ABSTRACT
Shewanella oneidensis strain MR-1 is a versatile bacterium capable of respiring extracellular, insoluble ferric oxide minerals under anaerobic conditions. The respiration of iron minerals results in the production of soluble ferrous ions, which at high concentrations are toxic to living organisms. It is not fully understood how Fe2+ is toxic to cells anaerobically, nor is it fully understood how S. oneidensis is able to resist high levels of Fe2+. Here we describe the results of a transposon mutant screen and subsequent deletion of the genes clpX and clpP in S. oneidensis, which demonstrate that the protease ClpXP is required for anaerobic Fe2+ resistance. Many cellular processes are known to be regulated by ClpXP, including entry into stationary phase, envelope stress response, and turnover of stalled ribosomes. However, none of these processes appears to be responsible for mediating anaerobic Fe2+ resistance in S. oneidensis. Protein trapping studies were performed to identify ClpXP targets in S. oneidensis under Fe2+ stress, implicating a wide variety of protein targets. Escherichia coli strains lacking clpX or clpP also display increased sensitivity to Fe2+ anaerobically, indicating Fe2+ resistance may be a conserved role for the ClpXP protease system. Hypotheses regarding the potential role(s) of ClpXP during periods of high Fe2+ are discussed. We speculate that metal-containing proteins are misfolded under conditions of high Fe2+ and that the ClpXP protease system is necessary for their turnover.
IMPORTANCE Prior to the evolution of cyanobacteria and oxygenic photosynthesis, life arose and flourished in iron-rich oceans. Today, aqueous iron-rich environments are less common, constrained to low-pH conditions and anaerobic systems such as stratified lakes and seas, digestive tracts, subsurface environments, and sediments. The latter two ecosystems often favor dissimilatory metal reduction, a process that produces soluble Fe2+ from iron oxide minerals. Dissimilatory metal-reducing bacteria must therefore have mechanisms to tolerate anaerobic Fe2+ stress, and studying resistance in these organisms may help elucidate the basis of toxicity. Shewanella oneidensis is a model dissimilatory metal-reducing bacterium isolated from metal-rich sediments. Here we demonstrate a role for ClpXP, a protease system widely conserved in bacteria, in anaerobic Fe2+ resistance in both S. oneidensis and Escherichia coli.
KEYWORDS: Shewanella, iron reduction, iron toxicity, proteases
INTRODUCTION
The bacterium Shewanella oneidensis MR-1 is a member of the gammaproteobacteria that resides in the oxic-anoxic transition zones of water columns and aquatic sediments (1–3). S. oneidensis is a facultative anaerobe able to utilize numerous compounds as terminal electron acceptors in the absence of oxygen, including nitrate, sulfite, trimethylamine N-oxide, fumarate (1, 4, 5), and metals such as iron and manganese (hydr)oxide minerals (1, 6), which are frequently abundant in aquatic sediments (7). The respiration and consequent change in oxidation state of a metal can influence its solubility. For example, ferric iron (Fe3+) is often found in sediments as insoluble iron oxides (8), but upon reduction, these minerals can dissolve and release soluble ferrous iron (Fe2+) (8, 9).
Like many transition metals, iron is required for numerous biological functions (10), but at higher concentrations, it becomes toxic to organisms (11–13). Fe2+ toxicity in aerobic conditions is believed to be due to oxidative stress resulting from the production of hydroxyl radicals (12, 14, 15), but the mechanism of anaerobic Fe2+ toxicity is not known. S. oneidensis is capable of tolerating millimolar levels of Fe2+ anaerobically (6), higher than many other bacterial species (13, 16, 17), consistent with an adaptation to metal-rich environments. S. oneidensis is able to limit the buildup of intracellular iron via the activities of the iron uptake regulator Fur, which suppresses the production of siderophores and iron import systems under iron-replete conditions (18–20). The inner membrane efflux protein FeoE removes excess Fe2+ from the cytoplasm produced during Fe3+ respiration and lowers Fe2+ sensitivity (21). To discover other Fe2+ resistance mechanisms encoded in the S. oneidensis genome and to uncover mechanisms of anoxic Fe2+ toxicity, we performed a transposon screen under excess-Fe2+ conditions. Here we present analysis of two genes that, upon inactivation, conferred a fitness defect in the presence of excess Fe2+: clpP and clpX.
clpP and clpX encode the AAA+ (ATPases associated with diverse cellular activities) cytoplasmic protease ClpXP. The ATP-dependent unfoldase ClpX recognizes substrate proteins (22) and feeds them into the serine protease ClpP, which degrades the unfolded target proteins into small peptides (23, 24). ClpA, a second unfoldase able to complex with ClpP in place of ClpX (24), targets a different but overlapping set of proteins for degradation by ClpP in Escherichia coli (22, 25). ClpXP is one of five AAA+ proteases encoded in the S. oneidensis genome, the other four being ClpAP, Lon, HslVU, and FtsH (26–30). ClpXP has several established roles in bacteria, including regulation of entry into stationary phase via degradation of the stress response regulator σS, degradation of cell division proteins, promoting release of the envelope stress response regulator σE, and turning over ribosomes by degrading proteins stalled during translation (25, 31–36). We demonstrate that the role of ClpXP in mediating resistance to anaerobic Fe2+ stress is independent of these previously established roles. A role for ClpXP in Fe2+ tolerance may extend beyond S. oneidensis, as clpPX genes from E. coli complement the Fe2+ toxicity phenotype of S. oneidensis clpPX mutants and E. coli strains defective in either clpX or clpP exhibit enhanced sensitivity to Fe2+ under anaerobic conditions.
RESULTS
Tn-Seq reveals genes required for anaerobic Fe2+ toxicity response.
To find genes involved in surviving high concentrations of Fe2+ anaerobically, transposon sequencing (Tn-Seq) was performed on wild-type and ΔfeoE mutant S. oneidensis libraries grown in the presence or absence of 0.8 mM FeCl2. Both wild-type and ΔfeoE strains were used in order to serve as replicates for the experiment while providing a means to discover genes that, upon deactivation, confer a stronger fitness defect in a strain with increased Fe2+ sensitivity (ΔfeoE mutant) (21). The results of the Tn-Seq screen are listed in Table S1 in the supplemental material. The fitness costs of genes required for anaerobic respiration or encoding proteins known to interact with Fe2+ were evaluated as controls to confirm the validity of the Tn-Seq results. Deactivation of genes encoding the fumarate reductase FccA, pyruvate formate-lyase PflB, and inner membrane tetraheme cytochrome CymA conferred strong defects for each strain under both outgrowth conditions (Table S1; see Materials and Methods for an explanation of fitness effect calculations), confirming that the Tn-Seq screen reflects genes required for growth under our conditions. Deactivation of feoE, which encodes an Fe2+ efflux pump (21), conferred a significant net fitness defect under high Fe2+ (−1.24) (Table S1) in the wild-type background. No reads were mapped to feoE in any of the ΔfeoE libraries, indicating that no cross-library contamination had occurred. Deactivation of feoB, which encodes an Fe2+ importer in E. coli (37), conferred a strong apparent net fitness benefit under high-Fe2+ conditions in both the wild-type and ΔfeoE mutant (+3.68 and +1.77, respectively) (Table S1). Under low-Fe2+ conditions, feoB mutations result in a strong fitness defect, which is partially rescued in the ΔfeoE background (−4.00 in the wild type and −2.54 in the ΔfeoE mutant) (Table S1), consistent with their opposing functions.
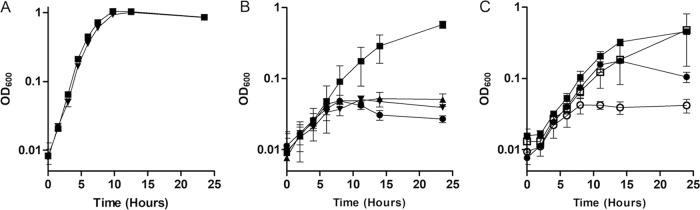
Two genes that, upon deactivation, produced some of the strongest net fitness defects under high Fe2+ were clpP (SO_1794) and clpX (SO_1795) (−1.09 and −1.55 in the wild-type background and −2.05 and −2.73 in the ΔfeoE mutant, respectively) (Table S1). Of the genes encoding the five AAA+ proteases in the S. oneidensis genome, only deactivation of clpX and clpP conferred a significant defect in the presence of excess Fe2+ in our transposon mutant screen. To confirm the Tn-Seq results for clpP and clpX, in-frame single and double deletions were made of clpP and clpX from the S. oneidensis genome. The ΔclpP, ΔclpX, and ΔclpPX mutants had strong growth defects compared to the wild type when grown anaerobically in LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2 but not when FeCl2 was omitted (doubling times without added Fe2+: ΔclpPX mutant, 1.23 ± 0.14 h; wild type, 1.22 ± 0.11 h) (Fig. 1A and B). Complementation with pBBR1MCS-2::clpPX restored the growth rate of the ΔclpPX mutant to that of the wild type (Fig. 1C).
FIG 1.
Growth of ΔclpP, ΔclpX, and ΔclpPX mutants with and without high FeCl2. (A) The rate of growth in anoxic LB supplemented with 20 mM lactate and 40 mM fumarate was measured for wild-type S. oneidensis (■) and the ΔclpPX mutant (▼). (B) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2 was measured for wild-type S. oneidensis (■) and the ΔclpPX (▼), ΔclpX (▲), and ΔclpP (●) mutants. (C) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2 was measured for wild-type S. oneidensis with empty pBBR1MCS-2 (□), wild-type S. oneidensis with pBBR1MCS-2::clpPX (■), the ΔclpPX mutant with empty pBBR1MCS-2 (○), and the ΔclpPX mutant with pBBR1MCS-2::clpPX (●). Results are the mean ± 1 standard deviation from three biological replicates.
ClpXP role in anaerobic Fe2+ resistance is unrelated to known proteolytic functions.
Much of the research into the function and structure of ClpXP has taken place in E. coli, although as mitochondria and additional bacterial species are analyzed, the known roles of ClpXP are expanding (reviewed in references 38 to 40). We focused the investigation of known ClpXP proteolysis targets to those annotated in the S. oneidensis genome. ClpXP targets the starvation and stationary-phase regulator sigma factor σS (31) and degrades proteins stalled in translation via recruitment and recognition by SspB, SmpB, and the SsrA transfer-mRNA tag (35, 41, 42). None of the fitness costs of deactivating any of these genes during growth in high Fe2+ met our threshold for significance in the Tn-Seq data set (rpoS, −0.08; sspB, −0.57; ssrA, +0.19; and smpB, +0.36 in the wild-type background; −0.77, −0.86, −0.53, and −1.65, respectively, in the ΔfeoE mutant) (Table S1). While smpB appears to have a significant result in the ΔfeoE background, the number of reads that mapped to smpB in the ΔfeoE background was low (50% less than the standard cutoff [Table S1]) and may therefore be an unreliable estimate of fitness.
A group of E. coli proteins regulated by ClpXP are the cell division proteins FtsZ, ZapC, FtsA, MinD, and SulA (25, 32, 33). Deactivation of neither zapA, zapB, zapC, ftsA, minD, nor sulA conferred a significant net fitness defect in high Fe2+ in our Tn-Seq data (−0.34, +0.33, +0.47, +0.85, −0.15, and +0.29, respectively, in the wild-type background; −0.57, −0.41, +0.53, −0.52, −0.22, and +0.35, respectively, in the ΔfeoE mutant) (Table S1). ftsZ appears to be an essential gene in S. oneidensis according to our Tn-Seq data, as no sequencing reads were mapped to the gene in any of the libraries (Table S1), consistent with previous observations (43).
ClpXP is involved in the release of the stress response sigma factor σE by degrading the cytoplasmic domain of the anti-sigma factor RseA (34). The proteases DegS and RseP are also required for degradation of RseA (44, 45), while RseB acts as a secondary regulator of σE (46). None of the genes encoding proteins involved in regulating σE had significant net fitness costs upon deactivation under high Fe2+ in our Tn-Seq data (+0.61, −0.42, +0.09, and +0.31 in the wild-type background, respectively; +0.41, −0.80, −0.65, and +0.64, respectively, in the ΔfeoE mutant) (Table S1). However, deactivation of rpoE itself, the gene encoding σE, did confer a net fitness defect near our significance threshold in high Fe2+ (−0.92 in the wild-type background, −1.06 in the ΔfeoE mutant). However, an in-frame rpoE deletion mutant was not significantly impaired compared to the wild type in either the presence or absence of excess Fe2+ (doubling times, respectively, of 3.13 ± 0.21 and 2.94 ± 0.21 h in excess Fe2+ and 1.38 ± 0.20 and 1.22 ± 0.11 h without excess Fe2+) (Fig. 2). Together, these data suggest that ClpXP plays a role in promoting survival during anaerobic Fe2+ stress, but this role is not related to previously described functions of ClpXP.
FIG 2.
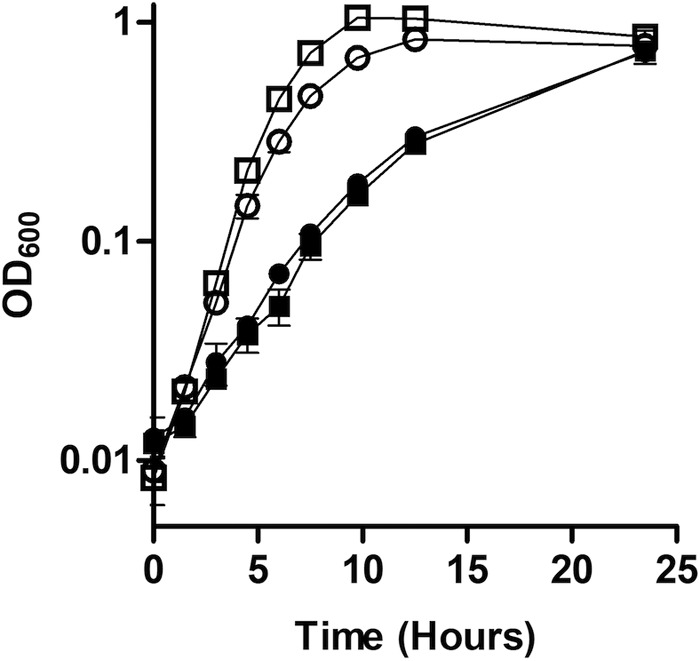
Growth of the ΔrpoE mutant with high FeCl2. The rate of growth in anoxic LB supplemented with 20 mM lactate and 40 mM fumarate, with (closed symbols) or without (open symbols) 2.5 mM FeCl2, was measured for wild-type S. oneidensis (□ and ■) and the ΔrpoE mutant (○ and ●). Results are the mean ± 1 standard deviation from three biological replicates.
ClpXP targets a distinct subset of proteins under high-Fe2+ conditions.
To determine the proteolysis targets of ClpXP under high-Fe2+ conditions, we set up a ClpP trapping experiment modified from that of Flynn et al. (25). The catalytic serine in the active site of ClpP was mutated (S106A) to eliminate proteolytic activity, and a His6-TEV-Myc3 affinity tag was attached to the C terminus, creating ClpPTrap. An allele encoding a protein without a mutated active site (S106) was also created, called ClpPTag, to confirm activity of the tagged protein. ClpPTrap folds correctly and continues to bind the ATPase subunits ClpA and ClpX (25), which feed protease targets into ClpPTrap. Substrate proteins are slowly released by ClpXPTrap or ClpAPTrap (47), allowing for the use of ClpPTrap, in conjunction with protein mass spectrometry, to detect which proteins are targeted for degradation by ClpXP or ClpAP.
A ΔsmpB ΔclpP background was used for all trapping strains, in order to remove SsrA-tagged proteins from protein analysis and to prevent degradation of ClpXP targets by an active ClpP protease (25). clpA and clpX deletions were created in this background to isolate proteins specifically targeted for degradation by either ClpXP or ClpAP. Additionally, a ΔclpA ΔclpX mutant was used to identify proteins that nonspecifically bind to ClpPTrap or the anti-Myc resin without being targeted by ClpX or ClpA. Each strain was transformed with pBBR1MCS-2::clpPTrap.
To confirm that deletion of smpB and clpA did not affect the growth of S. oneidensis under high-Fe2+ conditions, the wild-type and ΔsmpB and ΔclpA mutants were grown anaerobically in LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2. There was no difference in growth rates between the wild type and ΔsmpB and ΔclpA mutants (doubling times of 2.36 ± 0.22, 2.34 ± 0.35, and 2.52 ± 0.35 h, respectively) (Fig. 3A). The ΔsmpB ΔclpP ΔclpA mutant with pBBR1MCS-2::clpPTrap had the same growth defect as the ΔsmpB ΔclpP ΔclpA mutant with empty pBBR1MCS-2 when grown in anaerobic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2 (Fig. 3B), confirming that the S106A mutation mimics the phenotype of a clpP null mutant. The ΔsmpB ΔclpP ΔclpA mutant with pBBR1MCS-2::clpPTag had a higher growth rate than the ΔsmpB ΔclpP ΔclpA mutant with pBBR1MCS-2::clpPTrap (Fig. 3B), suggesting that removal of the propeptide sequence from and addition of the purification tag to ClpP did not interfere with its proper folding.
FIG 3.
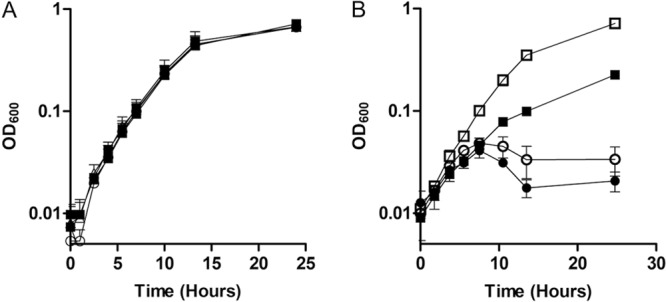
Growth of proteomic analysis strains. (A) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2 mM FeCl2 was measured for wild-type S. oneidensis (■) and the ΔsmpB (○) and ΔclpA (Δ) mutants. (B) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2.5 mM FeCl2 was measured for wild-type S. oneidensis with empty pBBR1MCS-2 (□), the ΔsmpB ΔclpA ΔclpP mutant with empty pBBR1MCS-2 (○), the ΔsmpB ΔclpA ΔclpP mutant with pBBR1MCS-2::clpPTag (■), and the ΔsmpB ΔclpA ΔclpP mutant with pBBR1MCS-2::clpPTrap (●). Results are the mean ± 1 standard deviation from three biological replicates.
The three ClpP-trapping strains were grown anaerobically in LB supplemented with 20 mM lactate and fumarate; the ClpX-only strain was also grown in anaerobic LB supplemented with 20 mM lactate, 40 mM fumarate, and 1.1 mM FeCl2. ClpP-trapped proteins were identified using mass spectrometry. The proteins listed in Table 1 were detected at least twice as frequently in the Fe2+ culture as in any of the others in at least two of three mass spectrometry runs. Proteins trapped by ClpXP that were enriched under high-Fe2+ conditions have disparate functions but frequently (10 of 11 proteins trapped) contain metal binding sites (Table 1; see Table S3 in the supplemental material), a higher proportion than the proteins trapped with ClpXP under lower-Fe2+ conditions (12 of 19), ClpAP (1 of 3), either ClpAP or ClpXP (17 of 36), or ClpP with no ATPase adapter (1 of 2). Additionally, a higher proportion of proteins (7 of 11 [Table 1; Table S3]) trapped with ClpXP in high Fe2+ utilize nucleotide-derived cofactors than those trapped by ClpXP under lower Fe2+ (8 of 19).
TABLE 1.
Proteins differentially trapped by ClpXP in high or low Fe2+
| Protein | Locus | Predicted function | Predicted features(s) or ligand(s)a |
|---|---|---|---|
| Trapped in high Fe2+ | |||
| RecA | SO_3430 | Recombinase A | ATP, Mg2+ |
| DeaD | SO_4034 | ATP-dependent RNA helicase | DEAD, ATP, Mg2+ |
| NrdA | SO_2415 | Aerobic ribonucleoside-diphosphate reductase alpha subunit | Tyrosyl radical, rNTP, ATP, Fe3+ |
| NrdD | SO_2834 | Anaerobic ribonucleoside-triphosphate reductase | Glycyl radical, rNTP, ATP, Zn2+ |
| GapA | SO_2345 | Glyceraldehyde-3-phosphate dehydrogenase | G3P, NAD(P) |
| TnpB_MuSo2 | SO_2655 | Mu phage transposase OrfB | ATPase, DNA, ATP, Mg2+ |
| CydA | SO_3286 | Cytochrome d ubiquinol oxidase subunit I | TM, UQ, heme |
| MgtE-1 | SO_1145 | Divalent metal transporter | TM, CBS, MgtE-N, Mg2+, Ca2+ |
| SO_0520 | Heavy metal efflux pump permease component CzcA family | TM, Cu2+ | |
| SO_1383 | ATP-dependent RNA helicase DEAD box family | DEAD, ATP, Mg2+ | |
| LepB | SO_1347 | Signal peptidase I | TM, S26, Mg2+, K+ |
| Trapped in low Fe2+ | |||
| AtpA | SO_4749 | ATP synthase subunit alpha | Mg2+, ADP, ATP |
| SecA | SO_4211 | Protein translocase subunit | Zn2+, ATP |
| ThrS | SO_2299 | Threonine-tRNA ligase | Zn2+, ATP, Thr, anticodon binding |
| GlyS | SO_0014 | Glycine-tRNA ligase beta subunit | ATP, Gly, anticodon binding |
| Fba | SO_0933 | Fructose-bisphosphate aldolase class II | Zn2+, Na+, DAP |
| SO_3743 | Transcriptional regulator TetR family | HTH | |
| DmsA | SO_1429 | Extracellular dimethyl sulfoxide/manganese oxide reductase molybdopterin-binding subunit A | Fe-S, MopB |
| AdhB | SO_1490 | Alcohol dehydrogenase II | Fe3+ |
| SO_0988 | Molybdopterin-binding oxidoreductase | Fe-S, Mo5+ | |
| DeoD2 | SO_1221 | Purine nucleoside phosphorylase DeoD-type 2 | Purine nucleoside, phosphate |
| ZapC | SO_2591 | Cell division protein | Z-ring binding |
| Ppc | SO_0274 | Phosphoenolpyruvate carboxylase | Mg2+ |
| HemB | SO_2587 | Delta-aminolevulinic acid dehydratase | Zn2+, Mg2+, Schiff base |
| SO_3363 | Transcriptional regulator LysR family | PBP, HTH | |
| NuoG | SO_1016 | NADH-quinone oxidoreductase subunit G | Fe-S, NADH, UQ |
| SO_1117 | Cytoplasmic leucyl metal-dependent aminopeptidase | Zn2+/Mn2+ | |
| NadB | SO_1341 | l-Aspartate oxidase | FAD, SQO |
| FdhD | SO_0107 | Sulfurtransferase | Mo-bisPGD |
| SO_3097 | Anti-sigma factor | TM; DUF3379 |
DEAD, DEAD box RNA helicase domain; TM, transmembrane domain; UQ, ubiquinone binding domain; CBS, cystathionine beta-synthase-like ligand binding domain; MgtE-N, Mg2+ transporter intracellular N domain; S26, S26 signal peptidase domain; DAP, dihydroxyacetone phosphate; HTH, helix-turn-helix DNA binding domain; Fe-S, iron-sulfur cluster; MopB, molybdopterin-binding domain; PBP, periplasmic effector binding pocket; FAD, flavin adenine dinucleotide; SQO, succinate:quinone oxidoreductase domain; Mo-bisPGD, molybdo-bis pyranopterin guanine dinucleotide.
Mg2+ concentration affects the growth of S. oneidensis in high Fe2+.
The protein trapping results indicated that a high intracellular ratio of Fe2+ to other metals might interfere with proper insertion of noniron metals into metalloproteins. To determine the effect of low Mg2+ concentration during growth in high Fe2+, wild-type S. oneidensis was grown in Shewanella basal medium (SBM) with either 475 or 119 μM MgSO4 (100% and 25% of the MgSO4 concentrations in SBM, respectively) and with or without 0.8 mM FeCl2. The lower concentration of Mg2+ did not affect the growth rate of S. oneidensis in the absence of Fe2+; however, the growth rate was lower in the combination of high Fe+ and low Mg2+ than in high Fe2+ alone (Fig. 4).
FIG 4.
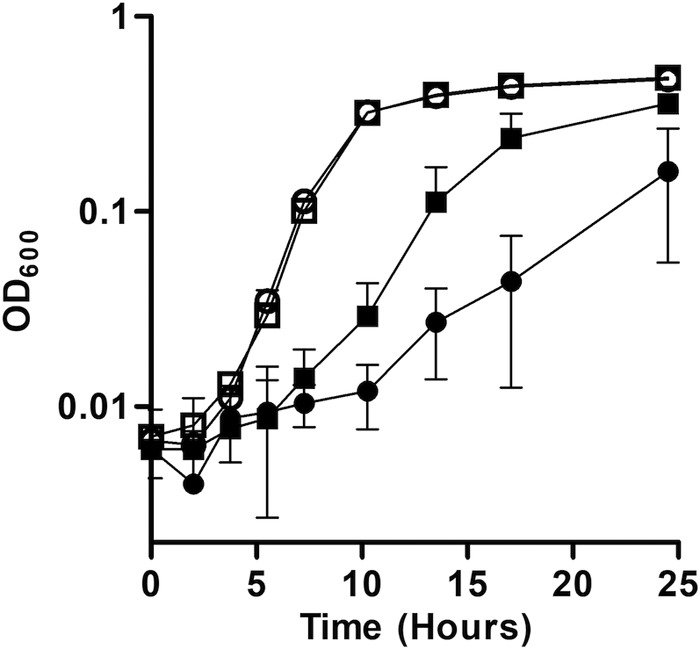
Growth of S. oneidensis with high Fe2+ and low Mg2+. The rate of growth in anoxic SBM supplemented with 20 mM lactate, 40 mM fumarate, and 475 μM MgSO4 and 0 mM FeCl2 (□), 119 μM MgSO4 and 0 mM FeCl2 (○), 475 μM MgSO4 and 0.8 mM FeCl2 (■), or 119 μM MgSO4 and 0.8 mM FeCl2 (●) was measured for wild-type S. oneidensis. Results are the mean ± 1 standard deviation from three biological replicates.
ClpXP is required for Fe2+ resistance in E. coli.
To determine whether ClpXP is involved in Fe2+ resistance beyond S. oneidensis, we grew S. oneidensis ΔclpX and ΔclpP mutants complemented with E. coli clpX and clpP genes, respectively, in anaerobic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2.5 mM FeCl2. The clpX and clpP genes from E. coli complemented the Fe2+ sensitivity phenotype of ΔclpX and ΔclpP S. oneidensis strains (Fig. 5A). E. coli strains with deletions of either clpX or clpP were grown in anaerobic LB supplemented with 20 mM lactate and 40 mM fumarate, with or without 6 mM FeCl2. This Fe2+ concentration was required to see impairment in the growth rate of E. coli in LB. While the ΔclpX and ΔclpP mutants grew at the same rate as the wild type without added Fe2+, both strains displayed a growth defect in the presence of excess Fe2+ (Fig. 5B).
FIG 5.
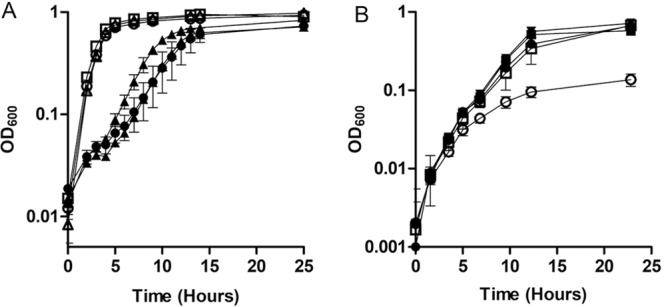
Growth of E. coli and S. oneidensis ΔclpX and ΔclpP mutant strains with and without high FeCl2. (A) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and with (closed symbols) or without (open symbols) 6 mM FeCl2 was measured for wild-type E. coli (□ and ■), the ΔclpX mutant (△ and ▲), and the ΔclpP mutant (○ and ●). (B) The rate of growth in anoxic LB supplemented with 20 mM lactate, 40 mM fumarate, and 2.5 mM FeCl2 was measured for wild-type S. oneidensis with empty pBBR1MCS-2 (□), the ΔclpPX mutant with empty pBBR1MCS-2 (○), the ΔclpPX mutant with pBBR1MCS-2::clpPXMR-1 (■), the ΔclpPX mutant with pBBR1MCS-2::clpPXE. coli (▼), and the ΔclpX mutant with pBBR1MCS-2::clpXE. coli (●). Results are the mean ± 1 standard deviation from three biological replicates.
DISCUSSION
AAA+ proteases have been implicated in numerous cellular processes in various bacterial species. Here we have provided evidence for a new function of the AAA+ protease ClpXP: resistance to Fe2+ toxicity. The loss of either clpP or clpX was shown to be detrimental to both S. oneidensis and E. coli growing under high-Fe2+ conditions but not at low Fe2+ concentrations (Fig. 1B and 5B; see Table S1 in the supplemental material). The loss of clpA, which encodes another ATP-dependent chaperone that complexes with ClpP (24), or clpS, which encodes an adaptor to the ClpAP complex (48), had no effect on the sensitivity of S. oneidensis to high Fe2+ concentrations (Fig. 3A; Table S1), indicating that the proteins specifically targeted by ClpX but not ClpA for degradation by ClpP are involved in Fe2+ sensitivity.
ClpXP regulates a number of cytoplasmic proteins and facilitates the turnover of stalled ribosomes (25, 31–34). None of these processes, however, appears to be involved in the resistance of S. oneidensis to high Fe2+ concentrations (Fig. 2 and 3; Table S1). Previous studies of σE in E. coli have indicated that rpoE is essential and that mutants can be made only in conjunction with suppressor mutations (49). Suppressor mutations could explain why we did not see an increase in Fe2+ sensitivity for our ΔrpoE strain; however, we believe that σE is neither essential in S. oneidensis nor critical for Fe2+ stress response. rpoE transposon and deletion mutants were readily generated (Fig. 2; Table S1), inconsistent with the requirement of a suppressor mutation. Additionally, we tested multiple ΔrpoE isolates, all of which displayed identical phenotypes regarding resistance to Fe2+ and sensitivity to other stressors (data not shown). Therefore, the role ClpXP plays in Fe2+ resistance does not appear to be mediated through the promotion of σE release or other previously described ClpXP targets.
As we could not identify the function of ClpXP in response to Fe2+ toxicity through Tn-Seq data or phenotypic tests with deletion mutants, we adapted a ClpP trapping method (25) to identify ClpXP proteolysis targets in S. oneidensis. While one-fourth to one-third of cellular proteins are believed to require a metal cofactor (81), nearly all proteins that were enriched for ClpXP trapping under high-Fe2+ conditions are predicted to contain metal-binding domains (91% [Table 1]), more than for the proteins trapped by ClpXP in lower Fe2+ (57% [Table 1; Table S3]). Additionally, 64% of the proteins enriched for trapping by ClpXP under high Fe2+ are predicted to utilize nucleotides or nucleotide derivatives as cofactors, a higher proportion than with proteins enriched for ClpXP trapping under lower Fe2+ (42% [Table 1; Table S3]). The mass spectrometry data provided here are meant to be qualitative and can be only suggestive of relative quantitation; truly quantitative measurements would need to be performed in future studies. That said, the protein trapping results were largely repeatable. Given the controls and conditions used here, the proteomics results suggest to us the hypothesis that ClpXP may target metalloproteins and/or proteins with nucleotide-derived cofactors in S. oneidensis under Fe2+ stress.
Fe2+ has an affinity for pyrophosphate and nucleotides (51, 52), where it is predicted that Fe2+ complexes with phosphate ester oxygens (52). It may be that free Fe2+ binds to the phosphate groups of nucleotide cofactors in proteins, perhaps forcing a change in conformation around the cofactor. Alternatively, high soluble Fe2+ may cause inappropriate phosphate hydrolysis and interfere with enzymatic function.
In addition to binding nucleotides, iron and other metals bind readily to hydrophilic regions within proteins (53). It is notable that many proteins apparently targeted by ClpPTrap under the high-Fe2+ condition bind Mg2+ (Table 1; Table S3). The Irving-Williams series places the order of metal affinity to proteins as Ca2+ < Mg2+ < Mn2+ < Fe2+ < Co2+ < Ni2+ < Cu2+ > Zn2+ (54), making Fe2+ likelier to bind proteins than Mg2+, Ca2+, Mn2+, and, under certain conditions, Zn2+. Cells have delivery systems to direct the insertion of most metals into their proper protein binding sites (50, 55–61). Proper insertion of metals at the lower end of the Irving-Williams series, such as Mn2+ and Mg2+, on the other hand, frequently simply depends upon high relative intracellular concentrations of those metals (62, 63).
The intracellular concentration of Mg2+ is commonly kept around 1 mM, the highest concentration of all metals that have been evaluated (64). Not coincidentally, the intracellular concentration of each metal in the Irving-Williams series is inversely correlated with its affinity for proteins (64), indicating that cells compensate for low binding affinity with high relative concentration. It is therefore not surprising that Mg2+-binding proteins appeared to be more frequently targeted by ClpXP under high Fe2+ in our study: as the concentration of Fe2+ rises, it may overwhelm the cells' iron storage and/or trafficking capacity and begin outcompeting Mg2+ and other metals less dependent on specific delivery systems for insertion into the correct protein binding sites.
Consistent with the hypothesis that high Fe2+ concentrations interfere with Mg2+ insertion into metalloproteins, lowering the Mg2+ concentration in the presence of high Fe2+ further decreased the growth rate of S. oneidensis beyond that in high Fe2+ alone (Fig. 4). Additionally, inactivation of the corA gene in our Tn-Seq screen caused a strong net fitness defect in both the wild type and ΔfeoE mutant under high Fe2+ (−1.23 and −1.70, respectively) (Table S1). The stronger defect in the ΔfeoE mutant indicates that the higher intracellular Fe2+ concentration in this mutant exacerbates the effect of corA mutation. CorA has a high affinity for Mg2+ (65, 66) and appears to be the primary Mg2+ importer in bacteria (67). Further lowering the ratio of Mg2+ to Fe2+ appears to increase sensitivity to Fe2+, which could be due to mismetallation of Mg2+-requiring proteins. Based on these results, we suggest that protein mismetallation may be at least one mechanism of Fe2+ toxicity to cells.
While Fe2+ is known to be toxic under aerobic conditions due to Fenton chemistry and the production of reactive oxygen species (12, 14, 15), the mechanism by which Fe2+ is toxic under anaerobic conditions has not been well understood. Previously published hypotheses about anoxic Fe2+ toxicity have included formation of organic radicals, inhibition of the F-ATPase (13), or reduction of Cu2+ to the more toxic Cu+ (68). Based on our findings in this study, we propose that mechanisms by which Fe2+ may be toxic anaerobically may include either binding nucleotide-derived cofactors or, as we believe to be more likely, overwhelming the normal mechanisms of proper metal insertion into metalloproteins, with Fe2+ replacing the required metal. Either of these events would interfere with proper enzyme activity, causing a buildup of multiple inactive cytoplasmic proteins that could overwhelm the cell if not properly turned over. This hypothesis is consistent with the growth behavior of the strains lacking ClpX, ClpP, and ClpXP, where the growth rate is identical to that of the wild type for the first ∼6 h before plateauing and beginning to decline (Fig. 1B). The growth pattern here is similar to that for temperature-sensitive mutations in the major chaperonin GroEL, which causes the accumulation of unfolded proteins (69). Interestingly, there was less overlap between our list of ClpXP-trapped proteins and those identified by Flynn et al. (25) than we expected. It may be that Fe2+ stress upends the usual set of ClpXP targets by making a certain subset of proteins more likely to become unstable or to partially unfold. If mismetallation of metalloproteins is a major mechanism behind Fe2+ toxicity, it is likely to be a major toxicity mechanism for other metals higher on the Irving-Williams series. In this case, ClpXP could target proteins with any misincorporated metal for degradation.
Here we have determined that ClpXP is an important factor in anaerobic Fe2+ resistance in both E. coli and S. oneidensis. Many bacterial species occasionally encounter high concentrations of metals, and consequently they have developed cellular mechanisms of metal toxicity resistance. S. oneidensis, for example, requires several mechanisms to resist the high Fe2+ concentrations that occur transiently and locally during the respiration of iron minerals. While the mechanism of ClpXP in resisting high Fe2+ needs to be confirmed with future work, the work presented here is consistent with our proposed model in which ClpXP is required to turn over misfolded proteins when cells experience anaerobic Fe2+ stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Table 2 lists the bacterial strains and plasmids used in this work. The chemicals used throughout were the highest purity available through Sigma-Aldrich, unless otherwise indicated. S. oneidensis MR-1 was isolated from Lake Oneida, NY (1). The E. coli strains for cloning (UQ950) and mating (WM3064) are described by Saltikov et al. (70). Overnight liquid Luria-Bertani (LB; BD Difco) cultures, supplemented with 50 μg/ml kanamycin when appropriate, were inoculated with isolated colonies from freshly streaked −80°C stocks. S. oneidensis and E. coli cultures were grown at 30 and 37°C, respectively. Cultures were grown in LB or Shewanella basal medium (SBM [71]) supplemented with 5 ml/liter vitamin solution (72), 5 ml/liter trace mineral solution (73), and 0.05% (wt/vol) Casamino Acids. Anaerobic cultures were sealed with butyl rubber stoppers, flushed with nitrogen gas, and supplemented with 20 mM sodium lactate and 40 mM sodium fumarate. Liquid cultures, except for 1-liter cultures prepared for protein purification, were shaken at 250 rpm.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| JG274 | S. oneidensis MR-1, wild type | 1 |
| JG2989 | JG274 ΔfeoE | 21 |
| JG3354 | JG274 ΔrpoE | This work |
| JG3355 | JG274 ΔclpPX | This work |
| JG3486 | JG274 ΔclpP | This work |
| JG3492 | JG274 ΔclpX | This work |
| JG3552 | JG274 ΔsmpB | This work |
| JG3556 | JG274 ΔclpA | This work |
| JG3560 | JG274 ΔsmpB ΔclpP ΔclpA | This work |
| JG3565 | JG274 ΔsmpB ΔclpPX | This work |
| JG3632 | JG274 ΔsmpB ΔclpPX ΔclpA | This work |
| JG168 | JG274 with empty pBBR1MCS-2, Kmr | 71 |
| JG3488 | JG3355 with empty pBBR1MCS-2, Kmr | This work |
| JG3549 | JG3486 with empty pBBR1MCS-2, Kmr | This work |
| JG3495 | JG3355 with pBBR1MCS-2::clpPXMR-1 | This work |
| JG3667 | JG3355 with pBBR1MCS-2::clpPXE. coli | This work |
| JG3668 | JG3492 with pBBR1MCS-2::clpXE. coli | This work |
| JG3570 | JG3560 with pBBR1MCS-2::clpPTag | This work |
| JG3599 | JG3560 with pBBR1MCS-2::clpPTrap | This work |
| JG3600 | JG3565 with pBBR1MCS-2::clpPTrap | This work |
| JG3635 | JG3632 with pBBR1MCS-2::clpPTrap | This work |
| UQ950 | E. coli DH5α λ(pir) cloning host; F− Δ(argF-lac)169 ϕ80dlacZ58(ΔM15) glnV44(AS) rfbD1 gyrA96(NalR) recA1 endA1 spoT1 thi-1 hsdR17 deoR λpir+ | 70 |
| WM3064 | E. coli conjugation strain; thrB1004 pro thi rpsL hsdS lacZΔM15 RP4–1360 Δ(araBAD)567 ΔdapA1341::[erm pir(wild type)] | 70 |
| MG1655 | E. coli K-12, wild type | Arkady Khodursky, University of Minnesota |
| JG3804 | MG1655 ΔclpP | This study |
| JG3805 | MG1655 ΔclpX | This study |
| Plasmids | ||
| pSMV3 | Deletion vector, Kmr sacB | 79 |
| pSMV3ΔclpP | pSMV3 with SO_1794 flanking sequences | This work |
| pSMV3ΔclpX | pSMV3 with SO_1795 flanking sequences | This work |
| pSMV3ΔclpPX | pSMV3 with SO_1794–5 flanking sequences | This work |
| pSMV3ΔrpoE | pSMV3 with SO_1342 flanking sequences | This work |
| pSMV3ΔclpA | pSMV3 with SO_2626 flanking sequences | This work |
| pSMV3ΔsmpB | pSMV3 with SO_1473 flanking sequences | This work |
| pSMV3ΔclpPEC | pSMV3 with b0437 flanking sequences | This work |
| pSMV3ΔclpXEC | pSMV3 with b0438 flanking sequences | This work |
| pBBR1MCS-2 | Broad-range cloning vector, Kmr | 80 |
| pBBR1MCS-2::clpPXMR-1 | SO_1794–5 (clpPX), 26 bp upstream, 8 bp downstream | This work |
| pBBR1MCS-2::clpPXE. coli | b0437–8 (clpPX), 22 bp upstream, 28 bp downstream | This work |
| pBBR1MCS-2::clpXE. coli | b0438 (clpX), 30 bp upstream, 28 bp downstream | This work |
| pBBR1MCS-2::clpPTag | SO_1794 (clpP) Δ2–9, downstream HIS6-TEV-MYC3 | This work |
| pBBR1MCS-2::clpPTrap | SO_1794 (clpP) Δ2–9, S106A, downstream HIS6-TEV-MYC3 | This work |
Creation and analysis of Tn-Seq mutant libraries.
Transposon library creation and selection were performed as previously described (43). Briefly, a delivery vector with MmeI restriction sites surrounding the MiniHimar transposon, which randomly inserts into the chromosome at TA sites (74), was transferred into wild-type and ΔfeoE mutant S. oneidensis strains via conjugation. Parent transposon libraries were outgrown for selection in anaerobic SBM with or without 0.8 mM FeCl2. The concentration of 0.8 mM FeCl2 was chosen to allow for an overall doubling time relatively close to that of unamended cultures while still inhibiting growth of Fe2+-sensitive mutants. Cultures were harvested and DNA extracted after approximately five doublings. Parent and outgrown DNA libraries were processed and sequenced as previously described (75). Adapters and primers used to prepare the DNA for sequencing have been published previously (76). Briefly, DNA was phenol-chloroform extracted and digested with MmeI. Adapters containing library-identifying barcodes were ligated to the digested DNA, and the transposon insertion sites were PCR amplified using primers containing Illumina-specific sequences. Single-read 50-bp sequence analysis was performed on an Illumina HiSeq250 at the University of Minnesota Genomics Center. Downstream sequence processing was performed using the Galaxy server maintained by the Minnesota Supercomputing Institute. Between 20 million and 33 million reads were mapped to the S. oneidensis chromosome and megaplasmid (accession no. NC_004347.2 and NC_004349.1, respectively) for each parent and outgrown library. Reads that did not match the genome sequence 100%, did not match uniquely to a gene, or fell in the first 1% or last 10% of a coding sequence were omitted from analysis. The number of reads for each gene was normalized to the total number of reads in each library.
Fitness effects of each gene under the outgrowth conditions were calculated by taking the natural log of the normalized number of reads in the outgrowth libraries divided by that in the parent library. Tn-Seq calculations and results are reported in Table S1 in the supplemental material. To exclude genes that confer a growth benefit or defect upon deactivation regardless of Fe2+ concentration, the net fitness effect of growing in excess Fe2+ for each gene was calculated by subtracting the fitness effect for the low-Fe2+ condition from that under the high-Fe2+ condition. A net fitness effect of ≥±1.0 was considered significant, a threshold that has been found in our hands to be a good predictor of whether a specific mutation has a significant effect on fitness. To avoid statistical anomalies, genes with fewer than 1,000 mapped reads to the parent library were omitted from consideration in this study (see the “Curated fitness values” tab in Table S1).
Plasmid and mutant construction.
Table 3 lists the primers used for construction of deletion and expression plasmids. In-frame deletion of genes was performed via homologous recombination as described in Saltikov et al. (70). Briefly, 1-kb upstream and downstream fragments for each gene were fused via a restriction site and inserted into the multiple-cloning site of pSMV3, which has kanamycin resistance and sacB cassettes. Complementation plasmids were created by cloning clpPX from the S. oneidensis genome and clpPX and clpX from the E. coli MG1655 genome and inserted into the multiple-cloning site of the constitutive expression vector pBBR1MCS-2 via BamHI and SpeI restriction sites. Tagged genes for protein purification were ordered as gBlocks from Integrated DNA Technologies and ligated into the expression vector pBBR1MCS-2 via EcoRI and BamHI restriction sites. Tagged alleles of clpP were created without the propeptide sequence (missing amino acids 2 to 9), with a C-terminal affinity (His6-TEV-Myc3) tag codon optimized for S. oneidensis, and with or without a deactivating point mutation in the active site (S106A), creating clpPTrap and clpPTag, respectively. clpPTag and clpPTrap sequences are listed in Table S2 in the supplemental material.
TABLE 3.
Primer sequences used for mutation and complementation in this work
| Primer | Sequence | Restriction site |
|---|---|---|
| S. oneidensis | ||
| rpoE deletion | ||
| 1342USF | GTACGGATCCCAATGCTTCGGTCAGCAG | BamHI |
| 1342USR | GTACACTAGTCTCATCCGAGCCGACTTC | SpeI |
| 1342DSF | GTACACTAGTGCCTTTGCTGGAAGAGTAAATTC | SpeI |
| 1342DSR | GTACGAGCTCCACCCTGAATATGATTAGAGAGG | SacI |
| clpP deletion | ||
| 1794USF | GTACGGATCCGATGTGGACAGCATGATTG | BamHI |
| 1794USR | GTACACTAGTGGCGAACTGCTAATCAAGTC | SpeI |
| 1794DSF | GTACACTAGTGATTTTTACTTTCGACTGGGC | SpeI |
| 1794DSR | GTACGAGCTCCTCAACTTGAGACAGGGTTTC | SacI |
| clpX deletion | ||
| 1795USF | GTACGGATCCCTCTATGGCTTCTGCTTACG | BamHI |
| 1795USR | GTACACTAGTGCCCATTAATTACCTCATTTGC | SpeI |
| 1795DSF | GTACACTAGTGGCGAGCAATAATTGTACAG | SpeI |
| 1795DSR | GTACGAGCTCCAGACATCGGTGACATCATG | SacI |
| clpA deletion | ||
| 2626USF | GTACGAGCTCCGCTAAACAAGCTATTGATTG | SacI |
| 2626USR | GTACGAATTCCAGATCTTTGTTCAGCATAAGC | EcoRI |
| 2626DSF | GTACGAATTCGCTTAACGCCAAGCTAATTTAC | EcoRI |
| 2626DSR | GCATACTAGTCTATTAGCCATAGGCTTTCG | SpeI |
| smpB deletion | ||
| 1473USF | GTACGAGCTCCTTCATCCTTGGCTTTATCAG | SacI |
| 1473USR | GTACGAATTCGTTTTTCTTTACCATAGTGGC | EcoRI |
| 1473DSF | GTACGAATTCGGATAATGAACAAACGATTGAAC | EcoRI |
| 1473DSR | GTACACTAGTGCAATCTGTGCTTCTCTATG | SpeI |
| clpPX expression | ||
| 1794F | GTACGGATCCGCCATTTTTATTTAGGGAAATG | BamHI |
| 1795R | GTACACTAGTCTGTACAATTATTGCTCGCC | SpeI |
| E. coli | ||
| clpP deletion | ||
| b0437USF | GTACACTAGTGAAGAATACCACGCAGAAAAC | SpeI |
| b0437USR | GTACGCGGCCGCCTGTATGACATTTCCGTCTCC | NotI |
| b0437DSF | GTACGCGGCCGCCATCGTAATTGATGCCAGAGG | NotI |
| b0437DSR | GTACGAGCTCCGGACTTCGCTTTTACCG | SacI |
| clpX deletion | ||
| b0438USF | GTACGAGCTCCCGTACCCATAACACAGG | SacI |
| b0438USR | GTACGCGGCCGCGCCATCTTTGCGTTTATC | NotI |
| b0438DSF | GTACGCGGCCGCGCATCTGGTGAATAATTAACC | NotI |
| b0438DSR | GTACACTAGTGCTCGTTCAGATAGTACTCAC | SpeI |
| clpPX and clpX expression | ||
| ECb0437F | GTACGGATCCCAATTTTATCCAGGAGACGG | BamHI |
| ECb0438F | GTACGGATCCGCACAAAGAACAAAGAAGAGG | BamHI |
| ECb0438R | GTACACTAGTGGTTAACTAATTGTATGGGAATGG | SpeI |
Growth curves.
Overnight liquid LB cultures were grown from freshly isolated colonies. Cells were pelleted by centrifugation, washed once, and resuspended in either LB or SBM, depending on the culture medium to be used. Fe2+ cultures were supplemented with 0.8 mM FeCl2 for SBM or either 2 or 2.5 mM FeCl2 for LB. A higher Fe2+ concentration was needed for growth curves in LB than in SBM to visualize the growth defects of mutants. Slightly different concentrations of FeCl2 were required in different batches of media to achieve a toxic effect while allowing for cell growth. Growth was measured by taking the optical density at 600 nm (OD600). Results are reported as the mean ± 1 standard deviation from three biological replicates. Growth curves were performed in triplicate at least twice; the results reported here are representative of each experiment.
ClpP trapping and protein purification.
A previously described ClpP trapping protocol (25) was adapted for S. oneidensis. Briefly, the ΔsmpB ΔclpP ΔclpA, and ΔsmpB ΔclpPX mutants and the ΔsmpB ΔclpPX ΔclpA mutant with pBBR1MCS-2::clpPTrap were grown anaerobically for 12 h in 1 liter anaerobic LB supplemented with 20 mM lactate and 40 mM fumarate; the ΔsmpB ΔclpP ΔclpA mutant with pBBR1MCS-2::clpPTrap was also grown for 12 h in in 3 liters anaerobic LB supplemented with 20 mM lactate, 40 mM fumarate, and 1.1 mM FeCl2. A 1.1 mM concentration of FeCl2 was chosen because clpP mutant strains are impaired but still able to grow at this concentration (data not shown). Cells were centrifuged 10 min at 5,000 rpm, resuspended in 40 ml Tris-buffered saline with 1 mM EDTA and 10 μM phenylmethylsulfonyl fluoride (pH 7.5), and centrifuged for 10 min at 5,000 rpm. Cell pellets were resuspended in 40 ml Tris-buffered saline and 10 μM phenylmethylsulfonyl fluoride (pH 8.0) and lysed by passing through a French press three times at 1,200 lb/in2. The lysate was centrifuged for 20 min at 10,000 rpm. The lysate supernatant was incubated with 4 ml anti-c-Myc–agarose (25% slurry; Thermo Scientific) for 5 h on a rocker at 4°C. The resin was collected on a 10-ml column and washed with 10 ml Tris-buffered saline with 0.5% Tween 20. The resin was eluted with 4 ml 50 mM NaOH, which was concentrated to 100 μl in a SpeedVac (Thermo Scientific).
Protein analysis.
Twenty micrograms of each protein elution was run into a Bio-Rad 8 to 16% Criterion precast polyacrylamide gel for 22 min at 25 mA. A single band for each sample containing total trapped protein was excised, digested in-gel with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on Thermo Orbitrap Velos and Orbitrap Fusion mass spectrometers. Detected peptides were mapped to S. oneidensis MR-1 protein and decoy (RefSeq Shewanella 70863) and common laboratory contaminant databases with Scaffold (Proteome Software, Inc.). Proteins with fewer than two spectra overall or more than one missed cleavage were excluded from analysis. The protein threshold was set at 99.0% minimum, a protein false-discovery rate (FDR) of 9%, a peptide threshold of 95.0% minimum, and a peptide FDR of 0.3%. The abundance of proteins trapped by ClpPTrap for each sample was quantified by evaluating both exclusive spectrum counts and the percentage of total spectra for each protein. Complete Scaffold results are listed in Table S3. Protein cofactors were predicted using Uniprot (77) and the Conserved Domain Database (78).
Supplementary Material
ACKNOWLEDGMENTS
We thank LeeAnn Higgins and Todd Markowski at the Center for Mass Spectrometry and Proteomics at the University of Minnesota for proteomics guidance and mass spectrometry analysis. We also thank Rebecca Maysonet for help with E. coli work.
This work was supported by the Office of Naval Research (N00014-13-1-0552 to J.A.G.). B.D.B. was supported in part by the University of Minnesota Biotechnology Training Grant Program through the National Institutes of Health. K.E.R. was supported by a MnDRIVE Seed Grant for Undergraduate Scholars.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00671-17.
REFERENCES
- 1.Myers CR, Nealson KH. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 2.Nealson KH, Myers CR, Wimpee BB. 1991. Isolation and identification of manganese-reducing bacteria and estimates of microbial Mn(IV)-reducing potential in the Black Sea. Deep Sea Res 38:S907–S920. doi: 10.1016/S0198-0149(10)80016-0. [DOI] [Google Scholar]
- 3.Brettar I, Höfle MG. 1993. Nitrous oxide producing heterotrophic bacteria from the water column of the central Baltic: abundance and molecular identification. Mar Ecol Prog Ser 94:253–265. doi: 10.3354/meps094253. [DOI] [Google Scholar]
- 4.Samuelsson MO. 1985. Dissimilatory nitrate reduction to nitrate, nitrous oxide, and ammonium by Pseudomonas putrefaciens. Appl Environ Microbiol 50:812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirodkar S, Reed S, Romine Saffarini MD. 2011. The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environ Microbiol 13:108–115. doi: 10.1111/j.1462-2920.2010.02313.x. [DOI] [PubMed] [Google Scholar]
- 6.Kostka JE, Nealson KH. 1995. Dissolution and reduction of magnetite by bacteria. Environ Sci Technol 29:2535–2540. doi: 10.1021/es00010a012. [DOI] [PubMed] [Google Scholar]
- 7.Canfield DE. 1989. Reactive iron in marine sediments. Geochim Cosmochim Acta 53:619–632. doi: 10.1016/0016-7037(89)90005-7. [DOI] [PubMed] [Google Scholar]
- 8.Schwertmann U. 1991. Solubility and dissolution of iron oxides. Plant Soil 130:1–25. doi: 10.1007/BF00011851. [DOI] [Google Scholar]
- 9.O'Reilly SE, Watkins J, Furukawa Y. 2005. Secondary mineral formation associated with respiration of nontronite, NAu-1 by iron reducing bacteria. Geochem Trans 6:67–76. doi: 10.1186/1467-4866-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riordan JF. 1977. The role of metals in enzyme activity. Ann Clin Lab Sci 7:119–129. [PubMed] [Google Scholar]
- 11.Moore B, Hawkes JL. 1908. An investigation of the toxic actions of dilute solutions of the salts of certain heavy metals (viz.: copper, iron, nickel, cobalt, manganese, zinc, silver, and lead) upon the Bacillus typhosus, with a view to practical application in the purification of shell-fish. Biochem J 3:313–345. doi: 10.1042/bj0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stohs SJ, Bagchi D. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med 18:321–336. doi: 10.1016/0891-5849(94)00159-H. [DOI] [PubMed] [Google Scholar]
- 13.Dunning JC, Ma Y, Marquis RE. 1998. Anaerobic killing of oral streptococci by reduced, transition metal cations. Appl Environ Microbiol 64:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton HC. 1985. Efficiency of chelated iron compounds as catalysts for the Haber-Weiss reaction. J Free Radic Biol Med 1:195–202. doi: 10.1016/0748-5514(85)90118-7. [DOI] [PubMed] [Google Scholar]
- 15.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol 177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman T, Kaplan B, Chava S, Parparova R, Nishri A. 1993. Effects of iron and chelation on Lake Kinneret bacteria. Microb Ecol 26:1–8. doi: 10.1007/BF00166024. [DOI] [PubMed] [Google Scholar]
- 17.Kersters I, Verstraete W. 1996. Inactivation of Aeromonas hydrophila by Fe(II)-related-radical generation in oxidizing groundwaters. Appl Environ Microbiol 62:3277–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hantke K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet 182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 19.Wan XF, Verberkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu L, Liu X, Yan T, Leaphart A, Hettich RL, Zhou J, Thompson DK. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J Bacteriol 186:8385–8400. doi: 10.1128/JB.186.24.8385-8400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Harris DP, Luo F, Wu L, Parsons AB, Palumbo AV, Zhou J. 2008. Characterization of the Shewanella oneidensis Fur gene: roles in iron and acid tolerance response. BMC Genomics 9(Suppl 1):S11. doi: 10.1186/1471-2164-9-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett BD, Brutinel ED, Gralnick JA. 2015. A ferrous iron exporter mediates iron resistance in Shewanella oneidensis MR-1. Appl Environ Microbiol 81:7938–7944. doi: 10.1128/AEM.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojtkowiak D, Georgopoulos C, Zylicz M. 1993. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem 268:22609–22617. [PubMed] [Google Scholar]
- 23.Hwang BJ, Park WJ, Chung CH, Goldberg AL. 1987. Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc Natl Acad Sci U S A 84:5550–5554. doi: 10.1073/pnas.84.16.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama-Fujimura Y, Gottesman S, Maurizi MR. 1987. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem 262:4477–4485. [PubMed] [Google Scholar]
- 25.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. 2003. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11:671–683. doi: 10.1016/S1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 26.Gottesman S, Clark WP, Maurizi MR. 1990. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate J Biol Chem 265:7886–7893. [PubMed] [Google Scholar]
- 27.Chung CH, Goldberg AL. 1981. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc Natl Acad Sci U S A 78:4931–4935. doi: 10.1073/pnas.78.8.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charette MF, Henderson GW, Markovitz A. 1981. ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc Natl Acad Sci U S A 78:4728–4732. doi: 10.1073/pnas.78.8.4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuang SE, Burland V, Plunkett G III, Daniels DL, Blattner FR. 1993. Sequence analysis of four new heat-shock genes constituting the hslTS/ibpAB and hslVU operons in Escherichia coli. Gene 134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 30.Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D'Ari R, Bouloc P. 1993. Cell growth and a phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci U S A 90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweder T, Lee KH, Lomovskaya O, Matin A. 1996. Regulation of Escherichia coli starvation sigma factor (σs) by ClpXP protease. J Bacteriol 178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neher SB, Villén J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, Baker TA. 2006. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol Cell 22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Buczek MS, Cardenas Arevalo AL, Janakiraman A. 2016. ClpXP and ClpAP control the Escherichia coli division protein ZapC by proteolysis. Microbiology 162:909–920. doi: 10.1099/mic.0.000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynn JM, Levchenko I, Sauer RT, Baker TA. 2004. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev 18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gottesman S, Roche E, Zhou Y, Sauer RT. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev 12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kammler M, Schön C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandu D, Nandi D. 2004. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res Microbiol 155:710–719. doi: 10.1016/j.resmic.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Frees D, Savijoki K, Varmanen P, Ingmer H. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol 63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- 40.Voos W, Jaworek W, Wilkening A, Bruderek M. 2016. Protein quality control at the mitochondrion. Essays Biochem 60:213–225. doi: 10.1042/EBC20160009. [DOI] [PubMed] [Google Scholar]
- 41.Karzai AW, Susskind MM, Sauer RT. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J 18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levchenko I, Seidel M, Sauer RT, Baker TA. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 43.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol Microbiol 86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 44.Ades SE, Connolly LE, Alba BM, Gross CA. 1999. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito A, Hizukuri Y, Matsuo E, Chiba S, Mori H, Nishimura O, Ito K, Akiyama Y. 2011. Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc Natl Acad Sci U S A 108:13740–13745. doi: 10.1073/pnas.1108376108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Las Peñas A, Connolly L, Gross CA. 1997. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol Microbiol 24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 47.Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci U S A 97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. 2006. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 49.De Las Peñas A, Connolly L, Gross CA. 1997. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol 179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia W, Li H, Sze KH, Sun H. 2009. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J Am Chem Soc 131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 51.Konopka K. 1978. Differential effects of metal-binding agents on the uptake of iron from transferrin by isolated rat liver mitochondria. FEBS Lett 92:308–312. doi: 10.1016/0014-5793(78)80776-5. [DOI] [PubMed] [Google Scholar]
- 52.Floyd RA, Lewis CA. 1983. Hydroxyl free radical formation from hydrogen peroxide by ferrous iron-nucleotide complexes. Biochemistry 22:2645–2649. doi: 10.1021/bi00280a008. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita MM, Wesson L, Eisenman G, Eisenberg D. 1990. Where metal ions bind in proteins. Proc Natl Acad Sci U S A 87:5648–5652. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irving H, Williams RJP. 1953. The stability of transition-metal complexes. J Chem Soc 1953:3192–3210. doi: 10.1039/jr9530003192. [DOI] [Google Scholar]
- 55.Leimkühler S, Wuebbens MM, Rajagopalan KV. 2011. The history of the discovery of the molybdenum cofactor and novel aspects of its biosynthesis in bacteria. Coord Chem Rev 255:1129–1144. doi: 10.1016/j.ccr.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Argüello JM, Raimunda D, Padilla-Benavides T. 2013. Mechanisms of copper homeostasis in bacteria. Front Cell Infect Microbiol 3:73. doi: 10.3389/fcimb.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira GC, Franco R, Lloyd SG, Moura I, Moura JJG, Huynh BH. 1995. Structure and function of ferrochelatase. J Bioenerg Biomembr 27:221–229. doi: 10.1007/BF02110037. [DOI] [PubMed] [Google Scholar]
- 58.Brayman TG, Hausinger RP. 1996. Purification, characterization, and functional analysis of a truncated Klebsiella aerogenes UreE urease accessory protein lacking the histidine-rich carboxyl terminus. J Bacteriol 178:5410–5416. doi: 10.1128/jb.178.18.5410-5416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song HK, Mulrooney SB, Huber R, Hausinger RP. 2001. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J Biol Chem 276:49359–49364. doi: 10.1074/jbc.M108619200. [DOI] [PubMed] [Google Scholar]
- 60.Raux E, Thermes C, Heathcote P, Rambach A, Warren MJ. 1997. A role for Salmonella typhimurium cbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J Bacteriol 179:3202–3212. doi: 10.1128/jb.179.10.3202-3212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raux E, Lanois A, Rambach A, Warren MJ, Thermes C. 1998. Cobalamin (vitamin B12) biosynthesis: functional characterization of the Bacillus megaterium cbi genes required to convert uroporphyrinogen III into cobyrinic acid a,c-diamide. Biochem J 335:167–173. doi: 10.1042/bj3350167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, Robinson NJ. 2008. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 63.Hung H-C, Chang G-G. 2001. Differentiation of the slow-binding mechanism for magnesium ion activation and zinc ion inhibition of human placental alkaline phosphatase. Protein Sci 10:34–45. doi: 10.1110/ps.35201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foster AW, Osman D, Robinson NJ. 2014. Metal preferences and metallation. J Biol Chem 289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hmiel SP, Snavely MD, Florer JB, Maguire ME, Miller CG. 1989. Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J Bacteriol 171:4742–4751. doi: 10.1128/jb.171.9.4742-4751.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Snavely MD, Florer JB, Miller CG, Maguire ME. 1989. Magnesium transport in Salmonella typhimurium: Mg2+ transport by the CorA, MgtA, and MgtB systems. J Bacteriol 171:4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niegowski D, Eshaghi S. 2007. The CorA family: structure and function revisited. Cell Mol Life Sci 64:2564–2574. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bird LJ, Coleman ML, Newman DK. 2013. Iron and copper act synergistically to delay anaerobic growth of bacteria. Appl Environ Microbiol 79:3619–3627. doi: 10.1128/AEM.03944-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chapman E, Farr GW, Usaite R, Furtak K, Fenton WA, Chaudhuri TK, Hondorp ER, Matthews RG, Wolf SG, Yates JR, Pypaert M, Horwich AL. 2006. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci U S A 103:15800–15805. doi: 10.1073/pnas.0607534103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hau HH, Gilbert A, Coursolle D, Gralnick JA. 2008. Mechanism and consequences of anaerobic respiration of cobalt by Shewanella oneidensis strain MR-1. Appl Environ Microbiol 74:6880–6886. doi: 10.1128/AEM.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. 1979. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci U S A 105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouhenni R, Gehrke A, Saffarini D. 2005. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Appl Environ Microbiol 71:4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Opijnen T, Camilli A. 2010. Genome-wide fitness and genetic interactions determined by Tn-Seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol Chapter 1:Unit1E.3. doi: 10.1002/9780471729259.mc01e03s19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43(Database issue):D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coursolle D, Gralnick JA. 2010. Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol Microbiol 77:995–1008. doi: 10.1111/j.1365-2958.2010.07266.x. [DOI] [PubMed] [Google Scholar]
- 80.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 81.Waldron KJ, Robinson NJ. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.