Abstract
Peptide stapling is a method for designing macrocyclic alpha-helical inhibitors of protein–protein interactions. However, obtaining a cell-active inhibitor can require significant optimization. We report a novel stapling technique based on a double strain-promoted azide–alkyne reaction, and exploit its biocompatibility to accelerate the discovery of cell-active stapled peptides. As a proof of concept, MDM2-binding peptides were stapled in parallel, directly in cell culture medium in 96-well plates, and simultaneously evaluated in a p53 reporter assay. This in situ stapling/screening process gave an optimal candidate that showed improved proteolytic stability and nanomolar binding to MDM2 in subsequent biophysical assays. α-Helicity was confirmed by a crystal structure of the MDM2-peptide complex. This work introduces in situ stapling as a versatile biocompatible technique with many other potential high-throughput biological applications.
Keywords: bioorthogonal chemistry, click chemistry, macrocycles, peptides, peptide stapling
Macrocyclization is an effective strategy for reinforcing peptides in stable secondary structures.[1] Whilst short peptides derived from proteins can lack a well-defined conformation when used in isolation, cyclisation can restore the native bioactive conformation and hence function of a peptide. By mimicking native binding motifs, cyclized peptides can competitively inhibit protein–protein interactions of clinical relevance, and they have received substantial attention as potential therapeutics.[1, 2] One of the most intensely studied targets is the oncogenic p53/MDM2 interaction, in which an N-terminal α-helix of the tumor suppressor p53 binds a hotspot on MDM2, an E3 ubiquitin ligase that downregulates p53 and is overexpressed in some cancers.[3, 4] Numerous peptide therapeutics have been developed to target this interaction, with some reaching as far as early-phase clinical trials.[5–10]
Stapled peptides are an important subset of macrocyclic peptides specific to α-helices.[11–15] Stapling involves two amino acids with non-native side chains that can be covalently linked, producing a cyclized peptide that may display enhanced proteolytic stability, binding affinity, and cellular uptake. Whilst the term “stapling” was originally coined by Verdine and Walensky for work on all-hydrocarbon linkers generated by metathesis (after Grubbs),[11, 12, 16] there is now great interest in alternative chemistries for side-chain cross-linking to give peptides with novel structures and biological properties. Of particular note are modular two-component strategies, where the staple linkage and peptide are separate moieties prior to cyclisation (Figure 1).[14] We recently developed a two-component stapling technique that makes use of double Cu-catalyzed azide–alkyne cycloaddition (CuAAC),[7, 17, 18] following related work by Bong[19] and other click systems.[20–22] Combining a two-component approach with click chemistry enables easy access to peptides bearing different functional staples. Whilst we appended cell-permeabilizing motifs to the staple, others have used a two-component approach to create photoswitchable,[23] reversible,[24] and dynamic linkers.[25]
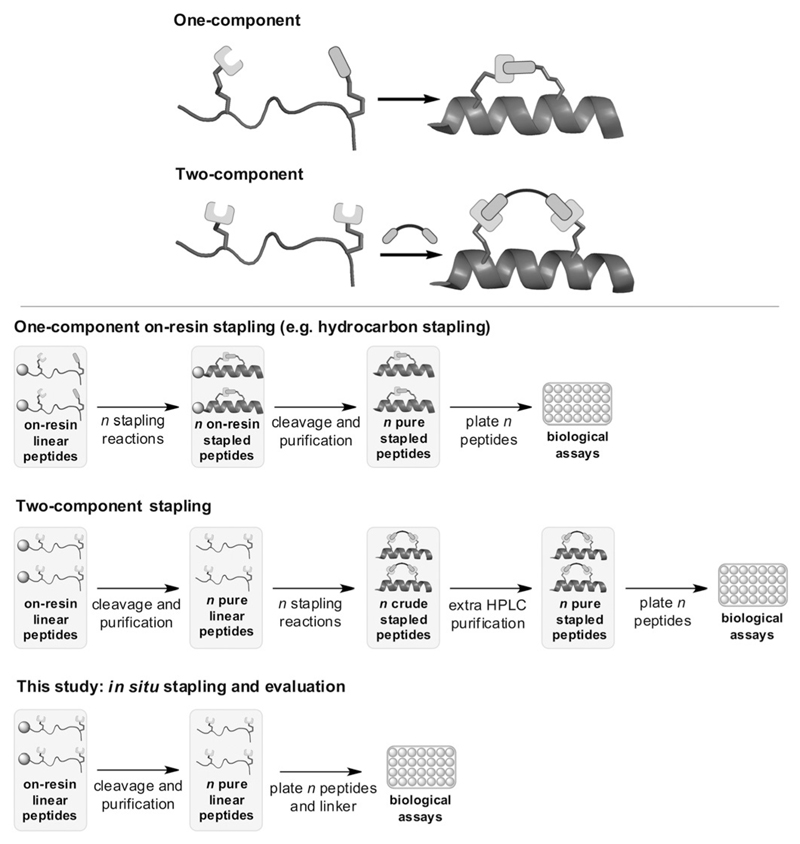
Figure 1.
Top: One- and two-component macrocyclization approaches. Bottom: Unlike classical one-component stapling[15] and existing two-component strategies such as our CuAAC method,[7, 18] the in situ strategy combines stapling and the primary biological assay in a single step.
When screening for an optimal inhibitor, stapling reactions are typically carried out on many peptide variants. A practical bottleneck for two-component strategies is that, unlike hydrocarbon stapling, cyclisation is typically done in solution to avoid on-resin site isolation.[7, 23, 25] Hence, from the pure unstapled peptide, extra purification is needed after cyclisation to remove reagents/catalyst prior to assays (Figure 1).
We decided to develop a stapling technique that would be biocompatible and so simple to conduct that it could be done in parallel on a large peptide library, even directly in the culture medium of a primary cell-based 96-well assay (Figure 1). This in situ approach would be faster than setting up a dedicated stapling reaction for each library variant, which is required in all current methods, and eliminates the extra purification step needed in other two-component strategies.
Inspired by the strain-promoted azide–alkyne cycloaddition reaction (Figure 2a),[26, 27] we report a catalyst-free double-click technique for parallel stapling in cell culture. This in situ strategy is the first example of stapling in a live biological context, since most techniques are incompatible owing to metal catalysis,[7, 19,20] inert atmosphere or protecting groups,[28, 29] or thiols[30–33]/amines[34] that might cross-react in biological systems. Whilst the oxime/hydrazone staples by Horne,[25] photocycloaddition staples by Lin,[35] and macrocyclic organo–peptide hybrids by Fasan have potential for in situ use,[10] we favored a non-dynamic system without UV irradiation or recombinant precursors.
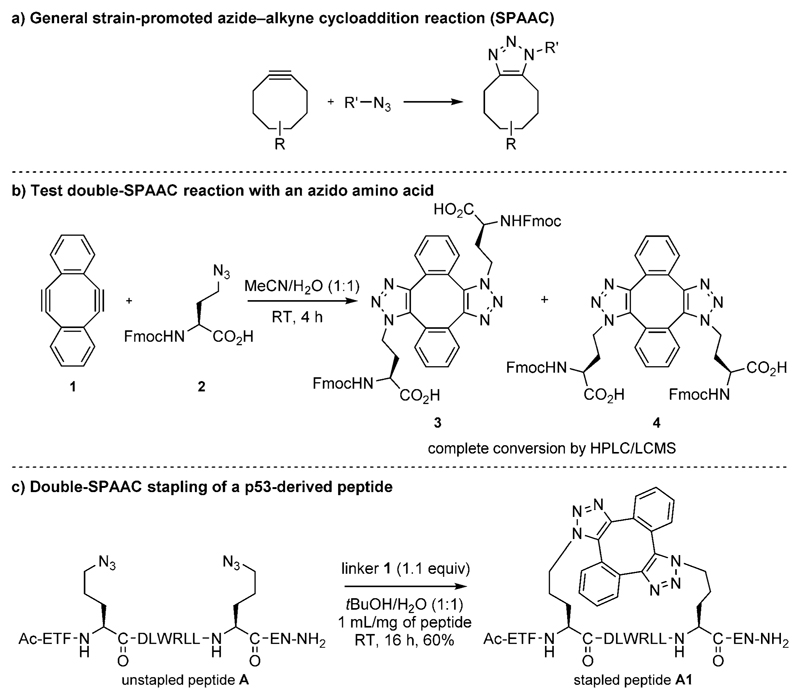
Figure 2.
a) Strain-promoted azide–alkyne cycloaddition (SPAAC).[26] b) Test double SPAAC on Fmoc-Aha-OH. c) Double-SPAAC stapling of p53-derived diazidopeptide A.
Strained diyne 1 was prepared according to the method of Orita et al.,[36] and a test reaction with Fmoc-Aha-OH 2 gave the expected bis(triazole) compounds 3 and 4 (Figure 2b and Figure S1.4.1 in the Supporting Information). Stapling of p53-derived diazidopeptide A (Figure 2c) with linker 1 in 1:1 H2O/tBuOH gave stapled peptide A1 in 60% yield (Figure 2c; assigned as the anti regioisomer, see later crystallography on an analogous peptide). Minor byproducts of the same mass were observed. These may be stable alternative conformations of the syn form, with MD simulations suggesting the possibility of at least two extra non-interchanging conformations (Figure S12.1.3). An excess of 1 did not affect the reaction, and comparable results were obtained when swapping tBuOH for MeOH, MeCN, or DMSO. Importantly, the reaction also proceeded in Dulbecco’s Modified Eagle’s medium (DMEM) with fetal calf serum and 1% DMSO.
The binding affinity of A1 for MDM2 was 3.1 ± 0.4 nm by competitive fluorescence polarization (FP),[7] which is more potent than that of wild-type p5317-29 (820 ± 60 nm) and unstapled A (16 ± 1 nm). Despite this encouraging in vitro result, up to 100 μm of A1 did not induce activation of p53 in an established T22 cell reporter assay,[9] which is in line with previous studies showing poor uptake of this sequence.[6, 7]
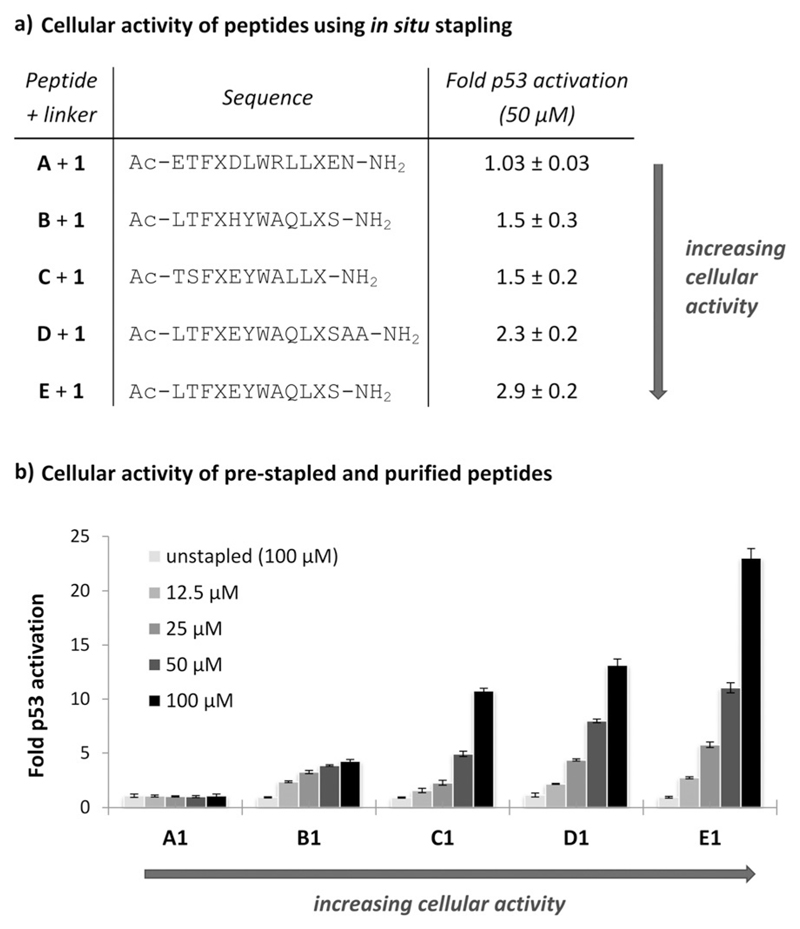
To test our in situ approach, we investigated sequence variants B–E, which are based on the phage-derived peptides PMI/PDI and were previously investigated for hydrocarbon stapling (Figure 3a).[5, 8, 9,37 To determine which variant would induce greatest p53 activation upon stapling with 1, we directly treated p53 reporter cells for the cell-based assay[9] in 96-well format with 0.5 mm 1 and 50 μm unstapled peptides A–E in DMEM. All five peptides were stapled in situ with similar yields (54–58%). After 18 h incubation, p53 activation was observed for B–E, whilst no activation was observed for A (Figure 3a). Cells treated with 1 or A–E only showed no p53 activation. From this in situ procedure, we were rapidly able to rank peptide activity, finding that E stapled with 1 (E + 1) was the most potent activator of p53.
Figure 3.
p53 activation in a cellular reporter assay for a) in situ stapling with peptides A–E (50 μm) and linker 1 (0.5 mm), and b) pre-stapled peptides A1–E1. Unstapled peptide controls A–E were tested at 100 μm (also see Figure S2.2.1). X = Orn(N3), data reported as fold activation over 1% DMSO. A is a p53-derived peptide (K24R) we previously used,[7] C is based on phage peptide PMI, and[8, 9] B, D, and E are based on phage peptide PDI.[5, 37]
We resynthesized and isolated stapled peptides A1–E1 to verify that the activity ranking from in situ stapling was consistent with standard testing of pure peptides.[7] Stapling in H2O/tBuOH proceeded with similar yield in each case, and the same activity ranking was observed for the purified peptides in the reporter assay, with E1 again inducing the greatest response (Figure 3b). An F3A control for E1 was found to have no cellular activity (Figure S11.2.1). The response using pure peptides was greater than in the in situ procedure, potentially due to gradual product formation over 18h and losses due to syn/anti byproducts. Despite these possible confounding factors, the reactivity of linker 1 was still sufficiently robust to rank peptide activity.
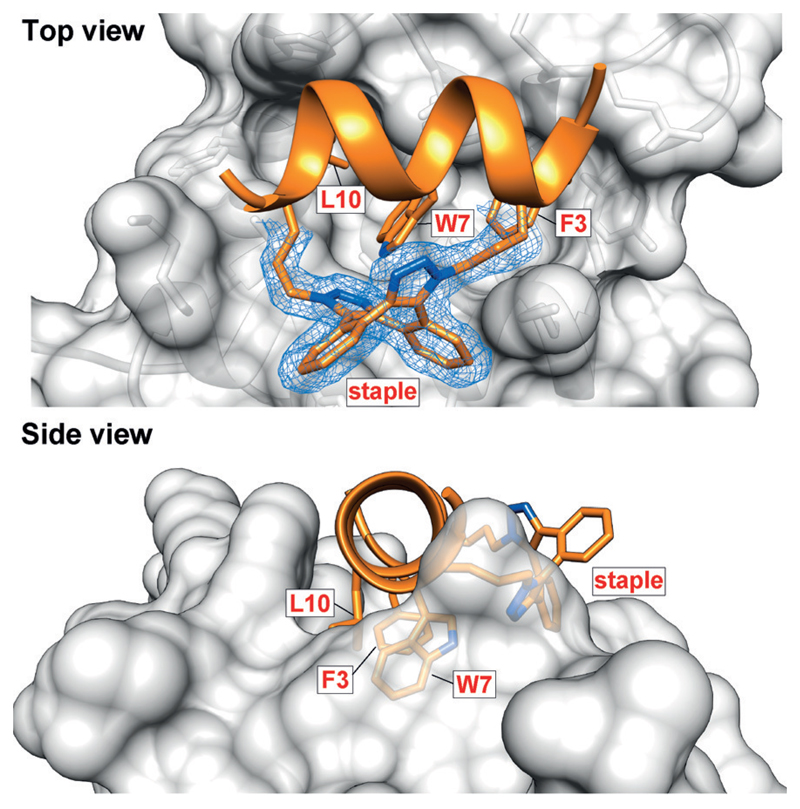
Taking E1 forward, we obtained its crystal structure in complex with MDM2 (17–108, E69A/K70A;[38] Figure 4). The structure shows E1 in α-helical conformation, placing the binding triad (F3,W7,L10) in the correct orientation for engaging the MDM2 hotspot. The bis(triazolyl) staple is found as the anti regioisomer and forms interactions with the protein, a phenomenon only previously seen with hydro-carbon-stapled peptides (Figure S5.3.2).[5, 39–41]
Figure 4.
Crystal structure of E1 bound to MDM2 at 1.9 Å resolution (PDB ID: 5afg), showing the α-helical conformation and the anti regioisomer of the staple. For clarity, only the staple and side chains of the three binding residues are shown. The 2Fo-Fc electron density map is contoured at 1σ.
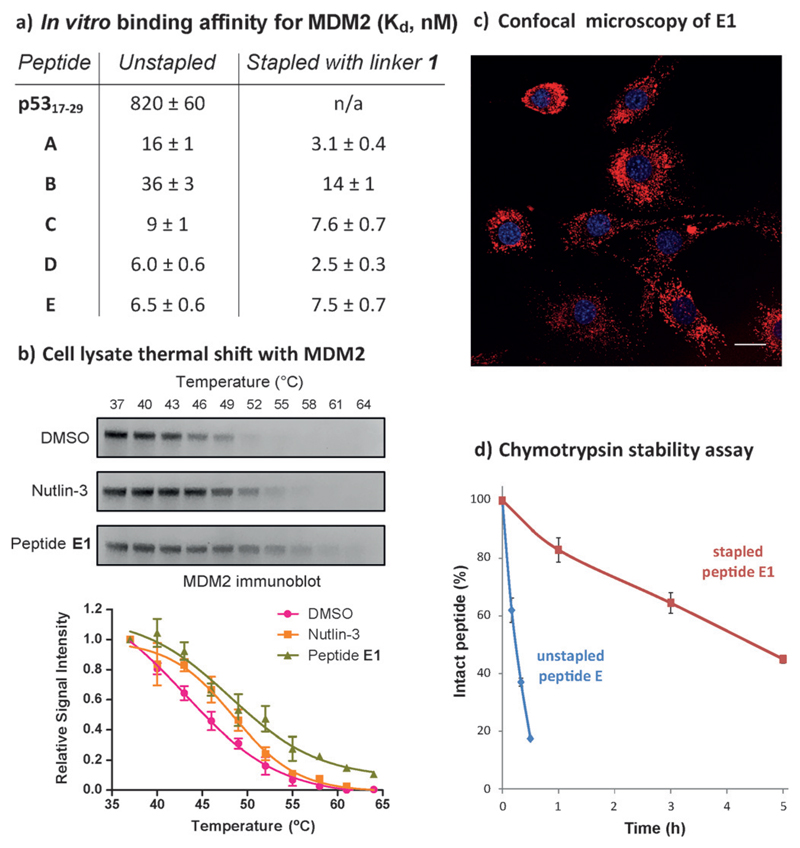
By FP,[7,9] A1–E1 all showed potent affinity for MDM2 (Figure 5a), thus exemplifying how in vitro binding does not always translate to cellular activity, owing to other factors such as uptake. A comparable binding affinity for E1 was obtained by isothermal calorimetry (12 ± 3 nm, Figure S4.2.1). Based on thermal shift in OCI/AML-2 lysate,[42,43] both E1 and control Nutlin-3 show increased stabilization of MDM2 (Figure 5b). Uptake of E1 was observed by confocal microscopy in T22 cells (Figure 5c). E1 did not exhibit non-specific toxicity in an LDH leakage assay (Figure S9.1.1), and was not observed to aggregate in solution by gel filtration and UV/Vis spectroscopy (Figures S10.1.1, S10.1.2). Finally, E1 showed significantly improved proteolytic stability in a chymotrypsin assay (Figure 5d).
Figure 5.
a) Binding affinities by fluorescence polarization (FP). b) MDM2 is stabilized by E1 as shown by cell lysate thermal shift. c) Confocal microscopy of TAMRA-E1 (20 μm) in live T22 cells, scale bar: 25 μm. Peptide shown in red, nuclei in blue. d) E1 shows increased stability to proteolysis by chymotrypsin.
In conclusion, this work introduces a new stapling technique with unique biocompatibility. Linker 1 was used for stapling in situ, leading to rapid selection of optimal candidate E1. Having established the chemistry, we now pave the way for applying in situ stapling to new biological targets. Only five peptides were screened in this study, since well-characterized sequences were already available. For new targets, more variants will be needed before finding a promising hit. A major advantage of our method is the potential to staple these variants with different strained linkers, efficiently covering a wider area of chemical space. Finally, different high-throughput assays can enable rapid evaluation of other properties, for example, high-content analysis of peptide uptake/localization, whilst stapling biosynthetic diazidopeptides could lead to vast screening libraries.
Supplementary Material
Acknowledgements
This work was supported by the EPSRC, BBSRC, MRC, Wellcome Trust (090340/Z/09/Z) and ERC (FP7/2007-2013; 279337/DOS). We thank Dr Judith Reeks for providing the MDM2 expression plasmid, and the Department of Biochemistry biophysics and X-ray facilities for crystallization and data collection equipment.
Footnotes
Supporting information and ORCID(s) from the author(s) for this article are available on the WWW under http://dx.doi.org/10.1002/anie.201508416.
Contributor Information
Dr. Maxim Rossmann, Department of Biochemistry, University of Cambridge, 80 Tennis Court Rd, Cambridge CB2 1GA (UK)
Dr. Ban Xiong Tan, p53Lab, A*STAR, 8A Biomedical Grove, #06-04/05 Neuros/Immunos, Singapore 138648 (Singapore)
Dr. Peterson de Andrade, Department of Chemistry, University of Cambridge, Lensfield Rd, Cambridge CB2 1EW (UK)
Dr. Yaw Sing Tan, Bioinformatics Institute, A*STAR, 30 Biopolis St, #07-01 Matrix, Singapore 138671 (Singapore)
Dr. Chandra Verma, Bioinformatics Institute, A*STAR, 30 Biopolis St, #07-01 Matrix, Singapore 138671 (Singapore); Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543; School of Biological Sciences, Nanyang Technological University, 50 Nanyang Drive, Singapore 637551
Dr. Marko Hyvönen, Department of Biochemistry, University of Cambridge, 80 Tennis Court Rd, Cambridge CB2 1GA (UK)
Prof. David R. Spring, Department of Chemistry, University of Cambridge, Lensfield Rd, Cambridge CB2 1EW (UK).
References
- [1].Pelay-Gimeno M, Glas A, Koch O, Grossmann TN. Angew Chem Int Ed. 2015;54:8896–8927. doi: 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2015;127:9022–9054. [Google Scholar]
- [2].Klein MA. ACS Med Chem Lett. 2014;5:838–839. doi: 10.1021/ml500235d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- [4].Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- [5].Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To K-H, Olson KA, Kesavan K, Gangurde P, Mukherjee A, et al. Proc Natl Acad Sci USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lau YH, de Andrade P, Quah S-T, Rossmann M, Laraia L, Sköld N, Sum TJ, Rowling PJE, Joseph TL, Verma C, et al. Chem Sci. 2014;5:1804–1809. [Google Scholar]
- [8].Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, et al. Proc Natl Acad Sci USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brown CJ, Quah ST, Jong J, Goh AM, Chiam PC, Khoo KH, Choong ML, Lee MA, Yurlova L, Zolghadr K, et al. ACS Chem Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- [10].Smith JM, Frost JR, Fasan R. Chem Commun. 2014;50:5027–5030. doi: 10.1039/c4cc01199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schafmeister CE, Po J, Verdine GL. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- [12].Blackwell HE, Grubbs RH. Angew Chem Int Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]; Angew Chem. 1998;110:3469–3472. [Google Scholar]
- [13].Azzarito V, Long K, Murphy NS, Wilson AJ. Nat Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- [14].Lau YH, de Andrade P, Wu Y, Spring DR. Chem Soc Rev. 2015;44:91–102. doi: 10.1039/c4cs00246f. [DOI] [PubMed] [Google Scholar]
- [15].Walensky LD, Bird GH. J Med Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lau YH, de Andrade P, Skold N, McKenzie GJ, Venkitaraman AR, Verma C, Lane DP, Spring DR. Org Biomol Chem. 2014;12:4074–4077. doi: 10.1039/c4ob00742e. [DOI] [PubMed] [Google Scholar]
- [18].Lau YH, Wu Y, de Andrade P, Galloway WRJD, Spring DR. Nat Protoc. 2015;10:585–594. doi: 10.1038/nprot.2015.033. [DOI] [PubMed] [Google Scholar]
- [19].Torres O, Yüksel D, Bernardina M, Kumar K, Bong D. ChemBioChem. 2008;9:1701–1705. doi: 10.1002/cbic.200800040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kawamoto SA, Coleska A, Ran X, Yi H, Yang C-Y, Wang S. J Med Chem. 2012;55:1137–1146. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ingale S, Dawson PE. Org Lett. 2011;13:2822–2825. doi: 10.1021/ol200775h. [DOI] [PubMed] [Google Scholar]
- [22].Scrima M, Le Chevalier-Isaad A, Rovero P, Papini AM, Chorev M, D’Ursi AM. Eur J Org Chem. 2010;2010:446–457. [Google Scholar]
- [23].Nevola L, Martín-Quirós A, Eckelt K, Camarero N, Tosi S, Llobet A, Giralt E, Gorostiza P. Angew Chem Int Ed. 2013;52:7704–7708. doi: 10.1002/anie.201303324. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2013;125:7858–7862. [Google Scholar]
- [24].Brown SP, Smith AB. J Am Chem Soc. 2015;137:4034–4037. doi: 10.1021/ja512880g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Haney CM, Loch MT, Horne WS. Chem Commun. 2011;47:10915–10917. doi: 10.1039/c1cc12010g. [DOI] [PubMed] [Google Scholar]
- [26].Agard NJ, Prescher JA, Bertozzi CR. J Am Chem Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- [27].Kii I, Shiraishi A, Hiramatsu T, Matsushita T, Uekusa H, Yoshida S, Yamamoto M, Kudo A, Hagiwara M, Hosoya T. Org Biomol Chem. 2010;8:4051–4055. doi: 10.1039/c0ob00003e. [DOI] [PubMed] [Google Scholar]
- [28].Kim Y-W, Grossmann TN, Verdine GL. Nat Protoc. 2011;6:761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- [29].Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. J Am Chem Soc. 2005;127:2974–2983. doi: 10.1021/ja0456003. [DOI] [PubMed] [Google Scholar]
- [30].Spokoyny AM, Zou Y, Ling JJ, Yu H, Lin Y-S, Pentelute BL. J Am Chem Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jo H, Meinhardt N, Wu Y, Kulkarni S, Hu X, Low KE, Davies PL, DeGrado WF, Greenbaum DC. J Am Chem Soc. 2012;134:17704–17713. doi: 10.1021/ja307599z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang F, Sadovski O, Xin SJ, Woolley GA. J Am Chem Soc. 2007;129:14154–14155. doi: 10.1021/ja075829t. [DOI] [PubMed] [Google Scholar]
- [33].Muppidi A, Doi K, Edwardraja S, Drake EJ, Gulick AM, Wang H-G, Lin Q. J Am Chem Soc. 2012;134:14734–14737. doi: 10.1021/ja306864v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fujimoto K, Kajino M, Inouye M. Chem Eur J. 2008;14:857–863. doi: 10.1002/chem.200700843. [DOI] [PubMed] [Google Scholar]
- [35].Madden MM, Rivera Vera CI, Song W, Lin Q. Chem Commun. 2009:5588–5590. doi: 10.1039/b912094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Orita A, Hasegawa D, Nakano T, Otera J. Chem Eur J. 2002;8:2000–2004. doi: 10.1002/1521-3765(20020503)8:9<2000::AID-CHEM2000>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- [37].Phan J, Li Z, Kasprzak A, Li B, Sebti S, Guida W, Schönbrunn E, Chen J. J Biol Chem. 2010;285:2174–2183. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Anil B, Riedinger C, Endicott JA, Noble MEM. Acta Crystallogr Sect D. 2013;69:1358–1366. doi: 10.1107/S0907444913004459. [DOI] [PubMed] [Google Scholar]
- [39].Stewart ML, Fire E, Keating AE, Walensky LD. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Phillips C, Roberts LR, Schade M, Bazin R, Bent A, Davies NL, Moore R, Pannifer AD, Pickford AR, Prior SH, et al. J Am Chem Soc. 2011;133:9696–9699. doi: 10.1021/ja202946k. [DOI] [PubMed] [Google Scholar]
- [41].Glas A, Bier D, Hahne G, Rademacher C, Ottmann C, Grossmann TN. Angew Chem Int Ed. 2014;53:2489–2493. doi: 10.1002/anie.201310082. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2014;126:2522–2526. [Google Scholar]
- [42].Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- [43].Tan BX, Brown CJ, Ferrer FJ, Yuen TY, Quah ST, Chan BH, Jansson AE, Teo HL, Nordlund P, Lane DP. Sci Rep. 2015;5:12116. doi: 10.1038/srep12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.