Abstract
One of the many biological functions of nitric oxide is the ability to protect cells from oxidative stress. To investigate the potential contribution of low steady state levels of nitric oxide generated by endothelial nitric oxide synthase (eNOS) and the mechanisms of protection against H2O2, spontaneously transformed human ECV304 cells, which normally do not express eNOS, were stably transfected with a green fluorescent-tagged eNOS cDNA. The eNOS-transfected cells were found to be resistant to injury and delayed death following a 2-h exposure to H2O2 (50–150 μM). Inhibition of nitric oxide synthesis abolished the protective effect against H2O2 exposure. The ability of nitric oxide to protect cells depended on the presence of respiring mitochondria as ECV304+eNOS cells with diminished mitochondria respiration (ρ−) are injured to the same extent as nontransfected ECV304 cells and recovery of mitochondrial respiration restores the ability of nitric oxide to protect against H2O2-induced death. Nitric oxide also found to have a profound effect in cell metabolism, because ECV304+eNOS cells had lower steady state levels of ATP and higher utilization of glucose via the glycolytic pathway than ECV304 cells. However, the protective effect of nitric oxide against H2O2 exposure is not reproduced in ECV304 cells after treatment with azide and oligomycin suggesting that the dynamic regulation of respiration by nitric oxide represent a critical and unrecognized primary line of defense against oxidative stress.
Under certain experimental conditions, nitric oxide has been shown to render cells resistant to oxidative stress (1–5). Multiple mechanisms have been proposed for the ability of nitric oxide to protect cells against oxidative stress, including biochemical reactions of nitric oxide and the induction of adaptive responses that require protein synthesis (1–6). Nitric oxide readily complexes with reduced iron, preventing the formation of strong oxidants (1–3). Similarly, kinetically fast reactions of nitric oxide with lipid and/or organic radicals result in termination of chain reactions and protect against membrane peroxidation and peroxidative chemistry-induced cell injury (1–3). Exposure to low, nonlethal doses of nitric oxide has been shown to induce adaptive responses that render cells resistant to lethal concentrations of nitric oxide and/or peroxides. These adaptive responses include the induction of hemoxygenase-1 (HO-1) and Mn superoxide dismutase (1, 6). The up-regulation of HO-1 was accompanied by an increase in ferritin to account for the release of iron from HO-1, indicating a role of both iron heme and nonheme iron for peroxide-mediated cellular injury (1, 3). Recent data has also revealed that nitric oxide, by regulating critical mitochondrial functions such as respiration, membrane potential, and release of cytochrome c, is able to trigger defense mechanisms against cell death induced by proapoptotic stimuli (7–14). Overall, a number of cellular pathways and mechanisms can be regulated by nitric oxide in a manner that confers protection against oxidative stress. However, with the exception of the studies of Lancaster and coworkers (1) that reported protection of hepatocytes from peroxides after cytokine-induced nitric oxide, the role of endogenous nitric oxide production against the deleterious effects of peroxides and oxidative stress remains largely unknown because the majority of the cellular systems use nitric oxide donors as sources of nitric oxide (2–6). Therefore, it remains unclear whether production of nitric oxide by the low-output endothelial NOS is capable of protecting cells against peroxides and whether regulation of mitochondrial function by low levels of endogenous nitric oxide can protect cells against oxidative stress.
To study the potential contribution and mechanisms of endogenous nitric oxide produced by the low-output endothelial NOS in protecting cells against peroxide stress, we utilize the ECV304 cell line. The ECV304 cells have been used to study endothelial nitric oxide synthase (eNOS) trafficking, lipoprotein metabolism, signal transduction events, expression of adhesion molecules, calcium metabolism and trafficking, and mechanisms of apoptosis and cell death from a variety of stimuli (13–20). ECV304 cells maintain typical cobblestone morphology, form lateral membrane interdigitations that prevent the diffusion of lanthanum, and respond to classical and selective endothelial stimulants such as ionomycin, ADP, vascular endothelial growth factor, bradykinin, angiotensin, and oxidized low density lipoprotein (13–20). The ECV304 cells express classical endothelial receptors such as thrombomodulin and vitronectin receptor CD51, and are capable of secreting plasminogen activator inhibitor-1 and endothelin (21). However, during transformation the cells lost the ability to generate nitric oxide and have acquired phenotypic expression of epithelial proteins such as E-cadherin and desmosomal proteins (21, 22). Therefore, although these cells express all of the classical endothelial markers in culture, they have differentiated toward an epithelium-like phenotype (22). Recently these cells were stably transfected with eNOS. The localization of eNOS after transfection is similar to that observed in endothelium of intact blood vessels, with perinuclear localization associated with Golgi markers and plasma localization with caveolin (20). The transfected cells generate sufficient nitric oxide to stimulate the production of smooth muscle cGMP in a coculture system. A basal stimulation of cGMP was observed that was increased by 10-fold after ionomycin stimulation. Both the basal and the inducible cGMP production in coculture studies were completely abrogated by the eNOS inhibitor NG-nitro-L-arginine methyl ester (L-NAME; ref. 20). We rationalize that this is a reasonable model to study the role and mechanisms of nitric oxide-mediated protection from oxidative stress because the production of NO recapitulates intracellular “low output” levels sufficient to stimulate guanylate cyclase in a coculture system in an L-NAME inhibitable manner.
Materials and Methods
Cell Culture, Exposure to H2O2, and Determination of Cell Viability.
Cells were cultured in 4.5 g/liter DMEM, with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. The medium of the eNOS-transfected cells was supplemented with 100 μg/ml geneticin (GIBCO/BRL) under the conditions described (20) and all experiments were performed in 6-well plates, using 1 × 106 cell per well. Reagent H2O2 was added as a bolus addition. During and after the 2-h H2O2 exposure, 2 μM YO-PRO1, a DNA-binding and membrane-impermeable dye that is used to determine membrane integrity and cell viability, is added. Cell viability was determined by counting the number of cells with and without YO-PRO1 fluorescence under an inverted fluorescence microscope as described (23).
ATP Levels and Glucose Utilization.
The steady state levels of ATP were determined by the bioluminescent somatic cell assay (Sigma). After a 1 h incubation with or without 100 μM L-NAME, adhered cells were washed twice to remove nonadhered and dead cells. The adhered cells were trypsinized, viability was confirmed with trypan blue exclusion, and aliquots were analyzed for ATP. Equivalent numbers of cells were incubated for 1 h with glucose that was labeled with 13C in the first carbon (13C-1 glucose) and glucose that was labeled with two deuterium ions in the sixth carbon (2H2-C6 glucose). The C1 of glucose is predominantly metabolized via the pentose phosphate pathway to CO2, whereas the remaining of the glucose molecule is oxidized by the glycolytic pathway to pyruvate. After a 1-h incubation with the labeled glucose mixture in rubber sealed plates to avoid loss of CO2, the cell extracts were treated with 3 ml NaOH to convert all CO2 to bicarbonate. Two milliliters of cell extract was reacted with acid under vacuum to convert all bicarbonate to CO2, and the label in the gas phase was detected by isotope ratio mass spectrometry by monitoring the m/z 45/44 ion ratio with a Sira 12 IR-MS (VG Instruments, Davers, MA). The remaining 1 ml of cell extract was divided into two 0.5-ml aliquots. One aliquot was used for the measurement of total lactate by the fluorimetric assay and the other was acidified, and then derivatized to yield the di-t-butyldimethylsilyl species. Label (atom percent excess) in the latter was determined with gas chromatography–mass spectrometry monitoring of the ion pair at m/z 276/274 with a Hewlett–Packard Mass Selective Detector. The product of the two lactate measurements represents the total lactate produced by the cells.
Determination of Enzymatic Activities.
Glucose-6-phosphate dehydrogenase (G6PDH) was assayed in 50 mM Tris⋅HCl (pH 7.8)/1 mM MgCl2/3 mM glucose-6-phosphate/20 μM NADP+. After the addition of 200 μg/ml cell lysate, formation of NADPH at 340 nm is proportional to the G6PDH activity as described (24). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity was determined in 50 mM Tris⋅HCl (pH 8.5)/0.1 mM diethylenetriamine pentaacetic acid (DTPA)/0.25 mM NAD+/0.5 mM glyceradehyde-3-phosphate/10 mM arsenic acid. GAPDH was assayed at 20°C by following the production of NADH at 340 nm during the first 5 min after the addition of 100 μg/ml of cell protein (25, 26). Glyceraldehyde-3-phosphate was obtained as barium diethylacetal-DL-glyceraldehyde-3-phosphate and converted to the free aldehyde as described (26). The activity of Mn superoxide dismutase was determined by the reduction of cytochrome c by superoxide generated from xanthine plus xanthine oxidase. To account for the contamination of mitochondrial fractions with Cu,Zn superoxide dismutase, the reduction of cytochrome c was assayed in the presence of freshly prepared 1 mM KCN (27). Catalase activity was measured by the initial rate of decomposition of 20 μM H2O2 at 240 nm (ɛ = 0.043 mM−1⋅cm−1). One unit of enzyme activity represents 1 μmol of substrate consumed for the first minute. Caspase-3 activity was determined as described (13, 23).
Cellular Concentration of Reduced Thiols.
After washing, the cells were harvested in 200 μl of buffer containing 20 mM Tris (pH 7.4), 150 mM NaCl, 10% glycerol, 1% Triton X-100, and 4 mM EGTA. An aliquot is used to quantify both protein and nonprotein thiols, using a thiol quantification kit (Molecular Probes) according to directions provided by the manufacturer. The total reduced thiol concentration is normalized to cellular protein.
Generation of Mitochondrial DNA Less ρ− Cells.
To generate cells without respiring mitochondria (ρ−), cells were grown in the presence of 50 ng/ml ethidium bromide for a total of 8 weeks, as described by King and Attardi (28, 29). Ethidium bromide selectively inhibits mitochondrial DNA replication without affecting nuclear DNA. Because mitochondrial DNA encodes for most but not all components of the respiratory chain, the ρ− cells have diminished respiratory function (Table 1) and rely for the most part on glycolysis for ATP synthesis (28, 29). The ρ− cells maintain their morphological appearance and have similar mitochondrial densities (Fig. 3) and intact cell membranes (Table 2) just as the parent cell lines. Moreover, removal of ethidium bromide form the media followed by a 2-week culturing in normal medium restores mitochondrial respiration (29) and the NO-mediated protection against H2O2 challenge (Table 2).
Table 1.
Oxygen consumption by mitochondrial respiratory components
| Substrate + inhibitor | ECV304 | ECV304 ρ− | ECV304+eNOS | ECV304+eNOS ρ− |
|---|---|---|---|---|
| Pyruvate + malate | 3.0 ± 0.7 | 0.3 ± 0.1* | 1.7 ± 0.8 | 1.2 ± 0.4 |
| Succinate + rotenone | 2.5 ± 0.5 | 0.4 ± 0.2* | 1.7 ± 1.0 | 0.4 ± 0.2** |
| Ascorbate/TMPD + antimycin | 5.9 ± 1.3 | 2.3 ± 1.0* | 3.6 ± 0.3 | 1.3 ± 0.4** |
ADP-stimulated oxygen consumption by mitochondrial complexes I, III, and IV was performed in digitonin-permeable cells with the addition of 10 mM pyruvate and 2 mM malate, and complexes II, III, and IV with 10 mM succinate plus 10 μM rotenone. Azide-sensitive cytochrome oxidase activity (complex IV) was measured by using 3 mM ascorbate plus 0.3 mM TMPD in the presence of 2 μM antimycin. Oxygen consumption is expressed as nAO/min/106 cells and the data represent means ± SD of 3–8 independent determinations.
, Statistically significant at P < 0.05 from ECV304;
, statistically significant at P < 0.05 from ECV304+eNOS.
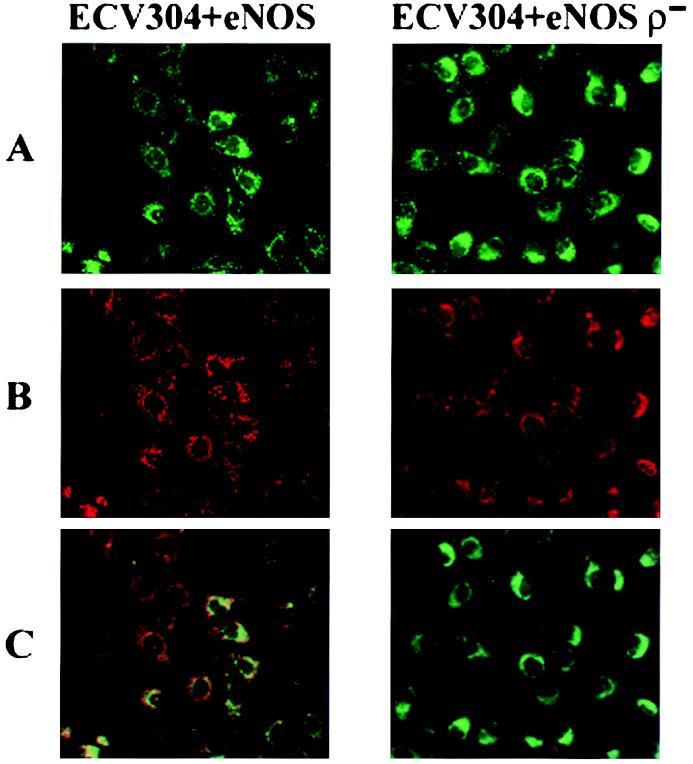
Figure 3.
Mitochondria imaging. Representative images of ECV304+eNOS and ECV304+eNOS ρ− cells loaded with MitoTracker Green to visualize cellular mitochondrial content (A) and with MitoTracker Red CM-H2XRos to visualize respiring mitochondria (B). In C, the fields in A and B were obtained under identical conditions and merged. (Magnification, ×38.)
Table 2.
Mitochondrial respiration and the protective effect of nitric oxide
| Cells YO-PRO1-labeled at the end of H2O2 exposure, % | Cells YO-PRO1-labeled 18 h after H2O2 exposure, % | |
|---|---|---|
| ECV304 | 9 ± 8 | 51 ± 17 |
| ECV304 ρ− | 5 ± 4 | 77 ± 6 |
| ECV304 post ρ− | 3 ± 3 | 53 ± 12 |
| ECV304+eNOS | 9 ± 8 | 9 ± 6 |
| ECV304+eNOS ρ− | 21 ± 12 | 62 ± 16 |
| ECV304+eNOS post ρ− | 2 ± 1 | 6 ± 3 |
ECV304 and ECV304+eNOS cells with or without respiring mitochondria (ρ−) and cells with recovered mitochondrial respiration (post ρ−) were exposed to 100 μM H2O2 and viability was determined. Data represent means ± SD of three independent experiments.
Oxygen Consumption.
Oxygen consumption in digitonin-permeabilized cells was performed with a Clark-type oxygen electrode in a magnetically stirred, thermostatically regulated chamber at 30°C. Cells were suspended in a total volume of 0.65 ml of air-saturated isotonic buffer composed of 220 mM mannitol, 70 mM sucrose, 10 mM Hepes (pH 7.2), 5 mM KH2PO4, and 1 mM EGTA. ADP-stimulated oxygen consumption by complexes I, III, and IV was performed after the addition of 10 mM glutamate plus 2 mM malate. ADP-stimulated oxygen consumption by complexes II, III, and IV was measured after the addition of 10 mM succinate and 10 μM rotenone. Azide-sensitive cytochrome oxidase activity (complex IV) was measured by using 3 mM ascorbate plus 0.3 mM N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) in the presence of 2 μM antimycin.
Protein, DNA Concentration, and Growth Rates.
ECV304, ECV304 ρ−, ECV304+eNOS, and ECV304+eNOS ρ− cells were grown in T175 flasks to a final density of ≈15.0 × 106 cells per flask. The cells were then trypsinized, counted with trypan blue exclusion assay, and aliquots of 2.0 × 106 alive cells were sonicated and the amount of protein determined by using the Bradford Protein Assay (Bio-Rad). DNA was purified by using a DNA purification kit (QIAamp DNA Mini Kit, Qiagen) and the concentration of the purified DNA was determined by using the PicoGreen double-stranded DNA Quantification Assay (Molecular Probes). The protein to DNA ratio for the ECV304, ECV304 ρ−, ECV304+eNOS, and ECV304+eNOS ρ− cells was 31.0 ± 2, 35.9 ± 9, 29.3 ± 0.4, and 39.6 ± 6, respectively (n = 3–5 independent cellular preparations). ECV304+eNOS and ECV304+eNOS ρ− cells were cultured in 35-mm tissue culture dishes by plating ≈80,000 cells in each well to measure the rate of proliferation. The cells were left to grow in the appropriate medium, and at various times after initiation of the culture cells were harvested and the total number of cells measured with the trypan blue exclusion assay. As expected from published data (28, 29) the cells grew slower in ethidium bromide, requiring 213 h to achieve 27% ± 1% confluence as compared with 116 h for the parent cell line ECV+eNOS cells. The ECV+eNOS cells were 100% confluent by 164 h, whereas 332 h were required for the ECV304+eNOS ρ− to reach 100% confluency.
Measurement of NO Metabolites.
Samples containing 2.0 × 106 cells were sonicated and the cell lysates were diluted 3-fold with cold ethanol, vortexed, and left on ice for 30 min. The diluted lysates were centrifuged at 14,000 rpm for 5 min, 60 μl of the supernatant was injected in a reaction chamber containing a VCl3/HCl mixture heated at 90°C, and nitric oxide was detected by chemiluminescence (Nitric Oxide Analyzer, Sievers Instruments, Boulder, CO). The standard curve was generated by injections of known concentrations of NaNO3. The nitrite plus nitrate levels in the lysates of the ECV+eNOS and ECV304+eNOS ρ− cells were similar: 598 ± 111 nM and 781 ± 85 nM, respectively. Moreover, confocal imaging of eNOS expression in the ECV304+eNOS ρ− cells were similar to the parent cell line ECV+eNOS (data not shown).
Mitochondrial Morphology.
ECV304+eNOS and ECV304+eNOS ρ− cells were cultured at a final density of ≈0.5 × 106 cells per well. The cells were washed with fresh medium, to remove nonadhered cells, and Mitotracker Green and Mitotracker Red CM-H2XROS (Molecular Probes), at final concentrations of 150 nM and 500 nM, respectively, were added in fresh medium as described (30, 31). MitoTracker Green (λex = 490 nm and λem = 516 nm), a cell-permeant, mitrochondrial-specific dye, is non fluorescence in aqueous solutions and becomes fluorescent only on sequestration and association with lipids within the mitochondria without requiring oxidation or reduction. The dye contains a mildly thiol-reactive chloromethyl moiety that keeps it inside the mitochondria and essentially measures cellular mitochondria content and distribution (30, 31). MitoTracker Red CM-H2XRos is a red fluorescence X-rosamine derivative (λex = 579 nm and λem = 599 nm) that is also cell-permeable and sequesters in the mitochondria, but does not emit fluorescence unless it is oxidized, implying that actively respiring mitochondria are required to obtain fluorescence (30, 31). The cells were incubated at 37°C for 30 min and then washed extensively with Dulbecco's PBS. Fluorescence was then visualized under an inverted microscope (X70, Olympus, New Hyde Park, NY).
Statistical Analysis.
Data were analyzed by one-way ANOVA using Tukey's post hoc test to determine statistical significance for all pairwise multiple comparison procedures and Dunnett's test for multiple comparisons against the control group. For all statistical analyses the SIGMASTAT program (Jandel, San Rafael, CA) was used. All of the data are expressed as mean ± SD and P < 0.05 was considered to be statistically significant.
Results
Effect of Nitric Oxide on Cell Viability After H2O2 Exposure.
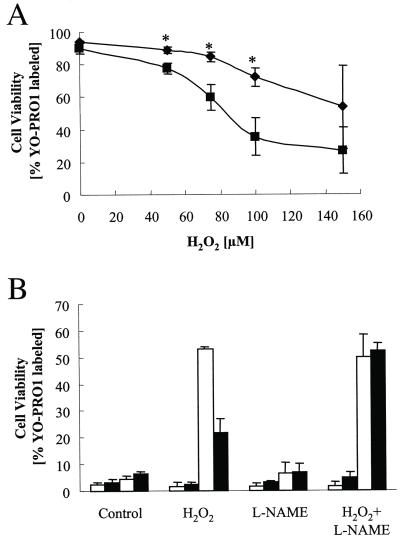
ECV304 and ECV304+eNOS cells were exposed to H2O2 at 0, 10, 25, 50, 100, and 150 μM for 2 h under the same conditions. Previously, we had determined that the consumption of exogenous added H2O2 by endothelial cells is on the order of 1.5–2 μM/min. Thus, we exposed the cells for 2 h to ensure complete consumption of the added H2O2. Cell injury was monitored over time by fluorescence microscopy and revealed that ECV304 and ECV304+eNOS cells, when exposed to 150 μM H2O2, showed evidence of injury immediately after the 2-h exposure. A delayed cell injury in ECV304 cells (≈18 h after exposure), but not in the eNOS transfected cells, was apparent in cells treated with up to 100 μM H2O2 (Fig. 1 A and B). To investigate whether eNOS-generated NO was responsible for the protective effect, the cells were incubated with the eNOS inhibitor L-NAME. As shown in Fig. 1B, L-NAME (added 1 h before H2O2 exposure and remaining in the solution for the duration of the experiment) was effective in abrogating the protective effect of nitric oxide. L-NAME had no effect in the nontransfected cells. These data indicated that endogenous nitric oxide production by the eNOS protects against peroxide-mediated delayed cell death, but not against acute cell death induced by relatively high H2O2 concentrations.
Figure 1.
(A) Nitric oxide-mediated protection against H2O2. After 2 h exposure to different concentrations of H2O2, cells were washed and fresh medium containing 2 μM YO-PRO1 was added. Cell injury measured as % cells with YO-PRO-1 labeling was determined 18 h after exposure. Data represent means ± SD of three to five independent experiments. There was significant (P < 0.05, Student's t test) protection in the ECV304-eNOS (diamonds) after exposure to 25, 50, and 100 μM H2O2, but not 150 μM H2O2. *, Statistically significant at P < 0.05 comparing ECV304 and ECV304+eNOS cells. (B) Viability was determined at the end of the 2-h exposure (first two bars) and 18 h after exposure to 100 μM H2O2. Open bars are ECV304 cells and filled bars ECV304+eNOS. l-NAME (100 μM) was added 1 h before the addition of H2O2 and it was present for the duration of the experiment. Data represent means ± SD of three to five independent experiments.
Effect of Nitric Oxide on Energy Metabolism.
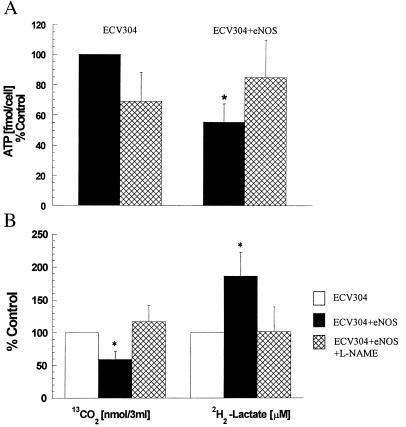
A potential mechanism for the protective role of nitric oxide may relate the nitric oxide-mediated regulation of cellular energy metabolism that is known to be perturbed by H2O2 exposure (32). Measurement of the steady state ATP levels (Fig. 2A) indicated that the eNOS transfected cells have nearly half the level of the nontransfected cells (P < 0.005, n = 4). The ATP levels in eNOS-transfected cells were the same as nontransfected cells after 1-h inhibition of nitric oxide synthesis with L-NAME, and addition of L-NAME to nontransfected cells did not alter ATP levels significantly. The differences in ATP production did not appear to be due to the differences in total glucose uptake by the cells. The rate of glucose uptake and flux through the two major glucose-using pathways, glycolysis, which accounts for nearly 80–90% of glucose breakdown, and the pentose phosphate pathway, was determined. Data in Fig. 2B indicate that the total glucose uptake in the transfected cells was the same as in the nontransfected cells, indicating that glucose uptake is not regulated by nitric oxide or transfection. However, the increase in the glycolytic utilization of glucose and the diminished flux via the pentose phosphate pathway in the eNOS transfected cells (both statistically significant, Student's t test, P < 0.001) was observed (Fig. 2B). The difference in the flux of glucose was not due to NO-mediated regulation of G6PDH or GAPDH activities (data not shown). Inhibition of eNOS with L-NAME abolished the changes in glucose utilization by the eNOS transfected cells. These data suggest that eNOS-derived nitric oxide regulates cellular metabolism, as indicated by the increased conversion of glucose to lactate at the expense of formation of reducing equivalents in the form of NADPH. However, the expected reduction in NADPH production is not sufficient to induce a decline in the total reduced thiol levels. Total (protein plus nonprotein) reduced thiol levels 1.1 ± 0.5 and 0.9 ± 0.2 nmol/mg protein (n = 3) in ECV304 and ECV304+eNOS, respectively, and it was not altered after exposure to H2O2, indicating that the glutathione peroxidase pathway and the total reductive capacity of the two cells was similar.
Figure 2.
(A) Effects of nitric oxide on ATP levels. The steady state levels of ATP were determined by the bioluminescent somatic cell assay after a 1 h incubation in the absence of l-NAME (solid bars) or presence of 100 μM l-NAME (hatched bars). Data represent means ± SD of four independent experiments. *, Statistically significant at P < 0.05. (B) Regulation of glucose utilization by nitric oxide. Glucose metabolism in ECV304+eNOS cells was measured as described in Material and Methods section after a 1 h incubation in the absence of l-NAME (solid bars) or presence of 100 μM l-NAME (hatched bars) and is expressed as % of ECV304 cells (open bars). Data represent means ± SD of four independent experiments (different days). Each independent experiment represents the average value from measurements performed in five different cell preparations. *, Statistically significant at P < 0.05.
Effects of Nitric Oxide on Mitochondrial Respiration and Cell Viability.
Another mechanism for the protective effect of nitric oxide may be the regulation of mitochondrial activities (7–14). To examine the critical role of mitochondria, three experimental approaches were used: measurement of cellular respiration, production of cells with the same mitochondrial mass but with reduced respiratory activity mitochondria (ρ− cells), and examination of whether the effect of NO can be substituted by inhibitors of mitochondrial function. The effect of nitric oxide on cellular respiration was determined by examining oxygen consumption. The cells were gently permeabilized with digitonin and the rate of oxygen consumption by the different components of the respiratory chain was determined by using appropriate substrates and specific electron transfer inhibitors. Consistent with the lower levels of ATP, oxygen consumption by complexes I and II and cytochrome oxidase were lower in eNOS-transfected cells as compared with control (Table 1).
To generate cells with the same mitochondrial mass, but without respiring mitochondria (ρ− cells), cells were grown in the presence of ethidium bromide for at least 8 weeks as described by King and Attardi (28, 29). Data in Table 1 indicate that respiration by complexes I and II and cytochrome oxidase is significantly declined in both the ECV304 and the ECV304+eNOS ρ− cells. To examine whether the ρ− contain the same density of mitochondria, the ECV304+eNOS and ECV+eNOS ρ− cells were labeled with a mitochondria-specific dye (MitoTracker Green) that becomes fluorescent on sequestration and association with lipids within the mitochondria without requiring oxidation or reduction. Staining of ECV304+eNOS and ECV304+eNOS ρ− cells with this dye revealed that both cells have equal distribution of fluorescence, implying similar content of mitochondria in these cells. Both cell types were also stained with a red fluorescence X-rosamine derivative (MitoTracker Red CM-H2XRos) that also sequesters in the mitochondria, but emits fluorescence only after it is oxidized, implying that actively respiring mitochondria are required to obtain fluorescence (30, 31). The images in Fig. 3B reveal that the ECV+eNOS ρ− cells have diminished red fluorescence, consistent with the lower oxygen consumption by the respiratory chain (Table 1). Moreover, overlay of the green and red fluorescence in the ECV+eNOS cells shows mostly orange and yellow, in contrast to the ECV+eNOS ρ− cells, which show predominantly green fluorescence. Collectively these data indicate that the ρ− cells have the same mitochondrial content, but diminished mitochondrial respiratory function. Exposure of ECV304+eNOS ρ− cells to H2O2 induced a significant amount of injury comparable with the nontransfected cells (Table 2), suggesting that the protective effect of nitric oxide depends on the presence of respiring mitochondria. Based on established protocols, ρ− cells can be reverted back to cells with normal mitochondrial function by removal of ethidium bromide (29). The ethidium bromide was removed and ECV304 and ECV+eNOS ρ− cells were allowed to replicate in normal culture medium for 2 weeks, at which time oxygen consumption by complexes I and II and cytochrome oxidase were restored to 85–90% of control (non-ethidium bromide-treated cells). Restoration of mitochondrial function restored the NO-mediated protection against exposure to100 μM H2O2 (Table 2).
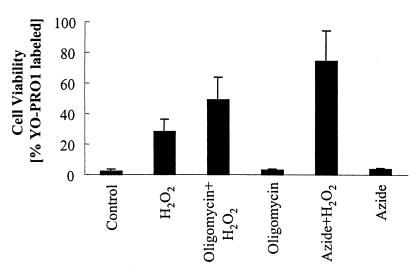
The protective effect of nitric oxide against H2O2 cannot be reproduced by azide, or by oligomycin. Treatment with 5 mM azide, which inhibits cytochrome oxidase, for 2 h does not reduce cellular levels of ATP (7.6 ± 0.8 control vs. 7.4 ± 1.0 fmol per cell, n = 4) and does not injure the cells (Fig. 4). However, addition of 100 μM H2O2 to azide-treated cells significantly augments cell death. Treatment of cells with oligomycin (2.5 μM for 2 h), which inhibits the mitochondrial ATPase, induces a significant decrease in ATP levels (7.6 ± 0.8 vs. 3.7 ± 1.0 fmol per cell, n = 4) and augments cell death on addition of 100 μM H2O2 (Fig. 4). Collectively, the data suggest that the presence of actively respiring mitochondria, not changes in cellular metabolism, is important for the protective effect of NO against H2O2-mediated delayed cell death.
Figure 4.
Effect of azide and oligomycin on cell viability. ECV304 cells were treated with 5 mM azide or 2.5 μM oligomycin for 2 h. Cell were then challenged with 100 μM H2O2 and viability was determined 18 h after exposure. Data represent means ± SD of four different experiments.
Antioxidant and Capsase-3 Enzymatic Activities.
It is possible that the protective effect of nitric oxide can be mediated by other cellular pathways. Critical among different enzymatic activities that can be regulated by nitric oxide and reported to protect cells against oxidative stress and cell death are antioxidant enzymes and caspase-3. Therefore, the activities of the antioxidant proteins catalase and Mn superoxide dismutase, and the activity of caspase-3, were determined (Table 3). However, the ECV304+eNOS cells appear to have significantly higher catalase and lower caspase-3 activities than ECV304 cells, and the increase in catalase activity may in part account for the increased resistance to H2O2. However, the activities of catalase and caspase-3 are not altered by the addition of L-NAME, and therefore may not account for the nitric oxide-mediated protection. Moreover, ECV+eNOS ρ− cells have similar catalase activity and even lower caspase-3 activity than ECV+eNOS cells, but are less resistant to H2O2 challenge, indicating that these enzymatic activities may not be critical for the nitric oxide-mediated protective effect.
Table 3.
Specific enzymatic activities
| Enzyme | ECV304 | ECV304 + l-NAME | ECV304 ρ− | ECV304+eNOS | ECV304+eNOS + l-NAME | ECV304+eNOS ρ− |
|---|---|---|---|---|---|---|
| Catalase (μmol H2O2/min/mg protein) | 9.5 ± 0.1 | 9.1 ± 0.3 | 14.2 ± 0.3* | 13.7 ± 0.2* | 13.6 ± 1.7* | 12.4 ± 0.3* |
| Mn superoxide dismutase (units/mg protein) | 7.6 ± 1.9 | 6.8 ± 1.2 | 9.5 ± 1.2 | 6.5 ± 0.7 | 5.9 ± 1.1 | 5.8 ± 0.8 |
| Caspase-3 (rfu/min/mg protein) | 23.8 ± 4.0 | 28.3 ± 2.9 | 16.9 ± 1.2* | 17.9 ± 2.3* | 17.7 ± 1.5* | 10.2 ± 0.5** |
Specific activities were determined as described in Materials and Methods and the data represent means ± SD of four different experiments.
, Statistically significant at P < 0.05 from ECV304;
, statistically significant at P < 0.05 from ECV304+eNOS.
Discussion
The present studies provides evidence that endogenous generation of low steady state levels of nitric oxide by eNOS dynamically regulates mitochondrial respiration, which provides protection against H2O2-mediated injury and death. Nitric oxide was effective in protecting cells exposed to concentrations of H2O2 (25–100 μM) that induced delayed cell injury, but not at concentrations (150 μM) that induce acute injury (Fig. 1). Although the nature of the delayed cell death was not investigated, the protective effect of nitric oxide appears to be critically dependent on mitochondrial respiratory activity. Data in Table 2 revealed that exposure to H2O2 effectively injures ECV304+eNOS cells with significantly reduced mitochondrial respiration. Restoration of mitochondrial respiration, however, also restored the ability of nitric oxide to protect the ECV304+eNOS cells from the H2O2-mediated delayed cell death. The mechanisms by which nitric oxide-mediated regulation of respiration protects cells from peroxide exposure are not completely understood, but certain possible pathways may be involved. Previous studies in cell models revealed that nitric oxide, supplied from nitric oxide donors or transfection with iNOS, regulates the mitochondrial membrane polarity (11, 14), mitochondrial permeability transition opening (11, 14), and cytochrome c release (11, 13). All of these functions are critical for cellular viability and all three, to a certain degree, depend on the presence of functionally respiring mitochondria. Recent data by Beltrán et al. (14) have indicated that the nitric oxide-mediated regulation of respiration induces a protective mechanism that maintains membrane potential and prevents apoptosis. It is possible that the same unidentified protective mechanism related to nitric oxide-mediated regulation of respiration and possibly membrane polarity is responsible for the protection against H2O2 stress.
In addition to respiration, the data presented in Fig. 2 indicated that nitric oxide also regulates glucose metabolism and cellular ATP levels. The total glucose uptake in the transfected cells is not regulated by nitric oxide or transfection, consistent with the observation that total glucose uptake was not altered in both the eNOS and nNOS knockout mice (33). However in the presence of nitric oxide, glucose is predominantly used by glycolysis as indicated by the increased conversion of glucose to lactate (Fig. 2) at the expense of the NADPH production by the G6PDH pathway. A nitric oxide-mediated decline in glucose utilization following IL-1 induction of iNOS in pancreatic islets has been reported (34), and a decrease in gluconeogenesis, which resulted in an increased flow through phosphofructokinase and pyruvate kinase, has been reported after lipopolysaccharide (LPS)-mediated induction of hepatocyte NOS (35). The decrease in glucose utilization by the G6PDH pathway does appear to compromise the levels of reduced thiols in the eNOS-transfected cells and is not due to an inhibition of G6PDH activity. Collectively these data suggest that nitric oxide dynamically regulates glucose utilization. However, in our paradigm this regulation does not appear to be causally related to the nitric oxide-mediated protection against H2O2 stress, because the protective effect of nitric oxide is not replicated by inhibition of complex IV with azide or by decreasing cellular ATP levels by inhibiting the mitochondrial ATPase activity with oligomycin (Fig. 4). Oxidative stress in the form of H2O2 has been shown to induce changes in the glycolytic pathway and disturbances to mitochondrial function, suggesting that mitochondrial energy production and metabolism are dynamically regulated under stress to meet energy demands (32). Although nitric oxide appears to regulate both mitochondrial bioenergetics and glucose metabolism, the first appears to be important for nitric oxide-dependent cell survival.
Recent evidence supports the presence of a mitochondrial form of NOS (36, 37). The nitric oxide produced by this NOS isoform has been suggested to be used for the regulation of oxygen consumption, thus modulating oxygen delivery to tissues, and also to modulate the mitochondrial production of reactive oxygen species (36, 37). Based on previously published data (14, 36, 37) and observations presented here, it is also reasonable to propose that endogenous nitric oxide released by eNOS or mitochondrial NOS by regulating respiration protects cells from oxidative stress and other stresses that induce cell death and apoptosis.
The role of nitric oxide in preconditioning may provide another physiological setting where the dynamic regulation of mitochondrial respiration by nitric oxide may be important. It is known that brief periods of ischemia or glucose withdrawal protect cells and organs from ischemia-reperfusion injuries and death (38). This preconditioning effect has been attributed to nitric oxide (38), although the precise mechanisms of this nitric oxide-mediated protection have not been elucidated. It is possible that the regulation of mitochondrial respiration and mitochondrial transmembrane potential by nitric oxide during the periods of preconditioning is able to protect cells against the subsequent, and otherwise lethal insults.
Overall, these observations suggest that nitric oxide-mediated regulation of mitochondrial respiration may represent an unrecognized primary line of defense against oxidative and other stresses.
Acknowledgments
This work was supported by National Institutes of Health Grants HD26979 and NS34900 (to M.Y.), HL61371 and HL64793 (to W.C.S.), and HL54926 and HL05926 (to H.I.). W.C.S. and H.I. are Established Investigators of the American Heart Association.
Abbreviations
- eNOS
endothelial nitric oxide synthase
- l-NAME
NG-nitro-l-arginine methyl ester
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kim Y-M, Bergonia H, Lancaster J R., Jr FEBS Lett. 1995;374:228–232. doi: 10.1016/0014-5793(95)01115-u. [DOI] [PubMed] [Google Scholar]
- 2.Wink D A, Hanbauer I, Krishna M, DeGraff W, Gamson J, Mitchell J B. Proc Natl Acad Sci USA. 1993;90:9813–9817. doi: 10.1073/pnas.90.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi M S, Ponthier J L, Lancaster J R., Jr Free Rad Biol Med. 1999;27:1357–1366. doi: 10.1016/s0891-5849(99)00179-3. [DOI] [PubMed] [Google Scholar]
- 4.Chang J, Rao N V, Markewitz B A, Hoidal J R, Michael J R. Am J Physiol. 1996;270:L931–L940. doi: 10.1152/ajplung.1996.270.6.L931. [DOI] [PubMed] [Google Scholar]
- 5.Herman C, Zeiher M, Dimmeler S. Arterioscler Thromb Vas Biol. 1997;17:3588–3592. doi: 10.1161/01.atv.17.12.3588. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Zulueta M, Ensz L M, Mukhina G, Lebovitz R M, Zwacka R M, Engelhardt J F, Oberley L W, Dawson V L, Dawson T M. J Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown G C. Biochim Biophys Acta. 2001;1504:46–57. doi: 10.1016/s0005-2728(00)00238-3. [DOI] [PubMed] [Google Scholar]
- 8.Shen W, Hintze T H, Wolin M S. Circulation. 1995;92:3505–3512. doi: 10.1161/01.cir.92.12.3505. [DOI] [PubMed] [Google Scholar]
- 9.Poderoso J J, Lisdero C, Schöpfer F, Riobó N, Carreras M C, Cadenas E, Boveris A. J Biol Chem. 1999;274:37709–37716. doi: 10.1074/jbc.274.53.37709. [DOI] [PubMed] [Google Scholar]
- 10.Clementi E, Brown G C, Foxwell N, Moncada S. Proc Natl Acad Sci USA. 1999;96:1559–1562. doi: 10.1073/pnas.96.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brookes P S, Salina E P, Darley-Usmar K, Eiserich J P, Freeman B A, Darley-Usmar V M, Anderson P G. J Biol Chem. 2000;275:20474–20479. doi: 10.1074/jbc.M001077200. [DOI] [PubMed] [Google Scholar]
- 12.Balakirev M Y, Khrantsov V V, Zimmer G. Eur J Biochem. 1997;256:710–718. doi: 10.1111/j.1432-1033.1997.00710.x. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Bombeck C A, Yang S, Kim Y-M, Billiar T M. J Biol Chem. 1999;274:17325–17333. doi: 10.1074/jbc.274.24.17325. [DOI] [PubMed] [Google Scholar]
- 14.Beltrán B, Mathur A, Duchen M R, Erusalimsky J D, Moncada S. Proc Natl Acad Sci USA. 2000;97:14602–14607. doi: 10.1073/pnas.97.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrie A M, Rizzuto R, Pozzan T, Simpson A W M. J Biol Chem. 1996;271:10753–10759. doi: 10.1074/jbc.271.18.10753. [DOI] [PubMed] [Google Scholar]
- 16.Abedi H, Zachary I. J Biol Chem. 1997;272:15442–15451. doi: 10.1074/jbc.272.24.15442. [DOI] [PubMed] [Google Scholar]
- 17.Bowie A G, Moynagh P N, O'Neill L A J. J Biol Chem. 1997;272:25941–25950. doi: 10.1074/jbc.272.41.25941. [DOI] [PubMed] [Google Scholar]
- 18.Escargueil-Blanc I, Andrieu-Abadie N, Caspar-Bauguil S, Brossmer R, Levade T, Negre-Salvayre A, Salvayre R. J Biol Chem. 1998;273:27389–27395. doi: 10.1074/jbc.273.42.27389. [DOI] [PubMed] [Google Scholar]
- 19.Winter M C, Kamath A M, Ries D R, Shasby S S, Chen Y T, Shasby D M. Am J Physiol. 1999;277:L988–L995. doi: 10.1152/ajplung.1999.277.5.L988. [DOI] [PubMed] [Google Scholar]
- 20.Sowa G, Liu J, Papapetropoulos A, Rex-Haffner M, Hughes T E, Sessa W C. J Biol Chem. 1999;274:22524–22531. doi: 10.1074/jbc.274.32.22524. [DOI] [PubMed] [Google Scholar]
- 21.Kiessling F, Kartenbeck J, Haller C. Cell Tissue Res. 1999;297:131–140. doi: 10.1007/s004410051340. [DOI] [PubMed] [Google Scholar]
- 22.Brown J, Reading S J, Jones S, Fitchett C J, Howl J, Martin A, Longland C L, Michelangeli F, Dubrova Y E, Brown C A. Lab Invest. 2000;80:37–45. doi: 10.1038/labinvest.3780006. [DOI] [PubMed] [Google Scholar]
- 23.Gow A, Chen Q, Gole M D, Themistocleous M, Lee M-Y V, Ischiropoulos H. Am J Physiol. 2000;278:C1099–C1107. doi: 10.1152/ajpcell.2000.278.6.C1099. [DOI] [PubMed] [Google Scholar]
- 24.Tian W-N, Braunstein L D, Apse K, Pang J, Rose M, Tian X, Stanton R C. Am J Physiol. 1999;276:C1131–C1131. doi: 10.1152/ajpcell.1999.276.5.C1121. [DOI] [PubMed] [Google Scholar]
- 25.Molina Y, Vedia L, McDonald B, Reep B, Brune B, Di Silvio M, Billiar T R, Lapetina E G. J Biol Chem. 1992;267:24929–24932. [PubMed] [Google Scholar]
- 26.Souza J M, Radi R. Arch Biochem Biophys. 1998;360:187–194. doi: 10.1006/abbi.1998.0932. [DOI] [PubMed] [Google Scholar]
- 27.Claiborne A. In: Handbook of Methods for Oxygen Radical Research. Greenwald R A, editor. Boca Baton, FL: CRC; 1985. pp. 181–188. [Google Scholar]
- 28.King M P, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 29.King M P, Attardi G. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 30.Ellerby H M, Arap W, Ellerby L M, Kain R, Andrusiak R, Del Rio G, Krajewski S, Lombardo C R, Rao R, Ruoslahti E, et al. Nat Med. 1999;5:1032–1038. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 31.Mathur A, Hong Y, Kemp B K, Barrientos A A, Erusalimsky J D. Cardiovasc Res. 2000;46:126–138. doi: 10.1016/s0008-6363(00)00002-x. [DOI] [PubMed] [Google Scholar]
- 32.Hyslop P A, Hinshaw D B, Halsey W A, Jr, Schraufstatter I U, Sauerheber R D, Spragg R G, Jackson J H, Cochrane C G. J Biol Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- 33.Browne S E, Ayata C, Huang P L, Moskowitz M A, Beal M F. J Cereb Blood Flow Metab. 1999;19:144–148. doi: 10.1097/00004647-199902000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Ma Z, Landt M, Bohrer A, Ramanadham S, Kipnis D M, Turk J. J Biol Chem. 1997;272:17827–17835. doi: 10.1074/jbc.272.28.17827. [DOI] [PubMed] [Google Scholar]
- 35.Horton R A, Knowles R G, Titheradge M A. Biochem Biophys Res Commun. 1994;204:659–665. doi: 10.1006/bbrc.1994.2510. [DOI] [PubMed] [Google Scholar]
- 36.Giulivi C, Poderoso J J, Boveris A. J Biol Chem. 1998;273:11038–11043. doi: 10.1074/jbc.273.18.11038. [DOI] [PubMed] [Google Scholar]
- 37.Ghafourifar P, Schenk U, Klein S D, Richter C. J Biol Chem. 1999;274:31185–31188. doi: 10.1074/jbc.274.44.31185. [DOI] [PubMed] [Google Scholar]
- 38.Guo Y, Jones W K, Xuan Y-T, Bao W, Wu W-J, Han H, Laubach V E, Ping P, Yang Z, Qiu Y, Bolli R. Proc Natl Acad Sci USA. 1999;96:11507–11512. doi: 10.1073/pnas.96.20.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]