ABSTRACT
Polyamides have been shown to bind double-stranded DNA by complementing the curvature of the minor groove and forming various hydrogen bonds with DNA. Several polyamide molecules have been found to have potent antiviral activities against papillomavirus, a double-stranded DNA virus. By analogy, we reason that polyamides may also interact with the structured RNA bound in the nucleocapsid of a negative-strand RNA virus. Vesicular stomatitis virus (VSV) was selected as a prototype virus to test this possibility since its genomic RNA encapsidated in the nucleocapsid forms a structure resembling one strand of an A-form RNA duplex. One polyamide molecule, UMSL1011, was found to inhibit infection of VSV. To confirm that the polyamide targeted the nucleocapsid, a nucleocapsid-like particle (NLP) was incubated with UMSL1011. The encapsidated RNA in the polyamide-treated NLP was protected from thermo-release and digestion by RNase A. UMSL1011 also inhibits viral RNA synthesis in the intracellular activity assay for the viral RNA-dependent RNA polymerase. The crystal structure revealed that UMSL1011 binds the structured RNA in the nucleocapsid. The conclusion of our studies is that the RNA in the nucleocapsid is a viable antiviral target of polyamides. Since the RNA structure in the nucleocapsid is similar in all negative-strand RNA viruses, polyamides may be optimized to target the specific RNA genome of a negative-strand RNA virus, such as respiratory syncytial virus and Ebola virus.
IMPORTANCE Negative-strand RNA viruses (NSVs) include several life-threatening pathogens, such as rabies virus, respiratory syncytial virus, and Ebola virus. There are no effective antiviral drugs against these viruses. Polyamides offer an exceptional opportunity because they may be optimized to target each NSV. Our studies on vesicular stomatitis virus, an NSV, demonstrated that a polyamide molecule could specifically target the viral RNA in the nucleocapsid and inhibit viral growth. The target specificity of the polyamide molecule was proved by its inhibition of thermo-release and RNA nuclease digestion of the RNA bound in a model nucleocapsid, and a crystal structure of the polyamide inside the nucleocapsid. This encouraging observation provided the proof-of-concept rationale for designing polyamides as antiviral drugs against NSVs.
KEYWORDS: RNA structure, antiviral agents, negative-strand RNA virus, nucleocapsid, viral RNA synthesis
INTRODUCTION
Many pathogenic viruses are negative-strand RNA viruses (NSVs), such as respiratory syncytial virus (RSV), and parainfluenza viruses (e.g., PIV3), which cause severe infections in children. For instance, RSV accounts for up to 80% of hospitalization due to acute respiratory infections in children (1). Treatment options are limited, and no effective vaccines are currently available, which creates an urgent unmet need for novel antiviral medications for respiratory illness in children and adults. Furthermore, deadlier NSVs include rabies virus and Ebola virus, for which no effective antiviral drugs are available (2–4).
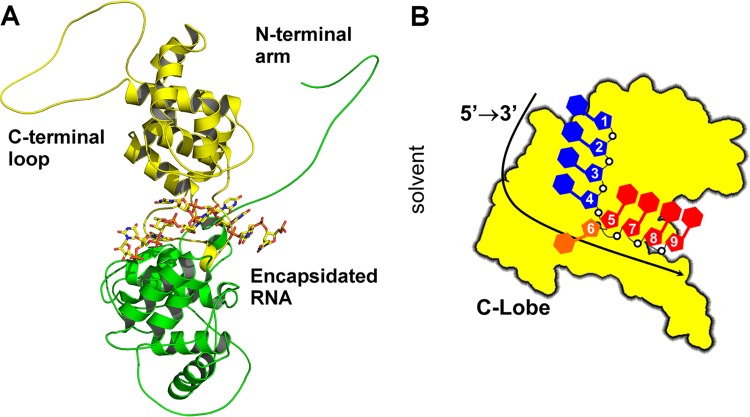
One common feature of all NSVs is that they encode their own RNA-dependent RNA polymerase (vRdRp) in the genome. During transcription/replication, the vRdRp must access the structured RNA genome sequestered in the nucleocapsid and will not recognize the free RNA genome as a template for RNA synthesis. This makes the nucleocapsid a prime target for broad antiviral therapies. The crystal structure of a nucleocapsid-like particle (NLP) was first solved for vesicular stomatitis virus (VSV) (5), revealing that the nucleocapsid protein (N) forms a polymer with extensive intermolecular interactions to package the genomic RNA (6). The N protein has a deep cleft between its N-terminal and C-terminal lobes where the genomic RNA is sequestered (Fig. 1A). When the genomic RNA is packaged in the nucleocapsid, it assumes a unique structure (Fig. 1B). If we count the nucleotides from the 5′ end to the 3′ end, nine nucleotides are covered by each N subunit. The bases of nucleotide 1 to 4 are stacked with the bases facing the exterior of the cleft, whereas bases of nucleotide 5 and nucleotides 7 to 9 are stacked with the bases facing the interior of the cleft. The base of nucleotide 6 does not stack with other bases. The base stacking of nucleotides 1 to 4, 5, and 7 to 9 is curved. The orientation of the ribose and the phosphate groups do not have a regular conformation either. The bases of VSV RNA are more parallel to each other in nucleotides 1 to 4 than in nucleotides 5 and 7 to 9.
FIG 1.
RNA structure in VSV NLP. (A) Ribbon and stick drawings of VSV N in complex with RNA. The N-terminal lobe is shown as ribbons in green, and the C-terminal lobe is indicated in yellow. The N-terminal arm and the C-terminal loop elements are labeled. The RNA structure in the center is shown as a stick model. (B) Cartoon diagram of a single subunit in the VSV NLP from the perspective of the N-lobe. This shows the RNA base stacking that occurs within the NLP. All drawings were created with PyMOL (51).
One such class of drugs that have been shown to target nucleotide moieties are polyamides. Several polyamides have been developed as inhibitors of gene expression and other biological effectors that bind double-stranded DNA in a sequence-specific manner (7–18), and other polyamides have also been developed as broad-spectrum antivirals for papillomaviruses and polyomaviruses precisely because they do not show sequence-specific binding properties (19–25; J. K. Bashkin et al., unpublished data). Others have also since discovered nonspecific polyamide effects (26, 27). Polyamides have not been cytotoxic to HeLa (28) or other mammalian cells in these reports, in one case showing selectivity index (SI) values of at least 2,000 before reaching compound solubility limits (22). Within the G nucleotides of a DNA helix, imidazole (Im) moieties of polyamides can act as an H-bond acceptor of one hydrogen from the exocyclic NH2, as predicted by Dickerson and coworkers (29). Furthermore, pyrrole (Py) residues and β-alanine residues (β) typically bind to A, T, and C through H-bond donation from amide bond NH groups to acceptor N or O groups in the minor groove.
Since polyamides have been shown to target structured nucleotide moieties and form hydrogen bonds, such as the structure found in the nucleocapsid-bound RNA of VSV, we tested a group of polyamides for possible antiviral activities against VSV (Table 1). One molecule, UMSL1011 (Fig. 2), was found to inhibit VSV replication. Further experiments found that UMSL1011 could protect the encapsidated RNA from thermo-release, digestion by RNase A, and inhibit vRdRp mRNA synthesis. Finally, the crystal structure of UMSL1011 bound to the VSV NLP was solved, and it was found to directly target the encapsidated RNA. Our results indicate a novel use of polyamides and can potentially be developed as antiviral drugs against other NSVs which cause widespread diseases. This opens a new avenue for drug discovery to effectively control NSV infection.
TABLE 1.
Polyamides used in antiviral screen, with their building blocks defineda
| Compound | N terminus | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | C terminus | Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UMSL1011 | dIm | Py | Py | Im | γ | Py | Py | Py | Py | β | Dp | Active |
| UMSL1013b | dIm | Py | Py | Py | γ | Py | Py | Im | Py | β | Dp | Inactive |
| UMSL1028c | dIm | Py | Py | β | Py | Py | Py | y | ||||
| Py | Py | β | Py | Py | Py | Py | β | Ta | Inactive | |||
| UMSL1055 | dIm | Im | Py | Py | γ | Py | Im | β | Py | β | Dp | Inactive |
| UMSL2115 | fPyd | Im | β | Im | γ | Py | Py | β | Py | β | Ta | Inactive |
| UMSL2082 | dIm | Im | Py | γ | Py | Py | Py | β | Ta | Inactive |
Im indicates an N-terminal n-methyl imidazole lacking an amino group (des-amino-N-methylimidazole-2-carboxylate). Py is derived from 4-amino-2-carboxy-N-methylpyrrole; Im is derived from 4-amino-2-carboxy-N-methylimidazole. β is derived from β-alanine; γ is derived from from γ-aminobutyric acid. Dp is derived from N,N-dimethylpropylamine; Ta is derived from bis(aminopropyl)methylamine.
UMSL1028 is a 16-residue polyamide active against HPV16, HPV18, and HPV31.
fPy is derived from 4-formamido-N-methylpyrrole-2-carboxylate.
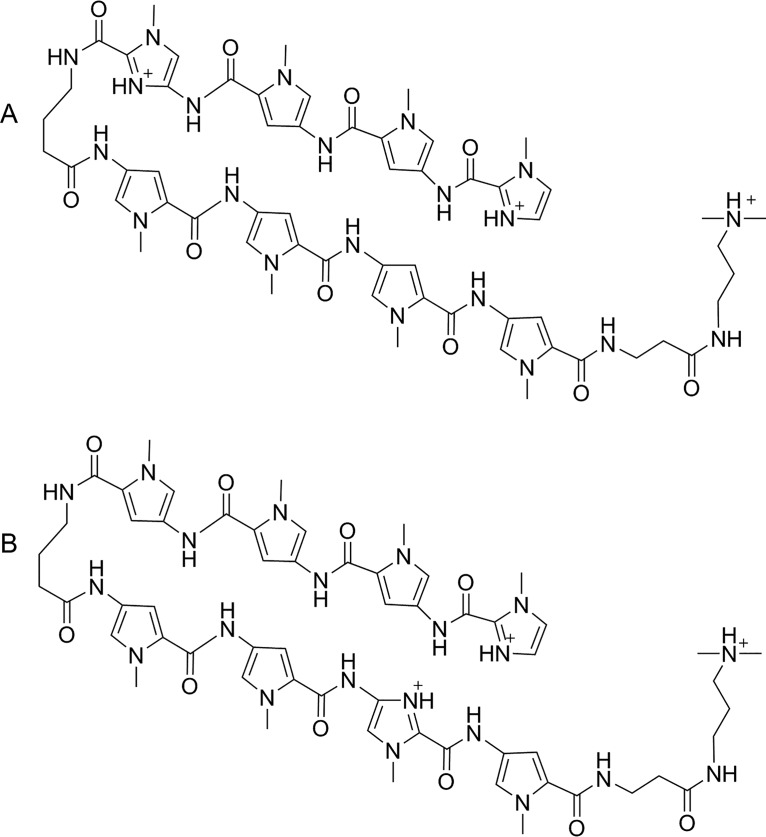
FIG 2.
(A) Cation of UMSL1011 isolated as a tris(TFA) salt. (B) Cation of UMSL1013 isolated as a tris(TFA) salt.
RESULTS
Antiviral screen.
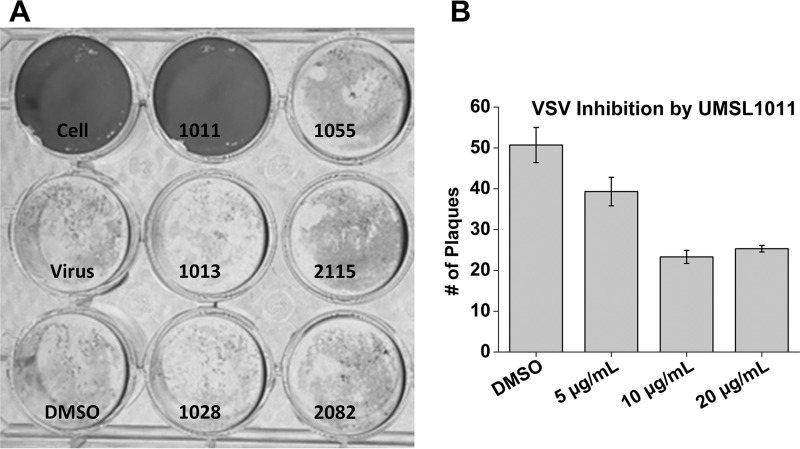
A plaque assay for VSV infection was set up for antiviral screening. Six different polyamide compounds (Table 1) prepared previously were included. When inhibitor solutions at a 20 μg/ml (130 μM) concentration of polyamides in 1% dimethyl sulfoxide (DMSO) were added, UMSL1011 inhibited VSV infection (Fig. 3). This result confirmed that a polyamide compound could inhibit infection by VSV. The antiviral activity by UMSL1011 is specific because other polyamide compounds did not inhibit VSV infection at this concentration. Inhibition of VSV infection by UMSL1011 was repeated with different concentrations of UMSL1011 (Fig. 3). At <10 μg/ml (<65 μM), UMSL1011 inhibits plaque formation by VSV infection.
FIG 3.
VSV plaque assays. (A) An initial screen of six polyamide compounds (20 μg/ml) was performed. Controls of uninfected cells, medium only, and 1% DMSO were included. The compound names were labeled for each well. (B) After virus adsorption for 1 h, UMSL1011 at different concentrations was added to the agarose overlap. DMSO was used as the solvent negative control. Experiments were performed in triplicate, and the plaques were counted at 18 h postinfection.
Melting curve analysis.
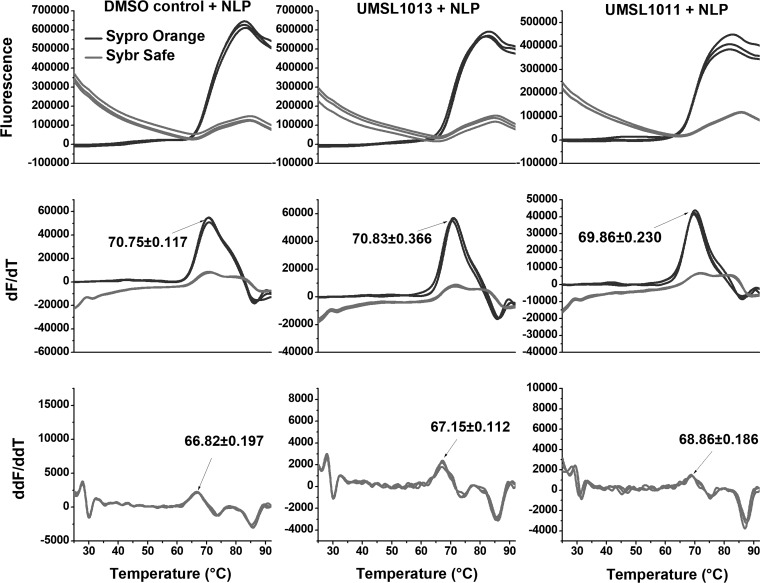
In order to verify that UMSL1011 is targeting the nucleocapsid, differential scanning fluorimetry (or a thermal shift assay [TSA]) was carried out. More specifically, we wanted to test whether UMSL1011 could delay the thermal release of the RNA from the NLP. Two different fluorescent dyes were selected to target the encapsidated RNA and protein, respectively. For RNA, SYBR Safe was used to stain DNA/RNA in electrophoresis gels because it works by intercalating DNA/RNA and then fluorescing. Since the bases of the RNA inside the NLP are stacked and the local RNA concentration is high inside the NLP as well, clear fluorescence was observed when SYBR Safe was incubated with the NLP (Fig. 4). To approximate the temperature when the RNA is dissociated from the NLP (Tfree), the first derivative graph was calculated for the rate of fluorescence changes. Most importantly, the peak in the second derivative of the melting curve defines the temperature point at which the rate change is at the maximum. We termed this temperature Tfree to indicate the RNA release from the NLP. To confirm our findings, SYPRO Orange was used to detect protein melting. The melting of the protein in the NLP is correlated with the release of encapsidated RNA because RNA can stabilize the protein in the NLP (6, 30). We have previously observed RNA release from the NLP when temperature was increased (30). Since the melting of the protein roughly follows the release of the RNA, as defined by Tfree, ΔTfree can be used to investigate the stability of the encapsidated RNA in the NLP.
FIG 4.
Melting curve analyses. VSV NLP was incubated with 200 μg/ml (1.3 mM) UMSL1011 (right column), with 200 μg/ml UMSL1013 (middle column), and in 1% DMSO (left column). NLP was at a concentration of 1.2 mg/ml in 50 mM Tris–300 mM NaCl (pH 7.5). Both SYBR Safe and SYPRO Orange were added to the NLP sample (light gray and dark gray traces, respectively). The temperature was scanned at 0.025°C/s using a QuantStudio 3 instrument. The first- and second-derivative graphs are presented. Tfree (68.9°C; equivalent to Tm) was determined from the second-derivative graph. The left column shows the results from the same experiment and presentation as the right column, except that the NLP was incubated with 1% DMSO. Tfree (66.8°C) was determined from the second-derivative graph. To obtain a negative control, the Tfree measurements were repeated with an inactive compound, UMSL1013 (central column). The Tfree value was essentially the same as that for the DMSO control. All errors are shown as standard deviations.
Using this assay, it was shown that when 200 μg/ml (1.3 mM) of UMSL1011 was incubated with NLP, Tfree was shifted upward by ∼2°C (68.86 versus 66.82°C) (Fig. 4). We interpreted this observation of the Tfree shift as UMSL1011 binding and stabilizing the RNA in the NLP. A shift of 2°C in melting temperature (Tm) is typical when an organic compound binds a protein in vitro (31). On the other hand, Tfree was shifted by only ∼0.3°C from an inactive polyamide molecule UMSL1013 (Fig. 4), suggesting that UMSL1013 does not stabilize the encapsidated RNA in the NLP. Interestingly, UMSL1011 has a slight effect on protein stability, showing a −0.9°C shift compared to the DMSO control (69.86°C with UMSL1011 and 70.75°C with DMSO alone) (red lines in Fig. 4). Furthermore, the unfolding of the protein begins after RNA is released from the NLP in all cases.
RNA protection.
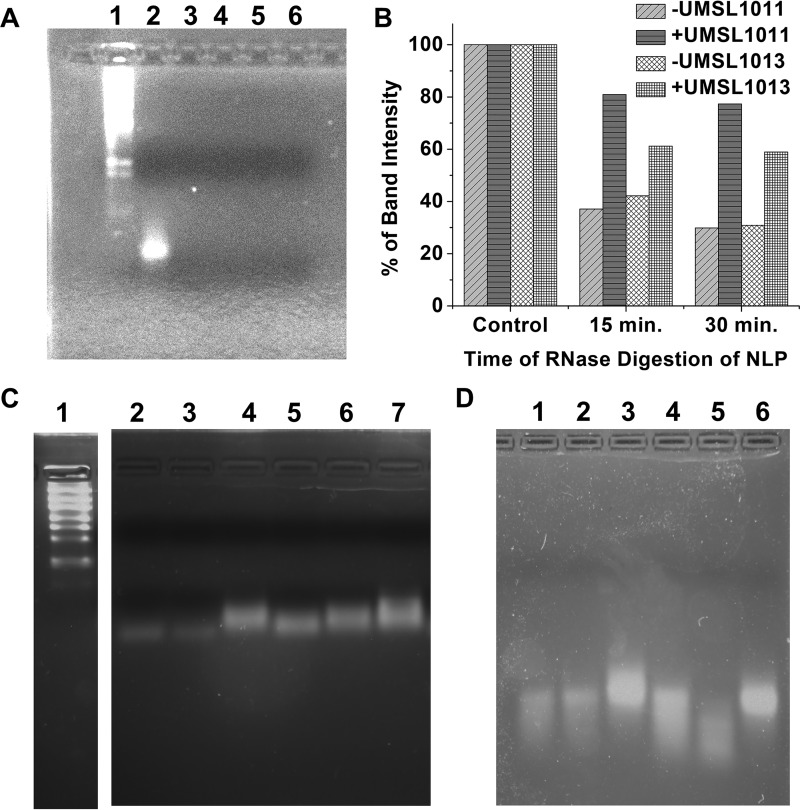
To further verify our melting curve analysis findings, RNA from the NLP was extracted by phenol extraction. The freed RNA was subjected to RNase A digestion in the absence or presence of 200 μg/ml (1.3 mM) UMSL1011 in 1% DMSO. There was no protection of the freed RNA from RNase A digestion by UMSL1011 (Fig. 5A). This observation is consistent with our hypothesis that UMSL1011 could bind only the structured RNA in the NLP. Digestion of RNA encapsidated by the NLP was also carried out. Previously, we have shown that RNA inside the NLP could be digested by RNase A (30). In this case, digestion of RNA in the NLP was protected by UMSL1011 (Fig. 5B), whereas the inactive UMSL1013 allowed for RNA digestion (Fig. 5B).
FIG 5.
RNA protection study. (A) RNA freed from NLP (Lane 2) was incubated with 15 μg/ml RNase A with (lanes 3 and 4) and without (lanes 5 and 6) 200 μg/ml (1.3 mM) UMSL1011 at 22°C for 30 min (lanes 2 and 4) and 60 min (lanes 3 and 5), respectively. RNA markers (lane 1) are included. (B) Bar graph quantifying the drop in intensity of each band in the gels shown in panels in C and D. Both + and −UMSL1011 and + and −UMSL1013 data are shown compared to each undigested control, which is set at 100%. The program GelAnalyzer 2010a was used to quantify the brightness and overall peak area of each gel, which were normalized on a percentage based on the control. (C) NLP was incubated with 1 mg/ml RNase A at 42°C without (lanes 2 to 4) or with (lanes 5 to 7) 200 μg/ml (1.3 mM) UMSL1011. Lanes 2 and 5, 30-min digestion; lanes 3 and 6, 15-min digestion. Lanes 4 and 7 had no RNase A. Lane 1, RNA markers (200 bases as the lowest band versus 90 bases of RNA from the NLP). (D) Control study with an inactive compound. UMSL1013 (lanes 4 to 6) at 200 μg/ml was incubated with NLP (lane 4, 15 min; lane 5, 30 min). Lane 6 has no RNase A. Lanes 1 to 3 correspond to the same reactions in the presence of 1% DMSO.
Evaluation of cytotoxicity.
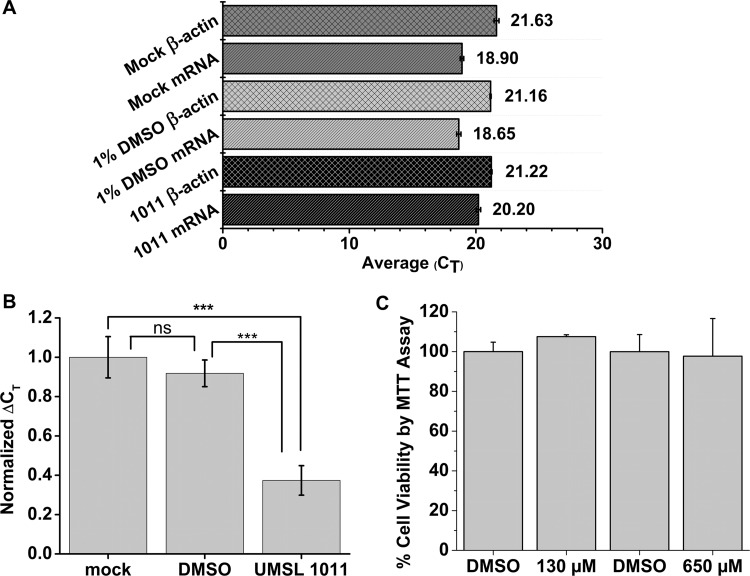
Previous studies showed that UMSL1011 and its analogs did not show cytotoxicity up to 200 μM (22). UMSL1011 was evaluated for cytotoxicity by MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assays using CellTiter 96 from Promega. Even at concentrations up to 650 μM, UMSL1011 exhibited very little cytotoxicity (Fig. 6C). This is indicative of a low probability of off-target effects and is consistent with previous data (21).
FIG 6.
Intracellular vRdRp assays and cytotoxicity study. (A) Raw CT values of mRNA transcripts from the minigenome assay and reporter gene, β-actin. Each CT value is taken as an average of three qPCRs, and error bars represent the standard deviations. (B) Normalized ΔCT values to the Mock, set as 1. Each ΔCT was calculated by subtracting the experimental CT value to its subsequent β-actin CT, and the error bars represent the standard deviations. This shows a significant decrease in mRNA transcripts when UMSL1011 is added. ***, P < 0.001; ns (not significant), P > 0.05. (C) Cytotoxicity studies. Monolayers of HeLa cells were incubated with 130 and 650 μM UMSL1011, respectively, for 25 h. We used 1% DMSO as the solvent control. Cell viability was measured using MTT assays. Experiments were carried out in triplicate, and error bars represent the standard deviations.
Minigenome assay.
To further test the specificity of UMSL1011 to inhibit VSV viral RNA synthesis, a commonly used minigenome assay/RT-qPCR was adapted to quantify mRNA transcripts (32–35). The N gene with three stop codons after the initiation Met codon was used as the reporter gene to encourage no bias. With this assay, UMSL1011 was shown to inhibit viral synthesis of mRNA for the reporter gene. The raw CT values for the qPCR assay are shown in Fig. 6A, and a normalized ΔCT value is shown in Fig. 6B. The ΔCT was calculated by comparing the housekeeping gene, β-actin, to the CT value of the mRNA. This was then normalized to the mock ΔCT values, allowing the calculation of a relative percent inhibition of transcription for UMSL1011. Compared to the DMSO control transfection, UMSL1011 at 20 μg/ml (130 μM) inhibited transcription of mRNA by 54.5%, whereas the DMSO treatment compared to the mock treatment (Mock) inhibited transcription of mRNA by about 8.2%. It is worth noting that a P value of >0.05 for comparing Mock to the DMSO control was not statistically significant. On the other hand, the P value for the UMSL1011 sample compared to the Mock/DMSO control was P < 0.001, showing a high level of confidence in statistical significance. Also, to corroborate our analysis with the well-known 2−ΔΔCT analysis, Table S1 in the supplemental material shows these values. These findings further confirm the pattern seen in our minigenome analysis.
Crystal structure.
In order to fully define the interactions of UMSL1011 with the structured RNA in the NLP, the crystal structure of UMSL1011-bound NLP was determined by X-ray crystallography (see Table 2 for statistics). The electron density corresponding to Py-3, Im-4, γ-5, and Py-6 of UMSL1011 (see Table 1 and Fig. 2) was found in the NLP (Fig. 7A). Due to resolution limitations and randomly encapsidated RNA when the NLP was purified from Escherichia coli, the RNA was first modeled as a poly(U) chain, as done previously (5, 30). In total, UMSL1011 forms seven hydrogen bonds with the RNA and two hydrogen bonds with the N protein (Fig. 7B). The electrostatic surface of UMSL1011 based on Adaptive Poisson-Boltzmann Solver software calculations confirmed the hydrogen bond potential of each UMSL1011 group (Fig. 7C). This confirmed that the methyl group in Im-4 is polarized, so its hydrogens may form hydrogen bonds with a proton acceptor, as found in previous studies (36–38). The amide nitrogen and the carbonyl group directly preceding the Im-4 each make a single hydrogen bond with the base of nucleotide 9. The three polarized methyl hydrogens coming off the Im-4 each form a hydrogen bond with the two riboses of nucleotides 7 and 8 and the phosphate between nucleotides 7 and 8. Im-4 also participates in weak π-π stacking with the base of nucleotide 9, further anchoring the Im-4 in place. Furthermore, the amide nitrogen and carbonyl group after Py-3 each forms a hydrogen bond with the bases on nucleotides 3 and 4, respectively. The carbonyl group directly preceding Py-6 forms a hydrogen bond with Arg-312, seemingly anchoring the γ-5 turn in place, and the methyl group on Py-6 interacts with the backbone carbonyl on Gln-318. The side of UMSL1011 that interacts with RNA is more positively charged, whereas the opposite side is mostly negatively charged. Unlike other polyamide interactions that would normally target the minor groove of DNA (39), UMSL1011 stretches across the structured RNA by making contacts with both the bases and the backbone (Fig. 7D). Interestingly, UMSL1011 fits in a pocket created by the interface between two N subunits and the encapsidated RNA (Fig. 7A).
TABLE 2.
Crystallographic statistics
| Parameter | Value(s) |
|---|---|
| Wavelength (Å) | 1 |
| Resolution range (Å) | 45.39–3.46 (3.584–3.46) |
| Space group | P2 21 21 |
| Unit cell dimensions | |
| a, b, c (Å) | 75.48, 165.48, 234.46 |
| α = β = γ (°) | 90 |
| Total no. of reflections | 168,290 (16,809) |
| No. of unique reflections | 38,347 (3,334) |
| Multiplicity | 4.4 (4.4) |
| Completeness (%) | 92.22 (86.26) |
| Mean I/σ〈I〉 | 6.79 (2.70) |
| Wilson B-factor | 100.3 |
| Rmerge | 0.1526 (0.6546) |
| Rmeas | 0.1718 (0.7406) |
| Rpim | 0.07654 (0.3363) |
| CC1/2 | 0.99 (0.742) |
| CC* | 0.997 (0.923) |
| No. of reflections: | |
| Used in refinement | 36,198 (3,334) |
| Used for Rfree | 1,999 (184) |
| Rwork | 0.2655 (0.3160) |
| Rfree | 0.3107 (0.3572) |
| CCwork | 0.912 (0.837) |
| CCfree | 0.868 (0.775) |
| No. of: | |
| Nonhydrogen atoms | 17,466 |
| Macromolecules | 17,417 |
| Ligands | 49 |
| Protein residues | 2,086 |
| RMS | |
| Bond length (Å) | 0.003 |
| Bond angles (°) | 0.76 |
| Ramachandran plot (%) | |
| Favored | 86.09 |
| Allowed | 9.71 |
| Outliers | 4.2 |
| Rotamer outliers (%) | 10.07 |
| Clashscore | 21.47 |
| Avg B-factor | 121.78 |
| Macromolecules | 121.77 |
| Ligands | 124.71 |
FIG 7.
Crystal structure of UMSL1011 bound to the NLP. (A) 2mFo-dFc map (shown in blue) at 1 sigma of UMSL1011 (shown in green) bound to the NLP. The magenta sticks are nearby amino acids of the NLP, while the RNA is modeled in cyan. The transparent surface reconstruction shows the pocket for which UMSL1011 binds. (B) Hydrogen bonds made by UMSL1011 (shown in green) to both the encapsidated RNA (cyan) and the protein (magenta). (C) Electrostatic surface potential of UMSL1011 calculated by APBS and visualized in Chimera (52, 53). The colored bar below shows the relative charge, with red indicating electron dense and blue indicating electron deficient. (D) Two-dimensional cartoon outlining the hydrogen bonds between UMSL1011 and the RNA[poly(rU)]/protein. Protein contacts are shown with the Gln318 amide carbonyl backbone and the Nε of Arg312.
For the genomic RNA in the VSV nucleocapsid, there could be any nucleotide sequence in each position that interacts with UMSL1011 (Table 3). To define the optimal sequence for binding with UMSL1011, we modeled each base at the RNA position in the UMSL1011-NLP complex. The possible hydrogen bonds between UMSL1011 and the NLP are summarized in Table 3. This model suggests that 5′-AXXCUXXGG-3′ may be the preferred sequence motif to bind with UMSL1011. The specific base is designated for that nucleotide position based on optimal hydrogen bond formation with the interacting moiety of UMSL1011. A survey of the genomic sequences found that there are 18 of these motifs in the negative strand and 6 in the positive strand, which represent all of the potential binding sites for UMSL1011.
TABLE 3.
UMSL1011 and RNA interactions based on nucleotide sequencea

In the UMSL1011-NLP structure, each base was modeled (A, G, U, and C) at each nucleotide position, and all possible hydrogen bonds were examined. The nucleotide position is numbred for each row, and each moiety of UMSL1011 is for each column. The interactions of each moiety of UMSL1011 with the respective base or the backbone are placed in each grid. The larger letters indicate possible selectivity for each nucleotide derived from optimal interactions, and the box highlighted in orange indicates interaction with the sugar/phosphate backbone.
To investigate whether UMSL1011 induces any structural changes, we aligned the A chain of the UMSL1011-bound structure with the A chain of PDB 2GIC, the original structure of the nucleocapsid. These crystals were grown under the same conditions as PDB 2GIC. There is almost no change in the N subunit structure where UMSL1011 is bound (Fig. 8A). However, globally, the overall NLP ring has tightened significantly. The diameter of the original NLP is 46.6 Å, measured from the C terminus to the C terminus of two opposite N subunits in the ring, whereas the UMSL1011-bound NLP has a diameter of 44.3 Å (Fig. 8B). This ring tightening of 2.3 Å can be corroborated with our TSA and RNA protection data.
FIG 8.
(A) Overlay of 2GIC and UMSL1011 bound crystal structure. UMSL1011 is indicated in green, while, as described above, magenta and cyan represent the protein and RNA, respectively. The 2GIC structure is yellow. There are few to no structural differences in the local area where UMSL1011 binds. (B) Reconstruction of the NLP ring. On the left-hand side is 2GIC (in yellow), and on the right-hand side is UMSL1011 structure (in cyan). An overall global tightening of the ring by 2.3 Å is observed when measuring the inner diameter from E-chain to E-chain, corresponding to the largest change in the ring diameter.
DISCUSSION
Many pathogens are negative-strand RNA viruses, including rabies virus, respiratory syncytial virus, and Ebola virus. Currently, there are no antiviral drugs targeting viral RNA synthesis by its unique viral RNA-dependent RNA polymerase (vRdRp). A large body of structural studies on the nucleocapsid of NSVs revealed a common mechanism of NSV viral RNA synthesis (40). During viral transcription and replication, vRdRp recognizes the nucleocapsid and gains access to the genomic RNA sequestered in the nucleocapsid to use it as the template. vRdRp must induce conformational changes in the nucleocapsid to release the genomic RNA for initiation and elongation during viral RNA synthesis. This provides a unique opportunity for antiviral inhibitors. Since the nucleocapsid is the template for viral RNA synthesis, viral replication will be blocked by a compound that binds the nucleocapsid.
By plaque assays, six different polyamide compounds were screened for inhibitory activities against VSV infection (Fig. 3). One compound, UMSL1011, was found to inhibit VSV infection. UMSL1011 was further confirmed to specifically inhibit viral RNA synthesis using an intracellular vRdRp minigenome assay (Fig. 6). It has been noted that UMSL1011 reduced viral plaques by about 50% at concentrations between 65 and 130 μM. This was further corroborated by the minigenome assay, which showed a reduction of 54.5% in the mRNA levels synthesized by the vRdRp when 130 μM UMSL1011 was added. This would indicate that UMSL1011 could not inhibit viral RNA synthesis very efficiently in this concentration range, allowing for a small amount of virus to continue through replication. Furthermore, the reduction in activity of viral RNA synthesis is not due to off-target effects of UMSL1011. Previously, it was shown that UMSL1011 had little effect on gene regulation in the apoptotic, DNA repair, and cell cycle pathways (21). This finding is important, since it allows one to use UMSL1011 as an existing model to design more active polyamide-based inhibitors.
To clearly define the target and reveal the mechanism of UMSL1011 inhibition, a melting curve and RNA protection studies were carried out on recombinantly expressed NLP. Due to the possible sequence-specific manner in which UMSL1011 interacts with the NLP and to account for the randomly encapsidated RNA from E. coli, the concentration of UMSL1011 was increased for all in vitro experiments on the NLP. By systematically increasing the temperature, structural changes are expected to occur in the nucleocapsid. Two fluorescent dyes were used to monitor the structural changes. One dye, SYBR Safe, intercalates between stacked bases of RNA. At the beginning, the fluorescence of SYBR Safe was high when incubated with NLP, suggesting tight binding to NLP. When the temperature continued to increase, the fluorescence of SYBR Safe started to drop, indicating a reduction in RNA binding, but the fluorescence of SYPRO Orange, the other dye we used to monitor protein unfolding, did not change very much. At a critical temperature, the fluorescence from the RNA released began to increase, indicating that that the RNA had already been released. To define a more common point after RNA release, the second derivative was utilized and named Tfree. The observation of thermo-release of RNA from the NLP was consistent with our previous studies of RNA digestion (30). We interpreted this observation as RNA sequestered in the NLP being thermally released, and we desgnated this temperature “Tfree.” Past this temperature, the fluorescence of SYPRO Orange began to increase rapidly, and a second Tm was observed, suggesting unfolding of the nucleocapsid protein. This indicates that the encapsidated RNA can stabilize NLP. When the melting curve measurement was repeated in the presence of UMSL1011, Tfree was increased by ∼2°C (Fig. 4), indicating that the binding of UMSL1011 stabilizes the NLP and prevents the thermo-release of sequestered RNA. When an inactive polyamide UMSL1013 was added, Tfree was not increased. Subsequently, an RNA protection assay was carried out to confirm that UMSL1011 binds only the RNA structure in the NLP (Fig. 5). If UMSL1011 was able to interact with the unstructured RNA extracted from the NLP, we would have seen distinct fragmentation of the RNA strand after RNase digestion. However, when the RNA in the NLP was extracted, it was completely digested by RNase A in the absence or presence of UMSL1011, suggesting no interactions between free RNA and UMSL1011. On the other hand, the RNA in the NLP may be digested by RNase A when the temperature was raised to release RNA (30). When the NLP was incubated with UMSL1011, the RNA in the NLP was protected from RNase A digestion, indicating that UMSL1011 binds the RNA in the NLP, further indicating that UMSL1011 stabilizes the NLP. The slight shift in the migration pattern is due to slight nibbling of exposed ends of the RNA by RNase. At the same time, the inactive UMSL1013 did not protect the RNA in NLP from RNase A digestion. The in vitro studies further argue that the polyamide UMSL1011 binds the RNA in the nucleocapsid to prevent it from serving as a template for RNA synthesis by vRdRp.
The cocrystal structure of UMSL1011 bound to NLP further validates the in vitro and biochemical results. Importantly, UMSL1011 was found to interact with both structured encapsidated RNA and nucleocapsid protein (Fig. 7). UMSL1011 binds the NLP in a manner not previously observed in polyamides. This unique binding pattern could be optimized to target more pathogenic NSVs. Not only is there an intricate hydrogen bonding network between UMSL1011 and the RNA/protein (Fig. 7B) but binding UMSL1011 also affects the NLP globally, causing a tightening of the NLP ring by 2.3 Å (Fig. 8B). Furthermore, a preferred UMSL1011 binding motif could be identified in the genomic RNA sequence (Table 2). Taking all of these observations into account, the inhibition of viral RNA synthesis by UMSL1011 could be a 2-fold process. The tightening of the nucleocapsid would make it harder for the polymerase to access the encapsidated genome, slowing the polymerase down or causing the complex to fall apart completely. Furthermore, UMSL1011 directly interacts with the RNA, causing an impassable blockage in the path of the polymerase complex.
To our knowledge, this is the first time that the encapsulated genome of an NSV has been directly targeted by an antiviral compound. Furthermore, this is the first evidence of a polyamide targeting and inhibiting the RNA synthesis of an NSV. Other NSVs exhibit similar RNA structures with stacked bases found inside the nucleocapsid, including rabies virus, respiratory syncytial virus, and measles virus (41–43). Such structures may also be expected in other NSVs, including Ebola virus and hantavirus, because the structure of their nucleocapsid proteins and the capsid assembly are very similar (44–46). Although UMSL1011 may be specific for VSV, it is possible that other polyamides can target other NSVs in a specific manner and should be the subject of future studies. Our proof-of-concept studies on nucleocapsid binding and the inhibition of VSV infection by UMSL1011 set a precedent for the design of polyamide molecules as antiviral agents for use against other NSVs.
MATERIALS AND METHODS
Antiviral screen.
VSV (Indiana strain) stocks were prepared in BHK-21 cells (30). Monolayer HeLa cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and grown to full confluence in 12-well plates. After washing the samples with Dulbecco's phosphate-buffered saline, VSV was dispensed over the cells and allowed to be absorbed for 1 h in 37°C and 5% CO2. The excess virus inoculum was washed away and the cells were overlaid with DMEM containing 0.8% agarose. After 48 h, the cells were fixed with 4% formaldehyde and the agarose overlay was removed. The plate was stained with 0.1% crystal violet, and the plaques were counted.
Thermal shift.
assay. Prior to carrying out the thermal shift assay (TSA), the isolation of recombinantly expressed NLP was purified according to a protocol previously established by our lab (47). All NLPs were checked by UV/Vis to verify the 260/280-nm wavelength ratio, ensuring RNA encapsidation, and SDS-page to verify protein purity. To test the efficacy of the compound in the TSA, a final concentration of 200 μg/ml (1.3 mM) UMSL1011 or UMSL1013, which corresponds to a final concentration of 1% DMSO, was incubated with 25 μM (1.2 mg/ml) NLP. To ensure binding to the NLP, three rounds of incubation at 42°C for 15 min, with subsequent incubation on ice for 15 min each round, were carried out; this was also done with the DMSO control. Stocks solutions 100× were prepared in the reaction buffer (50 mM Tris [pH 7.5], 300 mM NaCl) for the SYBR Safe and SYPRO Orange dyes. Each TSA reaction was carried out in 20-μl volumes that contained final concentrations of 20 μM NLP, 1% DMSO, or 180 μg/ml of UMSL1011/1013 dissolved in DMSO, as well as a 10× concentration of the dyes. The 20-μl reactions were prepared in triplicates and placed into a MicroAmp Fast 96-well plate. A QuantStudio3 was used to carry out the TSA with a gradient of 0.025°C/s from 25 to 95°C. The Savitzky-Golay method was used to smooth the raw data, and then first- and second-derivative analysis was performed to identify the Tm and Tfree, respectively.
RNA protection.
RNA was extracted using TRIzol reagent from Ambion by Life Technologies according to the manufacturer's protocol. After RNA extraction, the RNA was subjected to RNase treatment (final concentration of 15 μg/ml) both with 1% DMSO and 200 μg/ml (1.3 mM) of UMSL1011 for 30 min and 1 h, respectively. Samples were then loaded on a 2% agarose gel and visualized using SYBR Safe. The RNA encapsidated by the NLP was digested with 1 mg/ml of RNase for 15 or 30 min at 42°C. Prior to RNase digestion, 200 μg/ml of compounds and 1% DMSO was added to the NLP, and the cycling of temperature was carried out in the same manner as for the TSA. Each reaction was quenched by using TRIzol reagent and the subsequent extraction of the RNA. The samples were then loaded on a 2% agarose gel containing SYBR Safe and further visualized. Gel Analyzer 2010a was used to quantify the intensity of the bands on the gels for further analysis.
Evaluation of cytotoxicity.
HeLa cells were grown to confluence in 100 ml of DMEM supplemented with 10% FBS in 96-well. MTT assays were carried out according to the manufacturer's protocol (CellTiter 96; Promega, Madison, WI).
Minigenome assay.
A minigenome assay was designed and carried out as described previously (32–35). We constructed the minigenome to transcribe a nontranslatable N gene by adding three stop codons to the beginning of the gene. The minigenome and the helper plasmids were cotransfected, and either compound (final concentration, 20 μg/ml [130 μM]) or 1% DMSO was added to the media. RNA was extracted using an RNeasy RNA extraction kit (Qiagen) according to the manufacturer's protocol at 48 h posttransfection. Immediately after RNA extraction, reverse transcription was carried out to make the cDNA using the primer 5′-CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTCATTTGTCAAATTCTGACTTAG-3′ for the N mRNA transcripts and the primer 5′-AGCACTGTGTTGGCGTACAG-3′ for β-actin, using M-MLV reverse transcriptase from Invitrogen according to the manufacturer's protocol. After cDNA synthesis, qPCR was carried out with PowerUp SYBR green Mastermix according to the manufacturer's instructions for concentrations and cycling protocols. In brief, 10 ng of cDNA was used as a template, and 500 nM concentrations of both primers (5′-GTTGAATGGCTCGGATGGTTC-3′ and 5′-CCAGATCGTTCGAGTCGT-3′) were used for the mRNA transcript of N and both primers (5′-AGAGCTACGAGCTGCCTGAC-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′) were used for the β-actin transcript. A total of 40 cycles of PCR were carried out, and a subsequent melting curve was determined to check for transcript homogeneity. The primer-only (polymerase plus primers), no-primer (template plus polymerase), and no-reverse-transcriptase reactions showed no discernible CT values when we performed the experiment since the fluorescence was under the threshold to discern a CT value. The background fluorescence was taken into account when setting the baseline for the qPCRs, utilizing all of the controls as well; thus, only concentrations of the template yielding CT values of >15 were used in order to obtain an accurate baseline to set the CT value. Data were then analyzed using the ΔCT method for relative mRNA quantitation.
Structure.
Crystallization was carried out as previously described with minor modifications prior to crystallization to facilitate UMSL1011 binding (5). Otherwise, everything else was kept as described previously. Prior to crystallization, UMSL1011 (1 mg/ml [6.5 mM]) was incubated with the NLP in the same manner as for the TSA. In short, three rounds of incubation at 42°C for 15 min, each followed by a subsequent incubation on ice for 15 min, were carried out. Hanging drops were set up immediately after incubation, and crystals appeared after 7 to 10 days. Data were collected at the Advanced Photon Source on a SER-CAT 22-ID with an oscillation angle of 0.3°. Data were processed with the XDS package, and molecular replacement was carried out (with PDB 2GIC as a search model) using maximum-likelihood procedures in PHASER with the Phenix Suite (48, 49). The structure was then refined using phenix.refine in the Phenix Suite, and model building/ligand placement was performed in COOT (49, 50). The restraints for the UMSL1011 fragment were generated using eLBOW in the Phenix Suite from a SMILES string (49). All of the figures were prepared using PyMOL (51). Electrostatic surface potentials of UMSL1011 were generated using APBS and visualized using UCSF Chimera as described previously (52, 53).
Chemistry.
Polyamides were synthesized on ABI433A or CSBio 136XT peptide synthesizers using Boc methodology and extensive incorporation of dimer building blocks as previously described (23, 54–59). Boc-β-alanine-PAM resin was obtained from Peptides International, and building blocks were from A ChemTek or were synthesized in-house. Dimethylamine-free DMF was obtained from ABI (now Fisher). UMSL1028, UMSL1011, UMSL1055, and UMSL2115 were synthesized and purified as described previously (21, 22, 57). Compounds were cleaved off the resin with N,N-diaminopropylamine, leading to Dp derivatives, or with bis(aminopropyl)methylamine, leading to Ta derivatives. Preparative reversed-phase chromatography was carried out on a Phenomenex C18 Luna column in methanol-water with 0.1% trifluoroacetic acid (TFA), and the TFA salts were isolated by lyophilization. All compounds were analyzed for purity by two orthogonal high-pressure liquid chromatography (HPLC) and HPLC/mass spectrometry (MS) methods, using reversed-phase C12 on a Phenomenex Jupiter column in an acetonitrile-water gradient (HPLC/MS) and a Phenomenex Synergi Polar-RP column with ammonium formate running buffer for the orthogonal HPLC. Exact masses were determined by electrospray ionization, and 500- or 600 MHz 1H nuclear magnetic resonance (NMR) spectra were obtained. Typically, 13C and various two-dimensional NMR spectra were also obtained. All chemical characterizations are presented in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
R.H.G. thanks the Molecular Basis of Disease at Georgia State University for ongoing financial support. This study was supported in part by a National Institutes of Health grant (R01 AI106307) to M.L. We thank the University of Missouri Research Board for financial support to J.K.B. The UMSL Agilent 600-MHz NMR spectrometer was obtained using funds from the NSF (grant 0959360).
M.L. and R.H.G. thank the Southeast Regional Collaborative Access Team (SER-CAT) at Argonne National Laboratory for user support during remote data collection. We thank Todd J. Green for the drawing in Fig. 1B. J.K.B. thanks R. Luo for assistance with NMR and both B. J. Bythell (UMSL) and the Danforth Plant Sciences Center (NSF DBI 0922879) for HRMS.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00146-18.
REFERENCES
- 1.Bont L, Checchia PA, Fauroux B, Figueras-Aloy J, Manzoni P, Paes B, Simoes EA, Carbonell-Estrany X. 2016. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in western countries. Infect Dis Ther 5:271–298. doi: 10.1007/s40121-016-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appolinario CM, Jackson AC. 2015. Antiviral therapy for human rabies. Antiviral Ther 20:1–10. doi: 10.3851/IMP2851. [DOI] [PubMed] [Google Scholar]
- 3.Balmith M, Faya M, Soliman ME. 2017. Ebola virus: a gap in drug design and discovery—an experimental and computational perspective. Chem Biol Drug Design 89:297–308. doi: 10.1111/cbdd.12870. [DOI] [PubMed] [Google Scholar]
- 4.Wu W, Liu S. 2017. The drug targets and antiviral molecules for treatment of Ebola virus infection. Curr Top Med Chem 17:361–370. doi: 10.2174/1568026616666160829161318. [DOI] [PubMed] [Google Scholar]
- 5.Green TJ, Zhang X, Wertz GW, Luo M. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Green TJ, Tsao J, Qiu S, Luo M. 2008. Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J Virol 82:674–682. doi: 10.1128/JVI.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mysore VS, Szablowski J, Dervan PB, Frost PJ. 2016. A DNA-binding molecule targeting the adaptive hypoxic response in multiple myeloma has potent antitumor activity. Mol Cancer Res 14:253–266. doi: 10.1158/1541-7786.MCR-15-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiraoka K, Inoue T, Taylor RD, Watanabe T, Koshikawa N, Yoda H, Shinohara K-i, Takatori A, Sugimoto H, Maru Y, Denda T, Fujiwara K, Balmain A, Ozaki T, Bando T, Sugiyama H, Nagase H. 2015. Inhibition of KRAS codon 12 mutants using a novel DNA-alkylating pyrrole-imidazole polyamide conjugate. Nat Commun 6:6706. doi: 10.1038/ncomms7706. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto M, Bando T, Morinaga H, Kawamoto Y, Hashiya K, Sugiyama H. 2014. Sequence-specific DNA recognition by cyclic pyrrole-imidazole cysteine-derived polyamide dimers. Chem Eur J 20:752–759. doi: 10.1002/chem.201302482. [DOI] [PubMed] [Google Scholar]
- 10.Kang JS, Meier JL, Dervan PB. 2014. Design of sequence-specific DNA binding molecules for DNA methyltransferase inhibition. J Am Chem Soc 136:3687–3694. doi: 10.1021/ja500211z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floreancig PE, Swalley SE, Trauger JW, Dervan PB. 2000. Recognition of the minor groove of DNA by hairpin polyamides containing α-substituted-β-amino acids. J Am Chem Soc 122:6342–6350. doi: 10.1021/ja000509u. [DOI] [Google Scholar]
- 12.White S, Baird EE, Dervan PB. 1997. On the pairing rules for recognition in the minor groove of DNA by pyrrole-imidazole polyamides. Chem Biol 4:569–578. doi: 10.1016/S1074-5521(97)90243-X. [DOI] [PubMed] [Google Scholar]
- 13.Kopka ML, Goodsell DS, Han GW, Chiu TK, Lown JW, Dickerson RE. 1997. Defining GC-specificity in the minor groove: side-by-side binding of the di-imidazole lexitropsin to C-A-T-G-G-C-C-A-T-G. Structure 5:1033–1046. doi: 10.1016/S0969-2126(97)00255-4. [DOI] [PubMed] [Google Scholar]
- 14.White S, Baird EE, Dervan PB. 1996. Effects of the A.T/T.A degeneracy of pyrrole-imidazole polyamide recognition in the minor groove of DNA. Biochemistry 35:12532–12537. doi: 10.1021/bi960744i. [DOI] [PubMed] [Google Scholar]
- 15.Trauger JW, Baird EE, Mrksich M, Dervan PB. 1996. Extension of sequence-specific recognition in the minor groove of DNA by pyrrole-imidazole polyamides to 9-13 base pairs. J Am Chem Soc 118:6160–6166. doi: 10.1021/ja960726o. [DOI] [Google Scholar]
- 16.Trauger JW, Baird EE, Dervan PB. 1996. Extended hairpin polyamide motif for sequence-specific recognition in the minor groove of DNA. Chem Biol 3:369–377. doi: 10.1016/S1074-5521(96)90120-9. [DOI] [PubMed] [Google Scholar]
- 17.Pilch DS, Poklar N, Gelfand CA, Law SM, Breslauer KJ, Baird EE, Dervan PB. 1996. Binding of a hairpin polyamide in the minor groove of DNA: sequence-specific enthalpic discrimination. Proc Natl Acad Sci U S A 93:8306–8311. doi: 10.1073/pnas.93.16.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly JJ, Baird EE, Dervan PB. 1996. Binding site size limit of the 2:1 pyrrole-imidazole polyamide-DNA motif. Proc Natl Acad Sci U S A 93:6981–6985. doi: 10.1073/pnas.93.14.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasilieva E, Niederschulte J, Song Y, George Harris D J, Koeller KJ, Liao P, Bashkin JK, Dupureur C. 2016. Interactions of two large antiviral polyamides with the long control region of HPV16. Biochimie 127:103–114. doi: 10.1016/j.biochi.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He G, Vasilieva E, Harris GD Jr, Koeller KJ, Bashkin JK, Dupureur CM. 2014. Binding studies of a large antiviral polyamide to a natural HPV sequence. Biochimie 102:83–91. doi: 10.1016/j.biochi.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards TG, Vidmar TJ, Koeller K, Bashkin JK, Fisher C. 2013. DNA damage repair genes controlling human papillomavirus (HPV) episome levels under conditions of stability and extreme instability. PLoS One 8:e75406. doi: 10.1371/journal.pone.0075406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards TG, Koeller KJ, Slomczynska U, Fok K, Helmus M, Bashkin JK, Fisher C. 2011. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res 91:177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He G, Bashkin JK. 2015. What is the antiviral potential of pyrrole-imidazole polyamides? Future Med Chem 7:1953–1955. doi: 10.4155/fmc.15.120. [DOI] [PubMed] [Google Scholar]
- 24.Koeller KJ, Harris GD, Aston K, He G, Castaneda CH, Thornton MA, Edwards TG, Wang S, Nanjunda R, Wilson WD, Fisher C, Bashkin JK. 2014. DNA binding polyamides and the importance of DNA recognition in their use as gene-specific and antiviral agents. Med Chem 4:338–344. doi: 10.4172/2161-0444.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaneda CH, Jose' Scuderi M, Edwards TG, Harris GD, He G, Dupureur CM, Koeller K, Fisher C, Bashkin JK. 2016. Improved antiviral activity of a polyamide against high-risk human papillomavirus via N-terminal guanidinium substitution. MedChemComm 7:2076–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez TF, Phillips JW, Karanja KK, Polaczek P, Wang C-M, Li BC, Campbell JL, Dervan PB. 2014. Replication stress by Py–Im polyamides induces a non-canonical ATR-dependent checkpoint response. Nucleic Acids Res 42:11546–11559. doi: 10.1093/nar/gku866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao H, Ma C, Zhang X, Jing X, Li C, Zhao Y. 2015. Insight into DNA minor groove unspecific binding of pyrrole polyamide. Bioconjug Chem 26:2054–2061. doi: 10.1021/acs.bioconjchem.5b00309. [DOI] [PubMed] [Google Scholar]
- 28.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. 2004. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res 32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopka ML, Yoon C, Goodsell D, Pjura P, Dickerson RE. 1985. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A 82:1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green TJ, Rowse M, Tsao J, Kang J, Ge P, Zhou ZH, Luo M. 2011. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol 85:2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippova EV, Kieser KJ, Luan CH, Wawrzak Z, Kiryukhina O, Rubin EJ, Anderson WF. 2016. Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance. FEBS J 283:2206–2218. doi: 10.1111/febs.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nayak D, Panda D, Das SC, Luo M, Pattnaik AK. 2009. Single-amino-acid alterations in a highly conserved central region of vesicular stomatitis virus N protein differentially affect the viral nucleocapsid template functions. J Virol 83:5525–5534. doi: 10.1128/JVI.02289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, Ogino T, Banerjee AK. 2007. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol 81:13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filone CM, Hodges EN, Honeyman B, Bushkin GG, Boyd K, Platt A, Ni F, Strom K, Hensley L, Snyder JK, Connor JH. 2013. Identification of a broad-spectrum inhibitor of viral RNA synthesis: validation of a prototype virus-based approach. Chem Biol 20:424–433. doi: 10.1016/j.chembiol.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stillman EA, Rose JK, Whitt MA. 1995. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol 69:2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horowitz S, Yesselman JD, Al-Hashimi HM, Trievel RC. 2011. Direct evidence for methyl group coordination by carbon-oxygen hydrogen bonds in the lysine methyltransferase SET7/9. J Biol Chem 286:18658–18663. doi: 10.1074/jbc.M111.232876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yesselman JD, Horowitz S, Brooks CL III, Trievel RC. 2015. Frequent side chain methyl carbon-oxygen hydrogen bonding in proteins revealed by computational and stereochemical analysis of neutron structures. Proteins 83:403–410. doi: 10.1002/prot.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller G, Lutz M, Harder S. 1996. Methyl group conformation-determining intermolecular C-H…O hydrogen bonds: structure of N-methyl-2-pyrrolidone. Acta Crystallogr B 52:1014–1022. doi: 10.1107/S0108768196008300. [DOI] [Google Scholar]
- 39.Kielkopf CL, Baird EE, Dervan PB, Rees DC. 1998. Structural basis for G.C recognition in the DNA minor groove. Nat Struct Biol 5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 40.Luo M. 2012. The nucleocapsid of vesicular stomatitis virus. China Life Sci 55:291–300. doi: 10.1007/s11427-012-4307-x. [DOI] [PubMed] [Google Scholar]
- 41.Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 42.Tawar RG, Duquerroy S, Vonrhein C, Varela PF, Damier-Piolle L, Castagne N, MacLellan K, Bedouelle H, Bricogne G, Bhella D, Eleouet JF, Rey FA. 2009. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science 326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 43.Gutsche I, Desfosses A, Effantin G, Ling WL, Haupt M, Ruigrok RW, Sachse C, Schoehn G. 2015. Structural virology: near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science 348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 44.Guo Y, Wang W, Sun Y, Ma C, Wang X, Wang X, Liu P, Shen S, Li B, Lin J, Deng F, Wang H, Lou Z. 2015. Crystal structure of the core region of hantavirus nucleocapsid protein reveals the mechanism for ribonucleoprotein complex formation. J Virol 90:1048–1061. doi: 10.1128/JVI.02523-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchdoerfer RN, Abelson DM, Li S, Wood MR, Saphire EO. 2015. Assembly of the Ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep 12:140–149. doi: 10.1016/j.celrep.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong S, Yang P, Li G, Liu B, Wang W, Liu X, Xia B, Yang C, Lou Z, Guo Y, Rao Z. 2015. Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 Å resolution. Protein Cell 6:351–362. doi: 10.1007/s13238-015-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green TJ, Macpherson S, Qiu S, Lebowitz J, Wertz GW, Luo M. 2000. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J Virol 74:9515–9524. doi: 10.1128/JVI.74.20.9515-9524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Schrödinger. The PyMOL molecular graphics system, v1.3. Schrödinger, LLC, New York, NY. [Google Scholar]
- 52.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Baird EE, Dervan PB. 1996. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J Am Chem Soc 118:6141–6146. doi: 10.1021/ja960720z. [DOI] [PubMed] [Google Scholar]
- 55.Lacy ER, Le NM, Price CA, Lee M, Wilson WD. 2002. Influence of a terminal formamido group on the sequence recognition of DNA by polyamides. J Am Chem Soc 124:2153–2163. doi: 10.1021/ja016154b. [DOI] [PubMed] [Google Scholar]
- 56.Dupureur CM, Bashkin JK, Aston K, Koeller KJ, Gaston KR, He G. 2012. Fluorescence assay of polyamide-DNA interactions. Anal Biochem 423:178–183. doi: 10.1016/j.ab.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Nanjunda R, Aston K, Bashkin JK, Wilson WD. 2012. Correlation of local effects of DNA sequence and position of beta-alanine inserts with polyamide-DNA complex binding affinities and kinetics. Biochemistry 51:9796–9806. doi: 10.1021/bi301327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bashkin JK, Aston K, Ramos JP, Koeller KJ, Nanjunda R, He G, Dupureur CM, David Wilson W. 2013. Promoter scanning of the human COX-2 gene with 8-ring polyamides: unexpected weakening of polyamide-DNA binding and selectivity by replacing an internal N-Me-pyrrole with beta-alanine. Biochimie 95:271–279. doi: 10.1016/j.biochi.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang S, Aston K, Koeller KJ, Harris GD Jr, Rath NP, Bashkin JK, Wilson WD. 2014. Modulation of DNA-polyamide interaction by beta-alanine substitutions: a study of positional effects on binding affinity, kinetics and thermodynamics. Org Biomol Chem 12:7523–7536. doi: 10.1039/C4OB01456A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.