 % amino acid identity with NfrA (SA0367) in S. aureus (strain NCTC 8325), which has been proven to be involved in oxidative and disulfide stress responses [75]. Similarly, for B. subtilis, the expression of nfrA gene was induced upon heat shock and oxidative stress [53, 54]. NfsA (ECK0842) in E. coli shares 37 and 36.3 % amino acid identity with the NfrA protein of S. aureus NCTC 8325 and the putative NAD(P)H-flavin oxidoreductase (SAUSA300_0381), respectively. It is an oxygen-insensitive nitroreductase and acts as a lawsone-dependent azo reductase under anaerobic conditions [65]. Previous studies also have indicated that pollutant degrading bacteria might undergo the oxidative stress when exposed to pollutants [27, 29, 88]. Likewise, sulfonated azo dyes Reactive Orange 16 (RO16) and Reactive Red 120 (Red HE3B) generated oxidative stress when they were degraded by Lysinibacillus sp. RGS and bacterial consortium, respectively [3, 61]. Therefore, the high expression of the putative NAD(P)H-flavin oxidoreductase (SAUSA300_0381) might not only protect the cells of S. aureus from oxidative stress generated by sulfonated azo dyes Orange II but also has a probable role in decolorization of Orange II. mnhA encodes a Na+/H+ antiporter subunit A, which is widely distributed in cell membranes from bacteria to mammals. In S. aureus, the antiporter extrudes Na+ or Li+ in exchange for H+ and can make the cells survive under alkaline conditions, up to pH 9.5 [33]. Therefore, it seems that the alkaline pH tolerance of S. aureus will be decreased due to down-regulation of mnhA. In COG H (correspond to coenzyme transport and metabolism), SAUSA300_1585 and moaC (SAUSA300_2225) were found to be 1.52-fold up- and 1.53-fold down-regulated, respectively. Both of them were predicted to be located in cytoplasm. MoaC has an amino acid sequence identity of 50.31 and 53.66 % to E. coli and B. subtilis MoaC, respectively. This gene encodes a molybdenum cofactor (Moco) biosynthesis protein C (MoaC), which is involved in the first step of Moco biosynthesis [84]. Moco is considered to be an essential component that is required by large amounts of enzymes involved in global cycles of carbon, nitrogen and sulfur [38, 52]. acpP (SAUSA300_1125) encodes a cytoplasmic acyl carrier protein classified into COG I or Q (corresponding to lipid transport and metabolism and secondary metabolites biosynthesis, transport and catabolism, respectively). It was detected 1.74-fold down-regulated in the presence of Orange II. Acyl carrier protein (ACP) is the carrier of fatty acids during their synthesis and utilization, which has been found to play an essential part in a growing number of processes outside of fatty acids biosynthesis [36].
It is noteworthy that SAUSA_0113 encoding immuno-globulin G-binding protein A precursor in COG R (general function prediction only) was down-regulated. The protein was predicted to anchor cell wall and shares 98.3 % amino acid identity to SPA (staphylococcal protein A encoded by spa) of S. aureus strain NCTC 8325. Protein A from S. aureus has been suspected to play a role in the virulence of S. aureus [26, 46]. Therefore, it appears that Orange II can decrease the virulence of S. aureus as a pathogen. Up-regulated asp23 in COG S (function unknown), which might play a key role in alkaline pH tolerance of S. aureus [42] was relevant. Therefore, our results imply that the S. aureus has many interlaced mechanisms through a balance between up and down regulated gene expression for adaptation to Orange II. Additionally, it was of interest to find that all 4 differentially expressed genes encoding cytoplasmic membrane proteins were down-regulated in the presence of Orange II (Table 4).
Genes differentially expressed in the presence of Sudan III
Exposure of S. aureus cells to Sudan III revealed 57 differentially expressed genes, which amounted for 2.22 % of the total genome (Table 3). All these genes sorted by general COGs are shown in Table 5. Among these 57 genes, there were 3, 6, and 25 differentially expressed genes involved in message storage and processing (5.3 %), cellular processes and signaling (10.5 %) and metabolism (43.9 %), respectively. Unfortunately, there are still 11 and 12 differentially expressed genes, whose functions are poorly characterized (19.3 %) or not in COGs (21.1 %). Subcellular localizations of 57 differentially expressed gene products were predicted, of which 23 are cytoplasmic, 24 cytoplasmic membrane, 1 cell wall, 1 extracellular and 8 unknown (Table 5).
In message storage and processing functional category, 3 genes involved in transcription (COG K) and DNA replication, recombination, and repair (COG L) were up-regulated (Table 5). Among them, there are 2 genes encoding transcriptional regulators, including one with similarity to the multiple antibiotic resistance regulator (MarR) family (SAUSA300_0334, 1.55-fold) and the other with glutamine synthetase repressor (SAUSA300_1200, GlnR, 1.73-fold). MarR homologs regulate activity of genes involved in antibiotic resistance, stress responses, virulence or catabolism of aromatic compounds [13, 15, 50, 60]. The up-regulation of marR may be used by the bacterium for detoxification in response to Sudan III. In B. subtilis, GlnR plays a key role in regulation by directly controlling expression of glutamine synthetase as well as several other genes involved in nitrogen metabolism [69, 70, 80]. The up-regulation of glnR may relieve stress of nitrogen restriction caused by Sudan III in the bacterium.
In cellular processes and signaling functional category, there are four differentially expressed genes in COG M (cell wall/membrane/envelope biogenesis, Table 5). sgtB (SAUSA300_1855) and php3 (SAUSA300_1512) encoding monofunctional glycosyltransferase (MGT) and penicillin-binding protein 3 (PBP3) were up-regulated with 1.53- and 1.94-fold change, respectively. MGT has been demonstrated to play a key role in the elongation of peptidoglycan chains and PBPs also involve in the final stages of peptidoglycan biosynthesis in S. aureus [62, 78]. Both sgtB and pbp3 might involve in cell wall biosynthesis. It has been shown that over 60 % of the dyes (Sudan II or Sudan IV) penetrated into E. coli, and 90 % of these penetrated dyes remained on the membrane of the bacterium [43]. The accumulation of Sudan dyes on the membrane could cause damage to the bacterial cell walls. Therefore, the up-regulation of sgtB and pbp3 may be used by the bacterium for preventing the cell wall damage mediated by Sudan III. Whereas, another gene in COG M or G (carbohydrate transport and metabolism), SAUSA300_0394 encoding cytoplasmic FAD/NAD(P)-binding Rossmann fold Superfamily protein, which may function as oxidoreducatse [39], was down-regulated 1.98-fold. spsA (SAUSA300_0867), encoding cell wall anchor of signal peptidase IA in COG U (intracellular trafficking, secretion and vesicular transport), showed 1.65-fold up-regulated. In S. aureus, it was predicted that SpsA is an inactive signal peptidase homologue and may provide some other unknown functions for the bacterium [34]. msrA was also down-regulated with 1.55-fold in the presence of Sudan III exposure similar to Orange II. Studies on genotoxicity of Sudan I and Sudan IV have been showed that they can induce oxidative stress in HepG2 cells [2, 87]. So similar compound Sudan III may cause oxidative stress in S. aureus and inhibit the expression of msrA.
Twenty five differentially expressed genes within metabolism functional category were all up-regulated (fold change ranging from 1.51 to 3.14). They were grouped into 7 COG categories such as C, G, E, F, I, P and Q, which correspond to energy production/conversion, carbohydrate transport/metabolism, amino acid transport/metabolism, nucleotide transport/metabolism, lipid transport/metabolism, inorganic ion transport/metabolism, secondary metabolite biosynthesis/transport/catabolism, respectively.
SAUSA300_0170 and pckA (SAUSA300_1731) in COG C were significantly up-regulated with 1.88- and 2.31-fold, respectively (Table 5). SAUSA300_0170 encodes an aldehyde dehydrogenase involving in the catabolism of ethanol, which is oxidized via acetaldehyde into acetate. pckA encodes a phosphoenolpyruvate carboxykinase (PEPCK), which catalyzes the first committed step of gluconeogenesis pathway and likely plays a key role in the growth and survival of S. aureus in the absence of glucose [71]. gltT (SAUSA300_2329) also in COG C, encoding cytoplasmic membrane-bound proton/sodium-glutamate symport protein, showed a 1.67-fold increase. GltT in S. aureus is homologous to GltT in Bacillus stearothermophilus and Bacillus caldotenax (55.24 % identity for both) [76]. In bacteria, glutamate transporters participate in the nutrient uptake and are crucial to viability of cells [83]. So the binding of Sudan III to S. aureus cells might prevent the absorption of nutrition such as glucose and glutamate resulting in the up-regulation of pckA and gltT.
Four genes in COG G were up-regulated, of which 3 genes encode membrane transporter proteins, such as SAUSA300_2449 (encoding a putative transporter), mtlF (SAUSA300_2105, encoding mannitol-specificphos-photransferase system, IIBC component and glpT (SAUSA300_0337, encoding glycerol-3-phosphate transporter) with1.53-, 1.67- and 1.90-fold increases, respectively. It has been found that mtlF (SA1960) in S. aureus was up-regulated in late growth phase related to carbon metabolism under vancomycin stress conditions and in bio-film formation, respectively [4, 9, 51]. A glpT mutant of B. subtilis has been found to be defective in uptake of glycerol-3-phosphate [44]. Our results suggest that S. aureus could uptake other available carbon sources (like mannitol and glycerol-3-phosphate) after some of glucose prevented to into bacterium by the binding of Sudan III on the bacterial cell. Another up-regulated gene in COG G is zwf (SAUSA300_1454), which encodes cytoplasmic glucose-6-phosphate 1-dehydrogenase (G6PD) and catalyzs the oxidation of glucose 6-phosphate to gluconolactone 6-phosphate with either NADP+ or NAD+ as electron acceptor [66]. Up-regulation of zwf as a response to oxidative stress, recovery from heat stress, and even virulence has been demonstrated for gram-negative bacteria [30, 40, 47, 48]. The expression of zwf was found to be increased under oxidative stress conditions and acid-shock for Enterococcus mundtii CRL35 and S. aureus 50583, respectively [5, 66]. Similarly oxidative stress possibly generated by Sudan III could induce the up-regulation of zwf of S. aureus.
Eight genes identified as up-regulated belong to COG E. Especially, the products of SAUSA300_1231, 2538, 1808 and 0914 genes seem to function as amino acid transport. SAUSA300_1231 encodes a γ-aminobutyrate (GABA) permease, which has an amino acid sequence identity of 39.48 % to B. subtilis GabP protein involving not only uptakes the GABA as nitrogen source but also transports proline as a nutrient [25, 86]. It has been found that the expression of gabP in B. subtilis was induced during nitrogen-limited growth [7, 25]. An amino acid permease family protein is encoded by SAUSA300_2538, which shares 64.58 % amino acid sequence identity with BcaP in Lactococcus lactis [17]. BcaP has been confirmed to be the major branched-chain amino acids (BCAAs) carrier and the deletion of bcaP results in the loss of most of the BCAAs uptake [17]. It has been demonstrated that SAUSA300_1808 encoding an amino acid ABC transporter functions as an arogenate dehydratase, which converts L-arogenate to L-phenylalanine, and its activity is inhibited by phenylalanine (http://microcyc.genoscope.cns.fr/STAA3F1776/NEW-IMAGE?type=PATHWAY&object=PWY-3462). SAUSA300_0914 encodes a sodium/alanine symporter family protein. Three genes encoding cytoplasmic proteins in COG E involve in amino acid metabolism. Particularly, argG (SAUSA300_0864) encoding cytoplasmic argininosuccinate synthase and ilvD (SAUSA300_2006) encoding dihydroxy-acid dehydratase were observed to be significantly up-regulated (3.14 and 2.06-fold changes, respectively). These enzymes are involved in arginine biosynthesis II and BCAAs (isoleucine and valine) biosynthesis pathways, respectively. These results indicated that the binding of Sudan III to cell wall and membrane may induce amino-acids transport and biosynthesis for S. aureus growth.
gmk (SAUSA300_1102) was the only up-regulated gene (2.23-fold) classified into COG F. It encodes a cytoplasmic guanylate kinase involving in synthesis of nucleotide precursors and indirectly modulating the synthesis of DNA and RNA. Inhibition of Gmk would impact bacterial growth [22]. Additionally, Gmk is a potential target for novel antibacterial drugs in S. aureus [22]. This may indicate that excess expression of gmk in S. aureus may help the bacterium to overcome stress caused by Sudan III [57].
Four genes encoding proteins involved in lipid transport and metabolism (COG I) were up-regulated, namely acsA (encoding acetyl-coenzyme A synthetase, SAUSA300_1679), accB (encoding a biotin carboxyl carrier protein of acetyl-CoA carboxylase, SAUSA300_1476), SAUSA300_1673 (encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase) and SAUSA300_0229 (putative acyl-CoA transferase FadX). Except for SAUSA300_0229 with unknown location, the other 3 gene products were predicted in cytoplasm. AcsA in S. aureus has a high amino acid sequence identity of 69.7 % to AcsA in B. subtilis. The disruption of this gene in B. subtilis resulted in loss of the ability to use acetate for growth and sporulation [32]. SAUSA300_1673 is homologous to plsC in B. subtilis with 47.03 % amino acid sequence identity. PlsC depletion brought the cessation of phospholipids synthesis, but continued fatty acid synthesis in B. subtilis [59]. Altogether, it appears that up-regulation of fatty acid and phospholipids synthesis enzymes allowed S. aureus to overcome cell membrane damage caused by the binding of Sudan III.
Four genes (SAUSA300_2176, 1005, 1879 and 0619) grouped into COG P were all up-regulated (1.52–2.45 fold). Their gene products involve in inorganic ion transport rather than metabolism (two ABC transporters, one Mn2+/Fe2+ transporter and one sodium-dependent transporter) and are anchored in cytoplasmic membrane. In bacteria, ABC transporters play roles in nutrient uptake and in secretion of toxins and antimicrobial agents [16]. They exhibit specificity for different substrates: carbohydrates, amino acids, osmoprotectants, oligopeptides, inorganic ions, bacteriocins, and DNA [1]. They further confirmed that Sudan III leads to the damage of cell membrane. SAUSA300_1899, which encodes a conserved hypothetical cytoplasmic protein involved in secondary metabolite biosynthesis, transport and catabolism (COG Q), was also found to be 1.52-fold up-regulated.
All differentially expressed genes encoding cytoplasmic membrane proteins were identified to be up-regulated in the presence of Sudan III and most genes encode transporters. Their overexpression and functional enhancement seem to be an effective direct response to the unusual membrane environment caused by Sudan III accumulation, indicating that Sudan III is more toxic to the bacterium than Orange II which agreed with our previous study [57].
Validation of microarray data and functional identification of azoreductase by qRT-PCR
For validation of microarray data and functional identification of azoreductase, the expression of the genes, gyrA (SAUSA300_0006), an azoreductase gene (SAUSA300_0545, azo1), and randomly selected 11 genes of up or down-regulated in S. aureus in the presence of Orange II or Sudan III were further analyzed by qRT-PCR (Table 2). The gyrA gene did not show any variation in expression in the presence of Orange II and Sudan III, and therefore, was used for normalization. Consistent with the microarray data, the qRT-PCR experiment confirmed that the FMN-dependent NADPH-azoreductase Azo1 is constitutively expressed, indicating its functional responsibility for azoreduction. In addition, the expression patterns of the randomly selected genes measured by qRT-PCR were consistent with the expression values obtained by microarray analysis (data not shown).
Conclusions
In summary, this global transcriptome analysis showed that Orange II and Sudan III have apparent solubility-dependent impacts on the gene expression profiling of S. aureus. There were 21 (8 up- and 13 down-regulated) and 57 (51 up- and 6 down-regulated) genes differentially expressed after exposure to Orange II and Sudan III, respectively. In the presence of water-soluble Orange II, most differentially expressed genes with known functions were responsive to various stress factors, such as drugs (especially vancomycin), reactive oxygen species, and alkaline shock. However, in the presence of water-insoluble Sudan III, most differentially expressed genes were found to be up-regulated, especially genes encoding cytoplasmic membrane proteins and most of them involved in cell wall or membrane biogenesis, biosynthesis; nutrient (carbon, nitrogen, energy, inorganic ion) uptake, transport, metabolism; and stress responses. Conclusively, the azo dyes such as Orange II and Sudan III are stress-inducible xenobiotic compounds, and bacterial metabolic and stress responses depend mainly on the physicochemical properties of azo dyes and biodegradability.
Acknowledgments
We thank Drs. Steven L. Foley and Kidon Sung for their critical review of this manuscript. This study was funded by National Center for Toxicological Research, United States Food and Drug Administration, and supported in part by appointments (HP and JF) to the Postgraduate Research Fellowship Program by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. The views presented in this article do not necessarily reflect those of the Food and Drug Administration.
Contributor Information
Hongmiao Pan, Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Rd., Jefferson, AR 72079-9502, USA. Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China.
Joshua Xu, Division of Bioinformatics and Biostatistics, National Center for Toxicological Research, US Food and Drug Administration, Jefferson, AR 72079, USA.
Oh-Gew Kweon, Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Rd., Jefferson, AR 72079-9502, USA.
Wen Zou, Division of Bioinformatics and Biostatistics, National Center for Toxicological Research, US Food and Drug Administration, Jefferson, AR 72079, USA.
Jinhui Feng, Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Rd., Jefferson, AR 72079-9502, USA.
Gui-Xin He, Department of Clinical Laboratory and Nutritional Sciences, University of Massachusetts Lowell, Lowell, MA 01854, USA.
Carl E. Cerniglia, Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Rd., Jefferson, AR 72079-9502, USA
Huizhong Chen, Division of Microbiology, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Rd., Jefferson, AR 72079-9502, USA.
References
- 1.Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An Y, Jiang L, Cao J, Geng C, Zhong L. Sudan I induces genotoxic effects and oxidative DNA damage in HepG2 cells. Mut Res Genet Toxicol Environ Mutagen. 2007;627:164–170. doi: 10.1016/j.mrgentox.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Bedekar PA, Saratale RG, Saratale GD, Govindwar SP. Oxidative stress response in dye degrading bacterium Lysinibacillus sp. RGS exposed to Reactive Orange 16, degradation of RO16 and evaluation of toxicity. Environ Sci Pollut Res Int. 2014;21:11075–11085. doi: 10.1007/s11356-014-3041-2. [DOI] [PubMed] [Google Scholar]
- 4.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bore E, Langsrud S, Langsrud Ø, Rode TM, Holck A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology. 2007;153:2289–2303. doi: 10.1099/mic.0.2007/005942-0. [DOI] [PubMed] [Google Scholar]
- 6.Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim Biophys Acta. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Brechtel C, King S. 4-Aminobutyrate (GABA) transporters from the amine-polyamine-choline superfamily: substrate specificity and ligand recognition profile of the 4-aminobutyrate permease from Bacillus subtilis. Biochem J. 1998;333:565–571. doi: 10.1042/bj3330565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron DR, Ward DV, Kostoulias X, Howden BP, Moellering RC, Eliopoulos GM, Peleg AY. Serine/threonine phosphatase Stp1 contributes to reduced susceptibility to vancomycin and virulence in Staphylococcus aureus. J Infect Dis. 2012;205:1677–1687. doi: 10.1093/infdis/jis252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee I, Schmitt S, Batzilla CF, Engelmann S, Keller A, Ring MW, Kautenburger R, Ziebuhr W, Hecker M, Preissner KT. Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics. 2009;9:1152–1176. doi: 10.1002/pmic.200800586. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Hopper SL, Cerniglia CE. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology. 2005;151:1433–1441. doi: 10.1099/mic.0.27805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H. Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci. 2006;7:101–111. doi: 10.2174/138920306776359786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KC, Huang WT, Wu JY, Houng JY. Microbial decolorization of azo dyes by Proteus mirabilis. J Ind Microbiol Biotechnol. 1999;23:686–690. doi: 10.1038/sj.jim.2900689. [DOI] [PubMed] [Google Scholar]
- 13.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chengalroyen M, Dabbs E. The microbial degradation of azo dyes: minireview. World J Microbiol Biotechnol. 2013;29:389–399. doi: 10.1007/s11274-012-1198-8. [DOI] [PubMed] [Google Scholar]
- 15.Cohen SP, Hächler H, Levy S. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Ann Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 17.den Hengst CD, Groeneveld M, Kuipers OP, Kok J. Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA) J Bacteriol. 2006;188:3280–3289. doi: 10.1128/JB.188.9.3280-3289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 19.Dillon D, Combes R, Zeiger E. Activation by caecal reduction of the azo dye D & C red no. 9 to a bacterial mutagen. Mutagenesis. 1994;9:295–299. doi: 10.1093/mutage/9.4.295. [DOI] [PubMed] [Google Scholar]
- 20.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyl-transferases: a structural overview. Ann Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Hassouni M, Chambost JP, Expert D, Van Gijsegem F, Barras F. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Omari K, Dhaliwal B, Lockyer M, Charles I, Hawkins AR, Stammers DK. Structure of Staphylococcus aureus guanylate monophosphate kinase. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:949–953. doi: 10.1107/S174430910603613X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng J, Heinze M, Xu H, Cerniglia E, Chen H. Evidence for significantly enhancing reduction of azo dyes in Escherichia coli by expressed cytoplasmic Azoreductase (AzoA) of enterococcus faecalis. Protein Pept Lett. 2010;17:578–584. doi: 10.2174/092986610791112701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng J, Cerniglia CE, Chen H. Toxicological significance of azo dye metabolism by human intestinal microbiota. Front Biosci. 2012;4:568–586. doi: 10.2741/400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferson AE, Wray LV, Jr, Fisher SH. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 26.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frederick JR, Elkins JG, Bollinger N, Hassett DJ, McDermott TR. Factors affecting catalase expression in Pseudomonas aeruginosa biofilms and planktonic cells. Appl Environ Microbiol. 2001;67:1375–1379. doi: 10.1128/AEM.67.3.1375-1379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuller TE, Kennedy MJ, Lowery DE. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb Pathog. 2000;29:25–38. doi: 10.1006/mpat.2000.0365. [DOI] [PubMed] [Google Scholar]
- 29.Geckil H, Gencer S, Kahraman H, Erenler SO. Genetic engineering of Enterobacter aerogenes with the Vitreoscilla hemoglobin gene: cell growth, survival, and antioxidant enzyme status under oxidative stress. Res Microbiol. 2003;154:425–431. doi: 10.1016/S0923-2508(03)00083-4. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grice EA, Kong HH, Renaud G, Young AC, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 33.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J Bacteriol. 1998;180:6642–6648. doi: 10.1128/jb.180.24.6642-6648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 35.Kazakov AE, Vassieva O, Gelfand MS, Osterman A, Overbeek R. Bioinformatics classification and functional analysis of PhoH homologs. In silico Biol. 2003;3:3–15. [PubMed] [Google Scholar]
- 36.Keating DH, Carey MR, Cronan JE. The unmodified (apo) form of Escherichia coli acyl carrier protein is a potent inhibitor of cell growth. J Biol Chem. 1995;270:22229–22235. doi: 10.1074/jbc.270.38.22229. [DOI] [PubMed] [Google Scholar]
- 37.Khan MF, Wu X, Kaphalia BS, Boor PJ, Ansari GAS. Acute hematopoietic toxicity of aniline in rats. Toxicol Lett. 1997;92:31–37. doi: 10.1016/s0378-4274(97)00032-5. [DOI] [PubMed] [Google Scholar]
- 38.Kisker C, Schindelin H, Rees DC. Molybdenum-cofactor-containing enzymes: structure and mechanism. Ann Rev Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 39.Kleiger G, Eisenberg D. GXXXG and GXXXA motifs stabilize FAD and NAD(P)-binding Rossmann folds through C(alpha)-H… O hydrogen bonds and van der waals interactions. J Mol Biol. 2002;323:69–76. doi: 10.1016/s0022-2836(02)00885-9. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi H, Miyamoto T, Hashimoto Y, Kiriki M, Motomatsu A, Honjoh K, Iio M. Identification of factors involved in recovery of heat-injured Salmonella Enteritidis. J Food Prot. 2005;68:932–941. doi: 10.4315/0362-028x-68.5.932. [DOI] [PubMed] [Google Scholar]
- 41.Kuhn ML, Majorek KA, Minor W, Anderson WF. Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N-acetyltransferases with unknown substrate specificity. Protein Sci. 2013;22:222–230. doi: 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin-resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 43.Li L, Gao HW, Ren JR, Chen L, Li YC, Zhao JF, Zhao HP, Yuan Y. Binding of Sudan II and IV to lecithin liposomes and E. coli membranes: insights into the toxicity of hydrophobic azo dyes. BMC Struct Biol. 2007;7:16. doi: 10.1186/1472-6807-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindgren V. Mapping of a genetic locus that affects glycerol 3-phosphate transport in Bacillus subtilis. J Bacteriol. 1978;133:667–670. doi: 10.1128/jb.133.2.667-670.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo M, Bulach DM, Powell DR, Haake DA, Matsunaga J, Paustian ML, Zuerner RL, Adler B. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect Immun. 2006;74:5848–5859. doi: 10.1128/IAI.00755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loefdahl S, Guss B, Uhlen M, Philipson L, Lindberg M. Gene for staphylococcal protein A. Proc Natl Acad Sci. 1983;80:697–701. doi: 10.1073/pnas.80.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg BE, Wolf RE, Dinauer MC, Xu Y, Fang FC. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect Immun. 1999;67:436–438. doi: 10.1128/iai.67.1.436-438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J-F, Hager PW, Howell ML, Phibbs PV, Hassett DJ. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat) J Bacteriol. 1998;180:1741–1749. doi: 10.1128/jb.180.7.1741-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marmion DM. Handbook of US colorants: foods, drugs, cosmetics, and medical devices. Wiley; New York: 1991. [Google Scholar]
- 50.Martin RG, Rosner JL. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCallum N, Spehar G, Bischoff M, Berger-Bachi B. Strain dependence of the cell wall-damage induced stimulon in Staphylococcus aureus. Biochim Biophys Acta. 2006;1760:1475–1481. doi: 10.1016/j.bbagen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Mendel RR, Bittner F. Cell biology of molybdenum. Biochim Biophys Acta Mol Cell Res. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Moch C, Schrögel O, Allmansberger R. Transcription of the nfrA-ywcH operon from Bacillus subtilis is specifically induced in response to heat. J Bacteriol. 2000;182:4384–4393. doi: 10.1128/jb.182.16.4384-4393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mostertz J, Scharf C, Hecker M, Homuth G. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology. 2004;150:497–512. doi: 10.1099/mic.0.26665-0. [DOI] [PubMed] [Google Scholar]
- 55.Myslak ZW, Bolt HM. Occupational exposure to azo dyes and risk of bladder cancer. Zbl Arbeitsmed. 1998;38:310–321. [Google Scholar]
- 56.Novick RP, Geisinger E. Quorum sensing in staphylococci. Ann Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 57.Pan H, Feng J, Cerniglia CE, Chen H. Effects of Orange II and Sudan III azo dyes and their metabolites on Staphylococcus aureus. J Ind Microbiol Biotechnol. 2011;38:1729–1738. doi: 10.1007/s10295-011-0962-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan H, Feng J, He GX, Cerniglia CE, Chen H. Evaluation of impact of exposure of Sudan azo dyes and their metabolites on human intestinal bacteria. Anaerobe. 2012;18:445–453. doi: 10.1016/j.anaerobe.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paoletti L, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 61.Phugare SS, Kalyani DC, Patil AV, Jadhav JP. Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater. 2011;186:713–723. doi: 10.1016/j.jhazmat.2010.11.049. [DOI] [PubMed] [Google Scholar]
- 62.Pinho MG, de Lencastre H, Tomasz A. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J Bacteriol. 2000;182:1074–1079. doi: 10.1128/jb.182.4.1074-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu R, Pei W, Zhang L, Lin J, Ji G. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]
- 64.Rafii F, Hall JD, Cerniglia CE. Mutagenicity of azo dyes used in foods, drugs and cosmetics before and after reduction by Clostridium species from the human intestinal tract. Food Chem Toxicol. 1997;35:897–901. doi: 10.1016/s0278-6915(97)00060-4. [DOI] [PubMed] [Google Scholar]
- 65.Rau J, Stolz A. Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent azo reductases. Appl Environ Microbiol. 2003;69:3448–3455. doi: 10.1128/AEM.69.6.3448-3455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saavedra L, Sesma F. Atypical genetic locus associated with the zwf gene encoding the glucose 6-phosphate dehydrogenase from Enterococcus mundtii CRL35. Curr Microbiol. 2005;51:148–152. doi: 10.1007/s00284-004-4454-9. [DOI] [PubMed] [Google Scholar]
- 67.Sakoulas G, Eliopoulos GM, Moellering RC, Wennersten C, Venkataraman L, Novick RP, Gold HS. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sasindran SJ, Saikolappan S, Dhandayuthapani S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2007;2:619–630. doi: 10.2217/17460913.2.6.619. [DOI] [PubMed] [Google Scholar]
- 69.Schreier HJ, Brown SW, Hirschi KD, Nomellini JF, Sonenshein AL. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989;210:51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- 70.Schreier HJ, Caruso SM, Maier KC. Control of Bacillus subtilis glutamine synthetase expression by glnR from Staphylococcus aureus. Curr Microbiol. 2000;41:425–429. doi: 10.1007/s002840010162. [DOI] [PubMed] [Google Scholar]
- 71.Scovill WH, Schreier HJ, Bayles KW. Identification and characterization of the pckA gene from Staphylococcus aureus. J Bacteriol. 1996;178:3362–3364. doi: 10.1128/jb.178.11.3362-3364.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh VK, Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology. 2003;149:2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- 73.Stingley RL, Zou W, Heinze TM, Chen HZ, Cerniglia CE. Metabolism of azo dyes by human skin microbiota. J Med Microbiol. 2010;59:108–114. doi: 10.1099/jmm.0.012617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol. 2001;56:69–80. doi: 10.1007/s002530100686. [DOI] [PubMed] [Google Scholar]
- 75.Streker K, Freiberg C, Labischinski H, Hacker J, Ohlsen K. Staphylococcus aureus NfrA (SA0367) is a flavin mononucleotide-dependent NADPH oxidase involved in oxidative stress response. J Bacteriol. 2005;187:2249–2256. doi: 10.1128/JB.187.7.2249-2256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tolner B, Poolman B, Konings WN. Characterization and functional expression in Escherichia coli of the sodium/proton/glutamate symport proteins of Bacillus stearothermophilus and Bacillus caldotenax. Mol Microbiol. 1992;6:2845–2856. doi: 10.1111/j.1365-2958.1992.tb01464.x. [DOI] [PubMed] [Google Scholar]
- 77.Tong W, Cao X, Harris S, Sun H, Fang H, Fuscoe J, Harris A, Hong H, Xie Q, Perkins R. Arraytrack–supporting toxicogenomic research at the US Food and Drug Administration National Center for Toxicological Research. Environ Health Perspect. 2003;111:1819–1826. doi: 10.1289/ehp.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang QM, Peery RB, Johnson RB, Alborn WE, Yeh W-K, Skatrud PL. Identification and characterization of a monofunctional glycosyltransferase from Staphylococcus aureus. J Bacteriol. 2001;183:4779–4785. doi: 10.1128/JB.183.16.4779-4785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang RF, Chen H, Paine DD, Cerniglia CE. Microarray method to monitor 40 intestinal bacterial species in the study of azo dye reduction. Biosens Bioelectron. 2004;20:699–705. doi: 10.1016/j.bios.2004.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wray LV, Ferson AE, Rohrer K, Fisher SH. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu H, Heinze TM, Paine DD, Cerniglia CE, Chen H. Sudan azo dyes and Para Red degradation by prevalent bacteria of the human gastrointestinal tract. Anaerobe. 2010;16:114–119. doi: 10.1016/j.anaerobe.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15–e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yernool D, Boudker O, Folta-Stogniew E, Gouaux E. Trimeric subunit stoichiometry of the glutamate transporters from Bacillus caldotenax and Bacillus stearothermophilus. Biochemistry. 2003;42:12981–12988. doi: 10.1021/bi030161q. [DOI] [PubMed] [Google Scholar]
- 84.Yoshida H, Yamada M, Kuramitsu S, Kamitori S. Structure of a putative molybdenum-cofactor biosynthesis protein C (MoaC) from Sulfolobus tokodaii (ST0472) Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:589–592. doi: 10.1107/S174430910801590X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zaprasis A, Hoffmann T, Stannek L, Gunka K, Commichau FM, Bremer E. The γ-aminobutyrate permease GabP serves as the third proline transporter of bacillus subtilis. J Bacteriol. 2014;196:515–526. doi: 10.1128/JB.01128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, An Y, Jiang L, Geng C, Cao J, Jiang L, Zhong L. The role of oxidative stress in Sudan IV-induced DNA damage in human liver-derived HepG2 cells. Environ Toxicol. 2011;26:292–299. doi: 10.1002/tox.20558. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Meng D, Wang Z, Guo H, Wang Y, Wang X, Dong X. Oxidative stress response in atrazine-degrading bacteria exposed to atrazine. J Hazard Mater. 2012;229:434–438. doi: 10.1016/j.jhazmat.2012.05.054. [DOI] [PubMed] [Google Scholar]


