Abstract
Checkpoint Rad proteins function early in the DNA damage checkpoint signaling cascade to arrest cell cycle progression in response to DNA damage. This checkpoint ensures the transmission of an intact genetic complement to daughter cells. To learn about the damage sensor function of the human checkpoint Rad proteins, we purified a heteropentameric complex composed of hRad17-RFCp36-RFCp37-RFCp38-RFCp40 (hRad17-RFC) and a heterotrimeric complex composed of hRad9-hHus1-hRad1 (checkpoint 9-1-1 complex). hRad17-RFC binds to DNA, with a preference for primed DNA and possesses weak ATPase activity that is stimulated by primed DNA and single-stranded DNA. hRad17-RFC forms a complex with the 9-1-1 heterotrimer reminiscent of the replication factor C/proliferating cell nuclear antigen clamp loader/sliding clamp complex of the replication machinery. These findings constitute biochemical support for models regarding the roles of checkpoint Rads as damage sensors in the DNA damage checkpoint response of human cells.
DNA damage in human cells activates several distinct biochemical pathways that may eliminate the damage (DNA repair), arrest the cell cycle progression until the lesion is dealt with (DNA damage checkpoint), or carry out programmed cell death to eliminate seriously impaired cells (apoptosis). The molecular mechanisms of repair and apoptosis are fairly well understood; however, the DNA damage checkpoint response is, at present, biochemically ill-defined.
The DNA damage checkpoint response is the set of biochemical pathways that are activated by DNA damage to arrest cell cycle progression as long as the damage persists (1). The response, as revealed by genetic analyses in budding and fission yeasts, consists of damage sensor, signal transducer, and effector components that arrest the cell cycle at G1/S and G2/M (2–4). The signal transducers and effectors are protein kinases that phosphorylate the target molecules and halt cell cycle progression.
The least understood components of the checkpoint are the DNA damage sensors. Genetic analyses in Schizosaccharomyces pombe have identified six genes, Rad3, Rad17, Rad9, Rad1, Hus1, and Rad26, that are collectively referred to as the checkpoint Rad genes (5). These genes encode proteins thought to sense DNA damage and activate the signal transduction checkpoint-signaling cascade. The human homologs of the first five checkpoint Rad proteins are ATM/ATR, hRad17, hRad9, hRad1, and hHus1. ATM and ATR belong to the phosphatidylinositol 3-kinase-related protein kinase family of proteins and are thought to participate in both damage sensing and signal transduction (6). The remaining four checkpoint Rad proteins are thought to function primarily as damage sensors.
Recent in vivo biochemical studies in budding and fission yeasts and in human cells, as well as computational analyses of these proteins, have provided significant insights into possible mechanisms of action for the damage sensor checkpoint proteins. The Rad17 homologs exhibit sequence homology to all five subunits of the replication factor C (RFC) (7–9), which functions as a clamp loader. Evidence from budding and fission yeasts indicates that Rad17 interacts with the four small subunits of RFC (10–12), and thus, it has been proposed that Rad17 forms a complex with RFC proteins in which the large subunit of RFC (p140) is replaced by Rad17. Molecular modeling analyses of the rad checkpoint proteins Rad9, Rad1, and Hus1 have suggested structural similarities among these proteins and the sliding clamp, proliferating cell nuclear antigen (PCNA) (13–15). These observations have led to the proposal that these three rad proteins make a heterotrimeric complex with a PCNA-like structure possessing similar yet distinct functions as PCNA. In fact, immunoprecipitation and yeast two-hybrid analyses have provided experimental support for a PCNA-like Rad9-Hus1-Rad1 complex, termed the checkpoint 9-1-1 complex (14, 16–21). Collectively, these findings have led to the following model for the function of human checkpoint Rad proteins (2, 15): The primary DNA lesions or the special structures arising from processing these lesions by DNA repair or replication systems are recognized by the Rad17-RFC complex, which then acts as a molecular matchmaker (22) to recruit the checkpoint 9-1-1 complex and loads it onto the DNA, thus initiating the DNA damage checkpoint signaling. Although this is an attractive model, direct biochemical evidence in support of the model is lacking.
In this study, we have purified and biochemically characterized the hRad17-RFC and the checkpoint 9-1-1 complexes. Our results show that the hRad17-RFC/checkpoint 9-1-1 pair exhibits similarities to the RFC/PCNA pair in certain aspects but differs from the latter in certain key reactions. We have demonstrated that, in vivo, hRad17 forms a complex with the four RFC small subunits, and that, in vitro, hRad17 competes with RFC p140 for the RFC small subunits. Like p140, hRad17 depends on p38 for interaction with the three core RFC subunits. hRad17-RFC, like the classical RFC, binds to DNA, with a preference for primed DNA and possesses weak ATPase activity that is stimulated by primed DNA templates and single-stranded DNA. Unlike the classical RFC, hRad17-RFC ATPase activity was not stimulated by PCNA. Although we were unable to detect significant stimulation of hRad17-RFC ATPase activity by the checkpoint 9-1-1 complex, we did detect a stable interaction between the two complexes. This initial characterization of the biochemical properties of the checkpoint Rads provides the groundwork for future investigations regarding the roles of these proteins as damage sensors in the DNA damage checkpoint.
Materials and Methods
Plasmids.
hRad17 was amplified by PCR with pACT2-hRad17 [kind gift of Jorge Vialard, Janssen Research Foundation, Beerse, Belgium (9)] as the template. The PCR product was digested and ligated into pcDNA4 (Invitrogen) to generate pcDNA4-FlaghRad17 or ligated into pFastBacHTa (GIBCO/BRL) to generate pFast-His6-hRad17 and pFast-His6-Flag-hRad17. hRad9 was amplified by PCR with pcDNA3-AU1-hRAD9 [kind gift of Larry M. Karnitz, Mayo Foundation, Rochester, MN (18)] as a template, and the PCR product was digested and ligated into pFastBac1 to generate pFast-hRad9 and pFast-Flag-hRad9 that encode untagged and C-terminal Flag epitope-tagged hRad9, respectively. hHus1 was amplified by PCR from a HeLa cDNA library and cloned into pFastBac1 to generate pFast-hHus1 and pFast-Flag-hHus1 that encode untagged and C-terminal Flag epitope-tagged hHus1, respectively. hRad1 was amplified by PCR with pACT2-hRad1A as a template [kind gift of J. Vialard (9)]. The PCR product was digested and ligated into pFastBac1 to generate pFast-hRad1. All plasmids were sequenced to verify that no mutations were introduced during PCR and cloning.
Partial Purification of hRad17 from HeLa Cell-Free Extracts (CFE).
CFE from HeLa S3 cells were prepared as described (23). HeLa CFE were initially fractionated by an analytical velocity sedimentation as follows: 500 μg (100 μl) of HeLa CFE was applied to the top of a 12-ml gradient of 15–35% glycerol and centrifuged for 20 h at 37,000 rpm in a SW41 rotor (Beckman Instruments, Palo Alto, CA). Twenty-four 0.5-ml fractions were collected from the bottom to top and analyzed by SDS/PAGE and Western blotting (20 μl). The sedimentation position of standard proteins was determined by a gradient run in parallel.
For large-scale purification of hRad17, 1470 mg of HeLa CFE in buffer D (25 mM Hepes-KOH, pH 7.9/0.1 M KCl/12 mM MgCl2/0.5 mM EDTA/2 mM DTT/16% glycerol [vol/vol]) was loaded onto a 330-ml DEAE Sepharose column, eluted with a 2 liter 0.1–1 M KCl gradient, and 50-ml fractions were collected. The fractions were analyzed by Western blotting, and those containing hRad17 were pooled (eluting at 0.15 M KCl), resulting in a 7-fold purification. The pooled fractions, containing 225 mg of protein, were adjusted to 50 mM KCl, loaded onto a 300-ml SP Sepharose column, and eluted with a 2-liter 0.05–1 M KCl gradient, and 40-ml fractions were collected. The load and even fractions 16–52 (50 μl) were analyzed by SDS/PAGE and Western blotting. This column step resulted in a 12-fold purification of hRad17 (which peaked at 0.22 M KCl).
Expression and Purification of Recombinant Proteins.
Human embryonic kidney 293T cells were grown in DMEM (GIBCO/BRL) supplemented with 10% FBS and 100 units of penicillin and streptomycin/ml. Cells (3 × 106) were transfected with 17 μg of pcDNA4-FlaghRad17 plasmid by using the calcium phosphate transfection method (24). Forty-eight hours after transfection, cells were washed with PBS and lysed in 20 packed cell volumes of lysis buffer [50 mM Tris⋅HCl, pH 7.5/0.5% Nonidet P-40/protease inhibitors (Roche Molecular Biochemicals)] with 1 M NaCl. After a 15-min incubation on ice, the cell lysate was centrifuged for 30 min at 32,000 × g. The supernatant was incubated with anti-Flag agarose for 4 h at 4°C. The resin was then washed four times with lysis buffer and the protein was eluted with elution buffer [50 mM Tris-Cl, pH 7.5/0.05% Nonidet P-40/protease inhibitors/200 μg/ml Flag peptide Sigma)] with 1 M NaCl yielding ≈0.4 μg of protein.
Baculoviruses for expression of His6-hRad17, His6Flag-hRad17, hRad9, Flag-hRad9, hHus1, Flag-hHus1, and hRad1 were generated with the BAC-TO-BAC baculovirus expression system (GIBCO/BRL) and protocols suggested by the manufacturer. Baculoviruses for expression of RFC p40, His6-p38, p37, and p36 were described (25). The optimal multiplicity of infection was empirically determined for each recombinant virus. Expression of more than one protein at a time was accomplished by the infection of multiple viruses simultaneously (multiplicity of infection of five for each). Monolayer High Five (HF) insect cells (Invitrogen), grown in Grace's insect medium (GIBCO/BRL) supplemented with 10% FBS and 100 units of penicillin and streptomycin/ml, were infected with virus and then harvested after 48 h. The cells were lysed and centrifuged as described above for 293T cells, with the lysis buffer containing 0.3 M NaCl for the 9-1-1 complex and 1 M NaCl for the hRad17-RFC complex. The 9-1-1 complex was purified directly with anti-Flag agarose as described above for 293T, with the elution buffer containing 0.15 M NaCl. The hRad17-RFC complex, which contains His6-Flag-tagged hRad17 and His6-tagged p38, was first purified with Ni-NTA agarose (Qiagen, Chatsworth, CA), and the protein was eluted with 50 mM Tris⋅HCl (pH 7.5), 1 M NaCl, 0.5% Nonidet P-40, protease inhibitors, and 100 mM imidazole. The eluate was then purified with anti-Flag agarose as described above for 293T. The yield of 9-1-1 complex and hRad17-RFC from 109 of cells was 3 and 5.6 mg, respectively.
Antibodies.
hRad17 was detected with a rabbit polyclonal antibody raised against bacterially produced maltose-binding protein-tagged hRad17 (amino acids 1–170) and affinity-purified by using bacterially produced glutathione S-transferase-tagged hRad17 (amino acids 1–170) coupled to Pierce Amino Link resin according to the manufacturer's recommendations. The hRad1 rabbit polyclonal antibody was raised against a peptide corresponding to amino acids 9–26, and the serum was used for Western analysis. hRad9 was detected with affinity-purified rabbit polyclonal antibody M-389 (Santa Cruz Biotechnology). The anti-RFC p140 mAb was a kind gift of Bruce Stillman, Cold Spring Harbor Laboratory, Plainview, NY (26). The anti-RFC p37 polyclonal antibody has been described (27).
In Vitro Transcription-Translation of hRad17 and RFC Subunits.
Coupled in vitro transcription-translation reactions and immunoprecipitations were performed as described (27). Briefly, template DNAs (0.25 μg of each) that expressed hRad17 and the RFC subunits were added to a 50-μl TNT Quick transcription-translation mixture (Promega) containing 30 μCi (1 Ci = 37 GBq) of 35S-labeled methionine (1,175 Ci/mmol, NEN) and incubated at 30°C for 90 min. In reactions that contained the RFC complex, 0.5 μg of pET 16a-p140 was used to compensate for low p140 expression. After incubation, 50 units of DNaseI (Roche Molecular Biochemicals) was added, and the mixture was incubated for 10 min at 30°C to digest template DNAs. The labeled hRad17-RFC and RFC complexes from a 20-μl aliquot of the reaction mixture were immunoprecipitated as decribed (27). Ten percent of the reticulocyte lysate (load) and 50% of the immunoprecipitated materials were subjected to an SDS/12% PAGE, and labeled proteins were visualized by autoradiography. To determine the stoichiometry of the subunits in the hRad17-RFC complex, it was purified from reticulocyte lysate (250 μl scale) by phosphocellulose chromatography and sedimented twice in a 15–40% glycerol gradient as described (25, 27). After separation by SDS/PAGE, the 35S-labeled proteins in the peak glycerol gradient fraction were quantitated by using a PhosphorImager.
DNA Binding and ATPase Assays.
Nitrocellulose filter binding assays were performed as described (28). ATPase reaction mixtures (10 μl) contained 25 mM Tris⋅HCl (pH 7.5), 3 mM magnesium acetate, 2 mM DTT, 200 μg/ml BSA, 50 μM [γ32P]ATP (4,400 cpm/pmol), and DNA (type and amounts) as indicated. After 20 min at 37°C, aliquots (1 μl) were spotted on polyethyleneimine-cellulose thin layer plates that were developed in 1.0 M formic acid and 0.5 M LiCl at room temperature. 32Pi formation was quantitated by PhosphorImager analysis.
Phosphatase Treatment of the 9-1-1 Complex.
Three microliters (≈1 μg) of purified C-Flag-hRad 9-1-1 complex was incubated with the indicated amounts of λ phosphatase (Stratagene) in the absence or presence of 1 mM sodium orthovanadate at 30°C for 1 h.
Gel Filtration.
Purified C-Flag-hRad9 9-1-1 complex (100 μl; ≈20 μg) was loaded onto a Superdex 200 10/30 column (Amersham Pharmacia) and eluted with 50 mM Tris⋅HCl (pH 7.5) and 300 mM NaCl. The column was run at 0.3 ml/min, and 0.5-ml fractions were collected. Aliquots (100 μl) of the fractions were resolved by 12.5% SDS/PAGE and visualized by silver staining.
Purification of the Checkpoint Rad Complex.
HF cells were infected with baculoviruses expressing all eight subunits of the hRad17-RFC and 9-1-1 complexes, including Flag-hRad9, and the checkpoint complex was purified by affinity chomatography on anti-Flag resin (yielding 3.8 μg from 106 cells) and analyzed by SDS/PAGE and silver staining and Western blotting.
Results
Purification of the Rad17-RFC Complex.
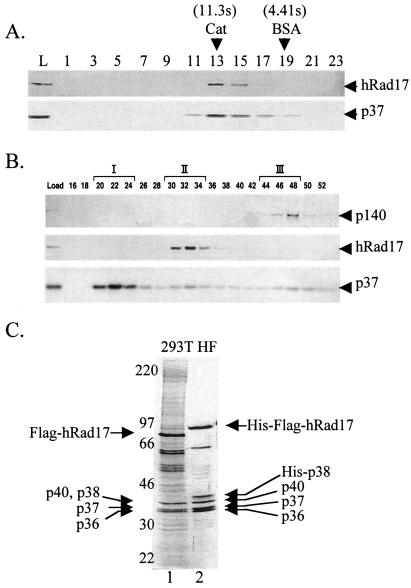
Currently, there is no in vitro assay available to assess enzymatic activities associated with either hRad17 or the hRad17-RFC complex. Therefore, we used immunoblotting to follow these proteins during purification of hRad17 from HeLa cells. Initially, we wanted to determine whether hRad17 was present in multiple forms such as a monomer or in association with the RFC small subunits. To this end, we separated HeLa CFE by glycerol density centrifugation and analyzed the fractions with antibodies against hRad17 and RFC p37. Fig. 1A shows that there is a single peak for both proteins. From this result, two conclusions can be made. First, there is only one species of hRad17. Second, this species has approximately the same molecular weight as RFC because the RFC p37 subunit, which represents all of the complexes formed with this and presumably the other RFC subunits, including classical RFC, comigrates with hRad17. Thus these data are consistent with the presence of a heteropentameric RFC complex containing hRad17 instead of the large RFC p140 subunit, as has been previously shown in yeast (10–12). This conclusion is strengthened by the following findings made during our attempts to purify hRad17 from HeLa cells.
Figure 1.
Isolation of hRad17-RFC. (A) Glycerol gradient sedimentation of hRad17 present in crude HeLa extracts. HeLa CFE was subjected to glycerol gradient centrifugation as described in Materials and Methods. Fractions were collected from the bottom of the tube and analyzed by Western blotting with anti-hRad17 and anti-RFCp37 antibodies. The sedimentation position of reference proteins in a parallel gradient is indicated. L, load. (B) Separation of at least three forms of RFC. Fractions from the SP Sepharose column (see Materials and Methods) were analyzed by Western blotting by using the indicated antibodies. The three RFC complexes are represented by I, II, and III. (C) Purification the hRad17-RFC complex from transfected human 293T cells and baculovirus-infected insect HF cells. 293T cells were transfected with pcDNA4-Flag-hRad17 and purified by using anti-Flag agarose (lane 1). The hRad17-RFC complex was reconstituted in insect cells by coinfection with five recombinant viruses capable of expressing His6-Flag-hRad17 and each RFC small subunit (lane 2). The p38 subunit contains a His6 tag. The complex was purified by chromatography with Ni-NTA and then anti-Flag agarose as described in Materials and Methods. The proteins were visualized after SDS/PAGE by silver staining. The amounts of protein loaded in lanes 1 and 2 were 1 and 0.5 μg, respectively.
Although immunoblots of HeLa CFE indicated that hRad17 is a relatively abundant protein (quantified by using recombinant hRad17 as the standard, data not shown), our repeated attempts to purify a complex containing hRad17 by using conventional chromatography failed because after 3–4 chromatographic steps the complex became unstable and the yield dropped drastically. However, these attempts revealed an interesting fact. As seen in Fig. 1B, at the second chromatographic step hRad17 eluted as a single peak, whereas p37, which represents all RFC-like complexes, eluted in three peaks. Of these, peak III contains RFC p140 and thus represents classical RFC; peak II contains hRad17 but no RFC p140 and thus represents the hRad17-RFC complex; peak I contains neither RFC p140 nor hRad17 and may represent other complexes containing RFC p37 such as the recently identified Ctf18-RFC complex (29, 30).
The purification profiles of hRad17 and the RFC subunits suggest that hRad17 exists in a heteropentameric RFC complex. We also obtained results from immunoprecipitation experiments that further support the existence of a hRad17-RFC complex (data not shown). In addition, we purified native hRad17 by affinity chromatography by using an epitope-tagged protein. Human 293T cells were transfected with a vector expressing the Flag-tagged protein, and hRad17 was purified by immunoaffinity chromatography. Fig. 1C (lane 1) shows that hRad17 purified in this manner contains additional proteins in the size range of 36–40 kDa, consistent with the size of RFC small subunits. Protein blotting confirmed the identity of these proteins as RFC p36, p37, p38, and p40 (data not shown). Thus, these findings show that hRad17 is indeed in an RFC-like complex, and in combination with data in Fig. 1A, the findings suggest that all of hRad17 is in this complex. The hRad17-containing RFC complex may be referred to as the checkpoint RFC to differentiate it from the classical RFC, which may be more appropriately referred to as the replicative RFC. However, to be more descriptive, the hRad17-containing complex will simply be called hRad17-RFC.
Although the Flag-tagged hRad17 expressed in human cells was useful in proving the existence of hRad17-RFC in these cells under nearly physiological conditions, the amount and purity of the protein purified by this method was insufficient for biochemical characterization. To obtain large quantities of hRad17-RFC, HF insect cells were coinfected with baculoviruses expressing hRad17, and the four small RFC subunits and the complex was purified by affinity chromatography with Ni-NTA agarose and subsequent immunoaffinity chromatography. This method enabled us to purify milligram amounts of the complex of >95% purity (Fig. 1C, lane 2). When necessary, hRad17-RFC was further purified by size exclusion chromatography or glycerol density gradient velocity sedimentation.
Order of Assembly of hRad17-RFC.
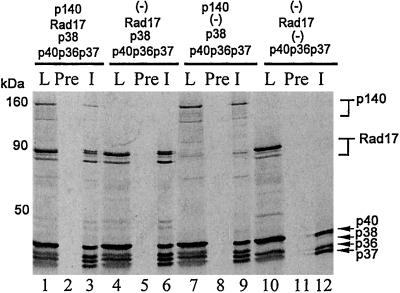
The classical human RFC can be reconstituted by expressing all five subunits in an in vitro transcription/translation system (27, 31) or in insect cells infected with recombinant baculoviruses expressing the individual subunits (25, 32, 33). These reconstitution studies suggested a cooperative mechanism of assembly, whereby formation of a stable core complex of p36, p37, and p40 is followed by cooperative binding of p38 and p140 to the core complex (27). Although a complex of the four small subunits has also been isolated (32, 33), its conversion to the five-subunit holoenzyme upon addition of p140 is unclear. Thus, it appears that p38 and p140 require each other for interaction with the core complex. We wanted to know whether hRad17-RFC was assembled by the same route with hRad17 replacing p140. The five RFC proteins and hRad17 were expressed in an in vitro transcription/translation system containing the genes in various combinations, and the complexes were immunoprecipitated with anti-p37 antibodies and analyzed by autoradiography. As apparent in Fig. 2, in this system both RFC and hRad17-RFC form efficiently (lanes 6 and 9), and when both p140 and hRad17 are present in the same mixture, they compete for assembly with the core complex (compare the p140 intensities in lanes 3 and 9). Importantly, p38 appears to be necessary for the association of hRad17 with the core p36-p37-p40 complex to form hRad17-RFC. Thus, we conclude that hRad17 behaves like p140 with regard to its interactions with the other RFC subunits. Along these lines the hRad17-RFC complex contains all five subunits in a one-to-one stoichiometry. When the in vitro translation products of a mixture of hRad17, RFC p36, p37, p38, and p40 were purified through phosphocellulose and then sedimented in a 15–40% glycerol gradient, the hRad17-RFC complex migrated at the same position as the classic RFC complex. Quantification of the 35S-labeled proteins in the peak fraction by PhosphorImaging gave the following ratios: 1 (hRad17):1.09, (p40):1.37, (p38):1.32, (p37):1.24 (p36).
Figure 2.
Order of assembly of hRad17-RFC. An in vitro transcription/translation system containing the indicated expression vectors was used to radiolabel the corresponding proteins. The products were immunoprecipitated with anti-p37 antibodies, separated on SDS/PAGE, and analyzed by autoradiography. L, load; Pre, preimmune serum, and I, immune (anti-p37) serum.
ATPase Activity of hRad17-RFC.
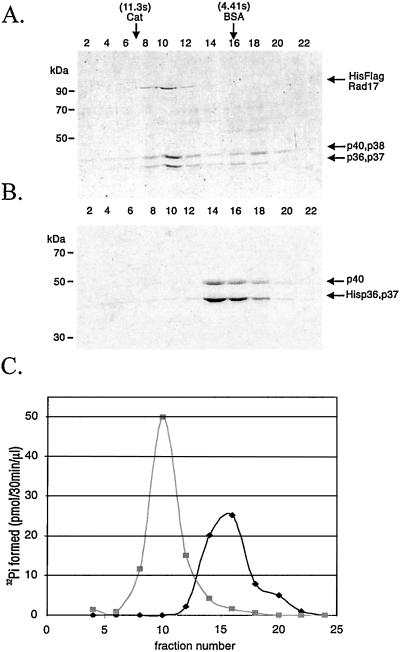
The classical RFC is an ATPase of moderate strength whose activity is stimulated by DNA and PCNA (33–36). Although the three-subunit RFC core complex exhibits weak ATPase activity that is modestly stimulated by DNA, the holoenzyme is more strongly stimulated by both DNA and PCNA (32, 34–37). We wanted to know whether hRad17-RFC had similar properties. The five-subunit hRad17-RFC and the three-subunit core complex were made in insect cells, isolated by affinity chromatography, and then further purified by glycerol density gradient sedimentation. The fractions were analyzed by SDS/PAGE and Coomassie blue staining (Fig. 3 A and B) and tested for ATPase activity (Fig. 3C). Densitometric analysis of the bands in the hRad17-RFC peak fractions showed that hRad17-RFC has a stoichiometry of 1.17 (hRad17):1.34 (p40 + p38):1 (p36 + p37) that along with the sedimentation data are consistent with a monomeric structure of the heteropentamer. Importantly, the peak fractions of hRad17-RFC contain more ATPase activity than the peak fractions of the core complex (Fig. 3C). More detailed analysis of the hRad17-RFC ATPase activity is summarized in Table 1. As is apparent, hRad17-RFC behaves similarly to the RFC holoenzyme: It is more active than the three-subunit core and is strongly stimulated by single-stranded DNA but not double-stranded (ds) DNA, although the level of stimulation is approximately one-half of that seen with RFC. Of importance, in contrast to RFC, hRad17-RFC ATPase is not stimulated by PCNA (data not shown). Of equal interest, the checkpoint 9-1-1 complex, which in current models is considered to be the PCNA counterpart of the checkpoint clamp loader/sliding clamp pair, also had little effect on hRad17-RFC in the presence or absence of DNAs used in Table 1. Whether this is a real difference between the two types of complexes (RFC/PCNA vs. hRad17-RFC/9-1-1 complex) or indicative of a unique DNA structure that might be necessary to observe the stimulatory effect of the checkpoint 9-1-1 complex on hRad17-RFC ATPase activity remains to be seen. It is clear, however, that hRad17-RFC is an RFC-like ATPase and its activity is stimulated by DNA.
Figure 3.
The copurification of ATPase activity with hRad17-RFC. (A and B) Analysis of glycerol gradient fractions of hRad17-RFC and p36-p37-p40 core RFC by SDS/PAGE and Coomassie blue staining. hRad17-RFC (0.2 ml, 0.73 mg/ml) or the p36-p37-p40 core RFC complex (0.2 ml, 1 mg/ml) was loaded onto a 5-ml 15–35% glycerol gradient and centrifuged for 20 h at 4° at 250,000 × g. Fractions were collected from the bottom of the gradients and 10 μl of each was loaded onto the gels. (C) ATPase activity of hRad17-RFC and core RFC. Each fraction (1 μl) was analyzed for ATPase activity in the presence of 12.5 μM (as nucleotides) poly dA4000:oligo dT12–18 as indicated in Materials and Methods. Peak fractions, fraction 10 for hRad17-RFC and fraction 16 for core RFC, contained 25 and 59 ng of protein/μl, respectively.
Table 1.
Comparison of ATPase activities of RFC, hRad17-RFC, and the 3-subunit complex
| DNA added | RFC | hRad17-RFC | 3S |
|---|---|---|---|
| None | 5.6 | 6.0 | 1.0 |
| Poly dA4000:Oligo dT12–18 | 24.1 | 15.2 | 2.5 |
| φXssDNA | 64.9 | 28.8 | 13.0 |
| φXRFI | 5.7 | 6.3 | 1.1 |
Reactions were carried out as described in Materials and Methods in the presence or absence of 25 μM of the indicated DNA. The values indicate pmol Pi/min per pmol of complex. The 3-subunit complex (3S) is composed of p40, p37, and p36.
DNA Binding of hRad17-RFC.
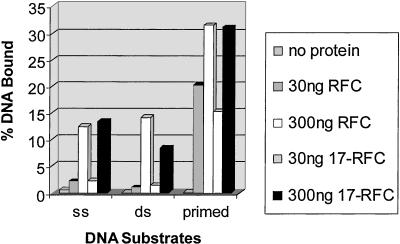
RFC binds to DNA with preference for primer-template-like structures (34). We tested hRad17-RFC for similar properties. As seen in Fig. 4, both RFC and hRad17-RFC have comparable affinities for single- and dsDNAs, and both protein complexes bind preferentially to primed DNA consistent with a role of a clamp loader at a primer terminus or a similar type structure.
Figure 4.
DNA-binding activity of hRad17-RFC. The indicated amounts of RFC or hRad17-RFC were incubated in a reaction mixture (30 μl) containing either 40 fmol of 32P-labeled 50-nt single-stranded (ss), 50-bp dsDNA or a primed 50-bp dsDNA with a 50-nt ssDNA (5′ overhang) in 25 mM Hepes (pH 7.5), 175 mM NaCl, 3 mM MgCl2, 1 mM DTT, and 0.1 mg/ml BSA for 30 min on ice. The fraction of DNA bound to the protein was quantitated by a nitrocellulose filter-binding assay. The details of the DNA substrates used were described (28).
Purification of the Checkpoint 9-1-1 Complex.
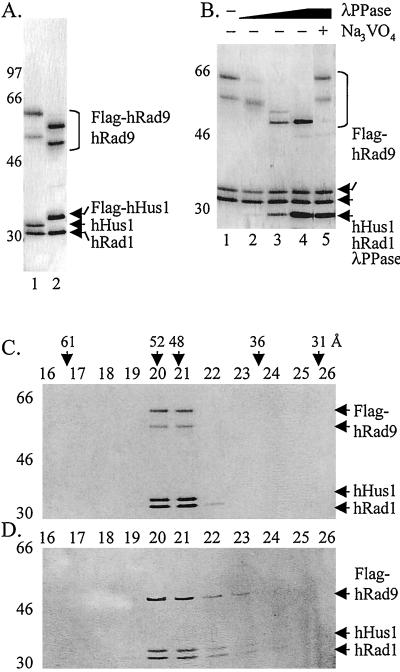
To isolate the hRad9-hHus1-hRad1 complex, we coinfected HF insect cells with baculoviruses expressing each of the subunits with either a Flag tag on the C terminus of hRad9 or hHus1. Immunoaffinity purification yielded the heterotrimeric complex with the three subunits, hRad9, hHus1, and hRad1, in a 1:1.2:1.2 stoichiometry as determined by densitometric scanning of a Coomassie blue-stained gel (Fig. 5A). Immunoblots of hRad9 from human cell extracts reveal multiple species ranging in size from 55 to 70 kDa, although hRad9 has a theoretical Mr of 45 kDa, and the fraction of heavily phosphorylated forms increase upon DNA damage (18, 38). It has been reported that only the phosphorylated forms of hRad9 associate with hRad1 and hHus1 as determined by coimmunoprecipitation experiments (18, 21). Therefore, it was of interest to determine the effect of phosphorylation on complex formation with recombinant proteins. As is apparent in Fig. 5A, hRad9 made in baculovirus-infected cells is mainly in the form of two species, which based on apparent molecular weights represent different levels of phosphorylation in insect cells. The stoichiometry of the subunits, and the elution pattern from gel exclusion chromatography (Fig. 5C), indicates that both of the forms of hRad9 are in a complex with hRad1 and hHus1. To investigate the role of phosphorylation in complex stability, checkpoint 9-1-1 was treated with λ phosphatase and analyzed by gel exclusion chromatography. Fig. 5B shows that with increasing concentrations of λ phosphatase there is a gradual conversion of hRad9 to the nonphosphorylated form, and interestingly during this conversion, discreet intermediate species are detected. When the maximally dephosphorylated form of hRad9 was analyzed by gel exclusion chromatography, the majority of the proteins migrated identically to the untreated 9-1-1 complex (Fig. 5 C and D). These results suggest that hyperphosphorylation of hRad9 is not required for the 9-1-1 complex stability. Whether or not any phosphorylation is required for the formation of the complex remains to be investigated.
Figure 5.
Purification and characterization of the checkpoint 9-1-1 complex. (A) Purification of the 9-1-1 complex. HF insect cells were infected with baculoviruses expressing all three subunits with tags where indicated, and the complex was purified by immunoaffinity chromatography as described in Materials and Methods. Lane 1, complex purified through the Flag tag on hRad9 (0.4 μg loaded) and lane 2, complex purified through the Flag tag on hHus1 (0.4 μg loaded). (B) Effect of λ phosphatase on the 9-1-1 complex. The complex was treated with 0, 3.2, 16, 80, and 80 units of phosphatase in lanes 1–5, respectively, under conditions described by the manufacturer. After 1-h incubation at 30°C, the products were analyzed by SDS/PAGE and silver staining. (C and D) Dephosphorylated hRad9 remains in the 9-1-1 complex. Checkpoint 9-1-1 complexes, before (C) or after (D) λ phosphatase treatment, were separated by gel filtration, and the fractions were analyzed by SDS/PAGE and silver staining.
Formation of the Checkpoint Rad Complex.
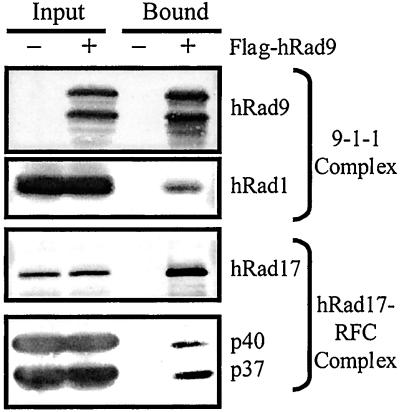
It is hypothesized that hRad17-RFC and the 9-1-1 complex function in a manner analogous to the conventional RFC/PCNA pair (3, 15). Complex formation between RFC and PCNA on and off DNA has been well documented (34); there is, however, no direct experimental evidence for a similar complex between hRad17-RFC and the checkpoint 9-1-1 complex. To find out whether such a complex exists, we infected HF insect cells with baculoviruses expressing the eight subunits of both complexes with the hRad9 containing a Flag tag. As a control, all of the subunits except hRad9 were expressed. The cell lysates were mixed with the anti-Flag resin and the bound material was analyzed by protein blotting for selected subunits of both complexes. As seen in Fig. 6, the hRad17-RFC is bound to the 9-1-1 complex. Similar results were obtained by mixing the separately purified hRad17-RFC and 9-1-1 complexes and analyzing the mixture by glycerol density gradient sedimentation (data not shown). Thus, hRad17-RFC and the 9-1-1 complex do form a RFC/PCNA-like complex with a special role in the DNA damage checkpoint. We propose the name checkpoint Rad complex to indicate its composition and unique function.
Figure 6.
Isolation of the checkpoint Rad complex. HF cells infected with either all of the subunits of the hRad17-RFC and 9-1-1 complexes, including Flag-tagged hRad9, or all of the subunits except hRad9 were lysed, and the complex was purified by immunoaffinity chromatography and analyzed by Western blotting as described in Materials and Methods.
In the RFC/PCNA pair, RFC binds to the primer terminus, recruits PCNA to the site, and opens the PCNA ring to clamp it onto DNA, thus providing the processivity factor for DNA polymerases δ and ɛ (see ref. 34). A standard assay for the molecular matchmaking activity of RFC is to measure RFC-dependent binding of PCNA to DNA; a nicked DNA circle is mixed with RFC and PCNA in the presence ATP, and then the free protein and DNA are separated by gel exclusion chromatography. Under these conditions, PCNA is found associated with the nicked DNA circle only in the presence of RFC and ATP (39). Current models predict a similar function for the checkpoint Rad complex. However, in repeated attempts with this experimental design, we were unable to detect the loading of the checkpoint 9-1-1 complex onto DNA by RFC or hRad17-RFC (data not shown). It is possible that different DNA structures, further posttranslational modifications of the checkpoint Rad complex, or additional factors are needed for loading of the checkpoint sliding clamp onto DNA.
Discussion
Recent computational and experimental studies have indicated that the human checkpoint Rad proteins make RFC-like and PCNA-like complexes with the distinct property of recognizing either primary or secondary DNA lesions and thereby initiating the DNA damage checkpoint cascade (3, 13). To learn about these proposed complexes and the features that differentiate them from the classical RFC and PCNA, we purified both hRad17-RFC, which is proposed to function as a clamp loader, and also the checkpoint 9-1-1 complex, which is predicted to be a checkpoint sliding clamp.
The purified hRad17-RFC has the predicted subunit composition of hRad17-RFCp36-RFCp37-RFCp38-RFCp40. Fractionation of CFE shows that virtually all of hRad17 exists in this complex. This finding contrasts with a report claiming that the majority of hRad17 is in the nucleolus and migrates out into the nucleoplasm to associate with other partners only upon DNA damage (40). Further studies are needed to understand the source of this discrepancy. In other aspects, hRad17-RFC exhibits the predicted RFC-like properties: It is a DNA-stimulated ATPase with a preference for primed DNA. In contrast with RFC, however, it does not associate with PCNA but instead specifically binds to the 9-1-1 complex.
The purified 9-1-1 complex also exhibited some of the predicted properties in terms of one-to-one stoichiometry of the three subunits and its specific interaction with hRad17-RFC but not with the classical RFC. We were unable to load the 9-1-1 complex onto primed DNA by either hRad17-RFC or RFC, and thus the checkpoint sliding complex function of the 9-1-1 complex remains to be demonstrated. There might be several reasons for our failure to load the 9-1-1 complex onto nicked DNA. Conceivably, more specialized DNA structures may be needed for loading the 9-1-1 complex. Exonuclease activity has been detected in both the hRad1 (41) and the hRad9 (42) subunits. It is possible that under appropriate conditions these nuclease activities aid in the loading process. The 9-1-1 complex itself has a weak nuclease activity (data not shown) that needs further characterization. Alternatively, it is possible that either posttranslational modification of the checkpoint Rad complex or its association with some yet to be identified accessory factors is necessary for the loading of the checkpoint sliding clamp. Indeed, it has been found that the Hus1 protein of S. pombe (16) and the hRad9 (18, 38) and hRad17 (43) are phosphorylated by the ATM/ATR family of kinases upon DNA damage. Furthermore, it has been reported that phosphorylation of hRad17 by ATR is required for high affinity interaction with hRad1, and presumably the entire 9-1-1 complex (43). Although in our in vitro system, we do detect a strong hRad17-RFC/9-1-1 complex interaction in the absence of ATR-induced hRad17 phosphorylation, it is possible that this interaction is stronger or qualitatively different with the ATR/ATM-phosphorylated hRad17, enabling the hRad17-RFC to function as a clamp loader of the 9-1-1 complex and activate the checkpoint-signaling cascade.
Acknowledgments
We thank L. Karnitz for the hRad9 cDNA clone, J. Vialard for the hRad1 and hRad17 cDNA clones, T. Bessho for the hHus1 cDNA clone, and B. Stillman for the anti-RFCp140 Abs. This work was supported by National Institutes of Health Grants GM322833 (to A.S.), GM38559 (to J.H.), and T32-CA09156 and F32-GM20830 (to L.A.L.-B.). J.H. is an American Cancer Society Professor.
Abbreviations
- RFC
replication factor C
- PCNA
proliferating cell nuclear antigen
- ds
double-stranded
- CFE
cell-free extracts
- HF
High Five insect cells
References
- 1.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 2.Lowndes N F, Murguia J R. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 3.O'Connell M J, Walworth N C, Carr A M. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhou B B, Elledge S J. Nature (London) 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 5.Rhind N, Russell P. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kastan M B, Lim D S. Nat Rev Mol Cell Biol. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths D J, Barbet N C, McCready S, Lehmann A R, Carr A M. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lydall D, Weinert T. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 9.Parker A E, Van de Weyer I, Laus M C, Verhasselt P, Luyten W H. J Biol Chem. 1998;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- 10.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 13.Thelen M P, Venclovas C, Fidelis K. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 14.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay H D, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr A M. Mol Cell Biol. 2000;20:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venclovas C, Thelen M P. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostrub C F, Knudsen K, Subramani S, Enoch T. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo T, Matsumoto K, Sugimoto K. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkmer E, Karnitz L M. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- 19.St. Onge R P, Udell C M, Casselman R, Davey S. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hang H, Lieberman H B. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 21.Burtelow M A, Roos-Mattjus P M, Rauen M, Babendure J R, Karnitz L M. J Biol Chem. 2001;4:4. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 22.Sancar A, Hearst J E. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 23.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan M, Schallhorn A, Wurm F M. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee C G, Phillips B, Finkelstein J, Yao N, O'Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunz F, Kobayashi R, Stillman B. Proc Natl Acad Sci USA. 1993;90:11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlmann F, Cai J, Flores-Rozas H, Dean F B, Finkelstein J, O'Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1996;93:6521–6526. doi: 10.1073/pnas.93.13.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J-K, Hurwitz J. J Biol Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 29.Hanna J S, Kroll E S, Lundblad V, Spencer F A. Mol Cell Biol. 2001;21:3144–3158. doi: 10.1128/MCB.21.9.3144-3158.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer M L, Gygi S P, Aebersold R, Hieter P. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- 31.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. EMBO J. 1999;21:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison V, Stillman B. J Biol Chem. 1998;273:5979–5987. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- 33.Podust V N, Fanning E. J Biol Chem. 1998;272:6303–6310. doi: 10.1074/jbc.272.10.6303. [DOI] [PubMed] [Google Scholar]
- 34.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 35.Tsurimoto T, Stilllman B. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 36.Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O'Donnell M, Hurwitz J. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 37.Podust V N, Tiwari N, Ott R, Fanning E. J Biol Chem. 1998;273:12935–12942. doi: 10.1074/jbc.273.21.12935. [DOI] [PubMed] [Google Scholar]
- 38.Chen M J, Lin Y T, Lieberman H B, Chen G, Lee E Y. J Biol Chem. 2001;276:16580–16586. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang M S, Sasaki H, Campbell M S, Kraeft S K, Sutherland R, Yang C Y, Liu Y, Auclair D, Hao L, Sonoda H, et al. J Biol Chem. 1999;274:36544–36549. doi: 10.1074/jbc.274.51.36544. [DOI] [PubMed] [Google Scholar]
- 41.Parker A E, Van de Weyer I, Laus M C, Oostveen I, Yon J, Verhasselt P, Luyten W H. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 42.Bessho T, Sancar A. J Biol Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 43.Bao S, Tibbetts R S, Brumbaugh K M, Fang Y, Richardson D A, Ali A, Chen S M, Abraham R T, Wang X F. Nature (London) 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]