Abstract
Motivation
5-Methylcytosine and 5-Hydroxymethylcytosine in DNA are major epigenetic modifications known to significantly alter mammalian gene expression. High-throughput assays to detect these modifications are expensive, labor-intensive, unfeasible in some contexts and leave a portion of the genome unqueried. Hence, we devised a novel, supervised, integrative learning framework to perform whole-genome methylation and hydroxymethylation predictions in CpG dinucleotides. Our framework can also perform imputation of missing or low quality data in existing sequencing datasets. Additionally, we developed infrastructure to perform in silico, high-throughput hypotheses testing on such predicted methylation or hydroxymethylation maps.
Results
We test our approach on H1 human embryonic stem cells and H1-derived neural progenitor cells. Our predictive model is comparable in accuracy to other state-of-the-art DNA methylation prediction algorithms. We are the first to predict hydroxymethylation in silico with high whole-genome accuracy, paving the way for large-scale reconstruction of hydroxymethylation maps in mammalian model systems. We designed a novel, beam-search driven feature selection algorithm to identify the most discriminative predictor variables, and developed a platform for performing integrative analysis and reconstruction of the epigenome. Our toolkit DIRECTION provides predictions at single nucleotide resolution and identifies relevant features based on resource availability. This offers enhanced biological interpretability of results potentially leading to a better understanding of epigenetic gene regulation.
Availability and implementation
http://www.pradiptaray.com/direction, under CC-by-SA license.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Transcriptional regulation is a complex, dynamic process established by regulatory pathways encompassing a variety of genetic and epigenetic mechanisms. 5-Methylcytosine (5-mC) and 5-Hydroxymethylcytosine (5-hmC) are major modifications to the cytosine base in the DNA, known to be correlated with gene expression (Hackett et al., 2013, Jones, 2012). The addition of a methyl group to cytosine creates the epigenetic modification 5-mC:- the most prevalent form of DNA methylation in mammals. 5-hmC is an oxidative derivative of 5-mC generated in a Ten-Eleven Translocation (TET) oxidase family mediated reaction (Yu et al., 2012). The role of 5-mC in transcriptional regulation is well understood, while the function of 5-hmC remains under investigation. 5-hmC is the intermediate step leading to demethylation of the cytosine (Hackett et al., 2013), known to closely associate with enhancers (Yu et al., 2012), exon-intron boundaries (Khare et al., 2012), elevated C-to-G transversion rates (Supek et al., 2014), labile nucleosomes and CTCF binding (Teif et al., 2014). Previous studies in mammalian systems have shown 5-hmC abundance across tissues to vary significantly, with neural tissue being 5-hmC enriched (Kim et al., 2014), and certain cancer tissues being 5-hmC depleted (Yang et al., 2013), suggesting a functional role of 5-hmC. The most accurate and comprehensive technique (Qu et al., 2013) for genome-wide methylation quantification is whole-genome sodium bisulfite treatment of DNA (Frommer et al., 1992) causing methylated cytosines to remain intact whilst unmethylated cytosines are deaminated to uracils (C-to-U conversion), followed by Polymerase Chain Reaction (PCR) amplification and shotgun sequencing. Whole-genome shotgun Bisulfite-sequencing (BS-seq) involves all PCR fragments genome-wide, while the Reduced Representation Bisulfite-sequencing (RRBS-seq) protocol leads to a small fraction of the fragments being selected (Gu et al., 2011). BS-seq experiments allow us to estimate a C-to-U conversion rate (CCR) or methylation level for each cytosine in the genome- an estimator of the degree of methylation. However, BS-seq does not differentiate between 5-mC and 5-hmC, hence the estimated methylation level is due to both 5-mC and 5-hmC. In order to quantify the degree of hydroxymethylation, alternate protocols like TET-Assisted BS-seq (TAB-seq) (Yu et al., 2012) and Oxidative BS-seq (oxBS-seq) (Booth et al., 2012) were developed. In this paper, we refer to detectable modifications from BS-seq experiments (yielding a summation of 5-mC and 5-hmC driven CCRs) as methylation, and genome-wide characterization of methylation as the methylome. We refer to detectable modifications from TAB-seq (yielding solely 5-hmC driven CCRs or 5-hmC levels) as hydroxymethylation, and corresponding genome-wide maps as the hydroxymethylome.
Importance of predicting methylation and hydroxymethylation: Our prediction framework, which can perform whole genome methylome or hydroxymethylome reconstruction as well as imputation of missing data in existing datasets, is important for several reasons. Despite the availability of high-throughput assays for querying DNA hydroxymethylation, there only exists a handful of publicly available TAB-seq or oxBS-seq datasets, and performing whole-genome BS-seq, oxBS-seq or TAB-seq requires significant expenditure and skilled labor. Sequencing (or hybridization) based assays are also invasive and destructive procedures that may be unfeasible in certain experimental setups. It is also impossible to set up high-throughput assays for all cell or tissue types and every developmental stage, physiological condition or perturbation, necessitating in silico prediction. In such situations, reconstruction of the whole epigenome predicated upon available data for correlated traits and a predictive model trained on a similar cell type is a practical, economical and efficient way to query methylation or hydroxymethylation. Additionally, DNA sequencing based protocols have amplification and fragment selection steps, effectively creating a biased sampling procedure that may cause a fraction of cytosines in the genome to be unrepresented or underrepresented in the survey. This is especially evident for protocols like RRBS-seq where only a small fraction of cytosines have reliable coverage for querying methylation (Gu et al., 2011). Our method can be used to predict such missing or low-quality data in imputation mode. Finally, inherent stochasticity of the sampling process makes it inevitable that some estimations of methylation levels using high coverage sequencing data can be potentially erroneous. However, in silico predictive models, trained using high-quality data with multiple input predictor variables, would be able to robustly predict DNA methylation.
We have devised a machine learning based integrative framework for high-accuracy, single-nucleotide resolution predictions of DNA methylation (either 5-mC or 5-hmC) and solely 5-hmC modifications in mammalian model system genomes. Our publicly available tool DIRECTION (Discriminative IntegRative whole Epigenome Classification at single nucleotide resoluTION) can be trained on shotgun sequencing-based mammalian methylation and hydroxymethylation datasets, by identifying and using available, correlated, high-throughput assays and genomic sequence-based traits as predictor variables. DIRECTION can be downloaded from http://www.pradiptaray.com/direction
Context in literature: Over the past decade, high-throughput assays and corresponding computational models have been actively pursued to annotate and predict the epigenome (Ernst and Kellis, 2012, 2015), including several approaches for predicting methylation as either a binary or continuous variable in CpG dinucleotides. Early models for DNA methylation prediction were based on Support Vector Machines (SVMs) and decision trees, which employed sequence and structure derived information (Bhasin et al., 2005, Bock et al., 2006, Das et al., 2006) to classify genes, CpG islands (CGI) or DNA fragments into hypermethylated versus hypomethylated classes. However, sequence-based prediction of methylation is limited in its ability to identify cell type, tissue, or condition-specific methylation patterns across datasets as underlying sequence features remain unchanged. Since such methylation patterns are of specific interest to biologists, several studies analyzed correlation between methylation and various assays profiling transcription factor (TF) binding or chromatin landscape (Wrzodek et al., 2012). Such knowledge has been leveraged to build explicit predictive models of DNA methylation based on histone modification, nucleosome positioning, chromatin accessibility and TF binding data, including several at single nucleotide or dinucleotide resolution. (Whitaker et al., 2015) uses discriminative sequence motifs for individual datasets to predict CpG methylation. (Ma et al., 2014) uses support vector regression to predict methylation as a continuous-valued response variable in CpG sites across tissues, and (Zhang et al., 2015) use Random Forests (RFs) on genome, epigenome and ChIP-seq derived traits and neighboring CpG methylation levels for imputing methylation arrays. (Yan et al., 2015) used RFs on sequence and epigenome-derived features training on BS-seq data, while (Wang et al., 2016) use SVMs and deep neural nets on topological domains and other features by training on RRBS-seq data. (Fan et al., 2016) predict stem cell CpG methylation for methylation arrays and BS-seq data (Supplementary Table T1 for a comprehensive survey updated from (Zhang et al., 2015)).
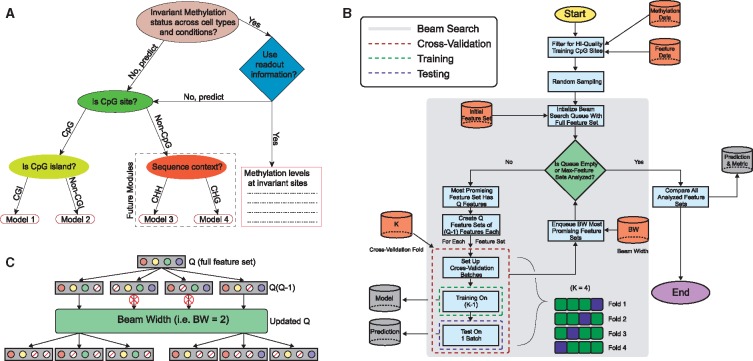
Uniqueness of approach: Firstly, DIRECTION is able to deconfound effects of 5-mC and 5-hmC modifications, as it can be separately trained on BS-seq and TAB-seq datasets for a given cell-type. This is the first time 5-hmC modifications have been predicted in silico (with a whole-genome accuracy of 0.82), allowing us to systematically reconstruct 5-hmC modification maps in different cell-types and tissues. Secondly, DIRECTION provides different usage modes (Supplementary Table T2) including imputation and whole methylome reconstruction (based on training a model in a related cell or tissue type). This is possible because we do not use predictor variables likely to be relevant only in specific cell-types (like DNA-binding motifs of cell-type restricted TFs), enabling transfer learning. Thirdly, DIRECTION is able to heuristically identify an optimal feature set (OFS) for predictions based on the set of available predictor variables (optionally using regional methylation patterns and methylation information from other cell types), allowing use in resource-poor scenarios and providing biologically interpretable results. Also, DIRECTION predicts 5-hmC modification at single nucleotide resolution (as opposed to CpG dinucleotide), since CpG dinucleotides may be asymmetrically modified for 5-hmC (Yu et al., 2012). Single nucleotide resolution allows us to collate predictions to any biologically relevant resolution (CpG dinucleotide, CGI, gene) for purposes of downstream functional analysis. We provide a novel framework for predicting the whole methylome, based on a decision tree topology (Fig. 1) with different classifiers corresponding to each leaf. This tree partitions the methylome by selecting the most appropriate classifier given the availability of predictor variables and their efficacy on the basis of biologically relevant methylation paradigms. Additionally, we identified CpG sites with invariant methylation by contrasting available reference methylomes, as an optional feature for methylation prediction.
Fig. 1.
DNA methylation reconstruction framework. (A) Decision Tree for partitioning methylome based on different prediction paradigms. (B) Schema of prediction framework outlining beam search (feature selection), training, testing and cross-validation modes. (C) Beam search algorithm feature set exploration shown for beam width = 2, for two levels of the search tree
2 Materials and methods
Bisulfite treatment protocols followed by short-read sequencing (BS-seq or TAB-seq) provide CCRs at single nucleotide resolution for cytosines ranging from 0 (unmethylated) to 1 (fully methylated). We formulate prediction of DNA methylation as a binary classification problem due to the bimodal nature of the distribution of CCRs in BS-seq experiments. Genome-wide empirical distributions of CCRs in mammalian reference methylomes (Kundaje et al., 2015) from inbred cell lines and sourced whole tissue (with low and high cellular heterogeneity respectively) show clear evidence of a bimodal distribution of CCRs (Supplementary Fig. 1A). This suggests that with respect to DNA methylation, cell-to-cell variation or within-cell heterogeneity across alleles at individual CpG sites are not prominent in mammalian cells and tissues.
5-hmC is an intermediate molecular state in the demethylation pathway, and TAB-seq CCRs tend to be significantly lower than BS-seq CCRs. Previous studies have shown that the vast majority of CpG sites are lowly hydroxymethylated and have a CCR of zero (Yu et al., 2012) and identified that significantly hydroxymethylated sites exhibit a unimodal distribution of CCRs peaking at 0.18 (Supplementary Fig. 1C). We thus also model 5-hmC prediction as a binary classification problem. 5-hmC has been shown to be a temporally stable (rather than transient) modification (Bachman et al., 2014), which is validated by concordance of TAB-seq levels across biological replicates of NPC (Supplementary Fig. 1D), and our BS-seq and TAB-seq datasets show good consistency between experiments. These evidences lend weight to the tractability of predicting 5-hmC modifications. Thus, we aim to learn a function that will map a set of input features {x1, x2, … xn} to binary class labels {low, high} for the purpose of reconstructing a discretized approximation of the BS-seq and TAB-seq CCRs at individual cytosines. The binary approximations of the BS-seq and TAB-seq CCRs are referred to as methylation and 5-hmC status respectively (Supplementary Text S1 for labeling classes, Supplementary Text S2 for feasibility of 5-hmC prediction).
Overall architecture: DIRECTION offers three primary modes of usage: for existing datasets, we can identify an OFS for predicting methylation and 5-hmC status based on available input feature sets, or impute low quality or missing data. Additionally, the toolkit allows us to perform whole methylome and hydroxymethylome reconstruction based on a user-provided feature set and SVM or RF model trained on a similar cell type or tissue. For other modes see Supplementary Table T2.
Machine learning based approaches, most prominently SVM and RF models have been successfully used to predict methylation in the past (Das et al., 2006, Zhang et al., 2015). Since we aim to perform genome-wide prediction, we chose not to use a single predictive model, but instead designed a scalable ensemble-learning framework that would be able to deconvolve multiple methylation paradigms that are at work in different regions of the genome. For this purpose, a decision tree with a biologically motivated topology is used (Fig. 1A), which partitions the methylome for methylation status prediction, based on available predictor variables and methylation paradigms. At each partition, we train separate predictive models predicated upon an SVM and RF, which exhibit comparable predictive accuracy. We also identified CpG sites with invariant methylation status across a set of high-quality reference methylomes, which can optionally be used as an additional feature to predict methylation status. With research on 5-hmC functionality currently underway, and due to a lack of reference hydroxymethylomes, we used a single predictive model (SVM or RF) to perform 5-hmC status prediction.
Model-based classification: SVMs typically seek to maximize the distance of training instances from the decision boundary in input space, using a kernel transformation to separate features in high dimensional space. We chose to use the popular Radial Basis Function (RBF) (Bishop, 2007), previously used to predict DNA methylation status (Das et al., 2006). RF is an ensemble-learning algorithm comprised of numerous decision trees, well known for high classification performance and resistance to overfitting. It averages predictions and feature weights across multiple decision trees and randomly samples subsets of features, subsequently separating class labels by splitting input features to optimize Gini Impurity or entropy (Breiman, 2001). SVMs and RFs were trained on balanced sets of both classes, and tested on both balanced sets (5-fold cross-validation) and on the whole genome (Supplementary Text S3). We include both models in our framework since they have differing strengths (eg. SVMs work well even with small training sets, RFs are naturally resistant to outliers), letting the user choose the model depending on the dataset, and they work with comparable efficiency for our data.
Evaluation of classification quality: For evaluating predictions on balanced sets, we used Precision and Recall, F-score and Area Under Curve (AUC). True Positive and True Negative Rates were used to evaluate whole genome predictions (Supplementary Text S4). Training and test set sizes were decided based on evaluation of metric stability (Supplementary Fig. 2A, B).
Beam search algorithm: All input features (listed in Supplementary Table T3) were first preprocessed for use in our predictive framework (Supplementary Text S5). Identifying OFSs for classification is computationally intractable for a large number of input features (Koller and Sahami, 1996), due to the curse of dimensionality. The problem is additionally complicated by the presence of noise in input features, label infidelity in the response variable, missing or low quality data for certain features, and high inter-feature correlation. While OFS selection and model training can be jointly performed (Nguyen and De la Torre, 2010), we heuristically identified an OFS using a recursive feature elimination strategy not limited to a specific learning algorithm, providing flexibility to choose a predictive model. Recursive feature elimination allows us to pick feature sets with fewer features that fit the data better in an iterative fashion, implicitly enforcing sparsity. We performed an initial feature elimination step based on inter-feature correlational redundancies (Supplementary Text S6, and S7 for recursive and initial feature elimination, respectively).
We then conducted recursive feature elimination on the remaining features by implementing the beam search algorithm (schema: Fig. 1B): a classical artificial intelligence search procedure, utilizing heuristic pruning rules to explore a graph with nodes corresponding to all possible feature sets (Zhang, 1998). Nodes (feature sets) are sorted in a queue according to classification evaluation metrics evaluated by 5-fold cross-validation, and the queued node having the highest metric is explored further by the algorithm until all nodes are evaluated or a maximum number of iterations are reached while simultaneously recoding the feature set with the optimum metric (e.g. Fig. 2E). The beam width parameter controls the number of nodes subject to further exploration and subsequent evaluation (Fig. 1C). Across different beam width values, we find that beam search exhibits stability since it generates similar results (Supplementary Table T4). The algorithm for identifying optimal feature sets is shown as pseudocode and a flowchart (Supplementary Text S6 and Supplementary Fig. 3D). While OFSs can be optimized for multiple classification evaluation metrics in our framework, in this paper ‘OFS’ typically refers to the feature set corresponding to highest F-score metric, unless otherwise mentioned explicitly. Finally, we examined contributions of individual features to the predictive ability of the OFS (Supplementary Text S8).
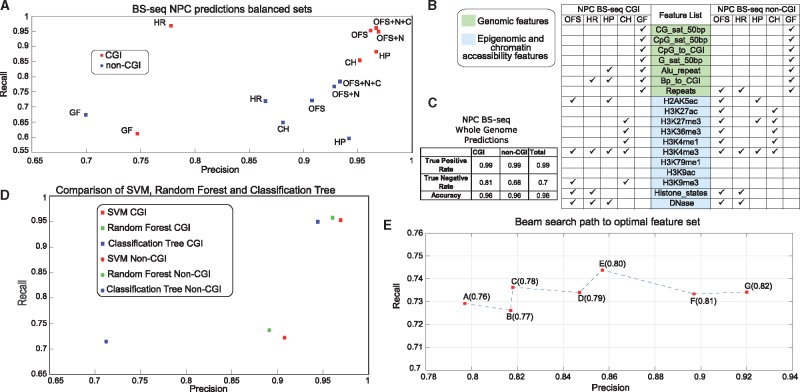
Fig. 2.
DNA methylation status prediction. (A) CGI and non-CGI SVM model predictions for GF (Genomic Features), CH (Chromatin Features), HR (Highest Recall Features), HP (Highest Precision Features), OFS (Highest F-Score Features), OFS + N (OFS + nearest neighbor), OFS + N + C (OFS + N + consensus reference methylome). (B) Feature sets for predicting NPC methylation status. (C) Whole-genome methylation status prediction performance in NPC. (D) Comparison of DIRECTION and classification tree for NPC methylation. (E) An example path traversed by beam search through Precision-Recall space, while optimizing F-score (in brackets) for H1 non-CGI SVM model [A and D: Supplementary Table T5, E: Supplementary Table T8]
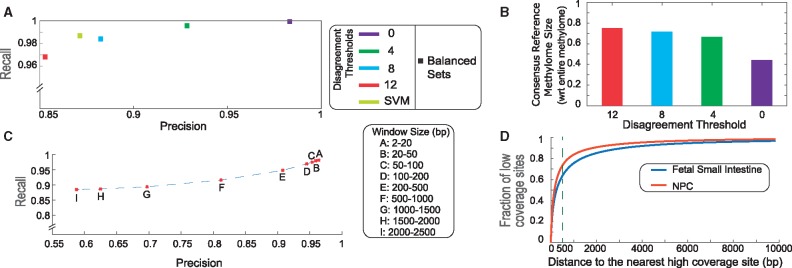
Exploiting correlation within datasets: Binding of DNMT1 to DNA results in a 6000 bp long random walk of an enzyme and subsequent methylation of 50 CpG sites on average, resulting in spatially contiguous stretches of 5-mC modified CpG sites seldom interrupted by lowly methylated CpGs. We engineered several predictor variables based on methylation status of neighboring CpG sites, previously used to impute methylation data (Zhang et al., 2015). Cytosines in CpG sites were divided into ‘high-coverage’ and ‘low-coverage’ sets (sequencing depth at CpG site in the dataset was ≥ or < 5) in NPC. To predict methylation status at each low-coverage cytosine, we compared predictive abilities of the methylation status of the three nearest high-coverage CpG sites to the CpG in question. We additionally contrasted another predictor constructed by using the most common methylation status (preforming a majority vote) across the three nearest high-coverage sites. We find that the precision of prediction drops from the nearest to furthest neighbor, and methylation status of the nearest neighbor’s predictive performance is comparable to the majority methylation status of the three nearest neighbors (Supplementary Fig. 3A). We analyzed the predictive quality of the nearest neighbor based on distance between the predicted CpG site and the nearest neighbor. As distance increases from contiguous up to 2500 bp, both precision and recall decrease (Fig. 3C), with a significant drop after 500 bp. Thus, methylation status of the nearest neighboring high-coverage CpG site within 500 bp was used as a discriminative predictor variable. Since a large fraction of CpG sites have a high coverage neighbor within 500 bp even for moderately sized BS-seq datasets (Fig. 3D), this feature was added to the beam search-identified OFS and the model was retrained for imputation (Supplementary Text S9).
Fig. 3.
DNA methylation predictions harnessing intra- and inter-methylome similarities. (A) Balanced sets predictions on methylation-invariant CpG sites using consensus reference methylome and SVM (Supplementary Table T5). (B) Consensus Reference Methylome size as fraction of total methylome for disagreement thresholds 0, 4, 8, 12 (Supplementary Table T9A). (C) Precision/Recall for methylation status imputation using methylation status of nearest neighboring CpG site as function of distance to nearest neighbor (Supplementary Table T5). (D) Cumulative Distribution Function of the fraction of low coverage CpG sites w.r.t. distance to the nearest high coverage site in a typical high-coverage and low-coverage BS-seq dataset (NPC and fetal small intestine respectively)
Identifying invariance in methylation across datasets: The underlying sequence composition of a genomic region has been documented to shape DNA methylation patterns locally (Yu et al., 2012). Accurate methylome predictions using sequence composition-derived features (Whitaker et al., 2015) suggest that a proportion of CpG sites have invariant methylation status across cell or tissue types and conditions. We identify such CpG sites and optionally use their methylation as an additional feature for performing whole methylome reconstruction or imputation in other datasets (Fig. 1A). Based on 25 high-quality reference human methylomes from the NIH Roadmap Epigenome consortium (Kundaje et al., 2015), we identified the majority methylation status for each CpG site with reliable sequencing depth across the 25 datasets. We refer to the set of cytosines and their corresponding majority methylation status as the consensus reference methylome. We systematically decrease the set of cytosines by additionally constraining that no more than 8, 4 or none out of the 25 reference methylomes could be different from the methylation status of the majority of the methylomes, referring to these variations as consensus reference methylome with disagreement threshold n. While determining methylation status in NPC using such consensus-based predictors, we identified a trade-off between accuracy and applicability. As we increase stringency of the disagreement criterion from 12 to 0, the prediction accuracy improves from 0.85 to 0.99 (on balanced test sets) (Fig. 3A), while the fraction of CpG sites in the genome that can be used to perform this prediction drops from 75% to 44% (Fig. 3B). Given high predictive ability of the consensus reference methylome with zero disagreement, we optionally use this dictionary driven approach as a predictor to reconstruct a portion of the methylome. Depending on the reconstructed methylome, the consensus reference methylome can be created using a different set of relevant reference methylomes, and can potentially provide insight into aberrant CpG methylation in perturbation or disease studies known to affect methylation (Supplementary Text S10).
3 Results
BS-seq and TAB-seq datasets from the NIH Roadmap Epigenome consortium (Kundaje et al., 2015) were used for training and testing our predictive model. Read counts for estimating CCRs in H1 human embryonic stem cell (ESC) line and H1-derived NPC neural progenitor BS-seq datasets (GEO GSE16256) were obtained from the uniformly processed data published by the Roadmap Epigenome consortium (Kundaje et al., 2015), while the BISMARK tool (Krueger and Andrews, 2011) was used for mapping and obtaining the CCRs for H1 (GEO GSE36173) and NPC (GEO GSM882245, GSM1463129) TAB-seq datasets (Supplementary Text S11). These cell types were chosen due to availability of BS-seq and TAB-seq data, and since previous studies performing functional enrichment and analysis of 5-hmC in human and mouse ESCs (Stroud et al., 2011, Wu et al., 2011, Yu et al., 2012, Zhang et al., 2016) and neural progenitors (Song et al., 2011, Tan et al., 2013, Wang et al., 2012), especially in neural development.
DNA methylation prediction: Since there is no precedent for in silico prediction of the 5-hmC modification, we first built a framework for conventional two-state classification of DNA methylation in CpG sites, supervised using BS-seq data. Since distributions and spatial contiguity patterns of highly and lowly methylated CpG sites vary between CGI and non-CGI regions, we trained two classifiers with separately inferred OFSs (Fig. 1A, Model 1, Model 2). Significant differences in prediction quality were observed among different feature sets (agreeing with previous studies (Das et al., 2006, Zhang et al., 2015)) suggesting the importance of feature set selection. We performed optimal feature selection using our beam search algorithm, and identified feature sets with the best precision, recall, and harmonic mean of the two (F-score) for training and testing balanced sets of both classes in H1 and NPC with minor performance differences (H1: Supplementary Fig. 2E, F, NPC: Fig. 2A). Whole genome predictions (Fig. 2C) were carried out subsequently (Supplementary Tables T5, T6 for results). The whole genome predictions were also used to assess the performance of DIRECTION across varying values of BS-seq CCRs (Supplementary Text S11).
Comparison with other DNA methylation prediction tools: Different methylation prediction algorithms work at differing genomic resolutions, on different datasets, using different predictor variables, to predict different response variables; making it challenging to set up unbiased comparisons between models. However, based on reported performances, DIRECTION is comparable to state-of-the-art high resolution methylation prediction algorithms (Whole-genome accuracy: DIRECTION: 0.96 versus (Zhang et al., 2015): 0.91, Supplementary Table T1). Also, under the constraint of the same predictor variable set, DIRECTION outperformed the well-established inbuilt MATLAB classification tree function (Fig. 2D).
OFS for DNA methylation prediction: The most discriminative features, contributing to high recall and precision, in DNA methylation predictions in NPC CGI regions were chromatin ‘states’ inferred by the ChromHMM model (Ernst and Kellis, 2012), and H2AK5ac histone modification (Fig. 2B and Supplementary Table ST7). The underlying biological interpretation of our findings is supported by published literature as H2AK5ac histone modification was shown to be enriched in regions of euchromatin and low methylation (Cuddapah et al., 2009). Also, the OFS for predicting DNA methylation in NPC CGI regions has only 5 features (Fig. 2B), including transcription activation (H3K4me3, H2AK5ac) and repression (H3K9me3) associated histone marks, and DNase hypersensitivity (discriminative with respect to underlying DNA methylation (Lazarovici et al., 2013). Contrasting CGI to non-CGI OFSs, we find several histone modification features (H3K27ac, H3K27me3, H3K36me3 and H3K4me1) in the non-CGI OFS, as opposed to H3K9me3 in the CGI OFS. The non-CGI OFS also contains the Repeat feature (repetitive elements), which is meaningful since repeat-containing retrotransposons in the human genome are silenced by methylation (Ooi et al., 2009). The most prominent changes in predictive ability are depicted by significantly different recall (Fig. 2A) and AUC (Supplementary Fig. 2G, H; Supplementary Text S12).
Transfer learning across cell types: Given that one of our goals is to perform whole-methylome reconstructions, we trained our classifier on H1 cells and tested its performance on NPC and vice versa. The results of the testing are only a few percentage points worse than the corresponding results in the same cell type (Supplementary Table T6), due to the fact that our approach relies on a minimal set of discriminative features (OFS) which are similar in H1 and NPC, and therefore has great promise for ‘transfer learning’ scenarios like de novo reconstruction of the methylome. In order to test the limits of such transfer learning, we used the NPC-trained SVM to perform whole methylome predictions in the totipotent Mesenchymal Stem Cells (MSC) and in the terminally differentiated fetal fibroblast cell line IMR90. Since loss of pluripotency is associated with epigenome reprogramming involving DNA methylation, we find that the NPC-trained SVM performs well on the MSC dataset, but performs only modestly in IMR90 (Supplementary Table ST6 and Supplementary Text S12).
Using neighboring CpG sites as predictor variables: For improving imputation, the methylation status of the nearest neighboring CpG site within 500bp was used to create an input feature. Our feature engineering analyses (see Methods, Fig. 3C) suggests that the predictive quality of the feature significantly decreases after 500 bp (a distance corresponding to the average size of CGIs (Kang et al., 2006), in agreement with findings that CGIs are typically consistently methylated or demethylated. We tested the ability of this feature to contribute to predictions in CGI and non-CGI SVM models by adding it to the beam search-identified OFS, followed by retraining the SVMs on balanced sets. It makes insignificant impact on the CGI SVM (where precision and recall are > 0.95) but strikingly improves recall of the non-CGI SVM from 0.72 to 0.77 (Fig. 2A), suggesting that even in non CpG-rich regions, spatial contiguity of methylation status is common.
Consensus reference methylome based predictions: Based on the 44% of CpG sites that are methylation-invariant in our reference, we compared our SVM prediction model to the prediction based on the consensus reference methylome. Both predictors were highly accurate and comparable on the set of cytosines underlying the consensus reference methylome with zero mismatches, and on balanced subsets, the precision of the SVM was 0.87, compared to 0.99 of the most stringent consensus-based predictor (Fig. 3A). We then incorporated the consensus reference methylome-based predictor into our ensemble-learning framework (Fig. 2A), testing the framework with and without the consensus reference methylome on balanced sets. The prediction metrics had incremental improvement in CGI regions, and significant improvement in non-CGI regions, suggesting that an ensemble prediction scheme is optimal. On whole genome datasets, we see incremental improvement in NPC methylation status prediction accuracy (0.97) as opposed to solely SVM or RF (0.96) (Supplementary Table T6).
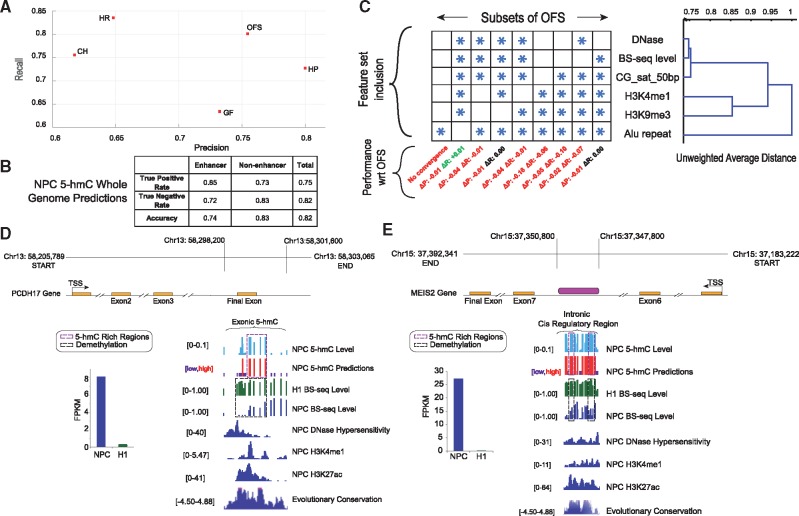
5-hmC status prediction: We performed 5-hmC status prediction using features from the initial feature set for methylation status prediction model, using methylation level as an additional feature (Supplementary Table T10). In order to identify the most discriminative features for 5-hmC status prediction, we ran our beam search algorithm and obtained discriminative feature sets. Based on the experimental design previously outlined, the performance of the OFS was compared against other biologically and statistically meaningful feature sets (NPC: Fig. 4A, F-score 0.78; H1: Supplementary Fig. 4B, F-score 0.7). The most distinguishing characteristic of assorted 5-hmC feature sets in both cell types was the profound presence of active enhancer histone modifications H3K4me1 and H3K27ac (Shlyueva et al., 2014), DNase and other genomic derived features including CpG content, and Alu repeats (Supplementary Table T11). Insightfully, a single addition to the OFS when our predictor was constrained to the enhancer regions was H3K27ac, suggesting biological interpretability of our results. The absence of H3K27ac from the 5-hmC OFS (when the predictor is not constrained to enhancer regions) can be explained by the presence of another enhancer chromatin mark (H3K4me1) in the OFS, and the relatively small size of enhancer regions compared to the non-enhancer portion of the genome. Unsurprisingly, we find the H3K4me1 enhancer mark being one of the most promising predictive features due to its presence in both the high recall and optimal feature sets. Significant depletion of 5-hmC in H3K9me3 rich heterochromatin regions, and its positive correlation with H3K4me3 active histone modification (Yamaguchi et al., 2013), clearly designates these chromatin marks as suitable candidates for the OFS. In order to show that the obtained OFS is discriminative towards 5-hmC signal, we predicted 5-hmC status across various TAB-seq level thresholds and noticed that the prediction metric grows slowly with the increase in threshold value (Supplementary Fig. 4G), and shows consistent AUC for a range of thresholds (Supplementary Fig. 4D, E). We performed whole-genome 5-hmC predictions in NPC and H1 and obtained 0.82 and 0.75 accuracy respectively (NPC: Fig. 4B; H1: Supplementary Fig. 4C). These results together suggest that 5-hmC status can be fairly accurately reconstructed in our datasets. Lower prediction accuracy in H1 can putatively be attributed to a lower coverage depth in the training data. Finally, we performed 5-hmC predictions restricted to cytosines with high BS-seq CCRs, yielding comparable results to our previous analyses, implying that the numerous public BS-seq datasets together with additional input features can be used to predict 5-hmC maps (Supplementary Text S13).
Fig. 4.
5-hmC status prediction in NPC. (A) 5-hmC balanced set prediction evaluation for SVM (Supplementary Table T10). (B) 5-hmC whole-genome prediction metrics. (C) OFS feature clustering. At each node, leaves (features) under it were removed from OFS to create new feature sets. For these, feature inclusion (starred) and resultant change in precision/recall w.r.t. OFS (by reclassifying dataset) characterize features’ contribution to classification quality. (D,E) Visualization of 5-hmC status prediction and discriminative input features in PCDH17 and MEIS2
5-hmC transfer learning across H1 and NPC: In analogous fashion to our methylation data, we trained our classifier on H1 cells and tested its performance on NPC and vice versa. The results of the testing suggest that transfer learning across H1 and NPC is feasible (Supplementary Table T10).
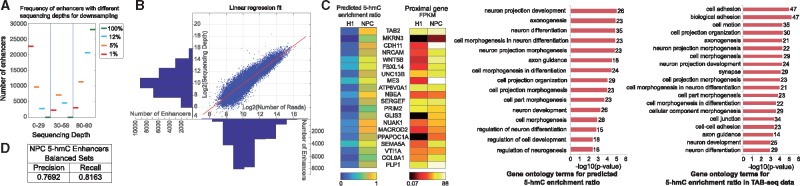
OFS feature contributions: We constructed a dendrogram (Fig. 4C) for the 5-hmC status prediction OFS, and eliminated subsets of features as described in Supplementary Text S8. The most notable changes to recall were observed upon elimination of the BS-seq CCR feature, while precision was affected by H3K4me1 and GC saturation removal, signifying the importance of these features to the prediction rate. Only four features (BS-seq CCR, GC saturation, DNase and Alu) are sufficient to capture the majority of TAB-seq signal by garnering 0.75 F-score in NPC (Fig. 4C). Several of these were identified in the literature to be enriched in regions of high hydroxymethylation (Yu et al., 2012). We show our 5-hmC prediction at work in two genomic regions proximal to PCDH17 and MEIS2 genes (Fig. 4D, E), previously implicated in synapse formation and interneuron development (Batista-Brito et al., 2009, Hoshina et al., 2013).
Overall prediction in enhancer regions: 5-hmC is differentially enriched in functionally important enhancers (Stroud et al., 2011). Thus, we trained and tested our model by restricting it only to NPC enhancers (identification of enhancers in Supplementary Text S14), obtaining 0.77 precision, 0.82 recall (Fig. 5D) and a high AUC (Supplementary Fig. 4F). The active enhancer mark H3K27ac was present in the OFS (Supplementary Table T11A) suggesting a correlation of 5-hmC with enhancer activation. A significant improvement in the maximum precision feature set (HP) was found in models constrained to enhancers (Supplementary Fig. 4A), due to 5-hmC overabundance in enhancers.
Fig. 5.
5-hmC status prediction in enhancers. (A) Sequencing depth across cytosines in enhancers after downsampling. (B) Log-log linear regression fit (mapped read count versus sequencing depth across cytosines) in NPC enhancers. (C) Heatmap of predicted 5-hmC enrichment ratio and proximal gene expression for enhancers with highest predicted gain in 5-hmC enrichment ratio (NPC versus H1). GO term enrichment for genes with highest 5-hmC enrichment ratio (NPC versus H1) using predictions and TAB-seq data. (D) 5-hmC status prediction in NPC enhancers
5-hmC prediction in small TAB-seq datasets: BS-seq and TAB-seq datasets require high sequencing depth to reliably determine CpG methylation and 5-hmC status across the genome, but as coverage decreases in smaller datasets, the ability to do so is diminished. The feasibility of training a model (like SVM) does not decrease proportionally to dataset size, as we can train SVMs with as few as 2000–2500 training examples (Supplementary Fig. 2A, Supplementary Table T12). We downsampled one of our NPC datasets to 12% (commensurate with RRBS-seq dataset sizes (Gu et al., 2011)) of the original number of reads, and predicted the corresponding sequencing depth in enhancer CpG cytosines (Fig. 5A, B). We find sufficient training examples (>2000) at resolutions of both whole enhancers and individual cytosines with sequencing depths suited for reliable CCR estimation in training SVMs, suggesting feasibility of robustly training 5-hmC status prediction models in enhancers for reduced representation TAB-seq data (Supplementary Text S15).
In silico framework for high throughput hypothesis-testing: Hypothesis testing using TAB-seq data to identify 5-hmC rich regions or differential 5-hmC enrichment across conditions, naturally leads to a feasibility study of performing such tests on in silico predictions. 5-hmC is an intermediate in the demethylation pathway and low DNA methylation levels are the hallmark of active enhancers (Yu et al., 2012). Thus, we hypothesized that increase in an enhancer’s 5-hmC enrichment (quantified as 5-hmC enrichment ratio, Supplementary Text S16) from H1 to NPC differentiation corresponded to changes in proximal gene expression, putatively indicative of functional differences between H1 and NPC. We identified enhancers with the largest changes in 5-hmC enrichment ratio using both experimental TAB-seq data and our 5-hmC predictions. Gene set enrichment analysis (Supplementary Text S17) on proximal genes to the identified enhancers reveal similar results for the two gene sets, enriched in neurodevelopmental processes. We find differential expression between H1 and NPC in the prediction-based gene set, suggesting our prediction-based functional study yields biologically relevant findings (Fig. 5C, Supplementary Data 1, Supplementary Data 2, Supplementary Data 3).
4 Discussion
Our work opens up new directions in DNA methylation studies. Discriminative feature sets for predicting 5-hmC status include features engineered to leverage idiosyncrasies of hydroxymethylation, like strand asymmetry, G-rich sequence bias, and enrichment in open chromatin and gene bodies. Such correlative descriptions of 5-hmC modification with respect to genomic and epigenomic features can help create fine-grained ‘epigenome states’ by integrating 5-mC and 5-hmC modifications with histone mark based chromatin states (Ernst and Kellis, 2012) in the future. For purposes of predicting BS-seq signal, we identified CpG sites that are methylation-invariant across reference human methylomes we analyzed, helping improve balanced set and whole-genome methylation status prediction (Supplementary Text S18 for strengths and limits of our framework). In the future, we aim to identify and characterize the correlational structure of reference methylomes across developmental linages and tissue types. Such studies can potentially yield insight into regulatory mechanisms, and identify aberrant methylation patterns in disease or perturbation models. DIRECTION is the first in-silico, whole-epigenome predictor of DNA methylation and 5-hmC status at single nucleotide resolution, with results comparable to state-of-the-art DNA methylation prediction tools. Our tool allows us to identify candidate genomic regions for differential hydroxymethylation as a first step in functional studies. Unlike previous feature-intensive approaches for predicting DNA methylation, our algorithm uses a sophisticated feature selection technique adopted from artificial intelligence and identifies a small subset of nonredundant, discriminative, predictive features. This allows for greater biological interpretability of generated results, superior performance in resource-scarce scenarios, making the model sparse without explicit regularization. DIRECTION is an open-source, agile, scalable ensemble predictor using biologically and practically motivated genome partitioning and training a predictive model per partition, allowing us to deconvolute inevitably mixed biological signals in whole-epigenome studies. In the future, we aim to extend DIRECTION by predicting DNA methylation and 5-hmC status in additional genomic contexts (like non-CpG cytosines), other methylation paradigms (like epigenetic reprogramming in gametes), and in non-mammalian species where methylation plays distinct functional roles.
Supplementary Material
Acknowledgements
PR designed search algorithm. PR & MP optimized SVM & RF. MP & PR wrote code. PR designed & MP performed experiments. PR & MP wrote manuscript, with input from MC & KP. MP & KP created figures. KP performed methylome clustering. AK performed feature engineering. PR supervised MP & AK. MC provided feedback and advised MP. MQZ supervised personnel, guided all research. All authors have read the manuscript and agree on its content.
Thanks: D. Tangellamudi for MRE coding, M. Guinn for figure help, Z. Xuan for enhancer annotation, T. Price & G. Dussor for feedback.
Funding
This work has been supported by a National Institutes of Health grant (Institute: NIEHS) U01 ES017166 to MQZ / San Diego Epigenome Institute and National Institutes of Health grant (Institute: NIAMS) K25 AR063761 to MC.
Conflict of Interest: none declared.
References
- Bachman M. et al. (2014) 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem., 6, 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R. et al. (2009) The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron, 63, 466–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin M. et al. (2005) Prediction of methylated CpGs in DNA sequences using a support vector machine. FEBS Lett., 579, 4302–4308. [DOI] [PubMed] [Google Scholar]
- Bishop C. (2007) Pattern Recognition and Machine Learning (Information Science and Statistics), 1st ed. 2006. corr. 2nd printing ed. Springer, New York. [Google Scholar]
- Bock C. et al. (2006) CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet., 2, e26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth M.J. et al. (2012) Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science, 336, 934–937. [DOI] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Cuddapah S. et al. (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res., 19, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. et al. (2006) Computational prediction of methylation status in human genomic sequences. Proc. Natl. Acad. Sci. U. S. A., 103, 10713–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M. (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M. (2015) Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol., 33, 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S. et al. (2016) Predicting CpG methylation levels by integrating Infinium HumanMethylation450 BeadChip array data. Genomics, 107, 132–137. [DOI] [PubMed] [Google Scholar]
- Frommer M. et al. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U. S. A., 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H. et al. (2011) Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc., 6, 468–481. [DOI] [PubMed] [Google Scholar]
- Hackett J.A. et al. (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science, 339, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshina N. et al. (2013) Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron, 78, 839–854. [DOI] [PubMed] [Google Scholar]
- Jones P.A. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet., 13, 484–492. [DOI] [PubMed] [Google Scholar]
- Kang M. et al. (2006) The length of CpG islands is associated with the distribution of Alu and L1 retroelements. Genomics, 87, 580–590. [DOI] [PubMed] [Google Scholar]
- Khare T. et al. (2012) 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat. Struct. Mol. Biol., 19, 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. et al. (2014) Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Hum. Mol. Genet., 23, 657–667. [DOI] [PubMed] [Google Scholar]
- Koller D., Sahami M. (1996) Toward optimal feature selection. In: Proceedings of 13th International Conference on Machine Learning, pp. 284–292.
- Krueger F., Andrews S.R. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A. et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarovici A. et al. (2013) Probing DNA shape and methylation state on a genomic scale with DNase I. Proc. Natl. Acad. Sci. U. S. A., 110, 6376–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B. et al. (2014) Predicting DNA methylation level across human tissues. Nucleic Acids Res., 42, 3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.H., De la Torre F. (2010) Optimal feature selection for support vector machines. Pattern Recognit., 43, 584–591. [Google Scholar]
- Ooi S.K. et al. (2009) Mammalian cytosine methylation at a glance. J. Cell. Sci., 122, 2787–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J. et al. (2013) MLML: consistent simultaneous estimates of DNA methylation and hydroxymethylation. Bioinformatics, 29, 2645–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyueva D. et al. (2014) Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet., 15, 272–286. [DOI] [PubMed] [Google Scholar]
- Song C. et al. (2011) Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol., 29, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H. et al. (2011) 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol., 12, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F. et al. (2014) Hydroxymethylated cytosines are associated with elevated C to G transversion rates. PLoS Genet., 10, e1004585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. et al. (2013) Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res., 41, e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teif V.B. et al. (2014) Nucleosome repositioning links DNA (de) methylation and differential CTCF binding during stem cell development. Genome Res., 24, 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2012) Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum. Mol. Genet., 21, 5500–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2016) Predicting DNA Methylation State of CpG Dinucleotide Using Genome Topological Features and Deep Networks. Sci. Rep., 6, 19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker J.W. et al. (2015) Predicting the human epigenome from DNA motifs. Nat. Methods, 12, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzodek C. et al. (2012) Linking the epigenome to the genome: correlation of different features to DNA methylation of CpG islands. PloS One, 7, e35327.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev., 25, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. et al. (2013) Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res., 23, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. et al. (2015) Chromatin modifications and genomic contexts linked to dynamic DNA methylation patterns across human cell types. Sci. Rep., 5, 8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. (2013) Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene, 32, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. et al. (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell, 149, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2015) Predicting genome-wide DNA methylation using methylation marks, genomic position, and DNA regulatory elements. Genome Biol., 16, 14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2016) Isoform Switch of TET1 Regulates DNA Demethylation and Mouse Development. Mol. Cell, 64, 1062–1073. [DOI] [PubMed] [Google Scholar]
- Zhang W. (1998) Complete anytime beam search. In: Proceedings of the Fifteenth National Conference on Artificial Intelligence (AAAI/IAAI), pp. 425–430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.