MtNFH1, a Medicago truncatula hydrolase cleaving nodulation signals of nitrogen-fixing rhizobia, possesses a symbiotic function in the nodule symbiosis.
Abstract
Establishment of symbiosis between legumes and nitrogen-fixing rhizobia depends on bacterial Nod factors (NFs) that trigger symbiosis-related NF signaling in host plants. NFs are modified oligosaccharides of chitin with a fatty acid moiety. NFs can be cleaved and inactivated by host enzymes, such as MtNFH1 (MEDICAGO TRUNCATULA NOD FACTOR HYDROLASE1). In contrast to related chitinases, MtNFH1 hydrolyzes neither chitin nor chitin fragments, indicating a high cleavage preference for NFs. Here, we provide evidence for a role of MtNFH1 in the symbiosis with Sinorhizobium meliloti. Upon rhizobial inoculation, MtNFH1 accumulated at the curled tip of root hairs, in the so-called infection chamber. Mutant analysis revealed that lack of MtNFH1 delayed rhizobial root hair infection, suggesting that excess amounts of NFs negatively affect the initiation of infection threads. MtNFH1 deficiency resulted in nodule hypertrophy and abnormal nodule branching of young nodules. Nodule branching was also stimulated in plants expressing MtNFH1 driven by a tandem CaMV 35S promoter and plants inoculated by a NF-overproducing S. meliloti strain. We suggest that fine-tuning of NF levels by MtNFH1 is necessary for optimal root hair infection as well as for NF-regulated growth of mature nodules.
INTRODUCTION
Rhizobia are symbiotic bacteria that fix nitrogen in symbiosomes of nodules formed on roots of legumes and the nonlegume Parasponia sp (Cannabaceae). Rhizobia typically enter legume roots via infection of root hairs. Rhizobia on root hair tips stimulate the reorientation of root hair growth, which usually results in curling and formation of an infection chamber containing entrapped bacteria. After penetration, bacteria within the root hair are surrounded by a host-derived infection thread. The infection thread, containing dividing bacteria, elongates and branches to reach cells of the root cortex where a nodule primordium is formed (Perret et al., 2000; Fournier et al., 2015). Nodules can be determinate or indeterminate. Determinate nodules are globose and possess a homogeneous population of nitrogen-fixing bacteroids, which turn synchronously senescent (Rolfe and Gresshoff, 1988). Indeterminate nodules contain a persistent meristem and show differentiation over a longitudinal gradient, which results in an elongate nodule shape. Mature indeterminate nodules consist of an apical meristematic zone, an infection zone (penetration zone), a zone with differentiating bacteroids, a nitrogen-fixing zone and a senescence zone (Vasse et al., 1990). Owing to meristem branching, indeterminate nodules can adopt bifurcate, palmate, or coralloid structures.
Establishment of symbiosis depends on signal exchange between rhizobia and the nodule-forming host legume. Flavonoids from host plants interact with rhizobial NodD proteins (LysR family transcriptional regulators) to activate expression of bacterial nodulation genes. As a result, most rhizobia synthesize strain-specific nodulation signals essential for infection and nodule formation, the so-called nodulation factors (Nod factors [NFs]). NFs are lipochitoligosaccharides, i.e., they are oligosaccharides of N-acetylglucosamine with a lipid modification at the nonreducing end (Perret et al., 2000). For example, Sinorhizobium (Ensifer) meliloti produces NFs (NodSm factors) with a tetrasaccharidic (IV) or pentasaccharidic (V) carbohydrate moiety, which is predominantly N-acylated by a C16:2 fatty acid (2E,9Z-hexadecadienoic acid). These NFs are decorated by a sulfate group (S) at the reducing end and may also carry an O-acetyl group (Ac) at the nonreducing end (Lerouge et al., 1990; Schultze et al., 1992). Host plants perceive NFs using NF receptors (LysM domain receptor kinases), which initiate NF signaling to express symbiosis-related genes required for bacterial infection and nodule formation (Oldroyd, 2013). Nodulation signaling in the model Medicago truncatula includes proteins such as the NF receptor proteins LYK3 (Limpens et al., 2003) and NFP (Amor et al., 2003; Arrighi et al., 2006), the ion channel DMI1 (Ané et al., 2004), the leucine-rich repeat receptor-like kinase DMI2/NORK (Endre et al., 2002), and the Ca2+/calmodulin-dependent protein kinase DMI3 (Lévy et al., 2004; Mitra et al., 2004). The role of these genes in formation of indeterminate nodules containing S. meliloti bacteroids has been well established.
Owing to the structural similarities between the carbohydrate moiety of NFs and chitin (polymer of N-acetylglucosamine), certain plant chitinases possess the capacity to hydrolyze β-1,4 glycosidic bonds in NFs. Structural modifications in NFs may confer resistance to cleavage by chitinases (Staehelin et al., 1994a, 1994b, Goormachtig et al., 1998, Minic et al., 1998; Schultze et al., 1998; Ovtsyna et al., 2000). Plants do not possess chitin and most chitinases appear to play a role in defense against pathogens or herbivores. For example, various plant chitinases inhibit growth of fungi by hydrolyzing their chitin-containing cell walls (Schlumbaum et al., 1986; Mauch et al., 1988). Moreover, specific endochitinases may release (or cleave) chitooligosaccharides from fungi to produce (or inactivate) elicitors of plant defense reactions. Notably, plant receptors perceiving chitooligosaccharides are LysM domain receptor kinases and thus are structurally related to the NF receptors of legumes (Liang et al., 2014). Plant chitinases are glycoside hydrolases (GHs) that are traditionally divided into different classes (GH family 18, class III and V; GH family 19, class I, II, and IV). In legumes, transcript levels and activities of specific chitinases are induced during symbiosis with rhizobia or in response to treatment with NFs (Staehelin et al., 1992; Goormachtig et al., 1998; Xie et al., 1999; Ovtsyna et al., 2000; Ovtsyna et al., 2005). NFs applied to the roots of Medicago sativa (Staehelin et al., 1994b, 1995), Vicia sativa (Heidstra et al., 1994), and Pisum sativum (Ovtsyna et al., 2000, 2005) are rapidly hydrolyzed, indicating that host enzymes limit NF levels. Hydrolyzed NFs applied to M. sativa or V. sativa roots show strongly reduced activity in inducing root hair deformations, indicating that action of NFs can be controlled by host plants (Staehelin et al., 1994b; Heidstra et al., 1994). In the rhizosphere of M. sativa seedlings pretreated with S. meliloti NFs, an enzyme activity rapidly releases lipodisaccharides from the NF substrates (Staehelin et al., 1995). In Sesbania rostrata, transcript levels of a gene encoding a class III chitinase were found to be strongly upregulated during symbiosis and the corresponding protein was able to hydrolyze NFs of Azorhizobium caulinodans in vitro (Goormachtig et al., 1998). Likewise, when P. sativum roots were treated with NFs of the microsymbiont Rhizobium leguminosarum bv viciae, an enzyme cleaving S. meliloti NFs was induced. In contrast to wild-type plants, such a stimulatory effect was absent in mutants deficient in NF signaling (Ovtsyna et al., 2005).
Recently, we identified MEDICAGO TRUNCATULA NOD FACTOR HYDROLASE1 (MtNFH1; formerly named MtChit5; Medtr4g116990.1) (Tian et al., 2013). MtNFH1 expressed in Escherichia coli can release lipodisaccharides from S. meliloti NFs with a C16:2 acyl chain, but does not cleave NFs with a C18:4 chain purified from R. leguminosarum bv viciae. Remarkably, MtNFH1 also lacks chitinase activity, i.e., it cleaves neither chitin nor nonmodified oligomers of chitin. MtNFH1 is most closely related to the class V chitinases MtCHIT5a and MtCHIT5b, which possess chitinase activity but lack the capacity to hydrolyze S. meliloti NFs. Activity of MtNFH1 depends on two loops that likely contribute to the formation of a fatty acid binding cleft for the NF substrate. Remarkably, MtCHIT5b with a single serine-to-proline substitution gains the capacity to hydrolyze NFs (Tian et al., 2013; Zhang et al., 2016). MtCHIT5b and MtNFH1 are located in tandem on chromosome 4 of M. truncatula, suggesting that MtNFH1 evolved from MtCHIT5b by gene duplication. The evolution of a NF hydrolase from a chitinase can be considered an example of symbiosis-related neofunctionalization (Zhang et al., 2016). Transcripts of MtNFH1 are elevated during symbiosis with S. meliloti, namely, in root hairs, but also at the stage of mature nodules (Salzer et al., 2004; He et al., 2009; Breakspear et al., 2014; Roux et al., 2014; Jardinaud et al., 2016). Application of S. meliloti NFs to roots results in increased MtNFH1 transcript levels (Salzer et al., 2004; Jardinaud et al., 2016). Likewise, expression of MtNFH1 is induced in response to mycorrhizal lipochitoligosaccharides in a DMI3-dependent manner (Camps et al., 2015). M. truncatula RNAi lines with reduced MtNFH1 transcripts show reduced hydrolytic activity when S. meliloti NFs are added to the rhizopshere (Tian et al., 2013).
Owing to the symbiosis-related gene expression and the particular substrate preference for NFs, we hypothesized that MtNFH1 must have a function in symbiosis. In this work, we report that MtNFH1 controls NF hydrolysis at the stage of root hair infection. Moreover, our results indicate that MtNFH1 influences growth and branching of mature nodules.
RESULTS
NF Hydrolysis in the Rhizosphere Depends on MtNFH1
A PCR screen of the Tnt1 insertion mutant collection at the Noble Research Institute resulted in three MtNFH1 mutant lines of M. truncatula ecotype R108 (NF16587, NF12841, and NF11260; subsequently named nfh1-1, nfh1-2, and nfh1-3). The Tnt1 insertions were located in the promoter region of MtNFH1 at −363, −305, and −148 bp upstream of the ATG start codon (Figure 1A). MtNFH1 expression was analyzed by RT-qPCR in plants that were incubated with (Figure 1B) or without (Supplemental Figure 1) NFs. The nfh1-1 and nfh1-2 mutants showed reduced MtNFH1 transcript levels in comparison with wild-type plants. Remarkably, MtNFH1 transcripts were not or barely detected in the nfh1-3 mutant.
Figure 1.
Characterization of nfh1 Mutants.
(A) Tnt1 insertion sites of the M. truncatula R108 mutants nfh1-1 (NF16587), nfh1-2 (NF12841), and nfh1-3 (NF11260). The box indicates the coding region of MtNFH1. The location of Tnt1 insertions in the promoter region are marked by indicated arrows.
(B) Analysis of MtNFH1 expression by RT-qPCR in wild-type and nfh1 mutant seedlings (nfh1-1, nfh1-2, and nfh1-3). Roots of seedlings were immersed in 1-mL syringes filled with Jensen medium containing 0.1 μM NodSm-IV(C16:2, Ac, S) for 18 h. Roots from 20 seedlings were used for each RNA extraction (3 RNA extractions per genotype; n = 3). Data indicate means ± se of normalized expression values (mean value of wild-type plants set to one). Statistically different transcript levels of mutants compared with wild-type plants are marked by asterisks (Student’s t test, P ≤ 0.05; Supplemental File 1).
(C) Formation of the lipodisaccharide NodSm-II(C16:2, Ac) released from NodSm-IV(C16:2, Ac, S) in the rhizosphere of wild-type and nfh1 mutant seedlings. Data indicate means ± se (1 plant per sample; n ≥ 9). Asterisks indicate significantly reduced hydrolytic activity in nfh1 mutants compared with wild-type plants (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
Tests with M. truncatula seedlings and purified NFs of S. meliloti were performed to determine the ability of intact roots to cleave NFs in the rhizosphere. Conversion of the O-acetylated tetrameric NodSm-IV(C16:2, Ac, S) into the O-acetylated lipodissaccharide NodSm-II(C16:2, Ac) was analyzed by HPLC. Assay conditions and representative HPLC chromatograms are shown in Supplemental Figure 2. Compared with wild-type plants, nfh1-1 and nfh1-2 exhibited reduced hydrolytic activity. The seedlings of nfh1-3 failed to cleave NodSm-IV(C16:2, Ac, S) under the used test conditions. These results indicate that the ability of intact M. truncatula roots to hydrolyze NodSm-IV(C16:2, Ac, S) in the rhizosphere can be completely attributed to MtNFH1 activity (Figure 1C). Likewise, tetrameric NodSm-IV(C16:2, S) lacking the O-acetyl group was not hydrolyzed by the nfh1-3 mutant. When NodSm-V(C16:2, S) was added to the rhizosphere of nfh1-3, however, the cleavage products NodSm-III(C16:2) and NodSm-II(C16:2) were formed, indicating the presence of another enzyme that hydrolyzes only pentameric NFs (Supplemental Figure 3).
MtNFH1 expression and MtNFH1 activity in the rhizosphere was further investigated in non-nodulating M. truncatula mutants that are deficient in NF signaling. When treated with NFs, the non-nodulating mutants showed considerably reduced MtNFH1 expression in comparison with wild-type plants (Figure 2A). By contrast, only small, but significant, differences in MtNFH1 expression were seen for wild-type and mutant seedlings that were not challenged with NFs (Supplemental Figure 4). These data suggest that wild-type seedlings under the test conditions showed constitutive activity of nodulation signaling genes that contributed to expression of MtNFH1 before NF treatment. NF hydrolysis tests indicated a relationship between MtNFH1 expression and enzyme activity. When pretreated with NFs, wild-type plants cleaved NodSm-IV(C16:2, Ac, S) more efficiently than the NF signaling mutants (Figure 2B). Low enzyme activity of the NF signaling mutants was also measured with the pentameric substrate NodSm-V(C16:2, S) in a similar experiment (Supplemental Figure 5). These findings indicate that activation of nodulation signaling in M. truncatula culminates in enhanced MtNFH1 expression and NF hydrolysis in the rhizosphere. Experiments with NF-induced and noninduced wild-type plants substantiated these findings. As determined with NodSm-IV(C16:2, S), NF-triggered stimulation of MtNFH1 expression and NF hydrolase activity required only a few hours (Figures 3A and 3B). Furthermore, transcript levels of MtNFH1 and NF hydrolase activity both depended on the NF concentration used for the pretreatment (Figures 3C and 3D).
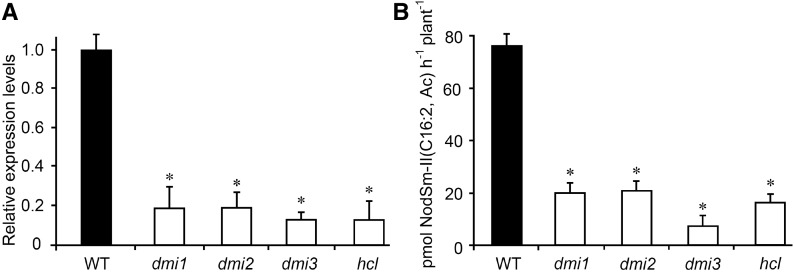
Figure 2.
MtNFH1 Transcript Levels and NF Hydrolysis Activity in the Rhizosphere of Nodulation Signaling Mutants.
(A) Analysis of MtNFH1 expression by RT-qPCR in M. truncatula Jemalong A17 wild-type plants and nodulation signaling mutants (dmi1, dmi2, dmi3, and hcl). Roots of seedlings were immersed in Jensen medium containing 0.1 μM NodSm-IV(C16:2, Ac, S) for 18 h. Roots from 20 plants were combined for each RNA extraction (3 RNA extractions per genotype; n = 3). Data indicate means ± se of normalized expression values (mean value of wild-type plants set to one). Statistically different transcript levels of mutants compared with wild-type plants are marked by asterisks (Student’s t test, P ≤ 0.05; Supplemental File 1).
(B) Corresponding NF hydrolysis tests: Roots of seedlings were pretreated with 0.1 μM NodSm-IV(C16:2, Ac, S) for 18 h and then incubated with 15 μM NodSm-IV(C16:2, Ac, S) for 3 h. The amounts of NodSm-II(C16:2, Ac) formed were deduced from HPLC chromatograms. Data indicate means ± se (1 plant per sample, n ≥ 9). Asterisks indicate significantly reduced hydrolytic activity in nodulation signaling mutants compared with wild-type plants (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
WT, M. truncatula wild-type (ecotype Jemalong A17); dmi1, dmi1-3 mutant (Y6); dmi2, dmi2-1 mutant (TR25); dmi3, dmi3-1 mutant (TRV25); hcl, hcl-1 mutant (lyk3, B56).
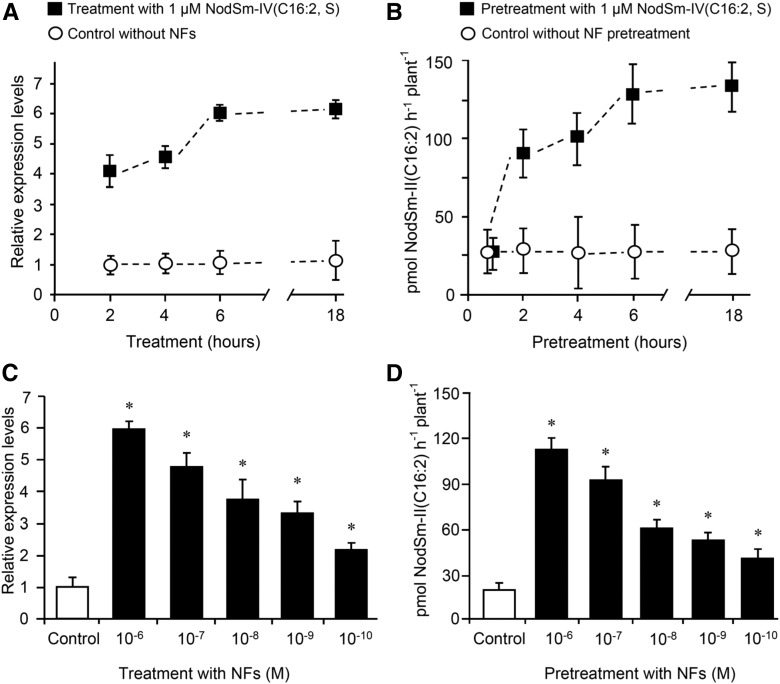
Figure 3.
Stimulation of MtNFH1 Expression and MtNFH1 Activity in Response to NF Treatments.
(A) Time-dependent stimulation of MtNFH1 expression by NFs: Roots of M. truncatula R108 seedlings were immersed in Jensen medium containing 1 μM NodSm-IV(C16:2, S) for the indicated time periods. Control plants without NF treatment were incubated under the same conditions. RNA from harvested roots (30 roots per RNA extraction; 3 RNA extractions; n = 3) was used for RT-qPCR analysis. Data indicate means ± se of normalized expression values (mean value of control plants at the 2-h time point set to one).
(B) Time-dependent stimulation of MtNFH1 activity by NFs: Roots of seedlings (Jemalong A17) were pretreated with 1 µM NodSm-IV(C16:2, S) for the indicated time periods. After pretreatment, plants were transferred to Jensen medium containing 5 µM NodSm-IV(C16:2, S) and incubated for 3 h. Plants without NF pretreatment were grown under the same conditions. The NodSm-II(C16:2) formed in samples (three to four plants per sample) was analyzed by reverse-phase HPLC. Data indicate means ± se from three independent experiments (n = 3).
(C) Concentration-dependent stimulation of MtNFH1 expression by NFs: Roots of R108 seedlings were immersed in Jensen medium containing NodSm-IV(C16:2, S) at indicated concentrations for 18 h. Plants without NF treatment served as a control. RNA was isolated from harvested roots (20 roots per RNA extraction; 3 RNA extractions; n = 3). Data indicate means ± se of normalized expression values (mean value of control plants set to one). MtNFH1 transcript levels of NF-treated plants significantly different from control plants are marked by asterisks (Student’s t test, P ≤ 0.05; Supplemental File 1).
(D) Concentration-dependent stimulation of MtNFH1 activity by NFs: Roots of R108 seedlings were pretreated with Jensen medium containing NodSm-IV(C16:2, S) at indicated concentrations for 18 h. MtNFH1 activity was then assayed with 5 μM NodSm-IV(C16:2, S) for 3 h. Formed NodSm-II(C16:2) (3 plants per sample) was analyzed by reverse-phase HPLC. Data indicate means ± se (3 samples; n = 3). Asterisks indicate a significant difference compared with the control (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
The NF hydrolase protein of M. sativa roots binds to the lectin concanavalin A (ConA) (Staehelin et al., 1995). We therefore expected that MtNFH1 would be an N-glycosylated protein and compared ConA binding protein fractions isolated from M. truncatula roots. NF cleavage was observed for wild-type plants but not for the nfh1-3 mutant (Supplemental Figure 6A). Gel filtration chromatography of ConA binding proteins from wild-type plants indicated that MtNFH1 is active as a monomeric protein with an apparent molecular weight of ∼40 kD (Supplemental Figure 6B).
Delayed Rhizobial Infection in nfh1-3 Mutant Plants
To identify any symbiotic role of MtNFH1 during rhizobial infection, wild-type and nfh1-3 mutant plants were compared. Seedlings were inoculated with a S. meliloti 2011 derivative constitutively expressing the galactosidase gene lacZ to visualize bacteria after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Infection threads containing bacteria were observed in the wild-type plants 3 d postinoculation (dpi). At the same time, only a few infection foci at tips of root hairs could be seen in the nfh1-3 mutant. At 5 dpi, the first nodule primordia were formed in the wild-type plants, while root hairs with infection threads were observed in the nfh1-3 mutant. At 7 dpi, nodule primordia, often clustered together, were also observed in the nfh1-3 mutant (Figures 4A to 4F). Hence, infection of the nfh1-3 mutant was delayed compared with wild-type plants.
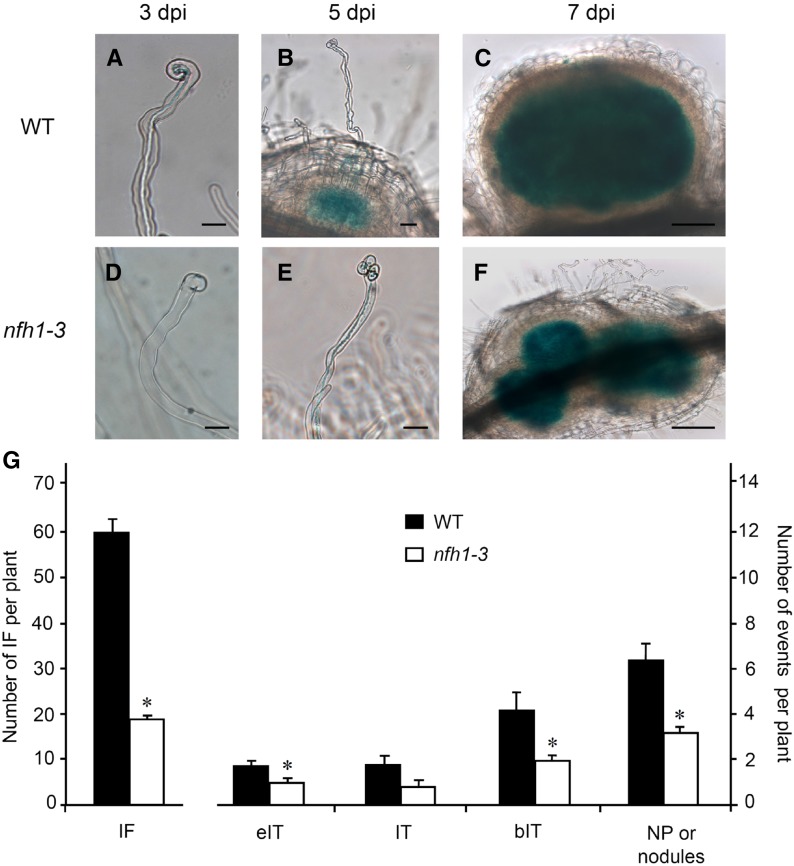
Figure 4.
Analysis of Early Symbiotic Stages in Wild-Type and nfh1-3 Mutant Plants.
Roots of seedlings inoculated with S. meliloti 2011 carrying pXLGD4 (lacZ) were harvested at indicated time points and bacteria visualized with X-Gal.
(A) to (C) Root hairs and formed nodule primordia of wild-type seedlings at 3 (A), 5 (B), and 7 (C) dpi. Bars = 20 µm in (A) and (B) and 100 µm in (C).
(D) to (F) Abnormal root hair deformation and formed nodule primordia of nfh1-3 mutant seedlings at 3 (D), 5 (E), and 7 (F) dpi. Bars = 20 µm in (D) and (E) and 100 µm in (F).
(G) Quantification of different symbiotic stages at 7 dpi. Data indicate means (±se) for 12 plants. Significant differences between wild-type and nfh1-3 mutant plants are marked with asterisks (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1). IF, infection foci; eIT, elongating infection thread in root hair; IT, fully elongated infection thread in root hair; bIT, branched infection thread in cortex; NP, nodule primordium.
To quantify these differences, plants harvested 7 dpi were analyzed for the frequency of infection events, namely, infection foci, elongating infection threads, fully elongated infection threads, and branched infection threads in the root cortex. Representative photographs for each category are shown in Supplemental Figure 7. Compared with wild-type plants, the number of infection events in the nfh1-3 mutant was considerably lower. Notably, a significant reduction of infection foci, fully elongated infection threads, branched infection threads, and nodule primordia (or young nodules) was observed in the mutant (Figure 4G). The nfh1-3 mutant often showed aberrant tip swelling without curling, multiple tip swelling or root hair branching (Figures 4D and 4E; Supplemental Figure 8).
MtNFH1 Accumulates in the Infection Chamber
To analyze MtNFH1 expression in M. truncatula root hairs, a 2051-bp promoter sequence upstream of the MtNFH1 coding region was used to construct a promoter-GUS (β-glucuronidase reporter gene) fusion. Transgenic M. truncatula roots were obtained after inoculation with Agrobacterium rhizogenes harboring a binary vector with this construct (MtNFH1pro-GUS). Roots were subsequently inoculated with S. meliloti Rm41, a strain that efficiently induces nodules on R108 roots. The harvested roots were stained for GUS activity with 5-bromo-4-chloro-3-indol-β-glucuronic acid (X-Gluc). GUS staining was clearly visible in the zone of infected root hairs close to the root tip. By contrast, no obvious blue coloration was observed in noninfected roots under the test conditions (plastic jar system). Hence, MtNFH1 expression was induced in root hairs in response to S. meliloti infection (Figures 5A to 5D).
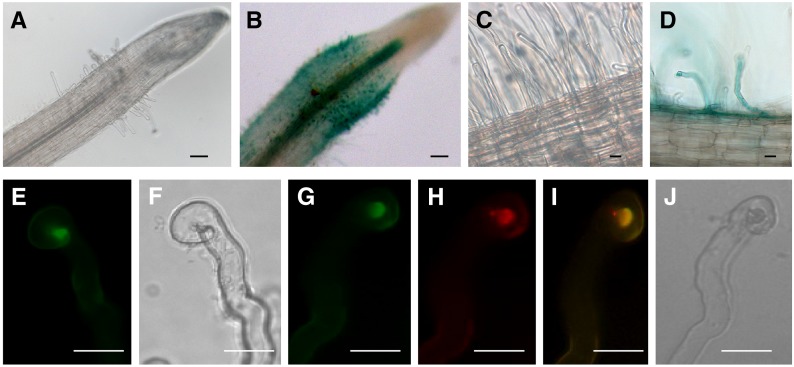
Figure 5.
Expression of MtNFH1 in Response to Rhizobial Inoculation and Accumulation of MtNFH1:GFP in the Infection Chamber.
(A) to (D) Analysis of M. truncatula R108 roots transformed with a MtNFH1pro-GUS construct. Plants were inoculated with S. meliloti Rm41 (3 dpi; [B] and [D]) or left noninoculated ([A] and [C]). Roots were stained with X-Gluc and then cleared with diluted NaClO solution. Bars = 50 µm in (A) and (B) and 20 µm in (C) and (D).
(E) to (J) Analysis of curled root hairs of R108 roots expressing MtNFH1:GFP driven by the MtNFH1 promoter (3 dpi) under green fluorescence ([E] and [G]), red fluorescence (H), and bright-field conditions ([F] and [J]). Roots were inoculated with S. meliloti Rm41 ([E] and [F]) or S. meliloti 1021 (pQDN03) constitutively expressing mCherry ([G] to [J]). GFP fluorescence signals reflecting the presence of MtNFH1:GFP protein are increased in the infection chamber. Colocalization with bacteria is indicated in yellow (merged image; [I]). Bars = 20 µm.
To examine MtNFH1 localization in M. truncatula root hairs, roots expressing MtNFH1 with a C-terminal GFP tag were constructed. The fusion protein construct contained the MtNFH1 promoter sequence as used before. Expression of this construct in nfh1-3 roots indicated that the MtNFH1:GFP fusion protein possesses NF-cleaving activity (Supplemental Figure 9). Microscopy analysis of transformed wild-type roots indicated that the MtNFH1:GFP protein accumulated in the apex of curled root hairs infected by S. meliloti Rm41 (Figures 5E and 5F). To visualize the bacteria, the transformed roots were inoculated with S. meliloti 1021 expressing mCherry fluorescent protein. Control roots (without MtNFH1:GFP) were also analyzed in these studies (Supplemental Figures 10A and 10B). The MtNFH1:GFP protein in root hairs colocalized with rhizobia at the site of bacterial entrapment, the so-called infection chamber (Figures 5G to 5J; Supplemental Figure 10C). However, GFP fluorescence signals reflecting the presence of MtNFH1:GFP were not detected in formed infection threads (Supplemental Figure 10D).
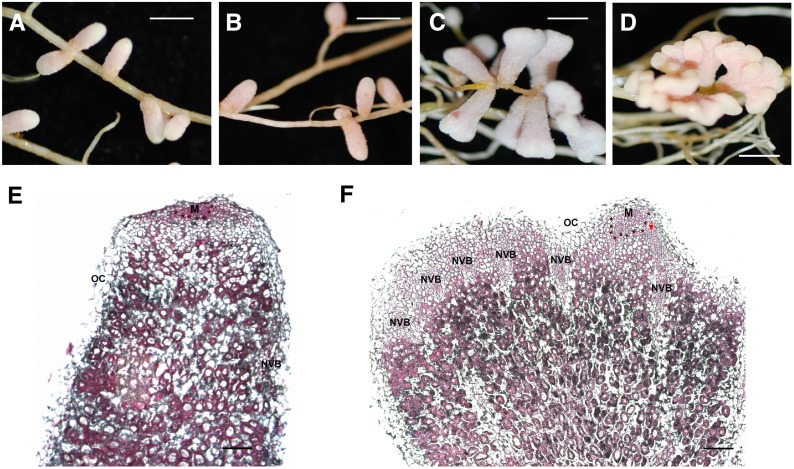
The nfh1-3 Mutant Shows Aberrant Nodule Branching
To investigate the role of MtNFH1 at a later symbiotic stage, nodulation tests were performed with M. truncatula wild-type and nfh1-3 (NF11260) mutant plants inoculated with S. meliloti Rm41. For comparison, we also analyzed sibling plants (from the original NF11260 population) that lacked a Tnt1 insertion in MtNFH1 and thus exhibited NF hydrolase activity. The nfh1-3 mutant plants did not phenotypically differ from wild-type or sibling plants with respect to the aerial part of the plant. However, nodules formed by the nfh1-3 mutant showed an altered shape. In contrast to the elongate nodules formed by wild-type or sibling plants (Figures 6A and 6B), nfh1-3 nodules were either bifurcate (dichotomously branched) or multiply branched resulting in palmate or even coralloid nodules. The distribution of the bifurcate and palmate-coralloid nodules in the root system was frequently clustered (Figures 6C and 6D). Micrographs illustrated the morphological differences between wild-type and nfh1-3 nodules. The nfh1-3 nodules possessed multiple meristems characterized by small dividing cells at the apex of each nodule lobe. Provascular meristems flanking the apical meristem and derived vascular bundles were observed at the periphery of each nodule lobe (Figures 6E and 6F). Expression of MtNFH1 in nodules was confirmed by A. rhizogenes-mediated transformation of wild-type roots with the MtNFH1pro-GUS construct (Supplemental Figure 11).
Figure 6.
Nodulation Phenotype of the nfh1-3 Mutant.
Plants were inoculated with S. meliloti Rm41.
(A) Unbranched nodules formed on M. truncatula R108 wild-type roots (20 dpi).
(B) Wild-type nodules formed on roots of a wild-type sibling line of nfh1-3 (20 dpi).
(C) and (D) Bifurcate (C) or palmate-coralloid (D) nodules formed on roots of the nfh1-3 mutant (20 dpi).
(E) and (F) Microscopy analysis of a wild-type (E) and nfh1-3 (F) nodule (20 dpi). Sections were stained with ruthenium red. M, meristem (indicated with asterisks); NVB, nodule vascular bundle; OC, outer cortex; PVM, provascular meristem (red arrow).
Bars = 2 mm in (A) to (D) and 200 µm in (E) and (F).
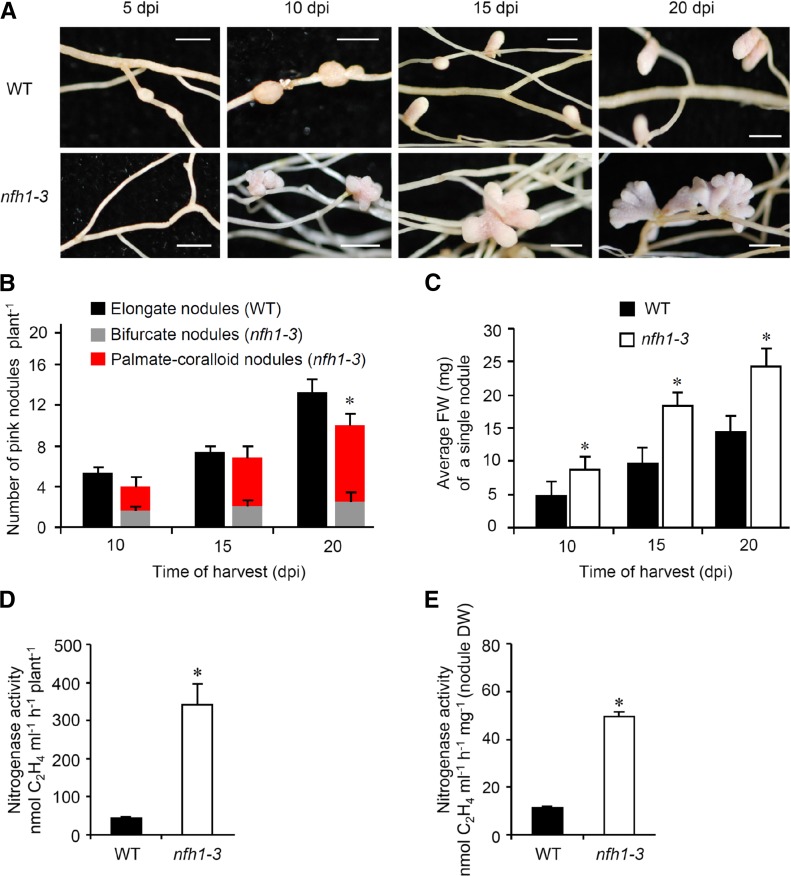
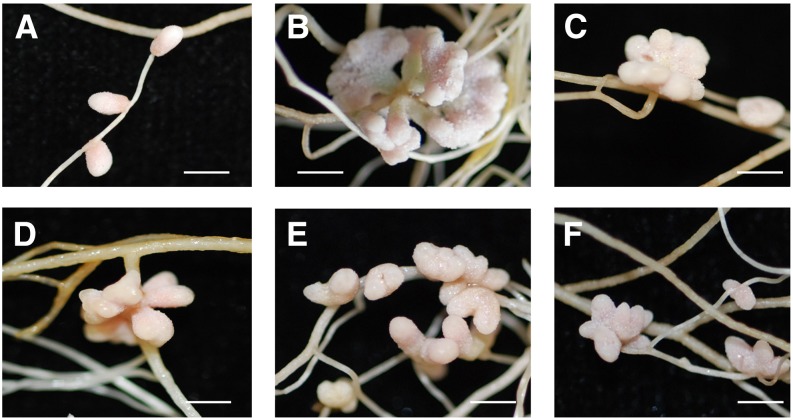
To further characterize the nfh1-3 mutant, plants inoculated with S. meliloti Rm41 were harvested at different time points. At 5 dpi, the first nodule primordia were observed on wild-type roots while the nfh1-3 mutant did not form nodule primordia at this stage. At 10 dpi, young nfh1-3 nodules were observed that were bifurcate or even palmate-coralloid (Figure 7A). The number of pink (leghemoglobin containing), mature nodules formed on nfh1-3 roots was lower than that on wild-type roots at 20 dpi (Figure 7B). However, the average fresh weight of an individual nfh1-3 nodule was higher than that of a wild-type nodule for all three time points (Figure 7C). Nitrogenase activity per plant or per nodule dry weight was also higher in nfh1-3 than in wild-type plants when measured at 20 dpi (Figures 7D and 7E). Wild-type plants harvested at 24 dpi or later time points started to form bifurcate and subsequently palmate nodules (data not shown).
Figure 7.
Time-Course Analysis of Nodule Formation and Nitrogenase Activity in Nodules.
M. truncatula R108 wild-type and nfh1-3 mutant plants were inoculated with S. meliloti Rm41 and harvested at indicated time points.
(A) Photographs of roots and nodules at the time of harvest. Bars = 2 mm.
(B) Quantification of different types of nodules. Wild-type plants formed elongate nodules while the nfh1-3 mutant formed either bifurcate or palmate-coralloid nodules. Data indicate means (±se) based on analysis of 10 plants of each genotype and for each time point. Compared with the wild type, a significantly reduced nodule number (marked by an asterisk) was observed for the nfh1-3 mutant at 20 dpi (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
(C) Quantification of the fresh weight of an individual nodule formed by wild-type and nfh1-3 roots. Data indicate means (±se) based from 10 plants of each genotype and for each time point. Asterisks indicate significantly increased values for the nfh1-3 mutant as compared with the wild-type (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
(D) and (E) Nitrogenase activity per plant (D) and per nodule biomass (dry weight) (E) at 20 dpi. Asterisks indicate significantly increased activities in nfh1-3 nodules as compared with wild-type nodules at this time point (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
As the nodule number of the nfh1-3 mutant was reduced at 20 dpi, we further asked whether nodule branching is a compensation reaction of plants that form fewer nodules. To test this, wild-type seedlings were inoculated with low inoculum doses to reach a lower nodule number. Wild-type plants formed elongate nodules at 20 dpi under these conditions, indicating that nodule branching was not necessarily stimulated in poorly nodulated plants (Supplemental Figure 12).
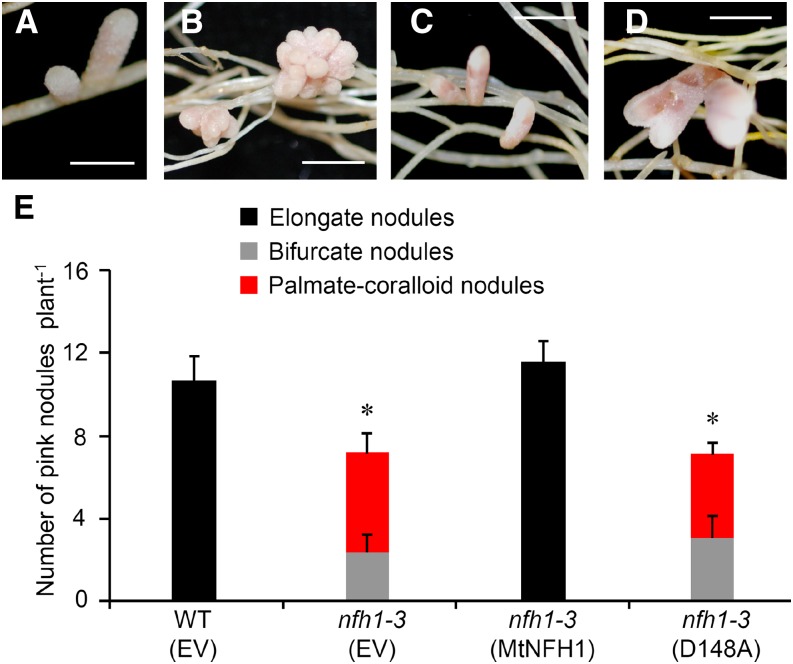
To confirm that the nodulation phenotype of the nfh1-3 mutant is indeed caused by MtNFH1 deficiency, we re-expressed MtNFH1 in the nfh1-3 mutant using the A. rhizogenes-mediated root transformation method. Expression of MtNFH1 was driven by its own promoter. Roots of the nfh1-3 mutant transformed with the MtNFH1pro-MtNFH1 construct developed elongate nodules that were morphologically not different from wild-type nodules. The number of pink nodules in roots re-expressing MtNFH1 was also similar to that in wild-type plants. These findings indicate that the wild-type phenotype could be restored. We also analyzed nodule formation on roots of the nfh1-3 mutant transformed with a mutated MtNFH1pro-MtNFH1(D148A) construct; MtNFH1(D148A) is an enzymatically inactive MtNFH1 variant (Tian et al., 2013). In this case, the nodules formed were morphologically identical to nfh1-3 nodules, indicating that the effect of MtNFH1 on nodule shape depends on its enzyme activity (Figure 8).
Figure 8.
Expression of MtNFH1 and MtNFH1(D148A) in the nfh1-3 Mutant.
Roots of the nfh1-3 mutant plants were transformed with the MtNFH1pro-MtNFH1 or MtNFH1pro-MtNFH1(D148A) construct. For comparison, wild-type (R108) plants and the nfh1-3 mutant were transformed with the empty vector pCAMBIA1302. Plants with transgenic roots were then inoculated with S. meliloti Rm41. Nodule formation was analyzed at 20 dpi.
(A) Elongate nodules formed on wild-type roots transformed with the empty vector.
(B) Palmate-coralloid nodules formed on nfh1-3 roots transformed with the empty vector.
(C) Elongate nodules formed on nfh1-3 roots transformed with MtNFH1pro-MtNFH1.
(D) Palmate-coralloid nodules formed on nfh1-3 roots transformed with MtNFH1pro-MtNFH1(D148A).
(E) Quantification of different types of nodules. Data from individually analyzed plants (n ≥ 12) indicate means (±se). The nfh1-3 mutant transformed with the empty vector or the MtNFH1pro-MtNFH1(D148A) construct formed fewer nodules than wild-type plants transformed with the empty vector (asterisks; Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
EV, empty vector; MtNFH1, transformed with MtNFH1pro-MtNFH1; D148A, transformed with MtNFH1pro-MtNFH1(D148A). Bars = 2 mm.
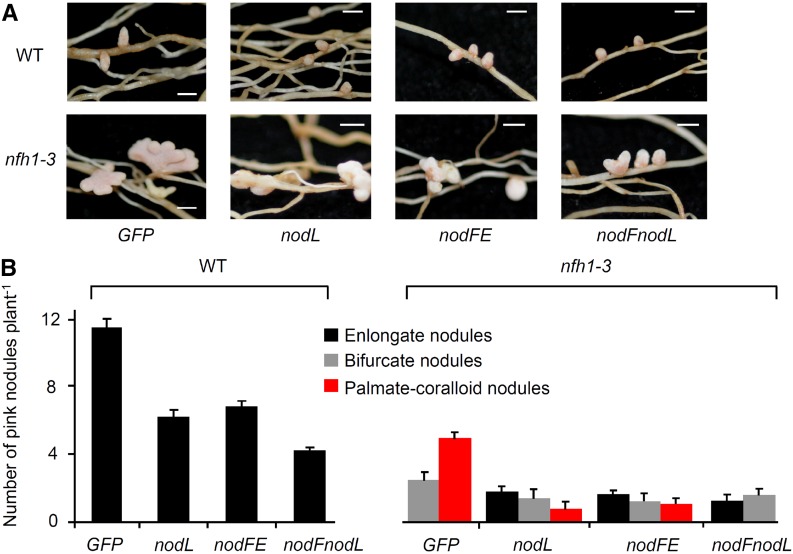
Next, we analyzed the branching of nfh1-3 nodules harboring S. meliloti 2011 mutants that synthesize structurally modified NFs. For comparison, wild-type plants were included into the experiment. Mutants deficient in the acetyl transferase NodL produce NFs that lack an O-acetyl group at the nonreducing end of the chitooligosaccharide moiety. As in wild-type M. truncatula plants (Ardourel et al., 1994; Limpens et al., 2003), delayed nodulation was observed for nfh1-3 plants inoculated with a nodL mutant compared with the parent strain 2011. Accordingly, nodules induced by the nodL mutant were smaller. Nevertheless, many nfh1-3 nodules were branched at 20 dpi (Figure 9). Similar data were also obtained for nodules harvested at 15 dpi (Supplemental Figure 13). Hence, branching of nfh1-3 nodules also occurred with bacteria that produce NFs without O-acetyl modification. S. meliloti mutants lacking nodFE or nodF do not synthesize NFs with C16 unsaturated fatty acids (16:1, 16:2, or 16:3) and instead produce C18:1 NFs with vaccenic acid (Demont et al., 1993). Nodules on nfh1-3 roots induced either by a nodFE mutant or a nodF/nodL double mutant were frequently branched when analyzed at 15 dpi (Supplemental Figure 13) and 20 dpi (Figure 9). These findings indicate that the nodule branching phenotype of nfh1-3 plants is not restricted to S. meliloti producing NFs with C16 unsaturated fatty acids.
Figure 9.
Nodules Induced by S. meliloti 2011 Mutants Producing Modified NFs (20 dpi).
M. truncatula R108 wild-type and nfh1-3 mutant plants were inoculated with S. meliloti strains that produce different NFs.
(A) Photographs of nodules at the time of harvest (20 dpi). Bars = 2 mm.
(B) Quantification of different types of nodules. Only elongate nodules were observed for wild-type plants. The nfh1-3 mutant formed elongate, bifurcate and palmate-coralloid nodules. Data indicate means (±se) based on analysis of 10 plants of each genotype.
GFP, strain 2011 carrying pHC60 constitutively expressing GFP; nodL, strain 2011nodL::Tn5-GFP producing NFs without O-acetyl group at the nonreducing end; nodFE, strain 2011ΔnodFE-GFP producing NFs with vaccenic acid; nodFnodL, strain ΔnodFnodL::Tn5-GFP producing NFs with vaccenic acid and lacking an O-acetyl group.
In addition to the nfh1-3 mutant, we examined plant lines with reduced MtNFH1 activity, namely, the nfh1-1 and nfh1-2 mutants (Figure 1C) and two RNAi lines constructed previously (L3 and L5; Tian et al., 2013). The distribution of nodules on the root system induced by S. meliloti Rm41 was sometimes clustered in these plant lines. Bifurcate nodules were observed at 20 dpi, particularly for the two RNAi lines. In contrast to the nfh1-3 mutant, however, nodules with a coralloid structure were not observed. In general, nodule branching of plants with reduced MtNFH1 activity was less pronounced as compared with the nfh1-3 mutant (Figure 10).
Figure 10.
Nodulation Phenotype of Various Genotypes with Reduced MtNFH1 Activity.
Plants (≥9 per genotype) were inoculated with S. meliloti Rm41 and harvested at 20 dpi. Bars = 2 mm.
(A) Nodules formed on wild-type R108 roots (normal MtNFH1 activity).
(B) Nodules formed on roots of the nfh1-3 mutant (no MtNFH1 activity).
(C) Nodules formed on roots of the nfh1-1 mutant (reduced MtNFH1 activity).
(D) Nodules formed on roots of the nfh1-2 mutant (reduced MtNFH1 activity).
(E) and (F) Nodules formed on roots of the RNAi lines L3 and L5 (reduced MtNFH1 activity; Tian et al., 2013).
Plants Expressing MtNFH1 Driven by a Tandem CaMV 35S Promoter Show Aberrant Nodule Branching
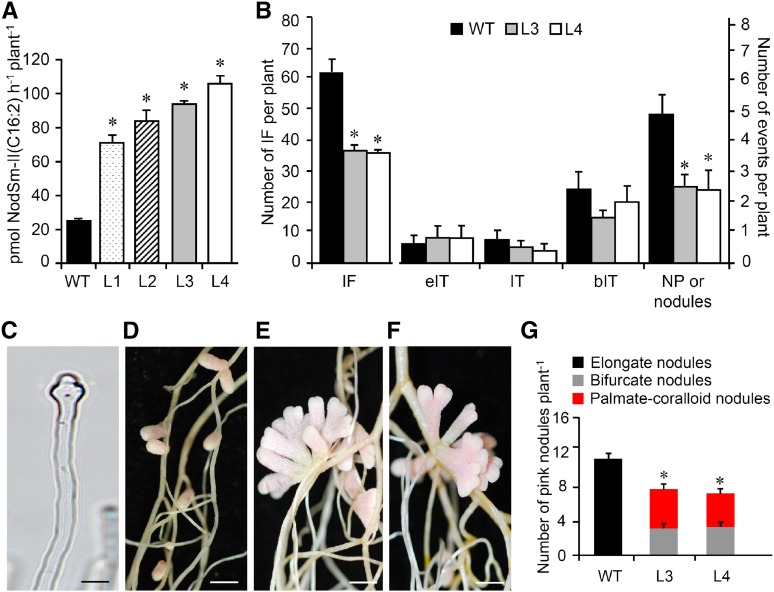
To study the effects of constitutive MtNFH1 expression, M. truncatula plants were stably transformed with A. tumefaciens carrying pISV-MtNFH1. The transgenic plants expressed MtNFH1 under the control of a tandem CaMV 35S promoter. NF hydrolysis tests showed that the MtNFH1 activity of obtained lines was increased compared with wild-type plants (Figure 11A). Two homozygous lines (L3 and L4) were further characterized. Root hair infection was frequently abnormal in these plants. In contrast to wild-type plants, root hairs showed often atypical swelling at the root hair tip as well as aberrant deformations such as root hair branching (Figure 11C; Supplemental Figure 14). We therefore expected impaired infection in these plants and quantitatively assessed the different infection stages. The number of infection foci in L3 and L4 plants was reduced in comparison with wild-type plants. Furthermore, nodule primordia (or young nodules) were frequently clustered together and their numbers were lower than in wild-type plants (Figure 11B). Nodules formed on L3 and L4 roots showed aberrant nodule branching. Nodules had a bifurcate or even a palmate-coralloid shape at 20 dpi, whereas wild-type plants only developed elongate nodules. The number of pink nodules formed on L3 and L4 lines roots was lower than that on wild-type roots at 20 dpi (Figures 11D to 11G). Hence, the L3 and L4 lines and the nfh1-3 mutant had similar symbiotic phenotypes.
Figure 11.
Symbiotic Phenotype of M. truncatula Plants Expressing MtNFH1 Driven by a Tandem CaMV 35S Promoter.
M. truncatula R108 plants were stably transformed with A. tumefaciens carrying pISV-MtNFH1. The symbiotic phenotype of two transgenic lines with increased NF-cleaving activity was compared with wild-type plants.
(A) Hydrolysis of NFs by intact roots of M. truncatula lines constitutively expressing MtNFH1. Roots of seedlings from wild-type R108 plants and four independent lines constitutively expressing MtNFH1 driven by a tandem CaMV 35S promoter (named L1 to L4; T4 generation) were first individually pretreated with 0.1 μM NodSm-IV(C16:2, S) for 24 h and then incubated with 15 μM NodSm-IV(C16:2, S) for 18 h. Formation of NodSm-II(C16:2) was analyzed by reverse-phase HPLC (1 plant per sample). Data indicate means ± se. In total, 36 transgenic and nine wild-type plants were analyzed. Hydrolysis of NFs by the four lines was significantly elevated compared with wild-type plants (Kruskal-Wallis test; P < 0.05; Supplemental File 1).
(B) Analysis of early symbiotic stages in wild-type, L3, and L4 lines inoculated with S. meliloti 2011 carrying pXLGD4 (lacZ). Roots were harvested at 7 dpi and stained with X-Gal to visualize bacteria. Data indicate means (±se) for 14 plants per genotype. Significant differences between the L3 or L4 lines and wild-type plants are marked with asterisks (Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1). IF, infection foci; eIT, elongating infection thread in root hair; IT, fully elongated infection thread in root hair; bIT, branched infection thread in cortex; NP, nodule primordium.
(C) Photograph of an abnormal root hair of L4 showing tip swelling induced by S. meliloti 2011 carrying pXLGD4 (3 dpi). Bar = 20 μm.
(D) to (F) Photographs of nodules induced by S. meliloti Rm41 harvested at 20 dpi. Wild-type plants (D) formed elongate nodules while the L3 (E) and L4 (F) lines formed bifurcate or palmate-coralloid nodules. Bars = 2 mm.
(G) Quantification of different types of nodules formed by wild-type, L3, and L4 plants. Data indicate means values (±se) from 10 plants per genotype. The number of nodules formed on L3 or L4 roots was significantly lower than on wild-type roots (differences marked by asterisks; Kruskal-Wallis test, P ≤ 0.05; Supplemental File 1).
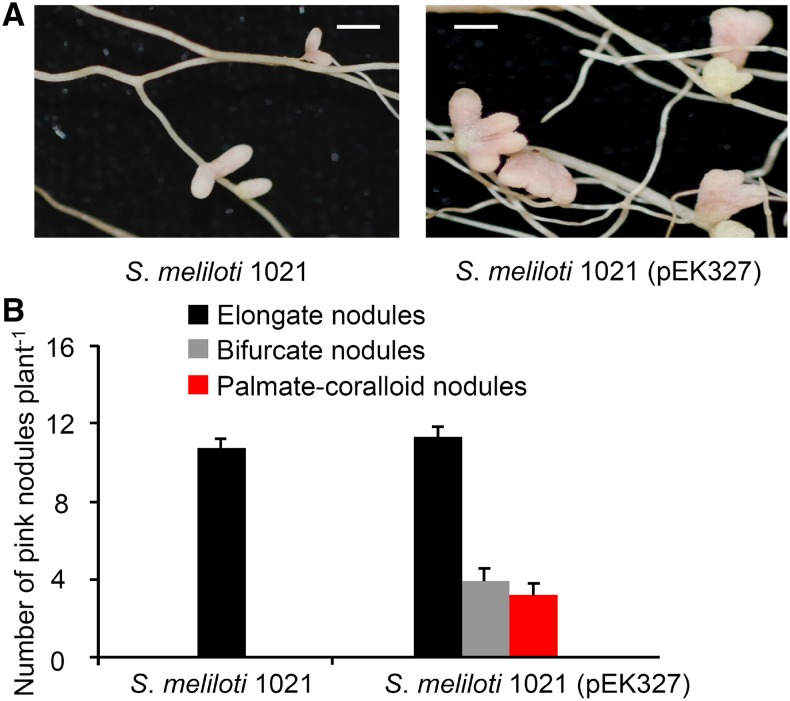
Overproduction of NFs Promotes Nodule Branching
To substantiate the role of NF levels in nodule branching, we inoculated M. truncatula R108 plants with S. meliloti 1021 (pEK327), a NF-overproducing strain carrying a plasmid with extra copies of nod genes (Schultze et al., 1992). The parent strain 1021 was used for comparison. As in previous experiments, plants inoculated with 1021 formed elongate nodules at 20 dpi. However, the NF-overproducing strain 1021 (pEK327) induced nodules that had an elongate, bifurcate, or even a palmate-coralloid shape. The total number of pink nodules was increased when plants were inoculated with the NF-overproducing strain (Figure 12). These results show that overproduction of NFs stimulates formation and branching of nodules.
Figure 12.
Nodule Shape of M. truncatula Plants Inoculated with a NF-Overproducing S. meliloti Strain.
(A) Photographs of nodules on M. truncatula R108 roots inoculated with S. meliloti 1021 or S. meliloti 1021 (pEK327). Plants were harvested at 20 dpi. Bars = 2 mm.
(B) Quantification of different types of nodules. Data indicate means (±se) from 12 plants per strain. S. meliloti 1021 (pEK327) induced significantly more nodules than the parent strain 1021 (Kruskal-Wallis test, P ≤ 0.05).
DISCUSSION
In this work, we found a symbiotic role for the lipodisaccharide-forming NF hydrolase protein MtNFH1. MtNFH1 promotes root hair infection of S. meliloti and formation of elongate nodules. Hydrolysis of NodSm-IV(C16:2, Ac, S) in the rhizosphere was abolished in the nfh1-3 mutant (Figure 1C). Accordingly, the ConA binding protein fraction from nfh1-3 roots did not show NF cleaving activity (Supplemental Figure 6A). These findings indicate that MtNFH1 is likely an N-glycosylated protein, a characteristic reported for MtNFH1-related class V chitinases in Nicotiana tabacum and Cycas revoluta (Melchers et al., 1994; Taira et al., 2009). The ConA binding properties of MtNFH1 are congruent with the previously characterized lipodisaccharide-forming activity of M. sativa (Staehelin et al., 1995).
NodSm-IV(Ac, S) was preferentially used in our hydrolysis tests because this NF is the most abundant NF of S. meliloti and efficiently cleaved by MtNFH1. Other NFs of S. meliloti, namely, NodSm-IV(C16:2, S) and NodSm-V(C16:2, S), are substrates for various plant chitinases in vitro (Staehelin et al., 1994b; Schultze et al., 1998; Minic et al., 1998; Ovtsyna et al., 2000). It is worth mentioning that nfh1-3 seedlings could hydrolyze pentameric NodSm-V(C16:2, S), indicating the presence of an additional uncharacterized enzyme in the rhizosphere (Supplemental Figure 3). However, an activity cleaving NodSm-IV(C16:2, Ac, S) into lipotrisaccharidic NodSm-III(C16:2, Ac), as previously described for M. sativa roots (Minic et al., 1998), was not detected in the nfh1-3 mutant under the used test conditions. The high stability of NFs in the rhizosphere of nfh1-3 therefore indicates that NF levels steadily increase over time when inoculated bacteria continuously produce NFs.
Analysis of the MtNFH1:GFP fusion protein in inoculated M. truncatula root hairs revealed strong fluorescence signals for the infection chamber of curled root hairs (Figure 5). These findings suggest a high turnover of NFs in the infection chamber. By contrast, MtNFH1:GFP was not detected in developed infection threads (Supplemental Figure 10D). Secretion of MtNFH1:GFP into the culture medium may result in fluorescence signals below the detection threshold. Localization of MtNFH1:GFP to the infection chamber suggests focal exocytosis of MtNFH1 at the tip of the root hair. Exocytosis of symbiosis-related proteins such as MtENOD11 has been proposed to play an important step in remodeling of the infection chamber, a prerequisite for subsequent infection thread initiation (Fournier et al., 2015). Like MtENOD11, expression of MtNFH1 in root hairs is strongly induced upon rhizobial inoculation (Figure 5). This finding is in accordance with expression data from transcriptomic studies (Breakspear et al., 2014; Jardinaud et al., 2016). Accordingly, a NF pretreatment of M. truncatula seedlings for 2 h was sufficient to stimulate MtNFH1 expression and NF hydrolysis in the rhizosphere (Figure 3). Such upregulation of MtNFH1 expression and NF hydrolysis was not detected in M. truncatula mutants deficient in NF signaling (Figure 2), indicating that MtNFH1 synthesis is regulated by NF signaling. Increased NF hydrolysis in response to applied S. meliloti NFs was previously observed for M. sativa seedlings. Remarkably, a concentration of 10−10 M NodSm-IV(Ac, S) was sufficient to obtain half-maximal stimulation of NF-cleaving activity in the rhizosphere (Staehelin et al., 1995). The NF pretreatment experiments of this study show similar results for MtNFH1 expression and NF hydrolysis in the rhizosphere of M. truncatula (Figure 3). Hence, high levels of NFs produced by root hair colonizing rhizobia are rapidly decreased by MtNFH1. The Michaelis-Menten constants of MtNFH1 for NFs are in the micromolar range (Tian et al., 2013); thus, NFs are efficiently hydrolyzed at high concentrations. By contrast, low NF concentrations only weakly stimulate MtNFH1 expression and rates of NF breakdown are low due to low enzyme amounts and low substrate concentrations.
Based on these considerations, we suggest that fine-tuning of NF levels promotes infection at the stage of infection thread initiation. In the case of the nfh1-3 mutant, too high levels of NFs likely resulted in infection defects as compared with wild-type plants. Excess amounts of NFs at infection foci apparently induced uncontrolled curling, swelling, or bending without the formation of a proper infection chamber (Figure 4; Supplemental Figure 8). NF levels at the stage of infection thread initiation appear to be also important for nodulation of pea roots (cv Afghanistan) by R. leguminosarum strain TOM. Increased NF production by expression of nodD in strain TOM strongly suppressed nodulation. The presence of R. leguminosarum strains producing high amounts of NFs and purified NFs applied to the roots also efficiently reduced nodulation by strain TOM (Hogg et al., 2002). However, levels of NFs below a certain threshold are certainly suboptimal for infection. M. truncatula plants expressing MtNFH1 driven by a tandem CaMV 35S promoter showed increased NF hydrolysis in the rhizopshere and the observed delay in infection thread initiation likely reflects spatiotemporal abnormalities in NF levels (Figure 11; Supplemental Figure 14).
It is worth noting in this context that a nodM (glucosamine synthase) mutant of S. meliloti produces only low amounts of NFs. When inoculated on M. sativa, delayed nodulation was observed as compared with the parental strain (Baev et al., 1991, 1992). Moreover, expression of a bacterial chitinase (chiB from Serratia marcescens) in S. meliloti RCR2011 and S. fredii USDA191 resulted in hydrolysis of produced NFs and delayed nodulation of M. sativa and soybean (Glycine max), respectively (Krishnan et al., 1999).
Activation of nod genes and synthesis of NFs in rhizobia is induced through secretion of flavonoids by the host plants. Once NF signaling is activated in the host legume, secretion of flavonoids is enhanced in many strain-host interactions, including M. sativa (so-called Ini response; Recourt et al., 1992; Dakora et al., 1993; Schmidt et al., 1994). This positive feedback response stimulates further NF synthesis. Activation of NF signaling in root hairs also results in MtNFH1 expression and reduced NF levels, indicating a negative feedback response. Hence, positive and negative feedback circuits guarantee that NF levels continuously remain at an optimal concentration during infection. Such NF homeostasis is reminiscent to nodule autoregulation, a regulatory feedback loop that restricts the number of formed nodules on the root through action of a shoot-derived inhibitor signal. Synthesis of this unknown inhibitor depends on CLE peptides, the ascending long-distance signals to the shoot. Expression of specific CLE genes in roots is mediated by activation of NF signaling (Staehelin et al., 2011; Hastwell et al., 2015). Formation of nodule primordia in the nfh1-3 mutant was frequently clustered in our study (Figure 4F), suggesting that NF levels influence distribution of nodules within the root system. Future work is required to assess the relationship between MtNFH1-mediated NF hydrolysis and autoregulation.
M. truncatula forms indeterminate nodules that typically consist of a persistent meristem at the nodule apex, an infection zone, a zone with nitrogen-fixing bacteroids, and a senescence zone. Young M. truncatula nodules have an elongate shape and nodules only start branching when they become older. Formation of bifurcate nodules is the result of apical meristem branching. The meristem splits into two parts when cells in the median part of the meristem stop dividing (Łotocka et al., 2012). A surprising finding of this work was that the nfh1-3 mutant formed nodules that exhibited strong branching and hypertrophy (Figures 6 and 7). Re-expression of MtNFH1 in nfh1-3 restored the wild-type phenotype. By contrast, MtNFH1(D148A) expression did not affect the nodule shape, indicating that MtNFH1(D148A), a MtNFH1 variant deficient in NF hydrolase activity (Tian et al., 2013), was symbiotically inactive (Figure 8). These findings indicate that the observed nodulation phenotype of nfh1-3 depends on the enzymatic activity of MtNFH1.
Nodules of the nfh1-3 mutant showed peripheral vascular bundles that branched to develop an appropriate vascular system for each nodule lobe. Thus, the peripheral vascular bundles in nfh1-3 nodules are wild-type-like and different from nodules with central vascular bundles formed by certain M. truncatula mutants (such as lin; Guan et al., 2013). Compared with wild-type plants, the multilobed nodules of nfh1-3 had a higher biomass and fixed more nitrogen (Figure 7), suggesting that accelerated nodule growth was leading to branching and hypertrophy. Nodules of wild-type plants induced by S. meliloti overproducing NFs showed a similar phenotype (Figure 12). These findings suggest that abnormally high levels of NFs in mature nodules trigger nodule branching and support the view that NFs are not only required for nodule initiation but also determine nodule shape. NF signaling in mature nodules has been poorly investigated because NF signaling mutants usually lack the capacity to form nodules. Nevertheless, infection defects in M. truncatula nodules were found for certain ipd3 (DMI3-interacting protein) mutants (Horváth et al., 2011; Ovchinnikova et al., 2011) and for nodules with an RNAi-silenced DMI2 (leucine-rich repeat receptor kinase) gene (Limpens et al., 2005). Additional evidence for NF signaling in M. truncatula nodules is provided by a recent study on localization of the NF receptor proteins LYK3 and NFP that are tightly regulated at the posttranslational level in the apical part of the nodule (Moling et al., 2014). Furthermore, many NF-responsive genes, including MtNFH1, are expressed in M. truncatula nodules (Roux et al., 2014; Supplemental Figure 11). NF signaling in mature nodules is also supported by immunolocalization of NFs in M. sativa nodules. Detection signals were mainly found in the infection zone but also appear to be present in the apical meristematic zone (Timmers et al., 1998), suggesting diffusion of NFs over a short distance.
Branching of nfh1-3 nodules and of nodules harboring NF-overproducing bacteria is likely associated with altered meristematic activity triggered by high NF levels. In this context, it is worth mentioning that M. truncatula and P. sativum plants with mutations in the NOOT (NODULE ROOT) and COCH (COCHLEATA) genes have been identified. These genes are related to BLADE-ON-PETIOLE transcriptional regulators of Arabidopsis thaliana. Mutations in NOOT/COCH caused various changes in plant development and perturbation of the nodule meristem. Nodules of these mutants are often branched and plants frequently form root-like structures (noot phenotype) or agravitropic roots (coch phenotype) emerging from the apical nodule meristem (Ferguson and Reid, 2005; Couzigou et al., 2012). Furthermore, similar to the nfh1-3 mutant, increased growth and nodule branching was observed for a cre1 (cytokinin receptor gene) mutant of M. truncatula. Older nodules often showed multiple lobes while wild-type nodules were mainly nonbranched. The authors of this study suggested that cytokinin in mature nodules regulates the balance between cell proliferation and differentiation (Plet et al., 2011). NF and cytokinin signaling pathways at early stages of symbiosis are directly linked and NF signaling results in accumulation of cytokinins in M. truncatula roots (van Zeijl et al., 2015; Boivin et al., 2016a; Miri et al., 2016). Based on the phenotypic similarities between the nfh1-3 and cre1 mutants, we hypothesize that NF levels in mature nodules control nodule branching in concert with cytokinin.
Moreover, auxin likely also affects growth and branching of nodules. Polar auxin fluxes play a crucial role in nodule initiation and auxin levels are expected to control cell divisions in indeterminate nodules (Breakspear et al., 2014; Ng et al., 2015; Laplaze et al., 2015; Boivin et al., 2016b). For example, nodules of Vicia hirsuta induced by an auxin-overproducing R. leguminosarum bv viciae strain showed an increased meristematic activity, resulting in enlarged and occasionally branched nodules (Camerini et al., 2008). Such nodule hypertrophy is reminiscent of the nodules formed by the nfh1-3 mutant. It is tempting to speculate that auxin levels or auxin fluxes in nodules are fine-tuned by levels of NFs and that MtNFH1 protects the meristematic zone from excess of NFs.
M. truncatula plants expressing MtNFH1 driven by a tandem CaMV 35S promoter showed nodule branching (Figure 11). In this context, it is worth noting that the CaMV 35S promoter activity is unequally distributed in nodules. Auriac and Timmers (2007) could not detect CaMV 35S promoter activity in the nodule meristem and in bacteroid-containing cells. Nevertheless, the nodule shape of our transgenic plants suggests spatiotemporal changes in NF levels that apparently caused misregulation of the apical meristem comparable to nodules formed by the nfh1-3 mutant. Formation of elongate nodules may depend on NFs that are active in a defined concentration range. NFs in the nodule apex could function as morphogens that trigger host gene expression in a concentration-dependent manner (Rogers and Schier, 2011). In M. truncatula nodules, the NF receptor proteins LYK3 and NFP localize in specific cell layers that form the border between the nodule meristem and the infection zone (Moling et al., 2014). Maintenance of a robust NF gradient from the infection zone (high NF concentrations) to the apical nodule meristem (reduced NF concentration) would be regulated by MtNFH1 that continuously inactivates the signal. Moderate concentrations of NFs are perhaps necessary for polarity of NF-exposed host cells and positional information directing cell divisions in a way that the meristem does not split into two parts. NF levels above or below thresholds would cause NF concentration gradients to flatten or disappear and consequently trigger nodule branching.
Taken together, this study demonstrates a role for MtNFH1 in the symbiosis between M. truncatula and S. meliloti. We suggest that MtNFH1 spatiotemporally limits NF levels to reach an optimal NF activity. In this view, NF hydrolysis can be considered as NF inactivation. In fact, hydrolyzed S. meliloti NFs applied to M. sativa roots showed reduced activity in stimulating root hair deformations and in inducing the lipodisaccharide-forming activity (Staehelin et al., 1994b, 1995). In addition, it could be hypothesized that the NF cleavage products represent secondary signals that influence processes related to root hair infection and nodule branching. Cleavage products could retain certain binding activity to NF receptors or even possess biological activity different from NFs. NF cleavage is probably a general phenomenon of nodule symbiosis. We suggest that different host plants have recruited various chitinases for symbiosis to hydrolyze the different types of NFs from their microsymbionts.
In conclusion, our data indicate that NF hydrolysis by specific hydrolases such as MtNFH1 play an important role in the fine-tuning of the symbiosis at two key points, namely, at the onset of infection (root hair curling and entry of bacteria) and in mature nodules.
METHODS
Medicago truncatula Genotypes and Bacterial Strains
M. truncatula ecotype R108, ecotype Jemalong A17, and cv Jester were used in this study. The mutants nfh1-1 (NF16587), nfh1-2 (NF12841), and nfh1-3 (NF11260) derived from M. truncatula R108 populations containing Tnt1 retrotransposon insertions (ďErfurth et al., 2003; Tadege et al., 2008; Pislariu et al., 2012). Clare Gough (CNRS, Toulouse, France) provided nodulation signaling mutants of M. truncatula Jemalong A17, namely, dmi1-3 (Y6; Catoira et al., 2000), dmi2-1 (TR25; Sagan et al., 1995), dmi3-1 (TRV25; Sagan et al., 1998), and hcl-1 (B56, lyk3; Catoira et al., 2001; Smit et al., 2007). Construction of RNAi-silenced R108 lines, showing reduced MtNFH1 transcript levels, has been reported previously (Tian et al., 2013). Construction of R108 lines constitutively expressing MtNFH1 is described below. M. truncatula R108 seeds were surface sterilized with diluted commercial NaClO for 20 min on a shaker (50 rpm). The seeds were washed with sterilized water three times and then incubated on 0.8% (w/v) agar plates in the dark at 4°C for 48 h. The seeds were left to germinate in the dark at 20°C for 18 h and then used for hydrolysis tests with Sinorhizobium meliloti NFs in 1-mL syringes.
For inoculation of M. truncatula plants, germinated seedlings were placed into 300-mL plastic jars, which had been filled with sterilized vermiculite and expanded clay (3:1 v/v) in the upper unit and B&D nutrient solution (Broughton and Dilworth, 1971) containing 1 mM KNO3 in the lower unit. Plants were kept in an air-conditioned growth room (16-h photoperiod; 2000 lux light intensity, Philips Lifemax TL-D 36W/54-765 and TL-D 36W/29-530 daylight fluorescent tubes at a ratio 3:1; 24°C). The following S. meliloti strains were used for inoculation: strain Rm41, strain 1021 (SU47 derivative), strain 1021 (pQDN03) constitutively expressing mCherry (Haney and Long, 2010), strain 1021 (pEK327) overexpressing NFs (Schultze et al., 1992), strain 2011 (SU47 derivative) carrying pXLGD4 with a hemApro-lacZ construct producing β-galactosidase (Ardourel et al., 1994), strain 2011 carrying pHC60 constitutively expressing GFP (Cheng and Walker, 1998), strain 2011nodL::Tn5-GFP (GMI6436 carrying pHC60) producing NFs without O-acetyl group at the nonreducing (Ardourel et al., 1994; Limpens et al., 2003), strain 2011ΔnodFE-GFP (GMI5622 carrying pHC60) producing NFs with vaccenic acid (Debellé et al., 1988; Demont et al., 1993; Limpens et al., 2003), and strain ΔnodFnodL::Tn5-GFP (GMI6628 carrying pHC60) producing NFs with vaccenic acid and lacking an O-acetyl group (Ardourel et al., 1994; Limpens et al., 2003). Agrobacterium tumefaciens, Agrobacterium rhizogenes, and Escherichia coli strains used for plasmid construction and plant transformation are listed in Supplemental Data Set 1.
Plasmid Construction
To estimate MtNFH1 promoter activity, a 2051-bp DNA fragment upstream of the start codon (GenBank accession no. KY751027) was PCR-amplified using genomic DNA of M. truncatula R108 and finally cloned into the binary vector pCAMBIA1305.1 digested with HindIII and NcoI. The constructed binary vector contains the MtNFH1 promoter with the GUS reporter gene (MtNFH1pro-GUS) and a RFP expression cassette (35Spro-RFP) inserted at the HindIII site.
To analyze localization of GFP-tagged MtNFH1 in M. truncatula root hairs, DNA consisting of the 2051-bp promoter sequence and the coding sequence of MtNFH1 was PCR-amplified from genomic DNA of M. truncatula R108 and inserted at the PstI site of the binary vector pCAMBIA1302 (yielding a MtNFH1pro-MtNFH1:GFP construct).
To rescue the nfh1-3 mutant phenotype by MtNFH1 re-expression, a similar DNA fragment [containing the 2051-bp the promoter sequence and the coding sequence of MtNFH1 or MtNFH1(D148A)] was inserted into pRT104 (Töpfer et al., 1987) and finally [with the poly(A) tail from pRT104] cloned into the binary vector pCAMBIA1302.
To create M. truncatula transformants constitutively expressing MtNFH1, the coding sequence of MtNFH1 was cloned as XbaI-XhoI fragment into pISV2678 (yielding pISV-MtNFH1; Supplemental Data Set 1). Transformants showed increased MtNFH1 expression driven by the used tandem CaMV 35S promoter.
All constructs were verified by DNA sequencing. Further information on the different plasmids is provided in Supplemental Data Set 1. Primer sequences for plasmid constructions are shown in Supplemental Data Set 2.
RT-qPCR
RT-qPCR was performed to measure transcript levels of MtNFH1 in M. truncatula wild-type and mutant plants. The tissue was extracted by using the RNAprep Pure Plant Kit (Tiangen Biotech). Each RNA sample was converted into cDNA by reverse transcription with the PrimeScript TM RT reagent Kit (TaKaRa). qPCR was performed with real-time PCR Master Mix (Faststart SYBR Green I; Roche) using a Roche LightCycler 480 System. Thermocycling conditions were as follows: (1) 95°C for 3 min; (2) 40 cycles: 95°C for 30 s, 60°C for 30 s, 72°C for 20 s; (3) 37°C for 4 min. All RNA extractions were performed in triplicate (three biological replicates) and each derived cDNA sample was three times PCR-analyzed (three technical replicates). Reactions with M. truncatula Ubiquitin gene primers served as internal reference to calibrate the value of transcript abundance among different samples. Threshold cycles (Ct values) were calculated using Roche LightCycler 480 software. Primers used for RT-qPCR are listed in Supplemental Data Set 2.
Identification of nfh1 Mutants
The M. truncatula Tnt1 retrotransposon insertion mutant collection, available at Noble Research Institute (ďErfurth et al., 2003; Tadege et al., 2008; Pislariu et al., 2012), was PCR-screened for nfh1 mutants. Genomic DNA samples of M. truncatula R108 were used for PCRs with Tnt1 primers and MtNFH1-specific primers listed in Supplemental Data Set 2. Obtained PCR products were sequenced to determine the Tnt1 insertion sites in the obtained mutants nfh1-1 (NF16587), nfh1-2 (NF12841), and nfh1-3 (NF11260). Details are shown in Figure 1. In addition, seedlings were analyzed for their capacity to hydrolyze NFs of S. meliloti.
NF Purification
NodSm-V(C16:2, S), NodSm-IV(C16:2, S), and NodSm-IV(C16:2, Ac, S) of S. meliloti strain 1021 (pEK327) (Schultze et al., 1992) were purified by reverse-phase HPLC as described (Staehelin et al., 1994b; Tian et al., 2013).
NF Hydrolysis Tests
NF hydrolysis with intact seedlings was performed as described previously (Staehelin et al., 1994b, 1995). Briefly, roots of germinated M. truncatula plants (wild-type, mutants, and lines constitutively expressing MtNFH1) were incubated in 1-mL plastic syringes, which were filled with 400 μL Jensen medium (Van Brussel et al., 1982) containing 0.5% (v/v) DMSO and the NF substrate at specified amounts (Supplemental Figure 15). Plants were incubated in the dark at 24°C for indicated time periods (3 h if not otherwise stated). Where indicated, seedlings were first pretreated with 0.1 or 1 μM NFs (24°C; 18 h if not otherwise mentioned) and then transferred to new 1-mL plastic syringes filled with 400 μL Jensen medium containing 0.5% (v/v) DMSO and the NF substrate. For HPLC analysis, nondegraded NFs and the acylated cleavage products from single or three to four seedlings were extracted with an equal volume of distilled n-butanol. This step was repeated for assays with NodSm-IV(C16:2, Ac, S). The samples were dried in a speed-vac evaporator, dissolved in 1 μL of DMSO, and analyzed by reverse-phase HPLC (Nova Pak C18; Waters) using 35% (v/v) acetonitrile/water containing 40 mM ammonium acetate as the mobile phase (Staehelin et al., 1994b).
To analyze the NF-cleaving activity isolated from M. truncatula roots, NFs were incubated with protein preparations in vitro as described (Tian et al., 2013).
Isolation of MtNFH1 from M. truncatula Roots
Proteins of M. truncatula R108 roots (wild type and nfh1-3) were extracted from roots inoculated with S. meliloti Rm41 and harvested at 20 dpi. N-glycosylated proteins were purified on a column containing ConA-agarose beads as described (Xiong et al., 2007). Proteins were eluted in 25 mM sodium acetate buffer (pH 5.0) containing 0.2 M methyl-α-d-mannopyranoside and used for NF hydrolysis tests. ConA binding proteins from M. truncatula cv Jester were obtained in a similar way and then subjected to gel filtration chromatography (fast protein liquid chromatography; gel filtration Superdex 75 10/300 GL column; 25 mM sodium acetate, pH 5.5, as running buffer).
A. rhizogenes-Mediated Transformation
Binary vectors with MtNFH1pro-GUS, MtNFH1pro-MtNFH1:GFP, MtNFH1pro-MtNFH1, and MtNFH1pro-MtNFH1(D148A) constructs were mobilized into A. rhizogenes LBA9402 by electroporation. Transgenic hairy roots were induced by A. rhizogenes LBA9402 carrying a given binary vector as described (Boisson-Dernier et al., 2001; Limpens et al., 2004).
Agrobacterium-Mediated Transformation
Agrobacterium strain EHA105 carrying pISV-MtNFH1 was used to create stable transformants constitutively expressing MtNFH1 under the control of a tandem CaMV 35S promoter. Leaf disk infiltration and regeneration of calli to whole M. truncatula R108 plants was performed according to published procedures (Hoffmann et al., 1997; Trinh et al., 1998). For selection, 6 mg L−1 glufosinate (phosphinothricin) was added to the agar plates. Regenerated plantlets were transferred into plastic jars containing vermiculite and expanded clay. After selfing, transformants were propagated and selected by spraying with 80 mg L−1 glufosinate. Transformants were confirmed by PCR analysis with primers listed in Supplemental Data Set 2. Plants from two homozygous lines (L3 and L4) with increased NF-cleaving activity were used for nodulation tests.
Histochemical Staining and Microscopic Analyses of Symbiotic Interactions
GUS activity in M. truncatula R108 roots and nodules containing the MtNFH1pro-GUS construct (inoculated with S. meliloti Rm41) were stained with X-Gluc solution (37°C for maximally 12 h). Tissues were then cleared with diluted NaClO and washed with water. Visualization of S. meliloti-2011-lacZ in wild type, nfh1-3 mutant plants, and lines constitutively expressing MtNFH1 was performed by staining tissues with X-Gal solution. Infection events were examined using bright-field microscopy. Analysis of transgenic root hairs expressing GFP-tagged MtNFH1 and inoculated with S. meliloti 1021 constitutively expressing mCherry was performed by fluorescence microscopy. Where indicated, nodule sections of M. truncatula R108 and nfh1-3 were prepared. Nodules were fixed overnight (4°C) with 4% (v/v) paraformaldehyde and 5% (v/v) glutaraldehyde in 0.1 mM phosphate buffer (pH 7.2), dehydrated in an ethanol series and finally embedded in paraffin. Sections (5 μm) were cut using a HM340 microtome (Microm) and stained in 0.1% (w/v) ruthenium red solution for 10 min.
For microscopy analysis, either a stereofluorescence microscope (Lumar.V12; Zeiss) or a Zeiss ImagerZ1 fluorescence microscope equipped with a CCD camera (Zeiss) were used.
Nodulation Tests and Determination of Nitrogenase Activity
M. truncatula R108 plants (wild-type plants, nfh1 mutants, nfh1-3 roots expressing MtNFH1, roots transformed with the MtNFH1pro-GUS construct, RNAi lines with reduced MtNFH1 activity, and lines constitutively expressing MtNFH1) were used for nodulation tests with a given S. meliloti strain. After germination, M. truncatula seedlings were transferred to 300-mL plastic jars and inoculated (∼108 bacteria per jar) 7 d later. Nodulation tests were performed with at least nine plants (one plant per jar). In an additional experiment, R108 wild-type plants were inoculated with a stepwise diluted inoculum suspension. Plants were harvested at indicated time points (20 dpi if not otherwise specified). Where indicated, nitrogenase activity was measured with the acetylene reduction method. Formed ethylene (2-h incubation, 28°C) was analyzed by gas chromatography [SP-2100 model; Beijing Beifen-Ruili Analytical Instrument (Group)].
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KY751027 (MtNFH1 promoter region) and KC833515.1 (MtNFH1; Medtr4g116990.1).
Supplemental Data
Supplemental Figure 1. MtNFH1 Transcript Levels of nfh1 Mutants in the Absence of NFs.
Supplemental Figure 2. HPLC Analysis Showing NodSm-IV(C16:2, Ac, S) Stability in the Rhizosphere of M. truncatula R108 Wild-Type and nfh1 Mutant Plants.
Supplemental Figure 3. Stability of NodSm-IV(C16:2, S) and NodSm-V(C16:2, S) in the Rhizosphere of the nfh1-3 Mutant.
Supplemental Figure 4. MtNFH1 Transcript Levels of Nodulation Signaling Mutants in the Absence of NFs.
Supplemental Figure 5. Hydrolysis of NodSm-V(C16:2, S) in the Rhizosphere of Nodulation Signaling Mutants.
Supplemental Figure 6. Analysis of NF Hydrolysis by ConA Binding Proteins Isolated from Wild-Type and nfh1-3 Mutant Plants.
Supplemental Figure 7. Different Infection Events Categorized in this Study.
Supplemental Figure 8. The nfh1-3 Mutant Shows Aberrant Root Hair Infections.
Supplemental Figure 9. Analysis of NF Hydrolysis by nfh1-3 Mutant Plants Expressing the MtNFH1:GFP Fusion Protein.
Supplemental Figure 10. MtNFH1:GFP Did Not Accumulate in Infection Threads.
Supplemental Figure 11. Expression of MtNFH1 in M. truncatula Nodules.
Supplemental Figure 12. Low Inoculum Doses Do Not Affect the Nodule Shape.
Supplemental Figure 13. Nodules Induced by S. meliloti 2011 Mutants Producing Modified NFs (15 dpi).
Supplemental Figure 14. Plants Constitutively Expressing MtNFH1 Frequently Show Aberrant Root Hair Deformations.
Supplemental Figure 15. Illustration of the NFs Hydrolysis Tests with Intact M. truncatula Seedlings.
Supplemental Data Set 1. Strains and Plasmids Used in This Study.
Supplemental Data Set 2. Primers Used in This Study.
Supplemental File 1. Statistical Analyses.
Acknowledgments
We thank Zhi-Yuan Tan (South China Agricultural University) for help with the nitrogenase activity measurements and Christian Wagner (Sun Yat-sen University) for helpful comments on this study. We thank Éva Kondorosi (BRC, Hungarian Academy of Sciences, Szeged, Hungary), Sharon R. Long (Stanford University, Stanford, CA), and Erik Limpens (Wageningen University, Wageningen, The Netherlands) for rhizobial strains. Clare Gough (CNRS, Toulouse, France) kindly provided M. truncatula NF signaling mutants. This work was supported by the National Natural Science Foundation of China (Grant 31670241), by the Department of Science and Technology of Guangdong Province, China (Grant 2016A030313299), by the Science Foundation of the State Key Laboratory of Biocontrol (Grants SKLBC 16A01 and SKLBC 322017A09), and by the Guangdong Key Laboratory of Plant Resources (Grant 2014B030301026). Generation of M. truncatula mutants was supported by National Science Foundation (Grants DBI 0703285 and IOS 1127155) and in part by the Noble Research Institute.
AUTHOR CONTRIBUTIONS
J.C., Z.-P.X., and C.S. conceived and designed the experiments. J.C., L.-Y.Z., W.L., Y.T., J.-S.X., Y.-H.W., R.-J.L., and H.-M.L. performed the experiments. J.W. and K.S.M. provided nfh mutants. J.C., L.-Y.Z., W.L., Y.T., J.-S.X., Z.-P.X., and C.S. analyzed the data. J.C., T.B., Z.-P.X., and C.S. wrote the article.
References
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auriac M.C., Timmers A.C.J. (2007). Nodulation studies in the model legume Medicago truncatula: advantages of using the constitutive EF1α promoter and limitations in detecting fluorescent reporter proteins in nodule tissues. Mol. Plant Microbe Interact. 20: 1040–1047. [DOI] [PubMed] [Google Scholar]
- Baev N., Endre G., Petrovics G., Banfalvi Z., Kondorosi A. (1991). Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol. Gen. Genet. 228: 113–124. [DOI] [PubMed] [Google Scholar]
- Baev N., Schultze M., Barlier I., Ha D.C., Virelizier H., Kondorosi E., Kondorosi A. (1992). Rhizobium nodM and nodN genes are common nod genes: nodM encodes functions for efficiency of nod signal production and bacteroid maturation. J. Bacteriol. 174: 7555–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34: 495–506. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garica F., Bécard G., Rosenberg C., Barker D.G. (2001). Hairy roots of Medicago truncatula as tools for studying nitrogen-fixing and endomycorrhizal symbioses. Mol. Plant Microbe Interact. 14: 693–700. [DOI] [PubMed] [Google Scholar]
- Boivin S., Kazmierczak T., Brault M., Wen J., Gamas P., Mysore K.S., Frugier F. (2016a). Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 39: 2198–2209. [DOI] [PubMed] [Google Scholar]
- Boivin S., Fonouni-Farde C., Frugier F. (2016b). How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 7: 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E.D., Downie J.A., Murray J.D. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini S., Senatore B., Lonardo E., Imperlini E., Bianco C., Moschetti G., Rotino G.L., Campion B., Defez R. (2008). Introduction of a novel pathway for IAA biosynthesis to rhizobia alters vetch root nodule development. Arch. Microbiol. 190: 67–77. [DOI] [PubMed] [Google Scholar]
- Camps C., Jardinaud M.F., Rengel D., Carrère S., Hervé C., Debellé F., Gamas P., Bensmihen S., Gough C. (2015). Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol. 208: 224–240. [DOI] [PubMed] [Google Scholar]
- Catoira R., Galera C., de Billy F., Penmetsa R.V., Journet E.P., Maillet F., Rosenberg C., Cook D., Gough C., Dénarié J. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12: 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R., Timmers A.C.J., Maillet F., Galera C., Penmetsa R.V., Cook D., Dénarié J., Gough C. (2001). The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development 128: 1507–1518. [DOI] [PubMed] [Google Scholar]
- Cheng H.P., Walker G.C. (1998). Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180: 5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J.M., et al. (2012). NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. Plant Cell 24: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakora F.D., Joseph C.M., Phillips D.A. (1993). Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101: 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Maillet F., Vasse J., Rosenberg C., de Billy F., Truchet G., Dénarié J., Ausubel F.M. (1988). Interference between Rhizobium meliloti and Rhizobium trifolii nodulation genes: genetic basis of R. meliloti dominance. J. Bacteriol. 170: 5718–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demont N., Debellé F., Aurelle H., Dénarié J., Promé J.C. (1993). Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J. Biol. Chem. 268: 20134–20142. [PubMed] [Google Scholar]
- d'Erfurth I., Cosson V., Eschstruth A., Luca H., Kondorosi Á., Ratet P. (2003). Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J. 34: 95–106. [DOI] [PubMed]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Reid J.B. (2005). Cochleata: getting to the root of legume nodules. Plant Cell Physiol. 46: 1583–1589. [DOI] [PubMed] [Google Scholar]
- Fournier J., Teillet A., Chabaud M., Ivanov S., Genre A., Limpens E., de Carvalho-Niebel F., Barker D.G. (2015). Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167: 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormachtig S., Lievens S., Van de Velde W., Van Montagu M., Holsters M. (1998). Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell 10: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D., et al. (2013). Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 162: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney C.H., Long S.R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 107: 478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell A.H., Gresshoff P.M., Ferguson B.J. (2015). Genome-wide annotation and characterization of CLAVATA/ESR (CLE) peptide hormones of soybean (Glycine max) and common bean (Phaseolus vulgaris), and their orthologues of Arabidopsis thaliana. J. Exp. Bot. 66: 5271–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Benedito V.A., Wang M., Murray J.D., Zhao P.X., Tang Y., Udvardi M.K. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R., Geurts R., Franssen H., Spaink H.P., Van Kammen A., Bisseling T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B., Trinh T.H., Leung J., Kondorosi Á., Kondorosi É. (1997). A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol. Plant Microbe Interact. 10: 307–315. [Google Scholar]
- Hogg B., Davies A.E., Wilson K.E., Bisseling T., Downie J.A. (2002). Competitive nodulation blocking of cv. Afghanistan pea is related to high levels of nodulation factors made by some strains of Rhizobium leguminosarum bv. viciae. Mol. Plant Microbe Interact. 15: 60–68. [DOI] [PubMed] [Google Scholar]
- Horváth B., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24: 1345–1358. [DOI] [PubMed] [Google Scholar]
- Jardinaud M.F., et al. (2016). A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol. 171: 2256–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H.B., Kim K.Y., Krishnan A.H. (1999). Expression of a Serratia marcescens chitinase gene in Sinorhizobium fredii USDA191 and Sinorhizobium meliloti RCR2011 impedes soybean and alfalfa nodulation. Mol. Plant Microbe Interact. 12: 748–751. [DOI] [PubMed] [Google Scholar]
- Laplaze L., Lucas M., Champion A. (2015). Rhizobial root hair infection requires auxin signaling. Trends Plant Sci. 20: 332–334. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Liang Y., Tóth K., Cao Y., Tanaka K., Espinoza C., Stacey G. (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol. 204: 289–296. [DOI] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633. [DOI] [PubMed] [Google Scholar]
- Limpens E., Ramos J., Franken C., Raz V., Compaan B., Franssen H., Bisseling T., Geurts R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55: 983–992. [DOI] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łotocka B., Kopcińska J., Skalniak M. (2012). Review article: The meristem in indeterminate root nodules of Faboideae. Symbiosis 58: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F., Mauch-Mani B., Boller T. (1988). Antifungal hydrolases in pea tissue. II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 88: 936–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers L.S., Apotheker-de Groot M., van der Knaap J.A., Ponstein A.S., Sela-Buurlage M.B., Bol J.F., Cornelissen B.J., van den Elzen P.J., Linthorst H.J. (1994). A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 5: 469–480. [DOI] [PubMed] [Google Scholar]
- Minic Z., Brown S., De Kouchkovsky Y., Schultze M., Staehelin C. (1998). Purification and characterization of a novel chitinase-lysozyme, of another chitinase, both hydrolysing Rhizobium meliloti Nod factors, and of a pathogenesis-related protein from Medicago sativa roots. Biochem. J. 332: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E.D., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri M., Janakirama P., Held M., Ross L., Szczyglowski K. (2016). Into the root: How cytokinin controls rhizobial infection. Trends Plant Sci. 21: 178–186. [DOI] [PubMed] [Google Scholar]
- Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., Gadella T.W.J., Bisseling T. (2014). Nod factor receptors form heteromeric complexes and are essential for intracellular infection in medicago nodules. Plant Cell 26: 4188–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.L.P., Hassan S., Truong T.T., Hocart C.H., Laffont C., Frugier F., Mathesius U. (2015). Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27: 2210–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E.D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E., et al. (2011). IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Mol. Plant Microbe Interact. 24: 1333–1344. [DOI] [PubMed] [Google Scholar]
- Ovtsyna A.O., Schultze M., Tikhonovich I.A., Spaink H.P., Kondorosi E., Kondorosi A., Staehelin C. (2000). Nod factors of Rhizobium leguminosarum bv. viciae and their fucosylated derivatives stimulate a nod factor cleaving activity in pea roots and are hydrolyzed in vitro by plant chitinases at different rates. Mol. Plant Microbe Interact. 13: 799–807. [DOI] [PubMed] [Google Scholar]
- Ovtsyna A.O., Dolgikh E.A., Kilanova A.S., Tsyganov V.E., Borisov A.Y., Tikhonovich I.A., Staehelin C. (2005). Nod factors induce Nod factor cleaving enzymes in pea roots. Genetic and pharmacological approaches indicate different activation mechanisms. Plant Physiol. 139: 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret X., Staehelin C., Broughton W.J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64: 180–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pislariu C.I., et al. (2012). A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiol. 159: 1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Recourt K., van Tunen A.J., Mur L.A., van Brussel A.A., Lugtenberg B.J., Kijne J.W. (1992). Activation of flavonoid biosynthesis in roots of Vicia sativa subsp. nigra plants by inoculation with Rhizobium leguminosarum biovar viciae. Plant Mol. Biol. 19: 411–420. [DOI] [PubMed] [Google Scholar]
- Rogers K.W., Schier A.F. (2011). Morphogen gradients: from generation to interpretation. Annu. Rev. Cell Dev. Biol. 27: 377–407. [DOI] [PubMed] [Google Scholar]
- Rolfe B.G., Gresshoff P.M. (1988). Genetic analysis of legume nodule initiation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 297–319. [Google Scholar]
- Roux B., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77: 817–837. [DOI] [PubMed] [Google Scholar]
- Sagan M., Morandi D., Tarenghi E., Duc G. (1995). Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn) after gamma-ray mutagenesis. Plant Sci. 111: 63–71. [Google Scholar]
- Sagan M., de Larambergue H., Morandi D. (1998). Genetic analysis of symbiosis mutants in Medicago truncatula. In Biological Nitrogen Fixation for the 21st Century, Elmerich C., Kondorosi Á., and Newton W.E., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 317–318. [Google Scholar]
- Salzer P., Feddermann N., Wiemken A., Boller T., Staehelin C. (2004). Sinorhizobium meliloti-induced chitinase gene expression in Medicago truncatula ecotype R108-1: a comparison between symbiosis-specific class V and defence-related class IV chitinases. Planta 219: 626–638. [DOI] [PubMed] [Google Scholar]
- Schlumbaum A., Mauch F., Vögeli U., Boller T. (1986). Plant chitinases are potent inhibitors of fungal growth. Nature 324: 365–367. [Google Scholar]
- Schmidt P.E., Broughton W.J., Werner D. (1994). Nod factors of Bradyrhizobium japonicum and Rhizobium sp. NGR234 induce flavonoid accumulation in soybean root exudate. Mol. Plant Microbe Interact. 7: 384–390. [Google Scholar]
- Schultze M., Quiclet-Sire B., Kondorosi E., Virelizer H., Glushka J.N., Endre G., Géro S.D., Kondorosi A. (1992). Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc. Natl. Acad. Sci. USA 89: 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M., Staehelin C., Brunner F., Genetet I., Legrand M., Fritig B., Kondorosi É., Kondorosi Á. (1998). Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide Nod signals. Plant J. 16: 571–580. [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C., Müller J., Mellor R.B., Wiemken A., Boller T. (1992). Chitinase and peroxidase in effective (fix+) and ineffective (fix-) soybean nodules. Planta 187: 295–300. [DOI] [PubMed] [Google Scholar]
- Staehelin C., Granado J., Müller J., Wiemken A., Mellor R.B., Felix G., Regenass M., Broughton W.J., Boller T. (1994a). Perception of Rhizobium nodulation factors by tomato cells and inactivation by root chitinases. Proc. Natl. Acad. Sci. USA 91: 2196–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C., Schultze M., Kondorosi É., Mellor R.B., Boller T., Kondorosi Á. (1994b). Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J. 5: 319–330. [Google Scholar]
- Staehelin C., Schultze M., Kondorosi E., Kondorosi A. (1995). Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 108: 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C., Xie Z.P., Illana A., Vierheilig H. (2011). Long-distance transport of signals during symbiosis: are nodule formation and mycorrhization autoregulated in a similar way? Plant Signal. Behav. 6: 372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., Ratet P., Mysore K.S. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54: 335–347. [DOI] [PubMed] [Google Scholar]
- Taira T., Hayashi H., Tajiri Y., Onaga S., Uechi G., Iwasaki H., Ohnuma T., Fukamizo T. (2009). A plant class V chitinase from a cycad (Cycas revoluta): biochemical characterization, cDNA isolation, and posttranslational modification. Glycobiology 19: 1452–1461. [DOI] [PubMed] [Google Scholar]
- Tian Y., Liu W., Cai J., Zhang L.Y., Wong K.B., Feddermann N., Boller T., Xie Z.P., Staehelin C. (2013). The nodulation factor hydrolase of Medicago truncatula: characterization of an enzyme specifically cleaving rhizobial nodulation signals. Plant Physiol. 163: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., de Billy F., Truchet G. (1998). Nod factor internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa. Development 125: 339–349. [DOI] [PubMed] [Google Scholar]
- Töpfer R., Matzeit V., Gronenborn B., Schell J., Steinbiss H.H. (1987). A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15: 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh T.H., Ratet P., Kondorosi É., Durand P., Kamaté K., Bauer P., Kondorosi Á. (1998). Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep. 17: 345–355. [DOI] [PubMed] [Google Scholar]
- Van Brussel A.A.N., Tak T., Wetselaar A., Pees E., Wijffelman C.A. (1982). Small Leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci. Lett. 27: 317–325. [Google Scholar]
- van Zeijl A., Op den Camp R.H., Deinum E.E., Charnikhova T., Franssen H., Op den Camp H.J., Bouwmeester H., Kohlen W., Bisseling T., Geurts R. (2015). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Vasse J., de Billy F., Camut S., Truchet G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172: 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.P., Staehelin C., Wiemken A., Broughton W.J., Müller J., Boller T. (1999). Symbiosis-stimulated chitinase isoenzymes of soybean (Glycine max (L.) Merr.). J. Exp. Bot. 50: 327–333. [Google Scholar]
- Xiong J.S., Balland-Vanney M., Xie Z.P., Schultze M., Kondorosi A., Kondorosi E., Staehelin C. (2007). Molecular cloning of a bifunctional β-xylosidase/α-L-arabinosidase from alfalfa roots: heterologous expression in Medicago truncatula and substrate specificity of the purified enzyme. J. Exp. Bot. 58: 2799–2810. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., Cai J., Li R.J., Liu W., Wagner C., Wong K.B., Xie Z.P., Staehelin C. (2016). A single amino acid substitution in a chitinase of the legume Medicago truncatula is sufficient to gain Nod-factor hydrolase activity. Open Biol. 6: 160061. [DOI] [PMC free article] [PubMed] [Google Scholar]