Abstract
We have shown previously that at physiologically relevant oxygen tension (pO2 ≈ 10 mmHg), NO S-nitrosylates 1 of ≈50 free cysteines per ryanodine receptor 1 (RyR1) subunit and transduces a calcium-sensitizing effect on the channel by means of calmodulin (CaM). It has been suggested that cysteine-3635 is part of a CaM-binding domain, and its reactivity is attenuated by CaM [Porter Moore, C., Zhang, J. Z., Hamilton, S. L. (1999) J. Biol. Chem. 274, 36831–36834]. Therefore, we tested the hypothesis that the effect of NO was mediated by C3635. The full-length RyR1 single-site C3635A mutant was generated and expressed in HEK293 cells. The mutation resulted in the loss of CaM-dependent NO modulation of channel activity and reduced S-nitrosylation by NO to background levels but did not affect NO-independent channel modulation by CaM or the redox sensitivity of the channel to O2 and glutathione. Our results reveal that different cysteines within the channel have been adapted to serve in nitrosative and oxidative responses, and that S-nitrosylation of the cysteine-containing CaM-binding domain underlies the mechanism of CaM-dependent regulation of RyR1 by NO.
The gaseous messenger nitric oxide (NO) has been proposed to exert regulatory effects on ion channels by modifying the redox state of critical thiols (1–3). NO-sensitive ion channels include the cardiac and skeletal muscle ryanodine receptors (RyRs; refs. 4–10), L-type Ca2+ channel (11), Na+ channel in baroreceptors (12), cyclic nucleotide-gated channel (13), Ca2+- or voltage-gated K+ channel (14, 15), and N-methyl-d-aspartate receptor channel complex (16, 17).
The skeletal muscle Ca2+ release channel/ryanodine receptor (RyR1) contains a large number of free thiols whose oxidation or nitrosylation influences channel function (9, 18–20). RyR1 redox state and activity depend on O2 tension (9), transmembrane glutathione redox potential (21), and effector molecules (Ca2+, Mg2+) that control RyR1 channel activity (22). Nearly half of the 404 cysteines within the tetrameric RyR1 channel complex are maintained in a reduced (free-thiol) state under redox conditions comparable to resting muscle (9). Changes in O2 tension dynamically oxidize/reduce as many as six to eight thiols per RyR1 subunit and, thereby, control channel responsiveness to physiological concentrations of NO. NO increases RyR1 activity by S-nitrosylation of one thiol per RyR1 subunit, and this effect is transduced by calmodulin (CaM; ref. 9).
CaM has a dual effect on skeletal muscle Ca2+ release channel activity. CaM activates the channel at Ca2+ concentrations below 1 μM, and inhibits the channel at Ca2+ concentrations above 1 μM (23). The skeletal muscle Ca2+ release channel binds one CaM per RyR1 subunit in the absence and presence of Ca2+, which suggests that both the Ca2+-free (apoCaM) and Ca2+-bound (CaCaM) forms of CaM bind to RyR1 (24, 25). Trypsin cleavage and analysis (24, 26) and site-directed mutagenesis (27) suggest that apoCaM and CaCaM have overlapping binding sites that mask C3635 from reaction with N-ethylmaleimide (24, 26) and that involve leucine-3624 (27). These data suggest that C3635 is part of the CaM-binding domain.
In the present study, we tested the hypothesis that C3635 is responsible for the CaM-dependent effects of NO on RyR1. We substituted an alanine residue for C3635 and expressed the mutant protein in HEK293 cells. Chemical analysis and immunoblotting with an anti-S-nitrosocysteine antibody revealed in vitro S-nitrosylation of wild-type (wt) RyR1 but not of the C3635A mutant. Moreover, functional studies with the mutant RyR showed a loss of NO responsiveness. These results indicate that NO mediates its effects on RyR1 by S-nitrosylation of C3635.
Experimental Procedures
Materials.
[35S]CaM (5 Ci/mmol; 1 Ci = 37 GBq) was prepared by using Tran35S-label (ICN) as described (25). [3H]Ryanodine (50–100 Ci/mmol) was purchased from Perkin–Elmer. Unlabeled ryanodine and myosin light chain kinase-derived CaM-binding peptide (CaMBP) were obtained from Calbiochem; CaM was obtained from Sigma; leupeptin, Pefabloc (a protease inhibitor), and 3,3-diaminobenzidine substrate kit were from Roche Molecular Biochemicals. NO gas (purity > 99%, AGA Specialty Gas, Maumee, OH) was scrubbed to remove O2 and nitrite by passing it through an argon-purged column filled with KOH pellets; the gas was then bubbled through a solution of NaOH. The concentration of NO dissolved in the argon-purged buffer was determined by using a hemoglobin titration assay (9). All other chemicals were of analytical grade.
Construction and Expression of C3635A Mutant.
The construction of the full-length rabbit RyR1 cDNA has been described (28). A 2,706-bp fragment (PvuI/NdeI, 8465/11171), excised from a 6.7-kb fragment of RyR1 in pBluescript II KS+ (PvuI-XbaI, 8465/11171) and cloned into pBluescript II KS+ vector, served as the template for mutagenesis. The C3635A mutation was prepared by introducing a double base change (TG to GC) by Pfu DNA polymerase-based chain reaction by using two mutagenic primers (forward primer: 5′-agccgtggtggccGCcttccgtatgacg-3′; reverse primer: 5′-cgtcatacggaagGCggccaccacggct-3′) and the QuickChange site-directed mutagenesis kit (Stratagene). The mutated sequence was confirmed by sequencing and reintroduced into the PvuI and XbaI sites of the 6.7-kb fragment in KS+. The mutated, full-length expression plasmid was prepared by the ligation of two fragments—ClaI/PvuI and PvuI/XbaI (containing the mutated sequence)—in the expression vector pCMV5 (ClaI/XbaI). wt and mutant RyR1 cDNAs were transiently expressed in HEK293 cells by using Effectene (Qiagen, Chatsworth, CA), according to the manufacturer's instructions. Cells were maintained in high-glucose DMEM containing 10% (vol/vol) FBS at 37°C under a 5% CO2/95% air atmosphere and plated the day before transfection. For each 10-cm tissue-culture dish, 6 μg of DNA was used at a DNA/Effectene ratio of 1:5. Cells were harvested 42–46 hr after transfection, and crude membrane fractions were prepared as described (29).
Isolation of Skeletal Sarcoplasmic Reticulum (SR) Vesicles.
SR membrane fractions enriched with RyR1 were prepared from rabbit skeletal muscle in the presence of protease inhibitors (100 nM aprotinin/1 μM leupeptin/1 μM pepstatin/0.2 mM phenylmethylsulfonyl fluoride/1 mM benzamidine) as described (30).
Quantification of RyR1 S-Nitrosothiol (SNO) Content.
The SNO content of the purified wt and C3635A mutant RyR1s was determined by a photolysis/chemiluminescence method as described (9).
Electrophoresis and Western Blotting.
All procedures were performed under nonreducing conditions. Proteins were denatured for 5 min at 95–100°C and separated on SDS/3–12% polyacrylamide gels and transferred to Immobilon-P membranes (28). Membranes were blocked in PBS buffer containing 0.05% Tween-20 and 5% nonfat milk, and transferred proteins were made to react with either anti-RyR1 monoclonal antibody D110 (29) or an anti-S-nitrosocysteine polyclonal antibody (1:200–1:500 dilution, Calbiochem). Bound antibodies were detected with goat anti-mouse or anti-rabbit IgG-horseradish peroxidase-linked antibody, respectively, with a 3,3-diaminobenzidine substrate kit.
[3H]Ryanodine Binding.
Unless otherwise indicated, skeletal SR vesicles (0.2 mg/ml) or microsomal membrane fractions containing the expressed RyR1s (0.4 mg/ml) were exposed to various concentrations of redox reagents and then incubated for 5 hr at 24°C with 5 nM [3H]ryanodine in media containing 0.125 M KCl, 20 mM imidazole (pH 7.0), 0.3 mM Pefabloc, 30 μM leupeptin, and the indicated concentrations of CaM and free Ca2+. Nonspecific binding was determined by using a 1,000-fold excess of unlabeled ryanodine. Aliquots of the samples were diluted with 20 volumes of ice-cold water and placed on Whatman GF/B filters soaked with 2% (wt/wt) polyethyleneimine. Filters were washed with three 5-ml vol of ice-cold 0.1 M KCl/1 mM KPipes (pH 7) buffer, and the radioactivity remaining on the filters was determined by liquid scintillation counting to obtain bound [3H]ryanodine.
[35S]CaM Binding.
Membrane fractions were incubated for 2 hr at 24°C with 5 or 15 nM [35S]CaM in 250 mM KCl/20 mM imidazole, pH 7.0/0.3 M sucrose/0.2 mg/ml BSA/5 mM reduced glutathione (GSH)/25 μM leupeptin/0.25 mM Pefabloc and either 5 mM EGTA (for apoCaM binding) or 100 μM free Ca2+ (for CaCaM binding). Aliquots were taken for determination of total radioactivity and centrifuged for 30 min at 30 psi (120,000 × g) in a Beckman Airfuge (Beckman Coulter) to obtain bound [35S]CaM. Nonspecific binding of [35S]CaM was determined by incubating equal amounts of membranes derived from vector or nontransfected HEK293 cells. In parallel experiments, Bmax values of [3H]ryanodine binding were determined by a 5-hr incubation of membrane fractions at 24°C with 30 nM [3H]ryanodine in 0.6 M KCl/20 mM imidazole, pH 7.0/25 μM leupeptin/0.25 mM Pefabloc/100 μM free Ca2+. Radioactivities were determined by scintillation counting.
Data Analysis.
Results are given as means ± SD. Significance of differences of data were analyzed with Student's t test. Differences were considered to be statistically significant at P < 0.05.
Results
C3635 Is Responsible for Modulation of RyR1 by NO.
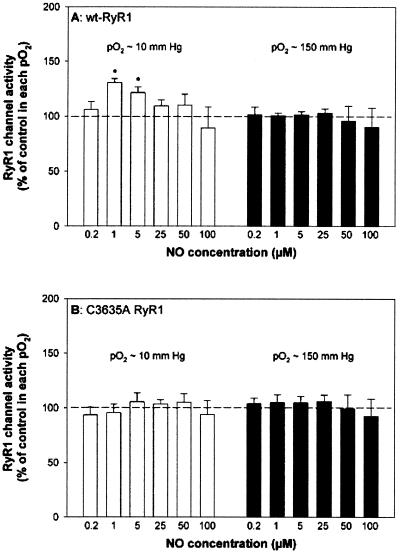
We have shown that at skeletal muscle O2 tension (pO2 ≈ 10 mmHg), submicromolar NO activates the native RyR1 by S-nitrosylation of one cysteine per RyR1 subunit, and that activation requires the presence of CaM. At ambient O2 tension (pO2 ≈ 150 mmHg), submicromolar NO neither S-nitrosylated nor activated the native RyR1 (9). The same strategy was used to explore the action of NO on the C3635A RyR1 mutant. wt and C3635A mutant RyR1s were expressed in HEK293, and the effects of a range of NO concentrations were tested by determining the [3H]ryanodine-binding levels of the expressed RyR1s at equilibrium in membrane fractions at pO2 ≈ 10 mmHg and pO2 ≈ 150 mmHg. Ryanodine is a highly specific plant alkaloid that is widely used as a probe of channel activity because of its preferential binding to the open RyR channel states (31, 32). As shown in Fig. 1A, wt RyR1 was activated by low concentrations of NO with a peak of [3H]ryanodine binding at 1 μM NO in pO2 ≈ 10 mmHg. In agreement with our previous study with the native RyR1 (9), NO concentrations greater than 5 μM reversed the activation of wt RyR1, indicating additional modifications by NO. No change in activity was observed when membranes were freed of endogenous CaM (see also Fig. 4) by pretreatment with 1 μM CaMBP followed by centrifugation to remove complexed CaMBP and CaM (25). Thus, wt RyR1 activation by physiological amounts of NO depended on CaM. wt RyR1 channel activity was not changed by NO at pO2 ≈ 150 mmHg, and in contrast to wt RyR1, the C3635A mutant did not respond to NO at either oxygen tension (Fig. 1B).
Figure 1.
C3635A RyR1 does not respond to NO. Specific [3H]ryanodine binding to membrane fractions prepared from HEK293 cells expressing wt-RyR1 (A) or mutant C3635A RyR1 (B) was determined in the presence of 10 μM free Ca2+ and increasing concentrations of NO in pO2 ≈ 10 mmHg (white bars) or pO2 ≈ 150 mmHg (black bars). [3H]Ryanodine-binding values, determined as described in Experimental Procedures, ranged from 0.05 to 0.15 pmol/mg protein for both wt-RyR1s and C3635A RyR1s. Normalized [3H]ryanodine binding data (100% in the absence of NO for each condition) are the means ± SD of five experiments. *, P < 0.05 when compared with controls in pO2 ≈ 10 mmHg and pO2 ≈ 150 mmHg.
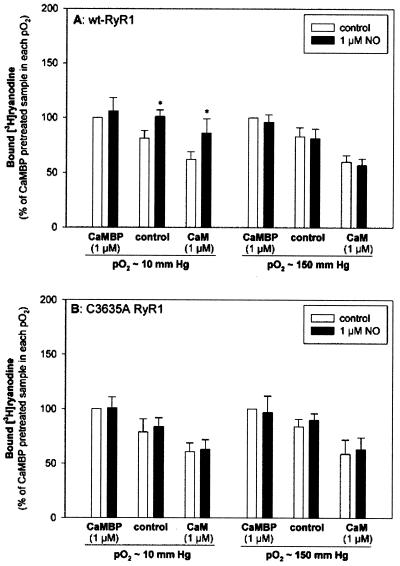
Figure 4.
Effects of CaM and NO on wt-RyR1 and C3635A RyR1 activities. Membrane fractions containing wt-RyR1 (A) or C3635A mutant RyR1 (B) (control) were preincubated with 1 μM CaMBP to sequester endogenous CaM (CaMBP) or were preincubated with 1 μM exogenous CaM (CaM). [3H]Ryanodine binding was assayed with 10 μM free Ca2+ in the absence (white bars) or the presence (black bars) of 1 μM NO in both pO2 ≈ 10 and pO2 ≈ 150 mmHg. Normalized [3H]ryanodine-binding data, expressed as percent of the binding values of membranes pretreated with CaMBP (NO), are the means ± SD of four to six experiments. *, P < 0.05 when compared with controls in pO2 ≈ 10 and pO2 ≈ 150 mmHg.
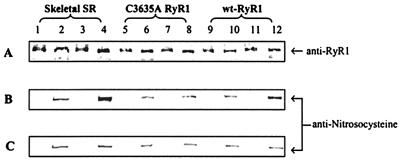
We used an anti-S-nitrosocysteine polyclonal antibody to determine whether NO S-nitrosylated the RyR1 C3635A mutant channel. The antibody detected a faint signal in the SR control sample (−NO) (Fig. 2 B and C, lane 2) in a region of the immunoblot containing the RyR1, as determined by an anti-RyR1 antibody (Fig. 2A). Specificity of the antibody for SNO was provided by treatment with HgCl2 that nearly eliminated the signal (Fig. 2, odd-numbered lanes). NO (1 μM) significantly increased the level of S-nitrosylation of the native RyR1 in pO2 ≈ 10 mmHg (Fig. 2B, lane 4) but not pO2 ≈ 150 mm (Fig. 2C, lane 4), which is in agreement with our previous study showing S-nitrosylation of the native RyR1 at low but not ambient oxygen tension (9). wt RyR1 showed a response to NO comparable to the native RyR1, which likewise exhibited an increased level of endogenous S-nitrosylation only in pO2 ≈ 10 mmHg (Fig. 2 B and C, lane 12). In contrast, NO did not increase noticeably the low levels of endogenous immunoreactivity of the C3635A RyR1 mutant at either low or high oxygen tension (Fig. 2 B and C, lanes 6 and 8).
Figure 2.
Immunoblots of nitrosocysteine. Skeletal SR vesicles (lanes 1–4, 10 μg each lane) and cell membrane fractions containing C3635A RyR1 (lanes 5–8, 40 μg each lane) or wt-RyR1 (lanes 9–12, 40 μg each lane) were incubated for 1 hr at 24°C in pO2 ≈ 10 mmHg or pO2 ≈ 150 mmHg in the presence of 1 μM NO. Anti-nitrosocysteine negative controls (treated with 0.1 mmHgCl2, odd-numbered lanes), samples without NO (lanes 2, 6, and 10), and samples treated with 1 μM NO (lanes 3 and 4, 7 and 8, 11 and 12) at pO2 ≈ 10 mmHg (B) and pO2 ≈ 150 mmHg (C) were separated by SDS/3–12% PAGE gradient. The proteins were transferred to Immobilon-P membranes overnight at 4°C. A polyclonal anti-nitrosocysteine antibody was used to detect an S-nitrosylation signal in the protein band region of RyR1 probed with D110 monoclonal anti-RyR1 (A). The data are representative of three experiments.
We also measured the extent of S-nitrosylation of the wt and C3635A mutant RyR1 channels with a photolysis/chemiluminescence method (9). wt and C3635A mutant RyR1 membrane fractions were treated with 1 μM NO at pO2 ≈ 10 mmHg. The receptors were solubilized in 1.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), without (controls), or after NO treatment for 1 hr at 24°C, and isolated by sucrose density-gradient centrifugation. Purified, control wt and C3635A mutant RyR1s showed essentially no signal (<0.01 SNO per RyR1 subunit). Purified, NO-treated wt and C3635A mutant RyR1s contained 1.15 ± 0.60 and 0.34 ± 0.25 SNO per subunit, respectively (n = 3), or a difference of ≈ 1 SNO per subunit.
CaM and Redox Modulation of C3635A.
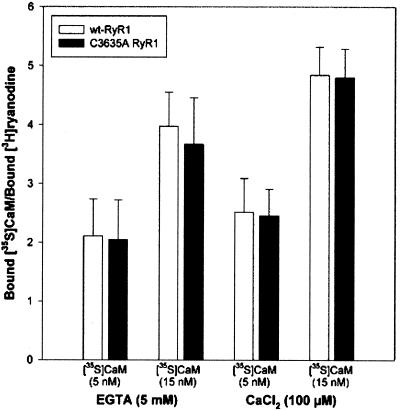
Recent studies have shown that NO activation of RyR1 depends on the presence of CaM (9). Moore et al. (26) showed that C3635 is alkylated by N-ethylmaleimide and that this reaction is blocked by CaM. In the present study, we examined whether the C3635-to-A substitution in RyR1 altered the channel's regulation by CaM and redox-active molecules. In Fig. 3, CaM binding to wt and binding to the C3635A mutant RyR1s are compared, by using [35S]CaM concentrations that are near and above the dissociation constants of apoCaM and CaCaM binding to the native, purified RyR1 (25). Cells expressing wt RyR1 bound ≈4 apo[35S]CaM and 5 Ca[35S]CaM per [3H]ryanodine-binding site. As there is only one high-affinity [3H]ryanodine-binding site per RyR1 tetramer, these ratios correspond to ≈1 apoCaM- and 1 CaCaM-binding site per RyR1 subunit. The binding of apo[35S]CaM and Ca[35S]CaM to C3635A was not different from wt RyR1, which indicates that C3635 is not crucial for conferring high-affinity apoCaM and CaCaM binding.
Figure 3.
[35S]CaM binding to wt-RyR and C3635A RyR. Membrane fractions prepared from HEK293 cells and expressing wt-RyR1 (white bars) or C3635A mutant RyR1 (black bars) were incubated with 5 or 15 nM [35S]CaM and either 5 mM EGTA or 100 μM Ca2+. The ratios of [35S]CaM-binding values to maximal binding values of [3H]ryanodine are shown. Maximal values of [3H]ryanodine binding were determined as described under Experimental Procedures. Data are the means ± SD of three experiments.
Next, we examined whether the C3635A mutation affected either CaM inhibition of [3H]ryanodine occupancy or CaM-dependent modulation of RyR1 activity by NO (Fig. 4). wt and C3635A mutant RyR1 membrane fractions were pretreated with 1 μM CaMBP to sequester endogenous CaM (25) or were preincubated with 1 μM exogenous CaCaM before being exposed to 1 μM NO in pO2 ≈ 10 or pO2 ≈ 150 mmHg. Pretreatment with CaMBP increased [3H]ryanodine binding, whereas the addition of exogenous CaCaM had an inhibitory effect. These results are in agreement with previous studies showing the inhibition of RyR1 activity by CaCaM (25). Fig. 4 further shows that the C3635A mutation did not eliminate inhibition of RyR1 activity by CaCaM. Fig. 4 also shows that in the CaMBP pretreated samples, 1 μM NO had no effect on [3H]ryanodine binding. In the presence of 1 μM CaM, however, NO increased wt RyR1 activity in pO2 ≈ 10 mmHg but not in pO2 ≈ 150 mmHg, which is in agreement with studies on the native RyR1 (9). NO did not activate significantly the mutant channel in the presence of CaM in either oxygen tension. Thus, C3635 is required for the CaM-dependent modulation of RyR1 by NO.
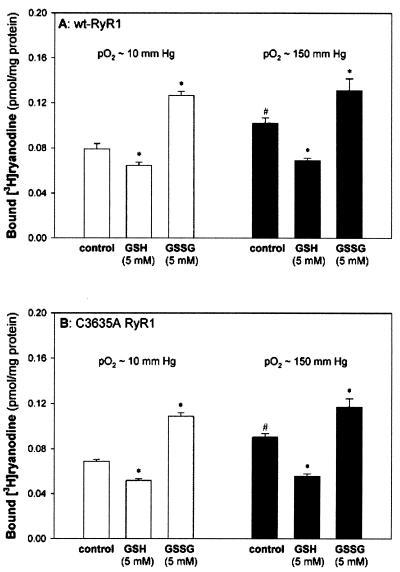
To address the question of whether C3635 is required for the general effects of redox-related molecules on the channel, we explored the effects of oxygen tension and GSH or oxidized glutathione (GSSG) on wt and C3635A mutant RyR1 activities (Fig. 5). We found that in both wt and mutant RyR, [3H]ryanodine binding was higher in ambient (pO2 ≈ 150 mmHg) than in biologically relevant muscle oxygen tension (pO2 ≈ 10 mmHg), and, likewise in both cases, [3H]ryanodine binding was decreased by GSH and increased by GSSG (irrespective of O2 tension). Similar data have been reported when SR vesicles were used (9, 20, 33). In other studies, we found that the mutant channel displayed a Ca2+ activation/inactivation profile (31, 32) that was essentially identical to wt RyR1 (data not shown). Taken together, the results of Figs. 4 and 5 suggest that the C3635A mutation does not eliminate the channel's response to physiological effectors, in particular, Ca2+, CaM, oxygen tension, and GSH or GSSG.
Figure 5.
Effect of oxygen tension and GSH/GSSG on wt-RyR1 and C3635A RyR1 activities. Specific [3H]ryanodine binding to membrane fractions prepared from HEK293 cells expressing wt-RyR1 (A) or C3635A RyR1 (B) was determined in 10 μM free Ca2+ medium in the absence or presence of 5 mM GSH or 5 mM GSSG and either in pO2 ≈ 10 mmHg (white bars) or pO2 ≈ 150 mmHg (black bars). [3H]Ryanodine-binding data are the means ± SD of three to six experiments. *, P < 0.05 compared with the controls in pO2 ≈ 10 mmHg and ≈ 150 mmHg, respectively. #, P < 0.05 compared with controls in pO2 ≈ 10 mmHg.
Discussion
We report that a single cysteine (C3635) is responsible for sensitizing the skeletal muscle Ca2+ release channel to NO. This finding is particularly remarkable given the unusually large number of reactive cysteines (≈50 free thiols per RyR1 subunit) in the channel. S-nitrosylation of a single cysteine also has been shown to be necessary and sufficient for NO responsiveness in cyclic nucleotide-gated channels (13), the N-methyl-d-aspartate receptor-channel complex (17), p21 ras (34), and hemoglobin (35). The site of posttranslational modification in these proteins is identified by an acid–base consensus motif, revealed in the primary sequence, i.e., cysteine is flanked on either side by lysine, arginine, histidine, aspartate, or glutamate [and, in the prototype case, is followed by an acidic amino acid residue, aspartate or glutamate, at the +1 (C-terminal) position] (13, 17, 36). The C3635 in RyR1 (AVVACF; refs. 26 and 37) lacks flanking acidic or basic residues. It has been shown recently, however, that the acid–base motif for S-nitrosylation may emerge from the tertiary structure, as illustrated in the cases of methionine adenosyltransferase (38) and caspase-3 (39) or even from protein–protein interactions. In addition, it has been suggested that hydrophobic compartments within or between proteins operationally define a “hydrophobic motif” for S-nitrosylation (40, 41); C3635 sits in such a hydrophobic pocket (41).
RyR1 was S-nitrosylated at low (10 mmHg) but not high (150 mmHg) O2 tension. We have shown that the O2 concentration determines the redox state of six to eight thiols per RyR subunit. Therefore, oxygen can be viewed as producing a preparatory modification of the channel that serves to facilitate S-nitrosylation. Our working model is that a redox-driven conformational change in the channel creates a local hydrophobic domain to concentrate NO/O2 for the purposes of generating nitrosylating equivalents. That is, the S-nitrosylation reaction likely occurs through intermediates formed by NO and O2 (42) and is accelerated in a hydrophobic compartment where NO and O2 concentrate (41–43).
S-nitrosylation of wt and C3635A mutant RyR1 was monitored with an anti-nitrosocysteine polyclonal antibody and by photolysis–chemiluminescence. Although the antibody provides a rapid and relatively sensitive means of detecting SNO groups in RyR1, we offer a word of caution because the antibody also detected a faint signal in the wt and C3635 control samples (−NO), although a more sensitive chemical analysis failed to confirm the presence of SNO groups. The finding that prior treatment with HgCl2 nearly eliminated these background signals suggests that the antibody also recognizes the unmodified thiol (and surrounding amino acid residues), although with much lower affinity.
The RyR1 C3635A mutation did not seem to introduce major global protein conformational changes, as the mutant channel showed a Ca2+ activation/inactivation profile comparable to wt RyR1. The functional consequences of the C3635A mutation also were assessed in CaM-binding studies and by the response to GSH/GSSG. CaM-binding studies with synthetic peptides (44) and site-directed mutagenic analysis (27) indicated that C3635 is a part of or near an RyR1 CaM-binding site that is partly shared by apoCaM and CaCaM. Moore et al. (26) found that N-[3H]ethylmaleimide labeling of C3635 is blocked by CaM, and conversely, that N-ethylmaleimide interferes with apo[35S]CaM binding, which suggested that C3635 may be important for CaM binding. Our data show that C3635A binds apo[35S]CaM and Ca[35S]CaM indistinguishably from wt RyR, and, therefore, indicate that the global conformation of the CaM-binding site is largely unchanged by mutagenesis, and that C3635 is not required for CaM binding. Also, we have shown that SNO modification, of what we now know to be C3635, does not displace CaM (9). A likely explanation for the difference between our results and those of Moore et al. (26) is that alkylation with N-ethylmaleimide introduces a bulky group at C3635, whereas mutagenesis and S-nitrosylation do not. Other studies have shown that RyR1 contains a large number of thiols whose redox state is controlled by oxygen tension and GSH/GSSG (9, 20–22, 33). We find that the C3635A mutation did not change the basic redox characteristics of RyR1 either; i.e., neither modulation by O2 tension nor modulation by glutathione is altered (Fig. 5). Taken together, these data suggest that C3635 does not contribute significantly to the redox modulation of RyR1 by glutathione or O2.
In conclusion, our studies show that a single amino acid substitution (C3635A) abolishes regulation of the skeletal muscle Ca2+-release channel by physiological concentrations of NO. C3635 is located within or near the CaM-binding domain of RyR1, which provides a mechanistic basis for the finding that NO regulation is CaM dependent and, more generally, implies that S-nitrosylation and CaM modulation of the RyR are irrevocably linked. Our data also rationalize the observation that S-nitrosylation of 1 of 50 thiols per RyR subunit can profoundly regulate the channel. It is intriguing to note that activation of constitutive NO synthase also depends on CaM. Thus, both the substrate of S-nitrosylation (RyR) and the catalytic source of S-nitrosylating equivalents (NO synthase) are regulated by the binding of CaM.
Acknowledgments
This work was supported by National Institutes of Health Grant HL04053 (to J.P.E), Grants HL52529 and HL59130 (to J.J.S), and Grants AR18687 and HL27430 (to G.M.).
Abbreviations
- RyR
ryanodine receptor
- RyR1
skeletal muscle isoform of RyR
- CaM
calmodulin
- apoCaM
Ca2+-free CaM
- CaCaM
Ca2+-bound CaM
- CaMBP
CaM-binding peptide
- wt
wild type
- SR
sarcoplasmic reticulum
- SNO
S-nitrosothiol
- GSH
reduced glutathione
- GSSG
oxidized glutathione
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Broillet M C. Cell Mol Life Sci. 1999;55:1036–1042. doi: 10.1007/s000180050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eu J P, Xu L, Stamler J S, Meissner G. Biochem Pharmacol. 1999;57:1079–1084. doi: 10.1016/s0006-2952(98)00360-8. [DOI] [PubMed] [Google Scholar]
- 3.Stamler J S, Meissner G. Physiol Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 4.Meszaros L G, Minarovic I, Zahradnikova A. FEBS Lett. 1996;380:49–52. doi: 10.1016/0014-5793(96)00003-8. [DOI] [PubMed] [Google Scholar]
- 5.Aghdasi B, Reid M B, Hamilton S L. J Biol Chem. 1997;272:25462–25467. doi: 10.1074/jbc.272.41.25462. [DOI] [PubMed] [Google Scholar]
- 6.Stoyanovsky D, Murphy T, Anno P R, Kim Y-M, Salama G. Cell Calcium. 1997;21:19–29. doi: 10.1016/s0143-4160(97)90093-2. [DOI] [PubMed] [Google Scholar]
- 7.Zahradnikova A, Minarovic I, Venema R C, Meszaros L G. Cell Calcium. 1997;22:447–453. doi: 10.1016/s0143-4160(97)90072-5. [DOI] [PubMed] [Google Scholar]
- 8.Xu L, Eu J P, Meissner G, Stamler J S. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 9.Eu J P, Sun J H, Xu L, Stamler J S, Meissner G. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 10.Hart J D E, Dulhunty A F. J Membr Biol. 2000;173:227–236. doi: 10.1007/s002320001022. [DOI] [PubMed] [Google Scholar]
- 11.Campbell D C, Stamler S, Strauss H C. J Gen Physiol. 1996;108:277–293. doi: 10.1085/jgp.108.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Chapleau M W, Bates J N, Bielefeldt K, Lee H C, Abboud F M. Neuron. 1998;20:1039–1049. doi: 10.1016/s0896-6273(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 13.Broillet M C. J Biol Chem. 2000;275:15135–15141. doi: 10.1074/jbc.275.20.15135. [DOI] [PubMed] [Google Scholar]
- 14.Bolotina V M, Najibi S, Palacino J, Pagano P, Cohen R A. Nature (London) 1994;368:850–853. doi: 10.1038/368850a0. [DOI] [PubMed] [Google Scholar]
- 15.Yuan X J, Tod M L, Rubin L J, Blaustein M P. Proc Natl Acad Sci USA. 1996;93:10489–10494. doi: 10.1073/pnas.93.19.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipton S A, Choi Y B, Pan Z H, Lei S Z, Sucher N J, Loscalzo J, Singel D J, Stamler J S. Nature (London) 1993;364:626–631. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y B, Tenneti L, Le D A, Ortiz J, Bai G, Chen H S V, Lipton S A. Nat Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- 18.Abramson J J, Salama G. J Bioenerg Biomembr. 1989;21:283–294. doi: 10.1007/BF00812073. [DOI] [PubMed] [Google Scholar]
- 19.Salama G, Menshikova E V, Abramson J J. Antioxid Redox Signal. 2000;2:5–16. doi: 10.1089/ars.2000.2.1-5. [DOI] [PubMed] [Google Scholar]
- 20.Sun J H, Xu L, Eu J P, Stamler J S, Meissner G. J Biol Chem. 2001;276:15625–15630. doi: 10.1074/jbc.M100083200. [DOI] [PubMed] [Google Scholar]
- 21.Feng W, Liu G, Allen P D, Pessah I N. J Biol Chem. 2000;275:35902–35907. doi: 10.1074/jbc.C000523200. [DOI] [PubMed] [Google Scholar]
- 22.Xia R H, Stangler T, Abramson J J. J Biol Chem. 2000;275:36556–36561. doi: 10.1074/jbc.M007613200. [DOI] [PubMed] [Google Scholar]
- 23.Tripathy A, Xu L, Mann G, Meissner G. Biophys J. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore C P, Rodney G, Zhang J Z, Santacruz-Toloza L, Strasburg G, Hamilton S L. Biochemistry. 1999;38:8532–8537. doi: 10.1021/bi9907431. [DOI] [PubMed] [Google Scholar]
- 25.Balshaw D M, Xu L, Yamaguchi N, Pasek D A, Meissner G. J Biol Chem. 2001;276:20144–20153. doi: 10.1074/jbc.M010771200. [DOI] [PubMed] [Google Scholar]
- 26.Moore C P, Zhang J Z, Hamilton S L. J Biol Chem. 1999;274:36831–36834. doi: 10.1074/jbc.274.52.36831. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi N, Xin C L, Meissner G. J Biol Chem. 2001;276:22579–22585. doi: 10.1074/jbc.M102729200. [DOI] [PubMed] [Google Scholar]
- 28.Gao L, Tripathy A, Lu X Y, Meissner G. FEBS Lett. 1997;412:223–226. doi: 10.1016/s0014-5793(97)00781-3. [DOI] [PubMed] [Google Scholar]
- 29.Gao L, Balshaw D B, Xu L, Tripathy A, Xin C L, Meissner G. Biophys J. 2000;79:828–840. doi: 10.1016/S0006-3495(00)76339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meissner G. J Biol Chem. 1984;259:2365–2374. [PubMed] [Google Scholar]
- 31.Meissner G. Annu Rev Physiol. 1994;56:485–508. doi: 10.1146/annurev.ph.56.030194.002413. [DOI] [PubMed] [Google Scholar]
- 32.Franzini-Armstrong C, Protasi F. Physiol Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 33.Zable A C, Favero T G, Abramson J J. J Biol Chem. 1997;272:7069–7077. doi: 10.1074/jbc.272.11.7069. [DOI] [PubMed] [Google Scholar]
- 34.Lander H M, Hajjar D P, Hempstead B L, Mirza U A, Chait B T, Campbell S, Quilliam L A. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 35.Gow A J, Stamler J S. Nature (London) 1998;391:169–173. doi: 10.1038/34402. [DOI] [PubMed] [Google Scholar]
- 36.Stamler J S, Toone E J, Lipton S A, Sucher N J. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 37.Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, Numa S. Nature (London) 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Mato I, Castro C, Ruiz F A, Corrales F J, Mato J M. J Biol Chem. 1999;274:17075–17079. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 39.Mannick J B, Hausladen A, Liu L, Hess D T, Zeng M, Miao Q X, Kane L S, Gow A J, Stamler J S. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 40.Nedospasov A, Rafikov R, Beda N, Nudler E. Proc Natl Acad Sci USA. 2000;97:13543–13548. doi: 10.1073/pnas.250398197. . (First Published November 28, 2000; 10.1073/pnas.250398197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess D T, Matsumoto A, Nudelman R, Stamler J S. Nat Cell Biol. 2001;3:E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- 42.Stamler J S, Hausladen A. Nat Struct Biol. 1998;5:247–249. doi: 10.1038/nsb0498-247. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Miller M J S, Joshi M S, Thomas D D, Lancaster J R., Jr Proc Natl Acad Sci USA. 1998;95:2175–2179. doi: 10.1073/pnas.95.5.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodney G G, Moore C P, Williams B Y, Zhang J Z, Krol J, Pedersen S E, Hamilton S L. J Biol Chem. 2001;276:2069–2074. doi: 10.1074/jbc.M008891200. [DOI] [PubMed] [Google Scholar]