Abstract
Introduction
Many lines of evidence indicate that low levels of HDL cholesterol increase the risk of cardiovascular disease (CVD). However, recent clinical studies of statin-treated subjects with established atherosclerosis cast doubt on the hypothesis that elevating HDL cholesterol levels reduces CVD risk.
Areas covered
It is critical to identify new HDL metrics that capture HDL’s proposed cardioprotective effects. One promising approach is quantitative MS/MS-based HDL proteomics. This article focuses on recent studies of the feasibility and challenges of using this strategy in translational studies. It also discusses how lipid-lowering therapy and renal disease alter HDL’s functions and proteome, and how HDL might serve as a platform for binding proteins with specific functional properties.
Expert commentary
It is clear that HDL has a diverse protein cargo and that its functions extend well beyond its classic role in lipid transport and reverse cholesterol transport. MS/MS analysis has demonstrated that HDL might contain >80 different proteins. Key challenges are demonstrating that these proteins truly associate with HDL, are functionally important, and that MS-based HDL proteomics can reproducibly detect biomarkers in translational studies of disease risk.
Keywords: Apolipoprotein A-I, biomarker, dysfunctional HDL, lipid-lowering therapy, parallel reaction monitoring, renal disease, selected reaction monitoring, shotgun proteomics
1. Atherosclerosis and high-density lipoprotein’s proposed cardioprotective properties
Atherosclerosis is the major cause of heart attacks and peripheral vascular disease in the industrialized world, and a key process in its pathogenesis is the accumulation of cholesterol-laden macrophages in the artery wall. Thus, one factor that greatly increases the risk for atherosclerotic cardiovascular disease (CVD) is an elevated level of low-density lipoprotein (LDL), which delivers cholesterol to macrophages [1]. In contrast, clinical, epidemiological, and animal studies have demonstrated a strong inverse relationship between high-density lipoprotein cholesterol (HDL-C) levels and risk for CVD [2–4], strongly supporting the proposal that HDL is antiatherogenic.
Several distinct pathways have been implicated in HDL’s cardioprotective effects [5–11]. One well-documented mechanism involves removal of excess cholesterol from artery wall macrophages, a process known as reverse cholesterol transport [6,8]. Importantly, HDL’s ability to remove cholesterol from cultured macrophages – termed cholesterol efflux capacity – strongly and negatively associates with CVD [12]. Moreover, HDL’s cholesterol efflux capacity is a stronger predictor of CVD than HDL-C [13,14], and HDL-C levels explain only a fraction (approximately one-third) of the variance in such functional assays [15].
HDL is a family of nanoparticles composed of non-covalently associated amphipathic lipids, neutral lipids, and proteins that undergo complex metabolic interactions with other classes of lipoproteins, cells, and plasma proteins [16]. Its major proteins, apolipoprotein A-I (APOA1) and APOA2, account for ~70% and ~20% of its protein mass [17,18] and over half the mass of the lipoprotein. However, it is not a homogeneous population of particles because the lipoproteins vary greatly in protein composition and size, consisting of multiple subspecies that vary from 7 to 14 nm in diameter. Moreover, shotgun proteomics has revealed that HDL carries a wide array of proteins linked to inflammation, complement regulation, and proteolysis inhibition (Figure 1) [19]. As HDL-C measurements do not reflect the abundance of such proteins [20], modulating HDL-C levels might have no impact on the properties of the particular particles responsible for atheroprotection [6,21]. Indeed, elevating HDL-C levels with drugs that act by different mechanisms has not lowered CVD risk in statin-treated humans with established atherosclerosis [22–24]. For example, torcetrapib, a potent cholesteryl ester transfer protein (CETP) inhibitor, inhibited the development of atherosclerosis in rabbits [25] and also increased HDL-C by 60–100% in early-phase human studies [26,27]. However, several large trials with torcetrapib showed no clinical benefits, as rates of CVD and total death were higher in the treated group than in the controls [22,28,29]. Another trial with a combination of niacin and statin also failed to reduce the risk of cardiovascular events due to no incremental clinical benefit and a twofold increase in stroke risk [30].
Figure 1.
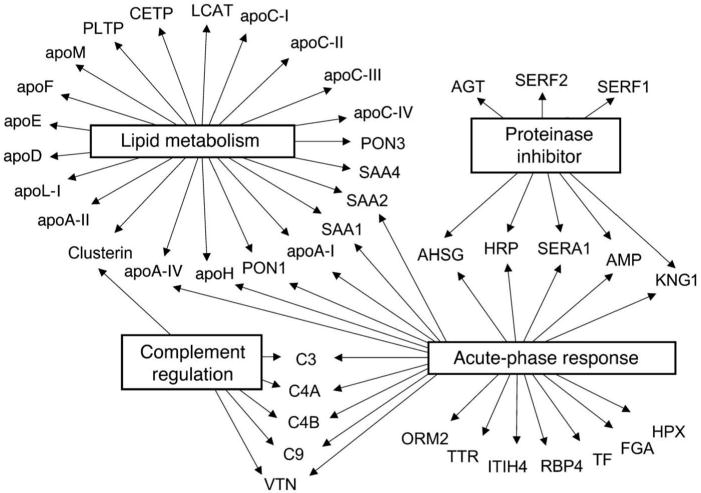
The proteomic diversity of HDL. Global view of biological processes and molecular functions of HDL proteins. A total of 48 proteins in total HDL and HDL3 were identified, using LC-ESI-MS/MS. Gene Ontology analysis demonstrated that 22 of the 48 HDL-associated proteins were linked to cholesterol and lipoprotein metabolism (P = 2 × 10−27). Twenty-three were acute-phase-response proteins (P = 1 × 10−18). Eight proteins were endopeptidase inhibitors (P = 2 × 10−6), and six were regulators of complement activation (P = 5 × 10−5). Collectively, these observations implicate HDL in the acute-phase response and innate immunity. They also strongly support the proposal that HDL carries proteins that may play previously unsuspected roles in regulating complement activation and inhibiting proteolysis. Reproduced with permission from [19].
To successfully implement CVD therapies that target HDL, it therefore will be critical to identify the HDL proteins, subspecies, and functions responsible for cardioprotection and ultimately to modulate those factors in a beneficial manner [6,21,31]. It will also be necessary to identify the proteins and other components that may cause HDL to become dysfunctional and lose its cardioprotective effects. For example, enrichment with inflammatory proteins, such as SAA1 and SAA2, or depletion of anti-atherosclerotic proteins, such as PON1, may generate dysfunctional HDL. Mouse genetic studies provide strong evidence that PON1, which associates exclusively with HDL in humans, is cardioprotective [32]. Moreover, the enzymatic activity of PON1 may predict CVD risk better than PON1 protein levels [33]. The significance of genetic polymorphisms in PON1 in the development of CVD remains controversial, however.
This review focuses on recent articles in four specific areas. First, we discuss how HDL can be isolated, how the isolation method affects its protein composition, and how to determine whether the numerous proteins that have been detected in HDL truly associate with HDL or are contaminants from plasma. Second, we review recent studies demonstrating the advantages of using parallel reaction monitoring (PRM) to identify potential protein biomarkers in HDL. Third, we highlight recent work addressing the role of lipids in HDL assembly and the functional properties of specific HDL-associated proteins affected by statin therapy. Finally, we discuss the potential strengths and limitations of mass spectrometry (MS)-based translational studies of HDL’s proteome and review recent studies of lipid-lowering therapy and renal disease subjects.
Other excellent new reviews summarize recent progress in our understanding of the diversity of the HDL proteome [34], the limitations of current laboratory technologies for separating and quantifying HDL [35], and proposed biological and cardioprotective effects of HDL [36,37].
2. Defining proteins that associate with HDL
HDL was defined more than 50 years ago by its density (1.063–1.21 g/ml) on ultracentrifugation [38]. However, it can now be isolated in many different ways, including density flotation, protein composition (e.g. affinity isolation of APOA1), molecular size, and electrophoretic migration [39]. In this review, we use the classic, density-based definition of HDL. It is important to note that this includes lipoproteins and lipid–protein complexes in the HDL density range that can lack apoA-I and contain other proteins such as apoE. Under this definition, lipid-free (or poorly lipidated) apoA-I, the major HDL protein, is not considered to be HDL because the particle does not float on ultracentrifugation.
A key issue in HDL analysis by MS/MS is the method used to isolate the lipoprotein. In many proteomic analyses of HDL, high levels of albumin and apoB-100 (LDL’s major protein) are detected, raising major questions about the purity of the HDL studied. Thus, different isolation techniques may significantly impact the results of HDL proteomic analysis. Most MS-based proteomic studies of HDL have used density gradient ultracentrifugation to isolate HDL. In our opinion, this is the ‘gold standard,’ both because it was originally used to define HDL and because it has been used for the vast majority of clinical and translational studies of the lipoprotein. However, the high shear forces and salt concentrations used in ultracentrifugation can disrupt protein–protein and protein–lipid interactions and therefore reduce the number of detectable proteins. For example, Kane and colleagues demonstrated more than 30 years ago that HDL isolated via affinity chromatography using polyclonal antibodies to apoA-I contained more total protein and minor apolipoproteins than HDL isolated by ultracentrifugation [40]. For example, immuno-isolated HDL, but not ultracentrifuge-isolated HDL, contains lecithin cholesterol acyl transferace (LCAT) and CETP – two key proteins in HDL metabolism [41].
To attempt to circumvent the limitations of ultracentrifugation for isolating lipoproteins, the Davidson group used high-resolution size-exclusion chromatography and phospholipid affinity chromatography to fractionate normal human plasma [42]. They identified 79 plasma proteins as associated with phospholipid. Subsequently, those investigators used three orthogonal separation techniques (gel filtration chromatography, ionic exchange chromatography, and preparative isoelectric focusing) and phospholipid affinity chromatography to isolate only those proteins associated with phospholipid in plasma [20]. This approach detected 76 lipid-associated proteins by mass spectrometry. A limitation of this strategy is that it was not possible to determine which proteins associated with specific lipoproteins as defined by ultracentrifugation. However, the authors were careful to note that ‘Given that most of the protein pairs identified are known to reside primarily in this [HDL] density range, our data challenge the notion that HDL is essentially a single entity with numerous interchanging protein components.’
An alternative analytical strategy is to use immunoaffinity methods to isolate HDL. Watanable et al. [43] employed immunocapture spin columns with chicken IgY antibody to isolate human HDL from plasma. The HDL proteins were then separated by electrophoresis followed by in-gel digestion and liquid chromatography tandem mass spectrometry (LC-MS/ MS) analysis. A total of 78 proteins were identified in HDL. It is noteworthy that many of the proteins identified in the above studies overlap with those identified in ultracentrifuge-isolated HDL, suggesting that most of these proteins are truly associated with HDL.
Another approach to isolating HDL combines ultracentrifugation with other isolation techniques. To generate ‘highly purified’ fractions of HDL2/3, Holzer et al. [44] isolated HDL by ultracentrifugation, followed by size-exclusion chromatography and shotgun proteomics. Surprisingly, they identified only nine proteins in the purified HDL2 and HDL3. The authors concluded that many of the proteins identified in HDL by other investigators were contaminants from plasma. However, the HDL they isolated by ultracentrifugation and used for additional purification contained only 26 proteins, suggesting that their MS analysis lacked adequate sensitivity or that their extensive isolation procedure caused the loss of proteins from the HDL preparation. In contrast to Holzer et al.’s work, Oberbach et al. [45] recently identified 494 distinct proteins in HDL from five controls and five chronic heart failure patients. Those investigators isolated HDL by sequential ultracentrifugation. The isolated HDL was digested with trypsin and the tryptic digests were then separated by a two-dimensional liquid chromatography followed by MS analysis. Compared with other proteomic studies, the very large number of proteins identified as HDL associated likely reflects in part the use of two-dimensional liquid chromatography to separate peptides and analysis with a high-resolution mass spectrometer. However, it is unclear whether some of the proteins might be derived from plasma or other sources distinct from HDL.
The Davidson and Shah groups (University of Cincinnati) maintain a website of HDL Proteome Watch [46] and LDL Proteome Watch [47] that catalogs proteins identified in HDL and LDL by both MS/MS and other analytical techniques. Importantly, they require any protein identified as lipoprotein associated to be identified by three separate reports from three independent laboratories. This approach greatly increased confidence in the identification of proteins as HDL associated. Using this criterion, HDL currently contains 95 associated proteins, while LDL contains only 22 proteins. These data clearly support the proposal that most of the proteomic diversity of lipoproteins is found in HDL and that a core group of proteins consistently resides in HDL.
Ultimately, the significance of HDL-associated proteins will be established by demonstrating their functional and/or translational importance in animal models and humans.
3. Quantitative protein analysis of HDL by PRM
Detection and quantification of proteins by MS depend on many technical factors, including the criteria used to identify peptides, the buffers and matrices used for protein isolation and MS analysis, and the MS instrumentation. Numerous articles and reviews address these important issues [48–51].
One key issue is the method used for protein quantification. Use of isotopically labeled peptides in selected reaction monitoring (SRM) is currently the standard approach in quantitative MS-based proteomics because of its sensitivity, specificity, and precision with complex biological samples [52]. This targeted approach permits multiplexed assays, with tryptic peptides as surrogates for the protein of interest. However, representative peptide candidates must be selected for each protein, and an isotope-labeled internal standard has to be synthesized for each selected peptide. Moreover, assay optimization requires peptides that are reproducibly generated during sample preparation, give a linear response in terms of protein concentration, and correlate with other candidate peptides for the same protein.
A disadvantage of quantifying peptides in candidate proteins together with stable isotope-labeled internal standards is that this approach fails to account for variability in the proteolytic step required before MS. Including isotope-labeled full-length proteins as standards overcomes this problem [53]. Importantly, use of isotope-labeled APOA1 rather than one labeled peptide or protein per precursor peptide of interest has been validated as a powerful method for quantifying HDL proteins [54].
PRM, which can quantify multiple peptides with high sensitivity and specificity, is an alternative to SRM that promises speedier assay development [55]. Because PRM does not require prior selection of target peptide transitions, it eliminates the time and effort required to optimize the best transitions. Therefore, it can involve much less effort during method development than the traditional SRM assay [56]. Moreover, PRM offers higher specificity than SRM on QqQ instruments because it simultaneously monitors product ions with high resolution and is less likely to be affected by interfering ions [57].
HDL is a particularly challenging biological substrate for proteomic analysis because two proteins, APOA1 and APOA2, account for ~90% of its protein mass [17,18], while at least 80 other proteins are detected in much lower abundance. We therefore compared PRM and SRM for their ability to quantify HDL proteins, using 15N-labeled APOA1 as the internal standard. PRM and SRM exhibited comparable linearity, dynamic range, precision, and repeatability for quantifying multiple proteins in HDL [52]. Moreover, for the quantification of three different proteins, the single internal standard protein performed as well as protein-specific internal standard peptides. PRM and SRM also yielded virtually identical quantitative results for 26 proteins in HDL isolated from 44 subjects. Because PRM requires less method development than SRM and is potentially more specific, our observations indicate that PRM in concert with a single isotope-labeled protein is a promising new strategy for quantifying HDL proteins in translational studies.
We used PRM with isotope dilution to test the hypothesis that HDL’s protein composition is abnormal in subjects with fenofibrate/rosiglitazone-induced hypoalphalipoproteinemia [58]. This disorder, a complication of fenofibrate therapy, causes a striking reduction in HDL-C and HDL’s major protein, apoA-I. HDL was isolated at baseline from plasma of subjects who received fenofibrate/rosiglitazone therapy in the ACCORD-Lipid study. One group subsequently developed hypoalphalipoproteinemia; in the other group of matched subjects, HDL levels either did not change or increased. After we applied stringent statistical criteria to control for multiple comparisons, our targeted PRM approach identified four HDL proteins – PON1, apoC-II, apoC-I, and apoH – that were relatively more abundant in HDL isolated from plasma of the subjects who subsequently developed fenofibrate/rosiglitazone-induced hypoalphalipoproteinemia. SRM analyses confirmed the relative increase in these proteins. In addition, using biochemical approaches, we confirmed our observations for PON1 and apoC-II. Quantifying PON1 by PRM correlated well with immunoblot analysis. For apoC-II, the PRM results correlated highly with ELISA data. These observations demonstrate that PRM is a powerful tool for quantifying proteins in translational studies of HDL.
4. Defining HDL and its protein partners by mass spectrometry in concert with biochemical fractionation and network analysis
HDL isolated by ultracentrifugation or non-denaturing gel electrophoresis consists of multiple populations of particles that differ in size and protein composition [59,60]. Moreover, it is well established that HDL contains defined molecular complexes with specific biological functions. For example, apoA-I, haptoglobin-related protein, and apoL-I form the trypanosome lytic complex [61,62], which plays an important role in host defenses against trypanosomes, the unicellular parasitic flagellates that cause sleeping sickness and Chagas disease. Indeed, HDL may play a key role in host defense mechanisms against these conditions, acting in a different capacity than when it promotes lipid transport [63].
These observations strongly suggest that HDL can serve as a platform for the assembly of molecular complexes of defined composition and function [19]. One possibility is that apoA-I forms a symmetrical cage-like structure covering most of HDL’s surface (Figure 2). The elegant models proposed by these investigators suggest that HDL particle size can be modulated by a twisting motion of the resident apoA-I molecules and that this in turn might affect the interaction of apoA-I with other proteins [64]. However, the factors that control the assembly of specific proteins on HDL and other lipoproteins remain poorly understood.
Figure 2.
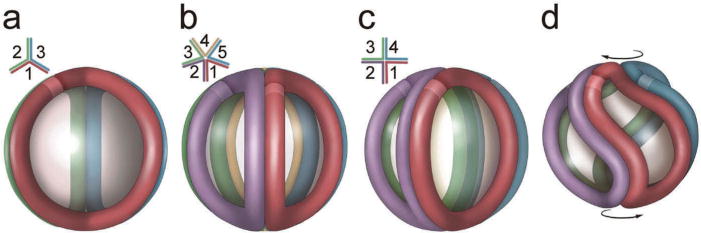
Proposed model of the structure of apolipoprotein A-I in HDL isolated from human plasma. Incorporation of additional apoA-I molecules into the trefoil model and adaptation of apoA-I to smaller HDL particle diameters. (a) Schematic representation of the three-molecule trefoil model as originally proposed, with each molecule of apoA-I shown in a different color. The lighter band on each molecule represents the N-terminus (residue 44, as the model was built in the absence of residues 1–43). The inset shows a schematic top view showing the bend angles of each apoA-I. (b) Pentameric complex proposed for the structure of LpA-I2b. (c) An idealized, fully extended tetrameric complex. (d) Twisted tetrameric complex with a reduced particle diameter as proposed for LpA-I2a. Reproduced with permission from [64]. Full color available online.
Gordon et al. tested the hypothesis that the phospholipid in lipoproteins serves as a platform that supports the assembly of specific protein complexes [42]. By using calcium silica hydrate to isolate any protein complexes containing phospholipid, they were able to use non-density-based methods to isolate HDL and the other classes of lipoproteins in plasma (LDL and very low density lipoprotein [VLDL]). After fractionating whole plasma with three techniques – size-exclusion chromatography, isoelectric focusing, or anion exchange – and then subjecting the preparation to phospholipid affinity chromatography and tandem MS analysis [20], they tracked the co-separation of 76 lipid-associated proteins. By applying a summed correlation analysis, they identified protein pairs that may co-reside on individual lipoproteins.
Gordon et al. [20] then quantified the correlations of relative protein abundance of all pairwise comparisons of the proteins detected in the three analyses, using a simple scoring system. The highest ranked pairs contained the various combinations of the fibrinogen α, β, and γ-chains; the very high scores indicated almost complete co-migration across the separations. Because fibrinogen is composed of two sets of three polypeptide subunits (α, β, and γ), identification of this well-established complex supported the strength of the analytical approach. The next highest scoring pair was apoA-I and apoA-II, HDL’s two major proteins. Moreover, trypanosome lytic factor was identified as co-migrating with apoL-I and haptoglobin-related protein. Collectively, these observations provide strong evidence that biochemical fractionation in concert with phospholipid affinity chromatography can identify biologically important protein–protein interactions in HDL and other lipoproteins.
Network analysis has provided additional important insights. For example, fibrinogen was found to preferentially associate with LDL, which is consistent with its presence in LDL that is precipitated by heparin [65]. Histidine-rich glycoprotein (HRG), whose multidomain structure interacts with many ligands [66], appeared to play previously unsuspected roles in organizing large numbers of other proteins [20]. These observations raise the possibility that HRG serves as a scaffold for assembling specific binding partners on HDL and other lipoproteins.
These observations suggest that individual lipoprotein species perform distinct biological functions. Because HDL is responsible for most of the proteomic diversity observed in lipoproteins [19,34], they further suggest that therapeutic interventions designed to mimic its cardioprotective function may need to elevate only specific subspecies of HDL to achieve benefits [67,68], particularly if altering other subspecies might have deleterious effects on other important functions, such as host defense mechanisms.
Another approach to isolating HDL uses immunoaffinity methods to select lipoprotein particles on the basis of protein composition. For example, phospholipid transfer protein (PLTP), which binds phospholipid and transfers it between lipoproteins, associates with apoA-I [69]. Isolation of PLTP by immunoaffinity chromatography revealed that the most abundant proteins were clusterin (apoJ), PLTP itself, coagulation factors, complement factors, and apoA-I [70]. Unexpectedly, lipids accounted for only 3% of the mass of the complexes. Collectively, these observations indicate that PLTP in human plasma resides on lipid-poor complexes dominated by clusterin and proteins implicated in host defense and inflammation. Network analysis and experiments with isolated complexes incubated with proteins found in the particles suggest that protein–protein interactions, not lipid–protein interactions, drive the formation of PLTP complexes in plasma.
5. Modulation of HDL’s protein composition and function by LDL-lowering therapy
The statin family of compounds lowers LDL-cholesterol levels by inhibiting a key early enzyme in cholesterol synthesis in the liver. Compelling evidence indicates that lowering LDL protects humans from myocardial infarction and other forms of atherosclerotic CVD. There is also strong evidence from animal studies that statins can inhibit atherosclerosis by mechanisms independent of LDL lowering [71], but the relevance of these findings to humans remains unclear.
A recent study tested the hypothesis that statin therapy might change HDL’s protein composition, thereby altering its potential cardioprotective effects [72]. These investigators isolated lipoproteins by size-exclusion chromatography from plasma of subjects before and on statin therapy and selectively recovered HDL from the appropriate fractions by phospholipid affinity chromatography. MS/MS analysis and protein quantification by spectral counting revealed striking alterations in the composition of the HDL proteome. Most notably, for large HDL particles, there was a greater than 20-fold elevation in α-1-antitrypsin (A1AT) spectral counts. These observations were confirmed by ELISA.
A1AT, an abundant member of the serine protease inhibitor (SERPIN) family of proteins, is an acute-phase reactive plasma protein. It is the primary physiological inhibitor of neutrophil elastase (NE), a serine protease secreted by activated neutrophils. NE is also produced by macrophages in atherosclerotic lesions, where it has been proposed to degrade extracellular matrix, which may be a key event in the rupture of atherosclerotic plaques, a common cause of myocardial infarction.
Modeling studies suggested that A1AT binds to HDL lipids via the two hydrophobic methionine amino acids in its reactive loop. Previous studies demonstrated that oxidative modification of either methionine residue causes A1AT to lose its anti-elastase activity [73]. In studies with reconstituted HDL, A1AT bound to the lipoprotein, impairing its ability to inhibit NE. Importantly, HDL-bound A1AT was protected from inactivation by hydrogen peroxide, suggesting that reduced activity might be offset by a beneficial stabilizing effect in a pro-oxidant environment such as the atherosclerotic lesion. Other studies demonstrated that HDL-bound A1AT reduced macrophage activation in vitro, suggesting that the complex might have multiple potential cardioprotective roles. These observations suggest that alterations in HDL’s protein composition caused by lipid-lowering therapy may increase the lipoprotein’s cardioprotective functions by mechanisms other than those that lower LDL.
6. Translational studies: HDL in renal disease subjects
Much of the interest in MS-based studies of HDL centers on the discovery of protein biomarkers that provide insights into disease risk. A plethora of recent papers has documented changes in the HDL proteome in disorders ranging from CVD and rheumatoid arthritis to diabetes and metabolic syndrome [74–81]. Many of these studies have centered on mechanistic implications of the observed changes in the HDL proteome. A key unresolved issue is whether any of the proposed alterations in HDL’s protein cargo are useful clinically.
Carr and colleagues noted six essential steps in demonstrating the clinical utility of a new biomarker [82]: candidate discovery, qualification, verification and research assay optimization, biomarker validation, and commercialization. Key criteria are that a biomarker should be reproducibly detected in different laboratories and should occur in large numbers of subjects (hundreds to thousands) in multiple populations. A major limitation of published MS-based HDL studies is the small number of subjects. It also remains unclear whether MS-based studies of HDL’s proteome are reproducible between different laboratories.
We review recent work on the HDL proteome in renal disease because several different laboratories have studied these subjects and because this work illustrates both the potential advantages of MS-based analysis of HDL for biomarker discovery and the challenges that confront investigators in this area.
The leading cause of death in subjects with end-stage renal disease (ESRD) is CVD, though the underlying mechanisms remain unclear. However, LDL-lowering therapies that markedly lower CVD risk in subjects with normal renal function appear much less effective in ESRD subjects, suggesting that alterations in lipoproteins distinct from LDL may be important in the pathogenesis of CVD. One important mechanism might be loss of HDL’s cardioprotective effects or even conversion of HDL into a pro-atherogenic form, perhaps through alterations in its proteome [83].
Early studies of ESRD subjects detected alterations in HDL’s protein cargo that might be relevant to CVD risk. For example, Holzer et al. found increased levels of SAA1 and apoC-III in HDL and showed that levels of these proteins associated inversely with cholesterol efflux capacity as assessed with RAW macrophages [84]. Weichhart et al. also found an elevated level of serum amyloid protein (SAA iso-form not specified) and showed that it both correlated inversely with the ‘anti-inflammatory potency’ of HDL and promoted the release of inflammatory cytokines from human monocytes [85]. Those workers also found increased levels of α-1-microglobulin/bikunin precursor (AMBP) but, in contrast to SAA, AMBP did not stimulate cytokine production by monocytes. Limitations of these studies include the small numbers of subjects and the semi-quantitative methods used to analyze HDL’s protein cargo.
To confirm and extend these observations in a larger number of subjects, we recently investigated alterations in the HDL proteome of ESRD subjects undergoing hemodialysis. HDL was isolated by ultracentrifugation from 20 control subjects and 40 ESRD subjects [86]. Using LC-MS/MS shotgun proteomic analysis and the criteria that at least two peptides unique to the protein of interest had to be detected in at least five subjects in any group, we identified 63 proteins in HDL. Among those, 35 proteins were observed in at least half the control subjects and 43 were present in at least half the ESRD subjects. This finding contrasts with other previous studies in which many proteins were detected in only one or a few subjects [84,85].
To quantitatively measure relative levels of candidate proteins in HDL isolated from our subjects, we used targeted proteomics with isotope-dilution SRM and [15N]apoA-I as the internal standard [86]. For most proteins, relative protein quantification by shotgun proteomics correlated very well with amounts quantified by SRM. This quantitative approach demonstrated that the relative abundance of 18 proteins was significantly higher in HDL isolated from the ESRD subjects than in HDL from the controls [86]. Moreover, our quantitative analysis of the HDL proteome identified a cluster of six proteins (AMBP, B2M, CFD, CST3, PTGDS, and RBP4) that were dramatically enriched in HDL from patients with the disorder (Figure 3). In contrast, 10 proteins were more abundant in HDL isolated from the controls. Importantly, levels of several antiatherogenic or atherogenic HDL proteins were altered in the ESRD subjects. For example, PON1 levels in HDL were significantly lower in the ESRD subjects than in the control group. PON1 is particularly interesting because its expression is antiatherogenic in multiple hypercholesterolemic mouse models. In contrast, levels of SAA1 in HDL were significantly higher in the ESRD subjects than in the control group, and recent studies have shown that SAA plays a critical role in remodeling the HDL proteome and impairing its efflux capacity during inflammation [87].
Figure 3.
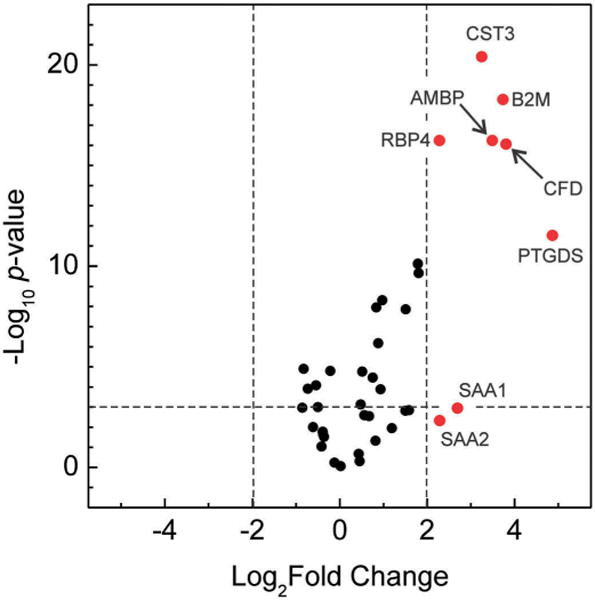
End stage renal disease remarkably remodels the HDL proteome. Volcano plot of HDL proteins: P value versus fold changes [end stage renal disease (ESRD) to control]. Relative levels of 37 proteins in HDL isolated from ESRD and control subjects were determined by isotope-dilution SRM analysis. The y axis is negative base 10 logarithm of the P values from unpaired Student’s t test for 37 proteins quantified with SRM. The × axis is base 2 logarithm of the fold change of the proteins (ratio of ESRD to control). The horizontal dotted line marks the threshold of the P value <0.001 (to the top). The right vertical dotted line marks the threshold of >4-fold increases in ESRD subjects than in control subjects (to the right). The eight proteins whose levels were >4-fold higher in ESRD subjects than in control subjects are shown in red. AMBP, α-1-microglobulin/bikunin precursor; B2M, beta-2-microglobulin; CFD, complement factor D; CST3, cystatin-C; PTGDS, prostaglandin-H2 D-isomerase; RBP4, retinol-binding protein 4; SAA1, serum amyloid A-1 protein; SAA2, serum amyloid A-2 protein. Reprinted with permission from [86]. Full color available online.
Collectively, these observations support the proposal that MS/MS-based proteomics of HDL can be quantitative, that similar results can be obtained when MS analysis is performed in different laboratories, and that the HDL proteome of ESRD subjects is markedly different from that of control subjects. Moreover, data from multiple investigators indicate that the HDL proteome is markedly remodeled in ESRD, supporting the proposal that HDL’s protein cargo can be a marker – and perhaps mediator – of renal disease and serve as a novel biomarker for ESRD status.
In future studies, it will be critical to extend these observations to larger numbers of subjects and to determine whether alterations in the HDL proteome predict incident CVD risk or other clinically important end points.
7. Expert commentary
It is clear that HDL’s protein cargo is diverse and that HDL’s functions extend well beyond its classic role in lipid transport and reverse cholesterol transport. Key challenges for the future are to identify the functional properties that account for HDL’s proposed cardioprotection in humans and to link specific components of HDL with these properties. One exciting possibility is that HDL’s protein composition is one key determinant of its cholesterol efflux capacity, as recently proposed for apoE in mouse HDL [88]. It is noteworthy that HDL is a sub-proteome of the plasma proteome, which simplifies MS/MS analysis and focuses the search for candidate biomarkers on proteins that are likely to be causally linked to CVD.
Different isolation techniques may significantly impact proteomic analyses of HDL. No matter which method is used; however, the key issue is to confirm that any predicted protein changes are functionally or clinically significant. For functional studies, this might include studies of HDL that is depleted or enriched in specific proteins. For clinical studies, it should include an adequate number of subjects, ideally from at least two different populations [82].
Most proteomic analyses of HDL have used shotgun proteomics to detect and quantify proteins. However, this approach is only semi-quantitative. To validate candidate proteins, it is important to use targeted methods for sensitive and specific protein quantification. Two MS-based targeted proteomic approaches are SRM and PRM, and both provide accurate, linear, and reproducible quantification of HDL proteins. However, PRM facilitates method development and biomarker validation because it does not require prior selection of target peptide transitions. A major advantage of targeted MS analysis with SRM or PRM is that multiple proteins can be readily quantified in a single analysis over a wide range of relative concentrations without cross-reacting interferences – a common problem in multiplexed immunoassays. Importantly, multiple studies have repeatedly demonstrated that results from SRM and PRM are consistent with other biochemical approaches, including antibody-based immunoassays. Therefore, MS-based targeted analysis could replace immunoassays.
A major focus of HDL proteomic studies is to determine whether HDL particles isolated from patients carry a unique cargo of proteins, perhaps with different cardioprotective functions. For example, the protein composition of HDL isolated from CVD patients is abnormal [19], but is normalized by aggressive lipid-lowering therapy [89]. These observations suggest that HDL contains proteins that may serve as markers, and perhaps mediators, of CVD risk. Again, a major limitation of these studies is the small number of subjects studied. In the future, it will be important to characterize differences in HDL’s proteome in large numbers of subjects and to determine whether such abnormalities predict incident CVD risk.
It will also be critical to identify and characterize the protein composition of specific HDL particles that may be cardioprotective because HDL-associated proteins are localized in distinct patterns across different subpopulations [90]. Importantly, Du et al. [67] provided strong evidence that small, dense HDL particles (HDL3b and HDL3c) are potent acceptors of cholesterol exported from macrophages by the ABCA1 pathway. Moreover, both the drugs that failed to reduce cardiac risk in the clinical trials increase the concentration of HDL2, the large form of HDL, but not the small, dense HDL3 [6]. Therefore, it will be critical to compare differences in protein cargo between HDL subpopulations (HDL3 or HDL2) isolated from diseased subjects and healthy controls and to determine whether alterations in the HDL proteome associate with impaired cholesterol efflux capacity or other HDL functions.
8. Five-year view
As highlighted in this review, strong evidence supports the proposal that HDL’s proteome is markedly altered in renal disease and statin-treated subjects. However, little is known about the HDL proteome and CVD risk. Although many translational studies have proposed that certain proteins in HDL are linked to CVD risk, most involved only a small number of subjects. Moreover, many of the observations were not replicated in a second set of subjects or in a different study, and both approaches are key to biomarker discovery and validation [82].
Recent advances, such as PRM with isotope dilution using 15N-labeled apoA-I, promise to greatly speed up the identification and quantification of HDL proteins that associate with CVD risk. PRM also enhances confidence in protein identification and is a powerful method for protein quantification. In concert with high-throughput isolation of HDL and automated data analysis, PRM provides a powerful pipeline for identifying candidate protein biomarkers in HDL. In future studies, it will be of great interest to apply non-MS multiplex assays to the analysis of HDL. For example, folded, single-stranded, anionic oligonucleotides can bind proteins with high specificity and affinity [91]. Assays such as these can be sensitive and amenable to high throughput, facilitating cost-effective analysis in translational and clinical studies.
The key challenge now is for investigators to quantify candidate HDL proteins in large clinical studies that focus on incident and prevalent CVD in high-risk subjects, ideally using two independent populations.
It will also be of great interest to associate specific HDL proteins with biological functions of HDL that are linked to cardioprotection. One promising area is cholesterol efflux capacity, which has been shown by multiple groups to predict both incident and prevalent CVD. To confirm the proposed function of a protein linked to HDL’s biological properties, it is possible, for example, to incorporate that protein into HDL. This approach has uncovered functional roles for SAA1 [87] and A1AT [72]. Recent studies have begun to explore this area with mouse models, which offer many advantages (e.g. deficiency of specific proteins, genetic homogeneity, and defined diets). A key challenge to translational investigators will be to extend these studies to humans. Demonstrating that specific proteins in HDL are linked to cardioprotection, atherosclerosis, and the functional properties of HDL would open up the exciting possibility that targeting those proteins pharmacologically could lower CVD risk in humans.
Key issues.
Atherosclerosis is the leading cause of cardiovascular disease (CVD) and death in industrialized nations.
High levels of LDL-cholesterol and low levels of HDL-cholesterol are strongly linked to CVD risk in clinical and epidemiological studies.
Lowering LDL-cholesterol clearly lowers the risk of heart attacks and other forms of atherosclerotic CVD. In contrast, therapies that raise HDL-cholesterol levels have not reduced CVD risk in statin-treated humans with established cardiovascular disease. It is therefore critical to identify new HDL metrics that capture HDL’s proposed cardioprotective effects.
HDL is a family of particles that vary markedly in size and protein composition. MS/MS analyses from multiple laboratories indicate that these particles might carry >80 distinct proteins. An unresolved issue is whether most of those proteins truly associate with HDL or if they are plasma contaminants that co-isolate with the lipoprotein.
Strong evidence indicates that LDL-lowering therapy with a statin and renal disease markedly affect the HDL proteome. Much less is known about alterations of the HDL proteome in subjects at increased risk of CVD.
Recent studies demonstrate the power of isotope dilution MS/MS for quantifying levels of HDL proteins in translational studies. A key challenge is to establish whether these methods can be used to reproducibly study large numbers of subjects.
Once a high-throughput proteomic pipeline is established and validated, it will be critical to identify proteins that indicate established CVD. A long-term goal will be to extend those observations to predict future CVD events in high-risk human populations, such as subjects with established CVD on statin therapy, ESRD subjects, and diabetic subjects.
Quantifying the HDL proteome may provide insights into the efficacy of lipid therapy, and it might identify different sub-fractions of HDL that have distinct biological functions.
Acknowledgments
Funding
This work was supported by grants from the National Institutes of Health: R01HL110418, R01HL108897, R01HL112625, P01HL092969, P01 HL128203 and DP3 DK108209; and the Diabetes Research Center, University of Washington: P30DK017047. Mass spectrometry experiments were performed in the Department of Medicine Mass Spectrometry Resource, Diabetes Research Center Quantitative and Functional Proteomics Core under grant: P30DK017047; and the Proteomics Resource, University of Washington grant: UWPR95794.
Footnotes
Declaration of interest
J. Heinecke is named as a co-inventor on patents from the US Patent Office on the use of HDL markers to predict the risk of cardiovascular disease. Jay Heinecke has served as a consultant for Merck, Amgen, Bristol Meyer Squibb, GlaxoSmithKline, and Pacific Biomarkers. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell. 2015 Mar 26;161(1):161–172. doi: 10.1016/j.cell.2015.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon DJ, Probstfield JL, Garrison RJ, et al. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation. 1989 Jan;79(1):8–15. doi: 10.1161/01.cir.79.1.8. [DOI] [PubMed] [Google Scholar]
- 3.Miller NE, Thelle DS, Forde OH, et al. The Tromso heart-study. High-density lipoprotein and coronary heart-disease: a prospective case-control study. Lancet. 1977 May 7;1(8019):965–968. doi: 10.1016/s0140-6736(77)92274-7. [DOI] [PubMed] [Google Scholar]
- 4.Moore RE, Kawashiri MA, Kitajima K, et al. Apolipoprotein A-I deficiency results in markedly increased atherosclerosis in mice lacking the LDL receptor. Arterioscler Thromb Vasc Biol. 2003 Oct 1;23(10):1914–1920. doi: 10.1161/01.ATV.0000092328.66882.F5. [DOI] [PubMed] [Google Scholar]
- 5.Oram JF, Heinecke JW. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol Rev [Review] 2005 Oct;85(4):1343–1372. doi: 10.1152/physrev.00005.2005. [DOI] [PubMed] [Google Scholar]
- 6.Rader DJ, Tall AR. The not-so-simple HDL story: is it time to revise the HDL cholesterol hypothesis? Nat Med. 2012 Sep;18(9):1344–1346. doi: 10.1038/nm.2937. [DOI] [PubMed] [Google Scholar]
- 7.Rye KA, Barter PJ. Cardioprotective functions of HDLs. J Lipid Res. 2014 Feb;55(2):168–179. doi: 10.1194/jlr.R039297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tall AR, Yvan-Charvet L, Westerterp M, et al. Cholesterol efflux: a novel regulator of myelopoiesis and atherogenesis. Arterioscler Thromb Vasc Biol. 2012 Nov;32(11):2547–2552. doi: 10.1161/ATVBAHA.112.300134. [DOI] [PubMed] [Google Scholar]
- 9.Vergeer M, Holleboom AG, Kastelein JJ, et al. The HDL hypothesis: does high-density lipoprotein protect from atherosclerosis? J Lipid Res. 2010 Aug;51(8):2058–2073. doi: 10.1194/jlr.R001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Rader DJ. Molecular regulation of macrophage reverse cholesterol transport. Curr Opin Cardiol. 2007 Jul;22(4):368–372. doi: 10.1097/HCO.0b013e3281ec5113. [DOI] [PubMed] [Google Scholar]
- 11.Sorci-Thomas MG, Thomas MJ. Microdomains, inflammation, and atherosclerosis. Circ Res. 2016 Feb 19;118(4):679–691. doi: 10.1161/CIRCRESAHA.115.306246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khera AV, Cuchel M, de la Llera-Moya M, et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N Engl J Med. 2011 Jan 13;364(2):127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Rohatgi A, Khera A, Berry JD, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014 Dec 18;371(25):2383–2393. doi: 10.1056/NEJMoa1409065. [Research Support, N.I.H., Extramural Research Support, Non-US Gov’t] This is the first report that serum cholesterol efflux capacity predicts cardiovascular events. Among the 2416 participants in the Dallas Heart Study, 132 had a primary atherosclerotic cardiovascular disease event. This report showed an inverse association of cholesterol efflux capacity and HDL particle concentration, but not HDL cholesterol concentration, with incident cardiovascular events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleheen D, Scott R, Javad S, et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: a prospective case-control study. Lancet Diabetes Endocrinol. 2015 Jul;3(7):507–513. doi: 10.1016/S2213-8587(15)00126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Llera-Moya M, Drazul-Schrader D, Asztalos BF, et al. The ability to promote efflux via ABCA1 determines the capacity of serum specimens with similar high-density lipoprotein cholesterol to remove cholesterol from macrophages. Arterioscler Thromb Vasc Biol. 2010 Apr;30(4):796–801. doi: 10.1161/ATVBAHA.109.199158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Rye KA, Barter PJ. Regulation of high-density lipoprotein metabolism. Circ Res. 2014 Jan 03;114(1):143–156. doi: 10.1161/CIRCRESAHA.114.300632. An excellent review of our current understanding of HDL metabolism. [DOI] [PubMed] [Google Scholar]
- 17.Rader DJ. Molecular regulation of HDL metabolism and function: implications for novel therapies. J Clin Invest. 2006 Dec;116(12):3090–3100. doi: 10.1172/JCI30163. [Research Support, N.I.H., Extramural Research Support, Non-US Gov’t Review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng W, Breslow JL. Dramatically decreased high density lipoprotein cholesterol, increased remnant clearance, and insulin hypersensitivity in apolipoprotein A-II knockout mice suggest a complex role for apolipoprotein A-II in atherosclerosis susceptibility. Proc Natl Acad Sci U S A. 1996 Dec 10;93(25):14788–14794. doi: 10.1073/pnas.93.25.14788. [Research Support, US Gov’t, P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaisar T, Pennathur S, Green PS, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007 Mar;117(3):746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Gordon SM, Deng J, Tomann AB, et al. Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Mol Cell Proteomics. 2013 Nov;12(11):3123–3134. doi: 10.1074/mcp.M113.028134. This proteomic analysis demonstrated that biochemical fractionation in concert with phospholipid affinity chromatography can identify biologically important protein–protein interactions in HDL and other lipoproteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinecke JW. The not-so-simple HDL story: a new era for quantifying HDL and cardiovascular risk? Nat Med. 2012 Sep;18(9):1346–1347. doi: 10.1038/nm.2930. [DOI] [PubMed] [Google Scholar]
- 22.Barter PJ, Caulfield M, Eriksson M, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007 Nov 22;357(21):2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer EJ. Effects of cholesteryl ester transfer protein inhibitors on human lipoprotein metabolism: why have they failed in lowering coronary heart disease risk? Curr Opin Lipidol. 2013 Jun;24(3):259–264. doi: 10.1097/MOL.0b013e3283612454. [DOI] [PubMed] [Google Scholar]
- 24.Fazio S, Linton MF. High-density lipoprotein therapeutics and cardiovascular prevention. J Clin Lipidol [Review] 2010 Sep-Oct;4(5):411–419. doi: 10.1016/j.jacl.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Morehouse LA, Sugarman ED, Bourassa PA, et al. Inhibition of CETP activity by torcetrapib reduces susceptibility to diet-induced atherosclerosis in New Zealand White rabbits. J Lipid Res. 2007 Jun;48(6):1263–1272. doi: 10.1194/jlr.M600332-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Brousseau ME, Schaefer EJ, Wolfe ML, et al. Effects of an inhibitor of cholesteryl ester transfer protein on HDL cholesterol. N Engl J Med. 2004 Apr 08;350(15):1505–1515. doi: 10.1056/NEJMoa031766. [DOI] [PubMed] [Google Scholar]
- 27.McKenney JM, Davidson MH, Shear CL, et al. Efficacy and safety of torcetrapib, a novel cholesteryl ester transfer protein inhibitor, in individuals with below-average high-density lipoprotein cholesterol levels on a background of atorvastatin. J Am Coll Cardiol. 2006 Nov 07;48(9):1782–1790. doi: 10.1016/j.jacc.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 28.Kastelein JJ, Van Leuven SI, Burgess L, et al. Effect of torcetrapib on carotid atherosclerosis in familial hypercholesterolemia. N Engl J Med. 2007 Apr 19;356(16):1620–1630. doi: 10.1056/NEJMoa071359. [DOI] [PubMed] [Google Scholar]
- 29.Bots ML, Visseren FL, Evans GW, et al. Torcetrapib and carotid intima-media thickness in mixed dyslipidaemia (RADIANCE 2 study): a randomised, double-blind trial. Lancet. 2007 Jul 14;370(9582):153–160. doi: 10.1016/S0140-6736(07)61088-5. [DOI] [PubMed] [Google Scholar]
- 30.Boden WE, Probstfield JL, Anderson T, et al. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N Engl J Med. 2011 Dec 15;365(24):2255–2267. doi: 10.1056/NEJMoa1107579. [DOI] [PubMed] [Google Scholar]
- 31.Pollard RD, Fulp B, Sorci-Thomas MG, et al. High-density lipoprotein biogenesis: defining the domains involved in human apolipoprotein A-I lipidation. Biochemistry. 2016 Sep 06;55(35):4971–4981. doi: 10.1021/acs.biochem.6b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih DM, Gu L, Xia YR, et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998 Jul 16;394(6690):284–287. doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- 33.Jarvik GP, Rozek LS, Brophy VH, et al. Paraoxonase (PON1) phenotype is a better predictor of vascular disease than is PON1(192) or PON1(55) genotype. Arterioscler Thromb Vasc Biol. 2000 Nov;20(11):2441–2447. doi: 10.1161/01.atv.20.11.2441. [DOI] [PubMed] [Google Scholar]
- 34••.Shah AS, Tan L, Long JL, et al. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J Lipid Res [Review] 2013 Oct;54(10):2575–2585. doi: 10.1194/jlr.R035725. An excellent review summarizing recent progress in our understanding of the diversity of the HDL proteome and its relationship with HDL functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Hafiane A, Genest J. High density lipoproteins: measurement techniques and potential biomarkers of cardiovascular risk. BBA Clin. 2015 Jun;3:175–188. doi: 10.1016/j.bbacli.2015.01.005. This review discusses the limitations of current laboratory technologies for methods that separate and quantify HDL and potential application to predict CVD, with an emphasis on emergent approaches as potential biomarkers in clinical practice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Karathanasis SK, Freeman LA, Gordon SM, et al. The changing face of HDL and the best way to measure it. Clin Chem. 2017 Jan;63(1):196–210. doi: 10.1373/clinchem.2016.257725. A thorough review on our current understanding of the formation and maturation of HDL particles via reverse cholesterol transport as well as other newly recognized roles of HDL. [DOI] [PubMed] [Google Scholar]
- 37.Sorci-Thomas MG, Thomas MJ. High density lipoprotein biogenesis, cholesterol efflux, and immune cell function. Arterioscler Thromb Vasc Biol. 2012 Nov;32(11):2561–2565. doi: 10.1161/ATVBAHA.112.300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havel RJ, Eder HA, Bragdon JH. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 1955 Sep;34(9):1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skinner ER. High-density lipoprotein subclasses. Curr Opin Lipidol. 1994 Jun;5(3):241–247. doi: 10.1097/00041433-199405030-00013. [DOI] [PubMed] [Google Scholar]
- 40.McVicar JP, Kunitake ST, Hamilton RL, et al. Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1356–1360. doi: 10.1073/pnas.81.5.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung MC, Wolf AC, Lum KD, et al. Distribution and localization of lecithin: cholesterolacyltransferase and cholesteryl ester transfer activity in A-I-containing lipoproteins. J Lipid Res. 1986 Nov;27(11):1135–1144. [PubMed] [Google Scholar]
- 42.Gordon SM, Deng J, Lu LJ, et al. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res. 2010 Oct 1;9(10):5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe J, Charles-Schoeman C, Miao Y, et al. Proteomic profiling following immunoaffinity capture of high-density lipoprotein: association of acute-phase proteins and complement factors with proinflammatory high-density lipoprotein in rheumatoid arthritis. Arthritis Rheum. 2012 Jun;64(6):1828–1837. doi: 10.1002/art.34363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holzer M, Kern S, Birner-Grunberger R, et al. Refined purification strategy for reliable proteomic profiling of HDL2/3: impact on proteomic complexity. Sci Rep. 2016 Dec 05;6:38533. doi: 10.1038/srep38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oberbach A, Adams V, Schlichting N, et al. Proteome profiles of HDL particles of patients with chronic heart failure are associated with immune response and also include bacteria proteins. Clin Chim Acta. 2016 Jan;30(453):114–122. doi: 10.1016/j.cca.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Davidson WS, Shah AS HDL Proteome Watch. [Internet] [updated Feb. 18 2015; cited 2017 Nov 1]; Available from: http://homepages.uc.edu/~davidswm/HDLproteome.html.
- 47.Davidson WS, Shah AS LDL Proteome Watch. [Internet]. [updated Feb. 18 2015; cited 2017 Nov 1]; Available from: http://homepages.uc.edu/~davidswm/LDLproteome.html.
- 48.Link AJ, Eng J, Schieltz DM, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999 Jul;17(7):676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 49.Mann M, Hendrickson RC, Pandey A. Analysis of proteins and proteomes by mass spectrometry. Annu Rev Biochem. 2001;70:437–473. doi: 10.1146/annurev.biochem.70.1.437. [Research Support, Non-US Gov’t Review] [DOI] [PubMed] [Google Scholar]
- 50.Peng J, Elias JE, Thoreen CC, et al. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003 Jan-Feb;2(1):43–50. doi: 10.1021/pr025556v. [Research Support, Non-US Gov’t Research Support, US Gov’t, PHS] [DOI] [PubMed] [Google Scholar]
- 51.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006 Apr 14;312(5771):212–217. doi: 10.1126/science.1124619. [Research Support, N.I.H., Extramural Review] [DOI] [PubMed] [Google Scholar]
- 52.Ronsein GE, Pamir N, Von Haller PD, et al. Parallel reaction monitoring (PRM) and selected reaction monitoring (SRM) exhibit comparable linearity, dynamic range and precision for targeted quantitative HDL proteomics. J Proteomics. 2015 Jan;15(113):388–399. doi: 10.1016/j.jprot.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brun V, Dupuis A, Adrait A, et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007 Dec;6(12):2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 54••.Hoofnagle AN, Becker JO, Oda MN, et al. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012 Apr;58(4):777–781. doi: 10.1373/clinchem.2011.173856. The first application of isotope dilution with SRM for quantifying HDL proteins. The authors demonstrated that the targeted proteomic technology can quantify HDL proteins as accurately as biochemical approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourmaud A, Gallien S, Domon B. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: principle and applications. Proteomics. 2016 Aug;16(15–16):2146–2159. doi: 10.1002/pmic.201500543. [DOI] [PubMed] [Google Scholar]
- 56.Domon B, Gallien S. Recent advances in targeted proteomics for clinical applications. Proteomics Clin Appl. 2015 Apr;9(3–4):423–431. doi: 10.1002/prca.201400136. [DOI] [PubMed] [Google Scholar]
- 57.Gallien S, Bourmaud A, Kim SY, et al. Technical considerations for large-scale parallel reaction monitoring analysis. J Proteomics. 2014 Apr;04(100):147–159. doi: 10.1016/j.jprot.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 58•.Ronsein GE, Reyes-Soffer G, He Y, et al. Targeted proteomics identifies paraoxonase/arylesterase 1 (PON1) and apolipoprotein CS as potential risk factors for hypoalphalipoproteinemia in diabetic subjects treated with fenofibrate and rosiglitazone. Mol Cell Proteomics. 2016 Mar;15(3):1083–1093. doi: 10.1074/mcp.M115.054528. PRM with isotope dilution was used to demonstrate marked changes in the HDL proteome of individuals enrolled in the ACCORD-Lipid study that developed hypoalphalipoproteinemia. The results were confirmed by SRM analysis and also by biochemical approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asztalos BF, Schaefer EJ. High-density lipoprotein subpopulations in pathologic conditions. Am J Cardiol. 2003 Apr 3;91(7A):12E–7E. doi: 10.1016/s0002-9149(02)03383-0. [DOI] [PubMed] [Google Scholar]
- 60.Asztalos BF, Tani M, Schaefer EJ. Metabolic and functional relevance of HDL subspecies. Curr Opin Lipidol. 2011 Jun;22(3):176–185. doi: 10.1097/MOL.0b013e3283468061. [Research Support, US Gov’t, Non-PHS Review] [DOI] [PubMed] [Google Scholar]
- 61.Raper J, Fung R, Ghiso J, et al. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. 1999 Apr;67(4):1910–1916. doi: 10.1128/iai.67.4.1910-1916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiflett AM, Bishop JR, Pahwa A, et al. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005 Sep 23;280(38):32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 63.Heinecke JW. The protein cargo of HDL: implications for vascular wall biology and therapeutics. J Clin Lipidol. 2010 Sep-Oct;4(5):371–375. doi: 10.1016/j.jacl.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang R, Silva RA, Jerome WG, et al. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat Struct Mol Biol. 2011 Apr;18(4):416–422. doi: 10.1038/nsmb.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaeger BR, Marx P, Pfefferkorn T, et al. Heparin-mediated extracorporeal LDL/fibrinogen precipitation–H.E.L.P.–in coronary and cerebral ischemia. Acta Neurochir Suppl. 1999;73:81–84. doi: 10.1007/978-3-7091-6391-7_13. [DOI] [PubMed] [Google Scholar]
- 66.Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: a novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol Cell Biol. 2005 Apr;83(2):106–118. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 67.Du XM, Kim MJ, Hou L, et al. HDL particle size is a critical determinant of ABCA1-mediated macrophage cellular cholesterol export. Circ Res. 2015 Mar 27;116(7):1133–1142. doi: 10.1161/CIRCRESAHA.116.305485. [DOI] [PubMed] [Google Scholar]
- 68.Ronsein GE, Hutchins PM, Isquith D, et al. Niacin therapy increases high-density lipoprotein particles and total cholesterol efflux capacity but not ABCA1-specific cholesterol efflux in statin-treated subjects. Arterioscler Thromb Vasc Biol. 2016 Feb;36(2):404–411. doi: 10.1161/ATVBAHA.115.306268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albers JJ, Vuletic S, Cheung MC. Role of plasma phospholipid transfer protein in lipid and lipoprotein metabolism. Biochim Biophys Acta. 2012 Mar;1821(3):345–357. doi: 10.1016/j.bbalip.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheung MC, Vaisar T, Han X, et al. Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation. Biochemistry. 2010 Aug 31;49(34):7314–7322. doi: 10.1021/bi100359f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becker L, Gharib SA, Irwin AD, et al. A macrophage sterol-responsive network linked to atherogenesis. Cell Metab. 2010 Feb 3;11(2):125–135. doi: 10.1016/j.cmet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Gordon SM, McKenzie B, Kemeh G, et al. Rosuvastatin alters the proteome of high density lipoproteins: generation of alpha-1-anti-trypsin enriched particles with anti-inflammatory properties. Mol Cell Proteomics. 2015 Dec;14(12):3247–3257. doi: 10.1074/mcp.M115.054031. A proteomics study demonstrated that α-1-antitrypsin (A1AT) was elevated 20-fold in large HDL particles after statin therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taggart C, Cervantes-Laurean D, Kim G, et al. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem. 2000 Sep 01;275(35):27258–27265. doi: 10.1074/jbc.M004850200. [DOI] [PubMed] [Google Scholar]
- 74.Yan LR, Wang DX, Liu H, et al. A pro-atherogenic HDL profile in coronary heart disease patients: an iTRAQ labelling-based proteomic approach. PLoS One. 2014;9(5):e98368. doi: 10.1371/journal.pone.0098368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaisar T, Mayer P, Nilsson E, et al. HDL in humans with cardiovascular disease exhibits a proteomic signature. Clin Chim Acta. 2010 Jul 04;411(13–14):972–979. doi: 10.1016/j.cca.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alwaili K, Bailey D, Awan Z, et al. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile. Biochimica Et Biophysica Acta. 2012 Mar;1821(3):405–415. doi: 10.1016/j.bbalip.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Riwanto M, Rohrer L, Roschitzki B, et al. Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling. Circulation. 2013 Feb 26;127(8):891–904. doi: 10.1161/CIRCULATIONAHA.112.108753. [DOI] [PubMed] [Google Scholar]
- 78.Holzer M, Wolf P, Curcic S, et al. Psoriasis alters HDL composition and cholesterol efflux capacity. J Lipid Res. 2012 Aug;53(8):1618–1624. doi: 10.1194/jlr.M027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manjunatha S, Distelmaier K, Dasari S, et al. Functional and proteomic alterations of plasma high density lipoproteins in type 1 diabetes mellitus. Metabolism. 2016 Sep;65(9):1421–1431. doi: 10.1016/j.metabol.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Raterman HG, Levels H, Voskuyl AE, et al. HDL protein composition alters from proatherogenic into less atherogenic and proinflammatory in rheumatoid arthritis patients responding to rituximab. Ann Rheum Dis. 2013 Apr;72(4):560–565. doi: 10.1136/annrheumdis-2011-201228. [DOI] [PubMed] [Google Scholar]
- 81.Hoofnagle AN, Wu M, Gosmanova AK, et al. Low clusterin levels in high-density lipoprotein associate with insulin resistance, obesity, and dyslipoproteinemia. Arterioscler Thromb Vasc Biol. 2010 Dec;30(12):2528–2534. doi: 10.1161/ATVBAHA.110.212894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006 Aug;24(8):971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 83.Rye KA. Biomarkers associated with high-density lipoproteins in atherosclerotic kidney disease. Clin Exp Nephrol. 2014 Apr;18(2):247–250. doi: 10.1007/s10157-013-0865-x. [DOI] [PubMed] [Google Scholar]
- 84.Holzer M, Birner-Gruenberger R, Stojakovic T, et al. Uremia alters HDL composition and function. J Am Soc Nephrol. 2011 Sep;22(9):1631–1641. doi: 10.1681/ASN.2010111144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weichhart T, Kopecky C, Kubicek M, et al. Serum amyloid A in uremic HDL promotes inflammation. J Am Soc Nephrol. 2012 May;23(5):934–947. doi: 10.1681/ASN.2011070668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Shao B, De Boer I, Tang C, et al. A cluster of proteins implicated in kidney disease is increased in high-density lipoprotein isolated from hemodialysis subjects. J Proteome Res. 2015 Jul 2;14(7):2792–2806. doi: 10.1021/acs.jproteome.5b00060. The first study of HDL in end-stage renal disease (ESRD) using targeted SRM with isotope dilution to quantify HDL proteins. The authors demonstrated that a cluster of six proteins are dramatically enriched in HDL from patients with ESRD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vaisar T, Tang C, Babenko I, et al. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. J Lipid Res. 2015 Aug;56(8):1519–1530. doi: 10.1194/jlr.M059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pamir N, Hutchins P, Ronsein G, et al. Proteomic analysis of HDL from inbred mouse strains implicates APOE associated with HDL in reduced cholesterol efflux capacity via the ABCA1 pathway. J Lipid Res. 2016 Feb;57(2):246–257. doi: 10.1194/jlr.M063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Green PS, Vaisar T, Pennathur S, et al. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation. 2008 Sep 16;118(12):1259–1267. doi: 10.1161/CIRCULATIONAHA.108.770669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davidson WS, Silva RA, Chantepie S, et al. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009 Jun;29(6):870–876. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010 Dec 07;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]


