Abstract
We owe the flexibility of our bodies to sophisticated articulations between bones. Establishment of these joints requires the integration of multiple tissue types: permanent cartilage that cushions the articulating bones, synovial membranes that enclose a lubricating fluid-filled cavity, and a fibrous capsule and ligaments that provide structural support. Positioning the prospective joint region involves establishment of an “interzone” region of joint progenitor cells within a nascent cartilage condensation, which is achieved through the interplay of activators and inhibitors of multiple developmental signaling pathways. Within the interzone, tight regulation of BMP and TGFβ signaling prevents the hypertrophic maturation of joint chondrocytes, in part through downstream transcriptional repressors and epigenetic modulators. Synovial cells then acquire further specializations through expression of genes that promote lubrication, as well as the formation of complex structures such as cavities and entheses. Whereas genetic investigations in mice and humans have uncovered a number of regulators of joint development and homeostasis, recent work in zebrafish offers a complementary reductionist approach toward understanding joint positioning and the regulation of chondrocyte fate at joints. The complexity of building and maintaining joints may help explain why there are still few treatments for osteoarthritis, one of the most common diseases in the human population. A major challenge will be to understand how developmental abnormalities in joint structure, as well as postnatal roles for developmental genes in joint homeostasis, contribute to birth defects and degenerative diseases of joints.
INTRODUCTION
Joints connect neighboring bones within the vertebrate skeleton. Fibrous joints called sutures separate the flat bones of the skull and allow for flexibility during childbirth (Figure 1(a)). Cartilaginous joints, for example between the vertebrae, provide limited mobility and act as shock absorbers (Figure 1(b)). The most sophisticated types of joints are the lubricated synovial articulations, such as those of the highly mobile limbs and jaw (Figure 1(c)). Despite their differing structures and extents of mobility, all three types of joints share key features. For example, developing joints are enriched for skeletal progenitor cells, which in the case of sutures provide a continuing supply of osteoblasts for bone growth.1–5 Conversely, each joint is characterized by its own specialized morphology and function, which is reflected by unique patterns of gene expression by their resident skeletal and supporting cells. A critical early event in the creation of all joints is the suppression of osteoblast or chondrocyte differentiation at particular sites within the developing skeleton. For many synovial joints, suppression of chondrocyte maturation occurs within a broad region called the interzone.6,7 Lineage analysis demonstrates that interzone cells contribute not only to the articular cartilage lining the joint cavity but also to the synovium, menisci, and ligaments.8,9 While much remains to be learned about how joints are positioned, the precise spatial deployment of activators and inhibitors of diverse signaling pathways is a recurrent theme. One consequence of such signaling is the induction of several types of transcription factors and chromatin modifiers, which function to inhibit cartilage maturation within the interzone. What is less clear is how later events in joint development are regulated, such as production of the lubricating protein PRG4/Lubricin by articular chondrocytes,10,11 creation of the joint cavity,12 and the local differentiation of interzone cells into menisci and ligament attachment points (entheses).
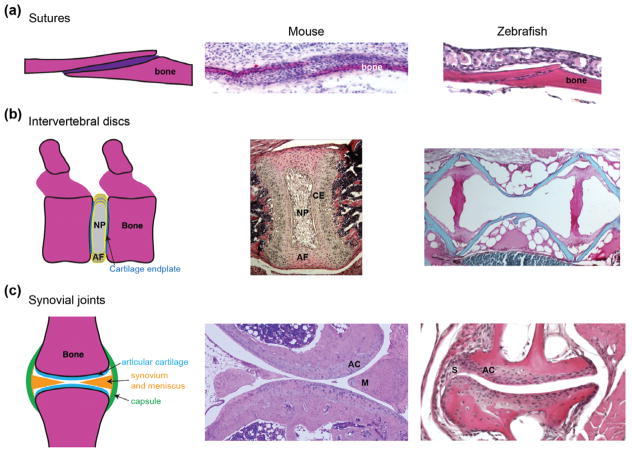
FIGURE 1.
Histological features of joints. (a) Sutures are a type of immoveable articulation between bones, with the suture mesenchyme housing progenitors for bone growth and repair. Hematoxylin and eosin staining shows comparable coronal sutures of an E16.5 mouse embryo and young adult zebrafish. (b) The intervertebral discs of mouse and zebrafish have a very different structure. In mammals (shown here for postnatal Day 15 mouse), a cartilage endplate (CE) covers each vertebra, with the disc consisting of a ring of annulus fibrosus (AF) tissue and a core of nucleus pulposus (NP) tissue. In adult zebrafish, vertebral bones (red) are separated by layers of fat that appear white upon sectioning. (c) Synovial joints are freely moveable articulations characterized by fluid-filled cavities lined by articular hyaline cartilage. Some synovial joints, as shown in a section of the knee joint from an adult mouse, include additional specializations such as menisci (M). The adult zebrafish jaw joint has a clear articular cartilage (AC) layer and synovium (S) but no mensicus. Photos courtesy of Camilla Teng (A), Jennifer Zieba (B, mouse), and Denis Evseenko (C, mouse).
While major congenital anomalies of joints are uncommon, the progressive loss of joint structure and function in osteoarthritis is the leading cause of disability in the United States.13 Genetic factors contribute substantially to osteoarthritis (e.g., estimated at 60% for the hip14), with recent studies beginning to uncover specific loci linked to arthritis predisposition.15 An emerging theme is that minor defects in developmental morphogenesis may lead to architectural defects in postnatal joints,16 which result in the increased wear-and-tear that predisposes to osteoarthritis. A key feature of osteoarthritis is the ectopic hypertrophic differentiation of articular chondrocytes at the joint surface, which erodes the cartilage cushions protecting the bones and eventually the bones themselves.17 This pathological observation suggests that a challenge for prolonged joint health is preventing joint-lining chondrocytes from undergoing hypertrophic maturation. Interestingly, many of the same signaling pathways and transcription factors that help to establish articular chondrocyte fate continue to be expressed at joints into adulthood, with postnatal genetic manipulations in some cases establishing continued requirements in maintaining articular fate.9,18 There are also potential roles for epigenetic modifications in establishing long-term repression of the hypertrophic program in articular chondrocytes,19 as well as other factors selectively required for the maintenance but not development of joints.
USE OF ZEBRAFISH FOR DEVELOPMENTAL STUDIES OF JOINTS
Although most studies on joints have been conducted in mammals, the zebrafish has recently emerged as an alternative model to investigate basic joint biology. Zebrafish have many of the same types of joints found in mammals (Figures 1 and 2). Sutures connect the skull bones in both fish and mammals, with undifferentiated mesenchyme residing between the interleaved bony plates20 (Figures 1(a) and 2(c) and (d)). Imaging studies in living zebrafish have revealed the stepwise formation of wild-type sutures, as well as abnormal suture development upon genetic or pharmacological perturbations (e.g., loss of retinoic acid signaling).21–23 Zebrafish also have intervertebral discs that are prone to degeneration upon aging.24 However, the discs of adult zebrafish differ in structure from those in mammals in lacking cartilage, with histology showing adipocytes between the vertebral bones rather than nucleus pulposus and annulus fibrosus structures as in mammals25 (Figures 1(b) and 2(c) and (d)). In the zebrafish fry, two cartilaginous joints in the head have been studied extensively: the jaw joint between the Meckel’s and palatoquadrate cartilages and the bipartite hyoid joint between the ceratohyal, interhyal, and hyosymplectic cartilages (Figure 2(a)). Studies of these joints have revealed conserved roles for GDF family ligands26 and new roles for Nkx3.227 and Irx family28 transcription factors. Contrary to previous dogma, a recent study also suggests that fishes have synovial-like lubricating joints, particularly in their jaw and fins.25 Building on older histological observations,29 this study showed that the jaw and fin joints of diverse fish species (zebrafish, stickleback, and spotted gar) have many of the features of mammalian synovial joints including articular cartilage, a joint cavity and synovium (Figures 1(c) and 2 (c) and (d)). Best illustrated in the much larger gar, the fish jaw joint also displays the same multilayered articular cartilage as in mammalian synovial joints (Figures 1(c) and 5(a)): a superficial zone of flattened chondrocytes, a deeper transitional zone of rounder chondrocytes, a radial zone of stacked chondrocytes, and finally calcified chondrocytes that integrate into the underlying bone (Figure 5(b)). Although it was not demonstrated that the synovial capsule is fully enclosed, the jaw joints of these fishes exhibited a clear synovial fibroblast layer at the periphery and an external fibrous capsule. At a molecular level, the jaw joints of all three fishes, as well as the pectoral fin joints of zebrafish (Figure 2(c)), express homologs of the lubricant proteoglycan PRG4/Lubricin,25 a prominent marker of superficial chondrocytes and synovium at mammalian joints.11 Furthermore, zebrafish lacking the Lubricin-encoding gene prg4b developed the same arthritic changes to their jaw and fin joints as observed in mice and human patients lacking Lubricin.11,25,30 These findings show that synovial joints likely evolved at least in the common ancestor of all bony vertebrates, and that zebrafish are a relevant model for the study of arthritis. While it remains unclear the extent to which the jaw and related joints of fishes have all the properties of mammalian synovial joints, future work in zebrafish has the potential to provide new insights into synovial joint development and disease. Where appropriate, we highlight recent contributions of the zebrafish model to our understanding of joint biology and pathology.
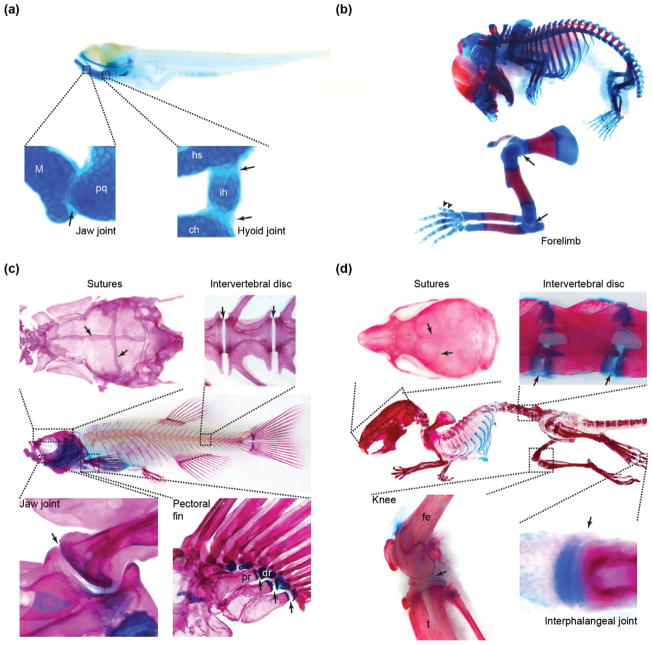
FIGURE 2.
Skeletal joints in zebrafish and mouse. (a) In zebrafish fry at 5 days postfertilization, well-studied cartilaginous joints (arrows) include the jaw joint between Meckel’s and palatoquadrate cartilages and the bipartite hyoid joint between hyosymplectic, interhyal, and ceratohyal cartilages. (b) In a mouse embryo at E17.5, representative joints include the shoulder and elbow joints (arrows) and interphalangeal joints (arrowheads). As in young zebrafish, these joints are largely cartilaginous at this stage. (c) The adult zebrafish skeleton is largely composed of bone and contains many types of joints (arrows), including sutures in the skull, intervertebral joints, and synovial joints in the jaw and pectoral fin. (d) The mouse skeleton at postnatal Day 21 has similar joints to zebrafish, including sutures, intervertebral discs, and synovial joints in the knee and digits (i.e., interphalangeal). Skeletons were stained with alcian blue for cartilage and alizarin red for bone. ch, ceratohyal; dr, distal radial; fe, femur; hs, hyosymplectic; ih, interhyal; M, Meckel’s; pq, palatoquadrate; pr, proximal radial; t, tibia.
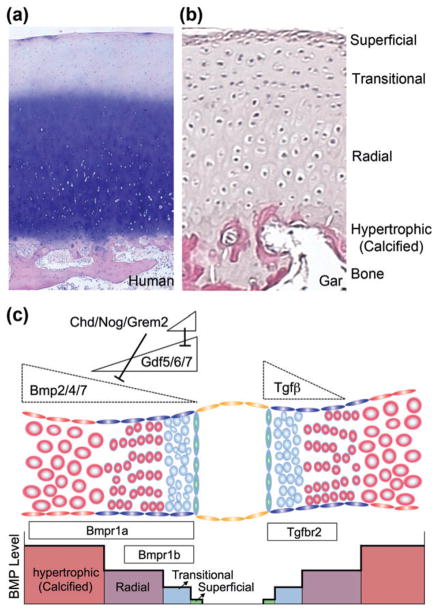
FIGURE 5.
Critical thresholds of Bmp and Tgfβ signaling in joint development. (a, b) Histological sections stained with hematoxylin and eosin show the different layers of joint cartilage in a human knee (16-year-old) and juvenile spotted gar jaw. (c) The levels, duration, and/or type of Bmp signaling help to specify the different types of chondrocytes at and around joints: hypertrophic, radial, transitional, and superficial. The Bmp ligands Gdf5/6/7 and antagonists Chordin (Chd), Noggin (Nog), and Gremlin2 (Grem2) are expressed in the developing interzone, and Bmp2/4/7 have been reported to be expressed either at a distance from the joint or at the joint itself. Variable diffusion of these ligands and antagonists may establish different levels of Bmp signaling, and signaling through Bmpr1a and Bmpr1b receptors could also influence joint fate. TGFβ signaling also has an important role to specify joint fates, with the Tgfbr2 receptor enriched at developing joints.
POSITIONING THE JOINT DOMAIN
The future joint domain could be determined in at least two major ways (Figure 3). First, an initial chondrogenic condensation can be split into two or more distinct zones by creation of a morphologically distinct interzone region, which is characterized by flattened and highly compacted cells relative to the adjacent growth plates. An example of this mechanism is the creation of the interphalangeal joints in the digits. Second, the prospective joint domain can be created by the appositional growth of two neighboring condensations, such as what occurs between the anlagen of the femur and pelvic bone31 and for the temporomandibular joint of the mammalian jaw.32 Interestingly, compromised condensation formation in barx1 zebrafish mutants can lead to ectopic joints, potentially through aberrant condensation splitting.33 It remains unclear, however, what effects the mode of joint specification have on later development. In principle, the preservation of progenitor zones at the leading edges of appositionally growing condensations may result in their later fusion into a structure closely resembling an interzone.
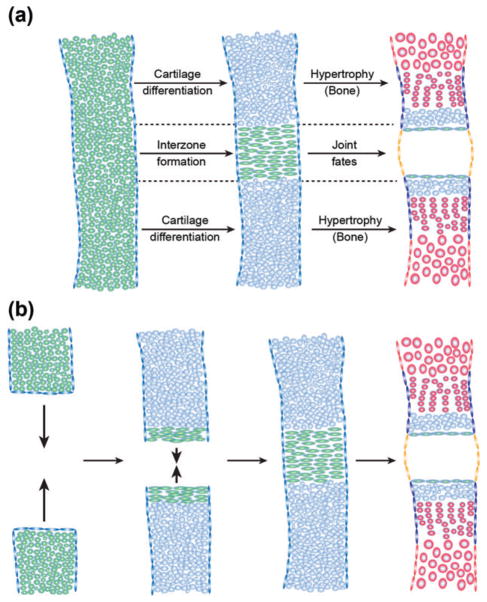
FIGURE 3.
Distinct modes of interzone formation. The joint interzone develops in the position of the presumptive joint and precedes articular cartilage differentiation and joint cavitation. The interzone can be generated from a single mesenchymal condensation (a, green) or through appositional growth of adjacent condensations (b). While cells outside the interzone undergo further cartilage differentiation (blue) and eventually hypertrophy and mineralize during endochondral bone development (red), cells within the interzone are maintained as immature chondrocytes at the articular surface (green flattened cells) and contribute to joint specializations such as the synovial membrane (orange). Also shown are chondrocyte progenitors within the perichondrium (blue flattened cells) and osteoblast progenitors within the periosteum (red flattened cells).
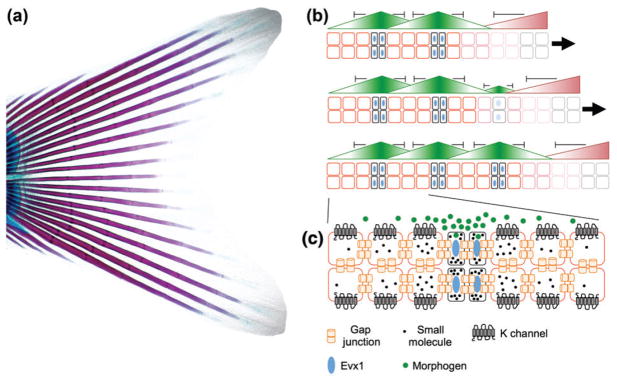
Reaction–Diffusion Mechanisms of Joint Spacing in Fin and Limb Skeletons
In many cases, joints develop in a periodic pattern, e.g., the interphalangeal joints of the digits. Studies of the ray joints of the zebrafish fins have begun to uncover potential mechanisms of iterative joint specification. The proximal portion of the fish fin is thought to be homologous to tetrapod limbs,34 with the more derived distal portion containing a series of bony rays segmented at regular intervals by fibrous joints (Figure 4). While these ray joints differ from the synovial-like joints at the base of the fin, they have been useful for understanding how repeated joints can be established. Loss-of-function mutations in the gap junction protein-encoding gene connexin43 (short-fin) result in joints spaced closer together, independent of effects on fin size, with Connexin43 overexpression eliminating joints.35,36 Gain-of-function mutations in the potassium channel-encoding gene kcnk5b (another-long-fin) also result in fins with irregularly spaced joints.37 In addition, the even-skipped transcription factor evx1 is essential for ray joint development in zebrafish.38–40 While it is unclear the extent to which iterative patterning of ray joints would be relevant to mammalian joint patterning, mesenchymal cells within the interzone of developing mammalian joints do express Connexins 32, 40, and 43,12,41 and Connexin40−/− mice display losses of specific joints in the distal limbs.42 Mutations of CONNEXIN43 in Oculodentodigital Dysplasia syndrome also result in skeletal defects, including missing and fused phalanges.43 The conserved requirement of Connexins in fish and mammals suggest that gap junctions could be required for the efficient spread of some unknown small molecule between cells in both species, although most growth factors would be assumed to be too large to pass through these channels. Nonetheless, modeling studies in zebrafish reveal how the expression and diffusion of just two factors is sufficient to explain the repeated spacing of joints within the fin.44 At the tip of the elongating fin, a growth factor would promote mesenchymal proliferation and inhibit joints, with joint formation being induced when the concentration of the growth factor falls below a certain threshold. At this position, a joint-promoting factor would be induced, which would secondarily inhibit other joints from forming nearby. Only in positions where the two factors were below a critical threshold would a new joint form (Figure 4(b)). It is tempting to speculate that a similar mechanism might occur for joints in the hands and feet of tetrapods, such as the periodically spaced Phalangeal joints of the digits. Good candidates for the limb/fin growth factor are FGFs, which are produced by the apical ectodermal ridge and required for limb/fin outgrowth.45 WNT9a/14 is a candidate joint factor that would inhibit additional joints at a distance, as its misexpression in mouse induces an ectopic joint while inhibiting development of the adjacent normal joint.46 Variations on the theme of diffusible activators and inhibitors generating repeated patterns are common in biological systems. Such reaction–diffusion mechanisms proposed by Turing47 have been used to explain the patterning of teeth,48 ectodermal appendages,49 and skin pigmentation.50 Future work is needed to explore the relative contribution of prepatterning versus iterative activator–inhibitor interactions for the positioning of different joints.
FIGURE 4.
Iterative segmentation of the bony rays of the zebrafish fin. (a) The tail fin from a 1-month-old zebrafish was stained with alizarin red and alcian blue to highlight bone and cartilage, respectively. In the bony rays, a series of joints form in a segmental pattern from the base of the fin to the tip (left to right in this image). (b) Modeling predicts that just two morphogens can generate the segmental pattern of joints in the fin rays. Bone cells are shown in red, joint cells in black with blue nuclei, and progenitors in gray. At forming joints, one morphogen (green) specifies joint fates while inhibiting neighboring cells from becoming joints. At the distal end, a growth factor (red) drives progenitor growth (right arrow) that lengthens the fin while inhibiting joint formation. Over time (top to bottom), new joints form where both morphogens fall below a critical threshold. (c) The gap junction protein Connexin43, the potassium channel Kcnk5b, and the transcription factor Evx1 have all been shown to be required for correct joint spacing in the fin rays. One possibility is that secreted morphogens, as well as the transport of small molecules and electrical signaling between neighboring cells, combine to regulate joint spacing.
Developmental Signaling to Establish the Interzone of the Future Jaw Joint
In contrast to the repeated joints in appendages, there are other contexts where the joint interzone appears to be prespecified by the convergence of multiple developmental signaling pathways. An illustrative example is the zebrafish jaw joint, which is positioned by integration of Edn1 and Bmp signaling within the neural crest-derived precursors of the jaw skeleton.51 In zebrafish, loss of the Edn1 ligand or its Ednra receptors results in defects in the lower jaw and jaw joint.52,53 The joint appears to be particularly dependent on optimal Edn1 signaling as partial reductions of the pathway in furina, plcb3, or mef2ca mutants have greater effects on joint versus lower jaw development.54–56 An important target of Edn1 signaling in joint formation is the transcription factor nkx3.2/bapx1, as antisense reduction in zebrafish results in specific loss of the jaw joint.27 In chick, ectopic expression of BMP4 and FGF8 abrogates Nkx3.2 expression and results in loss of joint structures such as the retroarticular process.57,58 In zebrafish, Bmp4 misexpression or reduction of the Bmp antagonist Gremlin2 disrupts jaw joint formation, with Edn1 and Jagged-Notch signaling functioning together to confine grem2b expression to nascent facial joints.28,59 Bmp signaling restricts the jaw joint-forming domain in part through induction of Hand2, which prevents expansion of nkx3.2 expression into the lower jaw.27 In humans, mutations in EDN1 or its signaling components such as PLCB4 and GNAI3 similarly result in abnormalities of the jaw joint in Auriculo-Condylar Syndrome.60–64 Absence of the jaw articulation is also seen in mice deleted for the Edn1 target gene Dlx5 and Dlx2.65 Somewhat differently, loss of Foxc1 results in pronounced bony fusions of the upper and lower jaw, a condition known as syngnathia.66 In addition to EDN1, BMP, and FGF signaling, WNT and HH signaling also play important roles in establishing joint domains.67–69 In mice, Wnt9a/Wnt14 is both necessary and sufficient to specify joint-forming domains in the limb,46,70 and Ihh is required for limb and jaw joint development.67,71 Conversely, HH activation has been found to inhibit joint development through repression of WNT signaling and Fgf18 expression.69 Going forward, it will be important to discern the extent to which these pathways establish early gene expression domains that prefigure joints versus more directly controlling chondrocyte and other cell fate at the joint (e.g., as discussed below for BMP signaling).
CONTROLLING CHONDROCYTE IDENTITY AT THE JOINT
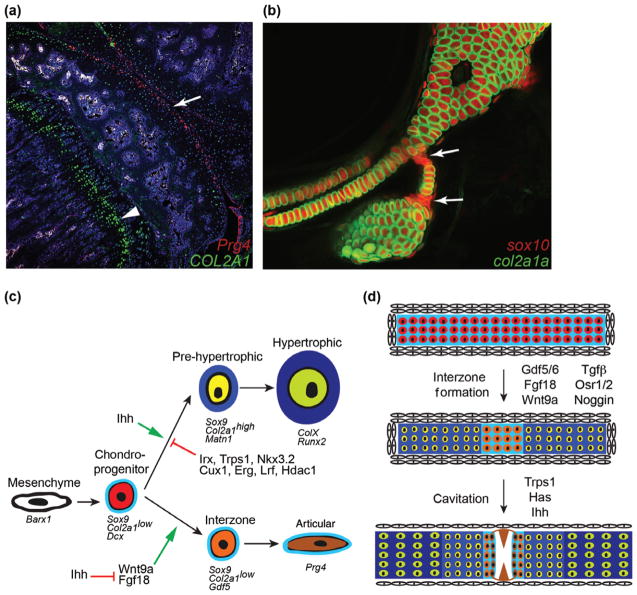
As the precartilage condensation forms, cells destined for both endochondral bone and the joint express a number of genes typical of mesenchymal progenitors, including Sox9 and low levels of Col2a1.72 In the anlagen of the endochondral bones, chondrocytes then upregulate cartilage matrix genes (e.g., Col2a1, Matrilin1, and Aggrecan) and stratify into proliferative, prehypertrophic, and hypertrophic zones.73 Hypertrophic chondrocytes express Col10a1 and low levels of genes in common with osteoblasts (e.g., Runx2 and Osterix), mineralize, and undergo apoptosis or transdifferentiation into long-lived osteocytes.74–77 In contrast, cells within the nascent interzone produce signaling molecules that help organize the joint and prevent further chondrocyte maturation. An ongoing debate is the extent to which joint chondrocytes arise from a separate lineage from those of endochondral bone. Lineage tracing studies based on Col2a1 and Dcx transgenes suggest that endochondral and joint chondrocytes arise from a common early field of cells in mice,78,79 and the observation that joint chondrocytes enter the endochondral program in certain zebrafish and mouse mutants would suggest that they share a common, albeit latent, potential with other chondrocytes in the condensation.4,28 On the other hand, there are reports of migratory cells from outside the interzone contributing to articular chondrocytes and the meniscus in mouse, which would imply an additional separate origin for at least some joint tissues.4,28,78,80 Notwithstanding, it is clear that a number of signaling pathways and transcriptional repressors converge to potently inhibit chondrocyte maturation at the developing joint surface.
Thresholds of BMP and TGFβ Signaling for Chondrocyte Fate
Complex regulation of BMP signaling at and around the joint suggests a central role of this pathway in joint progenitor specification.81 A variety of BMP ligands and antagonists are expressed in or around the developing interzone in mouse and zebrafish, including Bmp2, Bmp4, and Bmp7,82 Chordin,12,27 Noggin,83,84 and Gremlin2,59 and genetic evidence suggests that inhibiting Bmp signaling is critical for joint development. Transient misexpression of Bmp4 in zebrafish, at a stage after its jaw patterning role, blocks formation of facial joints,28 and loss of Noggin in the Col2a1-expressing lineage of mice disrupts interphalangeal joints.4 On the other hand, loss of Bmp2 and Bmp4 in limb mesenchyme blocks formation of the elbow joint.85 Of particular interest, the BMP ligands GDF5, GDF6, and GDF7 are prominently expressed within the joint interzone,26,27,86–89 with Gdf5-Cre activity extensively labeling joint tissues in mice.8 Gdf5 was first identified as the gene mutated in the brachypodism mouse,90 which displays limb and mild joint defects; mice doubly mutant for Gdf5 and Gdf6 show more extensive joint defects.87,88,90 GDF5 activity is not, however, sufficient to induce joints, as GDF5 misexpression promotes rather than inhibits cartilage maturation.91,92 One possibility is that high Bmp signaling further away from the joint promotes endochondral ossification, lower Gdf signaling near the joint promotes deeper articular chondrocyte fates, and a relative absence of Bmp/Gdf signaling at the joint surface preserves progenitors in an early chondrocyte/mesenchymal state (Figure 5). Members of the TGFβ signaling family are also critical for joint morphogenesis.93 The Tgfbr2 receptor is expressed in the joint interzone, with its loss in mice resulting in a near complete absence of the interphalangeal joints due to ectopic hypertrophy of articular chondrocytes.94–96 While differentiation of chondrocytes from human embryonic stem cells in the presence of BMP4 results in a hypertrophic identity, addition of TGFβ maintains chondrocytes in a nonhypertrophic state.97,98 Bmp/Gdf and TGFβ signaling are thought to signal through a mixture of shared and distinct SMAD effectors.81 Whereas competition for the common Smad4 effector is predicted to result in cross-inhibitory actions of these pathways, the finding that mice lacking Smad4 in the Col2a1 lineage have only mild joint defects indicates that the balance of Bmp/Gdf and TGFβ pathways may be more important than the absolute levels of either.99
Transcriptional Repression of Chondrocyte Maturation
Although lower levels of Bmp signaling are important for specifying joints, less is known about how such signaling is interpreted in the nucleus to repress the hypertrophic maturation of articular chondrocytes (Figure 6). Cux1100 and the C-1-1 isoform of the Ets factor Erg101 are both expressed at developing murine joints and can repress cartilage differentiation upon misexpression. Loss of Erg in Gdf5+ joint precursors did not, however, alter joint specification in mice, although it did lead to increased susceptibility to age- and injury-induced osteoarthritis of the knee.102 Recent work in zebrafish has found roles for the Iroquois family of transcriptional repressors in joint specification. At the hyoid joint in zebrafish, which functions in gill ventilation, prospective joint chondrocytes express SoxE factors but make only low levels of Col2a1a and Aggrecan. In animals lacking irx7 and irx5a, hyoid joint chondrocytes inappropriately mature into high Col2a1a-expressing chondrocytes, which reflects direct repression of a col2a1a enhancer by Iroquois proteins.28 In mammals, Irx1 and Irx2 are expressed in the developing interphalangeal joints,103 and either mouse IRX1 or zebrafish Irx7 can repress the chondrogenic differentiation of a murine chondrogenic cell line.28 In addition to Iroquois genes, the homolog of the Tricho-rhino-Phalangeal syndrome gene TRPS1 is expressed at joints in zebrafish,104 as well as in early chondrocytes of the murine growth plate.105,106 Loss of Trps1 results in precocious chondrocyte maturation in the condylar cartilage of the murine jaw joint,107 and cone-shaped epiphyses, joint hypermobility, and osteoarthritis-like changes to articular cartilage in humans.108 Interestingly, IRX5 and TRPS1 proteins form a complex during early neural crest development.109 Given their similar expression and requirement in early chondrocytes, it will be interesting to test whether a similar protein–protein complex functions to inhibit cartilage maturation at joints. As Trps1 and Iroquois genes are expressed in the perichondrium,28,110 they may also have requirements outside of joints to inhibit chondrogenic differentiation.
FIGURE 6.
Chondrocyte fate decisions. (a) Section of the knee joint from an 8-week-old mouse shows that Col2a1 expression (green) is stronger in growth plate chondrocytes (arrowhead) compared to articular chondrocytes (arrow). Superficial joint chondrocytes are labeled by treatment of Prg4-CreER; Rosa26:memTomato/memGFP mice with Tamoxifen three weeks earlier (anti-GFP antibody staining detects Prg4-CreER-converted cells in red). (c) Compared to the complex mammalian knee joint, the hyoid joint of 6-day-old zebrafish (arrows) provides a simplified model for understanding the specification of joint chondrocyte fate. In this example, transient chondrocytes express both a sox10:dsRed transgene (red) and a col2a1a:GFP transgene (green). In contrast, joint chondrocytes express sox10 but much lower levels of col2a1a, suggesting they are immature. (c) Cells within a mesenchymal condensation initially express Barx1 and then go on to express Sox9 (and in zebrafish also the related SoxE family member sox10), Dcx, and low levels of Col2a1 (in particular an A splice isoform). In the growth plate, these cells mature into prehypertrophic chondrocytes that express high levels of Col2a1 and Matn1 and then hypertrophic chondrocytes that express Col10a1, Runx2, and other genes associated with mineralization. In contrast, interzone cells differentiate into articular chondrocytes that maintain low Col2a1 and instead express Gdf5 and later Prg4. Although Wnt9a and Fgf18 promote a joint fate, Ihh signaling controls cartilage maturation. Specification of articular chondrocyte fate is promoted through inhibition of cartilage maturation by a number of transcription factors and chromatin remodelers, including Iroquois proteins (Irx), Trps1, Nkx3.2, Cux1, Erg, Lrf, and Hdac1. (d) Under the control of signaling factors including Gdf5/6, Wnt9a, Fgf18, TGFβ and Noggin, chondroprogenitor cells mature to form the different chondrocyte layers of the growth plate (darker blue) or remain relatively immature within the joint (orange cells in light blue matrix). Factors including Trps1, Has, and Ihh then drive interzone cells to cavitate as superficial zone articular chondrocytes (brown) begin to produce lubricating molecules such as Prg4.
Epigenetic Fixation of Articular Cartilage Identity
It is becoming increasingly appreciated that covalent modifications of DNA and their associated histones can have long-term consequences on gene expression. A salient feature of joint cartilage is that it must be maintained as permanent hyaline cartilage for the life of the animal. An attractive model then is that epigenetic silencing of the chondrocyte hypertrophy program that helps lock articular cells into a permanent cartilage identity.111 In cultured human chondrocytes, expression of COL10A1 involves active DNA demethylation of CpG islands in its promoter.112 In mice, loss of Histone Deacetylase 4 (Hdac4) results in accelerated hypertrophy and mineralization of cartilage, in part through increased histone acetylation and expression of Runx2.113 Furthermore, treatment of synovium-derived cells from pig with TGFβ1 induces chondrogenesis, with coexpression of Hdac1 blocking the hypertrophy of these chondrocytes.114 Similarly, the joint-promoting factor NKX3.2 has been shown to function as a transcriptional repressor in complex with HDAC1 and the common BMP/TGFβ effector, SMAD4, although the targets of NKX3.2 remain unclear.115 The transcriptional repressor LRF has also been shown to act together with HDAC1 to repress chondrogenic differentiation in rodents.116 These data suggest that DNA methylation and/or histone deacetylation of Col10a1, Runx2, and likely other genes associated with chondrocyte hypertrophy may be important for maintaining permanent cartilage at joints.
GENERATION OF SYNOVIAL SPECIALIZATIONS
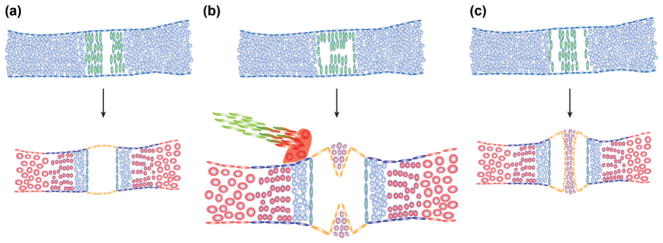
In addition to generating permanent articular cartilage, cells within the developing joint express a suite of genes involved in the creation of joint-specific structures (Figure 7). A prominent morphological event in synovial joints is the formation of a cavity between articular cartilage layers, which is accompanied by the loss of cell–cell contacts and production of molecular lubricants that promote joint function. In some joints, tissues with features in common with the articular cartilage layers can exist as menisci that project into the cavity (e.g., in the knee joint) or as discs between the articular surfaces (e.g., in the mammalian jaw joint). Supportive ligaments also attach to specific sites in the joint through a transitional structure, the enthesis. Joints are clearly complex organs that vary greatly depending on their location in the body, with their development relying on integration of common joint-promoting programs with unique positional specifiers.
FIGURE 7.
Synovial specializations. Distinct patterns of cavitation of the interzone (green) can generate simple synovial joints (a) or joints with specialized structures such as menisci (b) and articular discs (c). The synovium (orange) shares many properties with the fibroblasts ensheathing the menisci and disc (purple). Bony processes (red) act as attachment points for ligaments (green), with their connection point, or enthesis, consisting of a transitional cell type.
Formation of Lubricated Cavities
Cavitation of the developing joint separates the two articular surfaces, yet the cellular mechanisms underlying this morphogenetic event remain unresolved. Different joints fail to cavitate in a number of mouse mutants, including the temporomandibular joint in Trps1 mutants,107 the digit joints in Ihh mutants,67 and elbow and wrist joints in Osr1; Osr2 double mutants.117 In many of these cases, it remains unclear the extent to which lack of cavitation is secondary to earlier defects in joint specification. In one study, conditional deletion of the HH receptor Smo in Sox9+ chondroprogenitors resulted in a failure of the disc to separate from the mandibular condyle but did not affect earlier joint specification.32 Correlative evidence suggests that cavitation does not require extensive cell death within the developing interzone.118 Instead, asymmetric synthesis of hyularonan (HA), a major component of synovial fluid, may facilitate a decrease in cell–cell contacts that allows tissue separation.119 In the mouse limb, conditional inactivation of Has2, a member of the HA synthase family, using a Prrx1-Cre line that is active in early limb mesenchyme, inhibits cavitation of digit joints and leads to an accumulation of mesenchymal cells in the presumptive joint space.120 This effect is specific to Has2, as deletion of Has1 and/or Has3 does not affect cavitation.121
Synovial joints are also unique in producing lubricating fluid that reduces friction and protects joint integrity. A critical lubricating protein is the proteoglycan Lubricin, encoded by the Prg4 gene, which complexes with Aggrecan and HA in the synovial fluid.122 Within developing joints, Prg4 expression appears only after Gdf5 expression begins to subside, roughly correlating with initiation of cavitation.11 In addition to expression in the most superficial cells of articular cartilage, Prg4 expression can also be found in the synovium, meniscus, and ligaments of mammalian joints. Such shared expression may reflect combinatorial secretion of Lubricin from all cavity-lining tissues, as well as additional roles for Lubricin in the lubrication of nonjoint tissues such as ligaments.123 Although dispensable for joint cavitation, Lubricin is required for the maintenance of many types of joints with osteoarthritic phenotypes seen in human camptodactyly arthropathy-coxa vara-pericarditis (CACP) syndrome patients,124 mice lacking Prg4,11,125 and zebrafish prg4b mutants.25 The factors that induce Prg4 expression during later joint development remain unclear, although a recent study suggests a potential role for mechanical forces126 (see Box 1).
BOX 1. MECHANICAL FORCES IN JOINT DEVELOPMENTAL AND MAINTENANCE.
Although genetics clearly plays an important role in joint development, mechanical forces also help to refine joint structure. For example, experimental immobilization of the developing avian limb revealed a requirement for movement in the cavitation but not specification of joints.127,128 Mechanical motion has also been shown to stimulate production of the major lubricant of synovial cavities, PRG4.126 Imprecise joint architecture may also feed back through altered mechanosensation to locally modulate skeletal differentiation, in an attempt to correct joint structure. How cells in the joint sense mechanical forces and relay this information to modulate skeletal cell behavior remains largely a mystery. One clue is the identification of dominant mutations of TRPV4 in congenital arthropathy in humans.129 TRPV4 is a member of the Transient Receptor Potential class of gated calcium channels, and evidence suggests that it is a mechanosensitive channel that responds to compressive forces within the joint.130 Trpv4 is highly expressed by articular chondrocytes, and its loss in mice results in severe osteoarthritic changes in the knee joint.130,131 Similarly, the PIEZO1 and PIEZO2 channels have been shown to be required for calcium signaling within cultured articular chondrocytes in response to directly applied mechanical forces.132 What remains to be understood is how calcium signaling would result in changes in articular chondrocytes that maintain the health of the joint.
Integrating Ligaments and Structural Components of the Joint
Menisci and discs are similar to the articular surface in having Prg4-expressing flattened cells overlying rounder chondrocyte-like cells. These structures arise within the Gdf5-expressing interzone, with their complex structures influenced by varied patterns of cavitation (e.g., discs are likely generated by two separate and parallel cavitation events). There are also specialized endochondral bones at joints that act as ligament attachment sites. These endochondral bones often form as secondary cartilage outgrowths from the bone surface (e.g., fossa of the mammalian jaw joint).71 In zebrafish with reduced nkx3.2 or irx7 function, defects in articular cartilage are accompanied by loss of joint-associated endochondral bones at the jaw and hyoid joints, respectively, suggesting a high level of coordination of articular cartilage specification and joint-associated bone features.27,28 The enthesis is a specialized transitional tissue between the endochondral bone and ligament at the joint. A common feature of ligament, enthesis, bone, and cartilage progenitors is expression of Sox9.133,134 It is possible that there exists a common Sox9+ progenitor pool within the early interzone, with local induction of lineage-specific factors (e.g., Scleraxis for ligaments) resulting in precise arrays of cartilage, bone, and ligament cells within the joint organ. It remains unclear whether interzone cells constitute a homogenous cell population that later diverges into distinct lineages, or whether interzone cells are heterogenous and lineage biased from initial stages.
DEVELOPMENTAL PATHWAYS IN ADULT JOINT DISEASE
While genetic syndromes drastically affecting joint formation are rare, inherited susceptibility to joint degeneration (i.e., osteoarthritis) is common (Table 1). Osteoarthritis is thought to arise from accumulated “wear-and-tear” of joints. One possibility is that minor developmental defects lead to slightly abnormal joint architecture, which results in additive damage to the joint over time due to compromised function. Genetic programs that help specify articular at the expense of growth plate fates during development may also be required to continuously maintain the articular chondrocyte phenotype through adulthood. These developmental pathways may also serve to maintain a progenitor pool in the postnatal joint, which in mammals has a limited ability to replace damaged joint tissue. Genetic changes that affect progenitor maintenance or function could thus impact joint homeostasis and predispose to osteoarthritis. Understanding roles for developmental genes in postnatal joint function and maintenance may therefore offer new insights into the cause and possible prevention of joint diseases.
TABLE 1.
Selected Human Joint Syndrome Genes and Animal Models of Joint Development and Disease
| Gene Family | Genetic Models | Species | Human Syndrome | Joint Phenotype(s) | Selected References |
|---|---|---|---|---|---|
| A. Joint cartilage specification | |||||
| EDN1 |
Edn1−/− Ednra−/− |
h, z | Auriculo-Condylar Syndrome (ARCND3) (OMIM: 615706) | Abnormal TMJ and mandibular condyle, loss of jaw joint | 52–54 |
| Nkx3.2 | nkx3.2 morpholino | z | Loss of jaw joint | 27 | |
| Noggin | Col2a1-Cre; Nogfl/fl | h, m | Tarsal-carpal coalition syndrome (OMIM: 186570), proximal symphalangism (SYM1A) (OMIM: 185800) | Joint fusion (symphalangism), joint ankylosis, carpal and tarsal fusion, conductive deafness | 4,83,84 |
| FOXC1 | Foxc1−/− | m | Syngnathia (congenital adhesion of maxilla and mandible) | 66 | |
| IHH | Ihh−/− | m | Loss of limb joints, cavitation failure | 67,71 | |
| GDF | Gdf5−/−; Gdf6−/− | h, m | Proximal symphalangism (SYM1B) (OMIM: 615298) | Joint fusion at interphalangeal joints of fourth and fifth digit, digit joint fusion in mice | 87,88,90 |
| OSR1/OSR2 | Prrx1-Cre; Osr1fl/fl; Osr2−/− | m | Fusions at elbow joint, between the tibia and fibula, and carpal and tarsal joints; reduced Prg4 expression in articular cartilage | 117 | |
| TGFβ | Prrx1-Cre; Tgfbr2fl/fl | m | Interphalangeal joint fusion | 93,96 | |
| Connexin | connexin43−/− Connexin40−/− | h, m, z | Oculodentodigital dysplasia (OMIM: 164200) | Loss of distal limb joints, shorter interjoint spacing, missing and fused phalanges | 35,42,43 |
| Kcnk5b | kcnk5b GOF mutation | z | Longer and irregular spacing of fin ray joints | 37 | |
| Barx1 | barx1−/− | z | Ectopic joint in Meckel’s cartilage of the jaw | 33 | |
| BMP | Prrx1-Cre; Bmp2C/C; Bmp4C/C | m | Elbow joint fusion | 85 | |
| Irx | irx7−/−; irx5a−/− | z | Hyoid joint fusion | 28 | |
| B. Cavitation | |||||
| TRPS1 | Trps1−/− | h, m | Tricho-rhino-Phalangeal syndrome, type I (OMIM: 190350) | Temporomandibular joint cavitation defect, joint hypermobility | 107,108 |
| HAS | Prrx1-Cre; Has2fl/fl | m | Carpal, metacarpophalangeal, interphalangeal, elbow and shoulder cavitation defects | 120 | |
| C. Joint Cartilage Maintenance | |||||
| TRPV4 | TRPV4, Trpv4−/− | h, m | Familial digital arthropathy-brachydactyly (OMIM: 606835) | Defects of distal interphalangeal joints, early onset OA in fingers and toes, early onset OA of knee joints in mice | 125,135 |
| PRG4 | Prg4−/−, prg4b−/− | h, m, z | Camptodactyly arthropathy-coxa vara-pericarditis syndrome (OMIM: 208250) | Progressive failure of TMJ, ankle, knee, shoulder and elbow joints with age | 11,25,30,124,125 |
| PTHrP | Gdf5-Cre; PTHrPfl/fl | m | Increased OA susceptibility following knee DMM surgery | 133 | |
| Sirtuin | Sirt1−/−, Sirt6+/− | m | Arthritis progression in knee joints | 136,137 | |
| Notch | Prrx1-Cre; RBP-Jkfl/fl, Col2a1-CreER; RBP-Jkfl/fl | m | Cartilage degeneration in knee joints | 138–140 | |
| microRNA | miR-140−/− | m | Increased age-related and surgery-induced OA susceptibility in knee joints | 141 | |
h, human; m, mouse; z, zebrafish.
Do Subthreshold Developmental Defects Predispose to Osteoarthritis?
One theory is that minor alterations in patterning of joints during embryogenesis results in compromised postnatal joint structure and increased arthritis susceptibility.17 Changes in the proliferation, survival, and differentiation of cartilage and bone precursors in the developing joint could all lead to alterations in normal joint architectures. For example, polymorphisms in GDF5 are linked to early onset osteoarthritis in humans,142 and mice lacking Gdf5 have defects in joint formation, particularly in the digits.88 As Gdf5 expression peaks during initial joint formation and becomes much reduced by postnatal stages,11 it appears likely that the osteoarthritis susceptibility associated with GDF5 mutations is due to developmental defects at joints. Polymorphisms in other genes associated with the BMP, TGFβ, and WNT pathways have also been linked to osteoarthritis of the hip in humans, although causative effects of these mutations have not been shown in most cases. These include BMP5, the TGFβ inhibitor Asporin (ASPN), and the WNT inhibitor Frizzled-related protein (FRZB).16 The roles of these pathways in early chondrocyte differentiation suggest that setting the proper balance of endochondral bone to permanent articular cartilage may be critical for long-term joint maintenance. Definitive proof that the developmental roles ascribed to these genes are what predispose to osteoarthritis would require selectively removing gene function only during development.
Postnatal Maintenance of Permanent Articular Cartilage
Once established, permanent cartilage must be maintained at the joint surface through adulthood. The continued postnatal expression of many signaling components and transcription factors required for joint development suggest roles in joint maintenance, which in some cases is corroborated by osteoarthritic phenotypes upon postnatal gene deletion. Global expression profiling of bovine articular cartilage demonstrated decreased expression of TGFβ and BMP receptors with age,143 and slow-cycling TGFBR2-expressing cells have been suggested to be resident joint stem cells involved in homeostasis (see Box 2). When Bmpr1a is conditionally deleted from the Gdf5+ interzone in mice, joints of the ankle, knee, and digits display progressive loss of articular cartilage due to ectopic endochondral differentiation.9 Stronger evidence implicates TGFβ signaling in adult maintenance of joints, as postnatal deletion of Tgfbr2 in murine articular cartilage, using Col2a1-CreER, results in ectopic chondrocyte hypertrophy at the joint surface and osteoarthritis of the knee.144 Trps1 and Iroquois genes, which are required to inhibit articular cartilage maturation during development, also display continued expression at joints, at least in zebrafish.28 Similarly, Wnt9a expression is maintained in later joints72 and the zone of Parathyroid hormone-related protein (PTHrP) expression subadjacent to articular cartilage is maintained in adult mice.145 Deletion of PTHrP in Gdf5-Cre-expressing joint cells predisposes cartilage to arthritis of the knee following destabilization of the medial meniscus in mice.146 Furthermore, PTH analogs inhibit knee osteoarthritis progression in rats136 and mice.147 A failure of epigenetic maintenance of the permanent cartilage phenotype may also play a role in osteoarthritis progression. Genome-wide DNA methylation changes have been shown in hip and knee cartilage in human osteoarthritis patients137–139 and have been linked to differential expression of 70 genes, including the osteoarthritis-associated genes VIT, ROR2, and WLS.140 Altered activity of the Sirtuin family of histone deacetylases, in particular SIRT1, SIRT3, and SIRT6, has also been implicated in arthritis.19,148,149 In particular, increased histone acetylation within articular cartilage has been linked to ectopic hypertrophy in the mouse knee, including increased expression of matrix metalloproteases (e.g., MMP-13) that degrade joint cartilage.150
BOX 2. STEM CELLS IN THE POSTNATAL JOINT.
Researchers have sought to identify stem cells in the adult joint, both as potential endogenous therapeutic targets and as a source of transplantable cells for joint repair. Lineage tracing with tamoxifen-inducible Prg4-Cre revealed that Prg4+ joint cells drive the growth of articular cartilage through adulthood in mice.3 A subset of postnatal superficial joint cells also shows high colony-forming efficiency in culture.151 In a disease context, chondrogenic progenitor cells have been isolated from late-stage human osteoarthritic cartilage and display multipotency, clonogenicity, and migratory potential in vitro.152 BrdU labeling experiments have identified slow-cycling cells within the perichondrial zone of Ranvier at the periphery of the growth plate.141 Additionally, TGFBR2-expressing cells in the “synovio-entheseal-articular cartilage niche,” which includes the zone of Ranvier and a subset of the perichondrium, synovium, and articular cartilage, have been shown to be slow cycling, thus representing a potential stem cell population in the adult joint.153 When cultured in vitro, the synovium is expandable and chondrogenic,154,155 and synovial cells respond both to acute injury156 and joint disease124 with increased proliferation, which may reflect a progenitor response toward repairing the joint. Postnatal lineage tracing of these potential stem cell sources is needed to clarify their role in joint homeostasis and repair. In the future, targeted activation of these potential stem cell populations could be a potential strategy for reversing joint damage.
While continuity of developmental programs may play a role in joint maintenance, there also appear to be factors with specific postnatal requirements at the joint, including Notch signaling,135,157,158 PRG4,11,125 and ERRFI1.159 The Notch pathway has been implicated in progenitor maintenance in a variety of contexts, including a role for inhibiting cartilage differentiation in mice160 and zebrafish.161 Notch1 is expressed at postnatal joints in mice, and conditional deletion of the essential Notch effector RBP-Jk in either limb mesenchyme using Prx1-Cre or postnatal chondrocytes using Col2a1-CreER results in progressive joint cartilage degeneration and other osteoarthritic phenotypes.135,157,158 The miR-140 is the major miRNA expressed in articular cartilage of murine joints,162 and deletion of miR-140 predisposed mice to age-related osteoarthritis of the knee without affecting joint specification.163 Furthermore, miR-140 overexpression protects joints from injury-induced osteoarthritis.163 One mechanism of miR-140 function is to inhibit the expression of Adamts5,163,164 a metalloprotease that cleaves Aggrecan165 and whose deletion mitigates osteoarthritis in mice.166
CONCLUSION
The joint is a complex organ, composed of chondrocytes of different types and an array of associated connective tissues. Some of these connective tissues, in particular the synovium, may serve to not only encapsulate the joint cavity but also to act as a source of progenitors for new chondrocytes and joint tissues. A growing body of work is revealing how WNT, BMP/GDF, TGFβ, IHH, FGF, and other pathways are integrated in the developing joint interzone to control the expression of transcription factors and chromatin remodelers, which in turn locally specify cell fates. An important unanswered question is how members of the TGFβ superfamily—BMP, GDF, and TGFβ—are utilized to specify the different permanent cartilage zones at the articular surface versus the neighboring transient cartilage that converts into bone. Do the duration and/or relative levels of BMP versus TGFβ signaling drive different cartilage types? Or does signaling through distinct types of receptors and downstream effectors regulate different fates? A related question is how GDF and other signaling pathways regulate nonchondrocyte fates at joints. Chondrocytes, osteoblasts, and ligament cells all passage through a Gdf5+ interzone cell, with progenitors for each lineage also expressing Sox9. When do these lineages diverge from each other, and how are the major signaling pathways integrated differently to achieve these fates? An emerging theme is that there exists a continuum of cell fates throughout the joint, such as ligament cells that grade into osteoblasts in the enthesis, superficial chondrocytes that grade into synovial fibroblasts, and chondrocytes that grade into osteoblasts at the tidemark. Such graded fates may help link together the different components of the joint into a seamless organ.
A promising future research direction is to understand the extent to which the pathways that establish an articular cartilage fate function to maintain this fate, which in humans can be for over 100 years. An attractive but relatively unexplored possibility is that developmental pathways establish repressive DNA methylation and chromatin around genes associated with chondrocyte hypertrophy, thus locking articular chondrocytes into a permanent cartilage phenotype. Does the postnatal expression of transcriptional repressors such as Iroquois proteins and Trps1 indicate a requirement for continued reinforcement of silencing of chondrocyte hypertrophy at the joint surface? And how would injury and decreased lubrication (e.g., due to PRG4 loss) result in an override of epigenetic repression of chondrocyte hypertrophy? Relatedly, does failure to fully lock articular chondrocytes into a permanent cartilage identity during development predispose to later osteoarthritis? As such, new methods to assess chromatin status in joint tissue biopsies could result in the better diagnosis of susceptibility to arthritic changes, thus allowing for earlier detection and intervention before significant cartilage and bone erosion occurs.
While this review focuses on synovial joints, it will be interesting to understand the extent to which common pathways regulate cartilage and bone differentiation at other types of joints, such as intervertebral discs and sutures, as well as how their distinct utilization results in the wonderful diversity of joint architectures throughout the body. Are these differences created by the different deployment of similar pathways, or by additional joint-specific genetic programs? In addition, it will be interesting to explore whether core sets of anti-differentiation pathways are used in other contexts. Do the same pathways that inhibit cartilage maturation at the articular surface also function to maintain undifferentiated cartilage progenitors in the perichondrium, or to extinguish cartilage gene expression as hypertrophic chondrocytes transdifferentiate into osteocytes in the mature growth plate? A related issue is whether the synovium functions as a modified perichondrium that can generate limited numbers of new articular chondrocytes to maintain the joint. If so, osteoarthritis may reflect less a complete inability to replace articular chondrocytes in adults, and more the inability of a slow homeostasis program to keep pace with repeated injuries to the joint during wear-and-tear. An important question then is whether genes required to specify articular cartilage during embryogenesis also play a more limited role in regenerating articular chondrocytes in adults. Alternatively, as recently described for adult bone homeostasis,167 distinct progenitors may exist solely for postnatal joint homeostasis. In the future, a better understanding of how the different cell types of the joint are specified during development, then maintained and replaced as needed in the adult, will be critical for guiding new approaches toward both preventing and reversing osteoarthritis.
Footnotes
Conflict of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Gunnell LM, Jonason JH, Loiselle AE, Kohn A, Schwarz EM, Hilton MJ, O’Keefe RJ. TAK1 regulates cartilage and joint development via the MAPK and BMP signaling pathways. J Bone Miner Res. 2010;25:1784–1797. doi: 10.1002/jbmr.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn J, Shwartz Y, Blitz E, Krief S, Sharir A, Breitel DA, Rattenbach R, Relaix F, Maire P, Rountree RB, et al. Muscle contraction is necessary to maintain joint progenitor cell fate. Dev Cell. 2009;16:734–743. doi: 10.1016/j.devcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 3.Kozhemyakina E, Zhang M, Ionescu A, Ayturk UM, Ono N, Kobayashi A, Kronenberg H, Warman ML, Lassar AB. Identification of a Prg4-expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 2015;67:1261–1273. doi: 10.1002/art.39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray A, Singh PN, Sohaskey ML, Harland RM, Bandyopadhyay A. Precise spatial restriction of BMP signaling is essential for articular cartilage differentiation. Development. 2015;142:1169–1179. doi: 10.1242/dev.110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao H, Feng J, Ho TV, Grimes W, Urata M, Chai Y. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decker RS, Koyama E, Pacifici M. Genesis and morphogenesis of limb synovial joints and articular cartilage. Matrix Biol. 2014;39:5–10. doi: 10.1016/j.matbio.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longobardi L, Li T, Tagliafierro L, Temple JD, Willcockson HH, Ye P, Esposito A, Xu F, Spagnoli A. Synovial joints: from development to homeostasis. Curr Osteoporos Rep. 2015;13:41–51. doi: 10.1007/s11914-014-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, Okabe T, Ochiai T, Kamiya N, Rountree RB, et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316:62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, Kingsley DM. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2:e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet Cell Genet. 2000;90:291–297. doi: 10.1159/000056791. [DOI] [PubMed] [Google Scholar]
- 11.Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, Jay GD, Stewart M, Wang H, Warman ML, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer CW, Dowthwaite GP, Francis-West P. Development of synovial joints. Birth Defects Res C Embryo Today. 2003;69:144–155. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- 13.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, Liang MH, Kremers HM, Mayes MD, Merkel PA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58:15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 14.MacGregor AJ, Antoniades L, Matson M, Andrew T, Spector TD. The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum. 2000;43:2410–2416. doi: 10.1002/1529-0131(200011)43:11<2410::AID-ANR6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15.Consortium A. Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC, Boraska V, Esko T, Evangelou E, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker-LePain JC, Lane NE. Relationship between joint shape and the development of osteoarthritis. Curr Opin Rheumatol. 2010;22:538–543. doi: 10.1097/BOR.0b013e32833d20ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitsillides AA, Beier F. Cartilage biology in osteoarthritis—lessons from developmental biology. Nat Rev Rheumatol. 2011;7:654–663. doi: 10.1038/nrrheum.2011.129. [DOI] [PubMed] [Google Scholar]
- 18.Chen M, Zhu M, Awad H, Li TF, Sheu TJ, Boyce BF, Chen D, O’Keefe RJ. Inhibition of β-catenin signaling causes defects in postnatal cartilage development. J Cell Sci. 2008;121:1455–1465. doi: 10.1242/jcs.020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Wang J. Epigenetics and osteoarthritis. Genes Dis. 2015;2:69–75. doi: 10.1016/j.gendis.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quarto N, Longaker MT. The zebrafish (Danio rerio): a model system for cranial suture patterning. Cells Tissues Organs. 2005;181:109–118. doi: 10.1159/000091100. [DOI] [PubMed] [Google Scholar]
- 21.Jeradi S, Hammerschmidt M. Retinoic acid-induced premature osteoblast-to-preosteocyte transitioning has multiple effects on calvarial development. Development. 2016;143:1205–1216. doi: 10.1242/dev.129189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kague E, Roy P, Asselin G, Hu G, Simonet J, Stanley A, Albertson C, Fisher S. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev Biol. 2016;413:160–172. doi: 10.1016/j.ydbio.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laue K, Pogoda HM, Daniel PB, van Haeringen A, Alanay Y, von Ameln S, Rachwalski M, Morgan T, Gray MJ, Breuning MH, et al. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am J Hum Genet. 2011;89:595–606. doi: 10.1016/j.ajhg.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes AJ, Reynolds S, Nowell MA, Meakin LB, Habicher J, Ledin J, Bashford A, Caterson B, Hammond CL. Spinal deformity in aged zebrafish is accompanied by degenerative changes to their vertebrae that resemble osteoarthritis. PLoS One. 2013;8:e75787. doi: 10.1371/journal.pone.0075787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Askary A, Smeeton J, Paul S, Schindler S, Braasch I, Ellis NA, Postlethwait J, Miller CT, Crump JG. Ancient origin of lubricated joints in bony vertebrates. eLife. 2016;5:e16415. doi: 10.7554/eLife.16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed NP, Mortlock DP. Identification of a distant cis-regulatory element controlling pharyngeal arch-specific expression of zebrafish gdf6a/radar. Dev Dyn. 2010;239:1047–1060. doi: 10.1002/dvdy.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- 28.Askary A, Mork L, Paul S, He X, Izuhara AK, Gopalakrishnan S, Ichida JK, McMahon AP, Dabizljevic S, Dale R, et al. Iroquois proteins promote skeletal joint formation by maintaining chondrocytes in an immature state. Dev Cell. 2015;35:358–365. doi: 10.1016/j.devcel.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haines RW. Eudiarthrodial joints in fishes. J Anat. 1942;77:12–19. [PMC free article] [PubMed] [Google Scholar]
- 30.Hill A, Duran J, Purcell P. Lubricin protects the temporomandibular joint surfaces from degeneration. PLoS One. 2014;9:e106497. doi: 10.1371/journal.pone.0106497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capellini TD, Handschuh K, Quintana L, Ferretti E, Di Giacomo G, Fantini S, Vaccari G, Clarke SL, Wenger AM, Bejerano G, et al. Control of pelvic girdle development by genes of the Pbx family and Emx2. Dev Dyn. 2011;240:1173–1189. doi: 10.1002/dvdy.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell P, Joo BW, Hu JK, Tran PV, Calicchio ML, O’Connell DJ, Maas RL, Tabin CJ. Temporomandibular joint formation requires two distinct hedgehog-dependent steps. Proc Natl Acad Sci USA. 2009;106:18297–18302. doi: 10.1073/pnas.0908836106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140:2765–2775. doi: 10.1242/dev.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka M. Fins into limbs: autopod acquisition and anterior elements reduction by modifying gene networks involving 5′Hox, Gli3, and Shh. Dev Biol. 2016;413:1–7. doi: 10.1016/j.ydbio.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 35.Hoptak-Solga AD, Klein KA, DeRosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional inter-cellular communication. FEBS Lett. 2007;581:3297–3302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sims K, Jr, Eble DM, Iovine MK. Connexin43 regulates joint location in zebrafish fins. Dev Biol. 2009;327:410–418. doi: 10.1016/j.ydbio.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perathoner S, Daane JM, Henrion U, Seebohm G, Higdon CW, Johnson SL, Nusslein-Volhard C, Harris MP. Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 2014;10:e1004080. doi: 10.1371/journal.pgen.1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borday V, Thaeron C, Avaron F, Brulfert A, Casane D, Laurenti P, Geraudie J. evx1 transcription in bony fin rays segment boundaries leads to a reiterated pattern during zebrafish fin development and regeneration. Dev Dyn. 2001;220:91–98. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1091>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 39.Schulte CJ, Allen C, England SJ, Juarez-Morales JL, Lewis KE. Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev Dyn. 2011;240:1240–1248. doi: 10.1002/dvdy.22534. [DOI] [PubMed] [Google Scholar]
- 40.Ton QV, Iovine MK. Identification of an evx1-dependent joint-formation pathway during FIN regeneration. PLoS One. 2013;8:e81240. doi: 10.1371/journal.pone.0081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlegel W, Albrecht C, Eckl P, Freudenthaler H, Berger A, Vecsei V, Marlovits S. Dedifferentiation of human articular chondrocytes is associated with alterations in expression patterns of GDF-5 and its receptors. J Cell Mol Med. 2009;13:3398–3404. doi: 10.1111/j.1582-4934.2009.00953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pizard A, Burgon PG, Paul DL, Bruneau BG, Seidman CE, Seidman JG. Connexin 40, a target of transcription factor Tbx5, patterns wrist, digits, and sternum. Mol Cell Biol. 2005;25:5073–5083. doi: 10.1128/MCB.25.12.5073-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paznekas WA, Karczeski B, Vermeer S, Lowry RB, Delatycki M, Laurence F, Koivisto PA, Van Maldergem L, Boyadjiev SA, Bodurtha JN, et al. GJA1 mutations, variants, and connexin 43 dysfunction as it relates to the oculodentodigital dysplasia phenotype. Hum Mutat. 2009;30:724–733. doi: 10.1002/humu.20958. [DOI] [PubMed] [Google Scholar]
- 44.Rolland-Lagan AG, Paquette M, Tweedle V, Akimenko MA. Morphogen-based simulation model of ray growth and joint patterning during fin development and regeneration. Development. 2012;139:1188–1197. doi: 10.1242/dev.073452. [DOI] [PubMed] [Google Scholar]
- 45.Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 46.Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- 47.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation. Nature. 2010;464:583–586. doi: 10.1038/nature08838. [DOI] [PubMed] [Google Scholar]
- 49.Chuong CM, Yeh CY, Jiang TX, Widelitz R. Module-based complexity formation: periodic patterning in feathers and hairs. WIREs Dev Biol. 2013;2:97–112. doi: 10.1002/wdev.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondo S, Miura T. Reaction–diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 51.Medeiros DM, Crump JG. New perspectives on pharyngeal dorsoventral patterning in development and evolution of the vertebrate jaw. Dev Biol. 2012;371:121–135. doi: 10.1016/j.ydbio.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. Sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- 53.Nair S, Li W, Cornell R, Schilling TF. Requirements for Endothelin type-A receptors and Endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- 54.Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin-1 signaling in zebrafish. Dev Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker MB, Miller CT, Coffin Talbot J, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin-1 signaling in craniofacial patterning. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 56.Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. Phospholipase C, β 3 is required for Endothelin-1 regulation of pharyngeal arch patterning in zebrafish. Dev Biol. 2007;304:194–207. doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duprez D, Bell EJ, Richardson MK, Archer CW, Wolpert L, Brickell PM, Francis-West PH. Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech Dev. 1996;57:145–157. doi: 10.1016/0925-4773(96)00540-0. [DOI] [PubMed] [Google Scholar]
- 58.Wilson J, Tucker AS. Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev Biol. 2004;266:138–150. doi: 10.1016/j.ydbio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 59.Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon CT, Petit F, Kroisel PM, Jakobsen L, Zechi-Ceide RM, Oufadem M, Bole-Feysot C, Pruvost S, Masson C, Tores F, et al. Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. Am J Hum Genet. 2013;93:1118–1125. doi: 10.1016/j.ajhg.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon CT, Vuillot A, Marlin S, Gerkes E, Henderson A, AlKindy A, Holder-Espinasse M, Park SS, Omarjee A, Sanchis-Borja M, et al. Heterogeneity of mutational mechanisms and modes of inheritance in auriculocondylar syndrome. J Med Genet. 2013;50:174–186. doi: 10.1136/jmedgenet-2012-101331. [DOI] [PubMed] [Google Scholar]
- 62.Kido Y, Gordon CT, Sakazume S, Ben Bdira E, Dattani M, Wilson LC, Lyonnet S, Murakami N, Cunningham ML, Amiel J, et al. Further characterization of atypical features in auriculocondylar syndrome caused by recessive PLCB4 mutations. Am J Med Genet A. 2013;161A:2339–2346. doi: 10.1002/ajmg.a.36066. [DOI] [PubMed] [Google Scholar]
- 63.Leoni C, Gordon CT, Della Marca G, Giorgio V, Onesimo R, Perrino F, Cianfoni A, Cerchiari A, Amiel J, Zampino G. Respiratory and gastrointestinal dysfunctions associated with auriculocondylar syndrome and a homozygous PLCB4 loss-of-function mutation. Am J Med Genet A. 2016;170:1471–1478. doi: 10.1002/ajmg.a.37625. [DOI] [PubMed] [Google Scholar]
- 64.Rieder MJ, Green GE, Park SS, Stamper BD, Gordon CT, Johnson JM, Cunniff CM, Smith JD, Emery SB, Lyonnet S, et al. A human homeotic transformation resulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome. Am J Hum Genet. 2012;90:907–914. doi: 10.1016/j.ajhg.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inman KE, Purcell P, Kume T, Trainor PA. Interaction between Foxc1 and Fgf8 during mammalian jaw patterning and in the pathogenesis of syngnathia. PLoS Genet. 2013;9:e1003949. doi: 10.1371/journal.pgen.1003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koyama E, Ochiai T, Rountree RB, Kingsley DM, Enomoto-Iwamoto M, Iwamoto M, Pacifici M. Synovial joint formation during mouse limb skeletogenesis: roles of Indian hedgehog signaling. Ann N Y Acad Sci. 2007;1116:100–112. doi: 10.1196/annals.1402.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luyten FP, Tylzanowski P, Lories RJ. Wnt signaling and osteoarthritis. Bone. 2009;44:522–527. doi: 10.1016/j.bone.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 69.Rockel JS, Yu C, Whetstone H, Craft AM, Reilly K, Ma H, Tsushima H, Puviindran V, Al-Jazrawe M, Keller GM, et al. Hedgehog inhibits β-catenin activity in synovial joint development and osteoarthritis. J Clin Invest. 2016;126:1649–1663. doi: 10.1172/JCI80205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/β-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shibukawa Y, Young B, Wu C, Yamada S, Long F, Pacifici M, Koyama E. Temporomandibular joint formation and condyle growth require Indian hedgehog signaling. Dev Dyn. 2007;236:426–434. doi: 10.1002/dvdy.21036. [DOI] [PubMed] [Google Scholar]
- 72.Pitsillides AA, Ashhurst DE. A critical evaluation of specific aspects of joint development. Dev Dyn. 2008;237:2284–2294. doi: 10.1002/dvdy.21654. [DOI] [PubMed] [Google Scholar]
- 73.Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 2008;40:46–62. doi: 10.1016/j.biocel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 74.Mayne R, Vail MS, Mayne PM, Miller EJ. Changes in type of collagen synthesized as clones of chick chondrocytes grow and eventually lose division capacity. Proc Natl Acad Sci USA. 1976;73:1674–1678. doi: 10.1073/pnas.73.5.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von der Mark K, von der Mark H. The role of three genetically distinct collagen types in endochondral ossification and calcification of cartilage. J Bone Joint Surg Br. 1977;59-B:458–464. doi: 10.1302/0301-620X.59B4.72756. [DOI] [PubMed] [Google Scholar]
- 76.Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci USA. 2014;111:12097–12102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10:e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hyde G, Boot-Handford RP, Wallis GA. Col2a1 lineage tracing reveals that the meniscus of the knee joint has a complex cellular origin. J Anat. 2008;213:531–538. doi: 10.1111/j.1469-7580.2008.00966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Q, Cigan AD, Marrero L, Lopreore C, Liu S, Ge D, Savoie FH, You Z. Expression of doublecortin reveals articular chondrocyte lineage in mouse embryonic limbs. Genesis. 2011;49:75–82. doi: 10.1002/dvg.20702. [DOI] [PubMed] [Google Scholar]
- 80.Pacifici M, Koyama E, Shibukawa Y, Wu C, Tamamura Y, Enomoto-Iwamoto M, Iwamoto M. Cellular and molecular mechanisms of synovial joint and articular cartilage formation. Ann N Y Acad Sci. 2006;1068:74–86. doi: 10.1196/annals.1346.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 82.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 83.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 84.Stafford DA, Brunet LJ, Khokha MK, Economides AN, Harland RM. Cooperative activity of noggin and gremlin 1 in axial skeleton development. Development. 2011;138:1005–1014. doi: 10.1242/dev.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bruneau S, Mourrain P, Rosa FM. Expression of contact, a new zebrafish DVR member, marks mesenchymal cell lineages in the developing pectoral fins and head and is regulated by retinoic acid. Mech Dev. 1997;65:163–173. doi: 10.1016/s0925-4773(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 87.Settle SH, Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 88.Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- 89.Wolfman NM, Hattersley G, Cox K, Celeste AJ, Nelson R, Yamaji N, Dube JL, DiBlasio-Smith E, Nove J, Song JJ, et al. Ectopic induction of tendon and ligament in rats by growth and differentiation factors 5, 6, and 7, members of the TGF-β gene family. J Clin Invest. 1997;100:321–330. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee SJ. Limb alterations in brachypodism mice due to mutations in a new member of the TGF β-superfamily. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- 91.Coleman CM, Tuan RS. Growth/differentiation factor 5 enhances chondrocyte maturation. Dev Dyn. 2003;228:208–216. doi: 10.1002/dvdy.10369. [DOI] [PubMed] [Google Scholar]
- 92.Francis-West PH, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, Allen S, MacPherson S, Luyten FP, Archer CW. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 93.Spagnoli A, O’Rear L, Chandler RL, Granero-Molto F, Mortlock DP, Gorska AE, Weis JA, Longobardi L, Chytil A, Shimer K, et al. TGF-β signaling is essential for joint morphogenesis. J Cell Biol. 2007;177:1105–1117. doi: 10.1083/jcb.200611031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baffi MO, Slattery E, Sohn P, Moses HL, Chytil A, Serra R. Conditional deletion of the TGF-β type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Dev Biol. 2004;276:124–142. doi: 10.1016/j.ydbio.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 95.Longobardi L, Li T, Myers TJ, O’Rear L, Ozkan H, Li Y, Contaldo C, Spagnoli A. TGF-β type II receptor/MCP-5 axis: at the crossroad between joint and growth plate development. Dev Cell. 2012;23:71–81. doi: 10.1016/j.devcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Seo HS, Serra R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev Biol. 2007;310:304–316. doi: 10.1016/j.ydbio.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Craft AM, Rockel JS, Nartiss Y, Kandel RA, Alman BA, Keller GM. Generation of articular chondrocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:638–645. doi: 10.1038/nbt.3210. [DOI] [PubMed] [Google Scholar]
- 98.Wu L, Bluguermann C, Kyupelyan L, Latour B, Gonzalez S, Shah S, Galic Z, Ge S, Zhu Y, Petrigliano FA, et al. Human developmental chondrogenesis as a basis for engineering chondrocytes from pluripotent stem cells. Stem Cell Reports. 2013;1:575–589. doi: 10.1016/j.stemcr.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J, Tan X, Li W, Wang Y, Wang J, Cheng X, Yang X. Smad4 is required for the normal organization of the cartilage growth plate. Dev Biol. 2005;284:311–322. doi: 10.1016/j.ydbio.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 100.Lizarraga G, Lichtler A, Upholt WB, Kosher RA. Studies on the role of Cux1 in regulation of the onset of joint formation in the developing limb. Dev Biol. 2002;243:44–54. doi: 10.1006/dbio.2001.0559. [DOI] [PubMed] [Google Scholar]
- 101.Iwamoto M, Higuchi Y, Koyama E, Enomoto-Iwamoto M, Kurisu K, Yeh H, Abrams WR, Rosenbloom J, Pacifici M. Transcription factor ERG variants and functional diversification of chondrocytes during limb long bone development. J Cell Biol. 2000;150:27–40. doi: 10.1083/jcb.150.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohta Y, Okabe T, Larmour C, Di Rocco A, Maijenburg MW, Phillips A, Speck NA, Wakitani S, Nakamura T, Yamada Y, et al. Articular cartilage endurance and resistance to osteoarthritic changes require transcription factor Erg. Arthritis Rheumatol. 2015;67:2679–2690. doi: 10.1002/art.39243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zulch A, Becker MB, Gruss P. Expression pattern of Irx1 and Irx2 during mouse digit development. Mech Dev. 2001;106:159–162. doi: 10.1016/s0925-4773(01)00411-7. [DOI] [PubMed] [Google Scholar]
- 104.Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Singh PN, Ray A, Azad K, Bandyopadhyay A. A comprehensive mRNA expression analysis of developing chicken articular cartilage. Gene Expr Patterns. 2016;20:22–31. doi: 10.1016/j.gep.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 106.Kunath M, Ludecke HJ, Vortkamp A. Expression of Trps1 during mouse embryonic development. Mech Dev. 2002;119(Suppl 1):S117–S120. doi: 10.1016/s0925-4773(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 107.Michikami I, Fukushi T, Honma S, Yoshioka S, Itoh S, Muragaki Y, Kurisu K, Ooshima T, Wakisaka S, Abe M. Trps1 is necessary for normal temporomandibular joint development. Cell Tissue Res. 2012;348:131–140. doi: 10.1007/s00441-012-1372-1. [DOI] [PubMed] [Google Scholar]
- 108.Maas SM, Shaw AC, Bikker H, Ludecke HJ, van der Tuin K, Badura-Stronka M, Belligni E, Biamino E, Bonati MT, Carvalho DR, et al. Phenotype and genotype in 103 patients with tricho-rhino-Phalangeal syndrome. Eur J Med Genet. 2015;58:279–292. doi: 10.1016/j.ejmg.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 109.Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, Nelson SF, Ababneh OH, Uz E, Guran T, Kayserili H, et al. Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nat Genet. 2012;44:709–713. doi: 10.1038/ng.2259. [DOI] [PubMed] [Google Scholar]
- 110.McDonald LA, Gerrelli D, Fok Y, Hurst LD, Tickle C. Comparison of Iroquois gene expression in limbs/fins of vertebrate embryos. J Anat. 2010;216:683–691. doi: 10.1111/j.1469-7580.2010.01233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Furumatsu T, Ozaki T. Epigenetic regulation in chondrogenesis. Acta Med Okayama. 2010;64:155–161. doi: 10.18926/AMO/40007. [DOI] [PubMed] [Google Scholar]
- 112.Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S, Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum. 2008;58:2743–2753. doi: 10.1002/art.23736. [DOI] [PubMed] [Google Scholar]
- 113.Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 114.Pei M, Chen D, Li J, Wei L. Histone deacetylase 4 promotes TGF-β1-induced synovium-derived stem cell chondrogenesis but inhibits chondrogenically differentiated stem cell hypertrophy. Differentiation. 2009;78:260–268. doi: 10.1016/j.diff.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 115.Kim DW, Lassar AB. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3. 2. Mol Cell Biol. 2003;23:8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem. 2004;279:47081–47091. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 117.Gao Y, Lan Y, Liu H, Jiang R. The zinc finger transcription factors Osr1 and Osr2 control synovial joint formation. Dev Biol. 2011;352:83–91. doi: 10.1016/j.ydbio.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kavanagh E, Abiri M, Bland YS, Ashhurst DE. Division and death of cells in developing synovial joints and long bones. Cell Biol Int. 2002;26:679–688. doi: 10.1006/cbir.2002.0918. [DOI] [PubMed] [Google Scholar]
- 119.Craig FM, Bayliss MT, Bentley G, Archer CW. A role for hyaluronan in joint development. J Anat. 1990;171:17–23. [PMC free article] [PubMed] [Google Scholar]
- 120.Matsumoto K, Li Y, Jakuba C, Sugiyama Y, Sayo T, Okuno M, Dealy CN, Toole BP, Takeda J, Yamaguchi Y, et al. Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development. 2009;136:2825–2835. doi: 10.1242/dev.038505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jay GD, Waller KA. The biology of Lubricin: Near frictionless joint motion. Matrix Biol. 2014;39:17–24. doi: 10.1016/j.matbio.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 123.Taguchi M, Sun YL, Zhao C, Zobitz ME, Cha CJ, Jay GD, An KN, Amadio PC. Lubricin surface modification improves extrasynovial tendon gliding in a canine model in vitro. J Bone Joint Surg Am. 2008;90:129–135. doi: 10.2106/JBJS.G.00045. [DOI] [PubMed] [Google Scholar]
- 124.Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, Sood R, Makalowska I, Baxevanis A, Johnstone B, et al. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat Genet. 1999;23:319–322. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- 125.Koyama E, Saunders C, Salhab I, Decker RS, Chen I, Um H, Pacifici M, Nah HD. Lubricin is required for the structural integrity and post-natal maintenance of TMJ. J Dent Res. 2014;93:663–670. doi: 10.1177/0022034514535807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ogawa H, Kozhemyakina E, Hung HH, Grodzinsky AJ, Lassar AB. Mechanical motion promotes expression of Prg4 in articular cartilage via multiple CREB-dependent, fluid flow shear stress-induced signaling pathways. Genes Dev. 2014;28:127–139. doi: 10.1101/gad.231969.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dowthwaite GP, Flannery CR, Flannelly J, Lewthwaite JC, Archer CW, Pitsillides AA. A mechanism underlying the movement requirement for synovial joint cavitation. Matrix Biol. 2003;22:311–322. doi: 10.1016/s0945-053x(03)00037-4. [DOI] [PubMed] [Google Scholar]
- 128.Osborne AC, Lamb KJ, Lewthwaite JC, Dowthwaite GP, Pitsillides AA. Short-term rigid and flaccid paralyses diminish growth of embryonic chick limbs and abrogate joint cavity formation but differentially preserve pre-cavitated joints. J Musculoskelet Neuronal Interact. 2002;2:448–456. [PubMed] [Google Scholar]
- 129.Lamande SR, Yuan Y, Gresshoff IL, Rowley L, Belluoccio D, Kaluarachchi K, Little CB, Botzenhart E, Zerres K, Amor DJ, et al. Mutations in TRPV4 cause an inherited arthropathy of hands and feet. Nat Genet. 2011;43:1142–1146. doi: 10.1038/ng.945. [DOI] [PubMed] [Google Scholar]
- 130.O’Conor CJ, Leddy HA, Benefield HC, Liedtke WB, Guilak F. TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proc Natl Acad Sci USA. 2014;111:1316–1321. doi: 10.1073/pnas.1319569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clark AL, Votta BJ, Kumar S, Liedtke W, Guilak F. Chondroprotective role of the osmotically sensitive ion channel transient receptor potential vanilloid 4: age- and sex-dependent progression of osteoarthritis in Trpv4-deficient mice. Arthritis Rheum. 2010;62:2973–2983. doi: 10.1002/art.27624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee W, Leddy HA, Chen Y, Lee SH, Zelenski NA, McNulty AL, Wu J, Beicker KN, Coles J, Zauscher S, et al. Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proc Natl Acad Sci USA. 2014;111:E5114–E5122. doi: 10.1073/pnas.1414298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sugimoto Y, Takimoto A, Akiyama H, Kist R, Scherer G, Nakamura T, Hiraki Y, Shukunami C. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013;140:2280–2288. doi: 10.1242/dev.096354. [DOI] [PubMed] [Google Scholar]
- 135.Liu Z, Ren Y, Mirando AJ, Wang C, Zuscik MJ, O’Keefe RJ, Hilton MJ. Notch signaling in postnatal joint chondrocytes, but not subchondral osteoblasts, is required for articular cartilage and joint maintenance. Osteoarthritis Cartilage. 2016;24:740–751. doi: 10.1016/j.joca.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang JK, Chang LH, Hung SH, Wu SC, Lee HY, Lin YS, Chen CH, Fu YC, Wang GJ, Ho ML. Parathyroid hormone 1–34 inhibits terminal differentiation of human articular chondrocytes and osteoarthritis progression in rats. Arthritis Rheum. 2009;60:3049–3060. doi: 10.1002/art.24843. [DOI] [PubMed] [Google Scholar]
- 137.Bonin CA, Lewallen EA, Baheti S, Bradley EW, Stuart MJ, Berry DJ, van Wijnen AJ, Westendorf JJ. Identification of differentially methylated regions in new genes associated with knee osteoarthritis. Gene. 2016;576:312–318. doi: 10.1016/j.gene.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rushton MD, Reynard LN, Barter MJ, Refaie R, Rankin KS, Young DA, Loughlin J. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014;66:2450–2460. doi: 10.1002/art.38713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA, Sawalha AH. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis Rheumatol. 2014;66:2804–2815. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 140.den Hollander W, Ramos YF, Bomer N, Elzinga S, van der Breggen R, Lakenberg N, de Dijcker WJ, Suchiman HE, Duijnisveld BJ, Houwing-Duistermaat JJ, et al. Transcriptional associations of osteoarthritis-mediated loss of epigenetic control in articular cartilage. Arthritis Rheumatol. 2015;67:2108–2116. doi: 10.1002/art.39162. [DOI] [PubMed] [Google Scholar]
- 141.Karlsson C, Thornemo M, Henriksson HB, Lindahl A. Identification of a stem cell niche in the zone of Ranvier within the knee joint. J Anat. 2009;215:355–363. doi: 10.1111/j.1469-7580.2009.01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, Fujioka M, Sudo A, Uchida A, Yamamoto S, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–533. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 143.van Caam A, Madej W, Thijsen E, Garcia de Vinuesa A, van den Berg W, Goumans MJ, Ten Dijke P, Blaney DE, van der Kraan P. Expression of TGFβ-family signalling components in ageing cartilage: age-related loss of TGFβ and BMP receptors. Osteoarthritis Cartilage. 2016;24:1235–1245. doi: 10.1016/j.joca.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 144.Shen J, Li J, Wang B, Jin H, Wang M, Zhang Y, Yang Y, Im HJ, O’Keefe R, Chen D. Deletion of the transforming growth factor β receptor type II gene in articular chondrocytes leads to a progressive osteoarthritis-like phenotype in mice. Arthritis Rheum. 2013;65:3107–3119. doi: 10.1002/art.38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen X, Macica CM, Dreyer BE, Hammond VE, Hens JR, Philbrick WM, Broadus AE. Initial characterization of PTH-related protein gene-driven lacZ Expression in the mouse. J Bone Miner Res. 2005;21:113–123. doi: 10.1359/JBMR.051005. [DOI] [PubMed] [Google Scholar]
- 146.Macica C, Liang G, Nasiri A, Broadus AE. Genetic evidence of the regulatory role of parathyroid hormone-related protein in articular chondrocyte maintenance in an experimental mouse model. Arthritis Rheum. 2011;63:3333–3343. doi: 10.1002/art.30515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sampson ER, Hilton MJ, Tian Y, Chen D, Schwarz EM, Mooney RA, Bukata SV, O’Keefe RJ, Awad H, Puzas JE, et al. Teriparatide as a chondrore-generative therapy for injury-induced osteoarthritis. Sci Transl Med. 2011;3:101ra193, 101ra193. doi: 10.1126/scitranslmed.3002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ailixiding M, Aibibula Z, Iwata M, Piao J, Hara Y, Koga D, Okawa A, Morita S, Asou Y. Pivotal role of Sirt6 in the crosstalk among ageing, metabolic syndrome and osteoarthritis. Biochem Biophys Res Commun. 2015;466:319–326. doi: 10.1016/j.bbrc.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 149.Fu Y, Kinter M, Hudson J, Humphries KM, Lane RS, White JR, Hakim M, Pan Y, Verdin E, Griffin TM. Aging promotes SIRT3-dependent cartilage SOD2 acetylation and osteoarthritis. Arthritis Rheumatol. 2016 doi: 10.1002/art.39618. (Epub ahead of print; February 11, 2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gabay O, Zaal KJ, Sanchez C, Dvir-Ginzberg M, Gagarina V, Song Y, He XH, McBurney MW. Sirt1-deficient mice exhibit an altered cartilage phenotype. Joint Bone Spine. 2013;80:613–620. doi: 10.1016/j.jbspin.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJR, Haughton L, Bayram Z, Boyer S, Thomson B, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117:889–897. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 152.Koelling S, Kruegel J, Irmer M, Path JR, Sadowski B, Miro X, Miosge N. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4:324–335. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 153.Li T, Longobardi L, Myers TJ, Temple JD, Chandler RL, Ozkan H, Contaldo C, Spagnoli A. Joint TGF-β type II receptor-expressing cells: ontogeny and characterization as joint progenitors. Stem Cells Dev. 2013;22:1342–1359. doi: 10.1089/scd.2012.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Karystinou A, Dell’Accio F, Kurth TB, Wackerhage H, Khan IM, Archer CW, Jones EA, Mitsiadis TA, De Bari C. Distinct mesenchymal progenitor cell subsets in the adult human synovium. Rheumatology (Oxford) 2009;48:1057–1064. doi: 10.1093/rheumatology/kep192. [DOI] [PubMed] [Google Scholar]
- 155.Kurth T, Hedbom E, Shintani N, Sugimoto M, Chen FH, Haspl M, Martinovic S, Hunziker EB. Chondrogenic potential of human synovial mesenchymal stem cells in alginate. Osteoarthritis Cartilage. 2007;15:1178–1189. doi: 10.1016/j.joca.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 156.Kurth TB, Dell’accio F, Crouch V, Augello A, Sharpe PT, De Bari C. Functional mesenchymal stem cell niches in adult mouse knee joint synovium in vivo. Arthritis Rheum. 2011;63:1289–1300. doi: 10.1002/art.30234. [DOI] [PubMed] [Google Scholar]
- 157.Liu Z, Chen J, Mirando AJ, Wang C, Zuscik MJ, O’Keefe RJ, Hilton MJ. A dual role for NOTCH signaling in joint cartilage maintenance and osteoarthritis. Sci Signal. 2015;8:ra71. doi: 10.1126/scisignal.aaa3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mirando AJ, Liu Z, Moore T, Lang A, Kohn A, Osinski AM, O’Keefe RJ, Mooney RA, Zuscik MJ, Hilton MJ. RBP-Jk-dependent Notch signaling is required for murine articular cartilage and joint maintenance. Arthritis Rheum. 2013;65:2623–2633. doi: 10.1002/art.38076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Staal B, Williams BO, Beier F, Vande Woude GF, Zhang YW. Cartilage-specific deletion of Mig-6 results in osteoarthritis-like disorder with excessive articular chondrocyte proliferation. Proc Natl Acad Sci USA. 2014;111:2590–2595. doi: 10.1073/pnas.1400744111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, O’Keefe RJ, Hilton MJ. Cartilage-specific RBPjk-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–1212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barske L, Askary A, Zuniga E, Balczerski B, Bump P, Nichols JT, Crump JG. Competition between jagged-notch and endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genet. 2016;12:e1005967. doi: 10.1371/journal.pgen.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 163.Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis Rheum. 2009;60:2723–2730. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, Little CB, Last K, Farmer PJ, Campbell IK, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–652. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 166.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, Flannery CR, Peluso D, Kanki K, Yang Z, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 167.Zhou BO, Yue R, Murphy MM, Peyer JG, Morrison SJ. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]