Abstract
The structure of monomeric human chemokine IL-8 (residues 1–66) was determined in aqueous solution by NMR spectroscopy. The structure of the monomer is similar to that of each subunit in the dimeric full-length protein (residues 1–72), with the main differences being the location of the N-loop (residues 10–22) relative to the C-terminal α-helix and the position of the side chain of phenylalanine 65 near the truncated dimerization interface (residues 67–72). NMR was used to analyze the interactions of monomeric IL-8 (1–66) with ND-CXCR1 (residues 1–38), a soluble polypeptide corresponding to the N-terminal portion of the ligand binding site (Binding Site-I) of the chemokine receptor CXCR1 in aqueous solution, and with 1TM-CXCR1 (residues 1–72), a membrane-associated polypeptide that includes the same N-terminal portion of the binding site, the first trans-membrane helix, and the first intracellular loop of the receptor in nanodiscs. The presence of neither the first transmembrane helix of the receptor nor the lipid bilayer significantly affected the interactions of IL-8 with Binding Site-I of CXCR1.
Keywords: Chemokine, IL-8, CXCL8, GPCR, Nanodisc, Protein structure
Introduction
Chemokines are signaling proteins secreted by cells in response to infection or injury. They direct leukocytes, which contain chemokine receptors in their membranes, to the affected cells by providing a concentration gradient for chemotaxis, and activate them for killing the infectious agent through phagocytosis and other mechanisms (Rosenkilde and Schwartz 2004). Chemokines play roles in many diseases, including cancer, and various immune disorders (Bendall 2005). Humans have about fifty chemokines that interact with twenty-three different G protein-coupled receptors (GPCRs) (Alexander et al. 2015); the complexity of the system is witnessed by the ability of individual chemokines to interact with multiple receptors, and for individual receptors to interact with multiple chemokines.
Chemokines are small globular proteins typically containing less than one hundred residues. They are divided into four classes (Murphy et al. 2000), although members in different classes have limited sequence homology, the polypeptides share common features of secondary and tertiary structure. In particular, they have one or two disulfide linkages that stabilize the protein fold, and whose disposition in the sequence provides a classification system. The N-terminal residues are flexible; starting with the first cysteine residue, the protein is structured, with the long N-loop terminated by one turn of a 310-helix, which is followed by three β-strands connected by short loops, and a C-terminal α-helix (Grasberger et al. 1993). Interleukin-8 is a member of the CXC class of chemokines, some of which have a highly conserved glutamate-leucine-arginine (ELR) sequence motif that is crucial for biological activity (Bizzarri et al. 2006). The commonality of structural features suggests that findings about the structure, dynamics, and interactions of an individual chemokine are likely to apply to a broad array of chemokine—receptor structures and functions.
Interleukin-8 (IL-8, CXCL8) was the first chemokine to be discovered (Baggiolini 2015). It has 72 residues in its most common form and is a homodimer at high concentrations, although it is a monomer at low concentrations and interacts with its receptor in this form (Fernando et al. 2004; Rajarathnam et al. 2006; Ravindran et al. 2009). The structure of the wild-type IL-8 dimer was first determined by solution NMR and X-ray crystallography (Clore et al. 1990; Baldwin et al. 1991). Subsequently, its structure has been characterized under a range of conditions and with a number of mutations (Rajarathnam et al. 1995; Eigenbrot et al. 1997; Gerber et al. 2000).
Relatively minor amino acid sequence changes enable the preparation of IL-8 samples that exist as stable monomers or dimers in solution. Non-dissociating IL-8 dimers result from the introduction of a disulfide bond across the dimerization interface (Rajarathnam et al. 2006). IL-8 monomers have been produced in several ways; by replacing Leu25 at the dimerization interface with the unnatural amino acid N-methyl leucine, by mutating residues Leu25, Val27 and Glu29 to disrupt the hydrogen bonding network that stabilizes the dimeric form (Lowman et al. 1997; Joseph et al. 2013), and by removing the last six residues of the protein to disrupt the dimerization interface (Clark-Lewis et al. 1991; Rajarathnam et al. 1997), which is the monomeric form used here. The structures of each subunit of dimeric wild type IL-8 and of the monomeric IL-8 containing N-methyl leucine are similar, with some differences in the position of the first β-strand and the helical structure of residues 67–72 (Clore et al. 1990; Rajarathnam et al. 1995).
CXCR1 and CXCR2 were the first chemokine receptors to be discovered and have high sequence homology (Holmes et al. 1991; Murphy and Tiffany 1991). Of the forty GPCR structures that have been determined, six are chemokine receptors, including CXCR1 (Park et al. 2012), CXCR4 (Wu et al. 2010; Qin et al. 2015), CCR2 (Zheng et al. 2016), CCR9 (Oswald et al. 2016), CCR5 (Tan et al. 2013) and the viral receptor US28 (Burg et al. 2015). CXCR4 and US28 were co-crystallized with viral chemokines to obtain structural information about the complex (Qin et al. 2015), however, there are currently no structures available of IL-8 bound to a chemokine receptor. The available experimental data suggest that IL-8 interacts with two main binding sites that may be spatially contiguous on CXCR1. Binding Site-I is located within the extracellular 38-residue N-terminal region of the receptor. Binding Site-II includes Arg119, Arg203 in extracellular loop 2, and Asp265 in extracellular loop 3, among other residues (Leong et al. 1994; Barter and Stone 2012; Hébert et al. 1993; Suetomi et al. 2002); a synthetic peptide derived from these two extracellular loops connected by a linker was shown to bind IL-8 with a KD of 0.5 μM (Helmer et al. 2015).
There is evidence that wild-type IL-8(1–72) interacts with CXCR1 as a monomer under physiological conditions (Burrows et al. 1994). Monomeric IL-8(1–66) has been shown to have the same potency in neutrophil chemotaxis and Ca2+ mobilization assays (Clark-Lewis et al. 1991; Rajarathnam et al. 1997; Fernando et al. 2007).
The residues in Binding Site-I of chemokine receptors display high sequence diversity, and are likely responsible for the binding specificity of chemokines. Indeed, the sequences of CXCR1 and CXCR2 vary the most in this region, and swapping N-terminal residues between the two receptors alters their binding specificity (Barter and Stone 2012; Prado et al. 2007). The flexible N-terminal region appears to be a defining characteristic of chemokine receptors, and this has motivated the study of these residues in soluble peptides separate from the rest of the receptor. The affinity of IL-8 for various N-terminal peptides derived from CXCR1 is in the low micromolar range. The first atomic resolution insights came from NMR studies of IL-8 bound to a 21-residue peptide whose sequence corresponds to residues 9–29 of CXCR1 (Skelton et al. 1999). By itself, the peptide does not have a well-defined structure, but it becomes more ordered upon binding IL-8. Several specific residues clustered in the center of the peptide, including Pro21, Pro22, Asp24, Glu25, Asp26 and Pro29, were found to be involved in binding IL-8. More recently, we have shown (Park et al. 2011a, b) that additional residues near the N-terminus of CXCR1, including Asn16, Thr18 and Gly19, undergo large chemical shift perturbations upon interaction with IL-8. Moreover, (Rajagopalan and Rajarathnam 2004) have shown that IL-8 can bind CXCR1 N-terminal peptides of different lengths, with affinities that are increased by the presence of micelles, suggesting that lipids may influence the interactions of IL-8 with CXCR1. There is evidence that a Trp residue in a 34-mer peptide derived from the CXCR1 N-terminus interacts with DOPC lipid bilayers (Haldar et al. 2010). Additionally, we have shown that the N-terminus of CXCR1 interacts with the membrane surface in the absence of IL-8, but dissociates from the membrane in the presence of IL-8 (Park et al. 2011a, b).
IL-8 residues responsible for functions of CXCR1 have been identified. Although the characteristic ELR residues (Glu4, Leu5 and Arg6) have been shown to be essential for CXCR1 activation they are not involved in IL-8 binding to N-terminal peptides (Clark-Lewis et al. 1991; Hebert et al. 1991; Park et al. 2017). The main binding sites on IL-8 have been mapped to the N-loop (residues 12–18) and the 40 s loop and third β-strand (residues 44–51) and include both charged and hydrophobic residues (Fernando et al. 2004, 2007; Ravindran et al. 2009; Skelton et al. 1999; Park et al. 2011a, b; Clubb et al. 1994; Williams et al. 1996; Kendrick et al. 2014). Residues Tyr13, Lys15 and Phe21 of the N-loop appear to confer specificity for individual receptors (Hammond et al. 1996). Furthermore, it has been shown that IL-8 does not undergo a major conformational change upon interaction with the peptide corresponding to the N-terminus of CXCR1 (Skelton et al. 1999). There is also evidence that IL-8 dimers dissociate upon binding the N-terminal residues of CXCR1, and that there are some minor differences between a covalent IL-8 dimer binding and that of an obligatory IL-8 monomer (Fernando et al. 2004; Ravindran et al. 2009; Kendrick et al. 2014; Joseph et al. 2015).
Despite extensive studies (Kufareva et al. 2017) much remains to be learned about the molecular basis of chemokine-receptor interactions. Here we present the structure of monomeric IL-8(1–66) obtained by deleting the C-terminal residues 67–72, which contribute to the dimerization interface. We also describe binding studies of monomeric IL-8(1–66) with polypeptides corresponding to the N-terminal residues of CXCR1, ND-CXCR1(1–38) and 1TM-CXCR1(1–72), which includes the first transmembrane helix of CXCR1 in addition to the flexible N-terminal domain.
Materials and methods
Protein expression and purification
The gene for human IL-8(1–66) was cloned and expressed as described previously (Rajagopalan and Rajarathnam 2004). Cell lysis was achieved by sonication in lysis buffer (20 mM Tris, 500 mM NaCl, 20 mM imidazole, pH 7.4) in the presence of phenylmethane sulfonyl fluoride (MP Biomedicals) and lysozyme. Cell debris was removed by centrifugation, and the supernatant was incubated for 1 h on a Nickel-NTA column (Qiagen, Ni-NTA Superflow). The beads were washed with wash buffer 1 (20 mM Tris, 300 mM NaCl, 30 mM imidazole, pH 7.4) and wash buffer 2 (20 mM Tris, 150 mM NaCl, 30 mM imidazole, pH 7.4) before being eluted in 20 mM Tris, 150 mM NaCl with a gradient of 150–300 mM imidazole. The eluate was cleaved using factor Xa (New England BioLabs Inc) and purified by reverse-phase high-performance liquid chromatography on a C18 column (Waters DeltaPak 15 μm, 300 Å) using an acetonitrile gradient. The purified protein was stored as a lyophilized powder at −20°C. The polypeptide ND-CXCR1(1–38) containing the C30S mutation to enhance its stability was expressed and purified as described previously (Park et al. 2011; Casagrande et al. 2011). The gene for the polypeptide 1TM-CXCR1(1–72) including the C30S mutation and a C-terminal 5× His tag was also expressed and purified as described previously (Casagrande et al. 2011; Park et al. 2014). 1TM-CXCR1(1–72) from the final HPLC purification was lyophilized and stored at −20°C.
Nanodisc preparation
Nanodiscs were made with MSP1D1AH5 protein and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) (Hagn et al. 2013). MSP1D1AH5 was expressed and purified as described previously (Ritchie et al. 2009). The optimal molar ratio of 1TM-CXCR1: MSP1D1AH5: DMPC was found to be 0.1:1:43.3 by gel filtration chromatography screening. This ratio ensures that one nanodisc containing 1TM-CXCR1(1–72) is formed for every four nanodiscs that do not contain this polypeptide, which minimizes protein aggregation and oligomerization. Purified 1TM-CXCR1(1–72) and DMPC (Avanti Lipids) were solubilized in solutions of 0.5 and 1% SDS, respectively, and mixed. MSP1D1AH5 in 20 mM Hepes buffer was then added, and all three components incubated for 1 h with agitation. The detergent was slowly removed overnight by incubation with BioBeads SM-2 (Biorad). The absence of SDS was confirmed by testing with a colorimetric SDS-detection kit (G-Biosciences). Nanodiscs containing 1TM-CXCR1(1–72) were separated from empty nanodiscs by Ni-NTA chromatography exploiting the His-tag on the C-terminus of 1TM-CXCR1; the mixture was allowed to bind and the empty nanodiscs removed using a wash buffer (40 mM Tris 300 mM NaCl, pH 8). Nanodiscs containing 1TM-CXCR1(1–72) were eluted using elution buffer (20 mM Tris, 150 mM NaCl, 150 mM Imidazole pH 8). Finally, the remaining impurities were removed by size-exclusion chromatography (20 mM Tris, 50 mM NaCl, pH 7.4, Amersham Biosciences, HiLoad 16/60 Superdex 200).
NMR spectroscopy
Uniformly 13C, 15N-labeled IL-8(1–66) was dissolved in 20 mM Hepes pH 7.3 containing 10% D2O. The NMR experiments were performed on a 600 MHz Bruker AVANCE spectrometer equipped with a 5-mm triple-resonance cryoprobe (1H, 13C, and 15N) and z-axis pulsed gradient and on a Varian VS 800 MHz spectrometer equipped with a XYZ-gradient triple resonance cryoprobe (1H, 13C, and 15N). Triple-resonance HNCA, HNCO and HNCACB assignment experiments were performed on uniformly 13C, 15N labeled IL-8 (1–66) at 40°C. The side-chain assignments were made with HCC(CO) NH, CC(CO)NH, CβHδ and CβHε experiments. Distance restraints were obtained using 15N-edited NOESY-HSQC (mixing time, 200 ms) and 13C-edited NOESY-HSQC (mixing time, 120 and 80 ms) experiments. Heteronuclear 1H-15N NOEs were measured by comparing signal intensities obtained with and without 1H saturation during a relaxation delay of 6 s at 40 and 15°C. Hydrogen bond restraints were obtained from a two-dimensional 1H-15N HSQC spectrum of uniformly 15N labeled IL-8(1–66) in 90% D2O. The experiment was started following a 1-h incubation period. Residual dipolar couplings (RDCs) were measured with HSQC in-phase and anti-phase (IPAP) experiments on IL-8(1–66) in a solution of empty nanodiscs in the presence and absence of 13.5 mg/ml Y21M fd phage and 70 mM NaCl. The Y21M fd phage was produced using a previously described protocol. All NMR experiments were processed using NMRPipe and analyzed using SPARKY. The Chemical Shift Index was determined using TALOS. Chemical Shift Perturbations (CSP) were calculated using the following equation: .
Structure calculations
All calculations were performed with Xplor-NIH (Schwieters and Kuszewski 2006), using the EEFx energy function with the topology and parameter files protein_eefx2.top and protein_eefx2.par, and the statistical potential torsionDBPot to restrain dihedral angles. Experimental NMR restraints were implemented with the potentials: NOEPot, for distances derived from NOEs and hydrogen bonds; CDIH for dihedral angles; and RDCPot for RDCs. RDCs were used for cross-validation and not imposed as restraints. Each calculation was initiated from a fully extended polypeptide strand. Simulated annealing calculations were performed as described previously (Tian et al. 2015), using two protocols, the first to fold a total of 100 initial structures, and the second to refine the ten folded structures with lowest energy. A total of 100 structures were refined, and the ten structures with lowest energy were selected for the final ensemble. The coordinates were deposited in the protein data bank (PDB: 5WDZ) and the BMRB (ID: 30316).
Isothermal titration calorimetry (ITC)
The ITC experiments were performed at 23°C using a MicroCal iTC200 instrument (GE healthcare Life Sciences) in 20 mM Tris, 50 mM NaCl, pH 7.4 buffers. The concentrations of ND-CXCR1(1–38) in the cell were 65 and 50 μM, and that of 1TM-CXCR1(1–72) reconstituted in nanodiscs was 100 μm. IL-8 containing solutions present in the syringe had concentrations of 760, 600 and 985 μm. Experiments consisted of 19 injections of 2 μL aliquots separated by 150 s.
Results
Solution NMR structure of monomeric IL-8
As shown in Fig. 1a, both dimeric (Clore et al. 1990) and monomeric (Rajarathnam et al. 1995) forms of IL-8 yield well-resolved solution NMR spectra. Although many resonances have similar chemical shifts in both spectra, large differences are observed for selected signals, as marked in the Figure. The largest shifts are from residues Ile10, Lys15, Lys20, Lys23, Val27, Ser30, Asp52, Val61, Glu63, Lys64 and Phe65. These residues are located in the N-loop, the first β-strand, and the C-terminal α-helix, and the differences can be readily accounted for by the effects of truncation of six residues (67–72) at the C-terminus. For example, in the structure of the IL-8(1–72) dimer (PDB id: 1IL8), the side chains of Val27 and Ser30 in one of the monomers contact residues in the first β-strand and the α-helix of the other monomer, respectively.
Fig. 1.
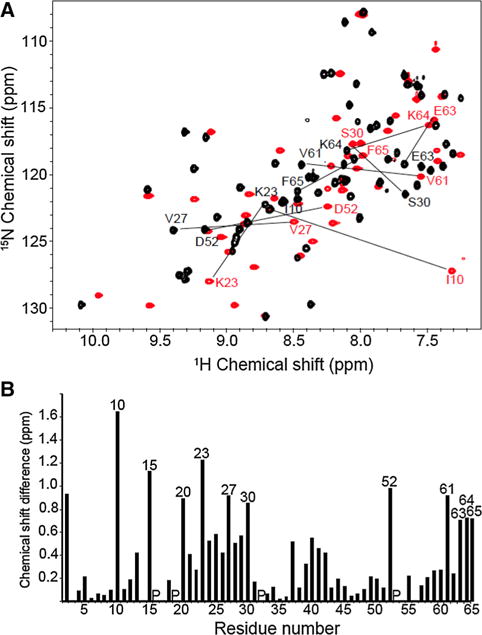
a Two-dimensional 1H/15N HSQC spectra of uniformly 15N labeled dimeric and monomeric IL-8 obtained under identical conditions in 20 mM HEPES buffer at pH 7.3 and temperature of 313 K. The spectra of dimeric full-length IL-8 (1–72) (in black) and of monomeric IL-8 (1–66) (in red) are superimposed with selected amide backbone resonances marked by their sequential assignments. b Bar graph of the weighted chemical shift differences observed between the resonances of dimeric IL-8 (1–72) and monomeric IL-8 (1–66) as a function of residue number; the resonances with the largest differences are marked by their sequential assignments
Samples of uniformly 13C, 15N labeled monomeric IL-8(1–66) enabled a variety of triple-resonance NMR experiments to be used to assign backbone and side chain resonances, to measure chemical shift index (CSI), residual dipolar coupling (RDC), and amide hydrogen exchange parameters as structural restraints, and to measure heteronuclear 1H/15N nuclear Overhauser enhancements (NOEs) as indicators of local dynamics. These data are summarized in Fig. 2 and Table 1.
Fig. 2.
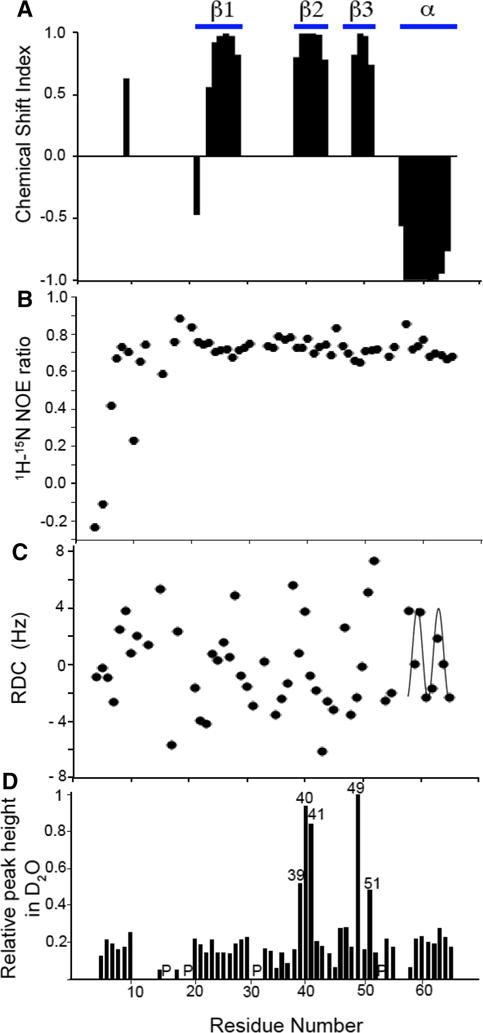
Plots of experimental data measured on monomeric IL-8(1–66) in solution as a function of residue number. a Chemical Shift Index. b 1H/15N heteronuclear NOE. c Residual Dipolar Couplings used to cross-validate the solution NMR structure of monomeric IL-8 (1–66). The helical region of the protein displays a characteristic Dipolar Wave pattern. d Resonance intensities after incubation of IL-8(1–66) in D2O for 1 h. Peak height was measured for all peaks in a 1H/15N HSQC spectrum and normalized to the peak height of residue Leu49. The locations of proline residues are indicated with a ‘P’
Table 1.
Restraint table to calculate the structure of monomeric IL-8 (1–66)
| NMR distance and dihedral constraints | |
| Number of unambiguous NOEs | 417 |
| Short-range | 204 |
| Medium-range | 159 |
| Long-range | 54 |
| Hydrogen bond restraints | 22 |
| Dihedral angle restraints (TALOS) | 107 |
| Structure statistics | |
| Molprobity analysis | |
| Molprobity score | 1.76 |
| Clashscore | 8.39 |
| Poor rotamer (%) | 2.90 |
| Ramachandran favored (%) | 99.68 |
| Ramachandran outliers (%) | 0.00 |
| Pairwise backbone RMSD (residues 7–65) | 0.97 Å |
| Heavy—atom RMSD (residues 7–65) | 1.83 Å |
The four types of NMR data displayed in Fig. 2 are aligned according to residue number. The major regions of secondary structure are delimited at the top of the Figure. Analysis of the CSI plot in Fig. 2a is consistent with the locations of the secondary structure elements established in previous studies of IL-8 (Clore et al. 1990; Baldwin et al. 1991; Rajarathnam et al. 1995; Eigenbrot et al. 1997; Gerber et al. 2000). Notably, monomeric IL-8(1–66) has three β-strands in the middle of the sequence and an α-helix located at the C-terminus. Although the C-terminus of the 72-residue full-length protein is shortened by six residues, the remaining helical residues (57–66) maintain a stable secondary structure as evidenced by the heteronuclear NOE values for these residues (Fig. 2b) and the ability to fit a Dipolar Wave to the RDCs for these residues (Fig. 2c) (Mesleh and Opella 2003).
Signals could not be observed for either residue 66, the C-terminal residue, or residue 1, the N-terminal residue, most likely due to local dynamics interfering with the NMR experiments. Signals from all of the other residues could be observed, even though the N-terminal residues of IL-8(1–66) show evidence of minimal structure and substantial local dynamics. Signals from residues 2–6 have negative 1H/15N heteronuclear NOEs (Fig. 2b) indicative of local motions, and the residues closest to the N-terminus have near-zero values for their 1H/15N RDCs (Fig. 2C) and only six observable long-range 1H–1H homonuclear NOEs, all of which connect this region to residues 32 and 33 in the 30 s loop (residues 29–37). Notably, the three conserved residues (4–6) of the ELR motif essential for activity show evidence of local motions. These results are consistent with those of previous solution NMR and X-ray crystallography studies of IL-8 where the positions of residues 1–6 could not be well defined (Clore et al. 1990; Baldwin et al. 1991; Rajarathnam et al. 1995).
The data in Fig. 2b show that residues 7–65 have 1H/15N heteronuclear NOE values of about 0.8, which indicates that, except for the N-terminal region, IL-8(1–66) has a stable backbone structure consistent with a well-folded protein. Using fd bacteriophage as the alignment medium, the measured 1H/15N RDCs plotted in Fig. 2c have values between 6.6 Hz and −6.2 Hz. As mentioned above, the RDCs for residues 58–65 were well fit to a Dipolar Wave with a periodicity of 3.6, which provide strong additional evidence that the C-terminal residues form a stable α-helix until the last residue of monomeric IL-8(1–66), despite the removal of the final six residues from the full-length wild-type protein. Hydrogen bonding restraints for structural analysis were obtained by measuring the loss of amide H–N resonance intensities in 1H/15N HSQC spectra of IL-8(1–66) following its transfer from H2O to D2O solution (Supplemental Fig. 1). These results are summarized in the plot of relative signal intensities as a function of residue number in Fig. 2d. Five particularly intense resonances are observed in spectra obtained after several hours of incubation in D2O, indicating the presence of stable hydrogen bonds in a number of residues located near the middles of the second and third β-strands.
We determined the structure of monomeric IL-8(1–66) in solution using NMR restraints summarized in Table 1 and methods described in the Experimental Section. As anticipated from the data in Fig. 2, the structure of IL-8(1–66) is well defined for residues 6–65, which contains three anti-parallel β-strands and the C-terminal α-helix, with a pair-wise RMSD of 0.97 A. Including the flexible N-terminal residues, the structure of residues 1–6 has an overall pairwise RMSD of 1.6 Å. This is illustrated graphically by the superimposed 10 lowest energy structures in Fig. 3a.
Fig. 3.
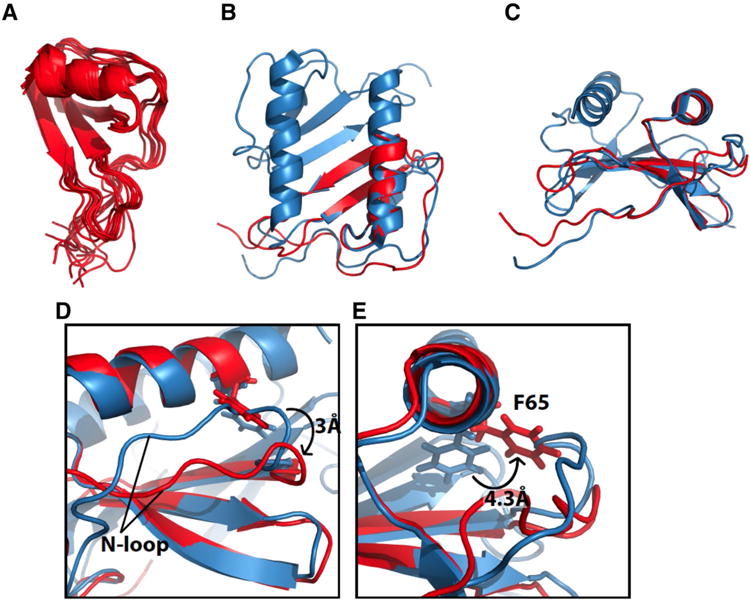
Solution NMR structure of monomeric IL-8 (1–66). a Overlay of the ten lowest energy structures of monomeric IL-8(1–66). b Structure of monomeric IL-8(1–66) in red overlayed with the solution NMR structure of full-length IL-8(1–72) in blue (PDB ID: 1IL8). c Same as in b only rotated 90°. d Expanded structural region of monomeric IL-8(1–66) in red and wild type IL-8 in blue. The change in position of the N-loop relative to the C-terminal helix between monomeric IL-8 and wild type IL-8 is shown. Differences in distances as measured between the amide nitrogen atoms of residues K20 and F65 e Expanded structural region of monomeric IL-8(1–66) in red and wild-type IL-8 in blue. The difference in position of the C5 atom of Phe65 between monomeric IL-8(1–66) and wild type IL-8(1–72) is shown
The overall structure of monomeric IL-8(1–66) (PDB: 5WDZ) shown in red in Fig. 3 is similar to that of each of the monomeric subunits of dimeric full-length IL-8(1–72) (PDB id: 1IL8) shown in blue. The principal differences in the structures are associated with the N-loop (residues 10–19) and the side chain of Phe65. In the previously determined structures of wild-type IL-8(1–72) and monomeric Leu25-NMe IL-8(1–72) (PDB id: 1IKL) the N-loop runs roughly parallel to the α-helix at a slight angle that places its N-terminal end further away from the α-helix than the C-terminal end (blue in Fig. 3d). By contrast, in the structure of monomeric IL-8(1–66) the N-loop and the α-helix are almost parallel (red in Fig. 3d). The C-terminal part of the N-loop in IL-8(1–66) shifts away from the α-helix by about 3 A compared to full-length IL-8, regardless of whether the protein is a monomer or a dimer. In the NMR structure of dimeric wild-type IL-8(1–72), the amide nitrogen atoms of Ser14 and Trp57, which are located in the N-terminal parts of the N-loop and the α-helix, respectively, are separated by 10.9 Å, and residues Lys20 and Phe65, which are in the C-terminal parts of the N-loop and α-helix, respectively, are separated by 7.5 Å. The same residues, Lys20 and Phe65, are separated by 10.52 and 10.46 A, respectively, in the structure of monomeric IL-8(1–66). This is shown in Fig. 3d. The difference in the relative position of the N-loop may be related to the change in position of the side chain of Phe65. In the structure of wild-type IL-8(1–72) the aromatic ring of Phe65 points away from the N-loop towards the dimerization interface; by contrast, in the structure of monomeric IL-8(1–66) the same side chain points towards the N-loop, in the space occupied by the N-loop in the other structures. This corresponds to a movement of 4.3 Å for the C82 atom, as illustrated in Fig. 1e.
Interaction of monomeric IL-8(1–66) with ND-CXCR1 and 1TM-CXCR1
To gain insight into how the chemokine IL-8 interacts with its receptor CXCR1 we expressed and purified polypeptides with sequences corresponding to residues 1–38 and 1–72 of CXCR1. There have been many studies of soluble peptides similar to ND-CXCR1(1–38) because the N-terminal domain (ND) contains the part of the agonist binding site referred to as Binding Site-I (Leong et al. 1994; Prado et al. 2007; Gayle et al. 1993; LaRosa et al. 1992; Lowman et al. 1997). The longer polypeptide 1TM-CXCR1(1–72) includes the complete N-terminal domain, the first transmembrane helix (1TM), and the first intracellular loop of CXCR1. This polypeptide is not soluble in aqueous solution and requires the presence of detergents or lipids for solubilization. The transmembrane helix ensures the proximity of Binding Site-I to the surface of the lipid bilayer when the polypeptide 1TM-CXCR1(1–72) is in nanodiscs.
The effects on residues of IL-8(1–66) by interaction with Binding Site-I of CXCR1 were determined using NMR titration experiments. Two-dimensional 1H/15N HSQC spectra were obtained from samples containing a fixed amount of uniformly 15N labeled IL-8(1–66) and varying amounts of unlabeled ND-CXCR1(1–38) or 1TM-CXCR1(1–72). Signals from IL-8 residues that occupy a different chemical environment upon addition of a receptor–derived polypeptide, either due to direct interaction or a local change in conformation, undergo changes in their 1H and/or 15N chemical shift frequencies. The expanded spectral regions in Fig. 4 compare the effects of IL-8(1–66) binding to ND-CXCR1(1–38) (panel A) and to 1TM-CXCR1(1–72) (panel D). The three main interaction regions of IL-8(1–66) can be discerned from the bar graphs of the chemical shift differences as a function of residue number in panels B and E. Signals from the N-loop (residues 10–19), the third β-strand (residues 47–51), and part of the C-terminal α-helix (residues 56–65) are affected by binding to the polypeptides containing Binding Site-I; in particular, the resonances from Thr12, Tyr13, Phe17, and His18 have the most strongly perturbed chemical shifts. No signal from Ser14 could be observed in any of the experiments, and the signal from Lys15 was broadened beyond detection upon interaction with both ND-CXCR1 and 1TM-CXCR1. No signal from residue 16 could be observed because proline lacks an amide N–H.
Fig. 4.
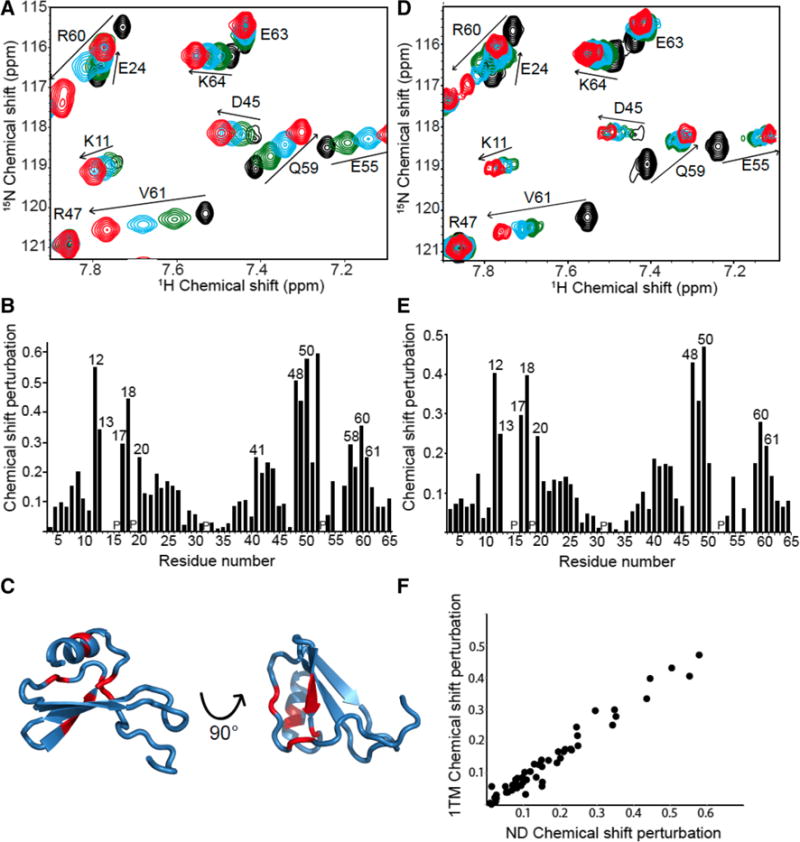
a. Selected region of solution 1H/15N HSQC spectra of uniformly 15N labeled monomeric IL-8(1–66) with increasing amounts of unlabeled ND-CXCR1(1–38) added. The colors represent the molar ratio ND-CXCR1:IL-8. Black is 0, green is 0.25, blue is 0.5, and red is 1. b Chemical Shift Perturbation plot of the data in (a). The locations of proline residues are indicated with a ‘P’ c Structure of monomeric IL-8(1–66) with red marking the residues with the largest chemical shift perturbation upon interaction with ND-CXCR1. d Selected region of a solution 1H/15N HSQC spectrum of uniformly 15N labeled monomeric IL-8(1–66) with increasing amounts of unlabeled 1TM-CXCR1 reconstituted in nanodiscs. The same color representation of molar ratios as used in Panel A. e Chemical shift perturbation plot of the data in (d). f Scatter plot showing the correlation between the chemical shift perturbations of monomeric IL-8(1–66) bound to ND-CXCR1(1–38) and to 1TM-CXCR1(1–72). R2 is 0.95
It has been proposed that residues in the N-loop confer receptor binding specificity (Ravindran et al. 2009). Evidence of interactions with the N-terminal part of the receptor by residues in the third β-sheet and part of the α-helix has been detected in studies of using similar constructs (Fernando et al. 2007; Skelton et al. 1999; Park et al. 2011a, b; Clubb et al. 1994; Kendrick et al. 2014; Hammond et al. 1996; Jiang et al. 2015). Both non-polar (Leu49, Val58, Val61) and charged (Glu48, Asp52, Arg60) residues of IL-8(1–66) are affected by binding to the polypeptides containing sequences from the N-terminal region of CXCR1. Chemical shift perturbations for Asp52 and Val58 could only be observed upon binding to ND-CXCR1, since the signals were broadened beyond detection upon interaction with 1TM-CXCR1 in nanodiscs.
The residues whose chemical shifts are affected by binding to the polypeptides containing the N-terminal domain of the receptor are marked by their residue numbers in Fig. 4b, e. They are located in three distinct regions of the sequence and are colored red in the structure of IL-8(1–66) shown in Fig. 4c where they are shown to be in close spatial proximity. Notably, signals from residues in the conserved ELR motif near the N-terminus and from the extreme C-terminus of the α-helix undergo only minor chemical shift perturbations upon binding.
Significantly, the chemical shift perturbation patterns for IL-8(1–66) binding to the polypeptides ND-CXCR1(1–38) and 1TM-CXCR1(1–72) are similar. The chemical shift perturbations resulting from binding to ND-CXCR1(1–38) are compared to those from binding to 1TM-CXCR1(1–72) in the scatter plot shown in Fig. 4f. The linear correlation has an R2 value of 0.95, and indicates that IL-8(1–66) binds similarly to the N-terminal domain of CXCR1 in the absence and presence of the first trans-membrane helix. Further, it suggests that the interactions between IL-8(1–66) and Binding Site-I of the receptor are not influenced by the presence of a lipid bilayer.
To ensure that the absence of spectroscopic differences in IL-8(1–66) binding to ND-CXCR1(1–38) and 1TM-CXCR1(1–72) are not due to different binding affinities or to the nanodiscs themselves, isothermal titration calorimetry experiments were performed (Supplemental Fig. 2). ND-CXCR1(1–38) and 1TM-CXCR1(1–72) reconstituted into nanodiscs were placed in the chamber and aliquots from a concentrated solution of IL-8(1–66) were added. The KD for IL-8(1–66) binding to ND-CXCR1(1–38) was determined to be 3.5 μM, and that for IL-8 binding to 1TM-CXCR1(1–72) was 12.5 μM. These results suggest that IL-8(1–66) has a slightly weaker interaction with 1TM-CXCR1(1–72) than it does with ND-CXCR1(1–38). The enthalpy and entropy changes associated with binding were also comparable; ΔH = −3.2 kcal/mol and ΔS = 14.1 cal/mol/deg for the interaction between IL-8(1–66) and ND-CXCR1 and ΔH = −3.1 kcal/mol and ΔS = 11.7 cal/mol/deg for the interaction between IL-8(1–66) and 1TM-CXCR1(1–72) in nanodiscs. No interactions were observed in control experiments performed with IL-8(1–66) titrated into blank buffer solution and into empty nanodiscs. An additional control experiment was performed by titrating IL-8 into a solution containing both ND-CXCR1(1–38) and empty nanodiscs. The KD for IL-8(1–66) binding to ND-CXCR1(1–38) in the presence of empty nanodiscs was determined to be 3.2 μM, very similar to the value found in the absence of nanodiscs, further demonstrating that the presence of a membrane does not affect the binding of IL-8(1–66) to Binding Site-I of CXCR1 (data not shown).
Discussion and conclusions
The chemokine IL-8 interacts with the chemokine receptor CXCR1. The structure of monomeric IL-8(1–66) is similar to that of full-length IL-8(1–72) in both monomeric and dimeric forms, which demonstrates that the truncation of six residues at the C-terminus, necessary to form a stable monomeric species, does not have a significant effect on the overall fold of the chemokine. IL-8(1–66) binds to two polypeptides with sequences corresponding to the N-terminal region of the CXCR1 receptor. ND-CXCR1(1–38) includes Binding Site-I and is water soluble, while 1TM-CXCR1(1–72) includes Binding Site-I and the first hydrophobic trans-membrane helix of CXCR1 and requires the presence of lipids.
Docking and molecular dynamics studies between IL-8 and CXCR1 have been performed using the solid-state NMR structure of CXCR1 in lipid bilayers (PDB id: 2LNL) supplemented with NMR data on N-terminal domain derived peptides and homology studies to other GPCRs (Park et al. 2012; Liou et al. 2014; Joseph and Rajarathnam 2015). In the resulting model, charged residues in the N-loop of IL-8 interact with residues in the N-terminal domain of CXCR1 (Binding Site-I), and hydrophobic residues in IL-8 contact hydrophobic residues in the N-terminus of CXCR1, which results in a rotation of IL-8. Finally, electrostatic interactions attract the ELR motif in IL-8 to the extracellular loops of CXCR1, which constitute Binding Site-II, triggering a conformational change that activates the receptor. A very similar series of events is expected to take place in other chemokine receptors (Kufareva et al. 2017).
The residues of IL-8(1–66) involved in the interactions with Site I of CXCR1 were identified by chemical shift perturbations. Three main regions of IL-8(1–66) were shown to interact with ND-CXCR1(1–38), the N-loop, the third β-sheet, and the N-terminal part of the α-helix. This result agrees with previous studies performed on wild-type IL-8 and similar soluble peptides with sequences derived from CXCR1 (Fernando et al. 2007; Skelton et al. 1999; Park et al. 2011a, b; Hebert et al. 1991; Clubb et al. 1994; Williams et al. 1996; Kendrick et al. 2014; Hammond et al. 1996; Joseph et al. 2015, 2010; Jiang et al. 2015; Monteclaro and Charo 1997). The studies of the interaction of IL-8 with soluble polypeptides containing Binding Site-I were extended by the preparation of a longer polypeptide that contains the N-terminal domain and the first trans-membrane helix of CXCR1. This insoluble polypeptide was reconstituted into nanodiscs, which provide a bilayer environment proximal to Binding Site-I. No significant differences were found in how IL-8(1–66) binds to the shorter construct ND-CXCR1(1–38) or the longer membrane— associated construct 1TM-CXCR1(1–72).
These results indicate that the presence of the first trans-membrane helix does not affect the interaction of IL-8 with Binding Site-I in the N-terminal domain of CXCR1. Similar conclusions have been reached based on experiments where the N-terminal domain of the chemokine receptor CCR2 was fused to the trans-membrane segment of an unrelated receptor; chemokine binding was not affected, showing that the N-terminal portion of a chemokine receptor is both necessary and sufficient for chemokine-receptor interactions (Chaudhuri et al. 2013).
It has been hypothesized that another possible difference between studies on the peptide and ligand binding in vivo could be the presence of the lipid bilayer (Rajagopalan and Rajarathnam 2004; Nasser et al. 2009). However, we do not observe significantly different chemical shift perturbations between IL-8(1–66) binding to ND-CXCR1(1–38) in an aqueous environment and to 1TM-CXCR1(1–72) in nanodiscs. This suggests that it is unlikely that the membrane influences the final conformation of ND-CXCR1 upon interaction with IL-8. However, the presence of the lipid bilayer may affect the conformation of the unbound N-terminal domain. Previously, we observed that ND-CXCR1 is immobilized in the presence of DMPC/DHPC bicelles, but appears to lose this interaction upon binding IL-8 (Park et al. 2011a, b; Haldar et al. 2010).
Activity and mutational studies have shown that residues Glu4, Leu5 and Arg6 are essential for receptor activation, but not for receptor interactions. The ELR motif is therefore thought to be important for the interaction with the second binding site associated with extracellular loops of CXCR1 and not involved in binding the N-terminal domain (Clark-Lewis et al. 1991; Hebert et al. 1991; Park et al. 2017; Williams et al. 1996). This is supported by the lack of chemical shift perturbations for these residues upon interaction with both ND-CXCR1(1–38) and 1TM-CXCR1(1–72). Only small chemical shift perturbations are observed at the C-terminus. This is the location of the α-helix and the site of the truncation to make this construct of IL-8(1–66) monomeric, confirming experimental data obtained by others that the truncation of the most C-terminal six amino acids has no effect on CXCR1 binding or activation (Clark-Lewis et al. 1991; Joseph et al. 2010; Krieger et al. 2004). However, these residues are thought to be important for interaction with glycosaminoglycans (Möbius et al. 2013).
IL-8(1–66) is a stable monomer because of the truncation of the last six residues, which participate in the dimer interface. This structure is very similar to those of dimeric and monomeric full-length IL-8(1–72), with the exception of the location of the N-loop relative to the α-helix and the side chain of phenylalanine 65. IL-8 binds to the residues in the N-terminal domain of CXCR1 that constitute Binding Site-I regardless of the presence of the first trans-membrane helix or a lipid membrane. This suggests that the presence of IL-8 the N-terminal domain of CXCR1 does not interact with the membrane. Thus, investigations of the interactions of IL-8 with Binding Site-II, located among the extracellular loops of CXCR1, are essential for understanding the how the agonist triggers the conformational changes and receptor activation in the membrane environment.
Supplementary Material
Acknowledgments
We thank Mitchell Zhao for his participation in the sample preparation and Dr. Andrey Bobkov for assistance with the ITC measurements. This research was supported by Grants P41EB002031, RO1GM066978, R35GM122501, and R35GM118186 from the National Institutes of Health and utilized the Biomedical Technology Resource Center for NMR Molecular Imaging of Proteins at the University of California, San Diego.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10858-017-0128-3) contains supplementary material, which is available to authorized users.
References
- Alexander SP, et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. Br J Pharmacol. 2015;172:5744–5869. doi: 10.1111/bph.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini M. CXCL8-the first chemokine. Front Immunol. 2015;6:258. doi: 10.3389/fimmu.2015.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin ET, et al. Crystal structure of interleukin 8: symbiosis of NMR and crystallography. Proc Natl Acad Sci USA. 1991;88:502–506. doi: 10.1073/pnas.88.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter EF, Stone MJ. Synergistic interactions between chemokine receptor elements in recognition of interleukin-8 by soluble receptor mimics. BioChemistry. 2012;51:1322–1331. doi: 10.1021/bi201615y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall L. Chemokines and their receptors in disease. Histol Histopathol. 2005;20:907–926. doi: 10.14670/HH-20.907. [DOI] [PubMed] [Google Scholar]
- Bizzarri C, et al. ELR + CXC chemokines and their receptors (CXC chemokine receptor 1 and CXC chemokine receptor 2) as new therapeutic targets. Pharmacol Ther. 2006;112:139–149. doi: 10.1016/j.pharmthera.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Burg JS, et al. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows SD, et al. Determination of the monomer-dimer equilibrium of interleukin-8 reveals it is a monomer at physiological concentrations. BioChemistry. 1994;33:12741–12745. doi: 10.1021/bi00209a002. [DOI] [PubMed] [Google Scholar]
- Casagrande F, Maier K, Kiefer H, Opella SJ, Park SH. Production of membrane proteins. Wiley; Weinheim: 2011. Expression and purification of G-protein coupled receptors for NMR structural studies. [Google Scholar]
- Chaudhuri A, et al. Organization and dynamics of the N-terminal domain of chemokine receptor CXCR1 in reverse micelles: effect of graded hydration. J Phys Chem B. 2013;117:1225–1233. doi: 10.1021/jp3095352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- Clore GM, Appella E, Yamada M, Matsushima K, Gronenborn AM. Three-dimensional structure of interleukin 8 in solution. BioChemistry. 1990;29:1689–1696. doi: 10.1021/bi00459a004. [DOI] [PubMed] [Google Scholar]
- Clubb RT, Omichinski JG, Clore GM, Gronenborn AM. Mapping the binding surface of interleukin-8 complexed with an N-terminal fragment of the type 1 human interleukin-8 receptor. FEBS Lett. 1994;338:93–97. doi: 10.1016/0014-5793(94)80123-1. [DOI] [PubMed] [Google Scholar]
- Eigenbrot C, Lowman HB, Chee L, Artis DR. Structural change and receptor binding in a chemokine mutant with a rearranged disulfide: X-ray structure of e38C/C50A IL-8 at 2 A resolution. Proteins. 1997;27:556–566. [PubMed] [Google Scholar]
- Fernando H, Chin C, Rosgen J, Rajarathnam K. Dimer dissociation is essential for interleukin-8 (IL-8) binding to CXCR1 receptor. J Biol Chem. 2004;279:36175–36178. doi: 10.1074/jbc.C400283200. [DOI] [PubMed] [Google Scholar]
- Fernando H, Nagle GT, Rajarathnam K. Thermodynamic characterization of interleukin-8 monomer binding to CXCR1 receptor N-terminal domain. FEBS J. 2007;274:241–251. doi: 10.1111/j.1742-4658.2006.05579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle RB, et al. Importance of the amino terminus of the interleukin-8 receptor in ligand interactions. J Biol Chem. 1993;268:7283–7289. [PubMed] [Google Scholar]
- Gerber N, Lowman H, Artis DR, Eigenbrot C. Receptor-binding conformation of the “ELR” motif of IL-8: X-ray structure of the L5C/H33C variant at 2.35 A resolution. Proteins. 2000;38:361–367. [PubMed] [Google Scholar]
- Grasberger BL, Gronenborn AM, Clore MG. Analysis of the backbone dynamics of interleukin-8 by 15N relaxation measurements. J Mol Biol. 1993;230:364–372. doi: 10.1006/jmbi.1993.1152. [DOI] [PubMed] [Google Scholar]
- Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized phospho-lipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar S, Raghuraman H, Namani T, Rajarathnam K, Chattopadhyay A. Membrane interaction of the N-terminal domain of chemokine receptor CXCR1. Biochim Biophys Acta. 2010;1798:1056–1061. doi: 10.1016/j.bbamem.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Hammond ME, et al. Receptor recognition and specificity of interleukin-8 is determined by residues that cluster near a surface-accessible hydrophobic pocket. J Biol Chem. 1996;271:8228–8235. doi: 10.1074/jbc.271.14.8228. [DOI] [PubMed] [Google Scholar]
- Hebert CA, Vitangcol RV, Baker JB. Scanning mutagenesis of interleukin-8 identifies a cluster of residues required for receptor binding. J Biol Chem. 1991;266:18989–18994. [PubMed] [Google Scholar]
- Hébert CA, et al. Partial functional mapping of the human interleukin-8 type A receptor. Identification of a major ligand binding domain. J Biol Chem. 1993;268:18549–18553. [PubMed] [Google Scholar]
- Helmer D, et al. Rational design of a peptide capture agent for CXCL8 based on a model of the CXCL8:CXCR1 complex. Rsc Adv. 2015;5:25657–25668. doi: 10.1039/c8ra90035c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Structure and functional expression of a human interleukin-8 receptor. Science. 1991;253:1278–1280. doi: 10.1126/science.1840701. [DOI] [PubMed] [Google Scholar]
- Jiang S-J, et al. Peptides derived from CXCL8 based on in silico analysis inhibit CXCL8 interactions with its receptor CXCR1. Scientific Rep. 2015;5:18638. doi: 10.1038/srep18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph PR, Rajarathnam K. Solution NMR characterization of WT CXCL8 monomer and dimer binding to CXCR1 N-terminal domain. Protein Sci. 2015;24:81–92. doi: 10.1002/pro.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph PR, et al. Probing the role of CXC motif in chemokine CXCL8 for high affinity binding and activation of CXCR1 and CXCR2 receptors. J Biol Chem. 2010;285:29262–29269. doi: 10.1074/jbc.M110.146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph PR, et al. Proline substitution of dimer interface β-strand residues as a strategy for the design of functional monomeric proteins. Biophys J. 2013;105:1491–1501. doi: 10.1016/j.bpj.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph PR, Mosier PD, Desai UR, Rajarathnam K. Solution NMR characterization of chemokine CXCL8/IL-8 monomer and dimer binding to glycosaminoglycans: structural plasticity mediates differential binding interactions. Biochem J. 2015;472:121–133. doi: 10.1042/BJ20150059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick AA, et al. The dynamics of interleukin-8 and its interaction with human CXC receptor I peptide. Protein Sci. 2014;23:464–480. doi: 10.1002/pro.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, et al. A structural and dynamic model for the interaction of interleukin-8 and glycosaminoglycans: support from isothermal fluorescence titrations. Proteins. 2004;54:768–775. doi: 10.1002/prot.10590. [DOI] [PubMed] [Google Scholar]
- Kufareva I, Gustavsson M, Zheng Y, Stephens BS, Handel TM. What do structures tell us about chemokine receptor function and antagonism? Annu Rev Biophys. 2017;46:175–198. doi: 10.1146/annurev-biophys-051013-022942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRosa GJ, et al. Amino terminus of the interleukin-8 receptor is a major determinant of receptor subtype specificity. J Biol Chem. 1992;267:25402–25406. [PubMed] [Google Scholar]
- Leong SR, Kabakoff RC, Hébert CA. Complete mutagenesis of the extracellular domain of interleukin-8 (IL-8) type A receptor identifies charged residues mediating IL-8 binding and signal transduction. J Biol Chem. 1994;269:19343–19348. [PubMed] [Google Scholar]
- Liou J-W, et al. In silico analysis reveals sequential interactions and protein conformational changes during the binding of chemokine CXCL-8 to its receptor CXCR1. PLoS ONE. 2014;9:e94178. doi: 10.1371/journal.pone.0094178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman HB, et al. Monomeric variants of IL-8: effects of side chain substitutions and solution conditions upon dimer formation. Protein Science. 1997;6:598–608. doi: 10.1002/pro.5560060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesleh MF, Opella SJ. Dipolar Waves as NMR maps of helices in proteins. J Magn Reson. 2003;163:288–299. doi: 10.1016/s1090-7807(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Möbius K, et al. Investigation of lysine side chain interactions of interleukin-8 with heparin and other glycosaminoglycans studied by a methylation-NMR approach. Glycobiology. 2013;23:1260–1269. doi: 10.1093/glycob/cwt062. [DOI] [PubMed] [Google Scholar]
- Monteclaro FS, Charo IF. The amino-terminal domain of CCR2 is both necessary and sufficient for high affinity binding of monocyte chemoattractant protein 1—Receptor activation by a pseudotethered ligand. J Biol Chem. 1997;272:23186–23190. doi: 10.1074/jbc.272.37.23186. [DOI] [PubMed] [Google Scholar]
- Murphy PM, Tiffany HL. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science. 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- Murphy PM, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- Nasser MW, et al. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 2009;183:3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald C, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. J Mol Biol. 2011a;414:194–203. doi: 10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, et al. Local and global dynamics of the G proteincoupled receptor CXCR1. BioChemistry. 2011b;50:2371–2380. doi: 10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, et al. Paramagnetic relaxation enhancement of membrane proteins by incorporation of the metal-chelating unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQA) J Biomol NMR. 2014;61(3–4):185–196. doi: 10.1007/s10858-014-9884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Berkamp S, Radoicic J, De Angelis AA, Opella SJ. Interaction of monomeric interleukin-8 with CXCR1 mapped by proton-detected fast MAS solid-state NMR and intermolecular paramagnetic relaxation enhancement. 2017 doi: 10.1016/j.bpj.2017.09.041. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado GN, et al. Chemokine signaling specificity: essential role for the N-terminal domain of chemokine receptors. BioChemistry. 2007;46:89618968. doi: 10.1021/bi7004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347:1117–1122. doi: 10.1126/science.1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan L, Rajarathnam K. Ligand selectivity and affinity of chemokine receptor CXCR1. Role of N-terminal domain. J Biol Chem. 2004;279:30000–30008. doi: 10.1074/jbc.M313883200. [DOI] [PubMed] [Google Scholar]
- Rajarathnam K, Clark-Lewis I, Sykes BD. 1H NMR solution structure of an active monomeric interleukin-8. BioChemistry. 1995;34:12983–12990. doi: 10.1021/bi00040a008. [DOI] [PubMed] [Google Scholar]
- Rajarathnam K, Kay CM, Clark-Lewis I, Sykes BD. Characterization of quaternary structure of interleukin-8 and functional implications. Methods Enzymol. 1997;287:89–105. doi: 10.1016/s0076-6879(97)87009-7. [DOI] [PubMed] [Google Scholar]
- Rajarathnam K, Prado GN, Fernando H, Clark-Lewis I, Navarro J. Probing receptor binding activity of interleukin-8 dimer using a disulfide trap. BioChemistry. 2006;45:7882–7888. doi: 10.1021/bi0605944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran A, Joseph PR, Rajarathnam K. Structural basis for differential binding of the interleukin-8 monomer and dimer to the CXCR1 N-domain: role of coupled interactions and dynamics. BioChemistry. 2009;48:8795–8805. doi: 10.1021/bi901194p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, et al. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkilde MM, Schwartz TW. The chemokine system—a major regulator of angiogenesis in health and disease. APMIS. 2004;112:481–495. doi: 10.1111/j.1600-0463.2004.apm11207-0808.x. [DOI] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Clore MG. Using Xplor,ÄìNIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- Skelton NJ, Quan C, Reilly D, Lowman H. Structure of a CXC chemokine-receptor fragment in complex with interleukin-8. Structure. 1999;7:157–168. doi: 10.1016/S0969-2126(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Suetomi K, Rojo D, Navarro J. Identification of a signal transduction switch in the chemokine receptor CXCR1. J Biol Chem. 2002;277:31563–31566. doi: 10.1074/jbc.M204713200. [DOI] [PubMed] [Google Scholar]
- Tan Q, et al. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Schwieters CD, Opella SJ, Marassi FM. A practical implicit membrane potential for NMR structure calculations of membrane proteins. Biophys J. 2015;109:574–585. doi: 10.1016/j.bpj.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G, et al. Mutagenesis studies of interleukin-8. Identification of a second epitope involved in receptor binding. J Biol Chem. 1996;271:9579–9586. doi: 10.1074/jbc.271.16.9579. [DOI] [PubMed] [Google Scholar]
- Wu L, et al. Discrete steps in binding and signaling of interleukin-8 with its receptor. J Biol Chem. 1996;271:31202–31209. doi: 10.1074/jbc.271.49.31202. [DOI] [PubMed] [Google Scholar]
- Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


