Abstract
Recently, haploinsufficiency of PURA has been identified as an essential cause of 5q31.3 microdeletion syndrome, which is characterized by severe psychomotor developmental delay, epilepsy, distinctive features, and delayed myelination. A new 5q31.2-q31.3 microdeletion that included PURA was identified in a patient with infantile spasms. Approximately 50% of patients with PURA-related neurodevelopmental disorders exhibited epilepsy regardless of whether they harbor a 5q31.3 deletion or PURA mutation. Patients with the 5q31.3 deletion or a PURA mutation should be carefully monitored for epileptic seizures.
In 2011, the first two patients with 5q31 microdeletion syndrome manifesting severe psychomotor development delay, epilepsy, distinctive features, and delayed myelination were reported by our group.1 Thereafter, several overlapping deletions have been identified in patients with similar clinical phenotypes.2–4 From these findings, the 5q31.3 microdeletion syndrome was established, and a gene responsible for these clinical features was suspected to be located in this region. Genotype–phenotype correlation using the data from these findings narrowed the region responsible for the common clinical features. Then the purine-rich element binding protein A gene (PURA, NM_005859) located on the shortest region of overlap was considered as a gene responsible for the 5q31 microdeletion syndrome. PURA is a small gene with only one exon with 322 amino acids. Detailed functions of this gene have not been recognized to date. In 2014, whole-exome sequencing finally revealed several de novo PURA mutations in patients with clinical features similar to the 5q31.3 microdeletion syndrome.5,6 Therefore, it was confirmed that PURA is a gene responsible for the 5q31.3 microdeletion syndrome. Recently, we experienced a new case of 5q31.3 microdeletion, including PURA, and the genotype–phenotype correlation was re-analyzed.
A 2-year-old boy was born with a birth weight of 3385 g (75–90th percentile), a length of 52.5 cm (>97th percentile), and occipito-frontal circumference of 33.0 cm (25–50th percentile) at 41 weeks and 5 days of gestation. Apnea attacks were often observed until 4 months. Given that electroencephalogram revealed no epileptic discharge, the apnea was considered to be the consequence of immature brain development. The patient was discharged from the hospital with oxygen supply. Muscular hypotonia was noted in early infancy. At 12 months, he started to exhibit spasms, and hypsarrhythmia was observed by electroencephalogram. After prescription of valproate acids, spasms disappeared. Hypsarrhythmia also disappeared, and no definite electroencephalogram abnormality was observed at that time. However, sporadic sharp waves were recorded in the left frontal area since 2 years.
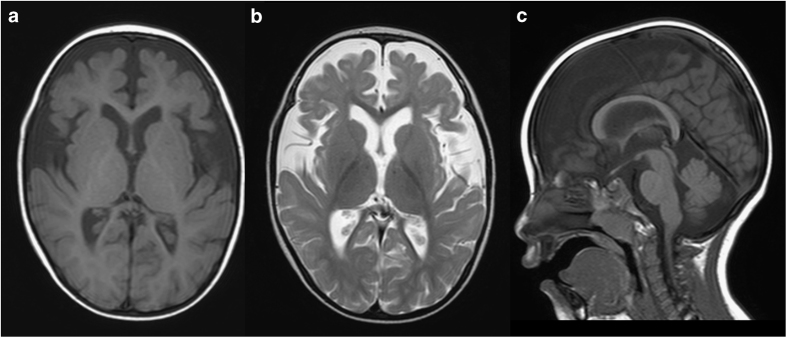
At present, his weight is 9.7 kg (<3rd percentile), length is 83.4 cm (3–10th percentile), and occipito-frontal circumference is 44.5 cm (<3rd percentile), indicating postnatal microcephaly. He exhibits a flat face, hypertelorism, short nose, thin upper lip, and down-turned corner of the mouth. His head is still not controlled. Rolling over has never been achieved. Brain magnetic resonance imaging examination at 2 years revealed cerebral volume loss in association with delayed myelination in the deep white matter (Figure 1).
Figure 1.
Brain magnetic resonance imaging examination at 2 years of age. Axial T1- and T2-weighted images (a and b, respectively) and a sagittal T1-weighted image (c). Decreased volume of the cerebrum (a, b) and delayed myelination in the deep white matter (b) are evident. The volume of the corpus callosum is also reduced (c).
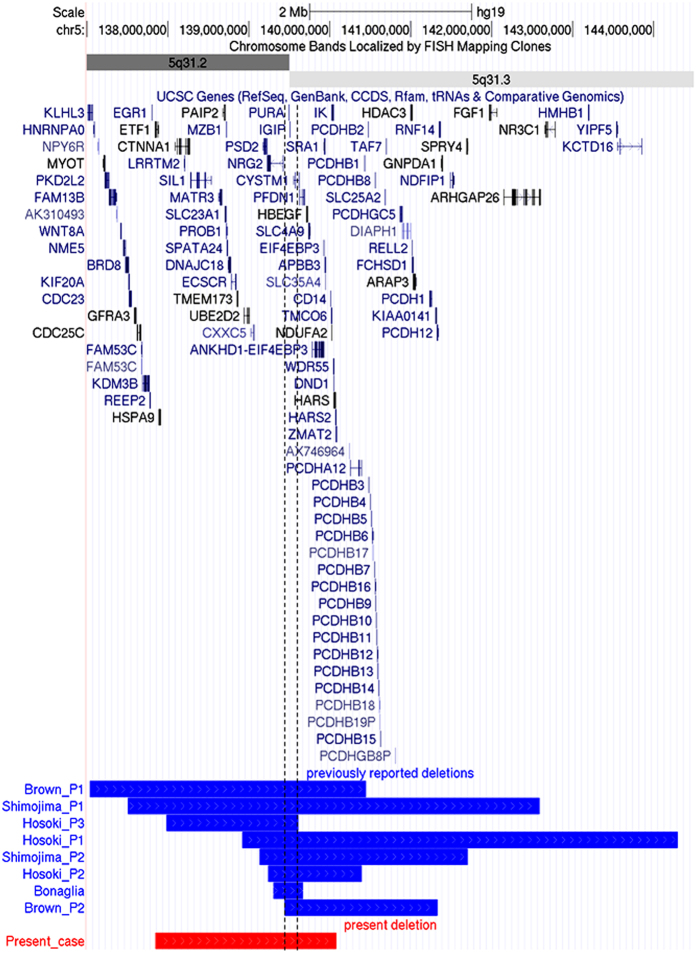
The study was performed in accordance with the principles in the Declaration of Helsinki and was approved by the ethics committee of Tokyo Women’s Medical University. Blood samples were obtained from the patient and his parents after receiving written informed consent. Chromosomal microarray testing using the Agilent 60 K Human Genome CGH Microarray platform (Agilent Technologies, Santa Clara, CA) was performed in accordance with previous descriptions of the method.7 Then a genomic copy number loss with a size of 2.2- Mb was identified and represented as arr[hg19] 5q31.2q31.3(137,851,716–140,085,772)X1 (Figure 2, Supplementary Figure S1). Subsequent fluorescence in situ hybridization analysis using BAC clones with RP11-678N8 (5q31.2:138,944,793–139,148,607) and RP11-78J2 (5p15.32:5,760,940–5,917,401) confirmed a deletion in the proband (Supplementary Figure S2). Given that parental fluorescence in situ hybridization analyses revealed no deletion, the microdeletion in the proband was confirmed as de novo.
Figure 2.
Genome map captured from the UCSC genome browser (https://genome.ucsc.edu/). The custom tracks reveal the deletion regions in the eight previously reported patients and the present patient depicted as blue lines and a red line, respectively. The shortest region of overlap is depicted as dotted lines for better understanding. All deletion regions are adapted to hg19.
Previously, chromosomal 5q31.3 deletions have been identified in eight patients (Figure 2, Supplementary Table S1). Almost all patients exhibited generalized hypotonia. Thus feeding difficulties and respiratory difficulties were also observed. Patients were nonverbal and not ambulatory due to severe psychomotor developmental delay. Distinctive facial features were also consistent. These findings are similar in patients derived from nucleotide alterations in PURA.8,9 This finding suggests that haploinsufficiency of PURA is an essential cause of 5q31.3 microdeletion syndrome. Now, the term “PURA-related neurodevelopmental disorders” is recommended to be used.10
In comparison, seizures and delayed myelination revealed by magnetic resonance imaging were observed in approximately 50% of patients. Although PURA is registered as MIM#616158 in Online Mendelian Inheritance in Man (OMIM; https://www.omim.org/) as a gene related to mental retardation, autosomal-dominant 31 (MIM#616158), approximately half of patients with PURA-related neurodevelopmental disorders exhibit epilepsy. In particular, the infantile spasms exhibited in the patient reported in this study have also been observed in several patients. Therefore, patients with PURA-related neurodevelopmental disorders should be carefully monitored for epileptic seizures.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We would like to express our gratitude to the patient and his family for their cooperation. This work is mainly supported by a Grant-in-Aid for Young Scientists (B) from Japan Society for the Promotion of Science (JSPS) (to KS). KS is grateful for a JSPS Research Fellowships for Young Scientists (Restart Postdoctoral Fellow; RPD) in association with a Grant-in-Aid for JSPS Research Fellow. This study is partially supported by a Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development (AMED), a grant from the Ministry of Health, Labour and Welfare, and a grant-in-aid for Scientific Research C from Japan Society for the Promotion of Science (JSPS) (all to TY).
Footnotes
Supplemental Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv).
The authors declare no conflict of interest.
References
- Shimojima K, Isidor B, Le Caignec C, Kondo A, Sakata S, Ohno K et al. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A 2011; 155 A: 732–736. [DOI] [PubMed] [Google Scholar]
- Hosoki K, Ohta T, Natsume J, Imai S, Okumura A, Matsui T et al. Clinical phenotype and candidate genes for the 5q31.3 microdeletion syndrome. Am J Med Genet A 2012; 158A: 1891–1896. [DOI] [PubMed] [Google Scholar]
- Brown N, Burgess T, Forbes R, McGillivray G, Kornberg A, Mandelstam S et al. 5q31.3 Microdeletion syndrome: clinical and molecular characterization of two further cases. Am J Med Genet A 2013; 161 A: 2604–2608. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Zanotta N, Giorda R, D'Angelo G, Zucca C. Long-term follow-up of a patient with 5q31.3 microdeletion syndrome and the smallest de novo 5q31.2q31.3 deletion involving PURA. Mol Cytogenet 2015; 8: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR et al. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet 2014; 95: 579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D et al. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet 2014; 51: 806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K et al. Subtelomeric deletions of 1q43q44 and severe brain impairment associated with delayed myelination. J Hum Genet 2012; 57: 593–600. [DOI] [PubMed] [Google Scholar]
- Okamoto N, Nakao H, Niihori T, Aoki Y. Patient with a novel purine-rich element binding protein A mutation. Congenit Anom (Kyoto) 2017; 57: 201–204. [DOI] [PubMed] [Google Scholar]
- Tanaka AJ et al. De novo mutations in PURA are associated with hypotonia and developmental delay. Cold Spring Harb Mol Case Stud 2015; 1: a000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnders MRF et al. PURA-related neurodevelopmental disorders. In: GeneReviews, Pagon RA et al. (eds). University of Washington: Seattle (WA) , USA, 2017. [PubMed]
Data Citations
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1848. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1767. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1770. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1893. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1776. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1779. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1782. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1785. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1896. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1791. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1794. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1797. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1899. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1803. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1806. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1809. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1902. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1815. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1818. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1821. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1905. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1827. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1830. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1833. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1908. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1839. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1842. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1845. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1851. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1857. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1860. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1863. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1911. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1869. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1872. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1875. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1878. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1881. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1884. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1887. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1848. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1767. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1770. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1893. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1776. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1779. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1782. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1785. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1896. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1791. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1794. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1797. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1899. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1803. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1806. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1809. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1902. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1815. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1818. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1821. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1905. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1827. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1830. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1833. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1908. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1839. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1842. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1845. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1851. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1857. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1860. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1863. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1911. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1869. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1872. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1875. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1878. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1881. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1884. [DOI]
- Yamamoto Toshiyuki.HGV Database. 2018. 10.6084/m9.figshare.hgv.1887. [DOI]