ABSTRACT
Commensal and beneficial microbes secrete myriad products which target the mammalian host and other microbes. These secreted substances aid in bacterial niche development, and select compounds beneficially modulate the host and promote health. Microbes produce unique compounds which can serve as signaling factors to the host, such as biogenic amine neuromodulators, or quorum-sensing molecules to facilitate inter-bacterial communication. Bacterial metabolites can also participate in functional enhancement of host metabolic capabilities, immunoregulation, and improvement of intestinal barrier function. Secreted products such as lactic acid, hydrogen peroxide, bacteriocins, and bacteriocin-like substances can also target the microbiome. Microbes differ greatly in their metabolic potential and subsequent host effects. As a result, knowledge about microbial metabolites will facilitate selection of next-generation probiotics and therapeutic compounds derived from the mammalian microbiome. In this article we describe prominent examples of microbial metabolites and their effects on microbial communities and the mammalian host.
BACKGROUND
The gastrointestinal tract (GIT) is a diverse and complex ecosystem shaped by continual interactions between host cells, nutrients, and the gut microbiota. The gut microbiome is estimated to contain approximately 1013 bacterial cells and is dominated by the major phyla Firmicutes, Bacteriodetes, Actinobacteria, Proteobacteria, and Verrucomicrobia (1, 2). Early colonizers of the GIT include bifidobacteria from the phylum Actinobacteria. These commensal microbes colonize immediately after birth and are speculated to prime the GIT and influence the gut-brain axis (3–5). The infant microbiota is considered to be relatively unstable. Despite dramatic changes in the microbiome structure during early life, the gut microbiota increases in diversity and stability over the first 3 years of life (6). Following this initial establishment, the microbiomes of children are generally enriched in Bifidobacterium spp., Faecalibacterium spp., and Lachnospiraceae compared to adults (7–9). During adulthood, the gut microbiome is considered to be stable and is dominated by the phyla Firmicutes and Bacteriodetes. While bacterial populations vary between individuals, the fecal microbiota of adults is highly stable through time (6). This stability is maintained until older age (>65), when the microbiome stability and function begin to decline (10, 11).
A mutualistic relationship exists between the gut microbiota and the host. Commensal microbes metabolize indigestible food components, produce vitamins, prime the immune system, modulate the enteric nervous system and potentially the central nervous system, contribute to intestinal architecture development, protect against colonization of opportunistic pathogens, and do more for the benefit of the host (12). Conversely, the host generates a stable ecosystem for the gut microbiota, providing nutrients and ecological niches. The gut selects resident commensal microbes based on their capacity to adapt to and colonize within the host.
Select commensal bacteria are able to positively alter the gut microbiome and/or host. Many of these commensal groups harbor strains that are considered to be probiotics. Probiotics are defined as microbes which confer advantages to the host and have not been implicated in conferring human disease (World Health Organization, http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf), and these well-characterized organisms are known to produce a number of beneficial products which promote host health. Certain strains of Lactobacillus, Bifidobacterium, Escherichia coli, Bacillus, and Propionibacterium spp. have been classically used as probiotics. The value of probiotics has been supported by multiple clinical trials, which demonstrate improvements for patients receiving probiotics in response to multiple pathologies including diarrhea, inflammatory bowel disease, allergic reactions, viral infections, cancer, and others. However, a number of commensal bacteria that are not defined as probiotics are also known to produce bioactive metabolites with health-promoting effects. These commensals include Ruminococcus, Eubacterium, Roseburia, Faecalibacterium, and Akkermansia spp. This review seeks to identify major pathways used by commensal bacteria to beneficially modulate the host, with the caveat that the majority of studies that have identified specific mechanisms tend to focus on probiotic strains.
Commensal and beneficial microbes have been hypothesized to promote health in a variety of ways: (i) exclusion of pathogens, (ii) immunomodulation, and (iii) enhancement of the intestinal barrier (Fig. 1). Several mechanisms have been proposed to explain the beneficial effects of key commensal microbes. One mechanism of great interest is the secretion of molecules which are capable of altering both the host and the microbiome (Fig. 2). Bacterial metabolites can be generated as intermediate or end products of bacterial metabolism. Secreted products can target the microbiome by acting as signaling molecules for intraspecies/strain communication (quorum sensing), for altering the intestinal environment, and for targeting certain microbes to control microbiota composition (antimicrobials). These molecules include lactic acid, hydrogen peroxide, and bacteriocins (Fig. 2). Likewise, bacterial molecules can modify the host. Metabolites can participate in functional complementation to the host metabolic capabilities, immune regulation, and improvement of intestinal barrier function. These factors include small molecule metabolites such as histamine, vitamins, short-chain fatty acids (SCFAs), polyunsaturated fatty acids, serpins, lactocepins, and secreted proteins (Fig. 2). Of note, the metabolic potential of microbes varies greatly among species and even among strains of the same species. Several studies have found that microbe-driven protective effects depend on the strains used (13–15). These differences could be due in part to the diversity of secreted metabolites. As a result, it is important to characterize the secreted products of individual microbes to identify which strains should be used for a specific disorder. Here we describe the most prominent examples of well-characterized secreted products and their documented effects on the microbiome and/or host function.
FIGURE 1.
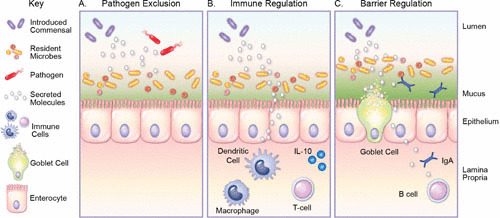
Methods utilized by commensal bacteria to beneficially modulate the intestinal environment. (A) Commensal bacteria secrete molecules which can alter the gut microbiota. By selectively inhibiting resident microbes, commensal bacteria establish an intestinal bacterial niche. Production of antimicrobial factors has also been shown to exclude pathogens. (B) Select commensal bacteria also secrete compounds which can modulate immune cells such as macrophages, dendritic cells, and lymphocytes such as T cells. These compounds decrease intestinal inflammation by dampening proinflammatory cytokines and promoting anti-inflammatory factors such as IL-10. (C) Commensal bacteria can secrete factors which modulate the functions of the epithelial barrier by enhancing the secretion of the protective mucus layer, upregulating tight junctions, and promoting secretion of molecules such as IgA.
FIGURE 2.
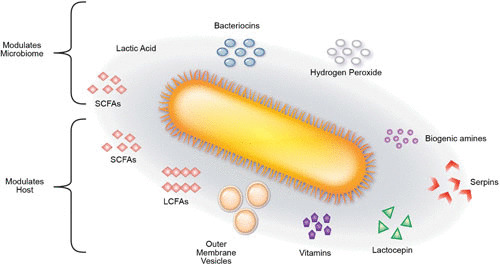
A depiction of secreted metabolites from commensal bacteria and their interactions with the microbiome or host. Lactic acid, hydrogen peroxide, short-chain fatty acids (SCFAs), and bacteriocins are all capable of serving as quorum-sensing molecules and/or directly modulating the composition of the microbiome. SCFAs, long-chain fatty acids (LCFAs), outer membrane vesicles, vitamins, lactocepins, serpins, and biogenic amines have all been demonstrated to beneficially modulate the host. Together, these bacterial products shape the intestinal environment and the host.
SIGNALING COMPOUNDS
Bacteria-Host Signaling Compounds
Biogenic amine neuromodulators
Select microbes are known to produce biologically active compounds that are associated with mammalian neurotransmission and behave as neuroactive compounds. These molecules include histamine, gamma-aminobutryic acid (GABA), and tryptophan metabolites. Microbe-generated neuromodulators in the intestinal lumen likely regulate signaling within the enteric nervous system and ultimately affect the gut-brain axis. Although bacterial products such as acetate (16) and peptidoglycan (17) have been found in the central nervous system, these neuroactive compounds primarily affect the enteric part of the peripheral nervous system and are thought to act in a local manner.
Biogenic amines (BAs) are low-molecular-weight organic bases generated through decarboxylation of specific free amino acids, reductive amination of aldehydes and ketones, transamination, or hydrolytic degradation of nitrogen compounds. BAs are ubiquitously present in both pro- and eukaryotes. In bacteria, the most common mechanism of BA synthesis is the microbial decarboxylation of amino acids catalyzed by amino acid decarboxylases (18). Certain bacteria use BA production to harness the proton gradient and generate energy and/or increase the cytoplasmic pH, thereby protecting cells against acid damage (19). BAs are separated into classes based on the number of amino groups present in the structure: monoamines, diamines, and polyamines (PAs) (18). BAs are known to be key regulators of host health, particularly when acting as hormones or neurotransmitters. BAs can be classified according to their chemical structure as aliphatic (putrescine, cadaverine, spermine, spermidine), aromatic (tyramine, β-phenylethylamine), or heterocyclic (histamine, tryptamine) (20, 21). Several BAs have been associated with promoting human health.
Histamine
Specific gut microbiota members have been reported to produce the BA histamine. Histamine is produced by decarboxylation of dietary l-histidine. Amino acid decarboxylation and BA synthesis maintain bacterial intracellular pH homeostasis (22) and can be used to generate energy using proton motive force (23). As a result, bacterial amino acid decarboxylase expression and activity are enhanced in acidic environments, leading to a locally increased pH and pH counterregulation. Bacterial amino acid decarboxylase expression is also regulated by fermentable carbohydrates, sodium chloride concentration, and oxygen saturation. Select Gram-negative and Gram-positive organisms generate histamine from histidine (24–28). The majority of histamine-producing strains belong to species of the genera Oenococcus, Lactobacillus, and Pediococcus (29, 30). These species harbor the gene encoding histidine decarboxylase (hdcA), which converts dietary histidine into histamine. Histamine is known to exert proinflammatory and anti-inflammatory effects on immunoregulatory processes (Fig. 3). The type of response is dependent on the type of histamine receptor (of four known histamine receptors) that is activated. Activation of histamine receptor type 1 (H1R) or 3 (H3R) has been associated with proinflammatory effects; in contrast, activation of H2R or H4R is associated with anti-inflammatory responses.
FIGURE 3.
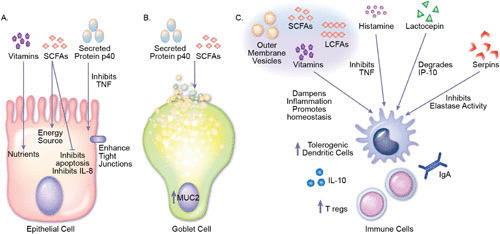
Mechanisms by which commensal secreted products beneficially modulate the host. (A) Epithelial cells. Vitamins produced by bacteria provide essential nutrients to the host. Likewise, short-chain fatty acids (SCFAs) such as butyrate are known to serve as energy sources for intestinal epithelial cells. The SCFA acetate has also been shown to inhibit IL-8 production and increase tubulin-α acetylation. Lactobacilli-produced p40 and p75 inhibit TNF-induced apoptosis and enhance tight junctions, which attenuates intestinal barrier disruption. (B) Goblet cells. p40 is known to transactivate the epidermal growth factor receptor, activating the downstream target Akt and stimulating Muc2 gene expression and mucin production. Acetate produced by bacteria has also been shown to increase goblet cell differentiation and expression of mucus-related genes. (C) Immune cells. Vitamins, outer membrane vesicles (OMVs), SCFAs, and long-chain fatty acids (LCFAs) are known to directly influence the development and function of immune cells. In general, these molecules modulate T cell and dendritic cell homeostasis and cytokine production, promoting production of anti-inflammatory IL-10 and inhibiting proinflammatory cytokines such as TNF. Biogenic amines such as histamine have also been shown to suppress proinflammatory cytokines such as TNF in immune cells, thereby ameliorating intestinal inflammation. Bacterial enzymes such as lactocepin selectively degrade lymphocyte-recruiting chemokine IP-10 and other proinflammatory chemokines such as I-TAC and eotaxin. The protease inhibitor serpin has been shown to suppress inflammatory responses by binding and inactivating neutrophil elastase. Using the highlighted mechanism, commensal bacteria produce signals that reduce intestinal inflammation and promote health.
While a number of species are capable of producing histamine, luminal histamine generated by Lactobacillus species has been documented to have beneficial anti-inflammatory properties. For example, several human-derived strains of L. reuteri contain the histidine decarboxylase gene cluster (hdcA, hdcB, hdcP, HisS), which is required for histamine production. L. reuteri ATTC PTA 6475-generated histamine suppressed the proinflammatory cytokine tumor necrosis factor (TNF) in Toll-like receptor 2 (TLR2)-activated human monocytoid cells (27). This suppression was driven by activation of the anti-inflammatory H2R and downregulation of mitogen-activated protein kinase activation. In vivo, histamine-producing L. reuteri strains ameliorate inflammation in vivo in a trinitrobenzene sulfonic acid-induced mouse model of acute colitis (26, 31). Likewise, histamine from Lactobacillus saerimneri strain 30a (ATCC 33222) significantly lowered NF-κB activation in human monocytoid cells and suppressed interleukin 17 (IL-17) and interferon-γ secretion in wild-type mice but not in H2R-deficient animals (32). Histamine has also been demonstrated to alter dendritic cell (DC) responses to microbial ligands (33). Histamine was found to suppress lipopolysaccharide (LPS; TLR-4 ligand)-driven proinflammatory cytokine secretion (TNF, IL-12, CXCL10) and Pam3Cys (TLR-2 ligand)-driven TNF production. Moreover, histamine increases production of the anti-inflammatory cytokine IL-10. These responses were driven by H2R signaling through cyclic AMP. In vivo addition of histamine-secreting Lactobacillus rhamnosus suppressed cytokine secretion (IL-2, IL-4, IL-5, IL-12, TNF-α, and granulocyte-macrophage colony-stimulating factor) secretion from Peyer’s patches in wild-type but not in H2R-deficient mice (33).These studies indicate that bacterial histamine exerts immunoregulatory effects in vitro and in vivo.
Amino acid neurotransmitters
GABA
GABA is a four-carbon amino acid which functions as an inhibitory neurotransmitter in the mammalian nervous system and mediates diverse functions with the host. GABA can be produced by select bacterial species from the decarboxylation of glutamic acid via glutamate decarboxylase (34). GABA production is highest in microbes when the environment is acidic. Decarboxylation of glutamate consumes a proton and results in the stoichiometric release of GABA. In this manner, GABA production increases the pH of the bacteria cytosol and allows cells to resist acid stress (34). Several microbes have been demonstrated to produce GABA (35–38). Lactic acid bacteria (LAB) strains are considered to be the major group of microbes responsible for GABA production. Some examples of known GABA-producing LAB include Lactobacillus paracasei NFRI 7415, Lactobacillus plantarum C48, L. paracasei PF6, Lactobacillus brevis PM17, Lactobacillus delbrueckii subsp. bulgaricus PR1, Lactococcus lactis PU1, and Bifidobacterium dentium, to name a few (39–41). GABA is known to elicit a number of beneficial effects on the host. As an inhibitory neurotransmitter, GABA lowers the blood pressure in vivo in animal models and human subjects (42–44). GABA is also known to have diuretic and antidiabetic effects (45–47). In the brain, GABA enhances plasma concentration, growth hormones, and protein synthesis (48). GABA intake can regulate sensations of pain and anxiety. Mice fed L. rhamnosus JB-1 were shown to have region-dependent alterations in GABA receptor mRNA in the brain when compared with control-fed mice (49). L. rhamnosus JB-1 also reduced stress-induced corticosterone and anxiety- and depression-related behavior in treated mice (49). In human subjects, administration of a 30-day course of Lactobacillus helveticus and Bifidobacterium longum led to decreased anxiety and depression (50). Other groups have found that daily oral administration of fermented products containing microbial GABA was effective in treating the neurological disorders of sleeplessness, depression, and autonomic disorder in female subjects (51). Together, these findings indicate that gut microbes participate in the bidirectional communication of the gut-brain axis via production of neurotransmitters such as GABA. Modulation of the gut-brain axis may prove to be beneficial for stress-related disorders such as anxiety and depression.
Polyamines
PAs are small aliphatic hydrocarbon molecules with two or more amino groups (-NH2) that have a net positive charge at physiological pH (52). PAs and inorganic cations such as magnesium and calcium play an essential role in maintaining optimal conformation of negatively charged nucleic acids. Moreover, PAs are essential factors for normal cell growth, cell differentiation, and the synthesis of DNA, RNA, and proteins (53). The majority of colonic PAs are derived from commensal gut microbiota (54–56). The main PAs are spermidine, putrescine, spermine, and cadaverine. These compounds are critical for the growth and multiplication of both prokaryotic and eukaryotic cells (52, 57). In the majority of bacteria, intracellular concentrations of spermidine (1 to 3 mM) are higher than putrescine (0.1 to 0.2 mM), with the known exception of E. coli, which has higher putrescine (10 to 30 mM) levels (57). Cadaverine is considered to be the least prevalent bacterial PA (52, 57). In contrast, spermine is primarily produced in the presence of specific dietary sources. Consumption of pectin has been shown to increase the cecal concentrations of multiple PAs, particularly spermine (56). This effect was shown to be gut-microbe dependent because consumption of pectin- or guar-containing diets altered the concentrations and the composition of cecal PAs in conventional rats but not in germ-free rats (56). The gut residents Bacteroides thetaiotaomicron and Fusobacterium varium are major contributors of spermine and spermidine in the gnotobiotic rat cecum, suggesting that commensal bacteria can modulate PA levels (55).
Commensal bacteria can both produce and be influenced by PAs. PAs are known to possess anti-inflammatory activities, which can benefit the host. They can decrease systemic inflammation by inhibiting proinflammatory cytokine production in macrophages and intestinal epithelial cells (58, 59). Additionally, PAs function as reactive oxygen species scavengers, chemical chaperones, positive regulators of stress genes, and antimutagenic agents (60, 61). Spermine possesses anti-inflammatory activity by inhibiting NF-κB activation (62) and inflammatory cytokine synthesis (58, 63) and by selectively activating T cell protein-tyrosine phosphatase (64). In mammals, systemic PA levels decrease with age (65, 66), and decreased PAs have been associated with intestinal barrier dysfunction (67). Supplementation of middle-aged (10-month-old) mice with Bifidobacterium animalis subsp. lactis LKM512 for 6 months increased fecal spermine concentrations (67). This spermine increase correlated with increased survival, reduced skin ulcers and tumors, improved colonic barrier function, downregulation of inflammation-associated genes, and alteration of the gut microbiota composition (67). In addition, supplementation of arginine to the diets of mice and rats increased colonic concentrations of spermine and putrescine (68). A combination of both arginine and B. animalis subsp. lactis LKM512 further suppressed inflammation, improved longevity, and provided protection from age-induced memory impairment in mice (68). Several of these findings have been mirrored in patients, because addition of B. animalis subsp. lactis LKM512 increased intestinal PAs (putrescine, spermidine, spermine, and cadaverine) (69, 70) and reduced the quantities of biomarkers of acute inflammation in hospitalized elderly patients (70).
Commensal bacteria also benefit from PA production because PAs can protect bacterial cells from reactive oxygen species (71) and mutagens (61, 72–75). PAs can also modulate gut community dynamics. Supplementation of PAs to a formula diet for neonatal BALB/c mice was found to regulate the concentrations of Akkermansia muciniphila, Lactobacillus, Bifidobacterium, Bacteroides-Prevotella, and Clostridium groups to levels found in the breast-fed group (76). Although PAs can be used by commensal bacteria, they can also be used by pathogens. Pathogens use PAs to promote toxin activity, bacteriocin production, biofilm formation, microbial carcinogenesis, and protection from oxidative and acid stress (77–85). While PA production and utilization are not unique to commensals, the highlighted studies provide evidence that PA synthesis by commensal bacteria may modulate the host immune response and improve patient well-being.
Tryptophan and indole
The essential amino acid l-tryptophan is used by the host to synthesize proteins and specialized molecules including the hormone and neurotransmitter serotonin (86). In addition to its use by the host, pathogens such as enteropathogenic E. coli and enterohemorrhagic E. coli can also utilize tryptophan as the sole source of carbon and nitrogen (87). As a result, conversion of tryptophan to its metabolites by commensal bacteria may effectively remove tryptophan from the amino acid pool available to pathogens. Luminal tryptophan can undergo bacterial degradation generating indole, indican, and indole acid derivatives (indolyl-3-acetic acid, indolyl-acetyl-glutamine, indolyl-propionic acid, indolyl-lactic acid, indolyl-acrylic acid, and indolyl-acryloyl-glycine). Indole is the main bacterial by-product of tryptophan. Several microbes are capable of producing indole and indole derivatives, including Bacteroides spp., Bifidobacterium spp., Clostridium spp., E. coli, Proteus vulgaris, Paracolobactrum coliforme, and Achromobacter liquefaciens (86, 88, 89). Bacterial tryptophanase converts tryptophan into indole, ammonia, and pyruvate. In many microbes, tryptophanase activity has been shown to be induced by tryptophan and repressible by glucose (89). Protein-rich diets have also been shown to induce bacterial tryptophanase activity (89).
Indole metabolites are noteworthy because they have been shown to provide several benefits to the host. Indole compounds secreted by commensal E. coli reduce attachment of pathogenic E. coli to epithelial cells (90). Of note, indole-3-carboxaldehyde downregulated production of pathogenic E. coli enterocyte effacement virulence locus and inhibited pedestal formation on mammalian cells (91). Oral administration of indole-3-carboxaldehyde was found to inhibit virulence and promote survival in a lethal mouse infection model of Citrobacter rodentium (91). Select indole derivatives also yielded bacteriostatic effects on Gram-negative enterobacteria, particularly pathogens such as Salmonella and Shigella (89).
In addition to the bacteriocidal effects of indole metabolites, indole itself may enhance the mucosal barrier by inducing tight junction-mediated trans-epithelial resistance and mucin production and by diminishing TNF-α-mediated activation of NF-κB (90). In vivo, oral administration of indole-containing capsules to germ-free mice resulted in increased quantities of tight junction- and adherens junction-associated proteins in colonic epithelial cells (92). Furthermore, germ-free mice supplemented with indole-containing capsules yielded greater resistance to dextran sodium sulfate-induced colitis (92). Indole compounds secreted by gut bacteria may be an important signal for the maintenance of intestinal epithelial homeostasis. Similar to other compounds produced by commensal microbes, indole is also utilized by several pathogens. Indole is known to regulate biofilm formation, virulence, and production of Shiga toxins in pathogenic E. coli strains, as well as virulence in the rodent attaching and effacing (A/E) pathogen C. rodentium, and Pseudomonas and Salmonella strains (91). However, since select indole metabolites synthesized by commensals inhibit these same species, future work may focus on selecting key commensals to combat pathogens via indole metabolism.
Bacteria-Bacteria Communication
Quorum-sensing molecules
In addition to the many routes of bacterial-host communication, bacteria themselves must communicate with each other to regulate cooperative activities (93). A number of bacteria release, sense, and respond to small diffusible signal molecules, known as quorum-sensing molecules, as a method of intra- and interspecies bacterial communication. In this manner, bacteria can behave as a collective unit (94–96). Quorum sensing regulates bacterial symbiosis, formation of spore or fruiting bodies, bacteriocin production, genetic competence, programmed cell death, virulence, and biofilm formation (93, 96–99). This behavior is thought to offer significant benefits to bacteria in terms of biofilm community structure, defense against competitors, adaptation to environmental changes, and overall host colonization (93, 96, 99, 100). In general, quorum sensing relies on diffusible signaling molecules and sensors or transcriptional activators which work in concert to promote gene expression (93, 96, 99, 101, 102). Quorum sensing can be divided into three classes: (i) LuxI/LuxR-type quorum sensing in Gram-negative bacteria, (ii) oligopeptide-two-component-type quorum sensing in Gram-positive bacteria, and (iii) luxS-encoded autoinducer 2 (AI-2) quorum sensing in both Gram-negative and Gram-positive bacteria (93).
In LuxI/LuxR-type quorum sensing, acyl-homoserine lactones (AHL) act as the signaling molecules (103, 104). In this system, AHL synthesis is dependent on a LuxI-like protein and AHLs increase in concentration in proportion to cell density. AHLs can freely diffuse across bacterial cell membranes where they are recognized and bound by the cognate LuxR-like protein, which subsequently binds specific promoter DNA elements and activates target genes. Hundreds of Gram-negative bacteria use the LuxI/LuxR-type quorum-sensing system, with each species producing a unique AHL. As a result, only similar species are able to recognize and respond to signals from their own kind (96, 99, 103, 105).
In contrast to the LuxI/LuxR system, the oligopeptide-two-component-type quorum-sensing system comprises three components: a signaling molecule and a two-component signal transduction system. The most common oligopeptide-two-component-type quorum-sensing system involves the autoinducer signaling peptide (AIP) and a corresponding two-component signal transduction system that specifically detects and responds to AIP (96, 102, 106–108). Unlike diffusible AHL signals, AIP must be transported by a dedicated oligopeptide transporter, typically an ABC transporter, to exit the cell (106–108). The AIP signal is then sensed by the two-component signal transduction system, which contains a membrane-associated, histidine kinase protein and a cytoplasmic response regulator protein, which translates the signal via regulation of target gene expression (106–108). Another type of quorum sensing identified in Gram-positive streptococci is the ComRS system, which involves a small double-tryptophan signal peptide pheromone, XIP. Similar to AIP, XIP is transported inside the cell via an oligopeptide ABC transport system (Opp/Ami). Inside the cell, XIP interacts with a transcriptional regulator, ComR or ComX, thereby activating competence genes for genetic transformation (109–111). Additionally, Streptococcus mutans contains ComCDE and ComRS quorum-sensing systems which regulate bacteriocin production (109).
Both Gram-negative and Gram-positive bacteria can utilize the AI-2 quorum-sensing system (93, 96, 99). In contrast to the LuxI/LuxR-type and oligopeptide-two-component-type quorum-sensing systems, which provide for intraspecies signaling, AI-2 quorum sensing allows for interspecies communication. As a result, AI-2 has been termed the “universal language” (101, 102). AI-2, a furanosyl borate diester, is produced and recognized by many Gram-negative and Gram-positive bacteria (96). AI-2 synthesis depends on a luxS encoded synthase: a metabolic enzyme which converts ribosyl-homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione (112). AI-2 is transported inside the bacteria by the Lsr ABC-type transporter in Enterobacteriaceae, Pasteurella, Photorhabdus, Haemophilus, and Bacillus (113). AI-2 is then phosphorylated by LsrK and subsequently binds the transcriptional repressor protein, LsrR. Binding of phosphor-AI-2 to LsrR releases the promoter/operator region of the lsr operon, thereby promoting transcription lsr genes.
Since AI-2 is produced and detected by a number of diverse bacteria, it has been speculated that AI-2 facilitates interspecies communication and social interactions. This compound has been postulated to be particularly relevant in the setting of biofilms. Biofilms are considered to be structured microbial communities organized within an extracellular matrix which harness symbiotic interactions for mutual benefit of the collective (94, 95). Microbes within biofilms have diverse characteristics compared with their free-living counterparts, including enhanced resistance to antibiotics and host immune responses (93, 95, 114). Biofilms are important for both virulent pathogens and complex commensal communities within the GIT (115). For commensal microbes within the GIT, colonization of the intestinal mucus layer is a critical step in establishing ecological niches. The intestinal epithelium is covered by a continuous layer of mucus secreted by goblet cells, chiefly composed of the mucin protein MUC2 (116, 117). Mucus provides a viscoeleastic gel barrier that prevents luminal contents and microbiota from interacting directly with the epithelium and immune cells of the lamina propria. The mucin proteins are heavily O-glycosylated, and these glycans can serve as adhesion sites for commensal bacteria (118–123). Several studies have shown that intestinal communities can be visualized as biofilms of sessile microorganisms within the mucus layer (116, 124–126). High bacterial cell density within biofilms favors a mode of communication using diffusible small molecules as a mechanism for regulating biofilm formation and for social interactions (97, 127). Several studies have shown that AI-2 signaling is important for the proper development of multispecies biofilms in natural ecosystems (128–130). This signaling pathway has been particularly well documented for the oral cavity. In mixed cultures of Actinomyces naeslundii T14V and Streptococcus oralis 34, AI-2 is essential for interdigitated biofilm growth where saliva is the primary nutrient source (129). Introduction of an S. oralis 34 luxS mutant with A. naeslundii T14V diminished mutualistic growth—a defect that could be rescued with the LuxS enzyme product 4,5-dihydroxy-2,3-pentanedione. Other groups have shown luxS-dependent biofilm formation in Streptococcus pneumoniae (131), clinical isolates of S. pneumoniae D39 (132), and L. reuteri (133).
In the intestine, AI-2 has also been shown to play a key role in community structure and colonization. Thompson and colleagues found that AI-2 can modulate the structure of the gut microbiota by using E. coli to manipulate signal levels (98). In this work, AI-2 influenced bacterial behaviors to restore the balance between the major phyla of the gut microbiota, Bacteroidetes and Firmicutes, following antibiotic treatment. Although few in vivo studies have been conducted thus far, existing data point to the critical role of quorum sensing in the establishment of commensal communities.
METABOLITES THAT BENEFIT THE HOST
Short-Chain Fatty Acids (SCFAs)
Several bacterial species in the GIT generate SCFAs as an end product of complex carbohydrate fermentation pathways in the intestine. SCFAs are composed of one to six carbons, with the most abundant and well-characterized SCFAs being acetate, propionate, and butyrate. The composition of SCFAs in the GIT depends on microbial composition and environmental conditions, including pH, hydrogen partial pressure, and host diet (134–137). SCFAs can reach local concentrations of approximately 13 mM in the terminal ileum, 130 mM in the cecum, and 150 mM in the descending colon, making them a class of abundant colonic anions (138, 139). Most SCFAs are absorbed by the host in exchange for bicarbonate. As a result, SCFAs gradually diminish in concentrations from the proximal to the distal colon, and the luminal pH correspondingly increases from cecum to rectum (138, 140, 141). The pH reduction may alter microbial composition of the gut and prevent overgrowth by pH-sensitive pathogenic bacteria such as certain strains of Enterobacteriaceae and Clostridium (142–146). SCFAs are absorbed by intestinal enterocytes via passive diffusion or carrier-mediated transportation through SMCT1 (SLC5a8) and MCT1 (SLC16a1) transporters (147, 148). SMCT1 acts as a sodium-coupled transporter, while MCT1 is a hydrogen-coupled transporter for SCFAs and related organic acids (147–149). These transporters are found on the apical membranes of intestinal enterocytes, dendritic cells, kidney cells, and brain cells.
In addition to intestinal absorption, SCFAs can activate several G-protein-coupled cell surface receptors. SCFAs bind to G protein-coupled receptors such as GPR41, GPR43, and GPR109A, on intestinal epithelial cells and enteroendocrine cells such as colonic L cells (150–154). In addition to intestinal cells, GPR41 is also expressed in enteric neuronal cells, adipocytes, renal smooth muscle cells, and pancreatic cells (155, 156), and GPR43 is expressed by granulocytes and some myeloid cells (157–159). GPR109a is also expressed by macrophages, dendritic cells, and adipocytes. As a result of the wide distribution of receptors, SCFAs have diverse effects on the host (Fig. 3). SCFAs can regulate different aspects of host physiology, including enhanced sodium uptake and subsequent water absorption, pH regulation, chloride and mucus secretion, immune regulation, decreased bioavailability of toxic amines, and remote effects on neural activity and development (160–162).
Acetate
Acetate is the most abundant SCFA and is generated as a fermentation end product produced by enteric bacteria, as well as a product of metabolism of H2, CO2, or formate by acetogenic bacteria (135). B. longum subsp. longum- and B. longum subsp. infantis-secreted products suppressed epithelial cell apoptosis, ameliorated intestinal inflammation, and inhibited translocation of the E. coli O157:H7 Shiga toxin, thereby protecting mice against infection by enterohemorrhagic E. coli O157 bacteria (163). This protection correlated with fecal acetate quantities. Non-acetate-producing Bifidobacteria strains as well as a B. longum mutant with reduced acetate production were unable to mimic the protective effects of the parental strain against E. coli O157. Moreover, administration of acetylated starch, which increased fecal acetate, improved the survival rate. These data indicate that bacteria-produced acetate protects the host against lethal infection.
In addition to epithelial cell effects, acetate also plays an important role in immune regulation. Acetate induces T regulatory cell proliferation and accumulation (160, 164–166) and inhibits T cell histone deacetylase 6, while increasing tubulin-α acetylation (167). Ishiguro and colleagues have demonstrated that in addition to immune cells, acetate inhibits IL-8 production and increases tubulin-α acetylation within intestinal epithelial cells (167). Inoculation of germ-free mice with the acetate producer B. thetaiotaomicron increased goblet cell differentiation and expression of mucus-related genes. These in vivo findings were confirmed in vitro using the mucus-producing cell line HT29-MTX, where acetate upregulated KLF4, a transcription factor involved in goblet cell differentiation (168). Mucus produced by goblet cells creates a protective barrier that prevents the epithelium and immune system from adhesion and invasion by pathogenic bacteria, microbial antigens, and other damaging agents present in the intestinal lumen (116, 127). Mucin glycans are also important sources of carbohydrate for saccharolytic bacteria, and the spatial organization and composition of mucosal communities may be influenced by variations in mucin production and glycan composition (118, 127). As a result, modulation of the mucus layer by microbial compounds such as acetate may serve to maintain a proper distance between the microbiota and host and potentially modulate the microbial community structure.
Acetate also inhibits the growth of several pathogens. Acetate alone inhibits the growth of Pseudomonas aeruginosa (169), while acetate in combination with propionate and butyrate inhibits the growth of pathogenic E. coli O157 (170), Proteus mirabilis, Klebsiella pneumoniae, and P. aeruginosa (169). Acetate and propionate acting via GPR43 participate in anti-inflammatory effects via the modulation of regulatory T cells (Tregs) (164, 171). In vivo, supplementation of acetate in germ-free mice was shown to be sufficient to ameliorate the dextran sodium sulfate-driven intestinal inflammation, an effect that was not observed in Gpr43–/– mice (171). Interestingly, De Vuyst and Leroy demonstrated the importance of acetate as a substrate for the production of butyrate by butyrate-producing bacteria (172), implicating a cross-talk of SCFAs in microbial metabolism.
Propionate
Propionate is primarily formed via the succinate pathway by the phyla Firmicutes (135, 173). Propionate is principally metabolized by the liver, while acetate is metabolized by peripheral tissues. Production of acetate by the commensal Akkermansia muciniphila modulates mouse gene expression, particularly Fiaf, Gpr43, histone deacetylases, and peroxisome proliferator-activated receptor gamma, which are important regulators of transcription factor regulation, cell cycle control, lipolysis, and satiety (174). In vitro propionate inhibits several pathogens including Salmonella enterica serovar Typhimurium (175–178), E. coli O157 (170), P. mirabilis, K. pneumoniae, and P. aeruginosa (169). Propionate and acetate generated by microbial species stimulated the cellular function of immune cells, specifically promoting neutrophil chemotaxis (157, 171, 179, 180). In addition, propionate has been utilized for its ability to release short-term modulators of satiation and satiety, including the anorectic gut hormones peptide YY and glucagon-like peptide-1, from intestinal enteroendocrine L cells (181–185). In humans, colonic delivery of inulin-propionate esters increased plasma peptide YY and glucagon-like peptide-1 and reduced energy intake, resulting in significantly reduced weight gain, intra-abdominal adipose tissue distribution, and intrahepatocellular lipid content after 24 weeks (184). These data suggest that colonic propionate may be an important microbial metabolite in the context of host body metabolism.
Butyrate
Butyrate is generated by microbes of the phylum Firmicutes (including Faecalibacterium prausnitzii, Roseburia spp., Eubacterium rectale, Eubacterium hallii, and Anaerostipes spp.) via the butyryl-CoA:acetate CoA-transferase enzyme or phosphotransbutyrylase and butyrate kinase pathway (135, 186–188). Butyrate is primarily metabolized in the colon as an energy source utilized by intestinal epithelial cells, and this SCFA yields immunoregulatory and cancer-protective effects in the GIT. Propionate and butyrate alter immune cell function. Both propionate and butyrate inhibit stimuli-induced expression of adhesion molecules and chemokine production and suppress monocyte/macrophage and neutrophil recruitment (179). Butyrate signaling via GPR109A regulates the differentiation of regulatory Treg and IL-10-producing T cells (162) and suppresses activation of NF-κB and induction of apoptosis (189). In vivo production of butyrate by clostridial species induces the differentiation of Treg cells and ameliorates development of colitis (160). Apart from the major effects of butyrate on immune cell populations, this SCFA can also serve as an energy source for colonic enterocytes. Furthermore, activation of GPR43 and GPR109A by butyrate and propionate mediates cancer-protective effects associated with high fiber intake (186, 190). Butyrate also participates in antitumorigenic properties by inhibiting proliferation and selectively inducing apoptosis of colorectal cancer cells (191–194). Intracellular butyrate and propionate, but not acetate, inhibit the activity of histone deacetylases in colonocytes and immune cells. Histone deacetylase inhibition promotes the hyperacetylation of histones, effectively downregulating proinflammatory cytokines such as IL-6 and IL-12 in colonic macrophages (191, 195, 196). Together, these studies demonstrate that commensal-produced SCFAs can elicit multiple advantages for the host through the stimulation of intestinal mucus production, inhibition of inflammation, modulation of immune cell populations, and inhibition of cancer proliferation.
Long-Chain Fatty Acids (LCFAs)
While SCFAs have been thoroughly characterized with respect to intestinal biology and human health, LCFAs are gaining attention for their health-promoting activities as well (197). Similar to SCFAs, commensal bacteria are also responsible for the composition and concentration of LCFAs and subsequently contribute to LCFA-induced signaling in host cells. LCFAs are produced when dietary polyunsaturated fatty acids such as linoleic acids are converted into conjugated linoleic acids and trans-fatty acids (198–200). Germ-free mice without a gut microbiota lack detectable LCFAs (201), while inoculation of mice with the commensal Bifidobacterium breve in combination with a linoleic acid-supplemented diet resulted in increased conjugated linoleic acids (202). Production of conjugated linoleic acid by bacteria reduced amounts of hepatic triacylglycerols and inhibited atherosclerosis (203, 204). Although it remains unclear if LCFAs regulate host immune functions, modified polyunsaturated fatty acids are potent agonists for peroxisome proliferator-activated receptor-γ and peroxisome proliferator-activated receptor-α, which are upregulated by commensal bacteria and implicated in attenuating inflammation (197, 205–208). The role of LCFAs in intestinal homeostasis was further confirmed in vivo in ethanol-fed mice. Exposure of mice to alcohol resulted in an altered gut microbiota and reduced synthesis of LCFAs (209). Relative abundances of Lactobacillus spp., known metabolizers of saturated LCFAs, were reduced in the feces of humans with active alcohol abuse (209). The authors hypothesized that targeted approaches to restore LCFA levels might reduce ethanol-induced liver injury and restore an intact community of gut microbiota.
Vitamins
Bacteria residing in mammals are able to produce vitamins which directly benefit the host (210). Metagenomic analyses of the human microbiota from the distal colon revealed the existence of diverse clustered orthologous groups which are involved in vitamin synthesis (211). Vitamins are essential organic micronutrients which are critical for cellular function and may not be synthesized by the host. Vitamins exist as precursors of intracellular coenzymes. The majority of vitamins are produced by bacteria via the 2-methyl-d-erythritol 4-phosphate pathway. Thirteen essential vitamins for human health include the water-soluble vitamins thiamine (B1), riboflavin (B2), niacin (B3), pyridoxine (B6), pantothenic acid (B5), biotin (B7 or H), folate (B11-B9 or M), and cobalamin (B12); vitamin C; and the fat-soluble vitamins A, D, E, and K (212). Vitamins generated by gut microbes are primarily absorbed in the colon, while dietary vitamins are absorbed mostly in the small intestine (213, 214). Data suggest that colonocytes absorb thiamine, folates, biotin, riboflavin, pantothenic acid, and menaquinones (213, 214). Vitamins provide essential nutrients to the host and directly influence the development and function of immune cells (215–218). In this manner, vitamins may promote host growth and immune homeostasis (Fig. 3).
Water-soluble vitamins
Riboflavin (vitamin B2)
Riboflavin, or vitamin B2, is a known component of cellular metabolism. Riboflavin is a precursor of the coenzymes flavin mononucleotide and flavin adenine dinucleotide (219). Flavin mononucleotide and flavin adenine dinucleotide are hydrogen carriers in cellular redox reactions, making B2 critical for host metabolism. B2 can exist in several active forms including riboflavin [7,8-dimethyl-10-(1′-d-ribityl) isoalloxazine], riboflavin-5′-phosphate (flavin mononucleotide), and riboflavin-5′-adenosyldiphosphate (flavin adenine dinucleotide). Riboflavin can be produced by both Gram-positive and Gram-negative bacteria and is well characterized in Bacillus subtilis and E. coli (220, 221). LAB strains including L. plantarum and Lb. lactis have also been identified as B2 producers (219, 222–225). B2 can be produced from the precursor guanosine triphosphate and d-ribulose 5-phosphate via seven enzymatic steps (220). Enhanced production of B2 has been shown in species that are capable of simultaneously expressing four biosynthetic genes (ribG, ribH, rib, and ribA) (223, 224). Deficiency in B2 levels leads to ariboflavinosis, which is associated with hyperemia, edema of oral and mucous membranes, cheilosis, and glossitis (226). In rats, supplementation of a fermented milk drink containing a genetically modified B2-producing Lb. lactis strain was effective in reversing ariboflavinosis in a riboflavin-deficiency model (223). In humans, daily consumption of a probiotic yogurt containing Streptococcus thermophilus, Lactobacillus bulgaricus, and Lactobacillus casei subsp. casei for 2 weeks contributed to the total intake of vitamin B2, as reflected by increased blood concentrations of plasma-free riboflavin in healthy women (227). These effects were ameliorated when subjects returned to their previous diet, implicating select supplemented probiotic bacteria in the enhanced production of vitamin B2 (227).
Folates (B11-B9 or M)
Folates are hydrophilic anionic molecules which are produced by a number of bacteria. The generic term “folate” is typically used to include all bacterially derived folate derivatives. Folates are involved in essential cellular metabolism functions including DNA replication, DNA repair, DNA methylation, and nucleotide synthesis. Folate deficiency has been linked to a large spectrum of disorders including colorectal cancer, osteoporosis, coronary heart disease, and Alzheimer’s disease, among others (228, 229). Although folate is primarily absorbed in the duodenum and jejunum, folate compounds generated by the mammalian microbiome in the colon represent a major source of host folate. Bacteria generate mono- and polyglutamylated folate, a form of folate which is easily absorbed by mammalian cells (230, 231). Multiple LAB strains such as L. reuteri, Lactobacillus acidophilus, L. plantarum, L. bulgaricus, Lactococcus lactis, Bifidobacterium adolescentis, B. animalis, and B. longum synthesize folate (228, 231–237). However, not all LAB strains are capable of generating folate. For example, Lactobacillus gasseri (236), Lactobacillus salivarius (238), and Lactobacillus johnsonii (239) do not contain folate biosynthesis genes and do not produce folate. Bacterial production of folate in the presence of existing folate appears to be species dependent, with select species producing folate solely in low-folate conditions, and others continually producing folate (240). Bacteria produce folate using the precursor 6-hydroxymethyl-7,8-dihydropterin pyrophosphate and para-aminobenzoic acid (pABA, vitamin B10) (25, 241).
All bacteria capable of producing folate contain the folC or homologous genes (231, 235). Additional genes involved in folate production include folKE genes, which encode 6-hydroxymethyl-dihydropterinpyrophosphokinase (folK), and guanosine triphosphate cyclohydrolase (folE) or pABA (242). Folate production relies on the combination of folate and pABA biosynthesis. Deletion of the pABA genes in Lb. lactis and L. reuteri resulted in a loss of folate production and inhibition of growth in the absence of purine nucleobases/nucleosides (242). Genetic manipulation of folate genes increased folate production in a number of species including Lb. lactis, L. gasseri, and L. reuteri (233, 236, 237, 242). In vivo addition of Lb. lactis overexpressing the folC, folKE, or folC-folKE genes improved folate status in a rat folate deficiency model (228). In L. reuteri ATCC PTA 6475, the gene folC2 is required for production of 5,10-methenyltetrahydrofolic acid (5,10-CH=THF) and folC participates in polyglutamylation of 5,10-CH=THF. Mutations in folC2 resulted in loss of 5,10-CH=THF and diminished the strain's ability to suppress TNF production by activated human monocytes (231). Additionally, the L. reuteri folC2 mutant was unable to suppress inflammation to the same degree as wild-type L. reuteri in a trinitrobenzene sulfonic acid-induced mouse model of acute colitis (231). These studies demonstrate that select folate-producing microbes can be utilized to improve folate status and modulate the immune system in animal models.
Vitamin B12
Vitamin B12, also known as cobalamin, exists in its natural form as 5′-deoxyadenosylcobalamin (coenzyme B12), methylcobalamin, or pseudocobalamin, and is a corrin ring or corrinoid compound. B12 is required for the metabolism of nucleic acids, amino acids, and fatty acids (243) and is primarily produced by anaerobic bacteria (244–246). Few bacteria are capable of producing vitamin B12 (247, 248). Typically, specialized bacteria found in food source animals produce vitamin B12. As a result, humans must absorb the coenzyme from animal sources such as meats, fish, and eggs. However, select lactobacilli have been demonstrated to produce vitamin B12. L. reuteri CRL1098 was found to produce a cobalamin-like compound, a form of B12 (249). L. reuteri DSM 20016 (237), JCM1112 (237), and CRL 1324 and 1327 (250) and Lactobacillus coryniformis (251) produced a cobalamin-type compound. These L. reuteri species contain an extensive cobalamin biosynthesis cluster, which is associated with the anaerobic catabolism of glycerol (or 1,2-propanediol) (252, 253). Vitamin B12 deficiency is associated with numerous hematopoietic, neurological, and cardiovascular pathologies. Pernicious anemia, a severe form of B12 deficiency, is the result of poor production of a gastric glycoprotein called intrinsic factor that facilitates the absorption of vitamin B12 in the small intestine (254). In vivo in a mouse model of vitamin B12-deficient animals, supplementation of L. reuteri CRL 1098 reversed vitamin B12 deficiency (255). Gut microbes may be important in maintaining adequate body concentrations of B complex vitamins, including vitamin B12.
Fat-soluble vitamins
Vitamin K
Vitamin K comprises a number of series of fat-soluble compounds which share a 2-methyl-1,4-naphthoquinone nucleus and different side chain structures at the 3-position. Vitamin K can be produced by both plants and microbes. In plants, vitamin K exists as phylloquinone (vitamin K1), which has a phytyl side chain. In contrast, bacteria generate a family of compounds known as menaquinones (vitamin K2). These compounds contain side chains based on repeating unsaturated 5-carbon (prenyl) units and are designated menaquinone-n (MK-n) according to the number (n) of prenyl units. Vitamin K is an essential cofactor in the formation of γ-carboxyglutamic acid residues in proteins which bind calcium ions. As a result, vitamin K serves a prominent role in bone formation, tissue calcification, kidney function, and blood clotting, to name a few functions (256, 257). Vitamin K deficiency has been implicated in osteoporosis-driven bone fracture and intracranial hemorrhage in newborns. The gut microbiota synthesizes large amounts of menaquinone K2, one of the forms of vitamin K (258). Quantitative measurements at different sites of the human intestine have demonstrated that most of these menaquinones are present in the distal colon (258). Specific microbes are capable of generating menaquinone K2, one of the forms of vitamin K. The genera Lactobacillus, Lactococcus, Enterococcus, Leuconostoc, and Streptococcus are known producers of K2 (259, 260). Other major menaquinone forms are produced by Bacteroides (MK-10, MK-11), Enterobacter (MK-8), Veillonella (MK-7), and Eubacterium lentum (MK-6). Vitamin K is predominantly absorbed in the terminal ileum of the intestine, a site where menaquinone-producing bacteria colonize as well. Collectively, the data acquired from all vitamin studies point to the selection of multivitamin-producing bacteria to compensate for common vitamin deficiencies and for promotion of gut homeostasis.
Outer Membrane Vesicles (OMVs)
Several bacteria are capable of releasing OMVs, which range in size from 20 to 300 nm in Gram-negative bacteria (261) to <20 nm in Gram-positive bacteria (262, 263). OMV production is considered to be a common feature of Gram-negative bacteria. They are generated by membrane remodeling which occurs when the outer membrane bulges and encapsulates periplasmic components (264, 265). As a result, OMVs contain a number of soluble proteins entrapped in the OMV periplasm and multiple proteins on the external surface. Secreted OMVs can disseminate compounds to distant sites. OMVs have yielded a wide range of biological functions, from delivery of enzymes to transport of toxins, transmission of communication signals, nutrient acquisition, and induction of commensal tolerance. One beneficial role of OMVs on host homeostasis is their ability to modulate innate and adaptive immune systems (266) (Fig. 3). Bacteroides fragilis OMV delivery of polysaccharide capsular antigen (PSA) yielded multiple immunomodulatory effects. Monocolonization of germ-free mice with B. fragilis was shown to modulate CD4+ T cell homeostasis and cytokine production (3). T cell modulation occurs in a PSA-dependent manner (3). OMV-delivered PSA stimulated TLR2 on Tregs and directly modulated DCs (267, 268). OMVs were internalized by DCs, and this interaction promoted tolerogenic DCs which produced IL-10 and stimulated regulatory Tregs. Furthermore, PSA production ameliorated intestinal inflammation (269), central nervous system inflammation (270, 271) and neurodegeneration (272). These studies demonstrate that OMV delivery of PSA is capable of promoting an anti-inflammatory profile which leads to tolerance and suppression of mucosal inflammation.
Serpin
Serpins are eukaryotic-type serine protease inhibitors. These molecules are synthesized by several commensal bacteria. Serpins are relatively large molecules consisting of approximately 330 to 500 amino acids (273). More than 70 serpin structures have been identified. These complex structures act as stoichiometric suicide inactivators and inhibit eukaryotic elastase-like serine proteases. Serpins are known to regulate a wide range of signaling pathways in eukaryotes. Select serpins have been shown to suppress inflammatory responses by inhibiting elastase activity (274) (Fig. 3). Several bifidobacterial species and subspecies (B. breve, B. longum subsp. infantis, B. longum subsp. longum, and B. dentium) are capable of producing serpins (275). B. longum NCC2705 was shown to secrete a serpin which binds and inactivates human neutrophil elastase, a product secreted by neutrophils during active inflammation (276). Moreover, bifidobacterial serpin-like proteins have been shown to reduce intestinal inflammation in a murine colitis model (277). Based on these findings, production of serpins which inhibit neutrophil elastases may be beneficial for reducing intestinal damage in the setting of overt inflammation.
Lactocepin
Lactocepins are bacterial enzymes which can degrade targeted bacteria of different genera via damage to prokaryotic cell membranes and induction of proinflammatory modulators (Fig. 3). Lactocepins can be cell wall associated or secreted, and the target specificity is strain specific (278–280). Lactocepins are mainly expressed by lactococci and lactobacilli. These enzymes are encoded by prtP, prtB, and/or prtH. The prtP genes are well documented for their caseinolytic properties. L. paracasei secreted prtP-encoded lactocepin, and this compound selectively degraded the lymphocyte-recruiting chemokine IP-10 and other proinflammatory chemokines such as I-TAC and eotaxin in vitro (281). Importantly, prtP-encoded lactocepin selectively degraded IP-10 in inflamed intestinal tissue and had no adverse effects on intestinal epithelial cell barrier function in vivo (281). This resulted in significantly reduced lymphocyte recruitment after intraperitoneal injection in an ileitis model. Another Lactobacillus strain, L. casei, was found to secrete lactocepin which degraded host IP-10 (282). In a murine colitis model (T cell transferred Rag2–/– mice), supplementation of an L. casei prtP-disruption mutant resulted in more IP-10, T cell infiltration, and inflammation in cecal tissue compared to the isogenic wild-type strain. Supplementation of the probiotic VSL#3, which contains Lactobacillus, Bifidobacterium, and Streptococcus, normalized intestinal levels of IP-10 in a murine colitis model and reduced inflammation in patients with inflammatory bowel disease (282). These studies support the role of lactocepin secreted by commensal microbes as an effective treatment for chemokine-mediated diseases such as inflammatory bowel disease.
Other Secreted Proteins Known To Enhance Host Health
In addition to all the secreted products highlighted by this review, select commensal bacteria generate putative proteins which modulate the host (Fig. 3). L. rhamnosus GG and L. casei secrete two proteins designated p40 and p75 (283, 284). Lactobacilli-produced p40 and p75 were found to inhibit TNF-induced apoptosis in the intestinal epithelium. The proteins signal via the antiapoptotic Akt kinase in a phosphoinositide 3-kinase-dependent manner likely via epidermal growth factor receptor activation (283, 285). The p40 and p75 proteins also enhance tight junctions and attenuate intestinal barrier disruption via protein kinase C and extracellular signal-regulated kinase 1. Furthermore, p40 was shown to transactivate the epidermal growth factor receptor, activating the downstream target Akt and stimulating mucus Muc2 gene expression and mucin production in the human goblet cell line LS174T and wild-type mice. Intestinal mucus is a critical component of the healthy intestinal barrier, particularly in the setting of infection and inflammation (122, 286–288). Stimulation of intestinal mucus by commensal bacteria likely enhances barrier function and promotes homeostasis. Other commensal bacteria produce putative proteins with immunoregulatory features. F. prausnitzii is known to produce a 15-kDa protein, termed MAM, with anti-inflammatory properties (289, 290). MAM inhibits the NF-κB pathway in intestinal epithelial cells and prevents colitis in vivo. Because subgroups of Crohn’s disease are associated with reduced relative abundances of F. prausnitzii, microbial and protein (e.g., MAM) supplementation has been hypothesized to alleviate intestinal inflammation in future clinical trials. These studies highlight the need to define secreted commensal products of the mammalian microbiome.
ANTIMICROBIAL COMPOUNDS
Lactic Acid
Commensal microbes may produce organic acids such as lactic acid that may reduce local pH and suppress the growth and survival of neighboring microbes. A group of microbes known as lactic acid bacteria (LAB) produce relatively large amounts of lactic acid as a major catabolic end product of glucose fermentation. In general, LAB consist of non-spore-forming Gram-positive bacteria with a DNA base composition of less than 53 mol% G+C (291). LAB members include Lactobacillus, Lactococcus, Leuconostoc, Enterococcus, Streptococcus, Pediococcus, Carnobacterium, Aerococcus, Oenococcus, Tetragenococcus, Vagococcus, and Weisella. The generation of lactic acid results in the recycling of electron acceptors for ATP generation by bacterial species. In addition to this role in the bacteria, lactic acid production benefits the host by reducing local pH and suppressing colonization and proliferation of potential pathogens. Lactic acid is readily miscible with water due to its low hydrophobicity and low acid dissociation constant. Lactic acid is effective against Gram-negative bacteria and to a lesser extent against Gram-positive bacteria (292, 293). As a liposoluble organic acid, lactic acid in its undissociated form can penetrate the bacterial cytoplasmic membrane (294). In Gram-negative bacteria, lactic acid transverses the outer membrane via water-filled porins and penetrates the cytoplasmic membrane. Additionally, in Gram-negative bacteria, lactic acid can act as a potent outer membrane-disintegrating agent, as evidenced by LPS release and sensitization of bacteria to detergents or lysozyme (293). Lactic acid-induced changes in cell membrane permeability can hinder substrate transport and promote further entrance of lactic acid into the cytoplasm, thereby effectively lowering the intracellular pH (293–295) (Fig. 3). Intracellular acidity can suppress NADH oxidation, altering the membrane electron transport system and transmembrane proton motive force (295). Malfunction of the electron transport system can lead to oxidative stress and generation of free radicals that damage DNA and proteins. These free radicals can then damage intracellular components as well as the extracellular membrane (296). Collectively, these mechanisms result in cell death to susceptible bacteria.
Lactic acid in sufficient quantities may inhibit the growth of a wide range of bacterial species (244, 297, 298). Addition of pure lactic acid in vitro has both inhibitory and biocidal effects against several pathogens, including the Gram-negative E. coli, P. mirabilis, Salmonella enteritidis, and P. aeruginosa and the Gram-positive Staphylococcus aureus, Enterococcus faecalis, Listeria monocytogenes, Bacillus cereus, and Bacillus megaterium and minimal fungicidal activity against the yeasts Rhodotorula sp., Saccharomyces cerevisiae, and Candida albicans (299). Generation of lactic acid by commensal bacteria also inhibits the growth of several pathogens including Helicobactor pylori, B. subtilis, B. cereus, Staphylococcus epidermidis, E. coli CB6, Klebsiella sp. strain CB2, Streptococcus pyogenes, P. aeruginosa, Salmonella enterica serovar Paratyphi, and Salmonella enterica serovar Typhimurium (298, 300–307). In these studies, lactic acid alters in vivo pH, inhibits pathogen urease activity, inhibits pathogen growth, and acts as a bactericidal agent. Additionally l-lactic acid suppresses immune cell-mediated proinflammatory responses (308). Modulation of the immune system and alteration of local GIT pH have been speculated to selectively manipulate the gut microbiota composition (120, 121, 309), which may further promote colonization resistance and inhibition of pathogens. In addition to the antimicrobial effects of lactic acid itself, several studies have demonstrated that unidentified bacterial substances, bacteriocins, and hydrogen peroxide act in concert with lactic acid to inhibit the growth of pathogens (300, 303, 307).
Hydrogen Peroxide
Bacterially produced hydrogen peroxide (H2O2) is known to act synergistically with l-lactic acid (310) (Fig. 3). H2O2 produced by certain microbes can damage bacterial nucleic acids by creating breaks in the carbon phosphate backbone of DNA, releasing nucleotides, and preventing chromosomal replication (311, 312). Additionally, hydroxyl radicals, which can be produced from the dissociation of H2O2, can attack the methyl group of thymine, resulting in damaged DNA (313, 314). Anaerobic bacteria are more sensitive to H2O2 because they do not produce catalase, which can break down H2O2. In general, Gram-negative bacteria are more sensitive than Gram-positive bacteria to H2O2. However, select Gram-negative bacteria have developed a mechanism to deal with H2O2. These microbes utilize an outer LPS layer which traps active molecular oxygen (315). Lactic acid disrupts the outer membrane of Gram-negative bacteria, releasing LPS and making cells sensitive to H2O2 and antimicrobial agents (293). Several microbes are known to produce H2O2, including lactobacilli and bifidobacteria (316, 317). Many studies have demonstrated that bacterially produced H2O2 inhibits the growth of pathogens such as S. aureus, S. enterica serovar Typhimurium, L. monocytogenes, E. faecalis, E. faecium, enterotoxigenic E. coli, E. coli CFT074, Listeria ivanovii, S. aureus, Yersinia enterocolitica, Aeromonas hydrophila, Gardnerella vaginalis DSM494, Neisseria gonorrhoeae, S. mutans, Bacteroides forsythus, Capnocytophaga sputigena, Eikenella corrodens, Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, and Wolinella recta (317–328). The effect of LAB-produced H2O2 on pathogen inhibition was found to be significantly enhanced by lactic acid (323), supporting the role of H2O2 and lactic acid acting in concert to shape microbial communities. However, commensal bacteria are not the sole producers of H2O2. The production of H2O2 by the pathogen S. pneumoniae inhibited growth of viral Haemophilus influenzae and fellow bacterial pathogens Moraxella catarrhalis, Neisseria meningitidis, and S. aureus (329, 330). Thus, it has been speculated that pathogens use H2O2 production to inhibit competing organisms and secure a niche.
Bacteriocins
The production of antimicrobial compounds by bacteria provides specific microorganisms with a competitive advantage for colonization. The production of bacteriocins is a nearly universal trait, because it is projected that the majority of bacteria and archaea produce at least one bacteriocin (331–333). The ubiquity of this trait implies that bacteriocins play an important role in vivo as colonizing peptides, as tools for inhibiting commensal or pathogen niche occupation, or as signaling peptides (333–338). Wide variation exists in the chemical composition and mechanisms of action of different bacteriocins. Bacteriocins can be classified based on the bacteria that secrete them (Gram-negative or Gram-positive). In general, bacteriocins produced by Gram-positive bacteria have antimicrobial effects on other Gram-positive bacteria (331). Production of bacteriocins by probiotic bacteria has been speculated to promote colonization and inhibit pathogens (161, 333). In contrast, production of bacteriocins by pathogenic bacteria has been postulated to provide a competitive edge for infection (339). Wide variation exists in the chemical composition and mechanism of action of different bacteriocins. Most bacteriocins target phosphate groups on bacterial cell membranes, deplete the transmembrane potential (Δψ) and/or the pH gradient, and form membrane pores, resulting in membrane disruption and cellular leakage (340–342) (Fig. 4). Similar to other compounds such as H2O2, bacteriocins may yield synergistic effects with lactic acid and exhibit greater antibacterial activities at lower pH values. To simplify our review, we have chosen to focus primarily on bacteriocins produced by commensal bacteria, as opposed to ones produced by pathogens.
FIGURE 4.
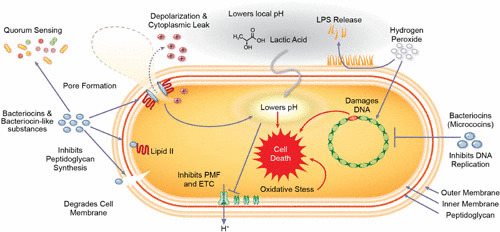
Schematic representation of the molecular mechanisms of commensal secreted products on a Gram-negative bacterium. Bacteriocins are classified based on their structure. Bacteriocins such as nisin bind to a peptidoglycan subunit transporter, thereby preventing cell wall synthesis and resulting in cell death. Furthermore, bacteriocins can initiate pore formation. Pore formation depletes the bacterial transmembrane potential (Δψ) and/or the pH gradient, resulting in membrane disruption and cellular leakage that lead to rapid cell death. Other bacteriocins insert themselves directly or degrade the target membrane, leading to depolarization and death. Bacteriocins have also been shown to serve as quorum-sensing molecules for other microbes. Lactic acid decreases local pH and suppresses the growth and survival of pathogens. Additionally, undissociated lactic acid can traverse the outer membrane via water-filled porins and penetrate the cytoplasmic membrane. This shift lowers the intracellular pH, disrupts the transmembrane proton motive force, and generates oxidative stress. Hydrogen peroxide and select bacteriocins such as microcins damage bacterial DNA and inhibit cell growth. Together, these compounds secreted by select members of the microbiota effectively target pathogens.
Bacteriocin diversity and classification
Gram-positive bacteriocins
Class I: the lantibiotics
Lantibiotics are small (<5 kDa) peptides characterized by the unusual amino acids lanthionine, α-methyllanthionine, dehydroalanine, and dehydrobutyrine. Within class I, molecules can be subgrouped into type A or type B according to their chemical structures and antimicrobial activities (343–345). Type A lantibiotics exhibit elongated screw-shaped peptides with a net positive charge. The shape and charge of type A molecules facilitate membrane pore formation and membrane depolarization in sensitive species. Type A molecules generally are 2 to 4 kDa in molecular weight. The best characterized of the type A lantibiotics is the Lc. lactis-produced nisin. Nisin inhibits the growth of a range of Gram-positive bacteria including L. monocytogenes, S. aureus, and B. cereus (346–349). Nisin also prevents spore germination by the pathogens Clostridium botulinum, Clostridium sporogenes, B. cereus, and Bacillus anthracis (341, 350–353). In vegetative cells, nisin binds to lipid II on targeted bacterial membranes. Nisin orients parallel to the surface of the target membrane and inserts the C terminus of the peptide into the phospholipids, thereby disrupting cell wall biosynthesis and creating a “wedge-like” pore (354). Pore formation causes a rapid nonspecific amino acid and cation efflux and subsequent cell membrane rupture and cell death (355, 356). In spores, nisin also utilizes lipid II binding and pore formation in germinated spores during outgrowth, leading to membrane disruption that inhibits spore development into vegetative cells. Type A lantibiotics include lacticin (Lc. lactis lacticin 3147 [357], Lc. lactis subsp. lactis lacticin 481 [358]), lactocin (L. rhamnosus lactocin 160 [342], Lactobacillus sake L45 lactocin S [359]), S. epidermidis epidermin (360), and Staphylococcus gallinarum gallidermin (361).
In comparison to type A molecules, type B lantibiotics are smaller (2 to 3 kDa) globular peptides with a negative or neutral charge. Also in contrast to type A lantibiotics, type B peptides exert their antimicrobial activity via cell lysis and inhibition of essential bacterial enzymes. Type B lantibiotics increase membrane permeability and reduce ATP-dependent protein transport and ATP-dependent calcium uptake in sensitive bacterial cells, resulting in cell lysis. Cell lysis was significantly reduced when the type B lantibiotics cinnamycin and duramycin were incubated with the phospholipid phosphatidylethanolamine (362–364). The data indicate that type B lantibiotics interact with phospholipid targets. In addition to membrane effects, type B lantibiotics are also known to inhibit bacterial enzymes such as phospholipase A and peptidoglycan synthesis. Examples of type B lantibiotics include Lactobacillus curvatus curvacin A, Streptomyces cinnamoneus cinnamycin, Streptomyces subsp. ancovenin (365), Streptoverticillium R2075 duramycins B and C, and Streptomyces griseoluteus (R2107) duramycins B and C (366).
Class II
Class II bacteriocins are small (<10 kDa), heat stable, nonlanthionine-containing membrane-active peptides. These peptides can be further divided into subgroups based on sequence and function: a, b, c, d, and e. The subgroup class IIa comprises pediocin-like peptides containing an N-terminal consensus sequence -Tyr-Gly-Asn-Gly-Val-Xaa-Cys. Class IIa bacteriocins are produced by food-associated bacterial strains and have garnered attention due to their anti-Listeria activity (367, 368). Similar to class I molecules, class IIa bacteriocins kill target cells by permeabilizing the cell membrane (340, 369). Class II molecules bind to the target membrane using the cationic N-terminal beta sheet domain of the peptide, while the C-terminal regions form a hairpin-like domain which penetrates into the target cell membrane. This penetration results in leakage of cytoplasmic components through the membrane, resulting in cell death. Bacteriocins belonging to the class IIa family include Lb. lactis lactococcin MMFII, Bifidobacterium bifidum NCFB bifidocin B, B. longum subsp. infantis bifidin I, Pediococcus acidilactici pediocin PA-1, Carnobacterium piscicola carnobacteriocin B2, Carnobacterium divergens divercin V41, Lactobacillus sakei sakacin P, L. sake sakacin A, Enterococcus faecium enterocin A, E. faecium enterocin P, Leuconostoc gelidum leucocin A, Leuconostoc mesenteroides mesentericinY105 (280, 370–378), and Pediococcus pentosaceus K23-2 (379). Class IIb contains bacteriocins that require two separate peptides for activity. Similar to class IIa they act as pore-forming peptides (380). Class IIb peptides include Lb. lactis lactococcin G and M, L. salivarius UCC118 Abp118, L. johnsonii lactacin F, and L. plantarum plantaricin A, S, E, F, and JK (338, 357, 381–388). The class IIc peptides have a wide range of effects on membrane permeability and cell wall formation. One of the well-documented class IIc molecules is E. faecalis bacteriocin AS-48 (389). Bacteriocin AS-48 consists of five alpha helices enclosing a hydrophobic core, creating a globular structure which creates membrane pores in susceptible Gram-positive or Gram-negative bacteria. Other examples include L. acidophilus acidocin B, C. piscicola carnobacteriocin A, C. divergens divergicin A, and E. faecium enterocin P and B (390–393). Class IId bacteriocins are linear, non-pediocin-like, single-peptide bacteriocins with similar antimicrobial activity. Class IId molecules include Lb. lactis lactococcin A, S. epidermidis epidermicin NI01, and Streptococcus cremoris diplococcin (386, 394, 395). Class IIe bacteriocins comprise nonribosomal siderophore-type posttranslational modifications at the serine-rich carboxy-terminal region of the peptide. Class IIe molecules include Klebsiella pneumoniae microcin E492 (396, 397).
Class III
Class III bacteriocins comprise large-molecular-weight (>30 kDa) heat-labile proteins. This class is further divided in two subclasses: IIIa and IIIb. Subclass IIIa, also known as bacteriolysins, encompasses peptides that degrade bacterial cell membranes, resulting in cell lysis and subsequent cell death. The most well-characterized members of subclass IIIa are Staphylococcus spp.-produced lysostaphin. Lysostaphin is a 27-kDa peptide that cleaves cell wall cross-linking pentaglycin bridges, thereby hydrolyzing susceptible staphylococci, including the pathogen S. aureus (398–400). Another member of the subclass IIIa group is E. faecium enterolysin (401). Subclass IIIb comprises peptides which disrupt target cell membrane potential, causing ATP efflux and death. In contrast to subclass IIIa, these peptides do not cause cell lysis. Other members of class III include bacteriocins from subclass IIIb such as L. casei caseicin 80 and L. helveticus helveticin J and V-1829 (402–404).
Class IV
Class IV bacteriocins were recently described and are defined as complex bacteriocins containing lipid or carbohydrate moities. The class IV bacteriocins are cyclical due to covalent bonding of the first and last amino acids and are considered to be S-linked glycopeptides with antimicrobial activity. Examples of class IV molecules include E. faecalis subsp. liquefaciens S-48 enterocin AS-48, E. faecalis F4-9 enterocin F4-9, B. subtilis 168 sublancin (an S-linked glycopeptide), and L. plantarum KW30 glycocin F (GccF) (405–408). Peptides from this class exhibit a wide pH range, variable resistance to heat, and loss of antimicrobial activity when exposed to proteolytic enzymes (408–410). The cationic charges of enterocin F4-9 and glycocin F were found to be essential for interactions with charged phospholipids in target bacterial cells. These interactions did not induce cell lysis, suggesting that these bacteriocins have a bacteriostatic effect. Enterocin F4-9 yielded antimicrobial activity against E. faecalis and E. coli JM109, but not E. faecium and other E. coli members (408). In contrast, glycocin F was found to have activity against lactobacilli (410), while sublancin 168 had activity against Gram-positive bacteria, specifically Bacillus spp., but not E. coli JM101 (409). These studies indicate that class IV bacteriocins target unique epitopes.
Gram-negative bacteriocins
Microcins
Microcins are low-molecular-weight (<10 kDa) hydrophobic antimicrobial peptides synthesized on bacterial ribosomes. These molecules are primarily produced by Gram-negative Enterobacteriaceae (phylum Proteobacteria). Microcins are generally heat-, extreme pH-, and protease-tolerant (411). They exhibit a wide range of structural diversity and antimicrobial mechanisms. Microcins target other bacterial cells via pore formation, DNAse or RNAse nuclease function, inhibition of protein synthesis, or inhibition of DNA replication (412). One of the more deceptive ways microcins exert their antibacterial activity is through a “Trojan horse” strategy whereby they imitate essential nutrients (411). Microcins mimic small, high-affinity iron-chelating compounds (iron-siderophore complexes). Because iron is required for bacterial DNA synthesis, microcins resembling iron-siderophores are able to bind to outer membrane receptors on target bacteria and translocate into the periplasmic space, where they can exert their antimicrobial activity.
Despite the wide diversity, these molecules are classified in two classes depending on their molecular masses, disulfide bonds, and posttranslational modifications: class I and II. Class I microcins are plasmid-encoded and low molecular weight (<5 kDa) and are posttranslationally modified. Examples of class I microcins include E. coli microcin B17, C7-C51, D93, and J25. Class II microcins are larger in molecular weight (5 to 10 kDa) and produced by and active against Enterobacteriaceae. Class II microcins are divided into two subclasses: IIa and IIb. Class IIa microcins do not undergo posttranslational modification, while class IIb microcins exhibit posttranslational modifications in the form of a salmochelin-like siderophore motif (413). Additionally, class IIa peptides typically contain one or two disulfide bond(s). Class IIa molecules include E. coli microcins L, V, S, and N, all of which require three different genes to synthesize the molecule. Class IIb microcins are differentiated from class IIa by their linear structure or lack of posttranslational modifications at the C terminus (412, 414). Examples of class IIb molecules include E. coli Nissle 1917 microcin M, E. coli Nissle 1917 microcin H47, and Klebsiella microcin E492.
Colicins
In contrast to low-molecular-weight microcins, colicins are high-molecular-weight (25 to 80 kDa) proteins with antimicrobial activity. Colicins act by binding to outer membrane receptors, penetrating the cytoplasmic membrane, and then causing cytotoxic effects, including cytoplasmic membrane depolarization, DNase activity, RNase activity, or murein synthesis inhibition. Colicins use a two-receptor system to target cells. First, colicins bind to outer membrane receptors such as the porins OmpF, FepA, BtuB, Cir, and FhuA, which are typically used for entry of specific nutrients into the bacterial cells. Next, colicins are translocated through the outer membrane and enter the cell cytoplasm by either the Tol or Ton system, where they can exert their cytotoxic effect (415). Colicins are divided into categories based on their outer membrane translocation system (either Tol or Ton). The Ton and Tol systems are two-protein arrangements which E. coli uses to transfer energy from the inner membrane to the outer membrane. The systems differ in their component proteins. The Ton system consists of TonB, ExbB, and ExbD, while the Tol system consists of TolA, TolQ, and TolR. In the Ton system, ExbB and ExbD proteins make up the energy-harvesting complex, which transfers energy to the protein TonB. This arrangement causes a conformational change in TonB, which subsequently delivers energy to the outer membrane. In the Tol system, TolQ and TolR proteins compose the energy-harvesting complex, which transfers energy to TolA, resulting in a conformational change and energy to the outer membrane. The Ton system is involved in transport of compounds across the membrane, while the Tol system is involved in outer membrane maintenance. Group A colicins use the Tol protein system to traverse the outer membrane of sensitive bacteria, while group B colicins use the Ton system.
Examples of group A colicins include colicins E1 to E9 and colicins A, K, and N, and examples of group B molecules include colicins 5, 10, B, D, M, V, Ia, and Ib (412, 416). Colicins can also be classified based on their mechanism of induced cell death: (i) pore-formation colicins, (ii) nuclease-type colicins, and (iii) peptidoglycanase-type colicins. Pore-formation colicins act by creating a pore within the bacterial cytoplasmic membrane, resulting in cytoplasmic content leakage, loss of electrochemical gradients, and cell death. Examples of pore-formation colicins include colicins A, B, E1, Ia, Ib, K, and N. Nuclease-type colicins utilize DNase, 16S rRNase, and tRNase to nonspecifically digest DNA and RNA of targeted bacteria. Examples of these molecules include colicins E2 to E9. Finally, peptidoglycanase-type colicins digest peptidoglycan precursors. This digestion prevents bacteria from synthesizing peptidoglycan, a key cell wall component, and results in bacterial death (417).
Bacteriocins in vivo
Bacteriocin-producing strains have been shown to have an ecological advantage over nonproducing strains in vivo, pointing to the role of bacteriocins in niche development. For example, E. coli BZB1011 strains which produced colicins (A, E1, E2, E7, K, and N) were found to be persistent over time in mice compared to non-colicin-producing isogenic strains (418). Bacteriocins, particularly the lantibiotics from Gram-positive bacteria, have been shown to be effective against a number of pathogenic strains in vivo. The lantibiotic nisin has been shown to inhibit the pathogens S. pneumoniae (419), S. aureus (420–422), and L. monocytogenes in mice (423). Furthermore, lantibiotic B-Ny266 was shown to inhibit S. aureus (424), and planosporicin inhibited S. pyogenes in mice (425). The class IIa bacteriocin peptides enterocin CRL35 and divercin V41 were also shown to inhibit L. monocytogenes (426, 427), while bacteriocin E50-52 inhibited Mycobacterium tuberculosis in mice (428). Intraperitoneal injection of microcin MccJ25 reduced Salmonella enterica serotype Newport quantities in the spleen and liver in mice (429). As a result of these in vivo studies, bacteriocins have been considered for the treatment of multi-drug-resistant pathogens.
Bacteriocins as quorum-sensing molecules
For many bacteria, bacteriocin production is regulated by quorum sensing (161, 430, 431). Quorum sensing, as discussed earlier, is a cell-density-dependent regulatory system in which autoinducing signals facilitate bacterial communication. This system can sense the number of cells of the same species and synchronize the expression of key genes. Bacteriocin peptides are sensed by membrane-located histidine kinases, which transmit a signal via an intracellular response regulator, thereby activating gene transcription of the inducer bacteriocin molecule (432). The two-component signal-transduction mechanism is a key step for transcription activation and production of several bacteriocins or bacteriocin-like peptides. These include the class I lantibiotics Lb. lactis nisin and B. subtilis subtilin, as well as the class II E. faecium enterocin A and L. salivarius UCC118 Abp118 (431, 433–437). Thus, the bacteriocin peptide itself functions as a pheromone to induce its own production. When bacterial cell density is high, an autoinduction loop is activated and bacteriocins are likewise produced at high concentrations. In this manner, bacteriocins are released to target similar species only when the bacterial levels are high enough to suppress the growth of competitive strains.
Bacteriocins within biofilms are known to play important roles in bacterial competition, ecological fitness, and overall community structure (93, 438, 439). In the naturally transformable streptococci, including S. mutans, Streptococcus gordonii, Streptococcus sanguinis, and Streptococcus mitis, bacteriocin production is tightly regulated by a quorum-sensing system that also regulates genetic competence and biofilm formation (93, 438, 439). S. mutans has served as a well-studied example of quorum sensing and bacteriocin production. S. mutans uses the ComCDE quorum-sensing system to connect to bacteriocin production, stress response, genetic competence, and biofilm formation (93, 342, 438, 440–442). S. mutans ComC mutants do not produce the signaling molecule CSP, and these biofilms exhibit reduced biomass (93). However, supplementation of cultures with synthetic CSP restores the wild-type biofilm phenotype. S. mutans CSP-induced bacteriocin (CipB) can also stimulate cell lysis (443, 444). As a result, bacteriocin production within biofilms is speculated to provide balanced competition and coexistence of multiple organisms within a microbial community (438, 439, 445).
Other Biogenic Substances
In addition to bacteriocins, select microbes are known to produce bacteriocin-like substances, which are used to describe compounds that have antimicrobial properties but are not ribosomally produced or gene-encoded precursor peptides. Examples of this class of compounds include reuterin and reutericyclin.
Reuterin
Select strains of L. reuteri generate reuterin (3-hydroxypropionaldehyde) during anaerobic fermentation of glycerol (446, 447). During this process glycerol is reduced to 1,3-propanediol, an event catalyzed by the coenzyme B12-dependent diol dehydratase, which regenerates NAD+ from NADH (448–450). Reuterin is water-soluble, active over a wide range of pH values, and bioactive against Gram-positive and Gram-negative bacteria, viruses, and fungi (451). The aldehyde group of reuterin is highly reactive with thiol groups and primary amines. As a result, reuterin inhibits bacterial growth by modifying protein thiol groups and inducing oxidative stress in target cells (449). Production of reuterin by L. reuteri was found to be enhanced by interactions with E. coli, indicating that cross-talk between members of the gut microbiota may regulate reuterin output (449). Reuterin has also been demonstrated to act synergistically with other bacteriocins. Nisin, lacticin 481, and enterocin AS-48 were found to enhance antimicrobial activity of reuterin against the pathogen L. monocytogenes (452). However, only nisin was found to increase antimicrobial activity of reuterin against S. aureus (452). These studies indicate that synergism between reuterin and bacteriocins is dependent on the bacteriocin and pathogen. Importantly, reuterin was found to be produced by L. reuteri in mono-associated germ-free mice, indicating that reuterin can be produced in vivo in the GIT (453).
Reutericyclin
Reutericyclin is a highly hydrophobic, negatively charged compound produced by certain L. reuteri strains (454, 455). Reutericyclin [(5R)-1-(2-decenoyl)-2-hydroxy-3-acetyl-5-isobutyl-Δ2-pyrroline-4-one] is an N-acylated tetramic acid (455), and its mode of action resembles that of weak organic acids. Reutericyclin targets the cytoplasmic membrane of sensitive bacteria by acting as a proton-ionophore. This action results in the translocation of protons across the cytoplasmic membrane and subsequent collapse of the transmembrane ΔpH (454). Reutericyclin exhibits a broad inhibitory spectrum against Gram-positive pathogens including E. faecalis, Listeria innocua, S. aureus, B. subtilis, and B. cereus (455). Reutericyclin has also been shown to inhibit germination of Bacillus spores (455). However, Gram-negative bacteria are resistant to reutericyclin due to the presence of LPS in their outer membrane. LPS is known to limit access of hydrophobic compounds, and the charged polysaccharide moieties of LPS have been proposed to bind reutericyclin, thereby rendering it inactive (454, 456).
SUMMARY AND CONCLUSIONS
This article highlighted multiple well-characterized systems that commensal bacteria use to communicate with other microbes and the mammalian host, to modify microbial communities, and to beneficially modulate the function of the mammalian host. These studies emphasize key roles of bacterial metabolites and secreted products in fine-tuning mammalian biology. Bacteria are capable of secreting multiple and chemically diverse host-modulating factors. This work has focused on commensal bacteria, but a number of these factors can also be used by pathogens. Future studies may begin to define the secretion of products in various environments and under different conditions (i.e., inflammation, altered pH, various dietary nutrients). As we begin to understand the health-promoting compounds and metabolites generated by commensal bacteria, we will be better able to identify individual species and strains which can be tailored for specific physiological or biochemical targets of interest. The work here highlights the importance of selecting well-characterized commensal or probiotic strains with known mechanisms to address specific disease processes. The recent development of the U.S. edition of the Clinical Guide to Probiotic Products reflects the growing need for evidence-based selection of probiotics. This online database provides guidelines for probiotic products and corresponding research studies supporting the use of each specific probiotic (http://usprobioticguide.com/). These tools will likely aid clinicians in selecting disease-relevant bacterial species.
Genetic engineering of probiotic strains has also been used as a method to enhance production of molecules of interest (124). In this system bacteria could be designed to have upregulated gene synthesis or to be constitutively active. Using this model, issues of whether a given product is being secreted in vivo under various conditions (pH, ion composition, changing osmolarity) could be minimized. Strains have also been engineered with genes from other bacteria to create organisms capable of secreting multiple beneficial factors. Microbial engineering may lead to the production of “super” strains with the ability to release multiple factors targeted at different purposes (i.e., immunomodulation, pathogen exclusion, enhancement of the mucus barrier). Additionally, genetic engineering has been used to facilitate probiotic selection and maintenance in the host (124). This method may ensure the transient introduction of strains which are not wanted to colonize the host in the long term. Currently, social acceptance of genetically modified foods and microbes is not universal, and thus these types of modifications will likely be closely monitored in the future. An alternative approach could be to isolate and purify valuable metabolites, which themselves could also be used directly as novel therapeutics for specific disorders.
Based on studies conducted thus far, we have strong evidence to pursue the selection of probiotics and examination of specific microbial genes, proteins, and metabolites of interest. In the future we hope to use this knowledge to thoughtfully select commensal strains for a given disease treatment or prevention. Overall, these studies contribute to the rapidly expanding body of evidence demonstrating that secreted microbial metabolites can beneficially influence host physiology. In addition to probiotics, future dietary and medicinal interventions may depend on the foundations of microbial metabolite and protein discovery in the context of microbiome science.
REFERENCES
- 1.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533 10.1371/journal.pbio.1002533. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenhower C, et al, Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 10.1038/nature11234. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118 10.1016/j.cell.2005.05.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, Deng Y, Blennerhassett PA, Fahnestock M, Moine D, Berger B, Huizinga JD, Kunze W, McLean PG, Bergonzelli GE, Collins SM, Verdu EF. 2011. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil 23:1132–1139 10.1111/j.1365-2982.2011.01796.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. 2010. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 170:1179–1188 10.1016/j.neuroscience.2010.08.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230 10.1038/nature11550. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson CL, Versalovic J. 2012. The human microbiome and its potential importance to pediatrics. Pediatrics 129:950–960 10.1542/peds.2011-2736. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, Raza S, Doddapaneni HV, Metcalf GA, Muzny DM, Gibbs RA, Petrosino JF, Shulman RJ, Versalovic J. 2015. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 3:36 10.1186/s40168-015-0101-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biagi E, Candela M, Turroni S, Garagnani P, Franceschi C, Brigidi P. 2013. Ageing and gut microbes: perspectives for health maintenance and longevity. Pharmacol Res 69:11–20 10.1016/j.phrs.2012.10.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, Brigidi P, De Vos W. 2010. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5:e10667. (Erratum, doi:10.1371/annotation/df45912f-d15c-44ab-8312-e7ec0607604d.) [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín R, Miquel S, Ulmer J, Kechaou N, Langella P, Bermúdez-Humarán LG. 2013. Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb Cell Fact 12:71 10.1186/1475-2859-12-71. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, Dewulf J, Brassart D, Mercenier A, Pot B. 2007. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol 13:236–243 10.3748/wjg.v13.i2.236. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marteau P, Lémann M, Seksik P, Laharie D, Colombel JF, Bouhnik Y, Cadiot G, Soulé JC, Bourreille A, Metman E, Lerebours E, Carbonnel F, Dupas JL, Veyrac M, Coffin B, Moreau J, Abitbol V, Blum-Sperisen S, Mary JY. 2006. Ineffectiveness of Lactobacillus johnsonii LA1 for prophylaxis of postoperative recurrence in Crohn’s disease: a randomised, double blind, placebo controlled GETAID trial. Gut 55:842–847 10.1136/gut.2005.076604. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maassen CB, van Holten-Neelen C, Balk F, Heijne den Bak-Glashouwer MJ, Leer RJ, Laman JD, Boersma WJ, Claassen E. 2000. Strain-dependent induction of cytokine profiles in the gut by orally administered Lactobacillus strains. Vaccine 18:2613–2623 10.1016/S0264-410X(99)00378-3. [DOI] [PubMed] [Google Scholar]
- 16.Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B, Cerdan S, Brody L, Anastasovska J, Ghourab S, Hankir M, Zhang S, Carling D, Swann JR, Gibson G, Viardot A, Morrison D, Louise Thomas E, Bell JD. 2014. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 5:3611 10.1038/ncomms4611. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schrijver IA, van Meurs M, Melief MJ, Wim Ang C, Buljevac D, Ravid R, Hazenberg MP, Laman JD. 2001. Bacterial peptidoglycan and immune reactivity in the central nervous system in multiple sclerosis. Brain 124:1544–1554 10.1093/brain/124.8.1544. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Bäumlisberger M, Moellecken U, König H, Claus H. 2015. The potential of the yeast Debaryomyces hansenii H525 to degrade biogenic amines in food. Microorganisms 3:839–850 10.3390/microorganisms3040839. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pessione A, Lamberti C, Pessione E. 2010. Proteomics as a tool for studying energy metabolism in lactic acid bacteria. Mol Biosyst 6:1419–1430 10.1039/c001948h. [PubMed] [DOI] [PubMed] [Google Scholar]
- 20.Bouchereau A, Guénot P, Larher F. 2000. Analysis of amines in plant materials. J Chromatogr B Biomed Sci Appl 747:49–67 10.1016/S0378-4347(00)00286-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 21.Suzzi G, Gardini F. 2003. Biogenic amines in dry fermented sausages: a review. Int J Food Microbiol 88:41–54 10.1016/S0168-1605(03)00080-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 22.Tabanelli G, Torriani S, Rossi F, Rizzotti L, Gardini F. 2012. Effect of chemico-physical parameters on the histidine decarboxylase (HdcA) enzymatic activity in Streptococcus thermophilus PRI60. J Food Sci 77:M231–M237 10.1111/j.1750-3841.2012.02628.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Molenaar D, Bosscher JS, ten Brink B, Driessen AJ, Konings WN. 1993. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J Bacteriol 175:2864–2870 10.1128/jb.175.10.2864-2870.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodwell AW. 1953. The histidine decarboxylase of a species of Lactobacillus; apparent dispensability of pyridoxal phosphate as coenzyme. J Gen Microbiol 8:233–237 10.1099/00221287-8-2-233. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Rossi F, Gardini F, Rizzotti L, La Gioia F, Tabanelli G, Torriani S. 2011. Quantitative analysis of histidine decarboxylase gene (hdcA) transcription and histamine production by Streptococcus thermophilus PRI60 under conditions relevant to cheese making. Appl Environ Microbiol 77:2817–2822 10.1128/AEM.02531-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemarajata P, Gao C, Pflughoeft KJ, Thomas CM, Saulnier DM, Spinler JK, Versalovic J. 2013. Lactobacillus reuteri-specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. J Bacteriol 195:5567–5576 10.1128/JB.00261-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. 2012. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One 7:e31951 10.1371/journal.pone.0031951. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessione E, Mazzoli R, Giuffrida MG, Lamberti C, Garcia-Moruno E, Barello C, Conti A, Giunta C. 2005. A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 5:687–698 10.1002/pmic.200401116. [PubMed] [DOI] [PubMed] [Google Scholar]
- 29.Lucas PM, Claisse O, Lonvaud-Funel A. 2008. High frequency of histamine-producing bacteria in the enological environment and instability of the histidine decarboxylase production phenotype. Appl Environ Microbiol 74:811–817 10.1128/AEM.01496-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izquierdo Cañas PM, Gómez Alonso S, Ruiz Pérez P, Seseña Prieto S, García Romero E, Palop Herreros ML. 2009. Biogenic amine production by Oenococcus oeni isolates from malolactic fermentation of Tempranillo wine. J Food Prot 72:907–910 10.4315/0362-028X-72.4.907. [PubMed] [DOI] [PubMed] [Google Scholar]
- 31.Gao C, Major A, Rendon D, Lugo M, Jackson V, Shi Z, Mori-Akiyama Y, Versalovic J. 2015. Histamine H2 receptor-mediated suppression of intestinal inflammation by probiotic Lactobacillus reuteri. MBio 6:e01358-15 10.1128/mBio.01358-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferstl R, Frei R, Schiavi E, Konieczna P, Barcik W, Ziegler M, Lauener RP, Chassard C, Lacroix C, Akdis CA, O’Mahony L. 2014. Histamine receptor 2 is a key influence in immune responses to intestinal histamine-secreting microbes. J Allergy Clin Immunol 134:744–746.e3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Frei R, Ferstl R, Konieczna P, Ziegler M, Simon T, Rugeles TM, Mailand S, Watanabe T, Lauener R, Akdis CA, O’Mahony L. 2013. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J Allergy Clin Immunol 132:194–204.e12 10.1016/j.jaci.2013.01.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 34.Dhakal R, Bajpai VK, Baek KH. 2012. Production of gaba (γ- aminobutyric acid) by microorganisms: a review. Braz J Microbiol 43:1230–1241 10.1590/S1517-83822012000400001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, Chen Z, Gu Z, Han Y. 2008. Isolation of γ-aminobutyric acid-producing bacteria and optimization of fermentative medium. Biochem Eng J 41:48–52. [Google Scholar]
- 36.Smith DK, Kassam T, Singh B, Elliott JF. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol 174:5820–5826 10.1128/jb.174.18.5820-5826.1992. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kono I, Himeno K. 2000. Changes in gamma-aminobutyric acid content during beni-koji making. Biosci Biotechnol Biochem 64:617–619 10.1271/bbb.64.617. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Barrett E, Ross RP, O’Toole PW, Fitzgerald GF, Stanton C. 2012. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol 113:411–417 10.1111/j.1365-2672.2012.05344.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 39.Komatsuzaki N, Shima J, Kawamoto S, Momose H, Kimura T. 2005. Production of y-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol 22:497–504 10.1016/j.fm.2005.01.002. [DOI] [Google Scholar]
- 40.Siragusa S, De Angelis M, Di Cagno R, Rizzello CG, Coda R, Gobbetti M. 2007. Synthesis of gamma-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl Environ Microbiol 73:7283–7290 10.1128/AEM.01064-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pokusaeva K, Johnson C, Luk B, Uribe G7, Fu Y, Oezguen N, Matsunami RK, Lugo M, Major A, Mori-Akiyama Y, Hollister EB, Dann SM, Shi XZ, Engler DA, Savidge T, Versalovic J. 2017. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil. [Epub ahead of print. 10.1111/nmo.12904.\] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayakawa K, Kimura M, Kasaha K, Matsumoto K, Sansawa H, Yamori Y. 2004. Effect of a gamma-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br J Nutr 92:411–417 10.1079/BJN20041221. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Kimura M, Hayakawa K, Sansawa H. 2002. Involvement of gamma-aminobutyric acid (GABA) B receptors in the hypotensive effect of systemically administered GABA in spontaneously hypertensive rats. Jpn J Pharmacol 89:388–394 10.1254/jjp.89.388. [PubMed] [DOI] [PubMed] [Google Scholar]
- 44.Izquierdo E, Marchioni E, Aoude-Werner D, Hasselmann C, Ennahar S. 2009. Smearing of soft cheese with Enterococcus faecium WHE 81, a multi-bacteriocin producer, against Listeria monocytogenes. Food Microbiol 26:16–20 10.1016/j.fm.2008.08.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 45.Adeghate E, Ponery AS. 2002. GABA in the endocrine pancreas: cellular localization and function in normal and diabetic rats. Tissue Cell 34:1–6 10.1054/tice.2002.0217. [PubMed] [DOI] [PubMed] [Google Scholar]
- 46.Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grütter MG. 2003. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J 22:4027–4037 10.1093/emboj/cdg403. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagiwara H, Seki T, Ariga T. 2004. The effect of pre-germinated brown rice intake on blood glucose and PAI-1 levels in streptozotocin-induced diabetic rats. Biosci Biotechnol Biochem 68:444–447 10.1271/bbb.68.444. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Cho YR, Chang JY, Chang HC. 2007. Production of gamma-aminobutyric acid (GABA) by Lactobacillus buchneri isolated from kimchi and its neuroprotective effect on neuronal cells. J Microbiol Biotechnol 17:104–109. [PubMed] [PubMed] [Google Scholar]
- 49.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. 2011. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 108:16050–16055 10.1073/pnas.1102999108. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, Bisson JF, Rougeot C, Pichelin M, Cazaubiel M, Cazaubiel JM. 2011. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr 105:755–764 10.1017/S0007114510004319. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Okada T, Sugishita T, Murakami T, Murai H, Saikusa T, Horino T, Onoda A, Kajimoto O, Takahashi R, Takahashi T. 2000. Effect of the defatted rice germ enriched with GABA for sleeplessness, depression, autonomic disorder by oral administration. Nippon Shokuhin Kagaku Kogaku Kaishi 47:596–603 10.3136/nskkk.47.596. [DOI] [Google Scholar]
- 52.Shah P, Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol 68:4–16 10.1111/j.1365-2958.2008.06126.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Pegg AE, McCann PP. 1982. Polyamine metabolism and function. Am J Physiol 243:C212–C221. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Milovic V. 2001. Polyamines in the gut lumen: bioavailability and biodistribution. Eur J Gastroenterol Hepatol 13:1021–1025 10.1097/00042737-200109000-00004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Noack J, Dongowski G, Hartmann L, Blaut M. 2000. The human gut bacteria Bacteroides thetaiotaomicron and Fusobacterium varium produce putrescine and spermidine in cecum of pectin-fed gnotobiotic rats. J Nutr 130:1225–1231. [PubMed] [DOI] [PubMed] [Google Scholar]
- 56.Noack J, Kleessen B, Proll J, Dongowski G, Blaut M. 1998. Dietary guar gum and pectin stimulate intestinal microbial polyamine synthesis in rats. J Nutr 128:1385–1391. [PubMed] [DOI] [PubMed] [Google Scholar]
- 57.Cohen SS. 1997. A Guide to the Polyamines. Oxford University Press, New York, NY. [Google Scholar]
- 58.Zhang M, Caragine T, Wang H, Cohen PS, Botchkina G, Soda K, Bianchi M, Ulrich P, Cerami A, Sherry B, Tracey KJ. 1997. Spermine inhibits proinflammatory cytokine synthesis in human mononuclear cells: a counterregulatory mechanism that restrains the immune response. J Exp Med 185:1759–1768 10.1084/jem.185.10.1759. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Rao JN, Bass BL, Wang JY. 2001. NF-kappaB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 280:G992–G1004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Rhee HJ, Kim EJ, Lee JK. 2007. Physiological polyamines: simple primordial stress molecules. J Cell Mol Med 11:685–703 10.1111/j.1582-4934.2007.00077.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pillai SP, Shankel DM. 1997. Polyamines and their potential to be antimutagens. Mutat Res 377:217–224 10.1016/S0027-5107(97)00075-4. [DOI] [PubMed] [Google Scholar]
- 62.Shah N, Thomas T, Shirahata A, Sigal LH, Thomas TJ. 1999. Activation of nuclear factor kappaB by polyamines in breast cancer cells. Biochemistry 38:14763–14774 10.1021/bi991291v. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Soda K, Kano Y, Nakamura T, Kasono K, Kawakami M, Konishi F. 2005. Spermine, a natural polyamine, suppresses LFA-1 expression on human lymphocyte. J Immunol 175:237–245 10.4049/jimmunol.175.1.237. [PubMed] [DOI] [PubMed] [Google Scholar]
- 64.Penrose HM, Marchelletta RR, Krishnan M, McCole DF. 2013. Spermidine stimulates T cell protein-tyrosine phosphatase-mediated protection of intestinal epithelial barrier function. J Biol Chem 288:32651–32662 10.1074/jbc.M113.475962. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Das R, Kanungo MS. 1982. Activity and modulation of ornithine decarboxylase and concentrations of polyamines in various tissues of rats as a function of age. Exp Gerontol 17:95–103 10.1016/0531-5565(82)90042-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 66.Matsumoto M, Benno Y. 2007. The relationship between microbiota and polyamine concentration in the human intestine: a pilot study. Microbiol Immunol 51:25–35 10.1111/j.1348-0421.2007.tb03887.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 67.Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y. 2011. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS One 6:e23652 10.1371/journal.pone.0023652. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kibe R, Kurihara S, Sakai Y, Suzuki H, Ooga T, Sawaki E, Muramatsu K, Nakamura A, Yamashita A, Kitada Y, Kakeyama M, Benno Y, Matsumoto M. 2014. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep 4:4548 10.1038/srep04548. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsumoto M, Aranami A, Ishige A, Watanabe K, Benno Y. 2007. LKM512 yogurt consumption improves the intestinal environment and induces the T-helper type 1 cytokine in adult patients with intractable atopic dermatitis. Clin Exp Allergy 37:358–370 10.1111/j.1365-2222.2007.02642.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Matsumoto M, Ohishi H, Benno Y. 2001. Impact of LKM512 yogurt on improvement of intestinal environment of the elderly. FEMS Immunol Med Microbiol 31:181–186 10.1111/j.1574-695X.2001.tb00518.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 71.Rider JE, Hacker A, Mackintosh CA, Pegg AE, Woster PM, Casero RA Jr. 2007. Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 33:231–240 10.1007/s00726-007-0513-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Clarke CH, Shankel DM. 1988. Antimutagens against spontaneous and induced reversion of a lacZ frameshift mutation in E. coli K-12 strain ND-160. Mutat Res 202:19–23 10.1016/0027-5107(88)90158-3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Clarke CH, Shankel DM. 1989. Antimutagenic specificity against spontaneous and nitrofurazone-induced mutations in Escherichia coli K12ND160. Mutagenesis 4:31–34 10.1093/mutage/4.1.31. [PubMed] [DOI] [PubMed] [Google Scholar]
- 74.Nestmann ER. 1977. Antimutagenic effects of spermine and guanosine in continuous cultures of Escherichia coli mutator strain mutH. Mol Gen Genet 152:109–110 10.1007/BF00264947. [PubMed] [DOI] [PubMed] [Google Scholar]
- 75.Lahue RS, Au KG, Modrich P. 1989. DNA mismatch correction in a defined system. Science 245:160–164 10.1126/science.2665076. [PubMed] [DOI] [PubMed] [Google Scholar]
- 76.Gómez-Gallego C, Collado MC, Pérez G, Ilo T, Jaakkola UM, Bernal MJ, Periago MJ, Frias R, Ros G, Salminen S. 2014. Resembling breast milk: influence of polyamine-supplemented formula on neonatal BALB/cOlaHsd mouse microbiota. Br J Nutr 111:1050–1058 10.1017/S0007114513003565. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Maurelli AT, Fernández RE, Bloch CA, Rode CK, Fasano A. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA 95:3943–3948 10.1073/pnas.95.7.3943. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldman ME, Cregar L, Nguyen D, Simo O, O’Malley S, Humphreys T. 2006. Cationic polyamines inhibit anthrax lethal factor protease. BMC Pharmacol 6:8 10.1186/1471-2210-6-8. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernandez IM, Silva M, Schuch R, Walker WA, Siber AM, Maurelli AT, McCormick BA. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J Infect Dis 184:743–753 10.1086/323035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 80.Torres AG, Vazquez-Juarez RC, Tutt CB, Garcia-Gallegos JG. 2005. Pathoadaptive mutation that mediates adherence of shiga toxin-producing Escherichia coli O111. Infect Immun 73:4766–4776 10.1128/IAI.73.8.4766-4776.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casero RA Jr, Marton LJ. 2007. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 6:373–390 10.1038/nrd2243. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Gerner EW, Meyskens FL Jr. 2004. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 4:781–792 10.1038/nrc1454. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Alam K, Arlow FL, Ma CK, Schubert TT. 1994. Decrease in ornithine decarboxylase activity after eradication of Helicobacter pylori. Am J Gastroenterol 89:888–893. [PubMed] [PubMed] [Google Scholar]
- 84.Patchett SE, Katelaris PH, Zhang ZW, Alstead EM, Domizio P, Farthing MJ. 1996. Ornithine decarboxylase activity is a marker of premalignancy in longstanding Helicobacter pylori infection. Gut 39:807–810 10.1136/gut.39.6.807. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu S, Ramanujam KS, Wong A, Fantry GT, Drachenberg CB, James SP, Meltzer SJ, Wilson KT. 1999. Increased expression and cellular localization of inducible nitric oxide synthase and cyclooxygenase 2 in Helicobacter pylori gastritis. Gastroenterology 116:1319–1329 10.1016/S0016-5085(99)70496-8. [DOI] [PubMed] [Google Scholar]
- 86.Keszthelyi D, Troost FJ, Masclee AA. 2009. Understanding the role of tryptophan and serotonin metabolism in gastrointestinal function. Neurogastroenterol Motil 21:1239–1249 10.1111/j.1365-2982.2009.01370.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Yanofsky C, Horn V, Gollnick P. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J Bacteriol 173:6009–6017 10.1128/jb.173.19.6009-6017.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aragozzini F, Ferrari A, Pacini N, Gualandris R. 1979. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl Environ Microbiol 38:544–546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith EA, Macfarlane GT. 1997. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol 33:180–188 10.1007/s002489900020. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA 107:228–233 10.1073/pnas.0906112107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bommarius B, Anyanful A, Izrayelit Y, Bhatt S, Cartwright E, Wang W, Swimm AI, Benian GM, Schroeder FC, Kalman D. 2013. A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PLoS One 8:e54456 10.1371/journal.pone.0054456. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. 2013. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604 10.1371/journal.pone.0080604. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li YH, Tian X. 2012. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel) 12:2519–2538 10.3390/s120302519. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Davey ME, O’Toole GA. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64:847–867 10.1128/MMBR.64.4.847-867.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watnick P, Kolter R. 2000. Biofilm, city of microbes. J Bacteriol 182:2675–2679 10.1128/JB.182.10.2675-2679.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199 10.1146/annurev.micro.55.1.165. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33 10.1016/j.tim.2004.11.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 98.Thompson JA, Oliveira RA, Xavier KB. 2016. Chemical conversations in the gut microbiota. Gut Microbes 7:163–170 10.1080/19490976.2016.1145374. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346 10.1146/annurev.cellbio.21.012704.131001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Cvitkovitch DG, Li YH, Ellen RP. 2003. Quorum sensing and biofilm formation in streptococcal infections. J Clin Invest 112:1626–1632 10.1172/JCI200320430. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Federle MJ, Bassler BL. 2003. Interspecies communication in bacteria. J Clin Invest 112:1291–1299 10.1172/JCI20195. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schauder S, Bassler BL. 2001. The languages of bacteria. Genes Dev 15:1468–1480 10.1101/gad.899601. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Fuqua C, Greenberg EP. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol 3:685–695 10.1038/nrm907. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Parsek MR, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP. 1999. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci USA 96:4360–4365 10.1073/pnas.96.8.4360. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Kievit TR, Iglewski BH. 2000. Bacterial quorum sensing in pathogenic relationships. Infect Immun 68:4839–4849 10.1128/IAI.68.9.4839-4849.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dunny GM, Leonard BA. 1997. Cell-cell communication in Gram-positive bacteria. Annu Rev Microbiol 51:527–564 10.1146/annurev.micro.51.1.527. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449 10.1046/j.1365-2958.2003.03526.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu Rev Microbiol 60:451–475 10.1146/annurev.micro.60.080805.142139. [PubMed] [DOI] [PubMed] [Google Scholar]
- 109.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606 10.1111/j.1365-2958.2010.07361.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol 80:1102–1119 10.1111/j.1365-2958.2011.07633.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192:1444–1454 10.1128/JB.01251-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549 10.1038/415545a. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Rezzonico F, Smits TH, Duffy B. 2012. Detection of AI-2 receptors in genomes of Enterobacteriaceae suggests a role of type-2 quorum sensing in closed ecosystems. Sensors (Basel) 12:6645–6665 10.3390/s120506645. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112:1466–1477 10.1172/JCI200320365. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.von Rosenvinge EC, O’May GA, Macfarlane S, Macfarlane GT, Shirtliff ME. 2013. Microbial biofilms and gastrointestinal diseases. Pathog Dis 67:25–38 10.1111/2049-632X.12020. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johansson ME, Larsson JM, Hansson GC. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA 108(Suppl 1):4659–4665 10.1073/pnas.1006451107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pullan RD, Thomas GA, Rhodes M, Newcombe RG, Williams GT, Allen A, Rhodes J. 1994. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 35:353–359 10.1136/gut.35.3.353. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Macfarlane S, Woodmansey EJ, Macfarlane GT. 2005. Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol 71:7483–7492 10.1128/AEM.71.11.7483-7492.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Holmén Larsson JM, Karlsson H, Sjövall H, Hansson GC. 2009. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology 19:756–766 10.1093/glycob/cwp048. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Engevik MA, Aihara E, Montrose MH, Shull GE, Hassett DJ, Worrell RT. 2013. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol 305:G697–G711 10.1152/ajpgi.00184.2013. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Engevik MA, Hickerson A, Shull GE, Worrell RT. 2013. Acidic conditions in the NHE2(-/-) mouse intestine result in an altered mucosa-associated bacterial population with changes in mucus oligosaccharides. Cell Physiol Biochem 32:111–128 10.1159/000356632. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Engevik MA, Yacyshyn MB, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol 308:G510–G524 10.1152/ajpgi.00091.2014. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. 2013. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23:1038–1046 10.1093/glycob/cwt040. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ahmed FE. 2003. Genetically modified probiotics in foods. Trends Biotechnol 21:491–497 10.1016/j.tibtech.2003.09.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis 38:1690–1699 10.1086/420823. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Lebeer S, Verhoeven TL, Claes IJ, De Hertogh G, Vermeire S, Buyse J, Van Immerseel F, Vanderleyden J, De Keersmaecker SC. 2011. FISH analysis of Lactobacillus biofilms in the gastrointestinal tract of different hosts. Lett Appl Microbiol 52:220–226 10.1111/j.1472-765X.2010.02994.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 127.Macfarlane S, Bahrami B, Macfarlane GT. 2011. Mucosal biofilm communities in the human intestinal tract. Adv Appl Microbiol 75:111–143 10.1016/B978-0-12-387046-9.00005-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224 10.1111/j.1574-6976.2008.00150.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 129.Rickard AH, Palmer RJ Jr, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol 60:1446–1456 10.1111/j.1365-2958.2006.05202.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Merritt J, Qi F, Goodman SD, Anderson MH, Shi W. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect Immun 71:1972–1979 10.1128/IAI.71.4.1972-1979.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. 2011. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun 79:4550–4558 10.1128/IAI.05644-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vidal JE, Ludewick HP, Kunkel RM, Zähner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060 10.1128/IAI.05186-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tannock GW, Ghazally S, Walter J, Loach D, Brooks H, Cook G, Surette M, Simmers C, Bremer P, Dal Bello F, Hertel C. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl Environ Microbiol 71:8419–8425 10.1128/AEM.71.12.8419-8425.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599 10.1128/AEM.72.5.3593-3599.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 186:2099–2106 10.1128/JB.186.7.2099-2106.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Macfarlane GT, Macfarlane S. 2012. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int 95:50–60 10.5740/jaoacint.SGE_Macfarlane. [PubMed] [DOI] [PubMed] [Google Scholar]
- 137.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:185 10.3389/fmicb.2016.00185. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28:1221–1227 10.1136/gut.28.10.1221. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim CH, Park J, Kim M. 2014. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw 14:277–288 10.4110/in.2014.14.6.277. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Annison G, Illman RJ, Topping DL. 2003. Acetylated, propionylated or butyrylated starches raise large bowel short-chain fatty acids preferentially when fed to rats. J Nutr 133:3523–3528. [PubMed] [DOI] [PubMed] [Google Scholar]
- 141.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. 2009. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 58:1509–1517 10.2337/db08-1637. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cherrington CA, Hinton M, Chopra I. 1990. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J Appl Bacteriol 68:69–74 10.1111/j.1365-2672.1990.tb02550.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 143.Prohászka L, Jayarao BM, Fábián A, Kovács S. 1990. The role of intestinal volatile fatty acids in the Salmonella shedding of pigs. Zentralbl Veterinarmed B 37:570–574. [DOI] [PubMed] [Google Scholar]
- 144.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68:5186–5190 10.1128/AEM.68.10.5186-5190.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, Flint HJ. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr 91:915–923 10.1079/BJN20041150. [PubMed] [DOI] [PubMed] [Google Scholar]
- 146.den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud DJ, Bakker BM. 2015. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 64:2398–2408 10.2337/db14-1213. [PubMed] [DOI] [PubMed] [Google Scholar]
- 147.Yanase H, Takebe K, Nio-Kobayashi J, Takahashi-Iwanaga H, Iwanaga T. 2008. Cellular expression of a sodium-dependent monocarboxylate transporter (Slc5a8) and the MCT family in the mouse kidney. Histochem Cell Biol 130:957–966 10.1007/s00418-008-0490-z. [PubMed] [DOI] [PubMed] [Google Scholar]
- 148.Miyauchi S, Gopal E, Babu E, Srinivas SR, Kubo Y, Umapathy NS, Thakkar SV, Ganapathy V, Prasad PD. 2010. Sodium-coupled electrogenic transport of pyroglutamate (5-oxoproline) via SLC5A8, a monocarboxylate transporter. Biochim Biophys Acta 1798:1164–1171 10.1016/j.bbamem.2010.03.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 149.Halestrap AP, Wilson MC. 2012. The monocarboxylate transporter family: role and regulation. IUBMB Life 64:109–119 10.1002/iub.572. [PubMed] [DOI] [PubMed] [Google Scholar]
- 150.Karaki S, Mitsui R, Hayashi H, Kato I, Sugiya H, Iwanaga T, Furness JB, Kuwahara A. 2006. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res 324:353–360 10.1007/s00441-005-0140-x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 151.Sleeth ML, Thompson EL, Ford HE, Zac-Varghese SE, Frost G. 2010. Free fatty acid receptor 2 and nutrient sensing: a proposed role for fibre, fermentable carbohydrates and short-chain fatty acids in appetite regulation. Nutr Res Rev 23:135–145 10.1017/S0954422410000089. [PubMed] [DOI] [PubMed] [Google Scholar]
- 152.Eberle JA, Widmayer P, Breer H. 2014. Receptors for short-chain fatty acids in brush cells at the “gastric groove”. Front Physiol 5:152 10.3389/fphys.2014.00152. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. 2008. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol 59(Suppl 2):251–262. [PubMed] [PubMed] [Google Scholar]
- 154.Nøhr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S, Jones RM, Offermanns S, Schwartz TW. 2013. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 154:3552–3564 10.1210/en.2013-1142. [PubMed] [DOI] [PubMed] [Google Scholar]
- 155.Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. 2004. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci USA 101:1045–1050 10.1073/pnas.2637002100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zaibi MS, Stocker CJ, O’Dowd J, Davies A, Bellahcene M, Cawthorne MA, Brown AJ, Smith DM, Arch JR. 2010. Roles of GPR41 and GPR43 in leptin secretory responses of murine adipocytes to short chain fatty acids. FEBS Lett 584:2381–2386 10.1016/j.febslet.2010.04.027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 157.Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, Franke A, Ott S, Häsler R, Nikolaus S, Fölsch UR, Rose-John S, Jiang HP, Li J, Schreiber S, Rosenstiel P. 2009. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 183:7514–7522 10.4049/jimmunol.0900063. [PubMed] [DOI] [PubMed] [Google Scholar]
- 158.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ. 2003. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 278:11312–11319 10.1074/jbc.M211609200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.Voltolini C, Battersby S, Etherington SL, Petraglia F, Norman JE, Jabbour HN. 2012. A novel antiinflammatory role for the short-chain fatty acids in human labor. Endocrinology 153:395–403 10.1210/en.2011-1457. [PubMed] [DOI] [PubMed] [Google Scholar]
- 160.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450 10.1038/nature12721. [PubMed] [DOI] [PubMed] [Google Scholar]
- 161.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476 10.1016/j.tim.2012.07.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 162.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40:128–139 10.1016/j.immuni.2013.12.007. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547 10.1038/nature09646. [PubMed] [DOI] [PubMed] [Google Scholar]
- 164.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573 10.1126/science.1241165. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455 10.1038/nature12726. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Arpaia N, Rudensky AY. 2014. Microbial metabolites control gut inflammatory responses. Proc Natl Acad Sci USA 111:2058–2059 10.1073/pnas.1323183111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ishiguro K, Ando T, Maeda O, Watanabe O, Goto H. 2011. Cutting edge: tubulin α functions as an adaptor in NFAT-importin β interaction. J Immunol 186:2710–2713 10.4049/jimmunol.1003322. [PubMed] [DOI] [PubMed] [Google Scholar]
- 168.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, Langella P, Thomas M. 2013. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11:61 10.1186/1741-7007-11-61. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Levison ME. 1973. Effect of colon flora and short-chain fatty acids on growth in vitro of Pseudomonas aeruginsoa and Enterobacteriaceae. Infect Immun 8:30–35. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shin R, Suzuki M, Morishita Y. 2002. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J Med Microbiol 51:201–206 10.1099/0022-1317-51-3-201. [PubMed] [DOI] [PubMed] [Google Scholar]
- 171.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286 10.1038/nature08530. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80 10.1016/j.ijfoodmicro.2011.03.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 173.Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. 2014. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8:1323–1335 10.1038/ismej.2014.14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. 2014. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 5:e01438-14 10.1128/mBio.01438-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. 2013. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87:1045–1060 10.1111/mmi.12149. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lawhon SD, Maurer R, Suyemoto M, Altier C. 2002. Intestinal short-chain fatty acids alter Salmonella Typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46:1451–1464 10.1046/j.1365-2958.2002.03268.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.Garner CD, Antonopoulos DA, Wagner B, Duhamel GE, Keresztes I, Ross DA, Young VB, Altier C. 2009. Perturbation of the small intestine microbial ecology by streptomycin alters pathology in a Salmonella enterica serovar Typhimurium murine model of infection. Infect Immun 77:2691–2702 10.1128/IAI.01570-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Durant JA, Corrier DE, Ricke SC. 2000. Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella Typhimurium. J Food Prot 63:573–578 10.4315/0362-028X-63.5.573. [PubMed] [DOI] [PubMed] [Google Scholar]
- 179.Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly-Y M, Stephens L, Hawkins PT, Curi R. 2011. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One 6:e21205 10.1371/journal.pone.0021205. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. 2011. Regulation of inflammation by short chain fatty acids. Nutrients 3:858–876 10.3390/nu3100858. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Anini Y, Fu-Cheng X, Cuber JC, Kervran A, Chariot J, Roz C. 1999. Comparison of the postprandial release of peptide YY and proglucagon-derived peptides in the rat. Pflugers Arch 438:299–306 10.1007/s004240050913. [PubMed] [DOI] [PubMed] [Google Scholar]
- 182.Cherbut C, Ferrier L, Rozé C, Anini Y, Blottière H, Lecannu G, Galmiche JP. 1998. Short-chain fatty acids modify colonic motility through nerves and polypeptide YY release in the rat. Am J Physiol 275:G1415–G1422. [PubMed] [DOI] [PubMed] [Google Scholar]
- 183.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. 2012. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61:364–371 10.2337/db11-1019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chambers ES, Viardot A, Psichas A, Morrison DJ, Murphy KG, Zac-Varghese SE, MacDougall K, Preston T, Tedford C, Finlayson GS, Blundell JE, Bell JD, Thomas EL, Mt-Isa S, Ashby D, Gibson GR, Kolida S, Dhillo WS, Bloom SR, Morley W, Clegg S, Frost G. 2015. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut 64:1744–1754 10.1136/gutjnl-2014-307913. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Murphy KG, Bloom SR. 2006. Gut hormones and the regulation of energy homeostasis. Nature 444:854–859 10.1038/nature05484. [PubMed] [DOI] [PubMed] [Google Scholar]
- 186.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672 10.1038/nrmicro3344. [PubMed] [DOI] [PubMed] [Google Scholar]
- 187.Louis P, Scott KP, Duncan SH, Flint HJ. 2007. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol 102:1197–1208 10.1111/j.1365-2672.2007.03322.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol 9:1101–1111 10.1111/j.1462-2920.2007.01281.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 189.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, Prasad PD, Ganapathy V. 2009. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res 69:2826–2832 10.1158/0008-5472.CAN-08-4466. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. 2013. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol 13:869–874 10.1016/j.coph.2013.08.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 191.Fung KY, Cosgrove L, Lockett T, Head R, Topping DL. 2012. A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br J Nutr 108:820–831 10.1017/S0007114512001948. [PubMed] [DOI] [PubMed] [Google Scholar]
- 192.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119 10.1111/j.1365-2036.2007.03562.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 193.Buda A, Qualtrough D, Jepson MA, Martines D, Paraskeva C, Pignatelli M. 2003. Butyrate downregulates alpha2beta1 integrin: a possible role in the induction of apoptosis in colorectal cancer cell lines. Gut 52:729–734 10.1136/gut.52.5.729. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clarke JM, Topping DL, Bird AR, Young GP, Cobiac L. 2008. Effects of high-amylose maize starch and butyrylated high-amylose maize starch on azoxymethane-induced intestinal cancer in rats. Carcinogenesis 29:2190–2194 10.1093/carcin/bgn192. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chang PV, Hao L, Offermanns S, Medzhitov R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA 111:2247–2252 10.1073/pnas.1322269111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wilson AJ, Chueh AC, Tögel L, Corner GA, Ahmed N, Goel S, Byun DS, Nasser S, Houston MA, Jhawer M, Smartt HJ, Murray LB, Nicholas C, Heerdt BG, Arango D, Augenlicht LH, Mariadason JM. 2010. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res 70:609–620 10.1158/0008-5472.CAN-09-2327. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shapiro H, Thaiss CA, Levy M, Elinav E. 2014. The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol 30:54–62 10.1016/j.coi.2014.07.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 198.Devillard E, McIntosh FM, Duncan SH, Wallace RJ. 2007. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol 189:2566–2570 10.1128/JB.01359-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.McIntosh FM, Shingfield KJ, Devillard E, Russell WR, Wallace RJ. 2009. Mechanism of conjugated linoleic acid and vaccenic acid formation in human faecal suspensions and pure cultures of intestinal bacteria. Microbiology 155:285–294 10.1099/mic.0.022921-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Gorissen L, Raes K, Weckx S, Dannenberger D, Leroy F, De Vuyst L, De Smet S. 2010. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl Microbiol Biotechnol 87:2257–2266 10.1007/s00253-010-2713-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 201.Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J, Kiyono H, Iwamoto R, Isobe Y, Arita M, Arai H, Ueda K, Shima J, Takahashi S, Yokozeki K, Shimizu S, Ogawa J. 2013. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA 110:17808–17813 10.1073/pnas.1312937110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wall R, Ross RP, Shanahan F, O’Mahony L, O’Mahony C, Coakley M, Hart O, Lawlor P, Quigley EM, Kiely B, Fitzgerald GF, Stanton C. 2009. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am J Clin Nutr 89:1393–1401 10.3945/ajcn.2008.27023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 203.Gudbrandsen OA, Rodríguez E, Wergedahl H, Mørk S, Reseland JE, Skorve J, Palou A, Berge RK. 2009. Trans-10, cis-12-conjugated linoleic acid reduces the hepatic triacylglycerol content and the leptin mRNA level in adipose tissue in obese Zucker fa/fa rats. Br J Nutr 102:803–815 10.1017/S0007114509297200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 204.Toomey S, Harhen B, Roche HM, Fitzgerald D, Belton O. 2006. Profound resolution of early atherosclerosis with conjugated linoleic acid. Atherosclerosis 187:40–49 10.1016/j.atherosclerosis.2005.08.024. [PubMed] [DOI] [PubMed] [Google Scholar]
- 205.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. 2004. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol 5:104–112 10.1038/ni1018. [PubMed] [DOI] [PubMed] [Google Scholar]
- 206.Are A, Aronsson L, Wang S, Greicius G, Lee YK, Gustafsson JA, Pettersson S, Arulampalam V. 2008. Enterococcus faecalis from newborn babies regulate endogenous PPARγ activity and IL-10 levels in colonic epithelial cells. Proc Natl Acad Sci USA 105:1943–1948 10.1073/pnas.0711734105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Moya-Camarena SY, Vanden Heuvel JP, Blanchard SG, Leesnitzer LA, Belury MA. 1999. Conjugated linoleic acid is a potent naturally occurring ligand and activator of PPARalpha. J Lipid Res 40:1426–1433. [PubMed] [PubMed] [Google Scholar]
- 208.Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, Nagy L, Yamamoto K, Schwabe JW. 2008. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat Struct Mol Biol 15:924–931 10.1038/nsmb.1474. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chen P, Torralba M, Tan J, Embree M, Zengler K, Starkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, Bajaj JS, Mutlu EA, Keshavarzian A, Tsukamoto H, Nelson KE, Fouts DE, Schnabl B. 2015. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology 148:203–214.e216. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hill MJ. 1997. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev 6(Suppl 1):S43–S45 10.1097/00008469-199703001-00009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 211.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 10.1126/science.1124234. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Brestoff JR, Artis D. 2013. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 14:676–684 10.1038/ni.2640. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Said HM, Mohammed ZM. 2006. Intestinal absorption of water-soluble vitamins: an update. Curr Opin Gastroenterol 22:140–146 10.1097/01.mog.0000203870.22706.52. [PubMed] [DOI] [PubMed] [Google Scholar]
- 214.Ichihashi T, Takagishi Y, Uchida K, Yamada H. 1992. Colonic absorption of menaquinone-4 and menaquinone-9 in rats. J Nutr 122:506–512. [PubMed] [DOI] [PubMed] [Google Scholar]
- 215.Bhaskaram P. 2002. Micronutrient malnutrition, infection, and immunity: an overview. Nutr Rev 60(suppl 5):S40–S45 10.1301/00296640260130722. [PubMed] [DOI] [PubMed] [Google Scholar]
- 216.Cheng CH, Chang SJ, Lee BJ, Lin KL, Huang YC. 2006. Vitamin B6 supplementation increases immune responses in critically ill patients. Eur J Clin Nutr 60:1207–1213 10.1038/sj.ejcn.1602439. [PubMed] [DOI] [PubMed] [Google Scholar]
- 217.Meydani SN, Meydani M, Blumberg JB, Leka LS, Siber G, Loszewski R, Thompson C, Pedrosa MC, Diamond RD, Stollar BD. 1997. Vitamin E supplementation and in vivo immune response in healthy elderly subjects. A randomized controlled trial. JAMA 277:1380–1386 10.1001/jama.1997.03540410058031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 218.Tamura J, Kubota K, Murakami H, Sawamura M, Matsushima T, Tamura T, Saitoh T, Kurabayshi H, Naruse T. 1999. Immunomodulation by vitamin B12: augmentation of CD8+ T lymphocytes and natural killer (NK) cell activity in vitamin B12-deficient patients by methyl-B12 treatment. Clin Exp Immunol 116:28–32 10.1046/j.1365-2249.1999.00870.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.LeBlanc JG, Laiño JE, del Valle MJ, Vannini V, van Sinderen D, Taranto MP, de Valdez GF, de Giori GS, Sesma F. 2011. B-group vitamin production by lactic acid bacteria: current knowledge and potential applications. J Appl Microbiol 111:1297–1309 10.1111/j.1365-2672.2011.05157.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 220.Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu Rev Nutr 20:153–167 10.1146/annurev.nutr.20.1.153. [PubMed] [DOI] [PubMed] [Google Scholar]
- 221.Bacher A, Fischer M, Kis K, Kugelbrey K, Mörtl S, Scheuring J, Weinkauf S, Eberhardt S, Schmidt-Bäse K, Huber R, Ritsert K, Cushman M, Ladenstein R. 1996. Biosynthesis of riboflavin: structure and mechanism of lumazine synthase. Biochem Soc Trans 24:89–94 10.1042/bst0240089. [PubMed] [DOI] [PubMed] [Google Scholar]
- 222.Capozzi V, Menga V, Digesu AM, De Vita P, van Sinderen D, Cattivelli L, Fares C, Spano G. 2011. Biotechnological production of vitamin B2-enriched bread and pasta. J Agric Food Chem 59:8013–8020 10.1021/jf201519h. [PubMed] [DOI] [PubMed] [Google Scholar]
- 223.LeBlanc JG, Burgess C, Sesma F, de Giori GS, van Sinderen D. 2005. Lactococcus lactis is capable of improving the riboflavin status in deficient rats. Br J Nutr 94:262–267 10.1079/BJN20051473. [PubMed] [DOI] [PubMed] [Google Scholar]
- 224.LeBlanc JG, Burgess C, Sesma F, Savoy de Giori G, van Sinderen D. 2005. Ingestion of milk fermented by genetically modified Lactococcus lactis improves the riboflavin status of deficient rats. J Dairy Sci 88:3435–3442 10.3168/jds.S0022-0302(05)73027-7. [DOI] [PubMed] [Google Scholar]
- 225.Burgess C, O’Connell-Motherway M, Sybesma W, Hugenholtz J, van Sinderen D. 2004. Riboflavin production in Lactococcus lactis: potential for in situ production of vitamin-enriched foods. Appl Environ Microbiol 70:5769–5777 10.1128/AEM.70.10.5769-5777.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sydenstricker VP. 1941. Clinical manifestations of ariboflavinosis. Am J Public Health Nations Health 31:344–350 10.2105/AJPH.31.4.344. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Fabian E, Majchrzak D, Dieminger B, Meyer E, Elmadfa I. 2008. Influence of probiotic and conventional yoghurt on the status of vitamins B1, B2 and B6 in young healthy women. Ann Nutr Metab 52:29–36 10.1159/000114408. [PubMed] [DOI] [PubMed] [Google Scholar]
- 228.LeBlanc JG, Sybesma W, Starrenburg M, Sesma F, de Vos WM, de Giori GS, Hugenholtz J. 2010. Supplementation with engineered Lactococcus lactis improves the folate status in deficient rats. Nutrition 26:835–841 10.1016/j.nut.2009.06.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.LeBlanc JG, Aubry C, Cortes-Perez NG, de Moreno de LeBlanc A, Vergnolle N, Langella P, Azevedo V, Chatel JM, Miyoshi A, Bermúdez-Humarán LG. 2013. Mucosal targeting of therapeutic molecules using genetically modified lactic acid bacteria: an update. FEMS Microbiol Lett 344:1–9 10.1111/1574-6968.12159. [PubMed] [DOI] [PubMed] [Google Scholar]
- 230.Kim TH, Yang J, Darling PB, O’Connor DL. 2004. A large pool of available folate exists in the large intestine of human infants and piglets. J Nutr 134:1389–1394. [PubMed] [DOI] [PubMed] [Google Scholar]
- 231.Thomas CM, Saulnier DM, Spinler JK, Hemarajata P, Gao C, Jones SE, Grimm A, Balderas MA, Burstein MD, Morra C, Roeth D, Kalkum M, Versalovic J. 2016. FolC2-mediated folate metabolism contributes to suppression of inflammation by probiotic Lactobacillus reuteri. MicrobiologyOpen 5:802–818 10.1002/mbo3.371. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Crittenden RG, Martinez NR, Playne MJ. 2003. Synthesis and utilisation of folate by yoghurt starter cultures and probiotic bacteria. Int J Food Microbiol 80:217–222 10.1016/S0168-1605(02)00170-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 233.Sybesma W, Starrenburg M, Kleerebezem M, Mierau I, de Vos WM, Hugenholtz J. 2003. Increased production of folate by metabolic engineering of Lactococcus lactis. Appl Environ Microbiol 69:3069–3076 10.1128/AEM.69.6.3069-3076.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sybesma W, Starrenburg M, Tijsseling L, Hoefnagel MH, Hugenholtz J. 2003. Effects of cultivation conditions on folate production by lactic acid bacteria. Appl Environ Microbiol 69:4542–4548 10.1128/AEM.69.8.4542-4548.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sybesma W, Van Den Born E, Starrenburg M, Mierau I, Kleerebezem M, De Vos WM, Hugenholtz J. 2003. Controlled modulation of folate polyglutamyl tail length by metabolic engineering of Lactococcus lactis. Appl Environ Microbiol 69:7101–7107 10.1128/AEM.69.12.7101-7107.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Wegkamp A, Starrenburg M, de Vos WM, Hugenholtz J, Sybesma W. 2004. Transformation of folate-consuming Lactobacillus gasseri into a folate producer. Appl Environ Microbiol 70:3146–3148 10.1128/AEM.70.5.3146-3148.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Santos F, Wegkamp A, de Vos WM, Smid EJ, Hugenholtz J. 2008. High-level folate production in fermented foods by the B12 producer Lactobacillus reuteri JCM1112. Appl Environ Microbiol 74:3291–3294 10.1128/AEM.02719-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren JP, Cerdeño-Tárraga AM, Parkhill J, Flynn S, O’Sullivan GC, Collins JK, Higgins D, Shanahan F, Fitzgerald GF, van Sinderen D, O’Toole PW. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci USA 103:6718–6723 10.1073/pnas.0511060103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A, Loux V, Dervyn R, Bossy R, Bolotin A, Batto JM, Walunas T, Gibrat JF, Bessières P, Weissenbach J, Ehrlich SD, Maguin E. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci USA 103:9274–9279 10.1073/pnas.0603024103. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pompei A, Cordisco L, Amaretti A, Zanoni S, Matteuzzi D, Rossi M. 2007. Folate production by bifidobacteria as a potential probiotic property. Appl Environ Microbiol 73:179–185 10.1128/AEM.01763-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rossi M, Amaretti A, Raimondi S. 2011. Folate production by probiotic bacteria. Nutrients 3:118–134 10.3390/nu3010118. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. 2007. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol 73:2673–2681 10.1128/AEM.02174-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Quesada-Chanto A, Afschar AS, Wagner F. 1994. Microbial production of propionic acid and vitamin B12 using molasses or sugar. Appl Microbiol Biotechnol 41:378–383. [DOI] [PubMed] [Google Scholar]
- 244.Roth LA, Keenan D. 1971. Acid injury of Escherichia coli. Can J Microbiol 17:1005–1008 10.1139/m71-160. [PubMed] [DOI] [PubMed] [Google Scholar]
- 245.Martens J-H, Barg H, Warren M, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285 10.1007/s00253-001-0902-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 246.Smith AD. 2007. Folic acid fortification: the good, the bad, and the puzzle of vitamin B-12. Am J Clin Nutr 85:3–5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 247.Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol 50:137–181 10.1146/annurev.micro.50.1.137. [PubMed] [DOI] [PubMed] [Google Scholar]
- 248.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem 278:41148–41159 10.1074/jbc.M305837200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 249.Taranto MP, Vera JL, Hugenholtz J, De Valdez GF, Sesma F. 2003. Lactobacillus reuteri CRL1098 produces cobalamin. J Bacteriol 185:5643–5647 10.1128/JB.185.18.5643-5647.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Vannini V, de Valdez G, Taranto MFS. 2008. Identification of new lactobacilli able to produce cobalamin (vitamin B12). Biocell 32:72. [Google Scholar]
- 251.Martin R, Olivares M, Marin ML, Fernandez L, Xaus J, Rodriguez JM. 2005. Probiotic potential of 3 lactobacilli strains isolated from breast milk. J Hum Lact 21:8–17; quiz 18–21, 41. [DOI] [PubMed] [Google Scholar]
- 252.Santos F, Vera JL, Lamosa P, de Valdez GF, de Vos WM, Santos H, Sesma F, Hugenholtz J. 2007. Pseudovitamin B(12) is the corrinoid produced by Lactobacillus reuteri CRL1098 under anaerobic conditions. FEBS Lett 581:4865–4870 10.1016/j.febslet.2007.09.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 253.Hüfner E, Britton RA, Roos S, Jonsson H, Hertel C. 2008. Global transcriptional response of Lactobacillus reuteri to the sourdough environment. Syst Appl Microbiol 31:323–338 10.1016/j.syapm.2008.06.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 254.Hunt A, Harrington D, Robinson S. 2014. Vitamin B12 deficiency. BMJ 349(sep04 1):g5226 10.1136/bmj.g5226. [DOI] [PubMed] [Google Scholar]
- 255.Molina VC, Médici M, Taranto MP, Font de Valdez G. 2009. Lactobacillus reuteri CRL 1098 prevents side effects produced by a nutritional vitamin B deficiency. J Appl Microbiol 106:467–473 10.1111/j.1365-2672.2008.04014.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 256.Olson RE. 1984. The function and metabolism of vitamin K. Annu Rev Nutr 4:281–337 10.1146/annurev.nu.04.070184.001433. [PubMed] [DOI] [PubMed] [Google Scholar]
- 257.Lippi G, Franchini M. 2011. Vitamin K in neonates: facts and myths. Blood Transfus 9:4–9. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Conly JM, Stein K. 1992. Quantitative and qualitative measurements of K vitamins in human intestinal contents. Am J Gastroenterol 87:311–316. [PubMed] [PubMed] [Google Scholar]
- 259.Cooke G, Behan J, Costello M. 2006. Newly identified vitamin K-producing bacteria isolated from the neonatal faecal flora. Microb Ecol Health Dis 18:133–138 10.1080/08910600601048894. [DOI] [Google Scholar]
- 260.Morishita T, Tamura N, Makino T, Kudo S. 1999. Production of menaquinones by lactic acid bacteria. J Dairy Sci 82:1897–1903 10.3168/jds.S0022-0302(99)75424-X. [PubMed] [DOI] [PubMed] [Google Scholar]
- 261.Olsen I, Amano A. 2015. Outer membrane vesicles: offensive weapons or good Samaritans? J Oral Microbiol 7:27468 10.3402/jom.v7.27468. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. 2011. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 6:e27958 10.1371/journal.pone.0027958. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Berleman J, Auer M. 2013. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ Microbiol 15:347–354 10.1111/1462-2920.12048. [PubMed] [DOI] [PubMed] [Google Scholar]
- 264.Mayrand D, Grenier D. 1989. Biological activities of outer membrane vesicles. Can J Microbiol 35:607–613 10.1139/m89-097. [PubMed] [DOI] [PubMed] [Google Scholar]
- 265.Kadurugamuwa JL, Beveridge TJ. 1995. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol 177:3998–4008 10.1128/jb.177.14.3998-4008.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Furuta N, Takeuchi H, Amano A. 2009. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun 77:4761–4770 10.1128/IAI.00841-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330:1768–1773 10.1126/science.1195568. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. 2012. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12:509–520 10.1016/j.chom.2012.08.004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Mazmanian SK, Round JL, Kasper DL. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453:620–625 10.1038/nature07008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 270.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 108(Suppl 1):4615–4622 10.1073/pnas.1000082107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. 2010. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol 185:4101–4108 10.4049/jimmunol.1001443. [PubMed] [DOI] [PubMed] [Google Scholar]
- 272.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155:1451–1463 10.1016/j.cell.2013.11.024. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Rühlmann A, Kukla D, Schwager P, Bartels K, Huber R. 1973. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor: crystal structure determination and stereochemistry of the contact region. J Mol Biol 77:417–436 10.1016/0022-2836(73)90448-8. [DOI] [PubMed] [Google Scholar]
- 274.Potempa J, Korzus E, Travis J. 1994. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem 269:15957–15960. [PubMed] [PubMed] [Google Scholar]
- 275.Turroni F, Foroni E, O’Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, Ventura M. 2010. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol 76:3206–3219 10.1128/AEM.02938-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA 99:14422–14427 10.1073/pnas.212527599. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ivanov D, Emonet C, Foata F, Affolter M, Delley M, Fisseha M, Blum-Sperisen S, Kochhar S, Arigoni F. 2006. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem 281:17246–17252 10.1074/jbc.M601678200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 278.Haandrikman AJ, Kok J, Laan H, Soemitro S, Ledeboer AM, Konings WN, Venema G. 1989. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol 171:2789–2794 10.1128/jb.171.5.2789-2794.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Haandrikman AJ, Kok J, Venema G. 1991. Lactococcal proteinase maturation protein PrtM is a lipoprotein. J Bacteriol 173:4517–4525 10.1128/jb.173.14.4517-4525.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Holck A, Axelsson L, Birkeland SE, Aukrust T, Blom H. 1992. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol 138:2715–2720 10.1099/00221287-138-12-2715. [PubMed] [DOI] [PubMed] [Google Scholar]
- 281.Hoermannsperger G, Clavel T, Hoffmann M, Reiff C, Kelly D, Loh G, Blaut M, Hölzlwimmer G, Laschinger M, Haller D. 2009. Post-translational inhibition of IP-10 secretion in IEC by probiotic bacteria: impact on chronic inflammation. PLoS One 4:e4365 10.1371/journal.pone.0004365. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.von Schillde MA, Hörmannsperger G, Weiher M, Alpert CA, Hahne H, Bäuerl C, van Huynegem K, Steidler L, Hrncir T, Pérez-Martínez G, Kuster B, Haller D. 2012. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11:387–396 10.1016/j.chom.2012.02.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 283.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132:562–575 10.1053/j.gastro.2006.11.022. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Bäuerl C, Pérez-Martínez G, Yan F, Polk DB, Monedero V. 2010. Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J Mol Microbiol Biotechnol 19:231–241 10.1159/000322233. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Yan F, Liu L, Dempsey PJ, Tsai YH, Raines EW, Wilson CL, Cao H, Cao Z, Liu L, Polk DB. 2013. A Lactobacillus rhamnosus GG-derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J Biol Chem 288:30742–30751 10.1074/jbc.M113.492397. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ganesh BP, Klopfleisch R, Loh G, Blaut M. 2013. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One 8:e74963 10.1371/journal.pone.0074963. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405 10.1371/journal.ppat.1004405. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, Sham HP, Ryz N, Huang T, Velcich A, Finlay BB, Chadee K, Vallance BA. 2010. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6:e1000902 10.1371/journal.ppat.1000902. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, Lequin O, Kharrat P, Thomas G, Rainteau D, Aubry C, Breyner N, Afonso C, Lavielle S, Grill JP, Chassaing G, Chatel JM, Trugnan G, Xavier R, Langella P, Sokol H, Seksik P. 2016. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65:415–425 10.1136/gutjnl-2014-307649. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Quévrain E, Maubert MA, Sokol H, Devreese B, Seksik P. 2016. The presence of the anti-inflammatory protein MAM, from Faecalibacterium prausnitzii, in the intestinal ecosystem. Gut 65:882 10.1136/gutjnl-2015-311094. [PubMed] [DOI] [PubMed] [Google Scholar]
- 291.Devi M, Rebecca LJ, Sumathy S. 2013. Bactericidal activity of the lactic acid bacteria Lactobacillus delbreukii. J Chem Pharm Res 5:176–180. [Google Scholar]
- 292.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656 10.1128/MMBR.67.4.593-656.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Alakomi HL, Skyttä E, Saarela M, Mattila-Sandholm T, Latva-Kala K, Helander IM. 2000. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl Environ Microbiol 66:2001–2005 10.1128/AEM.66.5.2001-2005.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ray B, Sandine WE. 1992. Acetic, Propionic, and Lactic Acids of Starter Culture Bacteria as Biopreservatives. CRC Press, Boca Raton, FL. [Google Scholar]
- 295.Kong Y-J, Park B-K, Oh D-H. 2001. Antimicrobial activity of Quercus mongolica leaf ethanol extract and organic acids against food-borne microorganisms. Korean J Food Sci Technol 33:178–183. [Google Scholar]
- 296.Mani-Lópeza E, Garcíaa HS, López-Malo A. 2012. Organic acids as antimicrobials to control Salmonella in meat and poultry products. Food Res Int 45:713–721 10.1016/j.foodres.2011.04.043. [DOI] [Google Scholar]
- 297.Östling CE, Lindgren SE. 1993. Inhibition of enterobacteria and Listeria growth by lactic, acetic and formic acids. J Appl Bacteriol 75:18–24 10.1111/j.1365-2672.1993.tb03402.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 298.Michetti P, Dorta G, Wiesel PH, Brassart D, Verdu E, Herranz M, Felley C, Porta N, Rouvet M, Blum AL, Corthésy-Theulaz I. 1999. Effect of whey-based culture supernatant of Lactobacillus acidophilus (johnsonii) La1 on Helicobacter pylori infection in humans. Digestion 60:203–209 10.1159/000007660. [PubMed] [DOI] [PubMed] [Google Scholar]
- 299.Stanojević-Nikolić S, Dimić G, Mojović L, Pejin J, Djukić-Vuković A, Kocić-Tanackov S. 2016. Antimicrobial activity of lactic acid against pathogen and spoilage microorganisms. J Food Process Preserv 40:990–998. [Google Scholar]
- 300.De Keersmaecker SC, Verhoeven TL, Desair J, Marchal K, Vanderleyden J, Nagy I. 2006. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol Lett 259:89–96 10.1111/j.1574-6968.2006.00250.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 301.Aiba Y, Suzuki N, Kabir AM, Takagi A, Koga Y. 1998. Lactic acid-mediated suppression of Helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am J Gastroenterol 93:2097–2101 10.1111/j.1572-0241.1998.00600.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 302.Lin WH, Lin CK, Sheu SJ, Hwang CF, Ye WT, Hwang WZ, Tsen HY. 2009. Antagonistic activity of spent culture supernatants of lactic acid bacteria against Helicobacter pylori growth and infection in human gastric epithelial AGS cells. J Food Sci 74:M225–M230 10.1111/j.1750-3841.2009.01194.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 303.Fayol-Messaoudi D, Berger CN, Coconnier-Polter MH, Liévin-Le Moal V, Servin AL. 2005. pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar Typhimurium. Appl Environ Microbiol 71:6008–6013 10.1128/AEM.71.10.6008-6013.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Adeniyi BA, Adetoye A, Ayeni FA. 2015. Antibacterial activities of lactic acid bacteria isolated from cow faeces against potential enteric pathogens. Afr Health Sci 15:888–895 10.4314/ahs.v15i3.24. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zheng W, Zhang Y, Lu HM, Li DT, Zhang ZL, Tang ZX, Shi LE. 2015. Antimicrobial activity and safety evaluation of Enterococcus faecium KQ 2.6 isolated from peacock feces. BMC Biotechnol 15:30 10.1186/s12896-015-0151-y. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Fujimura S, Watanabe A, Kimura K, Kaji M. 2012. Probiotic mechanism of Lactobacillus gasseri OLL2716 strain against Helicobacter pylori. J Clin Microbiol 50:1134–1136 10.1128/JCM.06262-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Lau AS, Liong MT. 2014. Lactic acid bacteria and bifidobacteria-inhibited Staphylococcus epidermidis. Wounds 26:121–131. [PubMed] [PubMed] [Google Scholar]
- 308.Watanabe T, Nishio H, Tanigawa T, Yamagami H, Okazaki H, Watanabe K, Tominaga K, Fujiwara Y, Oshitani N, Asahara T, Nomoto K, Higuchi K, Takeuchi K, Arakawa T. 2009. Probiotic Lactobacillus casei strain Shirota prevents indomethacin-induced small intestinal injury: involvement of lactic acid. Am J Physiol Gastrointest Liver Physiol 297:G506–G513 10.1152/ajpgi.90553.2008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 309.Engevik MA, Engevik KA, Yacyshyn MB, Wang J, Hassett DJ, Darien B, Yacyshyn BR, Worrell RT. 2015. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol 308:G497–G509 10.1152/ajpgi.00090.2014. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Niku-Paavola ML, Laitila A, Mattila-Sandholm T, Haikara A. 1999. New types of antimicrobial compounds produced by Lactobacillus plantarum. J Appl Microbiol 86:29–35 10.1046/j.1365-2672.1999.00632.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 311.Ananthaswamy HN, Eisenstark A. 1977. Repair of hydrogen peroxide-induced single-strand breaks in Escherichia coli deoxyribonucleic acid. J Bacteriol 130:187–191. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Freese EB, Gerson J, Taber H, Rhaese HJ, Freese E. 1967. Inactivating DNA alterations induced by peroxides and peroxide-producing agents. Mutat Res 4:517–531 10.1016/0027-5107(67)90038-3. [DOI] [PubMed] [Google Scholar]
- 313.Di Mascio P, Wefers H, Do-Thi HP, Lafleur MV, Sies H. 1989. Singlet molecular oxygen causes loss of biological activity in plasmid and bacteriophage DNA and induces single-strand breaks. Biochim Biophys Acta 1007:151–157 10.1016/0167-4781(89)90033-X. [DOI] [PubMed] [Google Scholar]
- 314.Florence TM. 1986. The production of hydroxyl radical from the reaction between hydrogen peroxide and NADH. J Inorg Biochem 28:33–37 10.1016/0162-0134(86)80021-6. [DOI] [PubMed] [Google Scholar]
- 315.Dahl TA, Midden WR, Hartman PE. 1989. Comparison of killing of Gram-negative and Gram-positive bacteria by pure singlet oxygen. J Bacteriol 171:2188–2194 10.1128/jb.171.4.2188-2194.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Servin AL. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev 28:405–440 10.1016/j.femsre.2004.01.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 317.Pridmore RD, Pittet AC, Praplan F, Cavadini C. 2008. Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti-Salmonella activity. FEMS Microbiol Lett 283:210–215 10.1111/j.1574-6968.2008.01176.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 318.Ito A, Sato Y, Kudo S, Sato S, Nakajima H, Toba T. 2003. The screening of hydrogen peroxide-producing lactic acid bacteria and their application to inactivating psychrotrophic food-borne pathogens. Curr Microbiol 47:231–236 10.1007/s00284-002-3993-1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 319.Siragusa GR, Johnson MG. 1989. Inhibition of Listeria monocytogenes growth by the lactoperoxidase-thiocyanate-H2O2 antimicrobial system. Appl Environ Microbiol 55:2802–2805. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Dahiya RS, Speck ML. 1968. Hydrogen peroxide formation by lactobacilli and its effect on Staphylococcus aureus. J Dairy Sci 51:1568–1572 10.3168/jds.S0022-0302(68)87232-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 321.Watson JA, Schubert J. 1969. Action of hydrogen peroxide on growth inhibition of Salmonella typhimurium. J Gen Microbiol 57:25–34 10.1099/00221287-57-1-25. [PubMed] [DOI] [PubMed] [Google Scholar]
- 322.Atassi F, Brassart D, Grob P, Graf F, Servin AL. 2006. In vitro antibacterial activity of Lactobacillus helveticus strain KS300 against diarrhoeagenic, uropathogenic and vaginosis-associated bacteria. J Appl Microbiol 101:647–654 10.1111/j.1365-2672.2006.02933.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 323.Atassi F, Servin AL. 2010. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol Lett 304:29–38 10.1111/j.1574-6968.2009.01887.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 324.Dubreuil D, Bisaillon JG, Beaudet R. 1984. Inhibition of Neisseria gonorrhoeae growth due to hydrogen peroxide production by urogenital streptococci. Microbios 39:159–167. [PubMed] [PubMed] [Google Scholar]
- 325.Holmberg K, Hallander HO. 1973. Production of bactericidal concentrations of hydrogen peroxide by Streptococcus sanguis. Arch Oral Biol 18:423–434 10.1016/0003-9969(73)90167-2. [DOI] [PubMed] [Google Scholar]
- 326.Hillman JD, Socransky SS, Shivers M. 1985. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol 30:791–795 10.1016/0003-9969(85)90133-5. [DOI] [PubMed] [Google Scholar]
- 327.Barnard JP, Stinson MW. 1996. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect Immun 64:3853–3857. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Barnard JP, Stinson MW. 1999. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect Immun 67:6558–6564. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997 10.1128/IAI.68.7.3990-3997.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001 10.1128/JB.00317-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Klaenhammer TR. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12:39–85 10.1111/j.1574-6976.1993.tb00012.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 332.Klaenhammer TR. 1988. Bacteriocins of lactic acid bacteria. Biochimie 70:337–349 10.1016/0300-9084(88)90206-4. [PubMed] [DOI] [PubMed] [Google Scholar]
- 333.Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl Environ Microbiol 78:1–6 10.1128/AEM.05576-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Czárán TL, Hoekstra RF, Pagie L. 2002. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA 99:786–790 10.1073/pnas.012399899. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Di Cagno R, De Angelis M, Limitone A, Minervini F, Simonetti MC, Buchin S, Gobbetti M. 2007. Cell-cell communication in sourdough lactic acid bacteria: a proteomic study in Lactobacillus sanfranciscensis CB1. Proteomics 7:2430–2446 10.1002/pmic.200700143. [PubMed] [DOI] [PubMed] [Google Scholar]
- 336.Gobbetti M, De Angelis M, Di Cagno R, Minervini F, Limitone A. 2007. Cell-cell communication in food related bacteria. Int J Food Microbiol 120:34–45 10.1016/j.ijfoodmicro.2007.06.012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 337.Majeed H, Gillor O, Kerr B, Riley MA. 2011. Competitive interactions in Escherichia coli populations: the role of bacteriocins. ISME J 5:71–81 10.1038/ismej.2010.90. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Riley MA, Wertz JE. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357–364 10.1016/S0300-9084(02)01421-9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 339.Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun 75:443–451 10.1128/IAI.01775-05. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Chen Y, Ludescher RD, Montville TJ. 1997. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol 63:4770–4777. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Gut IM, Blanke SR, van der Donk WA. 2011. Mechanism of inhibition of Bacillus anthracis spore outgrowth by the lantibiotic nisin. ACS Chem Biol 6:744–752 10.1021/cb1004178. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Li J, Aroutcheva AA, Faro S, Chikindas ML. 2005. Mode of action of lactocin 160, a bacteriocin from vaginal Lactobacillus rhamnosus. Infect Dis Obstet Gynecol 13:135–140 10.1080/10647440500148156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Moll GN, Konings WN, Driessen AJ. 1999. Bacteriocins: mechanism of membrane insertion and pore formation. Antonie van Leeuwenhoek 76:185–198 10.1023/A:1002002718501. [PubMed] [DOI] [PubMed] [Google Scholar]
- 344.van Kraaij C, de Vos WM, Siezen RJ, Kuipers OP, van Kraaij C, de Vos WM, Siezen RJ. 1999. Lantibiotics: biosynthesis, mode of action and applications. Nat Prod Rep 16:575–587 10.1039/a804531c. [PubMed] [DOI] [PubMed] [Google Scholar]
- 345.Guder A, Wiedemann I, Sahl HG. 2000. Posttranslationally modified bacteriocins: the lantibiotics. Biopolymers 55:62–73 . [PubMed] [DOI] [PubMed] [Google Scholar]
- 346.Beuchat LR, Clavero MR, Jaquette CB. 1997. Effects of nisin and temperature on survival, growth, and enterotoxin production characteristics of psychrotrophic Bacillus cereus in beef gravy. Appl Environ Microbiol 63:1953–1958. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Ryan MP, Rea MC, Hill C, Ross RP. 1996. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62:612–619. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Thomas LV, Wimpenny JW. 1996. Investigation of the effect of combined variations in temperature, pH, and NaCl concentration on nisin inhibition of Listeria monocytogenes and Staphylococcus aureus. Appl Environ Microbiol 62:2006–2012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zapico P, Medina M, Gaya P, Nuñez M. 1998. Synergistic effect of nisin and the lactoperoxidase system on Listeria monocytogenes in skim milk. Int J Food Microbiol 40:35–42 10.1016/S0168-1605(98)00008-7. [PubMed] [DOI] [PubMed] [Google Scholar]
- 350.Taylor LY, Cann DD, Welch BJ. 1990. Antibotulinal properties of nisin in fresh fish packaged in an atmosphere of carbon dioxide. J Food Prot 53:953–957 10.4315/0362-028X-53.11.953. [DOI] [PubMed] [Google Scholar]
- 351.Taylor SL, Somers EB, Krueger LA. 1985. Antibotulinal effectiveness of nisin-nitrite combinations in culture medium and chicken frankfurter emulsions. J Food Prot 48:234–239 10.4315/0362-028X-48.3.234. [DOI] [PubMed] [Google Scholar]
- 352.Wijnker JJ, Weerts EA, Breukink EJ, Houben JH, Lipman LJ. 2011. Reduction of Clostridium sporogenes spore outgrowth in natural sausage casings using nisin. Food Microbiol 28:974–979 10.1016/j.fm.2011.01.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 353.Vessoni Penna TC, Moraes DA, Fajardo DN. 2002. The effect of nisin on growth kinetics from activated Bacillus cereus spores in cooked rice and in milk. J Food Prot 65:419–422 10.4315/0362-028X-65.2.419. [PubMed] [DOI] [PubMed] [Google Scholar]
- 354.Beasley SS, Saris PE. 2004. Nisin-producing Lactococcus lactis strains isolated from human milk. Appl Environ Microbiol 70:5051–5053 10.1128/AEM.70.8.5051-5053.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ruhr E, Sahl HG. 1985. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob Agents Chemother 27:841–845 10.1128/AAC.27.5.841. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Gao FH, Abee T, Konings WN. 1991. Mechanism of action of the peptide antibiotic nisin in liposomes and cytochrome c oxidase-containing proteoliposomes. Appl Environ Microbiol 57:2164–2170. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.McAuliffe O, Ryan MP, Ross RP, Hill C, Breeuwer P, Abee T. 1998. Lacticin 3147, a broad-spectrum bacteriocin which selectively dissipates the membrane potential. Appl Environ Microbiol 64:439–445. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Piard JC, Kuipers OP, Rollema HS, Desmazeaud MJ, de Vos WM. 1993. Structure, organization, and expression of the lct gene for lacticin 481, a novel lantibiotic produced by Lactococcus lactis. J Biol Chem 268:16361–16368. [PubMed] [PubMed] [Google Scholar]
- 359.Mørtvedt CI, Nissen-Meyer J, Sletten K, Nes IF. 1991. Purification and amino acid sequence of lactocin S, a bacteriocin produced by Lactobacillus sake L45. Appl Environ Microbiol 57:1829–1834. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Allgaier H, Jung G, Werner RG, Schneider U, Zähner H. 1986. Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur J Biochem 160:9–22 10.1111/j.1432-1033.1986.tb09933.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 361.Kellner R, Jung G, Hörner T, Zähner H, Schnell N, Entian KD, Götz F. 1988. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem 177:53–59 10.1111/j.1432-1033.1988.tb14344.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 362.Choung SY, Kobayashi T, Inoue J, Takemoto K, Ishitsuka H, Inoue K. 1988. Hemolytic activity of a cyclic peptide Ro09-0198 isolated from Streptoverticillium. Biochim Biophys Acta 940:171–179 10.1016/0005-2736(88)90192-7. [DOI] [PubMed] [Google Scholar]
- 363.McAuliffe O, Ross RP, Hill C. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev 25:285–308 10.1111/j.1574-6976.2001.tb00579.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 364.Dunkley EA Jr, Clejan S, Guffanti AA, Krulwich TA. 1988. Large decreases in membrane phosphatidylethanolamine and diphosphatidylglycerol upon mutation to duramycin resistance do not change the protonophore resistance of Bacillus subtilis. Biochim Biophys Acta 943:13–18 10.1016/0005-2736(88)90341-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 365.Sahl HG, Jack RW, Bierbaum G. 1995. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem 230:827–853 10.1111/j.1432-1033.1995.tb20627.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 366.Fredenhagen A, Fendrich G, Märki F, Märki W, Gruner J, Raschdorf F, Peter HH. 1990. Duramycins B and C, two new lanthionine containing antibiotics as inhibitors of phospholipase A2. Structural revision of duramycin and cinnamycin. J Antibiot (Tokyo) 43:1403–1412 10.7164/antibiotics.43.1403. [DOI] [PubMed] [Google Scholar]
- 367.Ennahar S, Deschamps N, Richard J. 2000. Natural variation in susceptibility of Listeria strains to class IIa bacteriocins. Curr Microbiol 41:1–4 10.1007/s002840010081. [PubMed] [DOI] [PubMed] [Google Scholar]
- 368.Ennahar S, Sashihara T, Sonomoto K, Ishizaki A. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol Rev 24:85–106 10.1111/j.1574-6976.2000.tb00534.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 369.Rodríguez JM, Martínez MI, Horn N, Dodd HM. 2003. Heterologous production of bacteriocins by lactic acid bacteria. Int J Food Microbiol 80:101–116 10.1016/S0168-1605(02)00153-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 370.Tichaczek PS, Nissen-Meyer J, Nes IF, Vogel RF, Hammes WP. 1992. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst Appl Microbiol 15:460–468 10.1016/S0723-2020(11)80223-7. [DOI] [Google Scholar]
- 371.Henderson JT, Chopko AL, van Wassenaar PD. 1992. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys 295:5–12 10.1016/0003-9861(92)90480-K. [DOI] [PubMed] [Google Scholar]
- 372.Motlagh AM, Bhunia AK, Szostek F, Hansen TR, Johnson MC, Ray B. 1992. Nucleotide and amino acid sequence of pap-gene (pediocin AcH production) in Pediococcus acidilactici H. Lett Appl Microbiol 15:45–48 10.1111/j.1472-765X.1992.tb00721.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 373.Hastings JW, Sailer M, Johnson K, Roy KL, Vederas JC, Stiles ME. 1991. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol 173:7491–7500 10.1128/jb.173.23.7491-7500.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Héchard Y, Dérijard B, Letellier F, Cenatiempo Y. 1992. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J Gen Microbiol 138:2725–2731 10.1099/00221287-138-12-2725. [PubMed] [DOI] [PubMed] [Google Scholar]
- 375.Métivier A, Pilet MF, Dousset X, Sorokine O, Anglade P, Zagorec M, Piard JC, Marlon D, Cenatiempo Y, Fremaux C. 1998. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology 144:2837–2844 10.1099/00221287-144-10-2837. [PubMed] [DOI] [PubMed] [Google Scholar]
- 376.Aymerich T, Holo H, Håvarstein LS, Hugas M, Garriga M, Nes IF. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol 62:1676–1682. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ferchichi M, Frère J, Mabrouk K, Manai M. 2001. Lactococcin MMFII, a novel class IIa bacteriocin produced by Lactococcus lactis MMFII, isolated from a Tunisian dairy product. FEMS Microbiol Lett 205:49–55 10.1111/j.1574-6968.2001.tb10924.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 378.Yildirim Z, Winters DK, Johnson MG. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J Appl Microbiol 86:45–54 10.1046/j.1365-2672.1999.00629.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 379.Shin MS, Han SK, Ryu JS, Kim KS, Lee WK. 2008. Isolation and partial characterization of a bacteriocin produced by Pediococcus pentosaceus K23-2 isolated from kimchi. J Appl Microbiol 105:331–339 10.1111/j.1365-2672.2008.03770.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 380.Balla E, Dicks LM, Du Toit M, Van Der Merwe MJ, Holzapfel WH. 2000. Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl Environ Microbiol 66:1298–1304 10.1128/AEM.66.4.1298-1304.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Nissen-Meyer J, Håvarstein LS, Holo H, Sletten K, Nes IF. 1993. Association of the lactococcin A immunity factor with the cell membrane: purification and characterization of the immunity factor. J Gen Microbiol 139:1503–1509 10.1099/00221287-139-7-1503. [PubMed] [DOI] [PubMed] [Google Scholar]
- 382.van Belkum MJ, Kok J, Venema G, Holo H, Nes IF, Konings WN, Abee T. 1991. The bacteriocin lactococcin A specifically increases permeability of lactococcal cytoplasmic membranes in a voltage-independent, protein-mediated manner. J Bacteriol 173:7934–7941 10.1128/jb.173.24.7934-7941.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Allison GE, Fremaux C, Klaenhammer TR. 1994. Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J Bacteriol 176:2235–2241 10.1128/jb.176.8.2235-2241.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Jiménez-Díaz R, Ruiz-Barba JL, Cathcart DP, Holo H, Nes IF, Sletten KH, Warner PJ. 1995. Purification and partial amino acid sequence of plantaricin S, a bacteriocin produced by Lactobacillus plantarum LPCO10, the activity of which depends on the complementary action of two peptides. Appl Environ Microbiol 61:4459–4463. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Anderssen EL, Diep DB, Nes IF, Eijsink VG, Nissen-Meyer J. 1998. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl Environ Microbiol 64:2269–2272. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Davey GP, Richardson BC. 1981. Purification and some properties of diplococcin from Streptococcus cremoris 346. Appl Environ Microbiol 41:84–89. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Herranz C, Chen Y, Chung HJ, Cintas LM, Hernández PE, Montville TJ, Chikindas ML. 2001. Enterocin P selectively dissipates the membrane potential of Enterococcus faecium T136. Appl Environ Microbiol 67:1689–1692 10.1128/AEM.67.4.1689-1692.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Moll G, Ubbink-Kok T, Hildeng-Hauge H, Nissen-Meyer J, Nes IF, Konings WN, Driessen AJ. 1996. Lactococcin G is a potassium ion-conducting, two-component bacteriocin. J Bacteriol 178:600–605 10.1128/jb.178.3.600-605.1996. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.González C, Langdon GM, Bruix M, Gálvez A, Valdivia E, Maqueda M, Rico M. 2000. Bacteriocin AS-48, a microbial cyclic polypeptide structurally and functionally related to mammalian NK-lysin. Proc Natl Acad Sci USA 97:11221–11226 10.1073/pnas.210301097. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Leer RJ, van der Vossen JM, van Giezen M, van Noort JM, Pouwels PH. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629–1635 10.1099/13500872-141-7-1629. [PubMed] [DOI] [PubMed] [Google Scholar]
- 391.Worobo RW, Henkel T, Sailer M, Roy KL, Vederas JC, Stiles ME. 1994. Characteristics and genetic determinant of a hydrophobic peptide bacteriocin, carnobacteriocin A, produced by Carnobacterium piscicola LV17A. Microbiology 140:517–526 10.1099/00221287-140-3-517. [PubMed] [DOI] [PubMed] [Google Scholar]
- 392.Cintas LM, Casaus P, Håvarstein LS, Hernández PE, Nes IF. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol 63:4321–4330. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Nes IF, Diep DB, Håvarstein LS, Brurberg MB, Eijsink V, Holo H. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek 70:113–128 10.1007/BF00395929. [PubMed] [DOI] [PubMed] [Google Scholar]
- 394.Holo H, Nilssen O, Nes IF. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol 173:3879–3887 10.1128/jb.173.12.3879-3887.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Sandiford S, Upton M. 2012. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against staphylococci. Antimicrob Agents Chemother 56:1539–1547 10.1128/AAC.05397-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.de Lorenzo V. 1985. Factors affecting microcin E492 production. J Antibiot (Tokyo) 38:340–345 10.7164/antibiotics.38.340. [DOI] [PubMed] [Google Scholar]
- 397.de Lorenzo V, Pugsley AP. 1985. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob Agents Chemother 27:666–669 10.1128/AAC.27.4.666. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF. 2003. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother 47:3407–3414 10.1128/AAC.47.11.3407-3414.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Gründling A, Missiakas DM, Schneewind O. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol 188:6286–6297 10.1128/JB.00457-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Bastos MD, Coutinho BG, Coelho MLV. 2010. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals (Basel) 3:1139–1161 10.3390/ph3041139. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Nilsen T, Nes IF, Holo H. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984 10.1128/AEM.69.5.2975-2984.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Vaughan EE, Daly C, Fitzgerald GF. 1992. Identification and characterization of helveticin V-1829, a bacteriocin produced by Lactobacillus helveticus 1829. J Appl Bacteriol 73:299–308 10.1111/j.1365-2672.1992.tb04981.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 403.Ross RP, Morgan S, Hill C. 2002. Preservation and fermentation: past, present and future. Int J Food Microbiol 79:3–16 10.1016/S0168-1605(02)00174-5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 404.Müller E, Radler F. 1993. Caseicin, a bacteriocin from Lactobacillus casei. Folia Microbiol (Praha) 38:441–446 10.1007/BF02814392. [PubMed] [DOI] [PubMed] [Google Scholar]
- 405.Oman TJ, Boettcher JM, Wang H, Okalibe XN, van der Donk WA. 2011. Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat Chem Biol 7:78–80 10.1038/nchembio.509. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Stepper J, Shastri S, Loo TS, Preston JC, Novak P, Man P, Moore CH, Havlíček V, Patchett ML, Norris GE. 2011. Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett 585:645–650 10.1016/j.febslet.2011.01.023. [PubMed] [DOI] [PubMed] [Google Scholar]
- 407.Maqueda M, Gálvez A, Bueno MM, Sanchez-Barrena MJ, González C, Albert A, Rico M, Valdivia E. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr Protein Pept Sci 5:399–416 10.2174/1389203043379567. [PubMed] [DOI] [PubMed] [Google Scholar]
- 408.Maky MA, Ishibashi N, Zendo T, Perez RH, Doud JR, Karmi M, Sonomoto K. 2015. Enterocin F4-9, a novel O-linked glycosylated bacteriocin. Appl Environ Microbiol 81:4819–4826 10.1128/AEM.00940-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Paik SH, Chakicherla A, Hansen JN. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem 273:23134–23142 10.1074/jbc.273.36.23134. [PubMed] [DOI] [PubMed] [Google Scholar]
- 410.Kelly W, Asmundson R, Huang C. 1996. Characterization of plantaricin KW30, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol 81:657–662 10.1111/j.1365-2672.1996.tb01968.x. [DOI] [Google Scholar]
- 411.Rebuffat S. 2012. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans 40:1456–1462 10.1042/BST20120183. [PubMed] [DOI] [PubMed] [Google Scholar]
- 412.Yang SC, Lin CH, Sung CT, Fang JY. 2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol 5:241. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Thomas X, Destoumieux-Garzón D, Peduzzi J, Afonso C, Blond A, Birlirakis N, Goulard C, Dubost L, Thai R, Tabet JC, Rebuffat S. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J Biol Chem 279:28233–28242 10.1074/jbc.M400228200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 414.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. 2007. Low-molecular-weight post-translationally modified microcins. Mol Microbiol 65:1380–1394 10.1111/j.1365-2958.2007.05874.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 415.van den Elzen PJ, Walters HH, Veltkamp E, Nijkamp HJ. 1983. Molecular structure and function of the bacteriocin gene and bacteriocin protein of plasmid Clo DF13. Nucleic Acids Res 11:2465–2477 10.1093/nar/11.8.2465. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Kleanthous C. 2010. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol 8:843–848 10.1038/nrmicro2454. [PubMed] [DOI] [PubMed] [Google Scholar]
- 417.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229 10.1128/MMBR.00036-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Gillor O, Giladi I, Riley MA. 2009. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol 9:165 10.1186/1471-2180-9-165. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Goldstein BP, Wei J, Greenberg K, Novick R. 1998. Activity of nisin against Streptococcus pneumoniae, in vitro, and in a mouse infection model. J Antimicrob Chemother 42:277–278 10.1093/jac/42.2.277. [PubMed] [DOI] [PubMed] [Google Scholar]
- 420.van Staden AD, Brand AM, Dicks LM. 2012. Nisin F-loaded brushite bone cement prevented the growth of Staphylococcus aureusin vivo. J Appl Microbiol 112:831–840 10.1111/j.1365-2672.2012.05241.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 421.Brand AM, de Kwaadsteniet M, Dicks LM. 2010. The ability of nisin F to control Staphylococcus aureus infection in the peritoneal cavity, as studied in mice. Lett Appl Microbiol 51:645–649 10.1111/j.1472-765X.2010.02948.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 422.De Kwaadsteniet M, Doeschate KT, Dicks LM. 2009. Nisin F in the treatment of respiratory tract infections caused by Staphylococcus aureus. Lett Appl Microbiol 48:65–70 10.1111/j.1472-765X.2008.02488.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 423.Campion A, Casey PG, Field D, Cotter PD, Hill C, Ross RP. 2013. In vivo activity of nisin A and nisin V against Listeria monocytogenes in mice. BMC Microbiol 13:23 10.1186/1471-2180-13-23. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Mota-Meira M, Morency H, Lavoie MC. 2005. In vivo activity of mutacin B-Ny266. J Antimicrob Chemother 56:869–871 10.1093/jac/dki295. [PubMed] [DOI] [PubMed] [Google Scholar]
- 425.Castiglione F, Cavaletti L, Losi D, Lazzarini A, Carrano L, Feroggio M, Ciciliato I, Corti E, Candiani G, Marinelli F, Selva E. 2007. A novel lantibiotic acting on bacterial cell wall synthesis produced by the uncommon actinomycete Planomonospora sp. Biochemistry 46:5884–5895 10.1021/bi700131x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 426.Rihakova J, Cappelier JM, Hue I, Demnerova K, Fédérighi M, Prévost H, Drider D. 2010. In vivo activities of recombinant divercin V41 and its structural variants against Listeria monocytogenes. Antimicrob Agents Chemother 54:563–564 10.1128/AAC.00765-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Salvucci E, Saavedra L, Hebert EM, Haro C, Sesma F. 2012. Enterocin CRL35 inhibits Listeria monocytogenes in a murine model. Foodborne Pathog Dis 9:68–74 10.1089/fpd.2011.0972. [PubMed] [DOI] [PubMed] [Google Scholar]
- 428.Sosunov V, Mischenko V, Eruslanov B, Svetoch E, Shakina Y, Stern N, Majorov K, Sorokoumova G, Selishcheva A, Apt A. 2007. Antimycobacterial activity of bacteriocins and their complexes with liposomes. J Antimicrob Chemother 59:919–925 10.1093/jac/dkm053. [PubMed] [DOI] [PubMed] [Google Scholar]
- 429.Lopez FE, Vincent PA, Zenoff AM, Salomón RA, Farías RN. 2007. Efficacy of microcin J25 in biomatrices and in a mouse model of Salmonella infection. J Antimicrob Chemother 59:676–680 10.1093/jac/dkm009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 430.Wang WL, Liu J, Huo YB, Ling JQ. 2013. Bacteriocin immunity proteins play a role in quorum-sensing system regulated antimicrobial sensitivity of Streptococcus mutans UA159. Arch Oral Biol 58:384–390 10.1016/j.archoralbio.2012.09.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 431.Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CG. 2007. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA 104:7617–7621 10.1073/pnas.0700440104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 433.Kleerebezem M. 2004. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405–1414 10.1016/j.peptides.2003.10.021. [PubMed] [DOI] [PubMed] [Google Scholar]
- 434.Kleerebezem M, Kuipers OP, de Vos WM, Stiles ME, Quadri LE. 2001. A two-component signal-transduction cascade in Carnobacterium piscicola LV17B: two signaling peptides and one sensor-transmitter. Peptides 22:1597–1601 10.1016/S0196-9781(01)00494-6. [DOI] [PubMed] [Google Scholar]
- 435.Kleerebezem M, Quadri LE. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579–1596 10.1016/S0196-9781(01)00493-4. [DOI] [PubMed] [Google Scholar]
- 436.O’Keeffe T, Hill C, Ross RP. 1999. Characterization and heterologous expression of the genes encoding enterocin a production, immunity, and regulation in Enterococcus faecium DPC1146. Appl Environ Microbiol 65:1506–1515. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788 10.1038/nrmicro1273. [PubMed] [DOI] [PubMed] [Google Scholar]
- 438.van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol 187:3980–3989 10.1128/JB.187.12.3980-3989.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203 10.1128/JB.187.21.7193-7203.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Li YH, Hanna MN, Svensäter G, Ellen RP, Cvitkovitch DG. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol 183:6875–6884 10.1128/JB.183.23.6875-6884.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908 10.1128/JB.183.3.897-908.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Li YH, Tian XL, Layton G, Norgaard C, Sisson G. 2008. Additive attenuation of virulence and cariogenic potential of Streptococcus mutans by simultaneous inactivation of the ComCDE quorum-sensing system and HK/RR11 two-component regulatory system. Microbiology 154:3256–3265 10.1099/mic.0.2008/019455-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 443.Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. 2011. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol 193:6552–6559 10.1128/JB.05968-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917 10.1111/j.1365-2958.2009.06693.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev 71:653–670 10.1128/MMBR.00024-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Rodríguez E, Arqués JL, Rodríguez R, Nuñez M, Medina M. 2003. Reuterin production by lactobacilli isolated from pig faeces and evaluation of probiotic traits. Lett Appl Microbiol 37:259–263 10.1046/j.1472-765X.2003.01390.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 447.Talarico TL, Casas IA, Chung TC, Dobrogosz WJ. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother 32:1854–1858 10.1128/AAC.32.12.1854. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Lüthi-Peng Q, Dileme FB, Puhan Z. 2002. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl Microbiol Biotechnol 59:289–296 10.1007/s00253-002-1002-z. [PubMed] [DOI] [PubMed] [Google Scholar]
- 449.Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, Britton RA. 2010. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156:1589–1599 10.1099/mic.0.035642-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, Warren MJ, Prentice MB. 2008. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol 190:4559–4567 10.1128/JB.01535-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Cleusix V, Lacroix C, Vollenweider S, Duboux M, Le Blay G. 2007. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol 7:101 10.1186/1471-2180-7-101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Arqués JL, Rodríguez E, Nuñez M, Medina M. 2011. Combined effect of reuterin and lactic acid bacteria bacteriocins on the inactivation of food-borne pathogens in milk. Food Control 22:457–461 10.1016/j.foodcont.2010.09.027. [DOI] [Google Scholar]
- 453.Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T, Murakami M, Hisamatsu S, Kato Y, Takizawa T, Fukuoka H, Yoshimura T, Itoh K, O’Sullivan DJ, McKay LL, Ohno H, Kikuchi J, Masaoka T, Hattori M. 2008. Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res 15:151–161 10.1093/dnares/dsn009. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Gänzle MG. 2004. Reutericyclin: biological activity, mode of action, and potential applications. Appl Microbiol Biotechnol 64:326–332 10.1007/s00253-003-1536-8. [PubMed] [DOI] [PubMed] [Google Scholar]
- 455.Gänzle MG, Höltzel A, Walter J, Jung G, Hammes WP. 2000. Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl Environ Microbiol 66:4325–4333 10.1128/AEM.66.10.4325-4333.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Helander IM, Mattila-Sandholm T. 2000. Permeability barrier of the Gram-negative bacterial outer membrane with special reference to nisin. Int J Food Microbiol 60:153–161 10.1016/S0168-1605(00)00307-X. [DOI] [PubMed] [Google Scholar]


