Abstract
Motivation
Integrative approaches characterizing the interactions among different types of biological molecules have been demonstrated to be useful for revealing informative biological mechanisms. One such example is the interaction between microRNA (miRNA) and messenger RNA (mRNA), whose deregulation may be sensitive to environmental insult leading to altered phenotypes. The goal of this work is to develop an effective data integration method to characterize deregulation between miRNA and mRNA due to environmental toxicant exposures. We will use data from an animal experiment designed to investigate the effect of low-dose environmental chemical exposure on normal mammary gland development in rats to motivate and evaluate the proposed method.
Results
We propose a new network approach—integrative Joint Random Forest (iJRF), which characterizes the regulatory system between miRNAs and mRNAs using a network model. iJRF is designed to work under the high-dimension low-sample-size regime, and can borrow information across different treatment conditions to achieve more accurate network inference. It also effectively takes into account prior information of miRNA–mRNA regulatory relationships from existing databases. When iJRF is applied to the data from the environmental chemical exposure study, we detected a few important miRNAs that regulated a large number of mRNAs in the control group but not in the exposed groups, suggesting the disruption of miRNA activity due to chemical exposure. Effects of chemical exposure on two affected miRNAs were further validated using breast cancer human cell lines.
Availability and implementation
R package iJRF is available at CRAN.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
With the past decades rapid technological advances in genomic profiling, multiple types of genomic profiles can be collected on the same set of samples. Recently, we carried out an animal study to assess the effect of three environmental chemicals on miRNA and mRNA activity in mammary tissue (Gopalakrishnan et al., 2017). In this and similar studies, there is increasing interest to characterize changes in the regulatory patterns among different molecular types across different experimental conditions. Compared to commonly used marginal analyses, integrative approaches examining interactions often help to reveal more subtle yet biologically important mechanisms. For instance, it is well known that miRNAs drive the development of many diseases via the regulation of post-transcriptional gene expression (Jansson and Lund, 2012; Nogales-Cadenas et al., 2016). Thus it will be more powerful to characterize miRNA activities through monitoring the global regulatory system between miRNA and mRNA (Arner and Kulyté, 2015).
Multiple challenges arose during the construction of the high dimensional miRNA–mRNA interaction networks. First, such analysis involves thousands or tens of thousands of genes but a much smaller sample size (n = 20 in the motivating chemical exposure study). This problem, well known in statistics as the ‘large p, small n’ paradigm (Bernardo et al., 2003), arises in many biological applications (Kosorok et al., 2007). Second, samples under different conditions in the same study often share common properties, and borrowing information across conditions is crucial to maximize the power of the estimation process (Flutre et al., 2013; Li et al., 2011; Petretto et al., 2010). Third, properly incorporating information from existing databases in the analysis could greatly enhance the accuracy of the inference (Bernard and Hartemink, 2004; Petralia et al., 2015; Werhli et al., 2007; Yip et al., 2010; Zhu et al., 2008).
However, recent methodologies for integrating different genomic profiles (for example, Mo et al., 2013; Hwang et al., 2005; Ebrahim et al., 2016; Ritchie et al., 2015) either were not designed for characterizing interaction networks, or do not fully address the above challenges, especially the latter two. Recently, random forest based methods have been utilized to estimate multiple networks simultaneously (Petralia et al., 2016). In particular, we demonstrated the advantage of joint learning and showed the high performance of random-forest compared to Gaussian graphical models such as Danaher et al. (2014). In this work, we will extend these ideas to model the relationship of dependence among miRNAs and mRNAs. Moreover, in Petralia et al. (2015), we introduced a probability sampling scheme to effectively incorporate prior information in building random forest models. This framework can be effectively modified to incorporate existing knowledge on miRNA–mRNA regulatory relationships documented in miRNA–mRNA databases (Agarwal et al., 2015; Betel et al., 2010; Hsu et al., 2010; Kertesz et al., 2007).
Therefore, in this paper, we propose a new method—integrative Joint Random Forest (iJRF), which borrows information across multiple chemical exposure conditions and takes into account prior information from existing databases when inferring miRNA–mRNA interactions. iJRF is built upon our two previous random-forest based algorithms (Petralia et al., 2015, 2016). The advantages of our integrative framework are multiple. First, its ensemble nature allows the delivery of excellent performance with moderate sample size requirements. Second, treatment-specific tree ensembles are designed to share common structures, so that miRNAs regulations playing a crucial role in multiple conditions will be detected more accurately. Third, existing databases are utilized in order to prioritize miRNA–mRNA interactions. Specifically, TargetScan (Agarwal et al., 2015) was chosen over other databases (Betel et al., 2010; Kertesz et al., 2007) for its comprehensive list of predicted miRNA–mRNA interactions and, as mentioned in Lee et al. (2015), its consistency with databases containing experimentally validated targets (Farazi et al., 2014; Helwak et al., 2013).
Applying iJRF to the chemical exposure study, we simultaneously estimated miRNA–mRNA interaction networks for the control group and three chemical exposed groups: diethyl phthalate (DEP), methyl-paraben (MPB) and triclosan (TCS). We found that the interaction among miRNAs and mRNAs was greatly reduced in the chemical exposed groups compared to the control group. Among all chemicals, DEP exposure was associated with the highest loss of connectivity. iJRF also detected two important miRNAs: miR-200a and miR-375, that played crucial roles in the inferred regulatory network of the control group, but lost more than 90% connectivity in the network corresponding to the DEP exposure group. mRNAs connecting with miR-200a and miR-375 in control network only, were enriched for the ‘Mammary Gland Development’ and ‘Gland Morphogenesis’ pathways. This suggests a mediating role for these miRNAs associated with chemical exposure in mammary gland development. We then confirmed the effect of DEP on miR-375 and miR-200a using human breast cancer cell lines.
2 Materials and methods
2.1 Random forest for network construction
Random forest is a non-linear algorithm that models the response variable via a series of decision trees where each tree is constructed based on a random subset of samples (Breiman, 2001). At each node, a random subset of predictors is considered and the predictor maximizing a certain utility function (i.e. decrease in node impurity) is chosen to split observations into two subsets. Recently, Huynh-Thu et al. (2010) introduced GENIE3, a random forest based model for inferring gene regulatory networks (GRNs). In GENIE3, first, the expression of each target gene k is modeled as a function of the expression of all other genes via random forest, then, the regulatory events are ranked based on random forest importance scores. Specifically, the importance score is defined as the total decrease in node impurities from splitting on the jth predictor, averaged over all trees. Recently, Petralia et al. (2015) proposed iRafNet—a new random-forest based algorithm for network construction which can integrate prior information from database and independent datasets. According to iRafNet, potential regulators considered important by other datasets are prioritized and sampled more often within the random-forest framework. Petralia et al. (2016) extended the original random-forest algorithm to estimate multiple related networks (JRF). As shown by Petralia et al. (2016), borrowing information across multiple networks is crucial to accurately detect common mechanisms. In particular, information across different class of data is borrowed by using the same splitting variables for the tree construction. In this paper, JRF and iRafNet are combined to jointly estimate miRNA–mRNA interactions from different exposure conditions while integrating information from existing databases.
2.2 iJRF: integrative joint random forest
We are interested in inferring miRNA - mRNA interactions in tissues samples for control and three common environmental chemicals: diethyl phthalate (DEP), methyl paraben (MPB) and triclosan (TCS). Denote as the index over different treatment conditions. For each treatment condition g, we observe the expression of M miRNAs and p mRNAs for ng samples. Denote and as the expression of the kth mRNA and the jth miRNA for the ith sample exposed to the gth treatment.
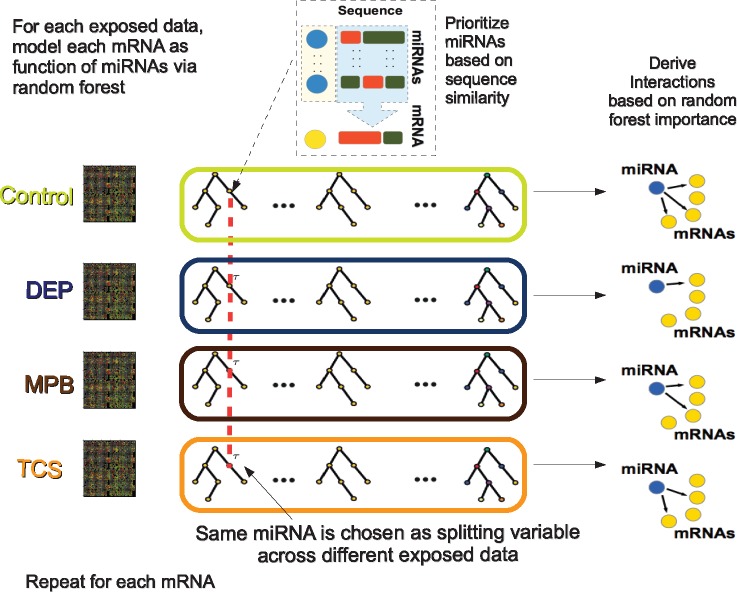
An overview of the proposed algorithm is shown in Figure 1. Specifically, for each treatment condition g, the expression of the kth mRNA is modeled as a function of the expression of miRNAs via random forest, i.e. . Therefore, for each target mRNA, G random forest models corresponding to G treatment conditions are constructed. The key idea of iJRF is to build G tree ensembles simultaneously. Let τg denote the current node in the gth tree model corresponding to the gth condition. As illustrated in Figure 1, the allocation processes for are performed simultaneously through the following steps:
Fig. 1.
Joint Random Forest with iRafNet sampling scheme. For each exposure condition, model the expression of mRNAs as function of the expression of miRNAs via random forest. At each node, sample miRNAs prioritizing those present in TargetScan (Agarwal et al., 2015). Following JRF model (Petralia et al., 2016), the four random forest tree ensembles (Control, DEP, MPB and TCS) use the same splitting variables (miRNAs) to build trees. In this way we achieve borrowing information across them. This procedure is repeated for each mRNA and, then, interactions are ranked based on random forest importance scores
- iRafNet Step (Petralia et al., 2015): For different tree ensembles corresponding to different treatments, the same set of predictors (miRNAs) are proposed for the splitting rule of nodes . This subset of predictors is selected by prioritizing miRNAs that have similar sequences to that of the target mRNA and, thus, are more likely to bind to the target mRNA. Specifically, we sample a set containing N miRNAs from the entire set of miRNAs with probabilities
where scores are derived based on prior information on miRNA–mRNA regulations from existing databases. In this study, TargetScan (Agarwal et al., 2015) was chosen over other databases given its comprehensive list of predicted miRNA–mRNA interactions. Also, as mentioned in Lee et al. (2015), TargetScan is consistent with databases containing experimentally validated targets (Farazi et al., 2014; Helwak et al., 2013). For each interaction , TargetScan provides a context score based on sequence similarity (Garcia et al., 2011). Context scores are non-positive with more negative values corresponding to more favorable sites. We then calculate: with and being the minimum context score. Scores take values in the interval [1, 2] and the probability to sample the miRNA with the most similar sequence to that of the target mRNA will be twice the probability of the least similar miRNA (context score equal to zero). This transformation was considered in order to not excessively penalize interactions that are not contained in prior databases.
- 2. JRF Step (Petralia et al., 2016): Among predictors contained in subset , the optimal splitting variable of nodes is the predictor maximizing the summation of the decrease in node impurity across different treatment conditions, i.e.,
with being the decrease in node impurity observed in the gth tree after splitting τg based on the jth predictor. Specifically, the decrease in node impurity is defined as where is the variance of observations allocated to set is the set of samples allocated to node τg in the gth tree ensemble; while sets and are respectively the sets of samples allocated to the left-child and right-child of node τg according to a splitting rule based on the jth predictor.
Once G random forest models are constructed for the kth mRNA, interactions between miRNAs and the kth mRNA under different conditions are ranked based on random forest importance scores. In order to derive the final unweighted networks, a proper cut-off value for importance scores needs to be chosen. Specifically, we follow the same permutation based procedure described in Petralia et al. (2016), whose details are provided in Supplementary Section 1.
The computational complexity of the proposed algorithm is in the same order as the complexity of JRF (Petralia et al., 2016), i.e. with T being the number of random forest trees. The computational burden can be greatly reduced by estimating in parallel random forest models corresponding to different target mRNAs (further information can be found in Supplementary Section 2.1). In practice, the number of trees (T) and the number of potential regulators to be sampled at each node (N) are parameters to be specified by the users. In this paper, we used the conventional choice of T = 1000 and with M being the number of predictors (Breiman, 2001).
3 Data
3.1 Data overview
Exposure to environmental chemicals, especially during mammary gland development, has been linked to breast cancer risk using animal models (Manservisi et al., 2015; Moral et al., 2008; Rudel et al., 2011; Russo et al., 2001). Increased understanding of the biological and genomic mechanisms mediating the effects of chemical exposures can lead to better prevention and treatment of the disease. In a recent pioneering study using a Sprague-Dawley (SD) rat model, animals were exposed chronically from birth through adulthood (postnatal day 146) to three environmental chemicals present in daily personal care products: diethyl phthalate (DEP), methyl paraben (MPB) and triclosan (TCS) at an exposure level comparable to those observed in human population (Teitelbaum et al., 2016). The study involved 20 rats in each of the following groups, control, DEP and MPB treatment groups, as well as 15 rats in the TCS treatment group. Gopalakrishnan et al. (2017) provides a detailed description of the animal study and the chemical treatment for MPB and TCS exposure. Likewise other chemicals, diethyl phthalate (DEP) (CAS 84-66-2, lot STBB0862V, 99% purity) was supplied in plastic containers (Sigma Aldrich, Italy). The experimental oral dose of DEP was 0.1735 mg/Kg/day, which represented of no observed adverse effect levels (NOAEL) of DEP (Brown et al., 1978; Moody and Reddy, 1978; Oishi and Hiraga, 1980).
3.2 Data processing
The expression matrices of both mRNAs and miRNAs can be found in the GEO database (ID: GSE72276) (Barrett et al., 2005). Quality control of.CEL files and preprocessing based on the robust multiarray average method (RMA) were done using the expression console software (Affymetrix, CA). For mRNA data, batch effects were removed using the ComBat package (Leek et al., 2012) available in R CRAN. We applied a signal intensity filter to retain only those probesets with high and stable expression (signal value > 30th percentile in at least 1 of the experimental groups). A variance-based filter was used to retain the top 50% of the probesets with high interquartile range. For miRNA data, 283 miRNAs were profiled and only miRNAs with variance different from zero were considered. The data were normalized using the package NanoStringNorm available in R CRAN (Waggott et al., 2012). The filtered data contained 7546 genes and 272 miRNAs which were used for downstream analyses. For both miRNA and mRNA data matrices, quantile normalization across samples was performed.
4 Results
4.1 Network estimation
For ease of explanation, we will refer to the network from control rats as Control-Net and networks from different chemical exposed rats as DEP-Net, MPB-Net and TCS-Net. For the analyses, we considered 7546 messenger RNAs and 272 miRNAs. As mentioned in section 3, the sample size was 20 each for the control, DEP and MPB treatment groups, and 15 for TCS. In order to implement the proposed algorithm, 1000 trees were considered and a total of miRNAs (predictors) were sampled at each node (Breiman, 2001). The four networks were estimated using iJRF and mRNA-miRNA interactions were derived using permutation techniques considering an FDR cut-off of 0.001 (further information can be found in Supplementary Section 1).
Table 1 shows the total number of interactions inferred for each network as well as the number of interactions shared across networks. As shown, all three chemicals result in a loss of interaction compared to Control-Net which involved 6829 edges linking 47 miRNAs and 2270 mRNAs. In particular, the total number of interactions in DEP-Net, MPB-Net and TCS-Net were respectively 44%, 84% and 52% of the total number of interactions in Control-Net. Therefore, DEP was the chemical exposure resulting in the most dramatic loss of interaction compared to control.
Table 1.
Number of interactions inferred in Control-Net, DEP-Net, MPB-Net and TCS-Net and number of interactions shared across networks
| Control | DEP | MPB | TCS | |
|---|---|---|---|---|
| Control | 6829 | 2079 | 3593 | 2374 |
| DEP | 3018 | 842 | 1630 | |
| MPB | 5743 | 3491 | ||
| TCS | 3557 |
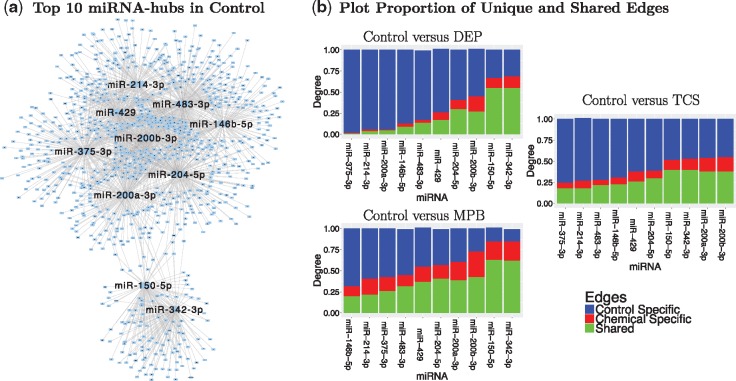
Figure 2(a) shows the top 10 hub-miRNAs in Control-Net, which were responsible for more than 85% of connecting edges in Control-Net. In particular, a substantial portion of those interactions () were not present in any of the chemical-networks. Figure 2(b) compares each chemical-Net and Control-Net showing, for each miRNA, the number of edges shared between chemical-Net and Control-Net (green bar), the number of control-specific edges (blue bar) and the number of chemical-specific edges (red bar). The three quantities have been normalized dividing them by the total number of connecting edges present in either Control-Net or chemical-networks. As shown, DEP-Net has miRNAs such as miR-375-3p, miR-200a-3p and miR-214-3p with remarkable loss in connectivity () compared to Control-Net. Given the dramatic loss in connectivity observed in DEP-Net, we decided to focus on this chemical and miRNAs miR-375-3p, miR-200a-3p and miR-214-3p for further investigation. Besides the consistent loss of connection in chemical-networks compared to Control-Net, differences among chemical-networks were also observed, as shown in Supplementary Section 2.2. Further investigation is needed to understand the biological implication of these differences across chemical-networks.
Fig. 2.
(a) Plot of top ten hub-miRNAs in Control-Net with interactions detected only in Control-Net (Shannon et al., 2003). These miRNAs were responsible for more than 85% of connecting edges in Control-Net. The total number of interactions detected in Control-Net was 6829. (b) For each miRNA, we show the number of edges shared by chemical and control (green bar), the number of control-specific edges (blue bar) and the number of chemical-specific edges (red bar). The three quantities have been normalized dividing them by the total number of connecting edges in either Control-Net or chemical-networks
It is important to note that, no significant results were detected by traditional univariate analysis (i.e. unpaired t-test, Wilcoxon test) when testing was conducted on one miRNA at-a-time, suggesting the advantage of the proposed network based approach (further information on univariate analysis based on the Wilcoxon test can be found in Supplementary Section 3.1). As a comparison, we also estimated miRNA–mRNA interactions under different treatment conditions via Pearson’s correlation test, a commonly used approach in the literature (Mertins et al., 2016; Zhang et al., 2014, 2016). As shown in Supplementary Section 3.2, when considering the same FDR cut-off (fdr = 0.001) as in the iJRF analysis, the correlation test detects far fewer edges than iJRF, and fails to reveal any informative hub structure or pathway enriched network module. We then relaxed the FDR cut-off to 0.01 for the correlation tests to obtain more connected networks, and tested for enriched GO terms. As shown in Supplementary Figure S6, the correlation test resulted in far fewer enriched GO terms compared to iJRF. This suggests that iJRF is more effective to detect biologically relevant interactions. Moreover, the percentage of shared edges across networks based on correlation tests is much smaller than that based on iJRF, which makes the detection of treatment-specific interactions particularly vulnerable to high false positive rates. This result is expected since iJRF, through joint learning, is more effective in detecting common associations than algorithms handling different treatment conditions separately (Petralia et al., 2016).
4.2 miR-375, miR-214 and miR-200a
Overview: One of the top hub miRNAs in Control-Net poorly connected in chemicals was miR-375-3p. In particular, the number of mRNAs interacting with miR-375-3p in DEP-Net, MPB-Net and TCS-Net was, respectively, 2%, 51% and 27% of the number of interacting mRNAs in Control-Net. Recent studies have investigated the role of miR-375 in breast cancer (Madhavan et al., 2016; Ward et al., 2013; Zehentmayr et al., 2016). In particular, Ward et al. (2013) reported a loss of miR-375 expression in drug-resistant breast cancer cells; while Madhavan et al. (2016) showed that miR-375 was significantly associated with breast cancer survival. As shown in Figure 2(b), other miRNAs poorly connected in DEP-Net were miR-214 and miR-200a. Their role in breast cancer has been investigated in several papers (Kalniete et al., 2015; Ming et al., 2015; Penna et al., 2015; Pieraccioli et al., 2013; Wang et al., 2015; Yao et al., 2014; Yu et al., 2015). In particular, miR-214 was shown to be associated with breast cancer survival (Kalniete et al., 2015) and drug sensitivity (Yu et al., 2015). On the other hand, miR-200a has been associated with survival in metastatic breast cancer (Madhavan et al., 2016) and its role in cell proliferation inhibition has been demonstrated (Yao et al., 2014).
Overlap with MirTarBase: Some Control-specific interactions detected by iJRF are contained in miRTarBase (Hsu et al., 2010)—the database of experimentally validated miRNA–mRNA interactions. Table 2 shows the list of interactions in Control-Net contained in miRTarBase for miR-375 and miR-200a (other interactions contained in miRTarBase can be found in Supplementary Table S2). As shown in Table 2, some of these interactions were contained in DEP-Net, MPB-Net and TCS-Net as well. Particularly interesting is the interaction of miR-375 with the human epidermal growth factor receptor 2 (HER2) (Pillai et al., 2014; Shen et al., 2014) only contained in Control-Net and MPB-Net. HER2 stimulates the growth of breast cancer cells and is one of the main targets for breast cancer survival and therapy. Another interesting interaction is miR-200a - ZEB2 which has been investigated in different ovarian and breast cancer studies (Ahmad et al., 2011; Bracken et al., 2008; Jang et al., 2014; Park et al., 2008; Truong et al., 2014; Wu et al., 2011). In particular, many articles have described ZEB2 as the crucial target of miR-200 family members (Burk et al., 2008; Christoffersen et al., 2007; Korpal et al., 2008). Brabletz and Brabletz (2010) showed that ZEB2 and miR-200a are involved in a ‘feedback loop’ which drives the progression of metastasis in breast cancer. As shown in Table 2, the interaction between ZEB2 and miR-200a is only contained in Control-Net, suggesting the impact of all chemicals on the regulatory mechanism between miR-200a and ZEB2.
Table 2.
List of interactions in Control-Net contained in miRTarBase for miR-200a and miR-375
| miRNAs | mRNAs | DEP | MPB | TCS |
|---|---|---|---|---|
| miR-375 | HER2, TMTC4, SFT2D2, KRT8 | x | ||
| miR-375 | PLAG1, CCDC88A, CELF2 | x | ||
| miR-375 | GATA6 | x | x | |
| miR-375 | CMTM4, FOLR1, CTSC | |||
| miR-200a | ZEB2 | |||
| miR-200a | HOXB5 | x | ||
| miR-200a | DLC1 | x |
For each interaction, we indicate if it was contained in other networks such as DEP-Net, MPB-Net and TCS-Net.
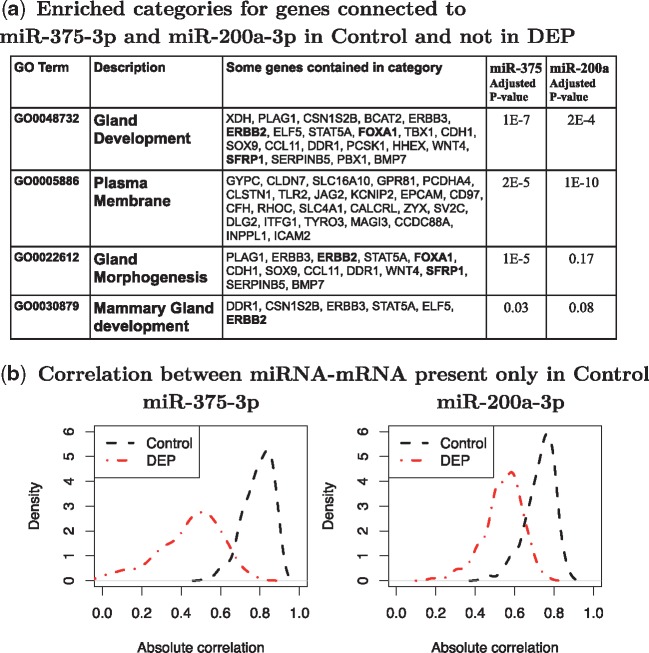
Enrichment Analysis: Among hub-miRNAs in Control-Net, interesting pathways were obtained for miR-375-3p and miR-200a-3p. Figure 3(a) shows some interesting enriched categories for mRNAs connected to miR-375-3p and miR-200a-3p in Control-Net but not in DEP-Net (enriched pathways for other hub-miRNAs in Control-Net can be found in Supplementary Table S3). The enrichment analysis was performed using David Tool 6.7 (Huang et al., 2008) and Benjamini adjusted p-values were reported in Figure 3(a). As shown, miR-375-3p and miR-200a-3p share some enriched categories such as ‘Mammary Gland Development’ with Benjamini adjusted p-values less than 0.1. This result is not surprising since the two miRNAs share some connections in Control-Net. Various studies have shown that chemical exposure can alter mammary gland development (Manservisi et al., 2015; Mandrup et al., 2015; Schwarzman et al., 2015). In this context, DEP exposure might alter the regulatory mechanism of miR-375-3p and miR-200a-3p and affect mammary gland development. As shown in Figure 3(a), enriched pathways include genes such as ERBB2 (HER2), FOXA1 and SFRP1 which play a crucial role in breast cancer. To further demonstrate the loss in connectivity in DEP-Net, Figure 3(b) shows the correlation density between each miRNA and mRNAs connected in Control-Net but not in DEP-Net. As expected, the correlation density for DEP exposure is shifted to the left compared to that of control revealing a loss of correlation. Supplementary Figure S7 shows that the loss of correlation observed in DEP-Net for both miR-375-3p and miR-200a-3p is significant.
Fig. 3.
(a) We consider genes connected to miR-375-3p and miR-200a-3p in Control-Net but not in DEP-Net and derived enriched categories using David Tools (Huang et al., 2008). Pathways ‘Gland Development’, ‘Plasma Membrane’ and ‘Mammary Gland Development’ were enriched for both miR-375-3p and miR-200a-3p with Benjamini adjusted p-values smaller than 0.10. Pathway ‘Gland Morphogenesis’ was enriched only for miR-375-3p. (b) Density of absolute correlation between miR-375-3p and miR-200a-3p with mRNAs connected only in Control-Net for DEP exposed data (red) and control data (black)
4.3 Validation via cell line experiments
In Section 4.1,we showed that DEP-Net resulted in a dramatic loss of interaction compared to Control-Net. In particular, the three hub- miRNAs poorly connected in DEP-Net were miR-375, miR-214 and miR-200a. To validate the effect of DEP, in vitro experiments of a human breast cancer cell line are utilized.
Experiments: MCF-7 cells were maintained in phenol-red-free DMEM (Gibco 11054) containing 5% (v/v) dextran-charcoal-stripped fetal calf serum, with M diethyl phthalate, for 4 weeks. These cells were subcultured every 3-4 days with a confluence level less than 70%. The total RNA was isolated from 10 cm Petri plate using the Promega Maxwell simplyRNA kit according to the manufacturer’s instructions. Reverse transcription was done using the Exiqon Universal cDNA Synthesis kit and the detection of qPCR was performed with the Exiqon ExiLENT SYBR Green master mix on Roche LightCycler 480 machine.
Results: We quantified the expression of miR-200a, miR-375 and miR-214 in MCF-7 cells. Unfortunately, miR-214 was not expressed in this cell line, thus only results for miR-200a and miR-375 are reported. As shown in Figure 4, the expression levels of both miR-375 and miR-200a are significantly different between control and DEP-exposed cells. This result suggests that the two miRNAs are affected by DEP exposure in breast cancer human cell lines. Further analyses are necessary to elucidate the role of these two miRNAs on mammary gland development.
Fig. 4.

Plot of cell line experiments. Expression levels of miR-375 and miR-200a for normal and DEP exposed cells. Red diamonds indicate average over the three replicates. Benjamini adjusted p-values from the unpaired t-test are reported
5 Discussion
In this paper, we focused on exposure to chemicals commonly used in personal care products during mammary gland development (birth to adulthood) and their effect on miRNA–mRNA interactions. Some challenges arose given the large number of miRNAs/genes involved and the limited sample size of our data. In order to overcome this problem, we proposed iJRF, an ensemble algorithm with small sample size requirement. The advantage of iJRF is dual. First, it is designed to borrow information across different treatment conditions so that associations shared across treatments can be detected more accurately. Second, iJRF can integrate information from existing miRNA–mRNA databases such as TargetScan.
Using our newly developed algorithm, we estimated miRNA–mRNA interaction in mammary tissues for control and chemical-exposed animals. All chemical-networks registered a loss in connectivity compared to control. In particular, DEP was the chemical registering the most dramatic loss of interaction compared to control. In fact, the total number of interactions in DEP-Net, MPB-Net and TCS-Net were respectively 44%, 84% and 52% of the total number of interactions in Control-Net.
Among the leading miRNAs in Control-Net, miR-200a, miR-214 and miR-375 lost more than 90% of connectivity in DEP-Net compared to Control-Net. Messenger RNAs connected to miR-200a and miR-375 in Control-Net but not in DEP-Net were enriched in ‘Gland Morphogenesis’ and ‘Mammary Gland Development’, indicating their potential involvement in mammary gland development mechanisms. Among genes in these pathways, we found targets in breast cancer such as ERBB2, FOXA1 and SFRP1. Recent studies have investigated the role of miR-375, miR-214 and miR-200a in breast cancer. For example, Ward et al. (2013) reported a loss of miR-375 expression in tamoxifen-resistant breast cancer cells; while Madhavan et al. (2016) reported a significant association between survival in metastatic breast cancer and both miR-375 and miR-200a. The expression of miR-214 was shown to be associated with breast cancer survival (Kalniete et al., 2015) and drug sensitivity (Yu et al., 2015); while Yao et al. (2014) demonstrated the role of miR-200a in the inhibition of cell proliferation in breast cancer.
Given the dramatic loss of connectivity observed in DEP-Net, we validated the effect using cell line experiments. Specifically, we focused on miRNAs with the highest loss in connectivity: miR-375, miR-214 and miR-200a. Using MCF-7 cells, we showed that the expression levels of both miR-375 and miR-200a were significantly different between the control and DEP exposed groups. In this study, we hypothesized that chemical exposures first affect miRNA activity, which then changes the regulatory pattern among miRNAs and mRNAs. In our validation experiments, we demonstrated the effects of chemical exposure on miRNA activity. Future research is warranted to further validate the detected changing of regulatory relationships among miRNAs and mRNAs.
In this paper we examined miRNA–mRNA networks for single exposure conditions. On the other hand, humans are exposed to a mixture of different chemicals with the interaction and combination of chemicals playing a crucial role. In order to deal with such complex data, as future work, we will design a model to estimate networks which vary across exposure conditions in a dynamic way. Once chemical-induced mechanisms will be identified, their association to breast cancer phenotypes will be assessed.
Finally, the proposed algorithm can be utilized in different biological applications. As an example, eQTL analysis might be performed for different tissues simultaneously while borrowing information from existing databases. It is well known that borrowing information across tissues is crucial to detect shared eQTLs more accurately (Flutre et al., 2013) and, iJRF is a non-parametric model that can be easily implemented for such analyses.
Supplementary Material
Acknowledgement
This work was supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Funding
F.P. and P.W. were supported by grant U24 CA210993, from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC) and NIH grants R01 GM108711 and R01 CA189532. The entire experiment was supported by NIH/NIEHS/NCI grant U01 ES019451. J.C., K. G, V. A. and S. T. were also supported by NIH/NCI grant R01 CA172460.
Conflict of Interest: none declared.
References
- Agarwal V. et al. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A. et al. (2011) Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by mir-200 in breast cancer cells. Cancer Res., 71, 3400–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner P., Kulyté A. (2015) MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol., 11, 276–288. [DOI] [PubMed] [Google Scholar]
- Barrett T. et al. (2005) Ncbi geo: mining millions of expression profilesdatabase and tools. Nucleic Acids Res., 33, D562–D566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A., Hartemink A.J. (2004). Informativestructure priors: joint learning of dynamic. Biocomputing, 2005, 459. [PubMed] [Google Scholar]
- Bernardo J. et al. (2003) Bayesian factor regression models in the large p, small n paradigm. Bayesian Stat., 7, 733–742. [Google Scholar]
- Betel D. et al. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol., 11, R90.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz S., Brabletz T. (2010) The zeb/mir-200 feedback loop a motor of cellular plasticity in development and cancer?. EMBO Rep., 11, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P. et al. (2008) A double-negative feedback loop between zeb1-sip1 and the microrna-200 family regulates epithelial-mesenchymal transition. Cancer Res., 68, 7846–7854. [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Brown D. et al. (1978) Short-term oral toxicity study of diethyl phthalate in the rat. Food Cosmetics Toxicol., 16, 415–422. [DOI] [PubMed] [Google Scholar]
- Burk U. et al. (2008) A reciprocal repression between zeb1 and members of the mir-200 family promotes EMT and invasion in cancer cells. EMBO Rep., 9, 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen N.R. et al. (2007) mir-200b mediates post-transcriptional repression of zfhx1b. RNA, 13, 1172–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danaher P. et al. (2014) The joint graphical lasso for inverse covariance estimation across multiple classes. J. R. Stat. Soc. Ser. B (Stat. Methodol.), 76, 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A. et al. (2016) Multi-omic data integration enables discovery of hidden biological regularities. Nat. Commun., 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi T.A. et al. (2014) Identification of distinct miRNA target regulation between breast cancer molecular subtypes using ago2-par-clip and patient datasets. Genome Biol., 15, R9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T. et al. (2013) A statistical framework for joint EQTL analysis in multiple tissues. PLoS Genet., 9, e1003486.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D.M. et al. (2011) Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol., 18, 1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan K. et al. (2017) Changes in mammary histology and transcriptome profiles by low-dose exposure to environmental phenols at critical windows of development. Environ. Res., 152, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A. et al. (2013) Mapping the human mirna interactome by clash reveals frequent noncanonical binding. Cell, 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-D. et al. (2010) mirtarbase: a database curates experimentally validated microRNA–target interactions. Nucleic Acids Res., gkq1107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W. et al. (2008) Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huynh-Thu V.A. et al. (2010) Inferring regulatory networks from expression data using tree-based methods. PloS One, 5, e12776.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D. et al. (2005) A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U. S. A., 102, 17296–17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K. et al. (2014) Loss of microrna-200a expression correlates with tumor progression in breast cancer. Transl. Res., 163, 242–251. [DOI] [PubMed] [Google Scholar]
- Jansson M.D., Lund A.H. (2012) MicroRNA and cancer. Mol. Oncol., 6, 590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalniete D. et al. (2015) High expression of mir-214 is associated with a worse disease-specific survival of the triple-negative breast cancer patients. Hered. Cancer Clin. Pract., 13, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M. et al. (2007) The role of site accessibility in microRNA target recognition. Nat. Genet., 39, 1278–1284. [DOI] [PubMed] [Google Scholar]
- Korpal M. et al. (2008) The mir-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of e-cadherin transcriptional repressors zeb1 and zeb2. J. Biol. Chem., 283, 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosorok M.R. et al. (2007) Marginal asymptotics for the large p, small n paradigm: with applications to microarray data. Ann. Stat., 35, 1456–1486. [Google Scholar]
- Lee E. et al. (2015) Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics, btv531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T. et al. (2012) The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. (2011) An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. Ann. Appl. Stat., 5, 994–1019. [Google Scholar]
- Madhavan D. et al. (2016) Circulating mirnas with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis, bgw008.. [DOI] [PubMed] [Google Scholar]
- Mandrup K.R. et al. (2015) Mixtures of environmentally relevant endocrine disrupting chemicals affect mammary gland development in female and male rats. Reprod. Toxicol., 54, 47–57. [DOI] [PubMed] [Google Scholar]
- Manservisi F. et al. (2015) Effect of maternal exposure to endocrine disrupting chemicals on reproduction and mammary gland development in female Sprague–Dawley rats. Reprod. Toxicol., 54, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P. et al. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature, 534, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming J. et al. (2015) Identification of mir-200a as a novel suppressor of connexin 43 in breast cancer cells. Biosci. Rep., 35, e00251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Q. et al. (2013) Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc. Natl. Acad. Sci. U. S. A., 110, 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody D.E., Reddy J.K. (1978) Hepatic peroxisome (microbody) proliferation in rats fed plasticizers and related compounds. Toxicol. Appl. Pharmacol., 45, 497–504. [DOI] [PubMed] [Google Scholar]
- Moral R. et al. (2008) Effect of prenatal exposure to the endocrine disruptor bisphenol a on mammary gland morphology and gene expression signature. J. Endocrinol., 196, 101–112. [DOI] [PubMed] [Google Scholar]
- Nogales-Cadenas R. et al. (2016) MicroRNA expression and gene regulation drive breast cancer progression and metastasis in PYMT mice. Breast Cancer Res., 18, 75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi S., Hiraga K. (1980) Testicular atrophy induced by phthalic acid esters: effect on testosterone and zinc concentrations. Toxicol. Appl. Pharmacol., 53, 35–41. [DOI] [PubMed] [Google Scholar]
- Park S.-M. et al. (2008) The mir-200 family determines the epithelial phenotype of cancer cells by targeting the e-cadherin repressors zeb1 and zeb2. Genes Dev., 22, 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna E. et al. (2015) mir-214 as a key hub that controls cancer networks: small player, multiple functions. J. Invest. Dermatol., 135, 960–969. [DOI] [PubMed] [Google Scholar]
- Petralia F. et al. (2015) Integrative random forest for gene regulatory network inference. Bioinformatics, 31, i197–i205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia F. et al. (2016) New method for joint network analysis reveals common and different coexpression patterns among genes and proteins in breast cancer. J. Proteome Res., 15, 743–754. PMID: 26733076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretto E. et al. (2010) New insights into the genetic control of gene expression using a Bayesian multi-tissue approach. PLoS Comput. Biol., 6, e1000737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieraccioli M. et al. (2013) Activation of mir200 by c-myb depends on zeb1 expression and mir200 promoter methylation. Cell Cycle, 12, 2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai M.M. et al. (2014) Hits-clip reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res. Treatment, 146, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.D. et al. (2015) Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet., 16, 85–97. [DOI] [PubMed] [Google Scholar]
- Rudel R.A. et al. (2011) Environmental exposures and mammary gland development: state of the science, public health implications, and research recommendations. Environ. Health Perspect., 119, 1053.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo J. et al. (2001) Mammary gland architecture as a determining factor in the susceptibility of the human breast to cancer. Breast J., 7, 278–291. [DOI] [PubMed] [Google Scholar]
- Schwarzman M.R. et al. (2015) Screening for chemical contributions to breast cancer risk: a case study for chemical safety evaluation. Environ. Health Perspect., 123, 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z.-Y. et al. (2014) mir-375 inhibits the proliferation of gastric cancer cells by repressing erbb2 expression. Exp. Therapeutic Med., 7, 1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum S.L. et al. (2016) Paired serum and urine concentrations of biomarkers of diethyl phthalate, methyl paraben, and triclosan in rats. Environ. Health Perspect., 124, 39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong H.H. et al. (2014) β1 integrin inhibition elicits a prometastatic switch through the tgfβ–mir-200–zeb network in e-cadherin–positive triple-negative breast cancer. Sci. Signal., 7, ra15–ra15. [DOI] [PubMed] [Google Scholar]
- Waggott D. et al. (2012) Nanostringnorm: an extensible r package for the pre-processing of nanostring mRNA and miRNA data. Bioinformatics, 28, 1546–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F. et al. (2015) microrna-214 enhances the invasion ability of breast cancer cells by targeting p53. Int. J. Mol. Med., 35, 1395–1402. [DOI] [PubMed] [Google Scholar]
- Ward A. et al. (2013) Re-expression of microrna-375 reverses both tamoxifen resistance and accompanying emt-like properties in breast cancer. Oncogene, 32, 1173–1182. [DOI] [PubMed] [Google Scholar]
- Werhli A.V. et al. (2007) Reconstructing gene regulatory networks with bayesian networks by combining expression data with multiple sources of prior knowledge. Stat. Appl. Genet. Mol. Biol., 6, 15.. [DOI] [PubMed] [Google Scholar]
- Wu Q. et al. (2011) Microrna-200a inhibits cd133/1+ ovarian cancer stem cells migration and invasion by targeting e-cadherin repressor zeb2. Gynecol. Oncol., 122, 149–154. [DOI] [PubMed] [Google Scholar]
- Yao J. et al. (2014) microRNA-200a inhibits cell proliferation by targeting mitochondrial transcription factor a in breast cancer. DNA Cell Biol., 33, 291–300. [DOI] [PubMed] [Google Scholar]
- Yip K.Y. et al. (2010) Improved reconstruction of in silico gene regulatory networks by integrating knockout and perturbation data. PloS One, 5, e8121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. et al. (2015) Mir-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Molecular Cancer, 14, 208–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehentmayr F. et al. (2016) Hsa-mir-375 is a predictor of local control in early stage breast cancer. Clin. Epigenet., 8, 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature, 513, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell, 166, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. et al. (2008) Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet., 40, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.