Abstract
Background
Inorganic arsenic (As) is methylated via one carbon metabolism (OCM) to mono- and dimethylated arsenicals (MMA and DMA), facilitating urinary excretion. Hyperhomocysteinemia (HHcys), a marker of impaired OCM, is a risk factor for As-induced skin lesions, but the influences of single nucleotide polymorphisms (SNPs) in OCM genes on Hcys, As metabolism and skin lesion risk is unclear.
Objectives
To (i) explore genetic sources of Hcys and the causal role of HHcys in As-induced skin lesion development using OCM genetic proxies for HHcys and (ii) identify OCM SNPs associated with urinary As metabolite proportions and/or skin lesion incidence.
Methods
We conducted a case-control study nested in the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh which 876 incident skin lesion cases were matched to controls on sex, age, and follow-up time. We measured serum Hcys, urinary As metabolites, and 26 SNPs in 13 OCM genes.
Results
Serum Hcys and urinary %DMA were independently associated with increased and decreased odds of skin lesions, respectively. The T allele of MTHFR 677 C→T (rs1801133) was associated with HHcys, higher %MMA, and lower %DMA, but not with skin lesions. Interactions between SNPs and water As on skin lesion risk were suggestive for three variants: the G allele of MTRR rs1801394 and T allele of FOLR1 rs1540087 were associated with lower odds of skin lesions with lower As (≤50 μg/L), and the T allele of TYMS rs1001761 was associated with higher odds of skin lesions with higher As.
Conclusions
While HHcys and decreased %DMA were associated with increased risk for skin lesions, and MTHFR 677 C→T was a strong predictor of HHcys, MTHFR 677 C→T was not associated with skin lesion risk. Future studies should explore (i) non-OCM and non-genetic determinants of Hcys and (ii) if genetic findings are replicated in other As-exposed populations, mechanisms by which OCM SNPs may influence the dose-dependent effects of As on skin lesion risk.
Keywords: arsenic, arsenic metabolism, one-carbon metabolism, gene-environment interaction, skin lesions, homocysteine
1. Introduction
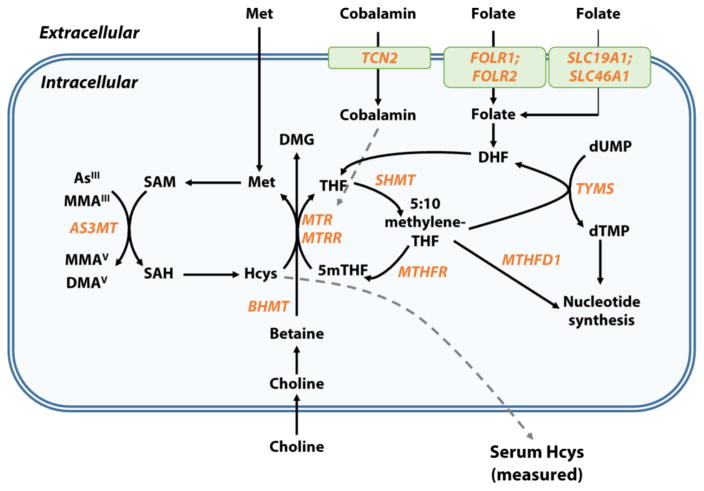
Elevated homocysteine (Hcys) in serum and/or plasma, a condition known as hyperhomocystenemia (HHcys), is a well-established risk factor for numerous health conditions, including cardiovascular disease (CVD) (Ganguly and Alam 2015), neurologic conditions (Ansari et al. 2014), and cancer (Wu and Wu 2002). Homocysteine is endogenously synthesized via B-vitamin-dependent one-carbon metabolism (OCM). As shown in Figure 1, the donation of a methyl group from S-adenosylmethionine (SAM) to various substrates yields the methylated product and S-adenosylhomocysteine, which is hydrolyzed to form Hcys (Scott and Weir 1998). HHcys is a sensitive indicator of B-vitamin deficiencies, especially for folate and cobalamin (Savage et al. 1994). Although Hcys induces oxidative stress and inflammation in experimental models (Dayal et al. 2004; Eberhardt et al. 2000), whether HHcys is a causative factor in human disease, or merely a biomarker of disease risk, remains under debate (Brattstrom and Wilcken 2000).
Figure 1. One-carbon metabolism, arsenic methylation, and homocysteine.
Folate enters the cell through receptor-mediated (folate receptor alpha [FRα], gene FOLR1; beta [FRβ], gene FOLR2; or gamma [FRγ]) or carrier-mediated (solute carrier family 19, member 1 [SLC19A1]) transport mechanisms. Folic acid is reduced to dihydrofolate (DHF) and tetrahydrofolate (THF). Serine hydroxymethyltransferase (SHMT) catalyzes the conversion of serine and THF to 5:10 methylene-THF, which can be used for nucleotide synthesis via methylenetetrahydrofolate dehydrogenase 1 (MTHFD) or thymidylate synthetase (TYMS). Alternatively, 5:10 methylene-THF can be converted to 5-methyl THF (5mTHF) by methylene tetrahydrofolate reductase (MTHFR). The methyl group from 5mTHF is transferred to Hcys via methionine synthetase (MTR), a cobalamin-dependent enzyme, which generates methionine (Met) and THF. Betaine can also serve as the methyl donor for the remethylation of Hcys to Met in a reaction catalyzed by betaine-homocysteine S-methyltransferase (BHMT). Met is activated to form S-adenosylmethionine (SAM), the methyl donor for the methylation of InAs and MMA, yielding MMA and DMA, respectively, and S-adenosylhomocysteine (SAH). SAH is hydrolyzed to regenerate Hcys, which can be remethylated to Met or directed toward the transsulfuration pathway by cystathionine-β-synthase (CBS). Excess intracellular Hcys can be exported extracellularly. Genes with SNPs examined in the current study are displayed in orange.
Chronic exposure to arsenic (As) is associated with increased all-cause mortality (Argos et al. 2010) and elevated risk for a variety of conditions, such as CVD (Moon et al. 2012), neurological deficits (Tyler and Allan 2014), and cancers of the skin, lung, bladder, liver, and kidney (Navarro Silvera and Rohan 2007). Inorganic As is metabolized through a series of methylation reactions to monomethyl (MMA) and dimethyl (DMA) species (Figure 1) by arsenite 3-methyltransferase (AS3MT), with methyl groups donated by SAM via OCM (Lin et al. 2002). An increased capacity to methylate As—as indicated by decreased proportions of inorganic As (InAs) and MMA and an increased proportion of DMA in urine—has been associated with reduced risk for several As-associated conditions (Steinmaus et al. 2007). Importantly, nutritional manipulation of OCM in randomized controlled trials (e.g., folic acid supplementation) has been shown to increase As methylation capacity, reduce total blood As concentrations, and reduce Hcys levels in As-exposed populations (Gamble et al. 2006; Gamble et al. 2007; Peters et al. 2015), suggesting the utility of public health interventions targeting OCM factors to reduce As toxicity.
A hallmark of chronic As exposure is the appearance of arsenical skin lesions, which are a sensitive indicator of increased risk for various As-related diseases (Karagas et al. 2015). As such, identifying risk factors for skin lesion development may provide insight into mechanisms of As toxicity and potential targets for intervention. Previously, our group found that HHcys and other factors associated with OCM status were associated with increased risk for As-induced skin lesions in Bangladesh (Pilsner et al. 2009), suggesting that compromised OCM might contribute to increased susceptibility to As toxicity, possibly by impacting As methylation capacity. Alternatively, HHcys may act as a biomarker reflecting dysregulation of other components of the OCM network that contribute to increased susceptibility to As, e.g., through nucleotide biosynthesis and its impact on DNA repair (Locasale 2013).
While Hcys levels are known to be influenced by single nucleotide polymorphisms (SNPs) in several genes involved in OCM, the most widely-studied variant is the nonsynonymous 677 C→T polymorphism (rs1801133) in methylenetetrahydrofolate reductase (MTHFR). MTHFR encodes for an enzyme that synthesizes 5-methyltetrahydrofolate (5mTHF), a substrate required for the remethylation of Hcys to methionine. MTHFR 677 C→T is a nonsynonymous SNP associated with reduced enzyme activity, thereby resulting in elevated Hcys, particularly in populations with low folate intakes (Jacques et al. 1996). Since the 677 C→T variant is causally associated with HHcys, it is commonly used as a genetic proxy for HHcys to examine the causal nature of HHcys-disease associations (Clarke et al. 2012; Wald et al. 2002).
The prevalence of HHcys is particularly high among individuals of South Asian descent, which has been hypothesized to reflect folate and cobalamin deficiencies and genetic factors (Chandalia et al. 2003; Senaratne et al. 2001). A previous cross-sectional survey from our group observed a high prevalence of HHcys in Bangladesh, and plasma folate and cobalamin was found to explain 15% and 5% of the variance in plasma Hcys, respectively (Gamble et al. 2005). It is unknown whether MTHFR 677 C→T and other one-carbon metabolism gene variants are associated with Hcys concentrations in Bangladesh: these SNPs have variable effects on Hcys concentrations across populations (Wald et al. 2012), and the associations of these genetic variants with Hcys have not been examined previously in this population.
Several studies have investigated the modulating effects of OCM gene variants on As-related health outcomes, including skin lesions (Ahsan et al. 2007; Seow et al. 2015), bladder cancer (Beebe-Dimmer et al. 2012; Chung et al. 2010; Karagas et al. 2005), breast cancer (Gamboa-Loira et al. 2017), elevated blood pressure (Farzan et al. 2015), and myelomeningocele (Mazumdar et al. 2015), though findings have been mixed. OCM gene variants have also been linked to altered uAs metabolite patterns in Argentina (Engstrom et al. 2011; Porter et al. 2010; Schlawicke Engstrom et al. 2009; Schläwicke Engström et al. 2007; Steinmaus et al. 2007), Bangladesh (Ahsan et al. 2007; Engstrom et al. 2011), Taiwan (Chung et al. 2010), China (Deng et al. 2007), and Eastern Europe (Lindberg et al. 2007). Collectively, these studies suggest that OCM may modify As-related health risks, possibly though altered As metabolism. However, only two of these studies (Ahsan et al. 2007; Chung et al. 2010) simultaneously examined OCM SNPs, As metabolites, and health risks.
Herein we hypothesized that an integrated assessment of OCM gene variants, Hcys, and uAs metabolites in As-exposed Bangladeshi adults would provide insight into the roles of HHcys and OCM in As-induced skin lesion pathogenesis. Thus, the objectives of our study were (1) to explore genetic sources of Hcys and the causal role of HHcys in skin lesion incidence using OCM genetic proxies and (2) to examine whether OCM gene variants are associated with urinary As metabolite proportions and/or As-induced skin lesion incidence. To address these objectives, we conducted a case-control study of As-induced skin lesions nested in the Health Effects of Arsenic Longitudinal Study (HEALS) cohort, a prospective cohort study in Araihazar, Bangladesh (Ahsan et al. 2006a).
2. Study Population and Methods
2.1. Eligibility criteria and study design
The Gene-Environment-Nutrition Interactions (GENI) study is a case-control study of incident As-induced skin lesions nested within the HEALS cohort, an ongoing prospective cohort study of As-exposed adults in Araihazar, Bangladesh (Ahsan et al. 2006a). GENI cases and controls were identified from the original cohort of 11,746 married men and women between 18 and 65 years, of which 9,727 participants completed the baseline physical examination, provided urine and blood samples, and were free of skin lesions. Initial HEALS recruitment was completed from October 2000 to May 2002, and active follow-up of the cohort was done every 2 years: the first follow-up was completed from September 2002 to May 2004; second follow-up from June 2004 to August 2006; and third follow-up from January 2007 to February 2009. At baseline and at every follow-up visit, trained physicians conducted comprehensive examinations to identify arsenicosis skin lesions (melanosis, leukomelanosis, and keratosis) across the entire body; the details of the structured examination protocol have been reported previously (Ahsan et al. 2006a; Ahsan et al. 2006b). Melanosis is characterized by skin hyperpigmentation over a wide surface area; leukomelanosis is characterized by both skin hyperpigmentation and skin hypopigmentation over a wide surface area; and keratosis is characterized by bilateral skin thickening of the palms of the hands and soles of the feet (Ahsan et al. 2000).
The GENI study includes 876 incident skin lesion cases identified through the end of the third follow-up period in HEALS (February 2009). The 876 skin lesion cases were individually matched to controls on gender, age (± 3 years), and follow-up time in the study (± 60 days). Since it was known a priori that some of the HEALS subjects did not have remaining serum and/or DNA samples from the baseline recruitment visit for analysis, we devised a matching scheme to maximize the number of controls with biological samples. Up to 3 possible controls were identified for each case, without prior knowledge of whether these controls had biological samples available. Sample availability (serum and/or DNA) was then determined for each possible control, and the control with the greatest number of biological samples in the freezers (serum and/or DNA) was preferentially selected for each case.
We had 703 cases and 705 controls with serum samples, and 701 matched pairs where both the case and control had a serum sample available. There were 732 successfully matched pairs in which a control was identified with a DNA and/or serum sample, and 608 matched pairs in which both the case and control have both DNA and serum samples.
Written informed consent was obtained by Bangladeshi field staff physicians upon enrollment to the HEALS cohort. This study was approved by the Institutional Review Boards of Columbia University Medical Center and the Bangladesh Medical Research Council.
2.2. Analytic techniques
2.2.1. Water As
Water samples were analyzed using graphite furnace atomic absorption (GFAA) spectrometry, with a detection limit of 5 μg/L; samples with non-detectable As using GFAA were analyzed with inductively coupled mass spectrometry, with a detection limit of 0.1 μg/L. Details about the sample analysis and quality control (QC) measures have been described previously (Van Geen et al. 2005; Zheng et al. 2004).
2.2.2. Urinary As
Total urinary As (uAs) was measured using graphite furnace atomic absorption (GFAA) spectrometry, as previously described (Nixon et al. 1991). Urinary As metabolites (arsenobetaine (AsB), arsenocholine (AsC), AsV, AsIII, MMA (MMAIII + MMAV), and DMA (DMAV)) were separated using high-performance liquid chromatography (HPLC), and metabolite concentrations were measured using ICP-MS. For total uAs and uAs metabolites, quality control (QC) samples were analyzed at the beginning of each daily batch and after every 10 samples to assess instrument drift; based on these QC samples, intra- and inter-precision CVs were 3.5 and 5.6%, respectively, for total uAs; 1.4 and 1.6%, respectively, for InAs (AsIII+AsV); 1.7 and 2.3%, respectively, for MMA; and 0.9 and 1.6%, respectively, for DMA. For uAs metabolites, 10% of study samples were run in duplicate on the same day, and 15% of study samples were repeated on a different day; intra- and inter-day CVs were 6.3 and 5.7%, respectively, for InAs (AsIII+AsV); 5.4 and 6.4%, respectively, for MMA; and 1.4 and 1.5%, respectively, for DMA. Urinary creatinine (uCr) was used to adjust for urine concentration, as previously described (Slot 1965).
2.2.3. Serum Hcys
Serum Hcys was measured by high performance liquid chromatography based on published methods (Pfeiffer et al. 1999), as previously described (Gamble et al. 2005). Serum samples in case-control pairs were assayed in a blinded manner in the same laboratory batch, and all samples were run in duplicate; samples were repeated if the CV within the duplicates was >10%. Matched pairs were randomized in a two-step process: first, the run order of the case-control pairs was randomized to ensure that sample pairs were analyzed in the same laboratory batch; second, the run order of the case and control samples within each matched pair. The intra- and inter-day CVs were 3 and 8%, respectively.
2.2.4. DNA extraction
DNA was extracted from blood clots using the Flexigene DNA kit (Qiagen) or whole blood as previously described (Pilsner et al. 2009), and the DNA concentration and quality (260/280 and 260/230 ratios) was determined using Nanodrop 1000. DNA samples were shipped to Columbia University on dry ice and stored at −80°C.
2.2.5. TaqMan OpenArray SNP genotyping
125 ng of DNA was processed with the TaqMan OpenArray Genotyping System (ThermoFisher Scientific, Carlsbad, California, USA) according to the manufacturer’s protocol using custom-designed chips with 64 SNPs in candidate genes involved in one-carbon metabolism, folate transport, and arsenic metabolism (full list of SNPs in Supplementary Table 1). DNA samples in case-control pairs were processed on the same array chip, and the run order was determined using the same two-step randomization process for serum Hcys, but with different randomization orders. Genotype calls were generated using the OpenArray SNP Genotyping analysis software 1.0.3.
A total of 1,383 samples were genotyped. For quality assurance, we excluded 11 SNPs with call rates < 90%, then removed samples with call rates on the remaining SNPs of < 80% (n = 56). Of the remaining 53 SNPs, 23 had minor allele frequencies (MAF) < 5% in our sample and were additionally excluded. Genotype frequencies for the 30 remaining SNPs were in accordance with Hardy-Weinberg equilibrium with the exception of 4 SNPs (p < 0.05 in controls), which were excluded from further analyses. The cleaned dataset contained 1,327 samples (694 cases, 633 controls) with a mean missing rate of 1.5% and 26 SNPs with a mean call rate of 98.9% (Supplementary Table 2). SNP alleles reported in the study reflect the alleles on the strand designated in the NCBI dbSNP Database (Sherry et al. 2001). Risk alleles were selected based on the literature; if a risk allele was not known for a SNP, the risk allele was selected as the minor allele (i.e., lowest frequency) among cases and controls.
2.3. Statistical methods
Statistical tests were conducted using RStudio version 1.0.143 (Team 2015); statistical tests were two sided with a significance level of 0.05. Descriptive statistics (mean and standard deviation [SD] for continuous variables, median and interquartile range [IQR] for continuous variables with skewed distributions, N [%] for categorical variables) were calculated separately for the cases and control groups. Differences in matched case-control pairs were examined using Wilcoxon signed-rank tests (continuous variables), McNemar’s tests (dichotomous variables), and Bowker’s tests (categorical variables). Accordance of genotype distributions with Hardy-Weinberg equilibrium (HWE) was conducted by comparing the expected vs. observed genotype frequencies in controls using Chi-square test with one degree of freedom. Calculation of HWE and other genetic statistics, such as pairwise linkage disequilibrium parameters (D′, r2) were conducted with the ‘genetics’ R package (Gregory Warnes 2013).
Urinary As metabolite concentrations below the limit of detection [LOD] were imputed with one half the minimum value for the corresponding metabolite in the full dataset (InAs, 1 observation, 0.4289/2; MMA, 2 observations, 0.4311/2). Urinary As metabolite proportions (%InAs [AsV + AsIII], %MMA, and %DMA) were calculated by dividing the metabolite concentration by the sum of all metabolite concentrations (excluding arsenocholine and arsenobetaine). Total uAs (as assessed by GFAA) was adjusted for uCr using the residual adjustment approach. Briefly, total uAs was adjusted for uCr by constructing a linear regression model with log-transformed uCr as the predictor of log-transformed uAs, adding the residuals from the model to the mean log-transformed uAs value, and exponentiating these values.
Associations between serum Hcys, wAs, and uAs metabolite percentages with skin lesion incidence was examined using conditional logistic regression in two models. The first model included each predictor, adjusted for wAs (categorical), age (continuous), and education (categorical), and the second model included predictors of wAs, (categorical), serum Hcys (categorical), urinary %DMA (categorical), adjusted for age (continuous) and education (categorical). Urinary metabolite percentage categories were determined based on tertile cutoffs in the control group. Both models were constructed using the 552 matched pairs with complete information on serum Hcys, uAs metabolites, and education.
For SNP analyses, an additive genetic effect (AA, Aa and aa coded as 0, 1, and 2, respectively) was of primary interest. Associations between SNPs with serum Hcys was examined with linear regression models adjusted for sex, age (continuous), wAs (continuous), and education (categorical). Beta regression (a model appropriate when the response has a beta distribution with values in the interval [0, 1]), using a logit link function of mean response, was used to examine associations between SNPs and uAs metabolites (urinary %InAs, %MMA and %DMA), adjusted for sex, age (continuous), wAs (log-transformed), and education (categorical) using the R package ‘betareg’ (Cribari-Neto and Zeileis 2010). The coefficient of a predictor in a beta regression model has the interpretation of a log odds ratio (Ferrari and Cribari-Neto 2004).
Associations between SNPs and skin lesions were examined in two conditional logistic regression models: the first model with the SNP predictor, adjusted for wAs (log-transformed), age, and education, and the second model additionally included a cross-product term (SNP × wAs) to examine SNP-As interactions. For SNPs with marginally-significant SNP by wAs interaction (p < 0.10), joint effects of SNPs and wAs were examined by creating categorical variables of genotypes with wAs (dichotomized at wAs > 50 μg/L, based on the median wAs in controls [51 μg/L] and the Bangladeshi standard for wAs in drinking water [50 μg/L]). Serum Hcys and wAs were log transformed to reduce right skewness. Associations were considered nominally-significant at a threshold of p < 0.05; to account for multiple tests, the Benjamini and Hochberg procedure (Benjamini and Hochberg 1995) was used to adjust p-values to control for the false discovery rate (FDR) considering all SNPs (26 tests) and LD blocks (15 tests, with the SNP with the lowest p-value considered within each block). FDRs for LD blocks are reported in the main manuscript, while FDRs for all SNPs are included in the Supplementary Materials.
3. Results
Demographic and clinical characteristics of the GENI study population are presented in Table 1. Controls were matched to skin lesion cases (876 matched pairs) on sex, age, and follow-up time. Biological samples were available for a majority of participants, with 701 matched pairs with serum Hcys measurements, 564 pairs with SNP genotyping data, and 547 pairs with both Hcys and SNP data. At baseline, skin lesion cases had a lower BMI, were more likely to report ever-use of cigarettes and betel nut, had fewer years of education, were less likely to own a television, and had higher As concentrations in water and urine. Cases had higher proportions of urinary InAs and lower proportions of urinary DMA, indicative of reduced As methylation capacity. Serum Hcys was higher in skin lesion cases, as was the proportion of individuals who were classified as having HHcys, defined here as serum Hcys > 15 uM.
Table 1.
Baseline descriptive and clinical characteristics.
| Variable | Controls (n = 876) | Cases (n = 876) | Pair diff. (p) |
|---|---|---|---|
| Demographic | |||
| Sex (male) | 622 (71.0)a | 622 (71.0) | 1.00 |
| Age (years) | 44.4 ± 9.3 (20 to 65)b | 44.5 ± 9.4 (20 to 65) | <0.0001 |
| BMI (kg/m2)d | 19.7 ± 3.2 (13.0 to 32.3) | 19.3 ± 2.8 (13.9 to 32.8) | 0.01 |
| Cigarette smoking (ever) | 507 (57.9) | 538 (61.4) | 0.05 |
| Betel nut use (ever)e | 434 (49.5) | 485 (55.5) | 0.07 |
| Educationf | 0.001 | ||
| 0 years | 374 (42.8) | 423 (48.3) | |
| 1–5 years | 236 (27.0) | 251 (28.7) | |
| > 5 years | 263 (30.1) | 201 (23.0) | |
| Own television (yes) | 336 (38.3) | 269 (30.7) | 0.009 |
|
| |||
| Arsenic exposure | |||
|
| |||
| Well water As (μg/L) | 84 ± 96 (0.1 to 571) | 140 ± 136 (0.1 to 790) | <0.0001 |
| Well water As category | <0.0001 | ||
| < 10 μg/L | 220 (25.1) | 135 (15.4) | |
| 10–50 μg/L | 210 (24.0) | 138 (15.8) | |
| 50–100 μg/L | 164 (18.7) | 154 (17.6) | |
| 100–200 μg/L | 178 (20.3) | 213 (24.3) | |
| >200 μg/L | 104 (11.9) | 236 (26.9) | |
| Urinary As (μg/L)g | 125 ± 125 (3.0 to 1,228) | 165 ± 176 (1.0 to 1,278) | <0.0001 |
| Urinary Cr (mg/dL) | 62.7 ± 46.2 (3.9 to 301) | 60.5 ± 46.8 (3.3 to 376) | 0.28 |
| Urinary As adj. for Cr (μg/L)h | 109 ± 91 (4.5 to 777) | 148 ± 132 (4.6 to 1,145) | <0.0001 |
| Urinary As metabolitesg | |||
| Urinary %InAs | 14.0 ± 6.1 (2.7 to 56.4) | 14.8 ± 5.8 (2.0 to 42.4) | 0.0009 |
| Urinary %MMA | 13.8 ± 5.0 (3.3 to 47.3) | 14.1 ± 5.2 (2.4 to 32.9) | 0.13 |
| Urinary %DMA | 72.3 ± 8.2 (35.0 to 92.2) | 71.1 ± 8.5 (38.6 to 90.5) | 0.003 |
|
| |||
| Homocysteine | |||
|
| |||
| Serum Hcys (μM)i | 13.1 ± 7.5 (3.1 to 108)c | 14.2 ± 8.0 (3.5 to 119.0)c | 0.003 |
| Serum Hcys categoryi | 0.04 | ||
| < 10 μM | 169 (24.0) | 133 (18.9) | |
| 10–15 μM | 264 (37.4) | 261 (37.1) | |
| 15–20 μM | 161 (22.8) | 159 (22.6) | |
| > 20 μM | 111 (15.7) | 150 (21.3) | |
N (%) (all such values);
mean ± SD (range) (all such values);
median ± IQR (range);
n = 875 controls, n = 868 cases, n = 867 pairs;
n = 876 controls, n = 874 cases, n = 874 pairs;
n = 873 controls, n = 875 cases, n = 872 pairs;
n =863 controls, n = 850 cases, n = 837 pairs;
n = 778 controls, n = 781 cases, n = 694 pairs;
n = 705 controls, n = 703 cases, n = 701 pairs
We used conditional logistic regression to estimate the odds ratios (ORs) for the skin lesions predictors, serum Hcys, uAs metabolite percentages, and wAs categories, shown in Table 2. We found that wAs was associated with a dose-dependent increase in odds for skin lesions across all wAs categories (compared to < 10 μg/L), including the wAs exposure comparison group (10–50 μg/L), which contained study participants who were exposed to wAs concentrations below the current Bangladeshi standard (50 μg/L). Consistent with our previous study, serum Hcys was associated with a dose-dependent increase in odds for incident skin lesions (Model 2): compared to Hcys < 10 μM, covariates-adjusted ORs for skin lesions for 10–15 μM, 15–20 μM, and > 20 μM were 1.53 (95% CI: 1.06, 2.20), 1.87 (95% CI: 1.24, 2.82), and 2.48 (95% CI: 1.57, 3.94), respectively. We also found that higher urinary %DMA was associated with decreased odds of skin lesions. Serum Hcys and urinary %DMA were significant predictors in a model containing both variables simultaneously (p = 0.0009 and p = 0.05, respectively, Wald test), suggesting that they are independently associated with skin lesion risk.
Table 2.
Odds ratios for serum homocysteine and arsenic exposure predicting arsenic-induced skin lesions in conditional logistic regression models (552 matched pairs).
| Predictor | Level/unit | Model 1a OR (95% CI) |
Model 2b OR (95% CI) |
|---|---|---|---|
| Serum Hcys | < 10 μM | ref. | ref. |
| 10–15 μM | 1.47 (1.03, 2.10) | 1.53 (1.06, 2.20) | |
| 15–20 μM | 1.82 (1.21, 2.73) | 1.87 (1.24, 2.82) | |
| > 20 μM | 2.38 (1.51, 3.76) | 2.48 (1.57, 3.94) | |
|
| |||
| Well water As | < 10 μg/L | ref. | ref. |
| 10–50 μg/L | 1.59 (1.03, 2.45) | 1.65 (1.06, 2.57) | |
| 50–100 μg/L | 2.06 (1.32, 3.21) | 2.08 (1.32, 3.28) | |
| 100–200 μg/L | 2.41 (1.58, 3.67) | 2.30 (1.49, 3.55) | |
| >200 μg/L | 4.35 (2.79, 6.79) | 4.35 (2.76, 6.85) | |
|
| |||
| Urinary InAs (%)c | < 11.1% | ref. | -- |
| 11.1 – 15.0% | 0.99 (0.72, 1.36) | -- | |
| > 15.0% | 1.34 (0.98, 1.83) | -- | |
|
| |||
| Urinary MMA (%)c | < 11.5% | ref. | -- |
| 11.5 – 15.4% | 0.85 (0.62, 1.17) | -- | |
| > 15.4% | 1.11 (0.81, 1.52) | -- | |
|
| |||
| Urinary DMA (%)c | < 69.6% | ref. | ref. |
| 69.6 – 76.3% | 0.79 (0.58, 1.07) | 0.78 (0.57, 1.06) | |
| > 76.3% | 0.72 (0.52, 0.98) | 0.68 (0.49, 0.94) | |
Model 1 included listed predictor, adjusted for wAs (categorical), age (continuous), and education (categorical);
Model 2 included predictors serum Hcys (categorical), wAs (categorical) and urinary %DMA (categorical), adjusted for age (continuous), and education (categorical);
Cutoffs based on tertiles in control group
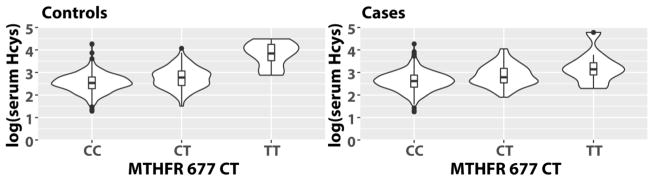
After quality control filtering, there were high-quality genotype data for 26 SNPs in 13 OCM genes (Table 3) that represented 15 independent linkage disequilibrium (LD) blocks (D′ > 0.8) (Supplementary File 1). Allele frequencies in cases and controls were similar to those reported for the Bengali in Bangladesh (BEB) population in the 1000 Genomes Database (The Genomes Project 2015). We first examined associations of the OCM variants with log-transformed serum Hcys (Supplementary Table 3). We found that the T allele of MTHFR 677 C→T (rs1801133) was strongly associated with higher serum Hcys (B [95% CI], 0.29 [0.25, 0.34], FDR = 5.1×10−31). In univariate analyses, geometric mean serum Hcys was 25% higher in CT heterozygotes (mean ± SD, 15.7 ± 1.6 μM) and 259% higher in TT homozygotes (45.2 ± 1.7 μM) compared to CC (12.6 ± 1.5 μM) in controls and 29% higher in CT (17.6 ± 1.6 μM) and 84% higher in TT (25.1 ± 2.0 μM) compared to CC (13.6 ± 1.5 μM) in cases (Figure 2).
Table 3.
Single nucleotide polymorphisms in one-carbon metabolism genes assessed in study.
| Gene | SNP | LD block | Type | Cases (n) | Controls (n) | Ref. allele | Risk allele | Risk allele freq, cases | Risk allele freq, controls | Risk allele freq, BEBa |
|---|---|---|---|---|---|---|---|---|---|---|
| AS3MT | rs10748835 | 1 | Intron | 666 | 617 | A | G | 0.53 | 0.54 | 0.47 |
| AS3MT | rs3740393 | 1 | Intron | 690 | 630 | G | C | 0.21 | 0.21 | 0.25 |
| BHMT | rs3733890 | 2 | Missense mutation (R239Q) | 688 | 624 | G | A | 0.28 | 0.29 | 0.28 |
| FOLR1 | rs1540087 | 3 | 5 prime UTR | 684 | 625 | C | T | 0.07 | 0.08 | 0.05 |
| FOLR1 | rs2071010 | 3 | Intron | 690 | 631 | G | A | 0.16 | 0.15 | 0.22 |
| FOLR1 | rs7109250 | 3 | Intron/5 prime UTR | 688 | 629 | C | T | 0.07 | 0.09 | 0.05 |
| FOLR2 | rs2298444 | 4 | Intron | 692 | 631 | A | G | 0.40 | 0.40 | 0.44 |
| FOLR2 | rs514933 | 5 | Intron | 674 | 615 | G | A | 0.62 | 0.59 | 0.65 |
| FOLR2 | rs651646 | 5 | Intron | 691 | 627 | A | T | 0.46 | 0.45 | 0.42 |
| FOLR2 | rs651933 | 5 | Upstream | 674 | 617 | A | G | 0.45 | 0.44 | 0.41 |
| MTHFD1 | rs11627387 | 6 | Intron | 679 | 621 | G | A | 0.28 | 0.26 | 0.24 |
| MTHFD1 | rs2236224 | 6 | Intron | 674 | 616 | C | T | 0.40 | 0.41 | 0.49 |
| MTHFD1 | rs2236225 | 6 | Missense mutation (R653Q) | 685 | 630 | C | T | 0.45 | 0.48 | 0.52 |
| MTHFD1 | rs1950902 | 7 | Missense mutation (K134R) | 693 | 633 | C | T | 0.13 | 0.10 | 0.12 |
| MTHFR | rs1476413 | 8 | Intron | 690 | 631 | G | A | 0.43 | 0.42 | 0.48 |
| MTHFR | rs1801131 | 8 | Missense mutation (E429A) | 669 | 615 | A | C | 0.38 | 0.38 | 0.41 |
| MTHFR | rs1801133 | 8 | Missense mutation (A222V) | 691 | 631 | C | T | 0.12 | 0.13 | 0.12 |
| MTR | rs1805087 | 9 | Missense mutation (D919G) | 690 | 631 | A | G | 0.27 | 0.27 | 0.33 |
| MTRR | rs1801394 | 10 | Missense mutation (I49M) | 686 | 631 | A | G | 0.50 | 0.54 | 0.54 |
| SHMT1 | rs1979277 | 11 | Missense mutation (L474F) | 675 | 625 | G | A | 0.17 | 0.17 | 0.16 |
| SLC19A1 | rs1888530 | 12 | Intron | 688 | 624 | T | C | 0.49 | 0.50 | 0.46 |
| SLC46A1 | rs1128162 | 13 | 3 prime UTR | 666 | 614 | T | G | 0.50 | 0.50 | 0.46 |
| TCN2 | rs1801198 | 14 | Missense mutation (R259P) | 686 | 632 | G | C | 0.39 | 0.38 | 0.45 |
| TYMS | rs1001761 | 15 | Intron | 685 | 625 | C | T | 0.54 | 0.53 | 0.62 |
| TYMS | rs2847149 | 15 | Intron | 693 | 627 | G | A | 0.53 | 0.52 | 0.60 |
| TYMS | rs502396 | 15 | Intron | 689 | 629 | T | C | 0.54 | 0.54 | 0.55 |
Allele frequencies for the Bengali in Bangladesh (BEB) population in the 1000 Genomes Project database
Figure 2.
Violin plots of log-transformed serum Hcys by MTHFR C677T genotype.
To explore the proportion of variation in log Hcys explained by MTHFR 677 C→T and other predictors, we constructed regression models separately in skin lesion cases and controls (Supplementary Table 4). In linear regression models, the change in R2 associated with 677 C→T genotype was 16.4% in controls, but only 7.4% in cases. Furthermore, the covariates-adjusted models explained a greater proportion of the variation in Hcys in controls (37.1% in controls, 30.9% in cases).
We also examined the associations of OCM SNPs with urinary %InAs, %MMA, and %DMA (Supplementary Table 3); nominally-significant SNPs (p < 0.05) are listed in Table 4. The T allele of MTHFR 677 C→T (rs1801133) was associated with reduced As methylation capacity, with increased %MMA (OR = 1.07, p = 0.002, FDR= 0.04) and decreased %DMA (OR = 0.94, p = 0.01, FDR = 0.16). For rs1805087, a missense mutation in the 5-methyltetrahydrofolate-homocysteine methyltransferase (MTR) gene, the G risk allele was associated with increased %InAs (OR = 1.04, p = 0.04, FDR = 0.30) and decreased %DMA (OR = 0.97, p = 0.049, FDR = 0.37). For rs1805087, a missense mutation in the serine hydroxymethyltransferase 1 (SHMT1) gene, the G allele was associated with increased %InAs (OR = 1.05, p = 0.03, FDR = 0.30). However, the associations for MTR and SHMT1 SNPs did not persist after correction for multiple testing (FDR ≥ 0.30).
Table 4.
Associations of OCM SNPs with urinary As metabolite percentages.
| Urinary %InAsa | Urinary %MMAa | Urinary %DMAa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Gene | SNP | Ref. allele | Risk allele | B (SE) | p | FDR | B (SE) | p | FDR | B (SE) | p | FDR |
| MTHFR | rs1801133 | C | T | 0.070 (0.023) | 0.002 | 0.04 | −0.060 (0.024) | 0.01 | 0.16 | |||
| MTR | rs1805087 | A | G | 0.039 (0.019) | 0.04 | 0.30 | −0.034 (0.017) | 0.05 | 0.37 | |||
| SHMT1 | rs1979277 | G | A | 0.050 (0.023) | 0.03 | 0.30 | ||||||
Beta regression model (with logit link) with SNP predictor (additive coding with AA, Aa, aa coded as 0, 1, and 2, respectively), adjusted for sex, age, log wAs, and education (categorical)
Finally, we examined associations of OCM SNPs with incident skin lesions in matched case-control pairs. Although 677 C→T was associated with elevated serum Hcys and decreased urinary %DMA in our sample, 677 C→T was not associated with skin lesion risk in a general genetic model (data not shown). Furthermore, there were no associations for the additive effects of 677 C→T and the other OCM SNPs in the main effect logistic models for skin lesions (Supplemental Table 5). In logistic models with interaction (top SNPs from each LD block listed in Table 5), there were three SNPs with suggestive SNP-wAs multiplicative interactions based on the SNP by wAs cross-product term (FOLR1 rs1540087, p = 0.03; MTRR rs1801394, p = 0.02; TYMS rs1001761, p = 0.06). However, no associations persisted after correction for multiple testing (FDR ≥ 0.15).
Table 5.
Interactions of OCM SNPs and water As on odds for incident skin lesions in conditional logistic regression models.
| Gene | SNP | Pairs (n) | Ref. allele | Risk allele | SNP B (95% CI)a |
Log wAs B (95% CI)a |
SNP*Log wAs B (95% CI)a |
pb | FDR |
|---|---|---|---|---|---|---|---|---|---|
| AS3MT | rs3740393 | 557 | G | C | −0.05 (−0.27, 0.18) | 0.19 (0.10, 0.27) | 0.07 (−0.05, 0.19) | 0.25 | 0.63 |
| BHMT | rs3733890 | 548 | G | A | −0.10 (−0.31, 0.10) | 0.23 (0.13, 0.32) | −0.01 (−0.11, 0.09) | 0.83 | 0.96 |
| FOLR1 | rs1540087 | 548 | C | T | −0.07 (−0.45, 0.30) | 0.19 (0.11, 0.26) | 0.25 (0.03, 0.47) | 0.02 | 0.15 |
| FOLR2 | rs2298444 | 557 | A | G | 0.11 (−0.08, 0.30) | 0.16 (0.05, 0.26) | 0.08 (−0.02, 0.18) | 0.10 | 0.38 |
| FOLR2 | rs651646 | 554 | A | T | −0.06 (−0.24, 0.13) | 0.21 (0.10, 0.32) | 0.02 (−0.08, 0.11) | 0.75 | 0.96 |
| MTHFD1 | rs11627387 | 538 | G | A | 0.15 (−0.05, 0.35) | 0.20 (0.10, 0.29) | 0.04 (−0.07, 0.14) | 0.47 | 0.88 |
| MTHFD1 | rs1950902 | 561 | C | T | 0.15 (−0.14, 0.44) | 0.22 (0.14, 0.30) | −0.02 (−0.16, 0.12) | 0.79 | 0.96 |
| MTHFR | rs1801133 | 556 | C | T | −0.17 (−0.44, 0.10) | 0.24 (0.16, 0.32) | −0.10 (−0.23, 0.04) | 0.16 | 0.48 |
| MTR | rs1805087 | 556 | A | G | −0.01 (−0.22, 0.19) | 0.22 (0.13, 0.31) | 0.00 (−0.11, 0.10) | 0.99 | 0.99 |
| MTRR | rs1801394 | 554 | A | G | −0.03 (−0.20, 0.15) | 0.10 (−0.02, 0.22) | 0.11 (0.02, 0.21) | 0.02 | 0.15 |
| SHMT1 | rs1979277 | 543 | G | A | 0.01 (−0.23, 0.24) | 0.22 (0.13, 0.30) | −0.02 (−0.14, 0.11) | 0.76 | 0.96 |
| SLC19A1 | rs1888530 | 546 | T | C | −0.07 (−0.25, 0.11) | 0.22 (0.10, 0.34) | 0.00 (−0.09, 0.09) | 0.92 | 0.99 |
| SLC46A1 | rs1128162 | 536 | T | G | −0.05 (−0.23, 0.13) | 0.25 (0.13, 0.37) | −0.03 (−0.12, 0.07) | 0.56 | 0.93 |
| TCN2 | rs1801198 | 557 | G | C | 0.13 (−0.06, 0.33) | 0.25 (0.14, 0.35) | −0.04 (−0.13, 0.05) | 0.44 | 0.88 |
| TYMS | rs1001761 | 550 | C | T | 0.11 (−0.08, 0.29) | 0.12 (0.00, 0.24) | 0.09 (0.00, 0.18) | 0.06 | 0.30 |
Conditional logistic regression model with predictors SNP (additive coding with AA, Aa, aa coded as 0, 1, and 2, respectively), log-transformed wAs centered at its median (log(71.5 μg/L)), and SNP × log-transformed wAs cross-product term, adjusted for age and education (categorical);
p-value and FDR for cross-product term
We explored the joint effects of the OCM SNPs and wAs exposure with cross-product terms with suggestive significance (p < 0.10) by constructing categorical variables for SNP genotypes and wAs, dichotomized at 50 μg/L (Table 6). For MTRR rs1801394 A→G, with lower wAs exposure, the GG genotype was associated with a decrease in skin lesion odds, indicating protective effects with lower wAs exposure (GG with wAs < 50 μg/L vs. AA with wAs < 50 μg/L, p = 0.02). A similar protective effect was observed for the T allele of FOLR1 rs1540087 C→T (CT/TT with wAs < 50 μg/L vs. CC with wAs < 50 μg/L, p = 0.04). For TYMS rs1001761 C→T, the T allele was associated with increased odds of skin lesions with high wAs exposures (CT with wAs ≥ 50 μg/L vs. CC with wAs < 50 μg/L, p = 0.009; CT with wAs ≥ 50 μg/L vs. CC with wAs < 50 μg/L, p = 0.003).
Table 6.
Joint effects of OCM genotypes with water As exposures on odds for incident skin lesions in conditional logistic regression models.
| Gene | SNP | Level | wAs < 50 μg/L OR (95% CI)a |
wAs ≥ 50 μg/L OR (95% CI)a |
|---|---|---|---|---|
| MTRR | rs1801394 | AA | ref. | 1.47 (0.87, 2.47) |
| AG | 0.76 (0.46, 1.26) | 1.44 (0.91, 2.29) | ||
| GG | 0.53 (0.30, 0.92) | 1.75 (1.06, 2.90) | ||
|
| ||||
| FOLR1 | rs1540087 | CC | ref. | 1.93 (1.46, 2.55) |
| CT/TT | 0.54 (0.29, 0.98) | 2.23 (1.34, 3.72) | ||
|
| ||||
| TYMS | rs1001761 | CC | ref. | 1.50 (0.89, 2.54) |
| CT | 0.88 (0.54, 1.43) | 1.85 (1.16, 2.94) | ||
| TT | 0.77 (0.45, 1.32) | 2.18 (1.31, 3.63) | ||
Conditional logistic regression model with listed categorical SNP/wAs predictor, adjusted for age and education
4. Discussion
In this nested case-control study of As-induced skin lesions in Bangladesh, we measured serum Hcys, uAs metabolites, and SNPs in OCM genes to address two primary objectives: first, to explore the sources and causal role of Hcys in the development of skin lesions using OCM genetic proxies; and second, to identify OCM genetic factors associated with uAs metabolite proportions and/or risk of skin lesions. First, we confirmed our group’s previous finding that serum HHcys was a risk factor for As-induced skin lesions (Pilsner et al. 2009). Additionally, we found decreased odds of skin lesions with higher proportions of DMA in urine independent of HHcys, suggesting that the involvement of Hcys in skin lesion development is independent of As methylation capacity. Next, we found that the risk allele for MTHFR 677 C→T, a nonsynonymous SNP known to influence Hcys concentrations in other populations, was associated with serum Hcys and higher urinary %MMA and nominally associated with lower %DMA, but was not associated with skin lesion risk. Suggestive gene-environment interactions were observed for three OCM SNPs and wAs for skin lesions: risk alleles for MTRR rs1801394 and FOLR1 rs1540087 were associated with lower odds for skin lesions with lower wAs exposures, and the risk allele for TYMS rs1001761 was associated with higher odds for skin lesions with higher wAs exposures, which must be replicated in future studies. Consistent with our 2009 study (Pilsner et al. 2009), elevated serum Hcys at baseline was predictive of the development of As-induced skin lesions. There was a marked dose-dependent association, where individuals with the highest Hcys levels (> 20 μM) were nearly 2.5 (95% CI: 1.6, 3.9) times as likely to develop skin lesions compared to individuals with the lowest Hcys levels (< 10 μM). While we initially hypothesized in 2009 that the link between Hcys and skin lesions might be attributable to a reduced capacity to methylate As (Pilsner et al. 2009), our results demonstrated that urinary As metabolite proportions and serum Hcys levels were independently associated with skin lesion incidence. To investigate other possible roles of Hcys, we explored the associations of OCM SNPs with Hcys levels. The MTHFR 677 C→T variant was strongly associated with elevated serum Hcys and explained a substantial proportion of the variance in Hcys levels. We failed to find an association between MTHFR 677 C→T and skin lesions, consistent with a previous case-control study of prevalent skin lesions in the HEALS cohort (Ahsan et al. 2007), which would seemingly contradict a causal role for Hcys in skin lesion pathogenesis. These observations are also consistent with a 2010 study of OCM SNPs and urothelial carcinoma (UC), which found that while MTHFR 677 C→T was associated with lower urinary %DMA and folate levels—which were both risk factors for UC—MTHFR 677 C→T was not associated with UC risk (Chung et al. 2010).
Although MTHFR SNPs have been used as proxies for HHcys in Mendelian randomization studies (Borges et al. 2016; Clarke et al. 2012), MTHFR is involved in numerous biologic processes that reflect other aspects of the OCM network that might influence As-induced skin lesion pathogenesis beyond the levels of Hcys. For example, 5:10-methylenetetrahydrofolate (5:10 methylene-THF), the substrate used by MTHFR for 5mTHF synthesis, is involved in the de novo production of thymidines (see Figure 1), and it has been hypothesized that 677 C→T variants, while having negative impacts on methylation capacity, might also have protective health effects by increasing DNA repair capacity and minimizing uracil-induced DNA double strand breaks (DSBs) (Blount et al. 1997; Ma et al. 1997). Given that AsIII is a direct inducer of DSBs in Saccharomyces cerevisiae (Litwin et al. 2013), it is plausible that the different 677 C→T genotypes might have both protective and deleterious effects on As-induced skin lesion risk. Further, since the biological influence of the 677 C→T variant is not exclusively through the modulation of Hcys levels, the null result for 677 C→T and skin lesions does not dismiss a possible causal role for Hcys.
MTHFR 677 C→T explained a higher proportion of the variance in baseline serum Hcys in healthy controls compared to individuals who went on to develop skin lesions (R2 = 15.8% vs. 7.0%, respectively), suggesting that there may be other important factors that explain serum Hcys levels among individuals at risk of skin lesions. Even in models containing additional covariates, the models explained 6% more of the variation in serum Hcys among controls. Our group’s 2009 study identified that the prevalence of folate deficiency two years prior to diagnosis was higher among incident skin lesion cases (Pilsner et al. 2009); given that dietary folate status is a major determinant of Hcys (Vollset et al. 2001), it is possible that among individuals who eventually go on to develop skin lesions, deficiencies in dietary folate and other important OCM nutrients (e.g., vitamin B6, riboflavin, cobalamin, betaine) play a larger role in determining Hcys levels than genetic factors. Alternatively, it is possible that there are other important genetic determinants of Hcys other than OCM genetic variants. Future studies should explore other nutritional, environmental, and genetic factors that explain the elevation in Hcys among individuals who are susceptible to developing skin lesions.
An increased capacity to fully methylate InAs to DMA (reflected by a higher urinary %DMA) was associated with a decreased risk of skin lesions. This finding is consistent with a Mendelian randomization study in the HEALS cohort that found strong evidence for a causal relationship between increased As methylation efficiency and decreased skin lesion risk (Pierce et al. 2013). The observed associations between SNPs and urinary As metabolites in the current study may provide insights into potential nutritional interventions to increase As methylation efficiency. The T allele of MTHFR 677 C→T was associated with higher urinary %MMA and lower urinary %DMA, confirming findings from studies in Eastern Europe (Lindberg et al. 2007), Argentina (Schläwicke Engström et al. 2007; Steinmaus et al. 2007), and China (Deng et al. 2007) and supporting a link between folate status and As methylation efficiency (Gamble et al. 2006; Peters et al. 2015). A missense variant in MTR (rs1805087, also known as 2756 A→G)—a gene encoding the cobalamin-dependent enzyme methionine reductase (MTR), which regenerates methionine (Met) from Hcys—was associated with reduced As methylation efficiency (higher urinary %InAs and lower urinary %DMA). Cobalamin (also known as vitamin B12) has been inconsistently linked to As metabolism in previous human studies (Gruber et al. 2012; Hall et al. 2009; Howe et al. 2014). We also identified a link between the A allele of rs1979277, a missense variant in SHMT1 (L474F), and higher urinary %InAs. The SHMT1 gene encodes for serine hydroxymethyltransferase (SHMT), a pyridoxal phosphate (Vitamin B6)-dependent enzyme that regulates the partitioning of one-carbon units between the synthesis of SAM (the substrate for As methylation) and the synthesis of thymidylate (Herbig et al. 2002). In vitro, the L474F variant blocks small ubiquitin-like modifier (SUMO)-ylation of SHMT, which may result in reduced nuclear translocation of SHMT and accumulation of SHMT in the cytoplasm (Woeller et al. 2007). Since cytoplasmic SHMT inhibits SAM synthesis by preferentially shunting one-carbon units toward thymidylate biosynthesis (Herbig et al. 2002), L474F might result in a reduction in SAM for As methylation.
Elevated Hcys, an indicator of OCM dysregulation, was associated with skin lesion incidence independently of urinary %DMA, and the association of TYMS rs1001761 with increased skin lesion risk highlights the potential role of OCM in As toxicity independent of As methylation efficiency. The TYMS gene encodes for thymidylate synthetase (TS), which uses the MTHFR substrate 5,10-methylenetetrahydrofolate for the methylation of 2-deoxy-uridine-5-monophosphate (dUMP) to 2-deoxy-thymidine-5-monophosphate (dTMP) (Carreras and Santi 1995) and is critical for DNA synthesis and repair (Boorstein and Pardee 1983; Johnston et al. 1995). Given that the rs1001761 risk allele is associated with increased odds for skin lesions with higher As exposures (> 50 μM), we hypothesize that DNA damage involving TS may be a mechanism of As toxicity at higher As concentrations. Consistent with this hypothesis, a recent study from the Stover group identified thymidylate biosynthesis as a sensitive target for As at levels observed in human populations (As trioxide (As2O3) at 0.5 μM, equivalent to 75 μg/L arsenic in water) (Kamynina et al. 2017). The study found that As exposure impaired folate-dependent dTMP biosynthesis, resulting in uracil misincorporation into DNA and genomic instability (Kamynina et al. 2017). Further, folate deficiency exacerbated the impact of As on uracil misincorporation and genomic instability (Kamynina et al. 2017), providing a potential mechanism linking elevated Hcys (a sensitive indicator of folate deficiency) and skin lesion risk. We caution that our TYMS finding did not persist after adjustment for multiple comparisons, but given the biological plausibility of the association, additional studies in human populations are warranted to replicate the link between TYMS and skin lesion development.
Additionally, we identified possible protective effects of variants in MTRR and FOLR1 with lower As exposures (< 50 μM) on risk of skin lesions. The missense variant rs1801394 in MTRR, also known as 66 A→G, is associated with reduced expression of methionine synthase reductase (MSR), which is required for MTR to convert Hcys to Met (Gaughan et al. 2001). The SNP rs7109250 is an intronic variant in the FOLR1 gene, which encodes for folate receptor alpha (FRα), a cellular folate transporter. These results were unexpected, and mechanisms to explain potential protective effects of these SNPs at low As exposures are unclear. These findings warrant additional genetic studies in humans and, if associations are replicated, further investigation in experimental models to explore pathogenic mechanisms.
To our knowledge, two previous candidate gene studies have explored the relationships among OCM gene variants and As-induced skin lesions. In a well-powered study with a discovery population and independent replication population in Pabna, Bangladesh, Seow and colleagues (2015) examined 25 SNPs in 23 genes involved in OCM, inflammation, and skin cancer, including three genes included in the current study (BHMT, MTHFD1, and TYMS). They observed SNP-wAs interactions for six SNPs in the discovery population, two of which were in OCM-related genes (dihydrofolate reductase [DHFR] and phosphatidylethanolamine N-methyltransferase [PEMT]) and one of which was significant in the replication population (rs1133400 in the skin cancer gene type I inositol-1,4,5-trisphosphate 5-phosphatase [INPP5A]). TYMS was not associated with skin lesion risk, though the TYMS SNP (rs699517) was different from the TYMS variants in the current study. A previous study in the HEALS population examined SNPs in MTHFR (rs1801131 and rs1801133 [677 C→T]) and did not find significant main effects or interactions with wAs on skin lesion risk (Ahsan et al. 2007), consistent with our findings.
Our study had several strengths, including the nested case-control design in a well-characterized longitudinal cohort and well-powered sample size. However, we also acknowledge several limitations. We did not have biologic samples (serum and/or DNA) for a small proportion of the cases and controls, which could potentially bias our results. In addition, we did not have serum and/or plasma samples that had not undergone a freeze-thaw cycle and were thus unable to measure plasma folate, which is a possible confounder in the relationships among Hcys, As metabolites, and skin lesions. Additionally, many of the SNP associations in our study did not persist after adjustment for multiple comparisons. It is thus imperative that the genetic findings are replicated in future studies before any mechanisms are explored experimentally. In future studies, we may follow up with these findings by assessing the effect of MTHFR 677 C→T on changes in folate, Hcys, and other biomarkers in our Folic Acid and Creatine Trial (FACT) (Peters et al. 2015), a randomized controlled trial of folic acid and creatine supplementation in the same region in Bangladesh.
CONCLUSIONS
We found that serum Hcys was a risk factor for As-induced skin lesions, and MTHFR 677 C→T was associated with elevated serum Hcys, increased urinary %MMA, and decreased urinary %DMA. However, MTHFR 677 C→T and other variants associated with urinary As metabolite proportions were not associated with skin lesion risk. There was a suggestive association between the T allele for TYMS rs1001761 and skin lesions with higher wAs exposures, consistent with recent experimental work indicating that the disruption of thymidylate biosynthesis may be a mechanism of As toxicity at higher exposures (Kamynina et al. 2017). Future studies should explore other determinants of HHcys, including non-genetic factors, to explain the link between HHcys and skin lesions in this population. Additionally, if the genetic findings are replicated in other populations, future studies should also explore how the roles of OCM in both As methylation and other processes might contribute to nuanced effects of OCM perturbations on the dose-dependent effects of As toxicity.
Supplementary Material
Highlights.
Arsenic (As) is methylated to MMA and DMA via one-carbon metabolism (OCM).
Elevated homocysteine (HHcys) is a marker of dysregulated OCM.
HHcys and lower %DMA were independent risk factors for As-induced skin lesions.
MTHFR C677T was associated with HHcys and %MMA, but not with skin lesions.
Dysregulated OCM may influence susceptibility to As toxicity.
Acknowledgments
Funding sources: This work was supported by the National Institutes of Health [grants R01 CA133595, R01 ES017875, P42 ES10349, P30 ES009089, and T32 CA009529-24]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health Sciences, the National Cancer Institute, or the National Institutes of Health.
Footnotes
Competing financial interests: The authors declare they have no actual or potential competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahsan H, Perrin M, Rahman A, Parvez F, Stute M, Zheng Y, et al. Associations between drinking water and urinary arsenic levels and skin lesions in bangladesh. Journal of occupational and environmental medicine. 2000;42:1195–1201. doi: 10.1097/00043764-200012000-00016. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Chen Y, Parvez F, Argos M, Hussain AI, Momotaj H, et al. Health effects of arsenic longitudinal study (heals): Description of a multidisciplinary epidemiologic investigation. Journal of exposure science & environmental epidemiology. 2006a;16:191–205. doi: 10.1038/sj.jea.7500449. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Chen Y, Parvez F, Zablotska L, Argos M, Hussain I, et al. Arsenic exposure from drinking water and risk of premalignant skin lesions in bangladesh: Baseline results from the health effects of arsenic longitudinal study. American journal of epidemiology. 2006b;163:1138–1148. doi: 10.1093/aje/kwj154. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Chen Y, Kibriya MG, Slavkovich V, Parvez F, Jasmine F, et al. Arsenic metabolism, genetic susceptibility, and risk of premalignant skin lesions in bangladesh. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:1270–1278. doi: 10.1158/1055-9965.EPI-06-0676. [DOI] [PubMed] [Google Scholar]
- Ansari R, Mahta A, Mallack E, Luo JJ. Hyperhomocysteinemia and neurologic disorders: A review. Journal of Clinical Neurology (Seoul, Korea) 2014;10:281–288. doi: 10.3988/jcn.2014.10.4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Kalra T, Rathouz PJ, Chen Y, Pierce B, Parvez F, et al. Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in bangladesh (heals): A prospective cohort study. Lancet (London, England) 2010;376:252–258. doi: 10.1016/S0140-6736(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe-Dimmer JL, Iyer PT, Nriagu JO, Keele GR, Mehta S, Meliker JR, et al. Genetic variation in glutathione s-transferase omega-1, arsenic methyltransferase and methylene-tetrahydrofolate reductase, arsenic exposure and bladder cancer: A case-control study. Environmental health: a global access science source. 2012;11:43. doi: 10.1186/1476-069X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein RJ, Pardee AB. Coordinate inhibition of DNA synthesis and thymidylate synthase activity following DNA damage and repair. Biochemical and biophysical research communications. 1983;117:30–36. doi: 10.1016/0006-291x(83)91536-x. [DOI] [PubMed] [Google Scholar]
- Borges MC, Hartwig FP, Oliveira IO, Horta BL. Is there a causal role for homocysteine concentration in blood pressure? A mendelian randomization study. The American journal of clinical nutrition. 2016;103:39–49. doi: 10.3945/ajcn.115.116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattstrom L, Wilcken DE. Homocysteine and cardiovascular disease: Cause or effect? The American journal of clinical nutrition. 2000;72:315–323. doi: 10.1093/ajcn/72.2.315. [DOI] [PubMed] [Google Scholar]
- Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annual review of biochemistry. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- Chandalia M, Abate N, Cabo-Chan AV, Jr, Devaraj S, Jialal I, Grundy SM. Hyperhomocysteinemia in asian indians living in the united states. The Journal of clinical endocrinology and metabolism. 2003;88:1089–1095. doi: 10.1210/jc.2002-021133. [DOI] [PubMed] [Google Scholar]
- Chung CJ, Pu YS, Su CT, Chen HW, Huang YK, Shiue HS, et al. Polymorphisms in one-carbon metabolism pathway genes, urinary arsenic profile, and urothelial carcinoma. Cancer causes & control: CCC. 2010;21:1605–1613. doi: 10.1007/s10552-010-9589-3. [DOI] [PubMed] [Google Scholar]
- Clarke R, Bennett DA, Parish S, Verhoef P, Dotsch-Klerk M, Lathrop M, et al. Homocysteine and coronary heart disease: Meta-analysis of mthfr case-control studies, avoiding publication bias. PLoS medicine. 2012;9:e1001177. doi: 10.1371/journal.pmed.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribari-Neto F, Zeileis A. Beta regression in r 2010. 2010;34:24. [Google Scholar]
- Dayal S, Arning E, Bottiglieri T, Boger RH, Sigmund CD, Faraci FM, et al. Cerebral vascular dysfunction mediated by superoxide in hyperhomocysteinemic mice. Stroke; a journal of cerebral circulation. 2004;35:1957–1962. doi: 10.1161/01.STR.0000131749.81508.18. [DOI] [PubMed] [Google Scholar]
- Deng FR, Guo XB, Chen L, Wang ZQ, Zhang K. relationship between 5,10-methylenetetrahydrofolate reductase genetic polymorphism and arsenic metabolism. Beijing da xue xue bao Yi xue ban = Journal of Peking University Health sciences. 2007;39:149–152. [PubMed] [Google Scholar]
- Eberhardt RT, Forgione MA, Cap A, Leopold JA, Rudd MA, Trolliet M, et al. Endothelial dysfunction in a murine model of mild hyperhomocyst(e)inemia. The Journal of clinical investigation. 2000;106:483–491. doi: 10.1172/JCI8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom K, Vahter M, Mlakar SJ, Concha G, Nermell B, Raqib R, et al. Polymorphisms in arsenic(+iii oxidation state) methyltransferase (as3mt) predict gene expression of as3mt as well as arsenic metabolism. Environmental health perspectives. 2011;119:182–188. doi: 10.1289/ehp.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan SF, Karagas MR, Jiang J, Wu F, Liu M, Newman JD, et al. Gene-arsenic interaction in longitudinal changes of blood pressure: Findings from the health effects of arsenic longitudinal study (heals) in bangladesh. Toxicol Appl Pharmacol. 2015;288:95–105. doi: 10.1016/j.taap.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Cribari-Neto F. Beta regression for modelling rates and proportions. Journal of Applied Statistics. 2004;31:799–815. [Google Scholar]
- Gamble MV, Ahsan H, Liu X, Factor-Litvak P, Ilievski V, Slavkovich V, et al. Folate and cobalamin deficiencies and hyperhomocysteinemia in bangladesh. The American journal of clinical nutrition. 2005;81:1372–1377. doi: 10.1093/ajcn/81.6.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MV, Liu X, Ahsan H, Pilsner JR, Ilievski V, Slavkovich V, et al. Folate and arsenic metabolism: A double-blind, placebo-controlled folic acid-supplementation trial in bangladesh. The American journal of clinical nutrition. 2006;84:1093–1101. doi: 10.1093/ajcn/84.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble MV, Liu X, Slavkovich V, Pilsner JR, Ilievski V, Factor-Litvak P, et al. Folic acid supplementation lowers blood arsenic. The American journal of clinical nutrition. 2007;86:1202–1209. doi: 10.1093/ajcn/86.4.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa-Loira B, Cebrian ME, Salinas-Rodriguez A, Lopez-Carrillo L. Genetic susceptibility to breast cancer risk associated with inorganic arsenic exposure. Environmental toxicology and pharmacology. 2017;56:106–113. doi: 10.1016/j.etap.2017.08.032. [DOI] [PubMed] [Google Scholar]
- Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutrition journal. 2015;14:6. doi: 10.1186/1475-2891-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan DJ, Kluijtmans LAJ, Barbaux S, McMaster D, Young IS, Yarnell JWG, et al. The methionine synthase reductase (mtrr) a66g polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. 2001;157:451–456. doi: 10.1016/s0021-9150(00)00739-5. [DOI] [PubMed] [Google Scholar]
- Gregory Warnes wcfGG, Friedrich Leisch and Michael Man. 2013. Genetics: Population genetics. Part R package version 1.3.8.1.
- Gruber JF, Karagas MR, Gilbert-Diamond D, Bagley PJ, Zens MS, Sayarath V, et al. Associations between toenail arsenic concentration and dietary factors in a new hampshire population. Nutrition journal. 2012;11:45. doi: 10.1186/1475-2891-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MN, Liu X, Slavkovich V, Ilievski V, Mi Z, Alam S, et al. Influence of cobalamin on arsenic metabolism in bangladesh. Environmental health perspectives. 2009;117:1724–1729. doi: 10.1289/ehp.0900734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang E-P, Lee L-R, Hills J, Shane B, Stover PJ. Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide ands-adenosylmethionine biosyntheses. Journal of Biological Chemistry. 2002;277:38381–38389. doi: 10.1074/jbc.M205000200. [DOI] [PubMed] [Google Scholar]
- Howe CG, Niedzwiecki MM, Hall MN, Liu X, Ilievski V, Slavkovich V, et al. Folate and cobalamin modify associations between s-adenosylmethionine and methylated arsenic metabolites in arsenic-exposed bangladeshi adults. The Journal of Nutrition. 2014;144:690–697. doi: 10.3945/jn.113.188789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, et al. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation. 1996;93:7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- Johnston PG, Lenz H-J, Leichman CG, Danenberg KD, Allegra CJ, Danenberg PV, et al. Thymidylate synthase gene and protein expression correlate and are associated with response to 5-fluorouracil in human colorectal and gastric tumors. Cancer Research. 1995;55:1407–1412. [PubMed] [Google Scholar]
- Kamynina E, Lachenauer ER, DiRisio AC, Liebenthal RP, Field MS, Stover PJ. Arsenic trioxide targets mthfd1 and sumo-dependent nuclear de novo thymidylate biosynthesis. Proceedings of the National Academy of Sciences. 2017;114:E2319–E2326. doi: 10.1073/pnas.1619745114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Park S, Nelson HH, Andrew AS, Mott L, Schned A, et al. Methylenetetrahydrofolate reductase (mthfr) variants and bladder cancer: A population-based case-control study. Int J Hyg Environ Health. 2005;208:321–327. doi: 10.1016/j.ijheh.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Karagas MR, Gossai A, Pierce B, Ahsan H. Drinking water arsenic contamination, skin lesions, and malignancies: A systematic review of the global evidence. Current environmental health reports. 2015;2:52–68. doi: 10.1007/s40572-014-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, et al. A novel s-adenosyl-l-methionine:Arsenic(iii) methyltransferase from rat liver cytosol. The Journal of biological chemistry. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Lindberg A-L, Kumar R, Goessler W, Thirumaran R, Gurzau E, Koppova K, et al. Metabolism of low-dose inorganic arsenic in a central european population: Influence of sex and genetic polymorphisms. Environmental health perspectives. 2007;115:1081–1086. doi: 10.1289/ehp.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin I, Bocer T, Dziadkowiec D, Wysocki R. Oxidative stress and replication-independent DNA breakage induced by arsenic in saccharomyces cerevisiae. PLoS genetics. 2013;9:e1003640. doi: 10.1371/journal.pgen.1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and the one-carbon cycle: Cancer metabolism in full circle. Nature reviews Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, Fuchs C, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Research. 1997;57:1098–1102. [PubMed] [Google Scholar]
- Mazumdar M, Valeri L, Rodrigues EG, Ibne Hasan MO, Hamid R, Paul L, et al. Polymorphisms in maternal folate pathway genes interact with arsenic in drinking water to influence risk of myelomeningocele. Birth defects research Part A, Clinical and molecular teratology. 2015;103:754–762. doi: 10.1002/bdra.23399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Guallar E, Navas-Acien A. Arsenic exposure and cardiovascular disease: An updated systematic review. Current atherosclerosis reports. 2012;14:542–555. doi: 10.1007/s11883-012-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Silvera SA, Rohan TE. Trace elements and cancer risk: A review of the epidemiologic evidence. Cancer causes & control: CCC. 2007;18:7–27. doi: 10.1007/s10552-006-0057-z. [DOI] [PubMed] [Google Scholar]
- Nixon DE, Mussmann GV, Eckdahl SJ, Moyer TP. Total arsenic in urine: Palladium-persulfate vs nickel as a matrix modifier for graphite furnace atomic absorption spectrophotometry. Clinical chemistry. 1991;37:1575–1579. [PubMed] [Google Scholar]
- Peters BA, Hall MN, Liu X, Parvez F, Sanchez TR, van Geen A, et al. Folic acid and creatine as therapeutic approaches to lower blood arsenic: A randomized controlled trial. Environmental health perspectives. 2015;123:1294–1301. doi: 10.1289/ehp.1409396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer CM, Huff DL, Gunter EW. Rapid and accurate hplc assay for plasma total homocysteine and cysteine in a clinical laboratory setting. Clinical chemistry. 1999;45:290–292. [PubMed] [Google Scholar]
- Pierce BL, Tong L, Argos M, Gao J, Jasmine F, Roy S, et al. Arsenic metabolism efficiency has a causal role in arsenic toxicity: Mendelian randomization and gene-environment interaction. International journal of epidemiology. 2013;42:1862–1872. doi: 10.1093/ije/dyt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, et al. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environmental health perspectives. 2009;117:254–260. doi: 10.1289/ehp.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KE, Basu A, Hubbard AE, Bates MN, Kalman D, Rey O, et al. Association of genetic variation in cystathionine-beta-synthase and arsenic metabolism. Environ Res. 2010;110:580–587. doi: 10.1016/j.envres.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DG, Lindenbaum J, Stabler SP, Allen RH. Sensitivity of serum methylmalonic acid and total homocysteine determinations for diagnosing cobalamin and folate deficiencies. The American journal of medicine. 1994;96:239–246. doi: 10.1016/0002-9343(94)90149-x. [DOI] [PubMed] [Google Scholar]
- Schlawicke Engstrom K, Nermell B, Concha G, Stromberg U, Vahter M, Broberg K. Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutation research. 2009;667:4–14. doi: 10.1016/j.mrfmmm.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schläwicke Engström K, Broberg K, Concha G, Nermell B, Warholm M, Vahter M. Genetic polymorphisms influencing arsenic metabolism: Evidence from argentina. Environmental health perspectives. 2007;115:599–605. doi: 10.1289/ehp.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JM, Weir DG. Folic acid, homocysteine and one-carbon metabolism: A review of the essential biochemistry. Journal of cardiovascular risk. 1998;5:223–227. [PubMed] [Google Scholar]
- Senaratne MP, MacDonald K, De Silva D. Possible ethnic differences in plasma homocysteine levels associated with coronary artery disease between south asian and east asian immigrants. Clinical cardiology. 2001;24:730–734. doi: 10.1002/clc.4960241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow WJ, Pan WC, Kile ML, Tong L, Baccarelli AA, Quamruzzaman Q, et al. A distinct and replicable variant of the squamous cell carcinoma gene inositol polyphosphate-5-phosphatase modifies the susceptibility of arsenic-associated skin lesions in bangladesh. Cancer. 2015;121:2222–2229. doi: 10.1002/cncr.29291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. Dbsnp: The ncbi database of genetic variation. Nucleic acids research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot C. Plasma creatinine determination. A new and specific jaffe reaction method. Scandinavian journal of clinical and laboratory investigation. 1965;17:381–387. doi: 10.3109/00365516509077065. [DOI] [PubMed] [Google Scholar]
- Steinmaus C, Moore LE, Shipp M, Kalman D, Rey OA, Biggs ML, et al. Genetic polymorphisms in mthfr 677 and 1298, gstm1 and t1, and metabolism of arsenic. Journal of Toxicology and Environmental Health, Part A. 2007;70:159–170. doi: 10.1080/15287390600755240. [DOI] [PubMed] [Google Scholar]
- Team R. Rstudio: Integrated development for r. Boston, MA: RStudio, Inc; 2015. [Google Scholar]
- The Genomes Project C. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler CR, Allan AM. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: A review. Current environmental health reports. 2014;1:132–147. doi: 10.1007/s40572-014-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Geen A, Cheng Z, Seddique AA, Hoque MA, Gelman A, Graziano JH, et al. Reliability of a commercial kit to test groundwater for arsenic in bangladesh. Environmental science & technology. 2005;39:299–303. [PubMed] [Google Scholar]
- Vollset SE, Refsum H, Ueland PM. Population determinants of homocysteine. The American journal of clinical nutrition. 2001;73:499–500. doi: 10.1093/ajcn/73.3.499. [DOI] [PubMed] [Google Scholar]
- Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: Evidence on causality from a meta-analysis. BMJ (Clinical research ed) 2002;325:1202. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald DS, Bestwick JP, Wald NJ. Homocysteine as a cause of ischemic heart disease: The door remains open. Clinical chemistry. 2012;58:1488–1490. doi: 10.1373/clinchem.2012.191791. [DOI] [PubMed] [Google Scholar]
- Woeller CF, Anderson DD, Szebenyi DM, Stover PJ. Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. The Journal of biological chemistry. 2007;282:17623–17631. doi: 10.1074/jbc.M702526200. [DOI] [PubMed] [Google Scholar]
- Wu LL, Wu JT. Hyperhomocysteinemia is a risk factor for cancer and a new potential tumor marker. Clinica chimica acta; international journal of clinical chemistry. 2002;322:21–28. doi: 10.1016/s0009-8981(02)00174-2. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Stute M, van Geen A, Gavrieli I, Dhar R, Simpson HJ, et al. Redox control of arsenic mobilization in bangladesh groundwater. Applied Geochemistry. 2004;19:201–214. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.