Summary
Dicer proteins are known to produce small RNAs (sRNAs) from long double-stranded RNA (dsRNA) templates. These sRNAs are bound by Argonaute proteins, which select the guide strand, often with a 5′ end sequence bias. However, Dicer proteins have never been shown to have sequence cleavage preferences. In Paramecium development, two classes of sRNAs that are required for DNA elimination are produced by three Dicer-like enzymes: Dcl2, Dcl3, and Dcl5. Through in vitro cleavage assays, we demonstrate that Dcl2 has a strict size preference for 25 nt and a sequence preference for 5′ U and 5′ AGA, while Dcl3 has a sequence preference for 5′ UNG. Dcl5, however, has cleavage preferences for 5′ UAG and 3′ CUAC/UN, which leads to the production of RNAs precisely matching short excised DNA elements with corresponding end base preferences. Thus, we characterize three Dicer-like enzymes that are involved in Paramecium development and propose a biological role for their sequence-biased cleavage products.
Keywords: Dicer-like enzymes, small RNAs, sequence specific cleavage, genome rearrangements, precise DNA elimination
Graphical Abstract
Highlights
-
•
Dicer-like proteins can cleave dsRNA in a sequence-specific manner
-
•
Dicer-like proteins produce small RNAs with 5′ and 3′ sequence bias
-
•
Small RNA sequence bias enables precise targeting of transposon-derived IESs
-
•
Dcl5 recognizes IES-IES junction sequence motifs
Like restriction enzymes and DNA, some Dicer-like proteins can cleave RNA with sequence selectivity.
Introduction
Dicer proteins are highly conserved dsRNA endoribonucleases with important roles in RNAi and post-transcriptional gene silencing processes (Kim et al., 2009, Carmell and Hannon, 2004, Hammond, 2005). Dicer proteins preferentially cleave dsRNA from their termini into small RNAs of distinct size (Zhang et al., 2002). The cleavage products are bound by Argonaute (AGO) proteins, which select the guide strand from the small RNA duplexes (Peters and Meister, 2007). For the strand selection, some AGO proteins show 5′ end nucleotide preference, which is typically a uridine (Mi et al., 2008). Dicer proteins typically contain an N-terminal helicase, DUF283 and a PAZ domain followed by two RNaseIII domains, and a double-stranded RNA (dsRNA)-binding domain (dsRBD) (MacRae and Doudna, 2007; Jinek and Doudna, 2009). However, many Dicer proteins lack some of these domains, such as the Dicer found in Giardia intestinalis that contains only the PAZ and RNaseIII domains (Macrae et al., 2006).
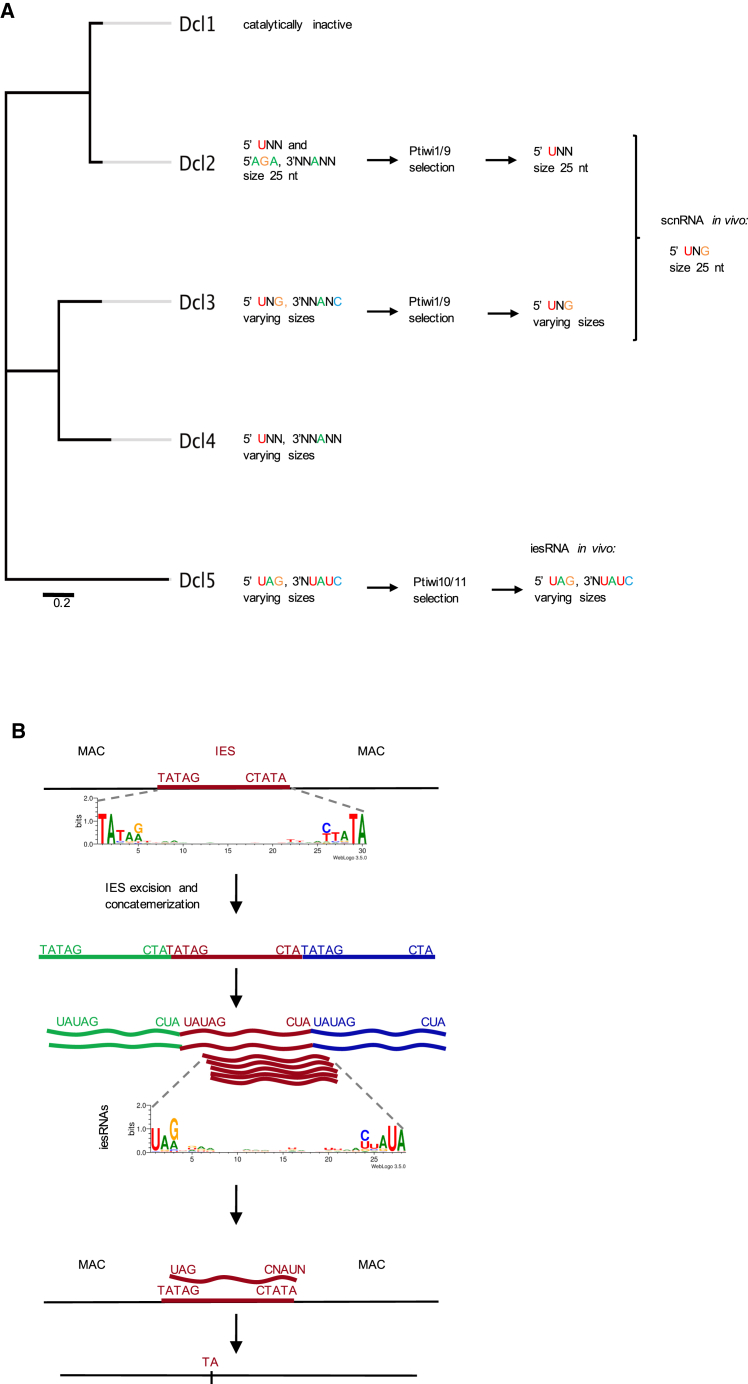
In Paramecium, eight genes encode for Dicer or Dicer-like enzymes. Dicer 1–3 (Dcr1–Dcr3) are canonical Dicer proteins, and it was shown that Dcr1 is responsible for small interfering RNA (siRNA) production. Additionally, the Paramecium genome encodes for five Dicer-like enzymes (Dcl1–Dcl5) (Lepère et al., 2009, Sandoval et al., 2014). These Dicer-like enzymes have similar protein domain architecture to the Dicer of Giardia intestinalis. They contain two RNaseIII domains and a highly diverged PAZ domain, but no helicase or DUF283 domain. Dcl5 also contains a dsRNA-binding domain (RBD) (Figure 1A). The three Dicer-like proteins, namely Dcl2, Dcl3, and Dcl5, produce two classes of small RNAs involved in a developmental genome rearrangement process (Lepère et al., 2009, Sandoval et al., 2014).
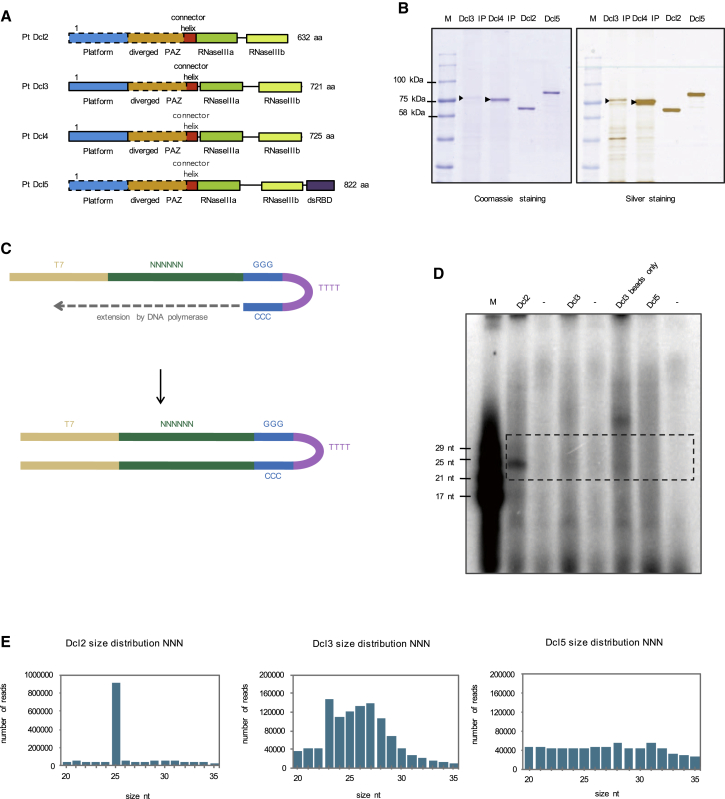
Figure 1.
In Vitro Cleavage Assays with Random Hairpin RNA
(A) Four Dicer-like proteins from Paramecium tetraurelia are shown schematically.
(B) SDS-PAGE analysis of immunopurified Dcl3 and Dcl4 proteins and purified recombinant Dcl2 and Dcl5 from insect cells. 500 ng of purified proteins and 20 μL beads were loaded on 8% SDS-PAGE gels and stained with either Coomassie or silver. Dcl3-3xFlagHA and Dcl4-3xFlagHA have a size of 88 kDa, Dcl2 has a size of 74 kDa, and Dcl5 has a size of 97 kDa.
(C) A schematic of DNA oligo used to generate a hairpin RNA with random nucleotides for in vitro cleavage assays.
(D) 15% polyacrylamide-urea gel with small RNAs 5′-labeled with 32P. M is the small RNA ladder. No enzyme controls are marked as (−). “Dcl3 beads only” is a no template RNA control for Dcl3.
(E) Size distribution graphs of Dcl2, Dcl3, and Dcl5 cleavage products based on sRNA sequencing.
See also Figure S1.
Ciliates, such as Paramecium, contain two kinds of nuclei: a somatic macronucleus and a germline micronucleus. The somatic nucleus is highly polyploid (∼800 n), transcriptionally active during the vegetative growth of the cell and does not contain any repetitive sequences such as transposons, transposon remnants or minisatellites. The germline genome is diploid and does contain these repetitive elements. During sexual development, these elements have to be excised either in a precise or imprecise manner from the zygotic nucleus to form a new functional somatic nucleus. Both small RNA classes guide the maternally inherited excision of transposon-derived sequences (internal eliminated sequences [IESs]) (Allen and Nowacki, 2017, Coyne et al., 2012). The first class, so-called scan RNAs (scnRNAs), are produced by Dcl2 and Dcl3 in the germline nuclei and are involved in the primary targeting of DNA excision (Lepère et al., 2009). The second class, iesRNAs, are produced in sexual progeny’s developing somatic nuclei from the excised, concatenated IESs and are involved in the amplification of the small RNA signal needed for complete DNA elimination (Sandoval et al., 2014, Allen et al., 2017). It is important for the small RNAs to target the precise excision of those DNA elements by helping the excision machinery. So far, it is not fully understood how the small RNAs target IESs for excision. The process needs to be highly precise, otherwise most open reading frames would be interrupted and/or frameshifted and the progeny would not be viable, which is what is observed upon depletion of scanRNAs (Lepère et al., 2009).
A hallmark of the excised elements is a weak 5′ consensus sequence of TAYAGYNR on both ends, with only one of the TA dinucleotides remaining in the macronuclear genome after elimination (Arnaiz et al., 2012). For shorter IESs (<30 base pairs [bp]), the consensus is TATAG (Swart et al., 2014). This size range between 26–30 bp of short IESs includes ∼35% of all IESs, and 93% of all IESs are shorter than 150 bp (Arnaiz et al., 2012). Interestingly, both classes of small RNAs contain specific 5′ and 3′ signature motifs. It was previously shown by small RNA sequencing that scnRNAs contain a 5′ UNG signature. Additionally, it was suggested that Dcl2 and Dcl3 have redundant functions because only the knock down of both enzymes led to unviable progeny and disappearance of scnRNAs (Lepère et al., 2009, Sandoval et al., 2014). However, small RNA sequencing after single gene knock downs showed indirectly that Dcl2 and Dcl3 have distinct functions during scnRNA production because DCL3KD shows sRNAs with a size of 25 nt but only a 5′U. DCL2KD, on the other hand, shows sRNAs of varying length but with the 5′UNG signature. In case of iesRNAs, it was shown that they contain a 5′UAG and a 3′ CNAUN signature motif (Sandoval et al., 2014).
In this study, we tested through in vitro assays whether the sequence specificity is a consequence of a specific Dicer cleavage, or as in other organisms, due to selection by Argonaute proteins. Here, we show that Paramecium Dicer-like proteins have sequence specificity for their target dsRNA, which contributes to the efficiency and precision of the DNA elimination machinery.
Results
Both developmental classes of small RNAs found in Paramecium contain end signature motifs. To analyze the basis for this sequence specificity and to elucidate the role of the Dicer-like enzymes, we performed in vitro processing assays. We purified recombinant Dcl2 and Dcl5 from insect cells and purified Dcl3-FlagHA-tagged protein directly from Paramecium cell lysates (Figure 1B). The purified proteins were incubated with different dsRNA templates. The processed RNA was 5′ end-labeled and analyzed on 15% denaturing PAGE gels and by deep sequencing.
Dicer-like Enzymes Cleave dsRNA with Preferences for Different Sizes
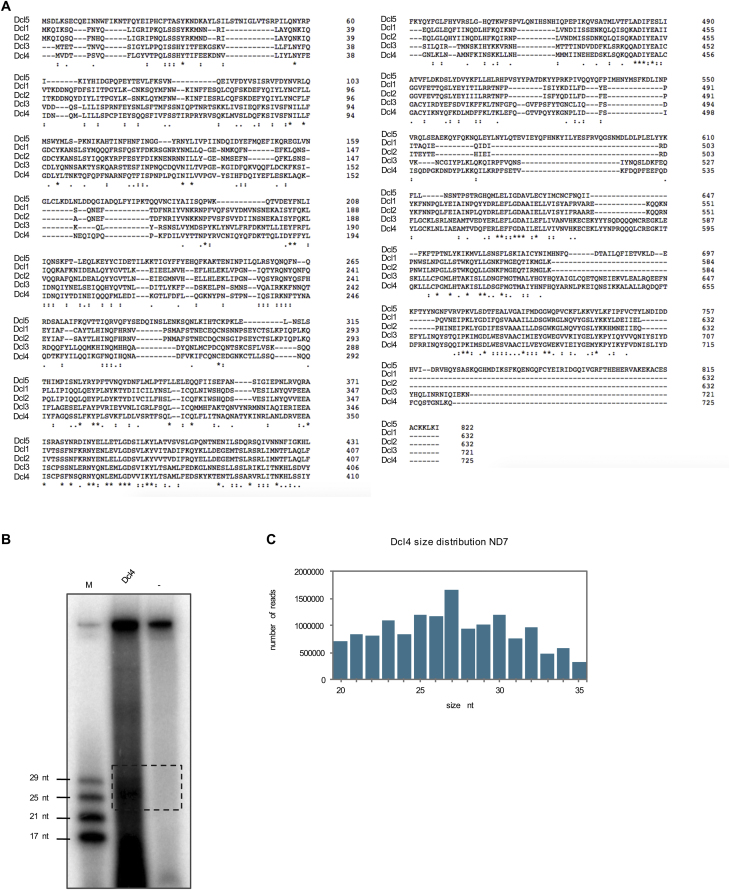
Because we were interested in the sequence preference of the Dicer-like enzymes in an unbiased way, we tested their cleavage activity with a random hairpin RNA substrate. We designed a DNA oligonucleotide containing a T7 promoter sequence on its 5′ end, followed by a stretch of 50 random nucleotides (50xN) and 12 Gs that loop and pair with 12 complementary Cs. We also added five Ts between the G and C stretch. This oligo was extended with the help of a DNA polymerase to form a DNA hairpin containing in its stem the random sequence. This DNA hairpin was used for in vitro transcription, using the T7 promoter, in order to form an RNA hairpin that could be used for the in vitro assays (Figure 1C). We incubated this RNA hairpin with the purified Dicer-like enzymes and analyzed the cleavage products both on acrylamide gels and with deep sequencing. The denaturing gel shows a specific band for Dcl2 around 25 nt. For Dcl3 and Dcl5, we did not observe discrete bands after the cleavage but a smear consisting of a varying range of sizes (Figure 1D). As a second cleavage template, we used a 500 bp long dsRNA fragment corresponding to the coding region of the non-essential ND7 gene, which is involved in a pathway leading to trichocyst discharge (Skouri and Cohen, 1997). Incubation of Dcl3 with the ND7 template gives a specific size pattern of cleavage products on the gel and after sequencing (Figure S1F). The same ND7 substrate is processed into 25 nt long sRNAs by Dcl2, as observed for the random templates (Figure S1F).
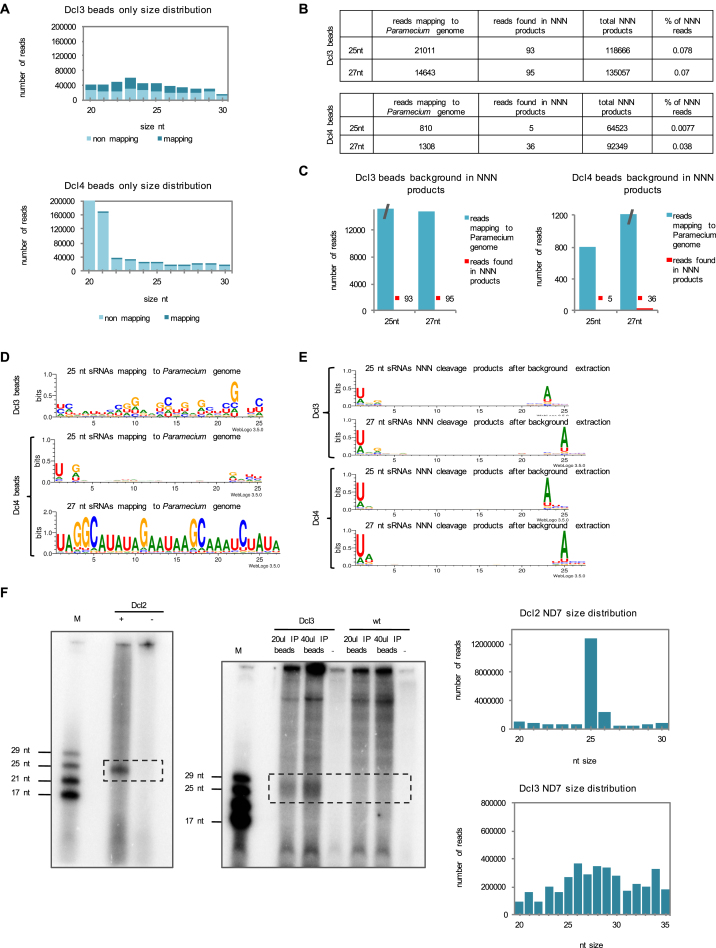
Figure S1.
Analysis of the Bead Contamination and Dcl2 and Dcl3 Results with ND7, Related to Figures 1 and 5
(A) Graphs of the mapping for Dcl3 beads control and Dcl4 beads control. y axis shows number of reads. x axis shows different nucleotides sizes. Different sRNAs mapped against the Paramecium MAC plus IES genome of strain 51. Number of reads mapped to the genome are shown in dark color. Non-mapping reads are shown in bright color.
(B) The table shows an overview of 25 nt and 27 nt reads of both bead controls mapping to the genome and how many of those reads are found in the NNN cleavage product reads to assess the background.
(C) Graphical representation of the table in (B).
(D) Sequence logos for both bead controls (Dcl3 and Dcl4) for either 25 nt reads or 27 nt reads.
(E) Sequence logos for 25 nt or 27 nt sRNA cleavage products with the NNN hairpin after extraction of the background reads.
(F) Results of the processing assay of Dcl2 and Dcl3 with the ND7 fragment RNA. For Dcl3 processing assay, wild-type lysates (wt) were used as a control. (-) indicates no enzyme controls. Size distribution graphs are shown for Dcl2 and Dcl3 deep sequencing after processing assay with ND7.
As controls, we used no enzyme samples (−) to visualize the unprocessed input RNA. For Dcl3, which we could not purify from insect cells, we additionally used immunoprecipitation (IP) bead samples incubated with wild-type (WT) lysates as a control (Figure S1F). The WT lysates do not contain the overexpressed Dcl3-FlagHA protein, and thus no protein should bind to the beads during IP. This control was used to assess the background RNA binding to the beads and to ensure that no non-specifically bound protein cleaves the dsRNA input. The hairpin RNA has a size of 175 nt and is therefore not able to migrate into the 15% denaturing gel, thus it is not visible on the gel. Additionally, for Dcl3 we used IP beads incubated without RNA as a control. This control was used to ensure that the cleavage products we detect are not cellular sRNAs already bound to the beads or the immunoprecipitated Dcl3. We do detect some RNA contamination in the beads-only control on the gel (Figure 1D). We think that the background level of RNA bound to the beads shows up after 5′ end labeling when there is no other RNA in the sample. This explanation is supported by the fact that in the beads-only control we see bands that are not present when the IP beads are incubated with template RNA. In order to make sure that this background RNA does not affect our cleavage preference results, we sequenced and analyzed the background RNA as well as our positive samples.
The sequencing for Dcl2, Dcl3, and Dcl5 shows different size distributions (Figure 1E). Dcl2, as already seen on the gel, has a distinct size preference for 25 nt. Canonical Dicer proteins generally bind their dsRNA target at the 3′ termini through the PAZ domain recognizing the 2 nt 3′ overhang, while the distance between the PAZ domain and the catalytic center determines the length of the cleavage product. Thus, canonical Dicers act as molecular rulers (Zhang et al., 2004, Macrae et al., 2006, Ma et al., 2004). However, Dicer proteins with no PAZ domain exist that still produce small RNAs of discrete sizes, such as the Dicer of S. pombe (Provost et al., 2002). In order to produce specific 25 nt long small RNAs, it is possible that Paramecium Dcl2 uses a similar mechanism to the Dicer of S. pombe, although structurally it is unclear how this works. Dcl3 and Dcl5, on the other hand, produce small RNAs of varying lengths.
Dcl2 and Dcl3 Cooperate to Produce Mature scnRNAs In Vivo
The analysis of the cleavage products shows different sequence specificities for Dcl2, Dcl3, and Dcl5. The abundance of each nucleotide at a certain position of the cleavage products was compared to the relative nucleotide abundance in the input RNA. For the random hairpin template, the input abundance is 25% for each nucleotide. This comparison is shown in the enrichment graphs. An enrichment factor of 1 for a specific nucleotide means that this nucleotide is no more abundant than in the input. A factor above 1 means the nucleotide is enriched (i.e., it is found more frequently at this position than would be expected by chance). The enrichment graph for Dcl2 shows a sequence preference for 5′U/AGA (Figure 2A). The same preference is observed in the weblogo analysis (Figure 2C). This 5′U/AGA signature for Dcl2 is not seen in RNA sequencing of DCL3KD cultures in vivo (Sandoval et al., 2014), which suggests a discrepancy between the Dcl2 cleavage products and the scnRNAs found in vivo, indicating that the Piwi proteins carry out a further selection on the RNAs in the cell. We further analyzed Dcl2 products by removing the passenger strands—identified as having 3′ NNU, assuming that the Piwi protein selects 5′ U—and analyzing the signature of the guide strands. The weblogo of the guide strands shows an even stronger bias toward 5′U (Figure 2C). We then also checked the signature for all the 25 nt reads that start with a U. The weblogo for those reads shows a much stronger 5′U signature (Figure 2C). These findings suggest that Dcl2 has a preference for either 5′UNN or 5′AGA. However, in vivo the Ago protein that binds Dcl2 products selects only the small RNAs starting with U. Therefore, in the small RNA sequencing of RNA extracted from cells, only the 5′U signature is seen.
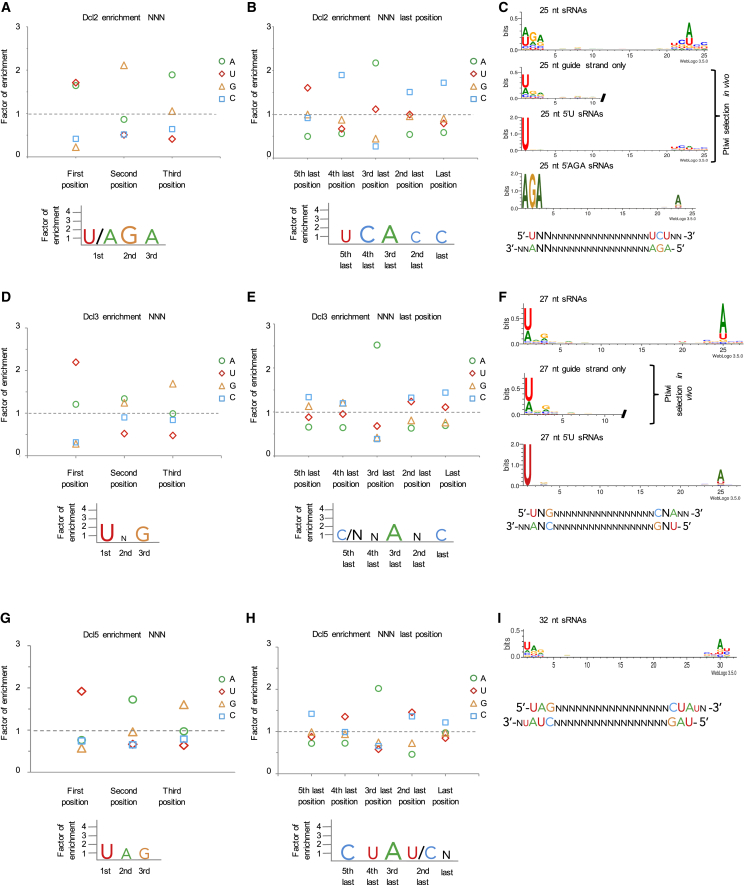
Figure 2.
Sequence Bias Analysis of Dcl Cleavage Products
(A) 5′ end nucleotide enrichment graph for Dcl2 cleavage products showing relative abundance of the first 3 nt. Enrichment factor of 1 (dashed line) represents expected frequency based on the nucleotide composition of the input dsRNA.
(B) 3′ end nucleotide enrichment graph for Dcl2 cleavage products.
(C) Sequence logos for 25 nt long Dcl2 cleavage products. A typical Dcl2 double-stranded cleavage product is shown schematically with end base signatures and the 2nt-3′ overhangs.
(D) 5′ end nucleotide enrichment graph for Dcl3 cleavage products.
(E) 3′ end nucleotide enrichment graph for Dcl3 cleavage products.
(F) Sequence logos for 27 nt long Dcl3 cleavage products. A typical Dcl3 double-stranded cleavage product is shown schematically.
(G–I) Enrichment graphs for Dcl5. Shown for the first three positions (G) and the last five positions (H). Sequence logo shown for 32 nt long sRNAs (I).
We also analyzed the nucleotide enrichment for the last few positions of the small RNA cleavage products. Dcl2 showed a 3′ UCACC signature, which is also visible in the weblogo of Dcl2 (Figures 2B and 2C). This 3′ UCACC is consistent with its corresponding to the passenger strand of the proposed 5′ UNN or 5′ AGA sequence preference of Dcl2, as Dicer products are double-stranded with a 2 nt 3′ overhang. The predicted complement of 5′ UNN is 3′ NNANN, including the 2 nt 3′ overhang, and the complement of 5′ AGA is 3′ UCUNN. Together, a mix of these two populations would appear as 3′ UCA/UNN on an enrichment graph or weblogo, which is what we observe, bar the 3′ most CC (Figures 2B and 2C). Strictly speaking, we cannot rule out Dcl2 having a 3′ sequence preference rather than a 5′ preference and the 5′ UNN or 5′ AGA coming from the complementary strand. Indeed, it is possible that Dcl2 recognizes both the 5′ and the 3′ ends of the RNA it is to cleave. If this is the case, it would explain the apparent dual specificity of the enzyme: if Dcl2 preferentially cleaves 5′ U and 3′UCUNN, then we would expect to see the enrichments found. In order to see whether the 5′ AGA could correspond to the complementary strand of a cleavage event where 5′ U and 3′ UCUNN were both selected for, we analyzed the weblogo of the RNAs that do contain 5′ AGA (Figure 2C). These RNAs have a preference for A at 3 nt from their 3′ end, which demonstrates that the 5′ A and the 3′ UCUNN are present on the same strand of RNA. This indicates that Dcl2 preferentially cleaves RNAs with 5′ U and 3′ UCUNN, as shown at the bottom of Figure 2C. Whether this preference results from a dual 5′ (or indeed 3′) specificity of the enzyme and the result of enzyme dimerization, or from both a 5′ and 3′ preference for the same strand, cannot be determined from this data.
The enrichment graph for Dcl3 shows a nucleotide preference of 5′UNG, which is also present in the weblogo (Figures 2D and 2F). The 5′UNG signature is even more dominant when we analyze the guide strand only. Moreover, the weblogo and the nucleotide enrichment for the last positions showed a 3′ C/NNANN motif (Figure 2E). Similarly to Dcl2, the enriched A is the expected complementary nucleotide to the 5′ U as shown in the schematic representation of the Dcl3 cleavage product (Figure 2F). The C is slightly enriched on the fifth last position. This C corresponds to the 5′ G on the third position of the passenger strand (Figure 2E).
Analysis of the beads-only control showed that the GC content of the reads is ∼50%, which is much higher than the GC content found in the Paramecium genome (GC 30%). Furthermore, only ∼50% of the IP bead reads mapped to the Paramecium genome (Figure S1A). The high GC content suggests that the non-mapping background mostly comes from bacterial contamination from the food of Paramecium. We analyzed the 25 nt and 27 nt reads in more detail and checked how many Paramecium-mapping background reads are found in our NNN hairpin cleavage reads for Dcl3 (Figures S1B and S1C). In other words, we analyzed how many reads in our sequencing results were actually not real Dicer products but background sRNAs from the beads, to ensure that the background does not affect our results. Of the 21,011 25 nt long reads from the IP beads mapping to the genome, only 93 reads were also found in the 25 nt long reads from the NNN hairpin sequencing. This is only 0.078% of the total 25 nt reads of the NNN hairpin reads. For the 27 nt long reads, only 0.07% of our NNN hairpin reads are background coming from the beads. We also analyzed whether the 25 nt reads from the IP beads mapping to the genome are scanRNAs. We checked the weblogo of the mapping reads (Figure S1D). The logo did not show the specific 5′UNG signature known for scnRNAs, which supports our finding that the background does not affect our cleavage preference results for Dcl3. Nevertheless, we extracted the background reads from our NNN reads and performed the weblogo analysis again to check if the logo changes. As expected, the weblogo still showed a 5′ UNG signature for Dcl3 after background extraction (Figure S1E). These findings suggest that there is some RNA contamination bound to the beads, which is visible on the gel, but this background RNA does not affect the results of the cleavage preferences of Dcl3.
Dcl5 Cleavage Products Have Specific 5′ and 3′ Signatures
Dcl5 seems to cleave the hairpin RNA less efficiently than Dcl2 or Dcl3, as can be seen on the gel and in the sequencing size distribution (Figures 1D and 1E). Nonetheless, analysis of the sequencing shows specific nucleotide preferences for Dcl5. The enrichment graph and the weblogo show a 5′UAG preference and a 3′ CUAU/CN (Figures 2G–2I). These 5′ and 3′ motifs are also seen in sRNA sequencing from total RNA extracts of the cell (Sandoval et al., 2014).
The 5′ and 3′ sequence cleavage preferences for Dcl5 are palindromic and could result from guide and passenger strands of the same cleaved duplex, as is shown by the complementary 5′ UAG and 3′ CUAU/CN, including a 3′ dinucleotide overhang. Thus, it is not possible to separate guide and passenger strands from each other to assess whether Dcl5 has a 3′ sequence preference as well as a 5′ cleavage preference, or whether the 3′ signature comes from the passenger strands. However, in the cellular small RNA sequencing (Sandoval et al., 2014) the same 5′ UAG and 3′ CUAU/CN are evident in the Dcl5 products, indicating that either the 3′ signature is a real feature of Dcl5 cleavage, or there is no guide/passenger strand selection and that all Dcl5 products are loaded onto Piwi proteins.
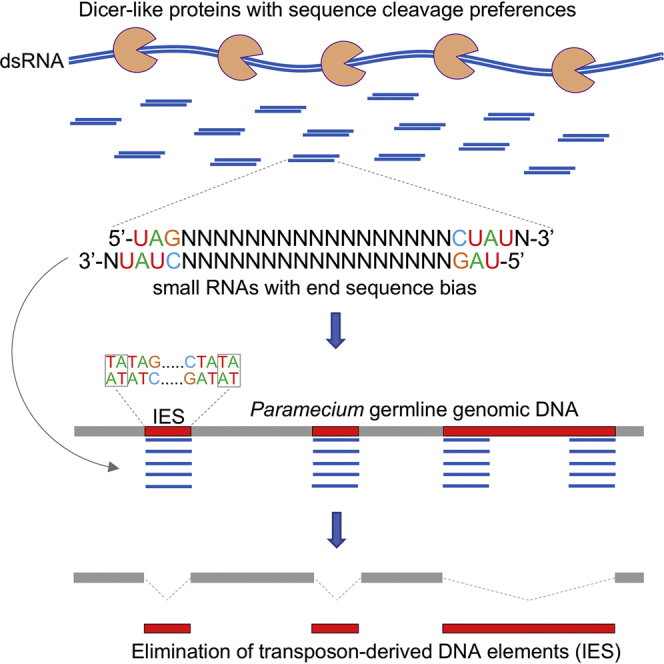
Dcl5 Recognizes and Preferentially Cleaves IES Sequences
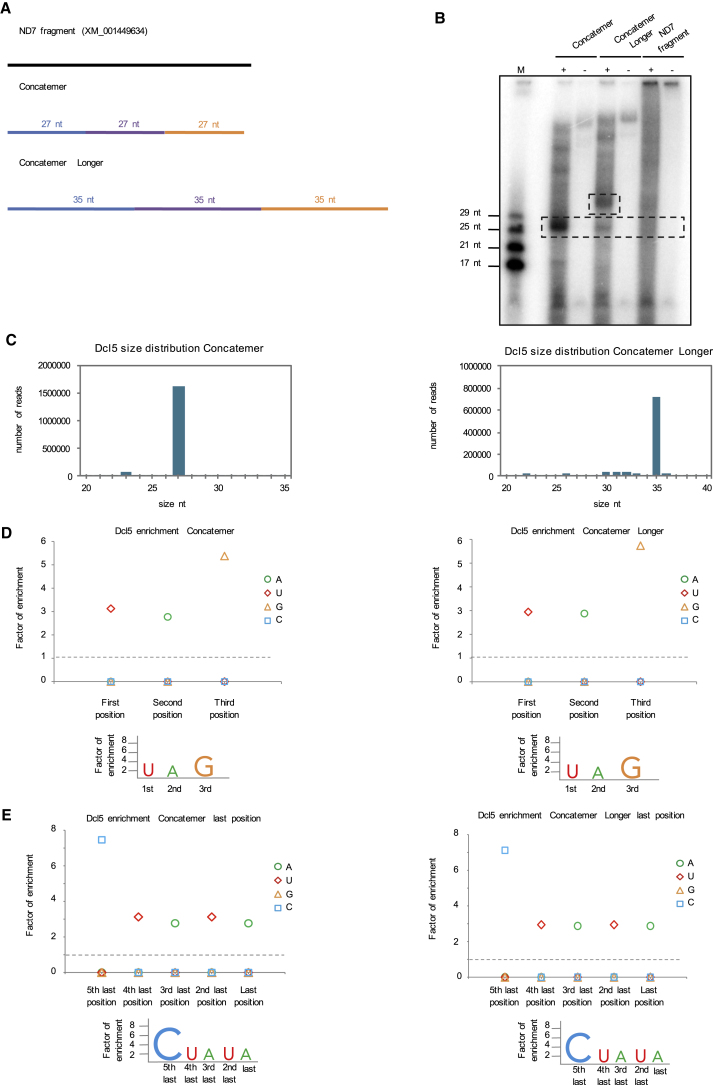
Because Dcl5 in vivo cleaves dsRNA templates composed of concatenated IESs (Allen et al., 2017), we decided to investigate its cleavage preferences on Paramecium sequences. We therefore performed in vitro assays for Dcl5 with several dsRNA templates (Figure 3A). We tested a 500 bp long dsRNA fragment of ND7 representing genomic DNA as well as dsRNA templates consisting of IESs. One template contained three different IESs, each 27 nt in length, joined together. IESs are mostly short sequences with a length mode of 27/28 bp (Arnaiz et al., 2012), therefore we chose the IES length in the concatemer to be 27 nt. This template was called ‘Concatemer’. Another template was derived from the same three IESs but their length was extended by adding 8 nt in the middle of each IES sequence resulting in 35 nt long IESs. This template is referred to as ‘Concatemer Longer’. The no enzyme controls (−) show the unprocessed RNA, which is visible for the two IES templates. The ND7 RNA is too large to migrate into the gel same as the hairpin NNN template. Dcl5 processes the Concatemer template into 27 nt products. The “Concatemer Longer” is cleaved into 35 nt dsRNAs, which were detected as a strong band on the gel. There is also a weaker 27 nt long band visible (Figure 3B). The size of the cleavage products reflects precisely the length of the IES sequences. These findings indicate that Dcl5 recognizes the IESs as discrete units. The ND7 fragment, similar to the NNN hairpin, does not get processed into products of distinct sizes (Figure 3B). We therefore postulate that Dcl5 has an ability to recognize and cleave IES ends. The size distribution observed on the gel for the “Concatemer” and “Concatemer Longer” templates is also found in the deep sequencing results, although some bands visible on the gel were not present in the sequencing data (Figure 3C). The discrepancy between what is seen on the gel and the corresponding sequencing data is not clear but it might come from RNAs that cannot be cloned during Illumina library preparation but can be 5′ end-labeled. We suggest that the additional products are cleaved off the ends of dsRNA templates RNAs when the Dicer-like protein recognizes the internal IES sequences. These leftovers are not true Dicer cleavage products (processed on both ends) and therefore do not contain 5′monophosphate required for adaptor ligation during library preparation. Both ends of the templates have 5′ triphosphates due to the T7 RNA synthesis and 3′OH. During library preparation, only one adaptor would be ligated to the 3′OH ends of those products that would lead to their absence in the sequencing libraries. This, however, would not affect the 5′ end labeling by T4 kinase exchange reaction.
Figure 3.
In Vitro dsRNA Processing by Dcl5 Using Templates Containing Paramecium MAC and IES Sequences
(A) dsRNA templates used for in vitro assays with Dcl5 are shown schematically: a 500 nt long fragment of ND7 coding sequence (black line) and two templates made of three concatenated IESs (blue, purple, and orange).
(B) 15% polyacrylamide-urea gel with small RNAs 5′-labeled with 32P. M is the small RNA ladder. (+) and (−) indicate reactions performed with or without the enzyme. Dashed boxes indicate the bands corresponding to the most abundant products after sRNA sequencing.
(C) Size distribution graphs of the cleavage products for Dcl5 with Concatemer and Concatemer Longer templates.
(D and E) Enrichment graphs for the 5′ (D) and 3′ (E) ends of Dcl5 cleavage products.
We analyzed the sequencing results for both concatemer templates processed by Dcl5. The sequencing data show a 5′ sequence preference of UAG, as these 3 nt were highly enriched at the first three positions compared to the general nucleotide composition of the template (Figure 3D). The sequencing analysis also shows nucleotide enrichment for 3′CUAUA (Figure 3E).
Dcl5 Specifically Recognizes IES Ends
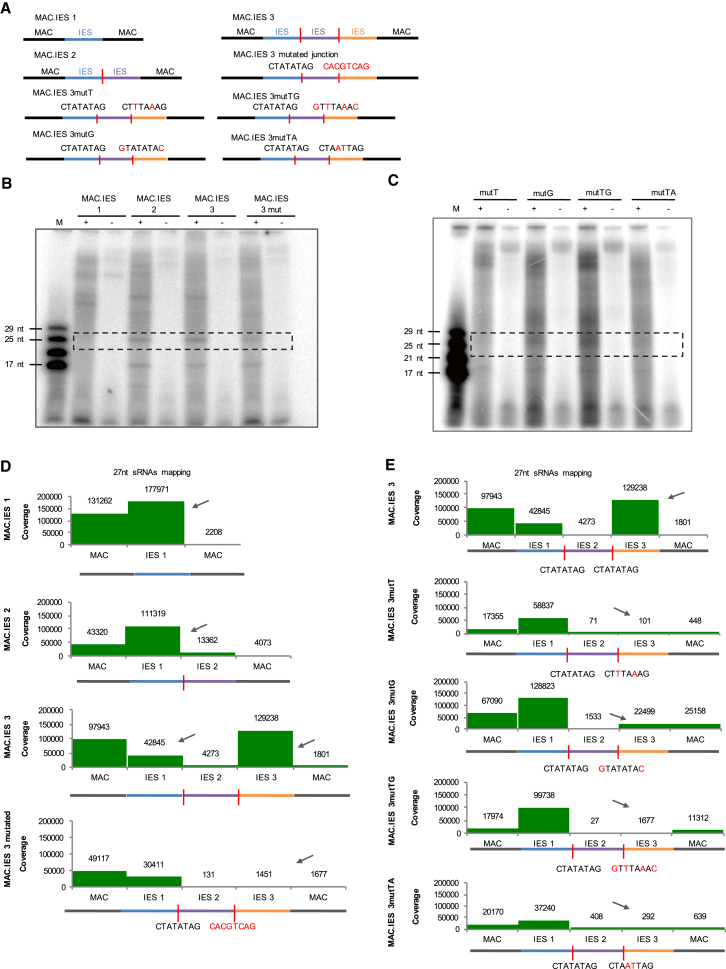
To assess whether Dcl5 has a preference for IES ends and/or IES-IES junctions, we performed processing assays with additional templates called MAC.IES 1, MAC.IES 2, or MAC.IES 3 (Figure 4A). These constructs contain 30 nt of a random sequence reflecting macronuclear (MAC) DNA nucleotide composition (30% CG content), flanking either one, two, or three concatenated IES sequences. Additionally, in one of the constructs, we introduced a mutation in one of the IES-IES junctions. It was previously shown that short IES ends show a consensus sequence of TATAG (Swart et al., 2014). Concatenation of IESs therefore results in an IES-IES junction of CTATATAG. In one of the junctions, we mutated this consensus to CACGTCAG. Nucleotide substitutions are shown in red (Figure 4A). All IESs in these templates are 27 nt long. The gel shows that 27 nt long products corresponding to the size of the IESs are present in all the experiments (Figure 4B). This was as expected because all the templates contain at least one IES sequence and should be processed by Dcl5 to 27 nt dsRNAs. The no enzyme control (−) shows the unprocessed input RNA (Figure 4B). The additional bands visible on the gel might again be cleaved off RNAs as described before. The size distribution of all the products based on sequencing indicates that Dcl5 produces mainly 27 nt sRNAs (Figures S2A and S2B). To investigate from which parts of the templates Dcl5 produces sRNAs, we mapped all 27 nt reads to the different templates. The mapping shows that Dcl5 preferably produces sRNAs from all the IES sequences (Figure 4D). The presence of sRNAs mapping to the flanking randomized MAC sequences could be due to Dcl5 recognizing and cleaving an IES end-like sequence within the flanking region. In addition, as soon as we introduce one IES-IES junction, sRNAs matching IESs are much more abundant than those matching the flanking region (see the difference between MAC.IES1 and MAC.IES2 templates in Figure 4D). Moreover, many sRNAs were produced from the third IES in the MAC.IES3 template and as soon as we introduce the mutated IES-IES junction sequence, nearly no sRNAs match the third IES (Figure 4D). Our in vitro cleavage assays indicate that the ability of Dcl5 to cleave dsRNA template depends on the presence of at least one IES end sequence. Mutation of IES2-IES3 junction reduces sRNA coverage of IES2 9.8-fold and IES3 26.7-fold (normalized to the total number of reads mapping to each of the templates) (Figure 4D).
Figure 4.
Dcl5 Cleavage of dsRNA Templates with and without Nucleotide Substitutions in IES-IES Junction Sequence
(A) dsRNA templates used for processing assays are shown schematically. Blue, purple, and orange represent different IES sequences. Red line indicates IES-IES junction. IES-IES junction sequence is shown in black font. Nucleotide substitutions are shown in red.
(B and C) 15% polyacrylamide-urea gel with small RNAs 5′-labeled with 32P for the cleavage with MAC.IES templates (B) and single mutated MAC.IES templates (C). M is the small RNA ladder. (+) and (−) indicate reactions performed with or without the enzyme. Dashed boxes indicate the bands corresponding to the most abundant products after sRNA sequencing (see Figure S2).
(D and E) Mapping of all 27 nt long sRNA reads to the MAC.IES templates (D) and to the single mutated MAC.IES templates (E) used for processing assays. Arrow indicates the sRNA peaks most affected upon changes in the template sequence.
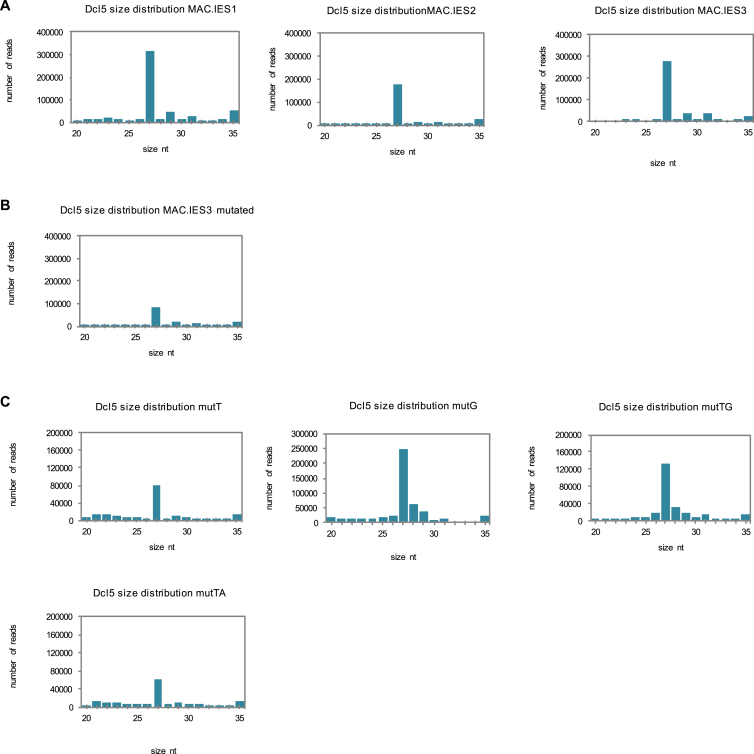
Figure S2.
Size Distributions of IES-IES Junction Templates and Mutated Templates, Related to Figure 4
(A–C) Size distribution graphs for all the different templates used for Dcl5 processing assays.
Each Nucleotide within IES End Sequence Is Important for Dcl5 Recognition
To determine which of the nucleotides at the IES-IES junction are important for Dcl5 recognition we performed an in vitro assay with the MAC.IES 3 template containing nucleotide substitutions within the CTATATAG IES-IES junction (Figure 4A). We used the following mutated junctions (nucleotide substitutions underlined): CTTTAAAG (MAC.IES 3mutT), GTATATAC (MAC.IES 3mutG), GTTTAAAC (MAC.IES 3mutTG), and CTAATTAG (MAC.IES 3mutTA). We performed an in vitro cleavage assay with Dcl5 as before and loaded the products on a gel (Figure 4C). The (−) shows the no enzyme control for each template. As for the other MAC.IES templates, we detect a 27 nt long cleavage product as well as additional non-specific uncloneable bands. sRNA sequencing revealed prominent 27 nt cleavage products in all the samples (Figure S2C). sRNA mapping shows that each of the mutated IES-IES junctions leads to a very strong reduction of Dcl cleavage (Figure 4E). This effect was slightly weaker in the case of the G to C mutation as seen in the mapping to the MAC.IES3 mutG template.
Dcl3 and Dcl4 Differ in Cleavage Preference
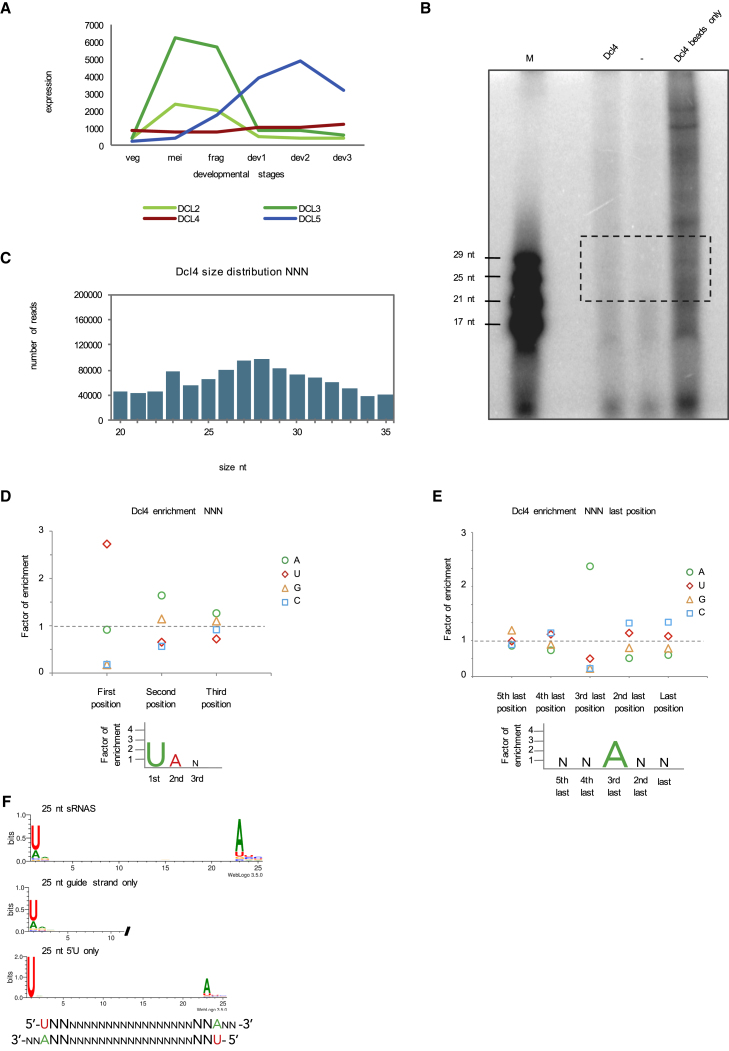
So far, we determined that Dcl2, Dcl3, and Dcl5 show different sequence cleavage specificities resulting in sRNA products carrying specific 5′ and 3′ signature motifs. To get an idea of which part of the protein may be responsible for the sequence recognition, we decided to analyze cleavage preference of Dcl4. Dcl4 is one of the two Dicer-like proteins in Paramecium that biological function is unknown (the other one being Dcl1). Dcl4 is not upregulated during the sexual reproduction (Figure 5A) and its knockdown does not affect Paramecium development (Lepère et al., 2009). Dcl4 is an ohnolog (paralog from the most recent whole genome duplication) of Dcl3, therefore their amino acid sequences are very similar (72% similar, 51% identical). For comparison, Dcl3 is only 46% similar and 27% identical to Dcl2. The ohnolog of Dcl2, Dcl1, carries mutations in the catalytic residues of the second RNaseIII domain thus is supposed to be catalytically inactive (Lepère et al., 2009). If Dcl4 and Dcl3 differ in cleavage preference, one could try to identify regions of the proteins responsible for the differences. We therefore cloned the coding sequence of FlagHA-DCL4 fusion under the regulatory regions of PTIWI09 in order to express it at a high level. We then purified the protein from Paramecium cell lysate. Dcl4 cleavage preference was tested using the random hairpin RNA and analyzed like the other Dcls on the gel and by deep sequencing of the cleavage products. Similar to Dcl3, the gel after the assay shows a smear (Figure 5B). This suggests that Dcl4 does not have a strict size preference for its products. We also performed the assay for Dcl4 with the ND7 dsRNA fragment that produces more abundant products between 25 nt and 30 nt (Figure S3B). The pattern observed on the gel with the ND7 RNA was also confirmed by sequencing (Figure S3C). We also loaded IP beads no template control to assess the background and, like in the case of Dcl3, we detect several bands of different sizes (Figure 5B). The most likely explanation for the presence of the background is, as mentioned earlier, that the cellular and bacterial contamination RNA becomes preferentially 5′-labeled when no input template is present in the sample. The cleavage products for the NNN hairpin template was also sequenced and the size distribution shows that Dcl4 produces small RNAs with a broad size distribution with 27–28 nt ones being most abundant (Figure 5C). Furthermore, we analyzed the sequencing data and the enrichment graph for Dcl4 and a weblogo shows a strong 5′ U preference (Figures 5D and 5F). The preference was confirmed by the enrichment analysis of the 3′ positions that shows an enrichment for 3′ A on the 3rd last position (Figure 5E). This A corresponds to the 5′ U of the passenger strand, showing that the analyzed reads are typical Dicer products. However, the 3′A is also present when only 5′U reads were selected (Figure 5F), suggesting that at least a fraction of the double stranded products have 5′U on both ends, which was also observed for Dcl3. The sequencing analysis of Dcl4 beads only control shows similar results as Dcl3 beads only control (Figures S1A–S1E). Most of the reads from the beads control do not map to the Paramecium genome and have high GC content that suggests a bacterial RNA contamination (Figure S1A). Out of the 25 nt and 27 nt long reads that map to the Paramecium genome, only a tiny fraction could be found among the NNN cleavage product reads (Figures S1B and S1C). The weblogo analysis of the Paramecium genome matching contaminant reads shows that the 25 nt sRNAs are scnRNAs with 5′UNG signature (Figure S1D). Overall, because the background level is very small, we conclude that it does not affect our results. This is supported by the fact that the 5′U preference of Dcl4 remains the same after extracting the beads contamination reads from the NNN cleavage product reads (Figure S1E). Our results show that Dcl4 and Dcl3 proteins, despite being very similar, have different cleavage preferences (5′U and 5′UNG). Differences in the amino acid sequences of those two proteins should be responsible for the different sequence preferences. The alignment of the proteins and the identification of the differences could be used to change amino acids among those two proteins in order to manipulate the proteins so that they would cleave as the other (Figure S3A).
Figure 5.
In Vitro dsRNA Processing by Dcl4
(A) Expression profiles of four Dicer-like enzymes according to microarray data. y axis shows the expression level. x axis shows the developmental stages.
(B) 15% polyacrylamide-urea gel with small RNAs 5′-labeled with 32P. M is the small RNA ladder. No enzyme controls are marked as (−). “Beads only” is a no template RNA control for Dcl4.
(C) Size distribution graphs of Dcl4 cleavage products based on sRNA sequencing.
(D–F) 5′ nucleotide enrichment (D) and 3′ end nucleotide enrichment graph (E) for Dcl4 cleavage products showing relative abundance of the nucleotides. The sequence logos are shown for 25 nt sRNAs (F). A typical Dcl4 double-stranded cleavage product is shown schematically (F).
See also Figures S1 and S3.
Figure S3.
Alignment of Dcl3 and Dcl4 and Results for Dcl4 Processing Assay with ND7, Related to Figure 5
(A) Protein alignment of all Dcls using the EMBOSS Needle Software.
(B) Gel of processing assay of Dcl4 with ND7. M is the marker. (−) is the no enzyme control.
(C) Size distribution graph for deep sequencing of Dcl4 processing assay with ND7.
Discussion
In this study, we demonstrate that Paramecium Dicer-like enzymes acquired unique sequence cleavage preferences, which allow them to produce different classes of sRNAs used in the process of developmental DNA elimination. Dcl2 produces 25 nt sRNAs containing a 5′U/5′AGA signature, whereas Dcl3 cleaves small RNAs of different sizes containing 5′UNG signature. Dcl5 also cleaves different sizes but the products contain 5′UAG signature and a 3′ CUAU/CN signature. In addition, the ohnolog of Dcl3, Dcl4, produces sRNAs with varying sizes with a 5′U (Figure 6A). So far, it has been shown that the structure of the RNA template influences the Dicer cleavage efficiencies and that certain sequences on the template act as anti-determinants for processing (Vermeulen et al., 2005, Calin-Jageman and Nicholson, 2003, Zhang and Nicholson, 1997). Moreover, a class 4 enzyme, the RNase-MiniIII from Bacillus subtilis, has a target consensus sequence of AGGU/ACCU. MiniIII enzymes, however, consist only of one single RNaseIII domain (Głów et al., 2015). Nevertheless, sequence cleavage preference has never been shown so far for any Dicer or Dicer-like enzyme. Our results identify sequence-specific Dicer-like proteins, which produce sRNAs with strong base bias that is independent of selection by Argonaute proteins. This feature makes Paramecium Dicers more similar to restriction enzymes, although their sequence requirements are not as stringent as in the case of bacterial restriction endonucleases cleaving dsDNA. It remains to be determined how the Paramecium Dicer-like enzymes recognize the dsRNA target in a sequence-specific manner. Compared to dsDNA, which most often is found in the more open B-form, dsRNA helix is present in the A-form. The A-form contains a wide and shallow minor groove and a deep, narrow major groove, where the access to the bases is difficult. Therefore, it was shown that most RNA-binding proteins access the dsRNA helix from the minor groove in a shape-specific manner (Masliah et al., 2017, Tian et al., 2004). Because the Paramecium Dicer-like proteins show sequence preference, they might be able to access the major groove so that they better reach base information. For example, it was shown that loops and bulges in the dsRNA helix open the major groove to be more accessible (Tian et al., 2004). Another possibility is that perhaps the proteins are able to interact with the bases in the minor groove to get sequence information. Some proteins containing dsRBD were shown to interact with dsRNA bases via the minor groove, such as the adenosine deaminase ADAR2 (Stefl et al., 2010). More structural data would be needed to identify the mechanism of sequence recognition.
Figure 6.
Paramecium Dicer-like Cleavage Preferences and the Role of sRNA End Sequence Bias during DNA Elimination Process
(A) A Neighbor-Joining phylogenetic tree of Paramecium Dicer-like enzymes and a summary of their sequence and length preferences.
(B) A schematic of iesRNA-guided IES excision. Sequence cleavage preference of Dcl5 leads to the production of iesRNAs, which match preferentially to IES ends, thus enabling their precise excision.
Furthermore, the cleavage of Paramecium Dicer-like proteins seems to be sequence-specific in order to facilitate the precise elimination of germline-specific DNA that ends have a strong sequence bias. This suggests that the Dicer-like proteins evolved together with the excision machinery to be able to efficiently eliminate those DNA elements from the genome. Analysis of developmental-specific sRNAs in Paramecium shows that the sRNAs map preferentially to the very ends of IESs (Sandoval et al., 2014). Dcl proteins have therefore evolved to produce sRNAs that help recognizing the ends of excised DNA. This mechanism will ensure that IES ends are marked precisely by small RNAs and target their elimination in a precise fashion (Figure 6B). It has been previously shown in vivo that although iesRNAs are produced from concatenated IESs, they do not map to the IES-IES junctions. They either delineate short IESs or map to the ends of long IESs (Allen et al., 2017, Sandoval et al., 2014). This is consistent with our finding that Dcl5′s sequence cleavage preference enables it to identify the boundaries of excised DNA. This allows to optimize the mechanism of RNA-guided DNA elimination by only producing sRNAs that are useful. More importantly, it ensures production of small RNAs marking the DNA ends very precisely, which may be essential, because most IESs are located within coding regions of the genome. For instance, presence of iesRNAs matching IES-IES junctions would possibly lead to imprecise DNA elimination if, by chance, a part if the sRNA matched the adjacent macronuclear sequences. Imprecise DNA elimination would certainly lead to ORF frameshifts and no viable progeny.
Our results for Dcl2 and Dcl3 confirm that both proteins have to cooperate to form mature scnRNAs in vivo, whereby Dcl2 is responsible for sRNA length and Dcl3 is responsible for 5′ sequence bias. It should be noted that, in this study, Dcl3 in vitro assay was performed using a FLAG-HA-tagged protein due to insolubility of the protein in insect cell extracts. It is, however, highly unlikely that the tiny FLAG-HA tag would confer sequence cleavage preference, especially because the untagged Dcl2 and Dcl5 do show a preference without the presence of the tag. The 5′UNG preference in vitro is the same as the sRNA base bias previously observed in vivo in DCL2 knockdown cells (Sandoval et al., 2014). In addition, we have performed a control experiment where we silenced DCL2 and DCL3 together (because only the double silencing gives a lethal phenotype) in cells expressing synthetic FLAG-HA-tagged Dcl2 and Dcl3. The presence of the tagged proteins was able to rescue the lethal phenotype, suggesting that the tag does not interfere with the protein function (data not shown).
The domain architecture also supports these findings, because neither Dcl2 nor Dcl3 contain a dsRNA-binding domain (dsRBD), similarly to the Giardia intestinalis Dicer (Figure 1A). Crystal structure of the Giardia Dicer showed that it is an elongated, flat enzyme that binds dsRNA on its flat surface (Macrae et al., 2006). This structure allows the binding of another Dicer protein to the opposite side of the dsRNA. In the case of Dcl2 and Dcl3, this is a potential way for the two enzymes to accommodate the same dsRNA template and cleave it to form mature scnRNAs. In vivo, scnRNAs produced by Dcl2/3 have a strong 5′UNG bias, which, like in the case of iesRNAs produced by Dcl5, makes them preferentially matching the ends of IESs. Because scnRNAs initiate IES excision, a lack of precision would have a negative consequence for the following steps that are IES concatenation and production of iesRNAs. In addition, the preference of Piwi proteins toward 5′U strengthens the sRNA sequence bias because the Dcl cleavage preference is not absolute. The effect of this 5′U preference is especially evident in the case of Dcl2 cleavage products that carry 5′U on one strand and 5′AGA on the opposite strand of sRNA duplexed (Figure 2C). In vivo, however, the 5′AGA strands are undetectable (Sandoval et al., 2014). Although our results support the cooperation of the two enzymes, it is not clear how these two proteins interact in detail to produce mature scnRNAs. It is also unclear how Dcl3, Dcl4, and Dcl5 are so flexible in terms of the sizes of their cleaved products. For instance, we show that Dcl5 is able to produce 27 nt or 35 nt sRNAs with similar efficiency (Figure 3). Structural studies will be needed to identify the domains and amino acids responsible for the cleavage preferences and to understand the mechanism of action of these Dicer-like proteins in Paramecium.
The ability of Dicer-like proteins to cleave at specific sequences within a long dsRNA template provides an interesting insight into the diversity of mechanisms involved in RNA processing and could provide an opportunity for future protein engineering to produce enzymes with different sequence and size preferences.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-HA (Y-11), rabbit polyclonal | Santa Cruz | Cat#sc-805; RRID: AB_631618 |
| Anti HA affinity matrix, Immobilized Anti-HA high-affinity rat monoclonal antibody | Roche | Cat#11 815 016 001 |
| goat anti-rabbit IgG-HRP | Santa Cruz | Cat#sc-2004; RRID: AB_631746 |
| Bacterial and Virus Strains | ||
| Endura competent E.Coli cells | Lucigen | Cat#60242-0 |
| Klebsiella pneumoniae non-virulent strain, food source for Paramecium | Gift from Eric Meyer (ENS,Paris) | N/A |
| Baculovirus | EMBL, Heidelberg | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Wheat Grass Powder | Pines International,Lawrence,KS | N/A |
| B-sitosterol | Calbiochem, Millipore | Cat#567152; CAS: 83-46-5 |
| Ribonucleoside vanadyl complexes | Sigma-Aldrich | Cat#R3380 |
| Complete protease inhibitor cocktail | Roche | Cat#11 836 145 001 |
| 30% acrylamid:bisacrylamid 19:1 | BioRad | Cat#161-0154 |
| Critical Commercial Assays | ||
| T4 Polynucleotide kinase | Thermo Fisher Scientific | Cat#EK0032 |
| MEGAscript T7 Transcription Kit | Thermo Fisher Scientific | Cat#AM1333 |
| GoTaq G2 DNA Polymerase | Promega | Cat#M8296 |
| Wizard SV Gel and PCR Clean-Up System | Promega | Cat#A9281 |
| pGem-T Easy Vector System | Promega | Cat#A3600 |
| Deposited Data | ||
| All sequencing data of in vitro cleavage assays | This paper | Accession number ENA: PRJEB20111 |
| Experimental Models: Cell Lines | ||
| Sf21 cells | EMBL, Heidelberg | N/A |
| Experimental Models: Organisms/Strains | ||
| Paramecium tetraurelia strain 51 | Gift from Eric Meyer (ENS,Paris) | N/A |
| Oligonucleotides | ||
| See Table S1 for complete list | This paper, Microsynth, Switzerland | N/A |
| Recombinant DNA | ||
| DCL2-His | This paper | N/A |
| DCL5-GST | This paper | N/A |
| DCL3-Ptiwi09 regulatory, FlagHA | This paper | N/A |
| Bac-to-Bac HT vector kit | Invitrogen | Cat#10584027 |
| pFastBac M30, GST tagged vector | From EMBL protein facility, Heidelberg | N/A |
| DCL4-Ptiwi09 regulatory, FlagHA | This paper | N/A |
| Software and Algorithms | ||
| Geneious R6.1.6 | Geneious | https://www.geneious.com |
| WebLogo 3 | WebLogo, Crooks et al., 2004 | http://weblogo.threeplusone.com/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mariusz Nowacki (mariusz.nowacki@izb.unibe.ch).
Experimental Model and Subject Details
In all experiments, Paramecium tetraurelia strain 51 was used. The cells were cultured in 1X Wheat Grass Powder (WGP; Pines International, Lawrence, KS). The media was bacterized with Klebsiella pneumoniae and supplemented with 0.8 mg/l B-sitosterol. The cells were cultured at 27°C. Autogamy was induced by starvation.
Method Details
Gene synthesis and cloning into expression vectors
Synthetic, insect optimized DCL2 (Genebank: CR932227) and DCL5 (Genebank: XM_001455443.1) genes (Genscript) were inserted into SacI and XbaI sites of the expression vectors pFastBac-HT (Invitrogen) or pFastBacM30 (EMBL protein facility, Heidelberg) containing either a N-terminal His-tag or GST-tag. DCL3 (Genebank: CR932226) and DCL4 (Genebank: CR932225) genes were tagged with 3xFlag-HA at the N-terminal end and cloned by PCR and ligation into pGem-T Easy Vector (Promega) containing the 5′ and 3′ regulatory sequences of PTIWI09 (Genebank: CR932246) to increase the expression level.
Protein expression and immunoprecipitation
Plasmids containing DCL3 and DCL4 Flag-HA were linearized and injected into the macronucleus of vegetative cells. 1.2x106 of cells (400 mL culture) were harvested at an early developmental stage and cell pellets were frozen in liquid nitrogen. Immunoprecipitation was performed as described in Reuter et al. (2009) and Cora et al., (2014). In detail, the cell pellets were resuspended in 2 mL fresh lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% sodium deoxycholate, 1% Triton X-100, 1X protease inhibitor complete tablet (Roche), and 10% glycerol) and homogenized until complete lysis. The cell lysates were centrifuged at 13000x g for 15 min at 4°C. 1 mL of the supernatant was incubated with 50 μL of Anti-HA affinity resin (Roche) overnight at 4°C while rotating. Beads were washed three times with 1 mL IP buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.01% NP-40, 1 mM MgCl2 1X protease inhibitor and 5% glycerol) before incubation. After overnight incubation, beads were washed five times with 1 mL IP buffer. Washed beads were resuspended in 100 μL IP buffer and aliquots of 20 μL were stored at −20°C until further use.
Recombinant protein expression and purification
Proteins were purified from 1x106 Sf21 cells infected with baculoviruses containing either DCL2-His or DCL5-GST constructs. After 72 h at 27°C cells were collected and lysed. For Dcl2, cells were lysed in running buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl and 20 mM imidazole). Dcl5 was lysed in 1X PBS. Lysis was supplied with DNase, 10 mM MgCl2 and 25X complete protease inhibitor. The cells were sonicated 3x for 30 s with 30 s break and centrifuged for 30 min @ 40000 rpm in Beckmann Ti45 rotor. For the purification either Ni/NTA columns or GST/4b columns were used. Dcl2 was eluted with the elution buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl and 300 mM imidazole). Dcl5 was eluted with 1X PBS supplied with 30 mM GSH. Both proteins were digested with either His3C or GST3C to remove the tag. Dcl2 was dialyzed in 1 L running buffer without imidazole overnight at 4°C. Dcl5 was dialyzed against 1X PBS. For both proteins, a second purification with the same conditions was performed. Dcl2 was dialyzed against the storage buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl and 5% glycerol) and stored at −80°C. Dcl5 was supplied with 5% glycerol and stored at −80°C.
SDS-PAGE analysis of purified proteins
Proteins were analyzed on 8% SDS-polyacrylamid gels and stained either with Coomassie Blue solution or silver staining.
RNA preparation
DsRNA substrates were produced by T7 in vitro transcription with the MEGAscript T7 transcription kit (Thermo Scientific). 200 ng of PCR purified DNA templates were used as input for in vitro transcription. After transcription, RNA was DNase treated and phenol purified. Annealing of two complement RNA strands was performed by mixing equal amounts of sense and anti-sense RNA with 10 mM Tris pH 7.4 and 20 mM NaCl. The reaction was heated to 95°C for 5 min and then slowly cooled down to room temperature. All dsRNA templates have blunt ends and are produced from PCR products of either genomic DNA fragments or DNA oligonucleotides (Microsynth).
For the random NNN hairpin template, 10 μg of the designed oligo was extended to a hairpin structure by GoTaq G2 DNA polymerase (Promega). The oligo was mixed with 10 mM dNTPs, 1X GoTaq G2 reaction buffer and 1.25 U of the polymerase. The reaction was denatured for 2 min at 95°C, followed by the annealing step of the CCC stretch to the GGG stretch for 55 s at 45°C. Extension was performed for 10 min at 72°C. This extended hairpin template was used as input DNA for in vitro transcription.
Sequences of DNA templates used for transcription
For ND7 (Genebank: XM_001449634) template we used coding region 777-1281 as in vitro template. The fragment was amplified by PCR (see Table S1 for primers). The other templates were amplified from oligonucleotides (see Table S1 for sequences) synthesized from Microsynth.
dsRNA cleavage assays
Cleavage assays were done as previously described (Zhang et al., 2002, Han et al., 2004). In detail, Dcl2 cleavage assays were performed in 1X dicing buffer (30 mM Tris pH 7.4, 50 mM NaCl and 3 mM MgCl2) containing 200 ng of Dcl2 and 0.6-3.2 μg of dsRNA in a total volume of 10 μl. Dcl5 reactions were done by mixing 2.7 μg protein with 10 mM MgCl2 and RNA in a final reaction volume of 20 μl. Dcl3 and Dcl4 processing assays were performed in 30 μL reaction volume containing 1X dicing buffer, 20 μL IP beads and dsRNA. The reactions were incubated at 27°C for 1 h. Cleavage products were 5′ end labeled with T4 Polynucleotide kinase (Thermo Scientific) and [γ-32P]ATP by exchange reaction and analyzed on 15% denaturing PAGE gels or sent for small RNA sequencing (Fasteris, Geneva, Switzerland).
Quantification and Statistical Analysis
Analysis of small RNA sequencing
Small RNAs were mapped to the dsRNA templates using Geneious 6.1.6 software. The reads mapping to the input RNA were used for the nucleotide enrichment analysis and the size distribution graphs. For the NNN hairpin RNA input, reads mapping to the T7 promoter sequence and to the GGGGGGGGGGGGTTTTTCCCCCCCCCCCC stretch were extracted. All other reads were coming from the random NNN sequences and were analyzed.
For the analysis of the 5′ and 3′ end nucleotide enrichments, the nucleotide abundance was calculated for each position of all reads for Dcl2 NNN hairpin, Dcl5 with the Concatemer template and Dcl5 with the Concatemer Longer template since all the products had the same size. When the Dicer-like proteins produced products of varying sizes as for Dcl3, Dcl4 and Dcl5 with the NNN hairpin, 3000 reads were extracted randomly per cleavage product size and the nucleotide abundance was analyzed. To obtain the nucleotide enrichment, the calculated nucleotide abundance in the cleavage products were divided by the general nucleotide composition of the input template. Enrichment factor of 1 represents expected frequency based on the nucleotide composition of the input dsRNA. Values above 1 were considered as enrichment. 3000 randomly extracted reads were used for sequence logo analysis using WebLogo3 tool (Crooks et al., 2004). GC content used for the analysis depends on the input dsRNA used for cleavage. For the NNN hairpin RNA, a GC content of 50% was used. For the IES containing templates, a GC content of 32% was used.
Data and Software Availability
The accession number for the sequencing data reported in this paper is European Nucleotide Archive: PRJEB20111. Individual accession numbers are:
ERS1985774 for Dcl4 with ND7, ERS19855784 for Dcl5 with MAC.IESmutTA, ERS19855783 for Dcl5 with MAC.IESmutTG, ERS19855782 for Dcl5 with MAC.IESmutG, ERS19855781 for Dcl5 with MAC.IESmutT, ERS19855780 for Dcl4 beads only, ERS19855779 for Dcl4 with NNN hairpin, ERS19855778 for Dcl5 with NNN hairpin, ERS19855777 for Dcl3 beads only, ERS19855776 for Dcl3 with NNN hairpin, ERS19855775 for Dcl2 with NNN hairpin, ERS1624022 for Dcl2 with ND7, ERS1624024 for Dcl3 with ND7, ERS1624025 for Dcl5 with Concatemer, ERS1624026 for Dcl5 with Concatemer Longer, ERS1624027 for Dcl5 with MAC.IES 1, ERS1624028 for Dcl5 with MAC.IES 2, ERS1624029 for Dcl5 with MAC.IES 3 and ERS1624030 for Dcl5 with MAC.IES 3 mutated.
Acknowledgments
We thank Nasikhat Stahlberger for technical support and the group of Ramesh Pillai for technical help with the immunoprecipitation. We also thank Stefanie Jonas and Sarah Allen for fruitful discussions. This research was supported by grants from the European Research Council (ERC) (260358 “EPIGENOME” and 681178 “G-EDIT”), the Swiss National Science Foundation (31003A_146257 and 31003A_166407), and from the National Center of Competence in Research (NCCR) RNA and Disease.
Author Contributions
C.H. and M.N. designed the experiments. C.H. and M.N. wrote the manuscript. C.H. performed most of the laboratory experiments. I.H. sub-cloned the Dicer-like genes into insect expression vectors. M.N. supervised the project.
Declaration of Interests
The authors declare no competing interests.
Published: March 15, 2018
Footnotes
Supplemental Information includes three figures and one table and can be found with this article online at https://doi.org/10.1016/j.cell.2018.02.029.
Supplemental Information
References
- Allen S.E., Nowacki M. Necessity is the mother of invention: ciliates, transposons, and transgenerational inheritance. Trends Genet. 2017;33:197–207. doi: 10.1016/j.tig.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Allen S.E., Hug I., Pabian S., Rzeszutek I., Hoehener C., Nowacki M. Circular concatemers of ultra-short DNA segments produce regulatory RNAs. Cell. 2017;168:990–999. doi: 10.1016/j.cell.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O., Mathy N., Baudry C., Malinsky S., Aury J.-M., Denby Wilkes C., Garnier O., Labadie K., Lauderdale B.E., Le Mouël A. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin-Jageman I., Nicholson A.W. RNA structure-dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2003;31:2381–2392. doi: 10.1093/nar/gkg329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Cora E., Pandey R.R., Xiol J., Taylor J., Sachidanandam R., Mccarthy A.A., Pillai R.S. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. RNA. 2014;20:773–781. doi: 10.1261/rna.044701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne R.S., Lhuillier-Akakpo M., Duharcourt S. RNA-guided DNA rearrangements in ciliates: is the best genome defence a good offence? Biol. Cell. 2012;104:309–325. doi: 10.1111/boc.201100057. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Głów D., Pianka D., Sulej A.A., Kozłowski L.P., Czarnecka J., Chojnowski G., Skowronek K.J., Bujnicki J.M. Sequence-specific cleavage of dsRNA by Mini-III RNase. Nucleic Acids Res. 2015;43:2864–2873. doi: 10.1093/nar/gkv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Doudna J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lepère G., Nowacki M., Serrano V., Gout J.-F., Guglielmi G., Duharcourt S., Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.-B., Ye K., Patel D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae I.J., Doudna J.A. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Masliah G., Barraud P., Allain F.H.T. RNA recognition by double stranded RNA binding domains : a matter of shape and sequence. Cell Mol. Life Sci. 2017;70:1875–1895. doi: 10.1007/s00018-012-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L., Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Provost P., Silverstein R.A., Dishart D., Walfridsson J., Djupedal I., Kniola B., Wright A., Samuelsson B., Rådmark O., Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc. Natl. Acad. Sci. USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Chuma S., Tanaka T., Franz T., Stark A., Pillai R.S. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Sandoval P.Y., Swart E.C., Arambasic M., Nowacki M. Functional diversification of Dicer-like proteins and small RNAs required for genome sculpting. Dev. Cell. 2014;28:174–188. doi: 10.1016/j.devcel.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Skouri F., Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B., Allain F.H. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart E.C., Wilkes C.D., Sandoval P.Y., Arambasic M., Sperling L., Nowacki M. Genome-wide analysis of genetic and epigenetic control of programmed DNA deletion. Nucleic Acids Res. 2014;42:8970–8983. doi: 10.1093/nar/gku619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Bevilacqua P.C., Diegelman-Parente A., Mathews M.B. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- Vermeulen A., Behlen L., Reynolds A., Wolfson A., Marshall W.S., Karpilow J., Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Nicholson A.W. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. USA. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.