ABSTRACT
The organohalide-respiring bacterium Sulfurospirillum multivorans produces a unique cobamide, namely, norpseudo-B12, which serves as cofactor of the tetrachloroethene (PCE) reductive dehalogenase (PceA). As previously reported, a replacement of the adeninyl moiety, the lower base of the cofactor, by exogenously applied 5,6-dimethylbenzimidazole led to inactive PceA. To explore the general effect of benzimidazoles on the PCE metabolism, the susceptibility of the organism for guided biosynthesis of various singly substituted benzimidazolyl-norcobamides was investigated, and their use as cofactor by PceA was analyzed. Exogenously applied 5-methylbenzimidazole (5-MeBza), 5-hydroxybenzimidazole (5-OHBza), and 5-methoxybenzimidazole (5-OMeBza) were found to be efficiently incorporated as lower bases into norcobamides (NCbas). Structural analysis of the NCbas by nuclear magnetic resonance spectroscopy uncovered a regioselectivity in the utilization of these precursors for NCba biosynthesis. When 5-MeBza was added, a mixture of 5-MeBza-norcobamide and 6-MeBza-norcobamide was formed, and the PceA enzyme activity was affected. In the presence of 5-OHBza, almost exclusively 6-OHBza-norcobamide was produced, while in the presence of 5-OMeBza, predominantly 5-OMeBza-norcobamide was detected. Both NCbas were incorporated into PceA, and no negative effect on the PceA activity was observed. In crystal structures of PceA, both NCbas were bound in the base-off mode with the 6-OHBza and 5-OMeBza lower bases accommodated by the same solvent-exposed hydrophilic pocket that harbors the adenine as the lower base of authentic norpseudo-B12. In this study, a selective production of different norcobamide isomers containing singly substituted benzimidazoles as lower bases is shown, and unique structural insights into their utilization as cofactors by a cobamide-containing enzyme are provided.
IMPORTANCE Guided biosynthesis of norcobamides containing singly substituted benzimidazoles as lower bases by the organohalide-respiring epsilonproteobacterium Sulfurospirillum multivorans is reported. An unprecedented specificity in the formation of norcobamide isomers containing hydroxylated or methoxylated benzimidazoles was observed that implicated a strict regioselectivity of the norcobamide biosynthesis in the organism. In contrast to 5,6-dimethylbenzimidazolyl-norcobamide, the incorporation of singly substituted benzimidazolyl-norcobamides as a cofactor into the tetrachloroethene reductive dehalogenase was not impaired. The enzyme was found to be functional with different isomers and not limited to the use of adeninyl-norcobamide. Structural analysis of the enzyme equipped with either adeninyl- or benzimidazolyl-norcobamide cofactors visualized for the first time structurally different cobamides bound in base-off conformation to the cofactor-binding site of a cobamide-containing enzyme.
KEYWORDS: benzimidazoles, corrinoid-containing enzymes, reductive dehalogenase, vitamin B12
INTRODUCTION
Prokaryotes produce structurally different cobamides (Cbas; analogs of vitamin B12), but little is known about their selective or nonselective utilization as cofactors in cobamide-dependent enzymes, such as the reductive dehalogenases (RDases) (1–3). Cbas are composed of a contracted tetrapyrrole ring system harboring a cobalt ion as central atom (4). In addition to the four equatorial nitrogens of the pyrrole ring system, the cobalt is bound by two axial ligands (“upper” and “lower” ligands). Cbas vary in their structure by harboring different lower bases, either purines or benzimidazoles, which can be replaced by phenols (5). Purines or benzimidazoles can serve as a lower ligand to the cobalt, while phenols cannot. A 5′-deoxyadenosyl moiety, a methyl group, a hydroxyl group, or a water molecule has been described as a natural upper ligand of Cbas (1–3).
Cobamide-containing RDases function as terminal reductases in the membrane-bound electron transport chain of different anaerobic bacteria, which obtain energy via organohalide respiration (6–8). The need for Cbas as RDase cofactors in the so-called organohalide-respiring bacteria is covered either by de novo biosynthesis or by salvaging of Cbas from the surroundings (summarized in reference 9). The norpseudo-B12 of the tetrachloroethene (PCE)-respiring bacterium Sulfurospirillum multivorans is synthesized de novo, whereby norcobamide (NCba) biosynthesis is induced in the presence of PCE (10–12). In comparison to the structure of vitamin B12, the norpseudovitamin B12 lacks the methyl group at C-176 in the linker moiety of the Cba (the prefix “nor-” refers to this fact) and contains an adenine moiety as a lower base (13). A specific need for the production of NCba proteins by Sulfurospirillum spp. (13, 14) has not been verified. The results from a previous study indicated that the structure of the PCE reductive dehalogenase (PceA) might be flexible and allow the incorporation of both NCbas and Cbas (15). The presence of mainly Cba instead of NCba (i.e., presence of a methyl group at C-176 of the linker moiety) did not completely diminish PceA activity in cell extracts. However, no evidence is available for the binding and function of a standard-type Cba containing the methyl group at C-176 in the PceA enzyme.
S. multivorans does not possess genes for benzimidazole biosynthesis (11), either for the direct formation of 5,6-dimethylbenzimidazole (DMB) from flavin in an oxygen-dependent reaction (16) or for the anoxic formation of DMB via 5-OHBza, 5-OMeBza, and 5-methoxy-6-methylbenzimidazole from 5-aminoimidazole ribotide (17), an intermediate of purine biosynthesis. However, exogenous DMB, which serves as a lower base in vitamin B12, is utilized for cobamide biosynthesis by the organism. When DMB was added to cultures of S. multivorans, it efficiently replaced the adenine in the NCba (10). In general, various benzimidazoles are activated by the nicotinate-nucleotide:benzimidazole/purine phosphoribosyltransferase CobT, which adds a phosphoribosyl moiety and forms α-ribotides, prior to incorporation into Cbas (18). The relaxed specificity of some CobT proteins allowed for the formation of uncommon 6-substituted benzimidazolyl-ribotides from singly substituted benzimidazoles, such as 5-hydroxybenzimidazole (5-OHBza) or 5-methoxybenzimidazole (5-OMeBza), which can lead to the formation of unusual Cba isomers (19). Besides the effect of exogenous DMB on the norcobamide biosynthesis in S. multivorans, its presence also influenced the organism's ability to perform organohalide respiration (10). The PCE-dependent growth, the PceA activity, and the PceA maturation were impaired when elevated concentrations of DMB (10 or 25 μM) were added to S. multivorans cultures. Such negative effects on enzymatic reductive dehalogenation caused by modification of the Cba structure have also been reported for Dehalococcoides mccartyi strains (20–23). The obligate organohalide-respiring D. mccartyi is a Cba auxotroph (24). The dechlorination of trichloroethene (TCE) by D. mccartyi strain 195 was dependent on the availability of Cbas with either DMB, 5-methylbenzimidazole (5-MeBza), or 5-OMeBza as a lower base. Cbas with alternative lower bases, such as 5-OHBza or benzimidazole (Bza), did not sustain TCE-dependent growth of the organism. A plausible explanation for the inhibitory effect might be the incompatibility of a specific lower base in the Cba cofactor with the Cba-binding site of a particular RDase.
The lower ligand base, which is part of the Cba nucleotide loop and thereby covalently bound to the cofactor, can be displaced from the cobalt (base-off) in the protein-bound state. Here, an alternative lower ligand, such as a histidine residue (base-off/His-on), often fulfills this function (25, 26). A lower ligand of the cobalt was not detected by structural analysis of Cba cofactors bound to RDases (27, 28). The lower base of the Cba is displaced. In the case of the two known RDase structures, coordinative binding of the lower base is prevented by steric hindrance. These observations were supported by spectroscopic analyses of different RDases using electron paramagnetic resonance spectroscopy, which also detected the Cba cofactor in the base-off state (28–30). The nucleotide loop of Cbas is varied in structure, and this diversity might interfere with the incorporation and correct positioning of the cofactor in Cba-dependent enzymes. This assumption is expected to be especially relevant for RDases, since these enzymes bind the cofactor with a network of hydrogen bonds deep inside the protein (27, 28). In order to shed light on the functionality of a single RDase with different Cba cofactors, we investigated the utilization of various benzimidazolyl-NCbas by PceA of S. multivorans and analyzed the similarities or variations in binding an NCba harboring either a purine or a benzimidazole as lower base on a structural level. Thereby, we identified PceA of S. multivorans as an NCba-containing protein functioning with both purinyl- and benzimidazolyl-NCbas but with limited ability to incorporate NCbas containing methylated benzimidazoles as a lower base. Furthermore, we found evidence for an unusually strict regioselectivity of the S. multivorans NCba biosynthetic machinery that is controlled by the type of substituent present in singly substituted benzimidazoles.
RESULTS AND DISCUSSION
Guided biosynthesis of benzimidazolyl-NCbas.
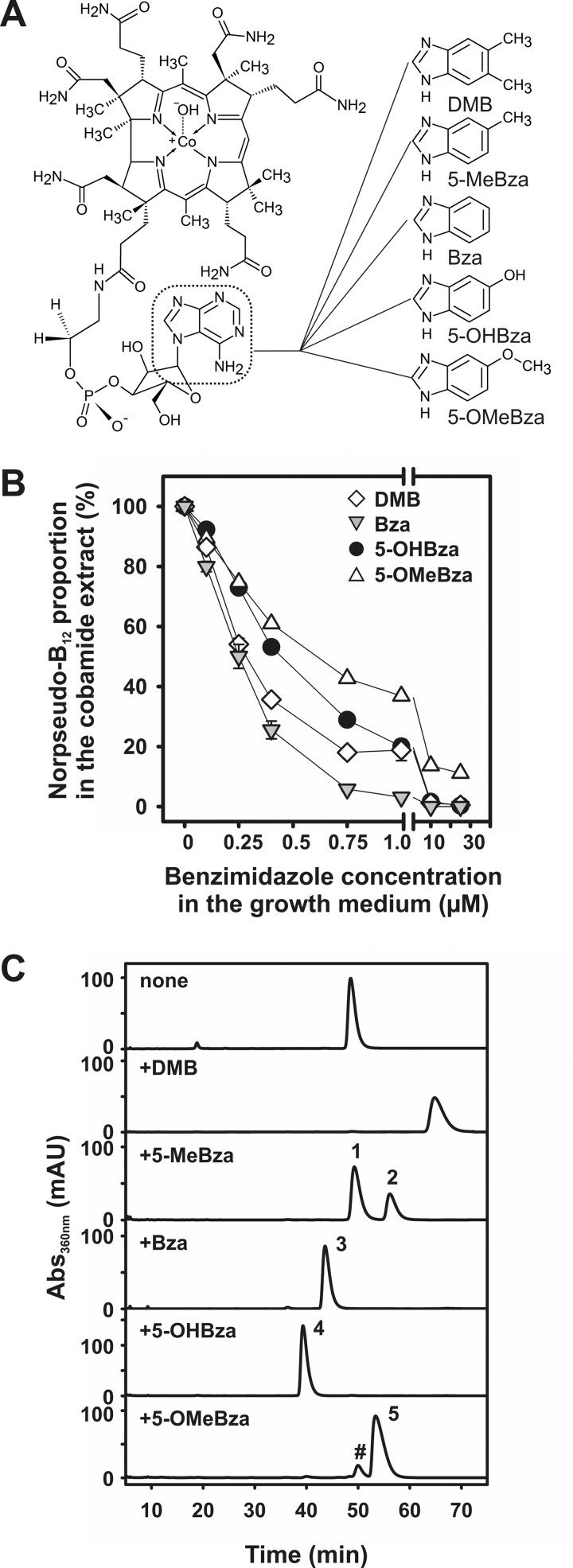
In order to test the utilization of various exogenous benzimidazoles as lower bases for the production of NCbas (Fig. 1A), S. multivorans was cultivated in the presence of DMB, 5-MeBza, Bza, 5-OHBza, or 5-OMeBza, and the NCbas were extracted. The incorporation efficiencies of all benzimidazoles were tested by analyzing the NCba fractions purified from cells cultivated in the presence of different concentrations of the lower-base precursors (Fig. 1B). The total amount of NCba extracted from cells cultivated in the presence of benzimidazoles was not altered in comparison to untreated cells. The absolute yield was about 1 μmol NCba per gram of cell protein used for the extraction in every case.
FIG 1.
Guided NCba biosynthesis in S. multivorans. (A) Norpseudo-B12 cofactor. A frame marks the adeninyl moiety in the nucleotide loop. The structures of the exogenous benzimidazoles, which were tested in the study, are depicted. (B) Ratio between norpseudo-B12 and various benzimidazolyl-NCbas formed in the presence of different concentrations of exogenous benzimidazoles. 5-MeBza was excluded from the analysis of the incorporation efficiency, since the norpseudo-B12 was still present in cells exposed to lower concentrations of exogenous 5-MeBza. The adeninyl-NCba has not been efficiently separated from the MeBza-NCba (peak 1 in panel C) by HPLC. This circumstance made a direct quantification of the portion of MeBza-NCba impossible. The data presented in panel B were obtained from two independent experiments. The maximal deviation is given by error bars. (C) HPLC analysis and mass determination of NCbas extracted from S. multivorans cells cultivated in the presence of different benzimidazoles (25 μM each). The pound sign marks the norpseudovitamin B12 in the NCba extract from cells treated with 5-OMeBza. Singly and doubly protonated ions were detected, which were assigned to the respective NCbas using UHPLC-ESI-HRMS (for details, see Fig. S1 and Table S1). mAU, milliabsorbance units.
The extracted NCbas were applied to high-performance liquid chromatography (HPLC) coupled to photometric detection in order to verify their homogeneity (Fig. 1C). The addition of DMB to the growth medium resulted in the formation of nor-B12, as has been reported previously (10). In order to identify the unknown NCbas, all samples were analyzed using ultrahigh-performance liquid chromatography coupled with electrospray ionization–high-resolution mass spectrometry (UHPLC-ESI-HRMS). By applying this technique, the efficient incorporation of all benzimidazoles as lower bases into the nucleotide loop of the NCbas produced by S. multivorans was confirmed, since the results are in agreement with the predicted masses (see Fig. S1 and Table S1 in the supplemental material). All singly substituted benzimidazoles gave rise to the production of new compounds. After three subsequent cultivations in the presence of 25 μM exogenously applied benzimidazoles, the amount of norpseudo-B12 in the cells was reduced to a nondetectable level (Fig. 1C). Exclusively in the case of 5-OMeBza, a residual amount (about 10%) of norpseudo-B12 was observed. In the case of the NCba extract obtained from cells cultivated in the presence of 5-MeBza, two NCba compounds were detected in a ratio of 2:1. The two NCba compounds showed an identical mass, which was attributed to MeBza-NCba. The difference in the retention times of the two isomers might be caused by a difference in the position of the methyl group of the MeBza moiety in the NCba structure.
Doubly substituted benzimidazoles different from DMB were also tested. While in cells cultivated in the presence of 25 μM 5,6-dinitrobenzimidazole (DNO2Bza) the norpseudo-B12 cofactor was nearly completely replaced, in cells cultivated in the presence of 25 μM 5,6-dimethoxybenzimidazole (DOMeBza), almost no production of the DOMeBza-NCba was detected. Most probably due to chemical transformations of the two NO2 groups, several NCbas were extracted from cells exposed to DNO2Bza. Mass determination revealed that DNO2Bza-NCba was not included, which hindered the evaluation of the effect of benzimidazoles harboring a substituent in positions 5 and 6.
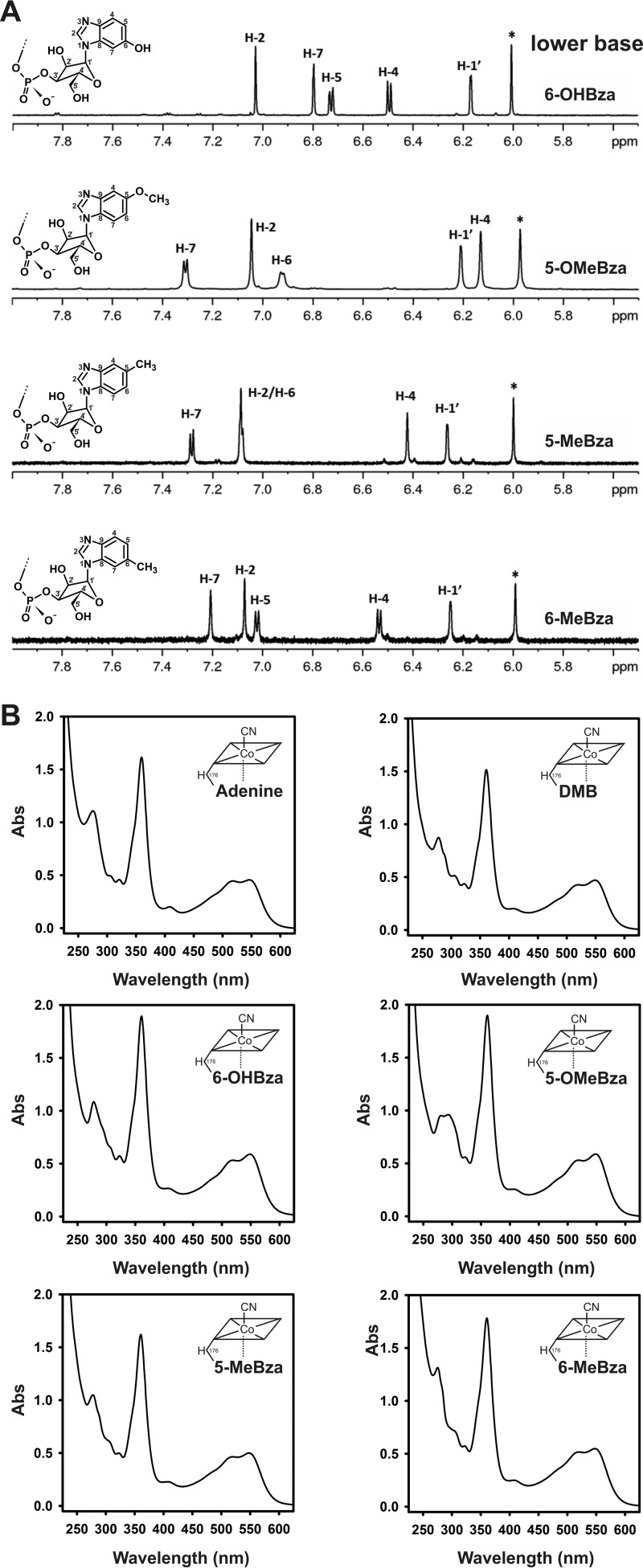
The purified NCbas formed in cells cultivated in the presence of the singly substituted benzimidazoles 5-MeBza, 5-OHBza, and 5-OMeBza were further analyzed by nuclear magnetic resonance (NMR) spectroscopy in order to identify the exact positioning of the substituents at the benzimidazoles in the NCbas. By applying NMR techniques, the incorporation of different benzimidazoles into the NCba structure was confirmed (Fig. 2A and S2 to S27). In theory, singly 5-substituted benzimidazoles can be incorporated in two different orientations into the cobamide structure, which gives rise to 5- or 6-substituted benzimidazolyl-Cbas. The determined chemical shifts in the NMR analysis obtained for the sample prepared from 5-OHBza-treated cells allowed for the identification of 6-OHBza-NCba, whereas in the case of the sample from 5-OMeBza-treated cells, the formation of 5-OMeBza-NCba was proven (Fig. 2A). The NMR data were in accordance with previously published results by Crofts and coworkers (19). Furthermore, the position of the substituent within the lower-base structure was determined in both NCbas obtained from 5-MeBza-treated cells, and the occurrence of two orientations of MeBza in the nucleotide loop was assigned (Fig. 2A). The structural differences of the four NCbas were reflected by characteristic absorbance spectra (Fig. 2B).
FIG 2.
Characterization of the isolated NCbas. (A) Low field range of 1H-NMR spectra of 6-OHBza-NCba, 5-OMeBza-NCba, 5-MeBza-NCba (peak 1 in Fig. 1C), and 6-MeBza-NCba (peak 2 in Fig. 1C). The depicted sections show the signals for the respective benzimidazolyl moieties and the signal for the anomeric position of the α-ribosyl unit. The signal marked with an asterisk belongs to position 10 of the corrin scaffold. (B) UV/Vis-absorbance spectra of the newly purified NCbas in comparison to those of the authentic adeninyl-NCba (norpseudovitamin B12) and the DMB-NCba (norvitamin B12).
The specificity in the formation of 6-OHBza-NCba from 5-OHBza and of 5-OMeBza-NCba from 5-OMeBza was an unexpected finding and adds a new facet to the unusual nucleotide loop assembly pathway in S. multivorans (31). Earlier, Friedrich and Bernhauer (32) reported the extraction and identification of 5-MeBza-Cba and 6-MeBza-Cba produced by Propionibacterium shermanii in the presence of 5-MeBza, albeit in a ratio of 96:4, which showed a clear preference for the formation of 5-MeBza-Cba in this organism. In Methanobacterium thermoautotrophicum, which has been shown to produce OHBza-Cba, approximately 10% of 6-OHBza-Cba was detected (33). A mixture of Cba isomers was found in Sinorhizobium meliloti and Veillonella parvula, both cultivated in the presence of 5-OMeBza or 5-OHBza (19, 34). However, the exclusive biosynthesis of benzimidazolyl-Cbas with a single substituent in position 6 of the benzimidazolyl-moiety, as shown here for S. multivorans, has not been described so far. Benzimidazoles are channeled into the Cba biosynthetic pathway by the activity of the phosphoribosyltransferase CobT. It is under investigation in our laboratory whether the regioselectivity of CobT from S. multivorans lays the foundation for the observed structural diversity in the NCbas containing OMeBza or OHBza in the organism.
Effect of singly substituted benzimidazoles on PceA production.
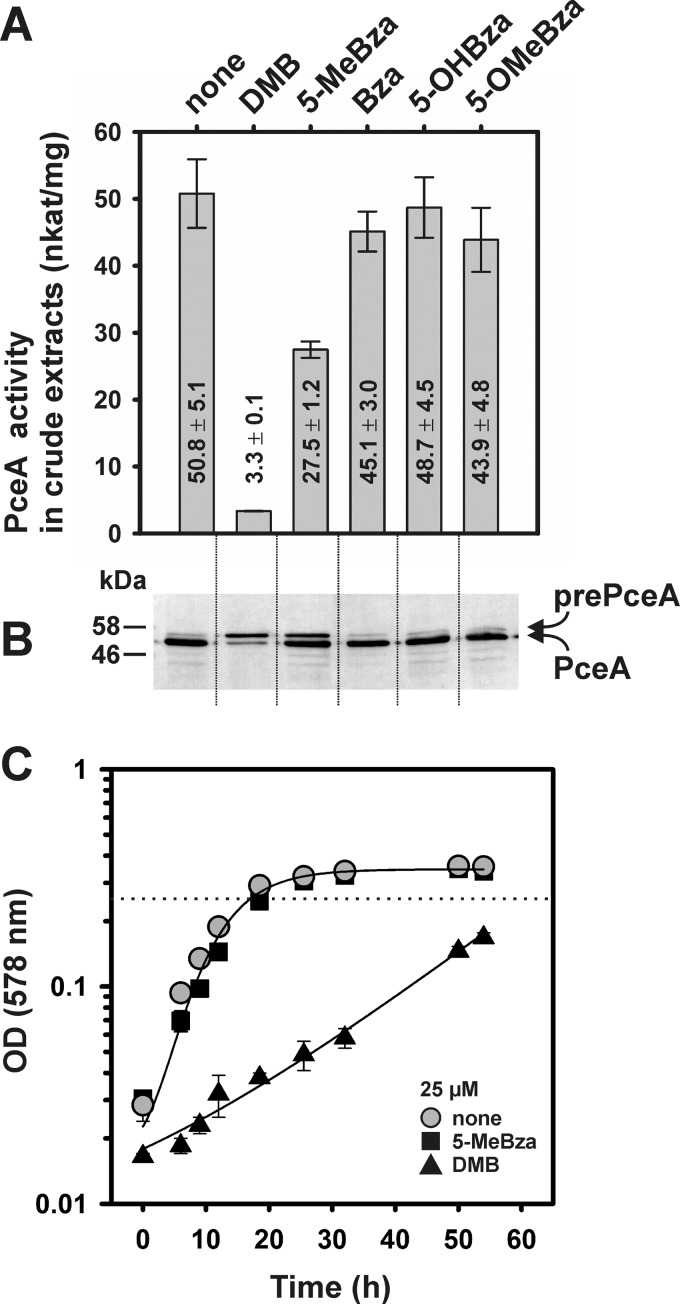
The exchange of the adeninyl moiety in the norpseudo-B12 cofactor by DMB caused a negative effect on the PCE-dependent growth and the biosynthesis of catalytically active PceA in S. multivorans (10). To date, no other benzimidazoles had been tested in functional assays. Thus, 5-MeBza, Bza, 5-OHBza, or 5-OMeBza (25 μM each) was added to cultures of S. multivorans that contained pyruvate and PCE as substrates. None of the singly substituted benzimidazoles tested in this study affected the PCE-dependent growth. In order to evaluate the impact on PceA function, the enzyme activity and protein level were tested in crude extracts of the cells supplemented with various benzimidazoles (Fig. 3A and B). Cultures of S. multivorans grown on PCE in the presence of the different benzimidazoles were harvested in the late exponential-growth phase, i.e., the protein concentration was 70 to 80 μg/ml, which was reached after 16 to 18 h of cultivation (Fig. 3C). In the case of the DMB-containing cultures, the same growth phase was reached after approximately 3 days of cultivation. Crude extracts were prepared, and the specific PceA activity was measured in a photometric assay using reduced methyl viologen as an artificial electron donor. The activity determined in cells cultivated in the presence of either Bza, 5-OHBza, or 5-OMeBza was similar to the PceA activity monitored in the nontreated S. multivorans cells (Fig. 3A). In contrast, the presence of 5-MeBza caused a substantial reduction in the enzyme activity (about 50%). This reduction was not as high as in DMB-treated cells (about 90%) and appeared to be sufficient for unaffected growth with PCE (Fig. 3C). It was observed before that the growth rate of S. multivorans with PCE was not severely reduced when the PceA activity in the cells was lowered by about 50% (15). Traces of the authentic norpseudo-B12 were not detected in cells cultivated in the presence of 25 μM 5-MeBza. Norpseudo-B12 was neither identified in an UHPLC-ESI-HRMS analysis of the extracted NCba fraction from such cells nor in the NMR analysis of the purified 5-MeBza-NCba. 5-MeBza-NCba showed a retention time similar to that of the adeninyl-NCba during separation (Fig. 1C).
FIG 3.
(A and B) PceA activity (A) and protein level (B) in crude extracts derived from cells grown with PCE in the presence of different benzimidazoles (25 μM). For the immunological analysis, 5 μg of protein was applied to each lane. The immunoblot was developed with an antiserum containing polyclonal antibodies against PceA. The data for the PceA activity were obtained from three different cultures. The standard deviation is given. prePceA, precursor with the N-terminal Tat signal peptide; PceA, processed form without the Tat signal peptide. (C) PCE-dependent growth of S. multivorans in the presence of 5-MeBza or DMB. Cells were harvested for functional analyses in the growth phase indicated by the dashed line.
The reduction in PceA activity in cells cultivated in the presence of 5-MeBza might be accompanied by a reduction in maturation and processing of the cytoplasmic PceA precursor (prePceA), as was previously observed for DMB (10). Thus, the presence of both forms of the enzyme was analyzed in crude extracts separated on an SDS-PAGE gel and applied to an immunoblot developed with PceA-specific antibodies (Fig. 3B). In comparison to the crude extract of nontreated cells, which displayed an intense band for the mature PceA enzyme (predicted molecular mass, 53.3 kDa) and only a thin band for the twin-arginine translocation (Tat) signal peptide-containing prePceA (predicted molecular mass, 57.1 kDa), the amount of prePceA was increased in 5-MeBza-treated cells. The total amount of the enzyme and its processing were apparently not affected in cells cultivated in the presence of Bza, 5-OHBza, and 5-OMeBza.
The quantification of the incorporation efficiency of the DMB-NCba into PceA was hindered in a previous study, since a sufficient amount of pure PceA enzyme was not obtained from such DMB-treated cells (10). This difficulty and the negative effect on the maturation of the PceA precursor under this condition indicated an improper incorporation of the DMB-NCba, containing the doubly substituted lower base, into the enzyme. A negative effect of DMB on pceA transcription in PCE-grown cells was not detected (Fig. S28). In contrast to the effects observed with DMB, the analysis of the PceA activity in cells treated with 5-OHBza or 5-OMeBza showed no decrease (Fig. 3). This result pointed toward an efficient utilization of both NCbas, 6-OHBza-NCba and 5-OMeBza-NCba, as cofactors of the PceA enzyme. Indeed, extraction of the NCbas from PceA isolated from the respective type of cells uncovered 100% saturation of the enzyme (1 mol cofactor per 1 mol PceA) in both cases, as well as in enzyme extracted from 5-MeBza-treated cells (>98%), and the elution profiles of the extracted cofactors obtained via HPLC separation (Fig. S29) mirrored the patterns observed earlier for the NCbas extracted from whole cells (Fig. 1C). For unraveling similarities or differences in the binding mode of the different NCba cofactors and for obtaining indications for the exclusion of the DMB-NCba, the different PceA enzymes were crystallized and subjected to structural analysis. For crystallization, PceA was purified from the membrane fraction that contained only the processed form of the enzyme without the Tat signal peptide. Crystals of PceA suitable for X-ray scattering and structural analysis were exclusively obtained for the enzyme purified from cells treated with 5-OHBza or 5-OMeBza. Two crystallographic data sets in high resolution (dmin = 1.6 Å) were recorded (Table S2). The inability of the PceA sample equipped with the mixture of the two MeBza-NCba isomers to form suitable crystals might indicate the presence of misfolded PceA. This assumption was supported by the fact that the specific activity of PceA purified from cells cultivated in the presence of 5-MeBza was about 50% lower than that of the PceA purified from cells treated with 5-OHBza or 5-OMeBza.
Structural details on NCba cofactor binding.
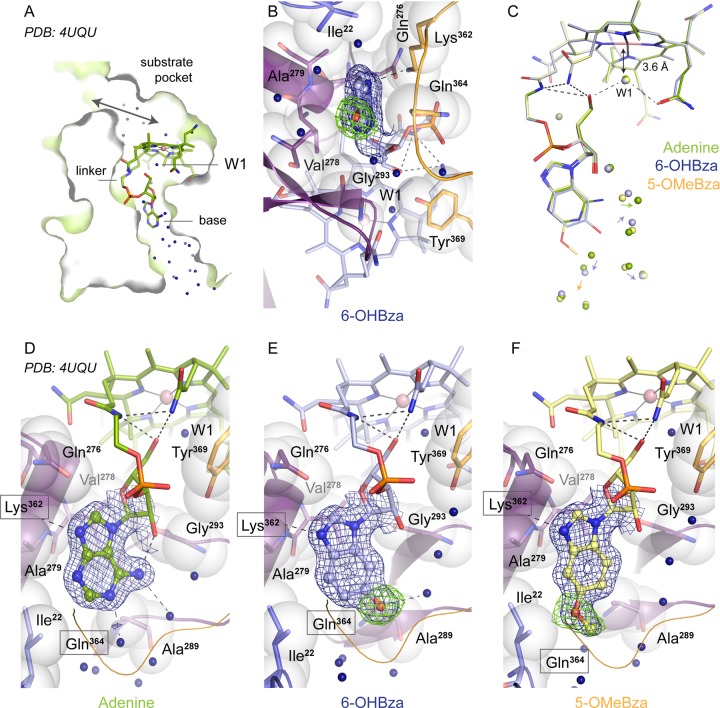
In both known RDase structures (27, 28), the Cba cofactor is deeply buried within the protein. Access to the upper (i.e., the β-face) of the corrin ring within the substrate-binding pocket of PceA is controlled by the protein structure. The nucleotide loop of the adeninyl-NCba (norpseudo-B12) in PceA is positioned in the base-off conformation (Fig. 4A). The unusual curled conformation of the tail is stabilized by intramolecular interactions in the form of hydrogen bonds between O-5 of the ribosyl moiety (for numbering of the atoms in norpseudo-B12, see reference 13), the linker amide, and the carboxamide side chain connected to ring C of the corrin core (27) (Fig. 4B to F). These stabilizing interactions are conserved in both the PceA equipped either with 6-OHBza-NCba or with 5-OMeBza-NCba. Another intramolecular hydrogen bond connects the amino group of the adenine and O-2 of the ribosyl moiety only in the adeninyl-NCba (Fig. 4D). The base is placed between the NCba-binding core of PceA (purple in Fig. 4) and a loop from the region binding the iron-sulfur clusters (27) (Fig. 4B). Lys362 from this loop is within hydrogen-bonding distance of both the base and the phosphate. Gln364 reaches across the base, and hydrogen bonds to O-2 of the sugar. Thus, intramolecular and protein interactions effectively lock the position of the base, and this is reflected by the conserved position observed for both benzimidazole-containing NCbas (Fig. 4C to F). In PceA but not in the structure of the RDase NpRdhA from Nitratireductor pacificus (28), a solvent channel reaches from the protein surface to the far end of the base. The amino group and N-1 of the adeninyl base are in hydrogen-bonding distance to water molecules (Fig. 4D). The three different substituents on the various bases, namely, the amino group of the adeninyl moiety (Fig. 4D), the hydroxyl group of the 6-OHBza moiety (Fig. 4E), and the methoxy group of the 5-OMeBza moiety (Fig. 4F), are accommodated by a change in the water structure rather than a change in the position of the base or a reorientation of surrounding side chains (Fig. 4C to F). The closest interactions between the new substituents were seen between Ala289 and the 6-OHBza moiety (3 Å) and between Ile22 of the N-terminal loop region and the 5-OMeBza moiety (3.5 Å). A substitution at a position other than C-5 or C-6 may not be possible without a change in the protein scaffold. Given the positioning of the 6-OHBza and 5-OMeBza moieties in PceA, no interference with the protein would be expected with a hypothetical MeBza or DMB as a lower base. Hence, the static folded structure cannot fully explain the apparent lack of functionality of the MeBza-NCba and the DMB-NCba. The unique feature of PceA is the solvent-accessible and water-bound base. Such solvation might play a role in cofactor acquisition and correct functional folding of both the PceA enzyme and cofactor.
FIG 4.
NCba cofactor binding in PceA. (A) Slice through a monomer of PceA showing the cofactor positioning. The upper side (β-face) of the corrin ring is accessible to solvent via the active-site pocket and the substrate channel. The lower side (α-face) of the corrin ring is isolated by the loop at Tyr 369 and Gly293 and faces the “curl” in the linker of the NCba cofactor (see panel C). The curl is stabilized by hydrogen bonds. The cavity formed by these structural elements contains a single water molecule (W1). The lower base of the NCba cofactor is displaced from the corrin ring and accessible to solvent. (B) View from below onto the 6-OHBza base. Residues from the B12-binding core of PceA (residues 139 to 163 and 216 to 323, purple) and a loop from the iron-sulfur cluster-binding unit (residues 324 to 394, orange) sandwich the base. The view is rotated compared to all other panels. (C) Overlay of the three PceA bound cofactors shown in panels D to F. The single water molecule near the lower face is within hydrogen-bonding distance to the ribose and a carboxamide side chain of the cofactor but not a ligand to Co (3.6-Å distance). Colored arrows indicate the displacement of water molecules by the different base substituents. (D to F) Structures of PceA equipped with NCbas containing either adenine (27), 6-OHBza, or 5-OMeBza as lower bases (ball and stick models). Lys362 and Gln364 side chains from the cluster-binding unit are in front of the view, where their position is indicated by boxes. Lys362 is within hydrogen-bonding distance of the imidazole moiety in all three bases (dashed lines). Structural changes in the base are accommodated by changes in the positioning of the surrounding water molecules. Electron density maps are shown for the base only (blue 1 σ 2Fo-Fc map and green 2.5 σ Fo-Fc difference maps calculated in the absence of the hydroxyl (E) or methoxy (F) group).
Since 5-OHBza and 5-OMeBza incorporated as lower bases in 6-OHBza-NCba or 5-OMeBza-NCba were functional in the enzyme, it is hard to conclude which isomer of MeBza-NCba causes PceA malfunction. In the case of the MeBza-NCbas, the incorporation did not appear to be hindered, since both isomers, the 5-MeBza- and the 6-MeBza-containing variant, were detected in the enzyme (Fig. S29), and the cofactor incorporation efficiency was nearly 100%. However, the enzyme activity of the pool of PceA molecules was reduced, and the difficulties observed in crystallization of purified PceA indicated the heterogeneity of the sample. Crystallization of 5-MeBza and 6-MeBza NCba-containing PceA resulted in a different crystal packing in which the base could not be fully resolved; thus, its detailed structure remains unknown. In the case of DMB, the respective NCba was most probably even excluded from the incorporation into PceA (see also reference 10). However, also the incompatibility of DMB as a lower base might not be due to steric hindrance inside the binding pocket. No structural incompatibilities between the substituents of the benzimidazole moiety and the protein environment were identified. Hence, it has to be assumed that the lower-base substitution pattern might interfere with the folding process rather than with the dimensions of the binding pocket present in the fully folded enzyme.
Whether the cofactor has to attain a specific conformation prior to the incorporation into the enzyme and whether this might be differentially affected in the various benzimidazolyl-NCbas tested here remain elusive. Here, the hydrophobic methyl groups in DMB-NCba and both MeBza-NCbas could interfere with the incorporation process. The lower base of the NCba cofactor in PceA is placed in a solvent-flooded cavity, which might be incompatible with the hydrophobic character of the methylated benzimidazoles. The DMB of the Cba cofactor in NpRdhA is protected from solvent (28), which could result in a different preference for alternative Cba cofactors.
Not all RDases seem to bind the Cba cofactor in the base-off mode. As reported for the reconstituted VcrA of D. mccartyi strain VS, the DMB-containing Cba cofactor showed that the lower ligand bound to the cobalt also in the enzyme-bound state (35). In addition, other RDases show different preferences in the type of lower base in the Cba cofactor from that of PceA of S. multivorans (20–23). It is conceivable that the evolution of RDases, especially those present in cobamide-auxotrophic organohalide-respiring bacteria, was influenced by the cobamides present in the surrounding environment. However, a general rule for the cofactor compatibility of RDases cannot be derived from the data available at the moment, and whether other proteins are involved in RDase cofactor selectivity is not known. The role of chaperones in the Cba cofactor incorporation has not been investigated so far. Specific chaperones, such as RdhT homologues (36, 37), are not found in S. multivorans and have not been identified in D. mccartyi. Padovani and coworkers reported the transfer of the Cba cofactor to the methylmalonyl coenzyme A (methylmalonyl-CoA) mutase to be mediated by the adenosyltransferase (ATR) (38). An ATR homologue is not present in S. multivorans. Since the NCba cofactor of PceA is catalytically active without the 5′-deoxyadenosyl-moiety, this gene function is dispensable in the organism.
It was observed earlier that the structure of the lower base has an effect on the ratio between the base-off and the base-on forms of Cbas in solution. This might influence the incorporation efficiency, especially when the Cba cofactor has to obtain its base-off conformation prior to the assembly with the RDase apoprotein. Fieber and coworkers showed the adenine-containing pseudocoenzyme B12 having a higher tendency to occur in base-off form in solution than that of the DMB-containing coenzyme B12 (39). Even between structural isomers, the tendency to obtain the base-off conformation differs. A lower pKa value for 5-OMeBza-Cba than that for 6-OMeBza-Cba was reported (19). However, a systematic analysis of the impact of all potential lower bases on the constitution of the cofactor under physiological conditions is not available. A cooperative process including PceA folding and structural adaptation of the NCba cofactor that lead to the final constitution of the enzyme-cofactor complex cannot be excluded. Reconstitution experiments with the apoprotein of PceA, which would help to analyze the compatibility of the unfolded protein with different NCba/Cba cofactors, were not successful or have not been tried with NCbas containing lower bases different from DMB. These efforts are going to be intensified in future studies.
MATERIALS AND METHODS
Cultivation of bacteria.
Sulfurospirillum multivorans DSMZ 12446 was cultivated as described previously by Keller et al. (10). Anoxic and sterile stock solutions of the different benzimidazoles (up to 4 mM) were prepared in ultrapure water and used for amendment of the media prior to the inoculation. All benzimidazole derivatives were purchased from Sigma-Aldrich (Munich, Germany), except for 5-OHBza, which was delivered by Combi-Blocks, Inc. (San Diego, CA), and DNO2Bza, which was purchased from Santa Cruz Biotechnology (Dallas, TX). Cells of S. multivorans were adapted to the different cultivation conditions in two subsequent precultures prior to the cultivation of cells for growth monitoring, PceA activity measurements, or cobamide extraction. The inoculum was 10%. The cells were transferred when the culture without the amendment of benzimidazoles reached the maximal optical density (OD). In every case, the first preculture was inoculated from a pyruvate-PCE-grown culture of S. multivorans routinely prepared for strain maintenance.
Measurement of PceA activity, immunoblot, cobamide extraction, and HPLC analysis.
The production of cell extracts and the determination of the PceA enzyme activity in crude extracts from S. multivorans cells were performed as described previously (10). This also applies to the immunological detection of PceA in cell extracts. The norcobamide (NCba) extraction and analysis were conducted in accordance to a protocol published earlier (10). The cells were cultivated in pyruvate/fumarate-containing medium. The mobile phases used for separation of the NCbas via high-performance liquid chromatography (HPLC) were 12% methanol–0.2% acetic acid (solvent A) and 99.8% methanol–0.2% acetic acid (solvent B). The flow rate was 0.5 ml/min, and the separation was performed isocratically at 30°C. The NCbas were purified finally via solid-phase extraction on a Chromabond HR-X column (3 ml, 200 mg; Macherey-Nagel, Düren, Germany), according to the manufacturer's instructions. The UV-Vis absorbance spectra of the purified NCbas were recorded with a Cary 100 UV-visible spectrophotometer (Agilent Technologies, Waldbronn, Germany).
UHPLC-ESI-HRMS.
For UHPLC, an Ultimate 3000 series rapid-separation liquid chromatography (RSLC; Dionex, Sunnyvale, CA) was used, applying an Acclaim C18 column (150 by 2.1 mm, 2.2-μm particles with 120-Å pore diameter; Dionex), with a flow rate of 300 μl/min in a binary solvent system of water (solvent A) and acetonitrile (hypergrade for LC-MS; Merck, Darmstadt, Germany) (solvent B), both containing 0.1% (vol/vol) formic acid (eluent additive for LC-MS; Sigma-Aldrich, Munich, Germany). Sample volumes were loaded onto the column and eluted by using a gradient as follows: linear increase from 0% B to 100% B within 15 min, to 100% B constant for 5 min, and to equilibration time at 0% B for 5 min. This system was coupled to a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Ionization was accomplished using electrospray ionization (ESI). ESI source parameters were set to 4 kV for spray voltage and 35 V for transfer capillary voltage at a capillary temperature of 275°C. The samples were measured in positive-ion mode in the mass range of m/z 100 to 2,000 using 30,000 m/Δm mass resolving power in the Orbitrap mass analyzer. The data were evaluated and interpreted using Xcalibur 3.0.63 (Thermo Fisher Scientific, Waltham, MA).
NMR measurements.
NMR spectra (1H NMR, 1H-1H COSY, 1H-1H ROESY, 1H-13C HSQC, and 1H-13C HMBC) were recorded on a Bruker Avance III HD 700 spectrometer, equipped with a CryoPlatform and a 1.7-mm TCI MicroCryoProbe (Bruker BioSpin GmbH, Rheinstetten, Germany). NMR tubes with 1.7-mm outer diameter were used for all measurements. All NMR spectra were recorded using D2O as a solvent. Accurate tuning of the spectrometer frequencies (700.45 MHz for 1H and 176.14 MHz for 13C) was accomplished prior to the experiments. Chemical shifts were left uncorrected. Data acquisition and processing were accomplished using TopSpin version 3.2 (Bruker BioSpin). Standard pulse programs as implemented in TopSpin were used for data acquisition.
Crystallization of PceA and structural analysis.
For the purification of PceA, the S. multivorans mutant strain GD21 was cultivated, as reported before (27, 40), in the presence of different benzimidazoles (see above). PceA was purified as described in previous reports (27, 40). PceA was crystallized and crystals flash-cooled under anoxic conditions in a glove box (model B; COY Laboratory Products, Grass Lake, MI) containing an atmosphere of 95% N2–5% H2 and less than 10 ppm oxygen. Crystals were grown by the sitting drop vapor diffusion method at room temperature. One microliter of 5 to 15 mg/ml PceA in 30 mM Tris-HCl (pH 7.5) and 5 mM Tris(2-carboxyethyl)phosphine (TCEP) was mixed with 1 μl of crystallization solution containing 12 to 17% (wt/vol) polyethylene glycol 3350 (PEG 3350) and 0.2 M sodium malonate, 2% benzamidine-HCl, and 50 mM Tris-HCl (pH 7.5). Crystals were flash-cooled in liquid nitrogen after protection in the crystallization solution supplemented with 20% (vol/vol) glycerol and 25% (final wt/vol) PEG 3350. Once plunged into liquid N2, crystals were removed from the anoxic atmosphere and thereafter stored and handled under liquid N2.
Diffraction data for PceA equipped with 5-OMeBza-NCba were collected on BL14.1 operated by the Helmholtz-Zentrum Berlin (HZB, Germany) at the BESSY II electron storage ring (Adlershof, Berlin, Germany) (41). Data for PceA equipped with 6-OHBza-NCba were collected at beamline P11 at PETRA-III (DESY, Hamburg, Germany) (42, 43). The data were indexed and integrated with the XDS package (44) and XDSAPP (45). Restraints for the base were generated by the Grade Server version 1.001 and used to modify the vitamin B12 restraint file posted by Oliver Smart (Global Phasing Ltd., Cambridge, UK). Models were fitted in COOT (46), refined with phenix.refine (47), and validated with MolProbity (48). Data collection and refinement statistics are summarized in the Table S2.
Accession number(s).
Model coordinates and structure factors for PceA equipped with different NCba proteins have been deposited in the Protein Data Bank (PDB) under accession numbers 5OBP (PceA with 6-OHBza-NCba cofactor) and 5OBI (PceA with 5-OMeBza-NCba cofactor).
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the German Research Foundation (DFG; grant SCHU 2605/1-1). The efforts of Cindy Kunze were funded by the DFG Research Unit FOR1530 and the Ernst Abbe Foundation. The efforts of Martin Bommer were financed by the SFB1078 (Protonation Dynamics in Protein Function).
We acknowledge access to beamlines of the BESSY II storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz-Zentrum Berlin für Materialien und Energie (HZB), the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück-Centrum, and the Leibniz-Institut für Molekulare Pharmakologie. Parts of this research were carried out at PETRA-III at DESY, a member of the Helmholtz Association (HGF). We thank Olga Lorbeer and Anja Burkhardt for assistance in using beamline P11. We are grateful to Gabriele Diekert for helpful discussions. We acknowledge the excellent technical assistance of Peggy Brand-Schön, and we are grateful to Inken Schulze-Hessing for performing the transcript-level analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00584-17.
REFERENCES
- 1.Banerjee R, Ragsdale SW. 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem 72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 2.Gruber K, Puffer B, Kräutler B. 2003. Vitamin B12-derivatives—enzyme cofactors and ligands of proteins and nucleic acids. Chem Soc Rev 40:4346–4363. doi: 10.1039/c1cs15118e. [DOI] [PubMed] [Google Scholar]
- 3.Bridwell-Rabb J, Drennan CL. 2017. Vitamin B12 in the spotlight again. Curr Opin Chem Biol 37:63–70. doi: 10.1016/j.cbpa.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenhert PG, Hodgkin DC. 1961. Structure of the 5,6-dimethyl-benzimidazolylcobamide coenzyme. Nature 192:937–938. doi: 10.1038/192937a0. [DOI] [PubMed] [Google Scholar]
- 5.Renz P. 1999. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids, p 557–576. In Banerjee R. (ed), Chemistry and biochemistry of B12. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 6.Holliger C, Wohlfarth G, Diekert G. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol Rev 22:383–398. doi: 10.1111/j.1574-6976.1998.tb00377.x. [DOI] [Google Scholar]
- 7.Jugder BE, Ertan H, Bohl S, Lee M, Marquis CP, Manefield M. 2016. Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation. Front Microbiol 7:249. doi: 10.3389/fmicb.2016.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fincker M, Spormann AM. 2017. Biochemistry of catabolic reductive dehalogenation. Annu Rev Biochem 86:357–386. doi: 10.1146/annurev-biochem-061516-044829. [DOI] [PubMed] [Google Scholar]
- 9.Moore TC, Escalante-Semerena JC. 2016. Corrinoid metabolism in dehalogenating pure cultures and microbial communities, p 455–484. In Adrian L, L̈offler FE (ed), Organohalide-respiring bacteria. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 10.Keller S, Ruetz M, Kunze C, Kräutler B, Diekert G, Schubert T. 2014. Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ Microbiol 16:3361–3369. doi: 10.1111/1462-2920.12268. [DOI] [PubMed] [Google Scholar]
- 11.Goris T, Schubert T, Gadkari J, Wubet T, Tarkka M, Buscot F, Adrian L, Diekert G. 2014. Insights into organohalide respiration and the versatile catabolism of Sulfurospirillum multivorans gained from comparative genomics and physiological studies. Environ Microbiol 16:3562–3580. doi: 10.1111/1462-2920.12589. [DOI] [PubMed] [Google Scholar]
- 12.Goris T, Schiffmann CL, Gadkari J, Schubert T, Seifert J, Jehmlich N, von Bergen M, Diekert G. 2015. Proteomics of the organohalide-respiring epsilonproteobacterium Sulfurospirillum multivorans adapted to tetrachloroethene and other energy substrates. Sci Rep 5:13794. doi: 10.1038/srep13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kräutler B, Fieber W, Ostermann S, Fasching M, Ongania KH, Gruber K, Kratky C, Mikl C, Siebert A, Diekert G. 2003. The cofactor of tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans is norpseudo-B12, a new type of a natural corrinoid. Helv Chim Acta 86:3698–3716. doi: 10.1002/hlca.200390313. [DOI] [Google Scholar]
- 14.Goris T, Schenz B, Zimmermann J, Lemos M, Hackermüller J, Schubert T, Diekert G. 2017. The complete genome of the tetrachloroethene-respiring epsilonproteobacterium Sulfurospirillum halorespirans. J Biotechnol 255:33–36. doi: 10.1016/j.jbiotec.2017.06.1197. [DOI] [PubMed] [Google Scholar]
- 15.Keller S, Treder A, von Reuss SH, Escalante-Semerena JC, Schubert T. 2016. The SMUL_1544 gene product governs norcobamide biosynthesis in the tetrachloroethene-respiring bacterium Sulfurospirillum multivorans. J Bacteriol 198:2236–2243. doi: 10.1128/JB.00289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazra AB, Han AW, Mehta AP, Mok KC, Osadchiy V, Begley TP, Taga ME. 2015. Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc Natl Acad Sci U S A 112:10792–10797. doi: 10.1073/pnas.1509132112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trzebiatowski JR, Escalante-Semerena JC. 1997. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella Typhimurium LT2. J Biol Chem 272:17662–17667. doi: 10.1074/jbc.272.28.17662. [DOI] [PubMed] [Google Scholar]
- 19.Crofts TS, Hazra AB, Tran JL, Sokolovskaya OM, Osadchiy V, Ad O, Pelton J, Bauer S, Taga ME. 2014. Regiospecific formation of cobamide isomers is directed by CobT. Biochemistry 53:7805–7815. doi: 10.1021/bi501147d. [DOI] [PubMed] [Google Scholar]
- 20.Yi S, Seth EC, Men YJ, Stabler SP, Allen RH, Alvarez-Cohen L, Taga ME. 2012. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl Environ Microbiol 78:7745–7752. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Im J, Yang Y, Löffler FE. 2013. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos Trans R Soc Lond B Biol Sci. 368:20120320. doi: 10.1098/rstb.2012.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl Environ Microbiol 78:6630–6636. doi: 10.1128/AEM.01535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan J, Şimşir B, Farmer AT, Bi M, Yang Y, Campagna SR, Löffler FE. 2016. The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J 10:1092–1101. doi: 10.1038/ismej.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig ML, Matthews RG. 1997. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem 66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 26.Buckel W, Golding BT. 2008. Chemistry of B12-dependent enzyme reactions, p 1–9. In Begley TP. (ed), Wiley encyclopedia of chemical biology. Wiley & Sons, New York, NY. [Google Scholar]
- 27.Bommer M, Kunze C, Fesseler J, Schubert T, Diekert G, Dobbek H. 2014. Structural basis for organohalide respiration. Science 346:455–458. doi: 10.1126/science.1258118. [DOI] [PubMed] [Google Scholar]
- 28.Payne KA, Quezada CP, Fisher K, Dunstan MS, Collins FA, Sjuts H, Levy C, Hay S, Rigby SE, Leys D. 2015. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517:513–516. doi: 10.1038/nature13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumacher W, Holliger C, Zehnder AJ, Hagen WR. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett 409:421–425. doi: 10.1016/S0014-5793(97)00520-6. [DOI] [PubMed] [Google Scholar]
- 30.van de Pas BA, Smidt H, Hagen WR, van der Oost J, Schraa G, Stams AJ, de Vos WM. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J Biol Chem 274:20287–20292. doi: 10.1074/jbc.274.29.20287. [DOI] [PubMed] [Google Scholar]
- 31.Schubert T. 2017. The organohalide-respiring bacterium Sulfurospirillum multivorans: a natural source for unusual cobamides. World J Microbiol Biotechnol 33:93. doi: 10.1007/s11274-017-2258-x. [DOI] [PubMed] [Google Scholar]
- 32.Friedrich W, Bernhauer K. 1958. Zur Chemie und Biochemie der “Cobalamine,” VIII. Über die 5- und 6-Methyl-Benzimidazol-Cobalamin-Analoga. Chemische Berichte 91:1665–1670. doi: 10.1002/cber.19580910815. [DOI] [Google Scholar]
- 33.Kräutler B, Moll J, Thauer RK. 1987. The corrinoid from Methanobacterium thermoautotrophicum (Marburg strain). Spectroscopic structure analysis and identification as Coβ-cyano-5′-hydroxybenzimidazolyl-cobamide (factor III). Eur J Biochem 162:275–278. [DOI] [PubMed] [Google Scholar]
- 34.Crofts TS, Seth EC, Hazra AB, Taga ME. 2013. Cobamide structure depends on both lower ligand availability and CobT substrate specificity. Chem Biol 20:1265–1274. doi: 10.1016/j.chembiol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Parthasarathy A, Stich TA, Lohner ST, Lesnefsky A, Britt RD, Spormann AM. 2016. Biochemical and EPR-spectroscopic investigation into heterologously expressed vinyl chloride reductive dehalogenase (VcrA) from Dehalococcoides mccartyi strain VS. J Am Chem Soc 137:3525–3532. doi: 10.1021/ja511653d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita Y, Futagami T, Goto M, Furukawa K. 2009. Functional characterization of the trigger factor protein PceT of tetrachloroethene-dechlorinating Desulfitobacterium hafniense Y51. Appl Microbiol Biotechnol 83:775–781. doi: 10.1007/s00253-009-1958-z. [DOI] [PubMed] [Google Scholar]
- 37.Maillard J, Genevaux P, Holliger C. 2011. Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157:2410–2421. doi: 10.1099/mic.0.050880-0. [DOI] [PubMed] [Google Scholar]
- 38.Padovani D, Labunska T, Palfey BA, Ballou DP, Banerjee R. 2008. Adenosyltransferase tailors and delivers coenzyme B12. Nat Chem Biol 4:194–196. doi: 10.1038/nchembio.67. [DOI] [PubMed] [Google Scholar]
- 39.Fieber W, Hoffmann B, Schmidt W, Stupperich E, Konrat R, Kräutler B. 2003. Pseudocoenzyme B12 and adenosyl-factor A: electrochemical synthesis and spectroscopic analysis of two natural B12 coenzymes with predominantly ‘base-off’ constitution. Helv Chim Acta 85:927–944. doi:. [DOI] [Google Scholar]
- 40.Kunze C, Bommer M, Hagen WR, Uksa M, Dobbek H, Schubert T, Diekert G. 2017. Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer. Nat Commun 8:15858. doi: 10.1038/ncomms15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mueller U, Darowski N, Fuchs MR, Förster R, Hellmi M, Paithankar KS, Pühringer S, Steffien M, Zocher G, Weiss MS. 2012. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat 19:442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meents A, Reime B, Stübe N, Fischer P, Warmer M, Göries D, Röver J, Meyer J, Fischer J, Burkhardt A, Vartiainen I, Karvinen P, David C. 2013. Development of an in-vacuum x-ray microscope with cryogenic sample cooling for beamline P11 at PETRA III. Proceedings of SPIE 8851, x-ray nanoimaging: instruments and methods, 88510K. doi: 10.1117/12.2027303. [DOI] [Google Scholar]
- 43.Burkhardt A, Pakendorf T, Reime B, Meyer J, Fischer P, Stübe N, Panneerselvam S, Lorbeer O, Stachnik K, Warmer M, Rödig P, Göries D, Meents A. 2016. Status of the crystallography beamlines at PETRA III. Eur Phys J Plus 131:56. doi: 10.1140/epjp/i2016-16056-0. [DOI] [Google Scholar]
- 44.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krug M, Weiss MS, Heinemann U, Mueller U. 2012. XDSAPP: a graphical user interface for the convenient processing of diffraction data using XDS. J Appl Cryst 45:568–572. doi: 10.1107/S0021889812011715. [DOI] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.