Abstract
Deadly infections from opportunistic fungi have risen in frequency, largely because of the at-risk immunocompromised population created by advances in modern medicine and the HIV/AIDS pandemic. This review focuses on dynamics of the fungal polysaccharide cell wall, which plays an outsized role in fungal pathogenesis and therapy because it acts as both an environmental barrier and as the major interface with the host immune system. Human fungal pathogens use architectural strategies to mask epitopes from the host and prevent immune surveillance, and recent work elucidates how biotic and abiotic stresses present during infection can either block or enhance masking. The signaling components implicated in regulating fungal immune recognition can teach us how cell wall dynamics are controlled, and represent potential targets for interventions designed to boost or dampen immunity.
Keywords: Fungi, cell wall, glucan, innate immunity, evasion
The fungal cell wall, an Achilles heel for a destructive class of pathogens
Recent years have seen the emergence of fungi as a major threat to public health and food security. The success of modern medical advancements such as chemotherapy and organ transplantation extend patient life, but leave immune compromised patients susceptible to invasive infections by opportunistic fungal pathogens like Candida albicans, Aspergillus fumigatus and Cryptococcus neoformans. These invasive fungal infections are accompanied by unacceptably high mortality rates, despite modern antifungal therapies. Candida, as an example, is the 4th most common cause of nosocomial bloodstream infections, thought to cause greater than 400,000 life-threatening infections annually [1]. Beyond being a threat to public health, fungi threaten agriculture, destroy crops, devastate amphibian and bat species, and are responsible for major extinctions caused by infectious disease [2]. Facing a global assault on plants, animals and our own health, we must devise novel strategies to address fungal infections.
Opportunistic fungi, like other pathogens in this class, are able to thrive in challenging and rapidly changing environments by using multiple pathways for stress adaptation [3]. For C. albicans, these stresses include environmental challenges, drug exposure and immune attack. Recent work has shown that the fungal responses to these stresses include altering their cell surfaces to enhance or limit immune recognition and responses, linking the two disparate fields of cell wall integrity (see Glossary) and immunity (Fig. 1, Key Figure). This new work highlights the translational importance of the cell wall, which serves as an effective drug target against both bacteria and fungi, and includes essential microbial components recognized by conserved innate immune pattern recognition receptors (PRRs). In this review, we summarize our current knowledge of fungal cell wall remodeling in response to the environment, with a focus on responses to stress relevant to infection. We also highlight the role of fungal cell wall remodeling in epitope masking.
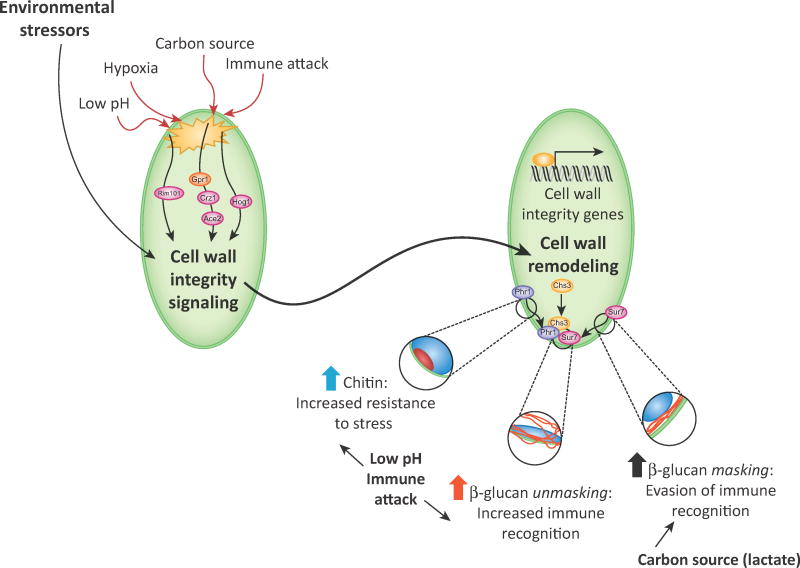
Figure 1. Stress-triggered cell wall remodeling affects immune recognition through epitope (un)masking.
Environmental stresses, including pH, hypoxia, altered carbon sources, and immune attack can trigger fungal response pathways that remodel the cell wall and alter PAMP availability. The roles of specific signaling proteins and pathways are detailed in Fig. 3 and Fig. 4. The effects on the cell wall lead to enhanced exposure (in the cases of low pH or immune attack) or increased masking of cell wall β-glucan and chitin (in the case of environmental lactate). The roles of specific cell wall remodeling enzymes in response to immune attack is detailed in Fig. 4. Changes in the availability of these PAMPs alter immune recognition and responses.
Adaptation to stresses during infection: fungal “feng shui”
The cell wall is the primary fungal barrier against the external environment, playing a central role in the maintenance of cellular integrity. The cell wall is also the initial point of contact between the host and fungus, where it initiates immune responses via immune receptors. To sustain cell integrity in the face of environmental stresses, the cell wall must adapt to external challenges that could compromise cell wall structure or change how it triggers immune responses.
Normal fungal cell wall architecture and stress responses
Most pathogenic fungi have an inner cell wall layer of chitin and β-glucan which provides cellular integrity, but the outer layer composition of the cell wall varies among fungal species and can cover inner polysaccharides to prevent immune availability (Fig. 2) [4]. In C. albicans, the outer layer consists of mannosylated glycoproteins connected to the inner layer by modified GPI anchors and (1,6)-β-glucan linkages to the inner core of β-glucan and chitin. However, the fungal cell wall is dynamic and varies between morphologies. For example, C. albicans hyphal cells have shorter, less dense mannan fibrils than yeast cells [5]. In A. fumigatus, conidia have a layer of α-glucan along with melanin and hydrophobic rodlets in their outer cell wall, while hyphae have α-glucan, galactomannan and galactosaminoglycan [4]. Similarly, Histoplasma capsulatumand other dimorphic fungi are covered with α-glucan during infection [4]. In contrast, C. neoformans has a thick outer capsule made largely of glucuronoxylomannan and galactoxylomannan attached to α-glucan [4].
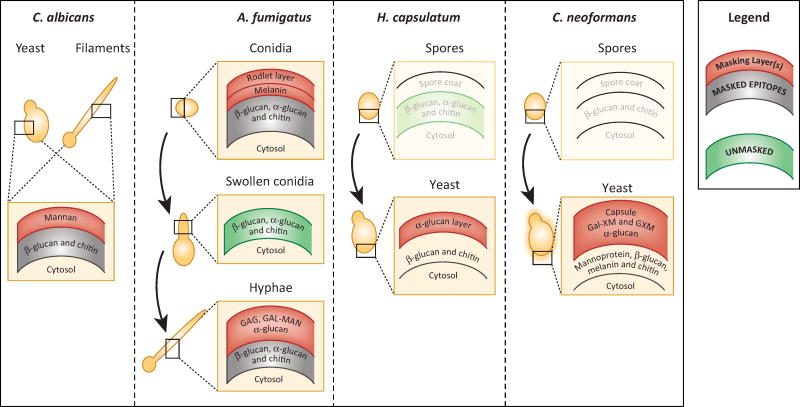
Figure 2. Fungi differentiate to change their function and form, and their cell wall architecture is designed to mask some epitopes from immune recognition.
The simplified cell wall architectures of select human pathogenic fungi are depicted schematically (adapted from [47]). The outer cell wall layers (shown in red) of Candida, Aspergillus, Histoplasma and Cryptococcus forms are generally capable of preventing pattern recognition receptors from binding to ligands that are buried within inner layers of the cell wall (shown in gray to indicate masking). The polysaccharide and protein components of the outer layers differ among pathogenic fungi. In some cases, such as A. fumigatus swollen conidia, rapid growth leads to the temporary unmasking of underlying epitopes, but in most cases during in vitro growth only small proportions of the cell wall are sufficiently unstructured to allow binding of immune receptors to inner polysaccharide molecules (shown in green to indicate surface exposure). Morphological transitions (indicated by curved arrows) that occur during infection by Aspergillus, Histoplasma and Cryptococcus are associated with cell wall changes that affect epitope exposure. Not much is known about the cell wall architecture of H. capsulatum or C. neoformans spores, so the layering is still unknown and these schematics are drawn in faded colors.
Although we know that the cell wall is dynamic, we still understand little about the pathways that regulate its architecture in pathogenic fungi. In the model fungus Saccharomyces cerevisiae, the cell wall integrity signaling network is able to sense stresses and transduce them into both transcriptional and post-transcriptional changes that modulate cell wall structure [6, 7]. This signaling network, which is evolutionarily conserved in C. albicans and other fungi, is initiated by stress sensors for pH, oxygen, carbon dioxide, shifts in carbon sources, osmotic shifts, reactive oxygen, nitrogen, and sulfur species, temperature, and direct cell wall perturbation [8–14]. Important parts of this network include cell wall sensors, two-component signaling proteins, MAPK signaling components, protein kinase C, calcineurin, transcription factors, and cytosolic and cell wall effectors [15]. Fungi use these signals to maintain “feng shui”, rapidly and accurately remodeling the architecture and composition of the cell wall to minimize the impact of these stressors in an ever-changing environment.
Cell wall stresses are felt during infection
A living host presents unique niches that require fungi to rapidly adapt their cell wall. Recent work has addressed how fungi respond to single and combinatorial stresses, and how cell wall integrity responses are activated in vivo. We know that changes in carbon availability, pH, oxygen availability and immune attack are all encountered in vivo [4, 16–18]. Rigorous in vitro experiments link these stresses to cell wall changes and altered immune recognition, and it is clear that immune attack also regulates cell wall remodeling and immune recognition in vivo [19–24].
In addition to natural environmental stresses, the echinocandin class of antifungal drugs imposes severe stress on the fungal cell wall by inhibiting β-glucan synthase. Exposure to echinocandins has been shown to induce numerous changes to the fungal cell wall, including lower β-glucan content, unmasking of cell wall β-glucan, and increased chitin synthesis and exposure [25–29]. Importantly, these responses are different in vivo and in vitro, indicating that signaling pathways may be activated or repressed by other cues within host niches [30].
The host environment challenges infecting organisms with multiple stressors at the same time. In vitro work on multiple concurrent stressors suggests that C. albicans uses microbial adaptive prediction, where exposure to an initial stress can influence survival when encountering a later stress [3]. While there is little discussion of adaptation prediction in A. fumigatus and C. neoformans in the literature, there is clear evidence that these fungal pathogens have evolved to integrate their responses to cell wall and other stresses. For example, multiple conserved pathways, including MAP kinase and protein kinase A signaling, coordinate the expression of cell wall genes with other stress genes in A. fumigatus, and signals other than cell wall stresses can trigger the cell wall integrity pathway and cell wall genes [31–33]. In C. neoformans, the transcription factorRim101 regulates cell wall gene expression as well as pH sensing pathways, and the cell wall integrity pathway coordinates capsule homeostasis as well as cell wall regulation [34, 35]. These data emphasize the limits of a reductionist approach and the necessity to study fungal physiology during infection.
Candida stress pathways are regulated during infection and important for pathogenesis
For C. albicans, the importance of many cell wall genes for fungal viability in the host has been confirmed through deletion mutants. For instance, conditional repression of the major β-glucan synthase FKS1 in a mouse model of disseminated candidiasis confirms the requirement for this enzyme in vivo [36]. PHR1, which remodels cell wall β-glucan and has been demonstrated to be recruited to sites of neutrophil attack, is required for virulence during disseminated but not vulvovaginal candidiasis [37]. The finding that some cell wall genes are only required in some tissues argues for the niche-specificity of cell wall requirements and virulence. Further support for the idea that virulence is relative to infection route was generated using competition assays of barcoded mutants in the gut and intravenous infection models [38]. More recently, similar in vivo competition experiments have demonstrated an unexpected connection between cell wall epitope exposure and gastrointestinal fitness, suggesting that cell wall architecture is especially important for commensal success [39].
Tools such as Nanostring profiling, RNAseq and cell intrinsic reporters have also enabled investigators to probe fungal physiology during infection [30, 40–44]. Imaging of single-cell responses using fluorescent reporters has also been useful in both endpoint and intravital studies using mice and zebrafish hosts [41, 43, 44]. Significantly, Nanostring profiling of fungal gene transcripts during drug treatment of disseminated candidiasis has demonstrated that the transcriptomic responses to caspofungin differ significantly between in vivo and in vitro conditions [30]. In vivo profiling has also revealed that transcripts related to the fungal cell wall and remodeling can differ between C. albicans strains and are impacted by the immune status of the host [27]. These studies emphasize that we now have the tools to translate in vitro findings into an understanding of cell wall remodeling during infection.
Pathogen recognition and epitope masking: wielding the cell wall as a mask
The discovery that the fungal cell wall is a dynamic structure that undergoes significant remodeling during morphogenesis prompts the idea that it may be a moving immunological target, enabling pathogenic fungi to exert some control over the host’s innate immune system. Recent work shows that pathogenic fungi can alter their cell wall structure in response to host cues in ways that could enable them to either evade and escape the immune system or to hyper-activate the immune system resulting in damage to the mucosa. The following sections review our current knowledge on this topic, focusing on C. albicans.
Fungal structures are recognized by pattern recognition receptors
The innate immune system is constantly on alert for microbial pathogens. To achieve this, the host uses a vast array of PRRs, which are capable of recognizing distinct components not normally found in the body. Upon binding these foreign ligands, called pathogen associated molecular patterns (PAMPs), PRRs initiate signaling cascades that activate downstream responses including phagocytosis, microbial killing mechanisms and cytokine production.
Fungal cell wall PAMPs include chitin, mannan and β-glucan, which are recognized by host innate immune PRRs that include multiple Toll-like receptors and C-type lectin receptors (CLRs) [45–47]. CLRs are particularly important for antifungal immunity, as evidenced by susceptibility to fungal infection in patients lacking effective CLR activity [48]. The cell wall, by controlling both fungal viability and host responses, therefore serves as a major focal point in determining the outcome of fungal infection.
Animal and plant pathogens mask epitopes from immune recognition
Manipulation of cell wall architecture to mask epitopes from host recognition is actually quite widespread and has been observed during fungal infection of plants and insects [49–52]. Importantly, epitope masking is also seen in multiple fungi capable of infecting humans (Fig. 2).
In particular, fungi seem to go to great lengths to mask β-glucans. H.capsulatum masks its β-glucan with α-glucan and Paracoccidioides brasiliensis actually converts its (1,3)-β-glucans to (1,3)-α-glucan in the host [53, 54]. A. fumigatus also masks β-glucan, though this PAMP becomes exposed during germination or in hypoxic host environments [23, 55]. This basal β-glucan masking in Aspergillus depends on cell wall architecture. In conidia, masking depends on the hydrophobin RodA, α-glucan, galactosaminogalactan and the pigment protein PskP [56–59], while epitope masking in hyphae depends on galactosaminogalactan [58]. Interestingly, A. fumigatus RodA also appears important for blocking both Dectin-1 and Dectin-2 mediated host responses, presumably to (1,3)-β-glucan and α-mannan epitopes [57]. Fonsecaea pedrosoi, a major agent of chromoblastomycosis, masks β-glucan in the cell wall of its sclerotic cells with a chitin-like component [60]. C. neoformansmasks PAMPs, including β-glucan, behind a thick capsule [61]. C. albicans also masks the vast majority of its β-glucan from Dectin-1, with exposure limited to places with active cell wall growth or bud scars. It is generally thought that the outer layer of mannan and mannoproteins in the cell wall shields the inner β-glucan from Dectin-1 recognition [4]. These diverse outer cell wall layers likely reflect species-specific evolutionary solutions to deal with environmental stresses, but they all function to restrict host access to the inner cell wall.
Maintenance of β-glucan masking by C. albicans requires a complex and diverse set of pathways in fungi. Disruptions of mannose structure, GPI anchor synthesis, phosphotidylserine biosynthesis, cell wall integrity signaling, β-glucan remodeling and β-glucan synthesis have all been shown to result in β-glucan unmasking [29, 62–68]. Atomic force microscopy demonstrates that β-glucan unmasking correlates with the increased surface roughness [69], while super-resolution microscopy has characterized the fine-scale architecture of caspofungin induced β-glucan unmasking in C. albicans [70]. Importantly, in each of these cases, the exposure of β-glucan drives enhanced Dectin-1 recognition and inflammatory responses against fungi. While epitope unmasking clearly impacts interactions with host immunity, the cell wall dynamics involved in the maintenance of epitope masking are only beginning to be understood.
Biotic and abiotic stresses cause epitope masking and unmasking: timely wearing the mask
Adaptation to the multitude of environmental cues in the host induces morphogenesis and activates stress response pathways. However, host-derived conditions, antifungals and host immune action have also been shown to induce considerable cell wall remodeling in fungi [71]. These changes in the fungal cell surface have profound effects on how the invading pathogen is perceived by the innate immune system. For instance, growth in the host triggers differentiation of Histoplasma capsulatum and stimulates deposition of the immunologically silent polysaccharide α-glucan on top of inflammatory β-glucan, thereby limiting inflammatory immune responses [54, 72]. Three recent studies in C. albicans now shed light on other ways in which fungal adaptation to the host niche influences the innate immune response.
Acidic pH enhances glucan unmasking
One key parameter C. albicans experiences during colonisation of the vaginal mucosa is low mucosal pH, with the vaginal mucosa being between pH 4–5, depending on age, ethnic origin and position in the menstrual cycle [73]. Investigation of the effect of pH adaptation on C. albicans identified that exposure to acidic environments induces the exposure of β-glucan and chitin, two cell wall carbohydrate PAMPs which are normally buried beneath the mannan fibrils [24] (Fig. 3). This exposure of underlying cell wall carbohydrates has been termed cell wall unmasking [29]. Both chitin and β-glucan are recognised by the innate immune system [74, 75], and C. albicans cells that have adapted to acidic conditions elicited a strong pro-inflammatory innate immune response, and enhanced neutrophil recruitment [24], immune responses characteristic of genital thrush.
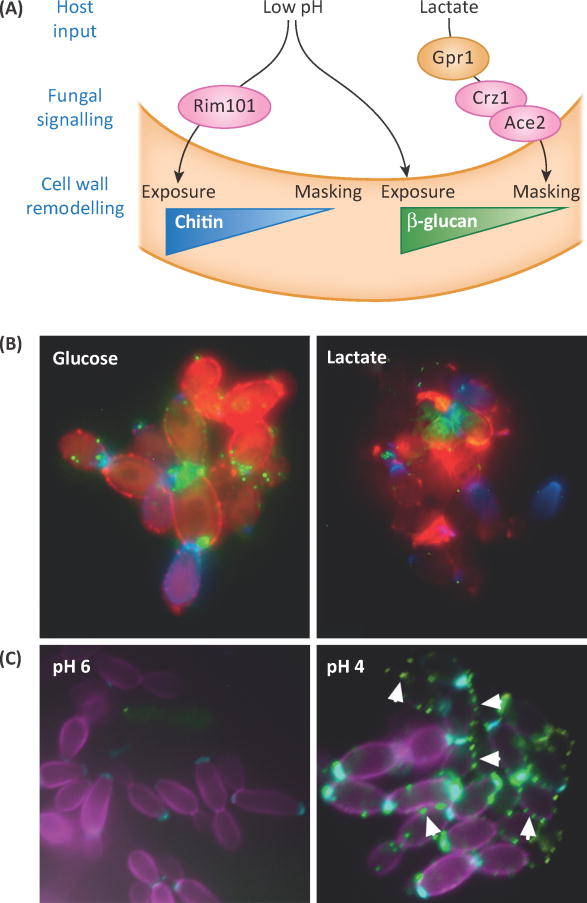
Figure 3. Abiotic environmental conditions can either enhance PAMP exposure or lead to greater masking.
(A) Schematic outlining how acidic pH and lactate signal C. albicans to alter availability of β-glucan and chitin for immune recognition, based on [24] and [19]. (B) C. albicanscells exposed to glucose or lactate were stained for exposure of β-glucan (Fc-Dectin-1, green), mannan (Concanavalin A, red) and chitin (wheat germ agglutinin, blue). Image generated by Gabriela Avelar, Aberdeen Fungal Group. (C) C. albicans cells exposed to growth media buffered to pH 6 or pH 4 were stained for exposure of β-glucan (Fc-Dectin1, green) chitin (wheat germ agglutinin, cyan) and total chitin (Calcofluor white, magenta). Arrowheads indicate points of β-glucan exposure.
Currently, the precise structural rearrangements that result in this cell wall unmasking are unknown. Visualization of the ultrastructure of C. albicans revealed that the mannan fibril layer was reduced upon adaptation to acidic conditions [24], which might indicate reduced mannan biosynthesis. Although the signaling mechanism regulating β-glucan unmasking remains to be elucidated, unmasking of chitin is regulated via the pH-dependent Rim101 signaling cascade [24] (Fig. 3). The Rim101 signaling cascade has been shown to be involved in the regulation of chitin exposure in other fungal pathogens [76], suggesting that this may be an evolutionary conserved process.
The impact of adaptation to acidic environments on cell wall remodeling in other pathogenic fungi has not been studied in detail to date. However, activation of cell wall salvage pathways in other fungi in response to various stress conditions also induces chitin deposition. This, in conjunction with the widely exploited fungal Rim101/PacC signaling pathway, suggests that other fungi may modulate chitin in response to environmental pH in a similar manner. As the mechanism underlying β-glucan exposure in response to environmental pH is still to be elucidated, it is hard to extrapolate whether mechanistic aspects of this phenomenon are conserved in other pathogenic fungi. However, exploration of pH dependent β-glucan exposure in other Candida species suggests that this phenomenon is species specific, with only C. albicans and Candida tropicalis exposing β-glucan at lower pH [24].
Lactate stimulates enhanced glucan masking
In contrast, it has become clear that host inputs can also stimulate epitope masking. This discovery emerged from the observation that growth of C. albicans cells on carbon sources such as amino, fatty or carboxylic acids, rather than sugars, leads to changes in their cell wall architecture and elasticity [71, 77] which affect host immune responses [21]. Further investigation revealed that C. albicans cells trigger masking of the PAMP β-glucan at their cell surface in response to physiological levels of lactate, generated either by host or bacterial cells [19] (Fig. 3). This lactate-induced β-glucan masking correlates with a reduction in neutrophil recruitment and an attenuation of inflammatory responses [19]. No doubt these effects contribute to the impact of carbon source adaptation upon the virulence of C. albicans during systemic and mucosal infection [71].
Lactate-induced β-glucan masking is mediated by a specific signaling pathway that appears to have recruited key components from other well-characterised signaling pathways. Lactate is detected by the receptor, Gpr1p [19], which lies upstream of the cyclic AMP-protein kinase A pathway that activates hyphal development [78] (Fig. 3). The lactate signal is then transduced to the transcription factor Crz1p [19]. Crz1p, which regulates cell wall remodeling, also responds to calcium ions via calcineurin signaling [79]. The mechanisms by which Gpr1p regulates Crz1p activity, and by which Crz1p drives β-glucan masking, are under investigation.
These findings in C. albicans have not yet been followed by work in other pathogenic fungi, but there are clear connections between carbon utilization and the cell wall in Aspergillus fumigatus. In this fungus, lowering the glucose concentration of the growth media to mimic the lung environment results in reduced cell wall β-glucan levels. This reduced β-glucan renders the fungus less susceptible to echinocandins, suggesting that environmental adaptation to carbon source may regulate therapeutic efficacy [20].
Immune attack triggers β-glucan unmasking
Clearly, C. albicans cells can tune the exposure of key epitopes at their cell surface to specific signals in host microenvironments. Interestingly, β-glucan becomes unmasked naturally over the course of disseminated C. albicans infection in vivo [29]. Recent work demonstrates that this PAMP unmasking is an active fungal response to attack from host neutrophils. Intriguingly, the fungal response also includes the deposition of chitin, which co-localizes with β-glucan unmasking [22] (Fig. 4). As expected, this altered pattern recognition following neutrophil extracellular trap (NET)-mediated cell wall changes enhances the subsequent responses of macrophages to damaged fungi. Importantly, neutrophils are required for the time-dependent unmasking of β-glucan in vivo, suggesting that these cell wall integrity pathways responding to immune attack in vitro are active during infection [22].
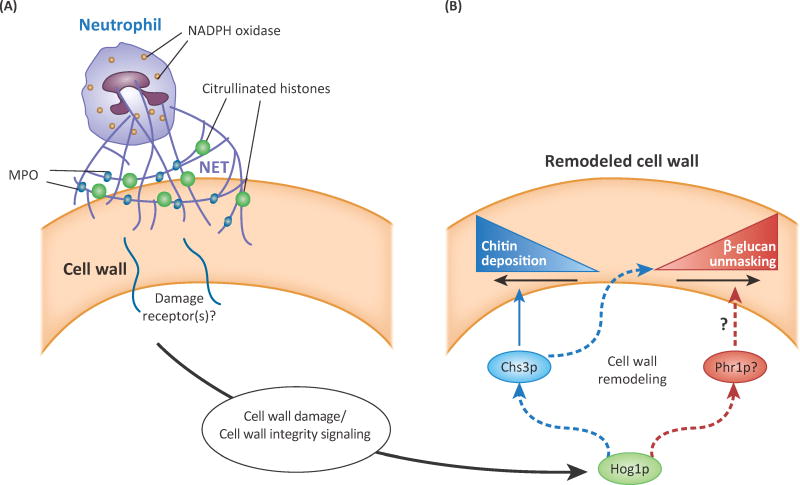
Figure 4. Immune-mediated attack of fungal hyphae triggers epitope exposure.
(A) Neutrophil attack through extracellular trap (NET) formation, including activity of phagocyte oxidase, myeloperoxidase (MPO), and reactive oxygen species (ROS) causes cell wall protein damage and provokes stress responses. (B) Cell wall remodeling triggered by NET attack includes localized Chs3p-mediated chitin deposition and enhanced β-glucan exposure. Unmasking of β-glucan is reduced or abolished in the absence of Hog1p or Chs3p activity. The Phr1p transglycosyltransferase localizes to sites of neutrophil attack and may actively remodel β-glucan in the cell wall. Indirect effects are indicated by lines with dotted outlines. Schematic based on [22].
The signaling and machinery involved in this fungal response have been partially elucidated (Fig. 4). Chitin deposition and β-glucan unmasking are both regulated by Hog1p signaling. Chitin synthase 3 (Chs3p) localizes to attack sites and is responsible for the majority of the chitin deposition [22]. In contrast, the stress-activated synthases Chs2p and Chs8p are not required for this immune-triggered remodeling, highlighting differences in responses to abiotic and biotic stress [80]. Two other proteins associated with cellular integrity and cell wall architecture, Phr1p and Sur7p, are recruited to immune attack sites with unique temporal dynamics, which suggests there is well-choreographed remodeling response to host immune action. These experiments thus give us a glimpse of key immune and cell wall integrity processes that are likely to act following immune attack during infection, and prompt further questions about whether immune cells trigger similar changes in other pathogenic fungi.
Concluding Remarks
Despite the foundation of knowledge outlined above, the role of fungal cell wall remodeling in adapting to the complex host environment and interactions with immunity remains poorly understood (see Outstanding Questions). Unmasking epitopes could be beneficial to the host, as it subverts attempts at immune evasion and enhances host responses directed against the pathogen. This appears to be the case for β-glucan [19, 22, 24, 39, 68] and chitin [27, 81]. Given that so many types of fungal pathogens mask β-glucan and other epitopes, there is a strong argument that blocking this masking benefits the host. However, it is also possible that, in other host contexts, dynamic fungal cell wall remodeling could instead directly benefit the pathogen by reinforcing the cell wall [22, 28, 82], exacerbating immunopathology [22, 25, 83–88], or affecting sensitivity to antifungal agent [28].
Outstanding questions box.
Can we enhance fungal clearance by targeting components of pathways that regulate epitope exposure?
Are stress-induced changes to the cell wall seen in murine infection and in vitro actually relevant for human infections? What is the end result when multiple stresses are present?
How relevant are the C. albicans stress response pathways for other human fungal pathogens such as Aspergillus fumigatus, Cryptococcus neoformans, Pneumocystisand dimorphic fungi?
Through which intermediates does stress sensing lead to changes in the cell wall?
How is cell wall remodeling spatially restricted to repair localized areas of damage?
What cell wall stresses are felt during infection in different tissues? Where and when are they felt? How are they sensed?
Fungal cell wall remodeling can have dramatic impacts on interactions with the host. Does epitope unmasking have beneficial or detrimental impacts on the host during infection?
Are responses to cell wall stresses dictated by adaptation to host niches of different fungal pathogens? Which fungal responses are conserved throughout the host and which are niche-specific?
Clearly, the fungal cell wall is a nexus of physiology, drug action and immunity. Therefore, an understanding of how the fungal cell wall senses, responds and remodels itself in response to stress can be leveraged on two fronts. First, this work will identify the fungal pathways critical for adapting and maintaining cell wall integrity during interactions with the host and immunity, potentially yielding novel targets for broad-spectrum antifungal drugs. Second, understanding how dynamic cell wall remodeling, especially epitope unmasking, influences host immune responses to fungi will open the door to designing novel immunotherapeutic strategies for patients with fungal infections.
Highlights.
The fungal cell wall is a dynamic organelle that provides structure, protects fungal viability and controls interactions with the host during infection.
Fungal cell wall architecture allows opportunistic fungal pathogens to evade innate immune recognition.
Immune attack and environmental stresses drive cell wall remodeling that unmasks or covers up cell wall epitopes and alters immunogenicity
Alternative carbon sources and acidic pH present in vulvovaginal candidiasis infections have distinct effects on epitope masking.
Neutrophil attack on C. albicans hyphae triggers chitin deposition and enhanced recognition of cell wall β-glucan.
Altered innate immune recognition due to stress during infection may affect immune and disease dynamics.
Acknowledgments
The authors wish to acknowledge the many other recent studies on cell wall dynamics and immune response that we did not have space to adequately include. The work was supported by funding from the following sources: Burroughs Wellcome Fund (RTW). AJPB was funded by the UK Biotechnology and Biological Research Council (BB/K017365/1), the UK Medical Research Council (MR/M026663/1); the Wellcome Trust (097377) and by the MRC Centre for Medical Mycology and the University of Aberdeen (MR/M026663/1).
Glossary
- Cell wall integrity
Cell wall integrity under stress is maintained by physiological responses to disruptions in the cell wall that generally lead to strengthening of the cell wall and adapting to changing conditions.
- Cell wall
layers of linked proteins, lipids, and polysaccharides that lie exterior to the plasma membrane and create a porous barrier to interface with the environment.
- Chs2, 3, and 8
chitin synthases that respond to stresses and synthesize new chitin
- Chitin
N-acetyl-glucosamine polymer present at low levels in fungal cell walls that provides crucial crosslinking strength to the cell wall. Chitin up-regulation is a common method for fungi to reinforce their cell walls when under stress.
- CLR
C-type lectin receptor. CLRs are one class of innate immune PRR, and recognize polysaccharide ligands.
- Crz1p
key protein in calcium sensing regulated by Gpr1 in the presence of lactate
- Dectin-1
a CLR that recognizes fungal β-glucan and mediates protective anti-fungal responses in innate immune phagocytes. Dectin-1 is required for anti-fungal immunity in mice and humans.
- Fks1p
(1,3)-β-glucan synthase, encoded by FKS1 gene, conserved among fungi. This is the target for the echinocandin class of anti-fungal drugs.
- β-glucan
refers to (1→3,1→6)-β-D-glucan, a highly crosslinked sugar in the fungal cell wall that provides strength and lies close to the plasma membrane
- Gpr1p
G protein-coupled receptor that senses presence of lactate, an alternative carbon source
- Hog1p
a mitogen-activated protein (MAP) kinase that regulates cell wall integrity and osmolarity sensing
- Nanostring profiling
a highly sensitive technique that directly detects and quantifies the copy number of selected RNA molecules
- NET
Neutrophil extracellular trap. A mix of DNA, hydrolases, antimicrobial peptides and myeloperoxidase released by neutrophils when attacking pathogens that can’t be engulfed.
- PAMP
pathogen-associated molecular pattern. These are non-self molecules such as (1→3,1→6)-β-D-glucan, mannan, and lipopolysaccharide that are conserved among classes of microbial pathogens.
- PRR
pattern recognition receptor. PRRs are evolutionarily conserved innate immune receptors that recognize non-self molecules that are shared among classes of microbial pathogens. Classes of PRRs include Toll-like receptors and C-type lectin receptors.
- Phr1p
transglycosylase that resides in the cell wall and is required for proper structure and cell wall integrity
- Rim101p
master regulator of pH-sensing in many fungi, including Candida species.
- RNAseq
high-throughput sequencing of RNA to determine the overall transcriptional profile of a group of cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown GD, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186–94. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AJ, et al. Stress adaptation in a pathogenic fungus. J Exp Biol. 2014;217(Pt 1):144–55. doi: 10.1242/jeb.088930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gow NAR, et al. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol Spectr. 2017;5(3) doi: 10.1128/microbiolspec.FUNK-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowman DW, et al. Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J Biol Chem. 2014;289(6):3432–43. doi: 10.1074/jbc.M113.529131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69(2):262–91. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189(4):1145–75. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso-Monge R, et al. Fungi sensing environmental stress. Clin Microbiol Infect. 2009;15(Suppl 1):17–9. doi: 10.1111/j.1469-0691.2008.02690.x. [DOI] [PubMed] [Google Scholar]
- 9.Arana DM, et al. The role of the cell wall in fungal pathogenesis. Microb Biotechnol. 2009;2(3):308–20. doi: 10.1111/j.1751-7915.2008.00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009;12(4):365–70. doi: 10.1016/j.mib.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Ernst JF, Pla J. Signaling the glycoshield: maintenance of the Candida albicans cell wall. Int J Med Microbiol. 2011;301(5):378–83. doi: 10.1016/j.ijmm.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Komalapriya C, et al. Integrative Model of Oxidative Stress Adaptation in the Fungal Pathogen Candida albicans. PLoS One. 2015;10(9):e0137750. doi: 10.1371/journal.pone.0137750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Meara TR, Cowen LE. Hsp90-dependent regulatory circuitry controlling temperature-dependent fungal development and virulence. Cell Microbiol. 2014;16(4):473–81. doi: 10.1111/cmi.12266. [DOI] [PubMed] [Google Scholar]
- 14.Smith DA, et al. Stress signalling to fungal stress-activated protein kinase pathways. FEMS Microbiol Lett. 2010;306(1):1–8. doi: 10.1111/j.1574-6968.2010.01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dichtl K, et al. Cell wall integrity signalling in human pathogenic fungi. Cell Microbiol. 2016;18(9):1228–38. doi: 10.1111/cmi.12612. [DOI] [PubMed] [Google Scholar]
- 16.Barelle CJ, et al. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8(6):961–71. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown AJ, et al. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014;22(11):614–22. doi: 10.1016/j.tim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grahl N, et al. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell. 2012;11(5):560–70. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballou ER, et al. Lactate signalling regulates fungal beta-glucan masking and immune evasion. Nat Microbiol. 2016;2:16238. doi: 10.1038/nmicrobiol.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clavaud C, et al. The composition of the culture medium influences the beta-1,3-glucan metabolism of Aspergillus fumigatus and the antifungal activity of inhibitors of beta-1,3-glucan synthesis. Antimicrob Agents Chemother. 2012;56(6):3428–31. doi: 10.1128/AAC.05661-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ene IV, et al. Growth of Candida albicans cells on the physiologically relevant carbon source lactate affects their recognition and phagocytosis by immune cells. Infect Immun. 2013;81(1):238–48. doi: 10.1128/IAI.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopke A, et al. Neutrophil Attack Triggers Extracellular Trap-Dependent Candida Cell Wall Remodeling and Altered Immune Recognition. PLoS Pathog. 2016;12(5):e1005644. doi: 10.1371/journal.ppat.1005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shepardson KM, et al. Hypoxia enhances innate immune activation to Aspergillus fumigatus through cell wall modulation. Microbes Infect. 2013;15(4):259–69. doi: 10.1016/j.micinf.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherrington SL, et al. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017;13(5):e1006403. doi: 10.1371/journal.ppat.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amarsaikhan N, et al. Caspofungin Increases Fungal Chitin and Eosinophil and gammadelta T Cell-Dependent Pathology in Invasive Aspergillosis. J Immunol. 2017;199(2):624–632. doi: 10.4049/jimmunol.1700078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamaris GA, et al. Caspofungin-mediated beta-glucan unmasking and enhancement of human polymorphonuclear neutrophil activity against Aspergillus and non-Aspergillus hyphae. J Infect Dis. 2008;198(2):186–92. doi: 10.1086/589305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marakalala MJ, et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013;9(4):e1003315. doi: 10.1371/journal.ppat.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker LA, et al. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4(4):e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler RT, et al. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 2008;4(12):e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu W, et al. Activation and alliance of regulatory pathways in C. albicans during mammalian infection. PLoS Biol. 2015;13(2):e1002076. doi: 10.1371/journal.pbio.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barker BM, et al. Transcriptomic and proteomic analyses of the Aspergillus fumigatus hypoxia response using an oxygen-controlled fermenter. BMC Genomics. 2012;13:62. doi: 10.1186/1471-2164-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown NA, Goldman GH. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J Microbiol. 2016;54(3):243–53. doi: 10.1007/s12275-016-5510-4. [DOI] [PubMed] [Google Scholar]
- 33.Valiante V, et al. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol. 2015;6:325. doi: 10.3389/fmicb.2015.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donlin MJ, et al. Cross talk between the cell wall integrity and cyclic AMP/protein kinase A pathways in Cryptococcus neoformans. MBio. 2014;5(4) doi: 10.1128/mBio.01573-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Meara TR, et al. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 2014;34(4):673–84. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker JM, et al. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A. 2010;107(51):22044–9. doi: 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Bernardis F, et al. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66(7):3317–25. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez JC, Johnson AD. Regulatory circuits that enable proliferation of the fungus Candida albicans in a mammalian host. PLoS Pathog. 2013;9(12):e1003780. doi: 10.1371/journal.ppat.1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sem X, et al. beta-glucan Exposure on the Fungal Cell Wall Tightly Correlates with Competitive Fitness of Candida Species in the Mouse Gastrointestinal Tract. Front Cell Infect Microbiol. 2016;6:186. doi: 10.3389/fcimb.2016.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allert S, et al. In Vivo Transcriptional Profiling of Human Pathogenic Fungi during Infection: Reflecting the Real Life? PLoS Pathog. 2016;12(4):e1005471. doi: 10.1371/journal.ppat.1005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brothers KM, et al. NADPH oxidase-driven phagocyte recruitment controls Candida albicans filamentous growth and prevents mortality. PLoS Pathog. 2013;9(10):e1003634. doi: 10.1371/journal.ppat.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruno VM, et al. Transcriptomic analysis of vulvovaginal candidiasis identifies a role for the NLRP3 inflammasome. MBio. 2015;6(2):e00182–15. doi: 10.1128/mBio.00182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enjalbert B, et al. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun. 2007;75(5):2143–51. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jimenez-Lopez C, et al. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell. 2013;12(1):91–100. doi: 10.1128/EC.00290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netea MG, et al. Immune defence against Candida fungal infections. Nat Rev Immunol. 2015;15(10):630–42. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 46.Kashem SW, Kaplan DH. Skin Immunity to Candida albicans. Trends Immunol. 2016;37(7):440–50. doi: 10.1016/j.it.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erwig LP, Gow NA. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol. 2016;14(3):163–76. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- 48.Lionakis MS, et al. Immunity against fungi. JCI Insight. 2017;2(11):93156. doi: 10.1172/jci.insight.93156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, St Leger RJ. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc Natl Acad Sci U S A. 2006;103(17):6647–52. doi: 10.1073/pnas.0601951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shinya T, et al. Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr Opin Plant Biol. 2015;26:64–71. doi: 10.1016/j.pbi.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 51.Fujikawa T, et al. Surface alpha-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 2012;8(8):e1002882. doi: 10.1371/journal.ppat.1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujikawa T, et al. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol Microbiol. 2009;73(4):553–70. doi: 10.1111/j.1365-2958.2009.06786.x. [DOI] [PubMed] [Google Scholar]
- 53.Borges-Walmsley MI, et al. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 2002;10(2):80–7. doi: 10.1016/s0966-842x(01)02292-2. [DOI] [PubMed] [Google Scholar]
- 54.Rappleye CA, et al. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A. 2007;104(4):1366–70. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steele C, et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005;1(4):e42. doi: 10.1371/journal.ppat.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beauvais A, et al. Deletion of the alpha-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 2013;9(11):e1003716. doi: 10.1371/journal.ppat.1003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Carrion SJ, et al. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. J Immunol. 2013;191(5):2581–8. doi: 10.4049/jimmunol.1300748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gravelat FN, et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog. 2013;9(8):e1003575. doi: 10.1371/journal.ppat.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luther K, et al. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 beta-glucan receptor and Toll-like receptor 2. Cell Microbiol. 2007;9(2):368–81. doi: 10.1111/j.1462-5822.2006.00796.x. [DOI] [PubMed] [Google Scholar]
- 60.Dong B, et al. A chitin-like component on sclerotic cells of Fonsecaea pedrosoi inhibits Dectin-1-mediated murine Th17 development by masking beta-glucans. PLoS One. 2014;9(12):e114113. doi: 10.1371/journal.pone.0114113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alspaugh JA. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol. 2015;78:55–8. doi: 10.1016/j.fgb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bain JM, et al. Candida albicans hypha formation and mannan masking of beta-glucan inhibit macrophage phagosome maturation. MBio. 2014;5(6):e01874. doi: 10.1128/mBio.01874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis SE, et al. Masking of beta(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect Immun. 2014;82(10):4405–13. doi: 10.1128/IAI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galan-Diez M, et al. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun. 2010;78(4):1426–36. doi: 10.1128/IAI.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall RA, Gow NA. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol. 2013;90(6):1147–61. doi: 10.1111/mmi.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLellan CA, et al. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem Biol. 2012;7(9):1520–8. doi: 10.1021/cb300235m. [DOI] [PubMed] [Google Scholar]
- 67.Shen H, et al. Abolishing Cell Wall Glycosylphosphatidylinositol-Anchored Proteins in Candida albicans Enhances Recognition by Host Dectin-1. Infect Immun. 2015;83(7):2694–704. doi: 10.1128/IAI.00097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wheeler RT, Fink GR. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006;2(4):e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hasim S, et al. beta-(1,3)-Glucan Unmasking in Some Candida albicans Mutants Correlates with Increases in Cell Wall Surface Roughness and Decreases in Cell Wall Elasticity. Infect Immun. 2017;85(1) doi: 10.1128/IAI.00601-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin J, et al. Nanoscopic cell-wall architecture of an immunogenic ligand in Candida albicans during antifungal drug treatment. Mol Biol Cell. 2016;27(6):1002–14. doi: 10.1091/mbc.E15-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ene IV, et al. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012;14(9):1319–35. doi: 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen Q, Rappleye CA. Differentiation of the fungus Histoplasma capsulatum into a pathogen of phagocytes. Curr Opin Microbiol. 2017;40:1–7. doi: 10.1016/j.mib.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Linhares IM, et al. Contemporary perspectives on vaginal pH and lactobacilli. Am J Obstet Gynecol. 2011;204(2):120e1–5. doi: 10.1016/j.ajog.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 74.Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001;413(6851):36–7. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 75.Wagener J, et al. Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 2014;10(4):e1004050. doi: 10.1371/journal.ppat.1004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ost KS, et al. Rim Pathway-Mediated Alterations in the Fungal Cell Wall Influence Immune Recognition and Inflammation. MBio. 2017;8(1):e02290–16. doi: 10.1128/mBio.02290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ene IV, et al. Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance. MBio. 2015;6(4):e00986. doi: 10.1128/mBio.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maidan MM, et al. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol Biol Cell. 2005;16(4):1971–86. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karababa M, et al. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59(5):1429–51. doi: 10.1111/j.1365-2958.2005.05037.x. [DOI] [PubMed] [Google Scholar]
- 80.Preechasuth K, et al. Cell wall protection by the Candida albicans class I chitin synthases. Fungal Genet Biol. 2015;82:264–76. doi: 10.1016/j.fgb.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dubey LK, et al. Induction of innate immunity by Aspergillus fumigatus cell wall polysaccharides is enhanced by the composite presentation of chitin and beta-glucan. Immunobiology. 2014;219(3):179–88. doi: 10.1016/j.imbio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 82.Heilmann CJ, et al. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot Cell. 2013;12(2):254–64. doi: 10.1128/EC.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.del Fresno C, et al. Interferon-beta production via Dectin-1-Syk-IRF5 signaling in dendritic cells is crucial for immunity to C. albicans. Immunity. 2013;38(6):1176–86. doi: 10.1016/j.immuni.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 84.Drummond RA, Brown GD. The role of Dectin-1 in the host defence against fungal infections. Curr Opin Microbiol. 2011;14(4):392–9. doi: 10.1016/j.mib.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 85.Lilly LM, et al. The beta-glucan receptor dectin-1 promotes lung immunopathology during fungal allergy via IL-22. J Immunol. 2012;189(7):3653–60. doi: 10.4049/jimmunol.1201797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lionakis MS, et al. Chemokine receptor Ccr1 drives neutrophil-mediated kidney immunopathology and mortality in invasive candidiasis. PLoS Pathog. 2012;8(8):e1002865. doi: 10.1371/journal.ppat.1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Majer O, et al. Type I interferons promote fatal immunopathology by regulating inflammatory monocytes and neutrophils during Candida infections. PLoS Pathog. 2012;8(7):e1002811. doi: 10.1371/journal.ppat.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smeekens SP, et al. An anti-inflammatory property of Candida albicans beta-glucan: Induction of high levels of interleukin-1 receptor antagonist via a Dectin-1/CR3 independent mechanism. Cytokine. 2015;71(2):215–22. doi: 10.1016/j.cyto.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]