Abstract
Prokaryotic Argonaute proteins (pAgos) have been proposed as an alternative to the CRISPR/Cas9 platform for gene editing. Although Argonaute from Natronobacterium gregoryi (NgAgo) was recently shown unable to cleave genomic DNA in mammalian cells, the utility of NgAgo or other pAgos as a targetable DNA-binding platform for epigenetic editing has not been explored. In this report, we evaluated the utility of two prokaryotic Argonautes (NgAgo and TtAgo) as DNA-guided DNA-binding proteins. NgAgo showed no meaningful binding to chromosomal targets, while TtAgo displayed seemingly non-specific binding to chromosomal DNA even in the absence of guide DNA. The observed lack of DNA-guided targeting and unexpected guide-independent genome sampling under the conditions in this study provide evidence that these pAgos might be suitable for neither gene nor epigenome editing in mammalian cells.
Introduction
Eukaryotic Argonaute proteins (eAgos) use small single-stranded RNA to target complimentary RNA sequences and play a key role in the RNA interference (RNAi) pathway [1]. On the other hand, recently discovered prokaryotic Argonaute proteins (pAgos) have been implicated in the targeting of foreign DNA for degradation [2]. Some pAgos have been shown to use small single-stranded DNA (ssDNA) to target and cleave double-stranded DNA and hence offer an intriguing possibility for gene and epigenome editing [3–6].
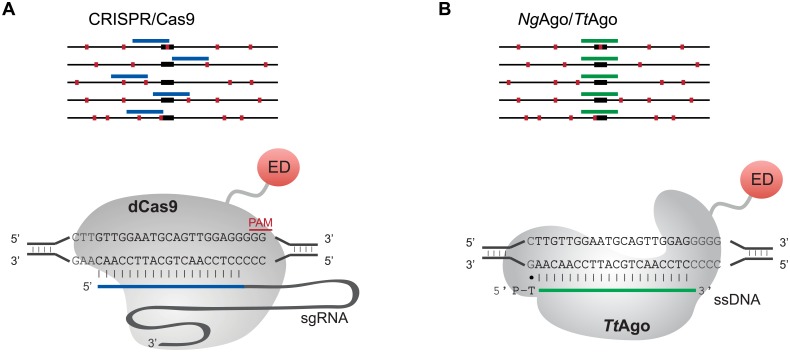
In principle, pAgos offer several advantages for DNA-guided site-specific binding in the mammalian genome, particularly because of their increased flexibility of targeting. While CRISPR/Cas9 is able to target near many genetic features, the necessity of a protospacer adjacent motif (PAM) in the target site often makes it impossible to design guide RNAs (gRNAs) to bind exactly at features such as SNPs, individual CpGs, intron/exon boundaries, or specific transcription factor binding sites (Fig 1A). Indeed this limitation has led to many efforts to expand the targeting lexicon using engineered or natural variant Cas9 proteins [7–10]. However, nearly all have been similarly or even more restricted than the 5’-NGG-3’ PAM site of the canonical SpCas9 [11]. pAgos do not require a PAM adjacent to their guide DNA (gDNA) target sites, raising the prospect of targeting without sequence restrictions (Fig 1B).
Fig 1. The potential for high-precision targeting by pAgos.
A. Targeting of CRISPR/Cas9 is limited by the requirement of a PAM site (green boxes). B. NgAgo or TtAgo might allow the precise targeting of features, such as transcription factor binding sites (black boxes). gRNA/gDNA, red lines.
An early report that the Argonaute from Natronobacterium gregoryi (NgAgo) could efficiently cleave genomic DNA in mammalian cells [12] was found to be irreproducible [13–17], eventually leading to its retraction [18]. The latter studies suggest that NgAgo will not be a useful tool for gene editing. However they did not rule out that NgAgo and other pAgos could be used as targetable DNA binding platforms, in analogy to catalytically inactive or dead Cas9 (dCas9) [19–21] or Cpf1 (dCpf1) [22, 23]. dCas9 has been widely used for applications such as gene activation or repression, epigenome editing, and imaging of DNA and RNA in living cells (reviewed in [24, 25]). Indeed, two recent studies found that NgAgo was able to inhibit gene expression even in the absence of DNA editing [15, 17], reminiscent of dCas9-mediated CRISPR interference (CRISPRi, [19]).
These observations motivated us to examine NgAgo as a PAM-less, DNA-guided DNA binding platform. In addition, pAgo from Thermus thermophilus (TtAgo) can be programmed to bind and cleave double stranded plasmid in a site-specific manner in vitro [4, 5]. The use of TtAgo as a tool for gene editing in mammalian cells is limited by its requirement for high temperatures to cleave double-stranded DNA. However, it has been shown to bind and cut single-stranded DNA at a physiological temperature of 37°C [5], making TtAgo an additional candidate as a programmable DNA-binding platform.
Results
Confirmation of lack of targeted DNA cleavage by hTtAgo and hNgAgo in the mammalian genome
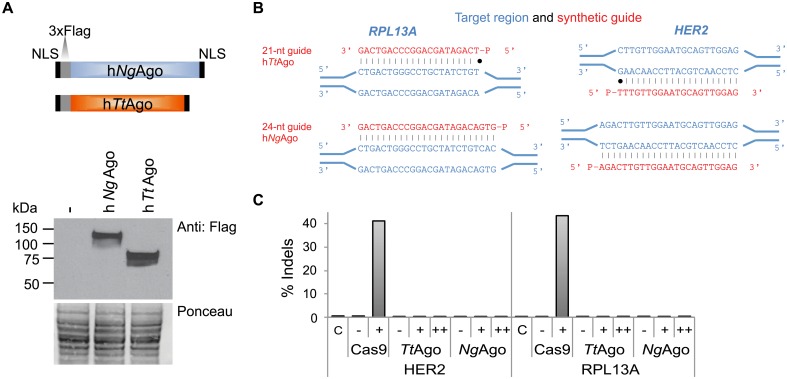
First we evaluated that TtAgo and NgAgo expressed in mammalian cells are in principle able to cleave DNA targets in vitro. HEK293 cells were transfected with plasmids expressing human codon optimized Argonautes (hNgAgo and hTtAgo) with an N-terminal 3xFlag epitope tag and nuclear localization signals both at the N and C termini (Fig 2A; protein sequence provided as S1 Fig). Western blot analysis confirmed hTtAgo and hNgAgo protein expression in HEK293 cells (Fig 2B). hTtAgo and hNgAgo proteins were purified from HEK293 cells (S2A Fig) and the nature of co-purified nucleic acids was evaluated (S2B Fig). The profile of nucleic acids co-purified with mammalian expressed hTtAgo recapitulates TtAgo binding characteristics observed in E. coli [5]. The majority of co-purified nucleic acids consisted of RNA, but in addition short DNAs were also observed. The ability of purified hTtAgo/hNgAgo to cleave single-stranded DNA templates was assessed in vitro (S2C Fig, S1 Table). As expected, hTtAgo cleaved a 98-nt single-stranded DNA target when provided with a 21-nt phosphorylated gDNA (previously described in [5]) at the high temperatures of 75°C and 55°C (S2D and S2E Fig). However, no cleavage activity was observed at the physiological temperature of 37°C (S2F Fig). It has recently been shown that bacterial expressed TtAgo has a preference for target sites with a G nucleotide in position 1 (t1G; [26]). Cleavage efficiency of mammalian-expressed hTtAgo at the RPL13A target site was only detectable when the target site had a G nucleotide in position 1 (t1G; S2E Fig). No DNA cleavage was observed for hNgAgo (S2D–S2F Fig).
Fig 2. hTtAgo and hNgAgo do not cleave genomic target sites.
A. Schematic of human codon-optimized TtAgo and NgAgo proteins containing two nuclear localization signals (NLSs) and a 3xFlag epitope tag. Western blot analysis of hTtAgo and hNgAgo proteins in HEK293 cells using an antibody against the 3xFlag tag. Untransfected cells serve as a negative control (-). Ponceau staining was used as a loading control. B. Diagram illustrating RPL13A and HER2 guide DNAs and genomic target site. Genomic target sites are indicated in blue and complementary ssDNA guides are indicated in red. C. Amplicon sequencing was carried out on HEK293 cells co-transfected with hTtAgo or hNgAgo expression plasmids with (+) or without (-) gDNAs to RPL13A and HER2. To increase the amount of gDNAs, cells were re-transfected with gDNAs 24 hours after the initial transfection (++). CRISPRESSO analysis confirms that hTtAgo and hNgAgo did not cause insertions or deletions (indels) under any of these conditions (S5 Table). As a control, HEK293 cells were co-transfected with Cas9 nuclease and gRNA expression plasmids targeting RPL13A and HER2. RNA-guided Cas9 displayed target site cleavage at the genomic RPL13A and HER2 target sites. The percentage of sequence reads containing indels relative to the total number of sequence reads is plotted on the y-axis.
As it is a prerequisite for transcriptional and epigenetic regulators to bind but not cleave their target sequence in vivo, we evaluated the cleavage ability of hTtAgo in HEK293 and HeLa cells at six genomic target sites (NFE2L1, NPAS1, RPL13A site1, RPL13A site2, RB1A site1, RB1A site2) using three gDNA configurations: 1) a single gDNA, 2) two complimentary gDNAs and 3) two gDNAs targeting the opposite DNA strand separated by a 15-nt spacer (S3 Fig, gDNAs are listed in S2 Table). An hTtAgo-expressing plasmid was transfected alone or in combination with different 5’-phosphorylated 21-nt gDNAs in HEK293 cells. Amplicon sequencing was used to capture even low frequency cleavage events. Genomic target regions were amplified by PCR (primer sequences are listed in S3 Table) and analyzed for the presence of insertions and deletions (indels) at the cleavage site using next-gen sequencing. We did not observe indel frequencies above background, or above the detection limit of 0.01%, at any of the regions targeted by hTtAgo (S4 Table). After adjusting for read depth, the number of substitutions were similar between untreated cells, cells that expressed hTtAgo only and cells that contained both hTtAgo and gDNAs. The same results were obtained when the 5’-phosphorylated gDNAs contained four phosphorothioate linkages to prevent degradation of the gDNAs (S5 Table).
Several studies have reported that NgAgo is not able to cleave genomic DNA in vivo [13–17]. When we expanded indel analysis to include hNgAgo at the HER2 and RPL13A loci in HEK293 cells, we did not observe cleavage by hTtAgo or hNgAgo in the presence of 21-nt or 24-nt gDNAs, respectively (S6 Table). To increase the amount of gDNAs present in the cell, we re-transfected cells with their respective DNA guides 24 hours after the initial co-transfection with pAgo and gDNAs. Increase of gDNAs in the cell did not result in DNA cleavage at the corresponding genomic target (Fig 2C). When HER2 and RPL13A sites were targeted with SpCas9-gRNA complexes, cleavage efficiencies of 40% and 43% were observed, respectively (Fig 2C).
5’ Phosphorylated single-strand DNA guides fail to guide hNgAgo and hTtAgo to specific genomic target sites
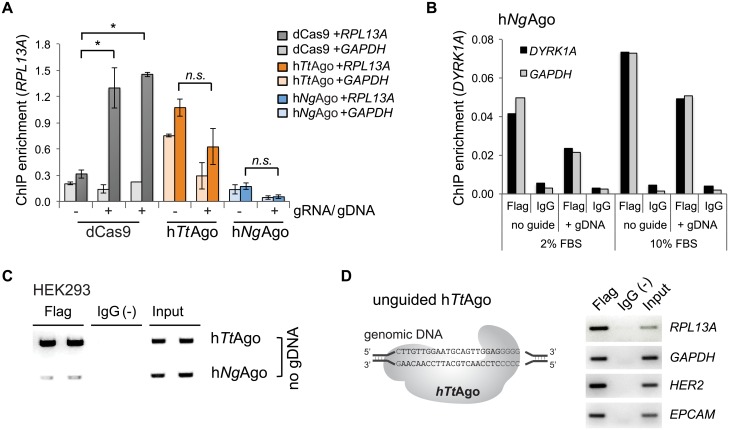
Having established that the pAgos did not cleave their genomic target sites, we next investigated if gDNAs can guide hTtAgo or hNgAgo to complementary genomic target sites in mammalian cells. We performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) using an antibody to the 3xFlag tag of hNgAgo or hTtAgo in the presence or absence of 5’ phosphorylated gDNA to the RPL13A locus in HEK293 cells. As a positive control, we used a catalytically inactive Cas9 (dead or dCas9) and a gRNA targeting RPL13A. As expected, dCas9 was able to bind the target locus in a gRNA-dependent manner, resulting in a 4-fold increase of dCas9 binding to the sense or anti-sense strand of the RPL13A locus in the presence of the gRNA (Fig 3A, Student’s t-test p<0.05). Binding of dCas9 was specific, since RPL13A gRNA did not increase dCas9 binding at the GAPDH control locus. However, the binding of hNgAgo to RPL13A was similar to the background level of Cas9 binding, and there was no preference for the target site compared to the non-target GAPDH control locus (Fig 3A). Similar results were observed when hNgAgo was targeted to DYRK1A using the G5 gDNA described in the original report by Gao et al. [12], in the presence of either 2% or 10% FBS-supplemented growth media (Fig 3B). ChIP enrichment for hNgAgo was always greater than for the IgG negative control, indicating some binding to chromosomal DNA (Fig 3B and 3C). However, the binding was weak, non-specific, and occurred even in the absence of a gDNA. In fact, the presence of a gDNA appeared to cause a slight decrease in DNA binding, although the trend did not reach the level of statistical significance. In contrast, hTtAgo displayed strongly enriched binding to chromosomal DNA in HEK293 cells (Fig 3A and 3C). However, there was no significant preference for the RPL13A target compared to the non-target GAPDH, and binding occurred in the absence of a gDNA (Fig 3A). The unexpected affinity of hTtAgo for untargeted genomic sites was further evaluated by ChIP-qPCR in HeLa cells at the RPL13A, EPCAM, HER2 and GAPDH loci. Strikingly, in the absence of any transfected gDNA, binding of hTtAgo was observed at all loci investigated (Fig 3D).
Fig 3. DNA guides do not facilitate binding of hTtAgo and hNgAgo to genomic target loci.
A. ChIP-qPCR enrichment to quantitate binding to RPL13A locus. HEK293 cells were transfected with hTtAgo- and hNgAgo-expression plasmids either with (+) or without (-) gDNAs. dCas9 and two different gRNA expressing plasmids were co-transfected as controls (Student two-sided T-test; *, p<0.05; n.s., not significant; n = 2 independent experiments; mean ± SEM). Binding to a non-target locus (GAPDH) was evaluated to interrogate binding specificity. B. Targeted binding of hNgAgo failed at the DYRK1A locus in HEK293 cells. HEK293 cells were co-transfected with hNgAgo expression plasmid together with 24-nt 5’-phosphorylated gDNAs (G5 gDNA) or without gDNA. ChIP assays were performed with an antibody to the 3xFlag tag of hNgAgo or with rabbit IgG as a negative control. Addition of G5 gDNA that was targeted to the DYRK1A locus did not increase hNgAgo binding above background level (no guide). There was no difference in binding to DYRK1A or to a control region (GAPDH). Two conditions (2% or 10% FBS in growth media) were tested. C. ChIP-PCR to measure unguided hTtAgo and hNgAgo binding to the RPL13A locus. ChIP assays were performed with two biological replicates. Flag, Flag ChIP; IgG(-), IgG negative control; Input, 0.1% chromatin input. D. hTtAgo binds multiple genomic regions without gDNA in HeLa cells. ChIP assays were performed in HeLa cells expressing hTtAgo without DNA guides. Standard PCR with locus-specific primers demonstrated hTtAgo binding to RPL13A, GAPDH, HER2 and EPCAM. Flag, Flag ChIP; IgG(-), IgG negative control; Input, 0.1% chromatin input.
Discussion
In this study, we evaluated the utility of pAgos as DNA-guided DNA-targeting tools using ChIP assays. Neither hNgAgo nor hTtAgo were able to bind to the mammalian genome in a targeted manner, but instead appeared to bind to essentially all loci examined in two human cell types, even in the absence of any gDNA. Overall our data demonstrate that single-stranded gDNAs do not efficiently guide hTtAgo or hNgAgo to specific genomic sites in the physiological context of a mammalian cell. We have demonstrated that the mammalian-expressed hTtAgo is functional in an in vitro cleavage assay. This is important to note since nuclear localization signals have been appended to the N- and C-terminal ends of hTtAgo. The C-terminal amino acid of hTtAgo and the Argonaute from Archaeoglobus fulgidus is structurally important for the binding of magnesium and the DNA or RNA guide [27, 28]. We were unable to observe evidence of cleavage using mammalian-expressed hNgAgo either in vitro or in vivo. By now, several groups have reported that NgAgo cannot cleave genomic DNA in vivo [13–17], but in vitro cleavage assays have not been reported. Our results lead us to conclude that hNgAgo and hTtAgo may not be suitable as targetable DNA-binding platforms for creating epigenetic editing tools. It is possible that some aspect of the experimental conditions prevented more favorable outcomes, such as unexpectedly poor transfection of guide DNAs. However, we note that the conditions used here did support binding of dSpCas9, and were the same used by others to show that NgAgo could not cleave DNA in vivo. Exploration of mutations or other variants of Argonaute proteins or covalently linking gDNAs to Argonaute proteins may provide a more positive outcome.
The mechanism for the dramatic unguided and seemingly non-specific binding is unclear. One explanation could be the sampling and cleavage of foreign DNA by pAgos that has been proposed as a means of host defense against mobile DNA from viruses or plasmids [5, 26, 29, 30]. The mechanism by which pAgos acquire guides to preferentially target foreign DNA remains elusive, but presumably could be operating in the mammalian cells to cause apparent non-specific binding to DNA sequences. The TtAgo double mutant (D478A/D546A) that is unable to bind small single-stranded DNA guides may be useful for testing such a hypothesis [5]. Another explanation could be that some pAgos simply have non-specific DNA binding activity (i.e., are sticky). In this regard, it is worth noting that hTtAgo, which showed the greatest binding activity, comes from a thermophilic species and may therefore have surface charges or other features that assist in its binding to DNA at high temperatures. These features might result in non-specific binding at lower temperatures. Alternatively, the pAgos might interact with unknown proteins or RNA in human cells.
Our findings would also seem to rule out a CRISPRi-like mechanism to explain the observed knock-down by NgAgo of fabp11a mRNA expression in zebrafish embryos [15] and hepatitis B virus pgRNA in human cells [17]. It is possible that NgAgo induced DNA-guided gene knockdown in these cases through a DNA-dependent RNA cleavage activity [31]. Prokaryotic Argonautes have been the subject of exuberant expectations and dramatic disappointments regarding their use as targeted tools to alter gene expression. However, continued careful study and characterization may yet provide the mechanistic insights required for their appropriate application.
Materials and methods
Construction of hTtAgo and hNgAgo expression plasmids
The human codon-optimized TtAgo was synthesized from GeneArt. The GeneArt plasmid was digested with SfiI restriction enzyme. pCDNA3-hTtAgo was created using Gibson Assembly (New England Biolabs) by inserting the hTtAgo fragment into the FseI and NheI digested ideal pCDNA3-dCas9 expression plasmid replacing the dCas9 sequence [32]. The human codon-optimized NgAgo was synthesized in two gBlocks (IDT) and was cloned into KpnI and NheI digested pCDNA3-hTtAgo by Gibson Assembly of the three fragments (New England Biolabs). The resulting pCDNA3-hTtAgo and pCDNA3-hNgAgo expression plasmids contain two nuclear localization signals (NLS) and a 3x Flag epitope tag (S1 Fig). Plasmids and annotated GenBank files are available through Addgene.
Design of gDNAs and gRNA expression vector
gDNAs and gRNAs were designed to the same genomic sequence. Single-stranded guide DNA sequences were 21 or 24 nucleotides in length for hTtAgo and hNgAgo, respectively and carried 5’-phosphorylation or an additional four phosphorothioate linkages (S1 and S2 Tables). Plasmids for gRNA expression were cloned as previously described [33]. Briefly, the empty gRNA cloning vector (Addgene plasmid # 41824) was linearized using the AflII restriction enzyme. 19-bp gRNA target sequences (19N) were selected and the G-19N sequence incorporated into two 60mer oligonucleotides that contained cloning vector overhangs for Gibson assembly. After annealing and extending the oligonucleotides to 100-bp, the PCR reaction was purified (PCR purification kit; QIAGEN) and dsDNA was Gibson assembled into the AflII linearized plasmid. Oligonucleotide sequences of gRNA target sites are listed in S2 Table.
Cell lines and transfection
The human cell lines HEK293-c18 (ATCC #CRL-10852) and HeLa (ATCC #CCL-2) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were maintained at 37°C and 5% CO2. Cells were transfected at ~70% confluency using Lipofectamine 3000 (Life Technologies) following the manufacturer’s instructions. For indel analysis, transfections were performed in 24-well dishes using 250 ng hTtAgo or hNgAgo expression plasmid and 1 μl of 100 μM gDNA per well. Alternatively, cells were co-transfected with 250 ng hCas9 nuclease (Addgene plasmid #41815) and 250 ng gRNA expressing plasmids. Cells were harvested 72 hours after transfection. For ChIP assays, transfections were carried out in 10-cm dishes using 2.5 μg pCDNA3-hTtAgo or pCDNA3-hNgAgo with or without 7.5 μl of 100 μM gDNA. Samples that were not co-transfected with gDNA, were instead co-transfected with 7.5 μg of plasmid pBABE-puro (Addgene plasmid #1764). Alternatively, cells were co-transfected with 2.5 μg pCDNA3-dCas9 and 7.5 μg of gRNA expressing plasmid.
NgAgo and TtAgo expression and purification from mammalian cells
HEK293-c18 cells were grown in 10 cm dishes and transfected with 10 μg hTtAgo or hNgAgo expression plasmid using Lipofectamine 3000 (Life Technologies) following the manufacturer’s instructions. 60 hours post transfection cells were rinsed once with PBS and protein was extracted using 1 mL 1xRIPA buffer (Millipore) supplemented with protease inhibitor cocktail (Roche). Anti-Flag M2 affinity gel (Sigma) was equilibrated by washing three times with 1mL TBS (50 mM Tris-HCl pH 7.5, 150 mM NaCl). 50 μl of equilibrated anti-Flag affinity gel was added to 1 mL RIPA lysate and incubated on a rotating platform for 2 hours at 4°C. Anti-Flag affinity gel was washed three times with 1mL TBS and bound protein was eluted with either 3XFlag peptide or 0.1 M glycine. Flag tagged protein was eluted in 100 μl TBS with 150 ng/μl 3X Flag peptide (SIGMA) by rocking at 4°C for 30 minutes and was then used for Western blot analysis and in vitro activity assay. For co-purification of nucleic acids, protein was eluted using 0.1 M glycine.
Extraction of nucleic acids co-purified with hTtAgo
100 μl purified hTtAgo obtained from one 10-cm dish of transfected HEK293 cells were incubated with 1 μl proteinase K for 1 hour at 37°C and separated into three reactions. Two samples were treated with RNase A or DNase I, respectively for 30 minutes at 37°C. The third sample was kept untreated. Nucleic acids were extracted using phenol/chloroform with an additional step of chloroform back extraction to increase recovery. Nucleic acids were precipitated (0.75M NH4OAc, 70% ethanol) and washed twice with 80% ethanol. The dried pellet was dissolved in TBE urea sample buffer (ThermoFisher) and heated for 5 minutes at 95°C. Nucleic acids were separated on a 15% Novex TBE-Urea Gel (ThermoFisher) and visualized with ethidium bromide staining.
Activity assay
15 μl purified hTtAgo or hNgAgo were mixed with 5 pmoles single-stranded DNA guide and 5 pmoles 98-nt ssDNA target (S6 Table) in assay buffer (20 mM Tris-HCl pH8, 250 mM NaCl supplemented with 5 mM MnCl2) and incubated for one hour at 75°C, 55°C or 37°C. The reaction mixture was treated with 10 μg RNase A. The reaction was stopped by addition of TBE urea sample buffer (ThermoFisher) and heated for 5 minutes at 95°C. Samples were resolved on a 15% Novex TBE-Urea Gel (ThermoFisher) and ssDNA was visualized by staining with ethidium bromide.
Amplicon sequencing
72 hours post-transfection with pCDNA3-hTtAgo and phosphorylated 21-nt gDNAs, genomic DNA was extracted using Quick-DNA Miniprep Kit (Zymo Research). Non-transfected cells and cells only transfected with pCDNA3-hTtAgo were used as a control. 100ng genomic DNA was used for PCR amplification with GoTaq polymerase (Promega). For the experiment with hTtAgo, oligonucleotide sequences amplifying 270-bp to 285-bp target regions are listed in S3 Table. PCR products were purified using the QIAquick PCR Purification (Qiagen) and six unique amplicons were mixed in equal amounts per amplicon pool. Illumina sequencing libraries were generated from individual amplicon pools (from different treatments or different cell types) by ligating barcoded adapters (Bioo Scientific) using T4 ligase (New England Biolabs) following the manufacturer’s instructions. Sequencing libraries were cleaned up using 0.8x Ampure XP beads (Agencourt) and library concentrations were determined with the Qubit™ dsDNA HS Assay kit (Invitrogen). Equal amounts of amplicon pools were mixed and sequenced in one lane of paired end 250 on the MiSeq (Illumina). Sequencing and demultiplexing of amplicons pools was performed at the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center. Results are summarized in S4 and S5 Tables. To compare cleavage efficiency of DNA-guided hTtAgo and hNgAgo (21-nt and 24-nt gDNAs, respectively) to that of RNA-guided SpCas9, genomic DNA was extracted 72 hours post-transfection using Quick-DNA Miniprep Kit (Zymo Research). 100ng genomic DNA was used for PCR amplification of ~200-bp fragments with Taq RED DNA Polymerase Master Mix (Apex) following the manufacturer’s recommendations. Forward primers contain a 5-nt barcode for multiplexing amplicons (S3 Table). All amplicons were purified using QIAQuick PCR Purification Kit (Qiagen) and pooled at equal concentrations for Illumina sequencing. Sequencing library preparation and amplicon sequencing were performed by the CCIB DNA Core Facility at Massachusetts General Hospital (Cambridge, MA). Results are summarized in S6 Table.
Amplicon sequencing data analysis
Sequencing data was processed using FLASH2 (https://github.com/dstreett/FLASH2) to overlap forward and reverse reads and merge paired reads into a single long read. For amplicons containing a 5-nt barcode, we demultiplexed merged reads with the FASTX barcode splitter by identifying barcodes at the beginning or the reverse complement barcodes at the end of sequence reads, allowing for one mismatch. Processed fastq files were analyzed with the CRISPResso online tool (http://crispresso.rocks) using default settings.
Chromatin immunoprecipitation (ChIP)
For ChIP assays transfected cells were cross-linked 48 hours post transfection by incubation with 1% formaldehyde solution for 10 min at room temperature and the reaction was stopped by the addition of glycine to a final concentration of 125 mM. Cross-linked cell pellets were stored at -80°C. Chromatin was extracted and ChIP performed using StaphA cells (Sigma-Aldrich, St. Louis, MO, USA) to collect the immunoprecipitates as previously described [33]. In summary, cells were lysed in Cell Lysis Buffer (5mM PIPES, 85mM KCl, 1% Igepal, 1x Protease Inhibitors; pH 8.0) on ice. Nuclei were pelleted and lysed for 30 minutes in Nuclear Lysis Buffer (50mM Tris-HCl, 10mM EDTA, 1% SDS, 1x Protease Inhibitors; pH 8.0) on ice. Chromatin was sheared to an average fragment size of 500-bp using a Bioruptor 2000 (Diagenode). Chromatin from an entire 10-cm dish was split into Flag and control ChIP assay and diluted with 3 volumes of IP Dilution Buffer (16.7mM Tris-HCl, 167mM NaCl, 1.2mM EDTA pH 8.0, 1.1% Triton X 100, 0.01% SDS, 1x Protease Inhibitors). ChIP enrichment was performed by incubation with 3 μg anti-Flag antibody (SIGMA M2 F1804) or 2 μg normal rabbit IgG (Abcam ab46540) for 16 h at 4 °C. Immuno complexes were captured with 3 μg rabbit anti mouse antibody for 1 hour at 4°C and were bound to StaphA cells for 15 minutes at room temperature. Chromatin-antibody complexes were pelleted and washed twice with Wash Buffer 1 (50mM Tris-HCl, 2mM EDTA pH 8.0, 0.2% Sarkosyl) and four times with Wash Buffer 2 (100mM Tri-HCl, 500mM LiCl, 1% Igepal, 1% Deoxycholic Acid; pH 8.0). Immunoprecipitated chromatin was eluted with 100 μl ChIP Elution Buffer (50mM NaHCO3, 1% SDS) by shaking at room temperature for 15 minutes. Crosslinks were reversed at 67°C overnight after addition of 12 μl 5M NaCl. Finally, immunoprecipitated DNA was treated with 10 μg RNase A at 37°C for 20 minutes, and purified using the QIAQuick PCR Purification Kit (Qiagen). ChIP-DNA and diluted Input control were used for subsequent qPCR or standard PCR reactions (Primers are listed in S3 Table). qPCR was carried out with 2x SYBR FAST mastermix (KAPA Biosystems) according to the manufacturer’s recommendations using the CFX384 Real-Time System C1000 Touch Thermo Cycler (BioRad). ChIP enrichment was calculated relative to input samples using the dCq method (dCq = Cq[ChIP]-Cq[input]). We applied the Student’s paired t-test with a two-tailed distribution to determine statistical significance for ChIP enrichment without or with gDNA/gRNA. Standard PCR was performed using GoTaq (Promega) DNA polymerase (2 min at 95°C; 30 sec at 95°C, 30 s at 55°C, 30 s at 72°C, 35 cycles; 5 min at 72°C). PCR products were separated on a 1.5% agarose gel and visualized using the Gel Doc™ XR+ System (BioRad).
Western blot analysis
HEK293-c18 cells were transfected in 6-well dishes with 2.5 μg pCDNA3-hTtAgo or pCDNA3-hNgAgo and lysed 48 hours post transfection in 1xRIPA buffer (Millipore) supplemented with protease inhibitor cocktail (Roche). Protein concentrations were determined by Bradford assay (BioRad) and 20 μg protein was separated on a 10–20% NuPAGE Bis-Tris gel (Thermo Fisher Scientific) in MOPS buffer and transferred onto nitrocellulose membranes. Protein loading was evaluated by Ponceau S stain. After rinsing the membrane with deionized water, non-specific antigen binding was blocked in TBST (50 mM Tris, 150 mM NaCl and 0.1% Tween-20) with 5% nonfat dry milk (Cell Signaling). Membranes were incubated with monoclonal antibody against Flag (1:1500; SIGMA M2 F1804) in blocking solution at 4°C over night. Membranes were washed with TBST three times for 10 minutes before incubation with anti-mouse HRP-conjugated antibody at room temperature. After 45 minutes, the membrane was washed three times in TBST and proteins were visualized with Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare) and autoradiobiography film.
Supporting information
(PDF)
A. hTtAgo was expressed in HEK293 cells and purified by immunoprecipitation (IP) with Flag antibody. A control IP was performed using rabbit IgG. Protein extract from transfected cells was loaded as a control. B. Copurification of nucleic acids with hTtAgo in HEK293 cells. Nucleic acids were resolved on a 15% TBE-urea gel. Nucleic acids were not treated (lane 1) or treated with RNaseA (lane 2) or DNaseI (lane 3). C. 21-nucleotide (nt) DNA guides are complimentary to 98-nt single stranded DNA target. Predicted cleavage sites are indicated by a black triangle. D-F. Purified hTtAgo or hNgAgo were incubated with 21-nt or 24-nt guides, respectively to cleave a 98-nt ssDNA target and run on a 15% TBE-urea gel. Cleavage assays were carried out at 75°C (A), 55°C (B) and 37°C (C). Control Flag-IPs were performed in untransfected HEK293 cells. hTtAgo was incubated with the a DNA guide complimentary to a G nucleotide in position 1 on the target strand (t1G) as indicated.
(PDF)
Diagram illustrating the three different gDNA constellations that were tested: forward (FW) guide only, forward (FW) and reverse (RV) guides or forward (FW) and reverse (RV) guides separated by a 15-nt spacer sequence. The genomic RPL13A target site is indicated in blue and complementary ssDNA guides are indicated in red.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
This work was supported by a University of California, Davis, Academic Senate Faculty Research Grant. C.R. was supported by the China State-Sponsored Postgraduates Study Abroad Program provided by the Chinese Scholarship Council.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a University of California, Davis, Academic Senate Faculty Research Grant. C.R. was supported by the China State-Sponsored Postgraduates Study Abroad Program provided by the Chinese Scholarship Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual review of biophysics. 2013;42:217–39. Epub 2013/05/10. doi: 10.1146/annurev-biophys-083012-130404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J. Biochemistry. A bacterial seek-and-destroy system for foreign DNA. Science. 2014;344(6187):972–3. Epub 2014/05/31. doi: 10.1126/science.1252962 . [DOI] [PubMed] [Google Scholar]
- 3.Hegge JW, Swarts DC, van der Oost J. Prokaryotic Argonaute proteins: novel genome-editing tools? Nature reviews Microbiology. 2017. Epub 2017/07/25. doi: 10.1038/nrmicro.2017.73 . [DOI] [PubMed] [Google Scholar]
- 4.Swarts DC, Hegge JW, Hinojo I, Shiimori M, Ellis MA, Dumrongkulraksa J, et al. Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic acids research. 2015;43(10):5120–9. Epub 2015/05/01. doi: 10.1093/nar/gkv415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507(7491):258–61. Epub 2014/02/18. doi: 10.1038/nature12971 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaya E, Doxzen KW, Knoll KR, Wilson RC, Strutt SC, Kranzusch PJ, et al. A bacterial Argonaute with noncanonical guide RNA specificity. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(15):4057–62. Epub 2016/04/02. doi: 10.1073/pnas.1524385113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. Epub 2015/06/23. doi: 10.1038/nature14592 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinstiver BP, Prew MS, Tsai SQ, Nguyen NT, Topkar VV, Zheng Z, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nature biotechnology. 2015;33(12):1293–8. Epub 2015/11/03. doi: 10.1038/nbt.3404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, et al. Engineered Cpf1 variants with altered PAM specificities. Nature biotechnology. 2017. Epub 2017/06/06. doi: 10.1038/nbt.3900 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brocken DJW, Tark-Dame M, Dame RT. dCas9: A Versatile Tool for Epigenome Editing. Current issues in molecular biology. 2017;26:15–32. Epub 2017/09/08. doi: 10.21775/cimb.026.015 . [DOI] [PubMed] [Google Scholar]
- 11.Braff JL, Yaung SJ, Esvelt KM, Church GM. Characterization of Cas9-Guide RNA Orthologs. Cold Spring Harbor protocols. 2016;2016(5): pdb top086793. Epub 2016/05/04. doi: 10.1101/pdb.top086793 . [DOI] [PubMed] [Google Scholar]
- 12.Gao F, Shen XZ, Jiang F, Wu Y, Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016;34(7):768–73. doi: 10.1038/nbt.3547 . [DOI] [PubMed] [Google Scholar]
- 13.Burgess S, Cheng L, Gu F, Huang J, Huang Z, Lin S, et al. Questions about NgAgo. Protein Cell. 2016;7(12):913–5. doi: 10.1007/s13238-016-0343-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javidi-Parsijani P, Niu G, Davis M, Lu P, Atala A, Lu B. No evidence of genome editing activity from Natronobacterium gregoryi Argonaute (NgAgo) in human cells. PloS one. 2017;12(5):e0177444 doi: 10.1371/journal.pone.0177444 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi J, Dong Z, Shi Y, Wang X, Qin Y, Wang Y, et al. NgAgo-based fabp11a gene knockdown causes eye developmental defects in zebrafish. Cell research. 2016;26(12):1349–52. doi: 10.1038/cr.2016.134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khin NC, Lowe JL, Jensen LM, Burgio G. No evidence for genome editing in mouse zygotes and HEK293T human cell line using the DNA-guided Natronobacterium gregoryi Argonaute (NgAgo). PloS one. 2017;12(6):e0178768 doi: 10.1371/journal.pone.0178768 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Z, Tan S, Xu L, Gao L, Zhu H, Ma C, et al. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017;145:20–3. doi: 10.1016/j.antiviral.2017.07.005 . [DOI] [PubMed] [Google Scholar]
- 18.Editors. Time for the data to speak. Nature biotechnology. 2017;35(8):689 doi: 10.1038/nbt.3938 . [DOI] [PubMed] [Google Scholar]
- 19.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–83. Epub 2013/03/05. doi: 10.1016/j.cell.2013.02.022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013;31(9):833–8. doi: 10.1038/nbt.2675 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51. doi: 10.1016/j.cell.2013.06.044 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tak YE, Kleinstiver BP, Nunez JK, Hsu JY, Horng JE, Gong J, et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nature methods. 2017. Epub 2017/10/31. doi: 10.1038/nmeth.4483 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamano T, Nishimasu H, Zetsche B, Hirano H, Slaymaker IM, Li Y, et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell. 2016;165(4):949–62. Epub 2016/04/27. doi: 10.1016/j.cell.2016.04.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17(1):5–15. doi: 10.1038/nrm.2015.2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–64. doi: 10.1146/annurev-biochem-060815-014607 . [DOI] [PubMed] [Google Scholar]
- 26.Swarts DC, Szczepaniak M, Sheng G, Chandradoss SD, Zhu Y, Timmers EM, et al. Autonomous Generation and Loading of DNA Guides by Bacterial Argonaute. Molecular cell. 2017;65(6):985–98 e6. Epub 2017/03/07. doi: 10.1016/j.molcel.2017.01.033 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434(7033):666–70. Epub 2005/04/01. doi: 10.1038/nature03514 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456(7219):209–13. Epub 2008/08/30. doi: 10.1038/nature07315 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swarts DC, Koehorst JJ, Westra ER, Schaap PJ, van der Oost J. Effects of Argonaute on Gene Expression in Thermus thermophilus. PloS one. 2015;10(4):e0124880 Epub 2015/04/23. doi: 10.1371/journal.pone.0124880 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Molecular cell. 2013;51(5):594–605. Epub 2013/09/17. doi: 10.1016/j.molcel.2013.08.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye S, Bae T, Kim K, Omer H, Lee SH, Kim YY, et al. DNA-dependent RNA cleavage by the Natronobacterium gregoryi Argonaute. BioRxiv. 2017; preprint: https://doi.org/10.1101/101923. [Google Scholar]
- 32.O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, et al. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression Nucleic acids research. 2017;45(17):9901–16. doi: 10.1093/nar/gkx578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic acids research. 2015;43(6):3389–404. Epub 2015/02/26. doi: 10.1093/nar/gkv137 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A. hTtAgo was expressed in HEK293 cells and purified by immunoprecipitation (IP) with Flag antibody. A control IP was performed using rabbit IgG. Protein extract from transfected cells was loaded as a control. B. Copurification of nucleic acids with hTtAgo in HEK293 cells. Nucleic acids were resolved on a 15% TBE-urea gel. Nucleic acids were not treated (lane 1) or treated with RNaseA (lane 2) or DNaseI (lane 3). C. 21-nucleotide (nt) DNA guides are complimentary to 98-nt single stranded DNA target. Predicted cleavage sites are indicated by a black triangle. D-F. Purified hTtAgo or hNgAgo were incubated with 21-nt or 24-nt guides, respectively to cleave a 98-nt ssDNA target and run on a 15% TBE-urea gel. Cleavage assays were carried out at 75°C (A), 55°C (B) and 37°C (C). Control Flag-IPs were performed in untransfected HEK293 cells. hTtAgo was incubated with the a DNA guide complimentary to a G nucleotide in position 1 on the target strand (t1G) as indicated.
(PDF)
Diagram illustrating the three different gDNA constellations that were tested: forward (FW) guide only, forward (FW) and reverse (RV) guides or forward (FW) and reverse (RV) guides separated by a 15-nt spacer sequence. The genomic RPL13A target site is indicated in blue and complementary ssDNA guides are indicated in red.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.