Abstract
A new isolate (Mg-mungbean-1) of yellow mosaic virus (YMV) was identified and characterized from mungbean growing in mid-hill condition of Meghalaya, India. Full genome of components (DNA A and DNA B; NCBI accessions number KU95030 and KU95031, respectively) of the virus were amplified through rolling circle amplification and sequenced. Both, DNA A and DNA B shared a common region (CR) with 90.4% similarity. The DNA A of Mg-mungbean-1 showed maximum (97.59%) nucleotide identity with mungbean yellow mosaic India virus (MYMIV) isolate (HF922628) reported from West Bengal, India and DNA B showed ~ 96% nucleotide identity with mungbean yellow mosaic virus (MYMV) isolates having variant DNA B. Phylogenetic tree of DNA A also identified Mg-mungbean-1 as a MYMIV. Based on DNA B the current isolate grouped with the variant Indian MYMV isolates. One recombination event in the CR of DNA B of Mg-mungbean-1 was detected, where MYMV:India:clonePB1 and MYMIV:India:cloneMBB-B31 have been identified as major and minor parents, respectively. Overall, the current study indicated occurrence of an isolate of MYMIV with a recombinant DNA B component on mungbean from mid-hills of Meghalaya, India. To the best of our knowledge this is the first molecular characterization of YMV from northeast India.
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0429-5) contains supplementary material, which is available to authorized users.
Keywords: Northeast India, MYMIV, MYMV, Mungbean, Genome, Recombination
Introduction
The yellow mosaic disease (YMD) of legumes is a major constraint for pulse production across the Indian sub-continent [28]. It affects five major crops such as mungbean, urdbean, french bean, pigeon pea and soybean along with other legumes. YMD symptoms largely depend on host species and susceptibility. On mungbean plants, it initially appears as small scattered chlorotic specks on young leaves. Later these specks coalesce to give mosaic pattern followed by complete chlorosis and necrosis. Infected plant bears few flowers and deformed pods with some poorly filled shriveled seeds. Thus, the disease affects both quality and quantity of the seed yield. Depending on the severity of the disease infection, the yield penalty may reach up to 85% [9].
At present, four distinct bipartite begomoviruses are recognized as causal agent of YMD of legumes in southern Asia [20] which are collectively known as the yellow mosaic viruses (YMVs): mungbean yellow mosaic virus (MYMV), mungbean yellow mosaic India virus (MYMIV), horsegram yellow mosaic virus (HgYMV) and dolichos yellow mosaic virus (DoYMV). The virus species belong to the genus Begomovirus in the family Geminiviridae [11] and transmitted by whitefly (Bemisia tabaci) in a persistent (circulative) manner. The members of Begomovirus has bipartite, circular single stranded DNA (cssDNA) genome encapsidated in geminate icosahedral particles (18–20 × 30 nm size) and replicate by rolling circle mechanism. The genome comprises of two components known as DNA A and DNA B, which are about 2800 nucleotides in length. The two components have a highly conserved intergenic common region (CR) containing a stem-loop structure with the invariant nonanucleotide motif (TAATATTAC) that marks the origin of virion-strand DNA replication. The larger component i.e., DNA A has seven genes or open reading frames (ORFs) which encode proteins required for encapsidation, replication and transcription regulation. The DNA B component has two ORFs that encode protein for inter and intracellular movement of the virus. Apart from it, satellite DNA molecule (~ 1300 bp length) has been found associated with the virus which is responsible for causing typical symptoms in the primary host [3]. Nucleotide sequence identity < 89% is generally considered indicative of a distinct species within the genus Begomovirus [11].
The YMVs have a very narrow host range within legumes and causes biologically indistinguishable symptoms, making specific identification of the viruses difficult [19]. They are closely related and have distinct but overlapping host ranges. In India, MYMV and MYMIV are most important as they infect large number of legumes [21]. Among them MYMIV is predominant in northern, central and eastern regions of India [27] whereas, MYMV is more ubiquitous in the southern and western regions [6, 10]. Incidence of HgYMV [1] and DoYMV has also been reported along with some reports on mixed infection from India [16].
Breeding for resistance to YMV(s) is largely advocated as the key scheme for efficient management of YMD. Thus, it is crucial to understand the relationship between the YMV(s) affecting different legumes, as well as, to consider the diversity among the various isolates of the virus too. The incidence and diversity of YMVs has been well documented from plains of India. The north- eastern region of India is a bio-diversity hotspot and possesses diverse germplasm of black gram, cow pea and rice bean. Earlier report suggested mungbean yellow mosaic virus disease as the main constraint for mungbean cultivation in Assam [17]. However, the molecular evidence on actual YMV species affecting various legumes in this region is not well documented. In the current study, the complete genome of a new YMV isolate from mungbean grown under mid-hill of Meghalaya, India has been characterized. The nucleotide identity and phylogeny of the current isolate was assessed with those of earlier reported MYMV, MYMIV, HgYMV and DoYMV. We have also performed recombination analyses for better understanding of genetic distinctiveness of DNA B component.
Materials and methods
Virus source and sequencing of viral genome
Typical YMD-like symptoms were observed on mungbean [Vigna radiata (L.) Wilczek] plants gown in the experimental field (mid-hill condition) of ICAR Research Complex for NEH Region, Umiam, Ri-Bhoi district, Meghalaya during pre-kharif (March–June) season in the year 2015. Total genomic DNA from the leaf samples was extracted using DNeasy Plant Mini Kit (Qiagen, Germany). Full length viral genomes (DNA A and DNA B) were amplified through rolling circle amplification (RCA) mechanism using REPLI-g® minikit (Qiagen, Germany). Aliquots of RCA product were digested with a range of restriction endonucleases (BamHI, BglI, EcoRI, EcoRV, HindIII, PstI, SalI, SacI and XbaI) to identify single cutting enzyme giving linear products of ~ 2700 nucleotide length. The full-length DNA A (produced using restriction enzyme EcoRV) and DNA B (produced using restriction enzyme BglI) were gel purified (Gel extraction kit, Qiagen, Germany) and sequenced bi-directionally (Xcelris Genomics, India).
Sequence analysis
Sequence data were assembled and analyzed using Bioedit (V7.2). The open reading frames (ORFs) were determined by ORF finder available at NCBI (www.ncbi.nlm.nih.gov/gorf/gorf.html). The identity and homology of the sequences were first evaluated using the BLASTN program. The full-length DNA A and DNA B sequences of Meghalaya isolate (further stated as Mg-mungbean-1) were deposited in NCBI GenBank with the accession numbers KU950430 and KU950431, respectively. For further analysis, reference sequences (DNA A and DNA B) of MYMIV, MYMV, HgYMV, DoYMV were downloaded from NCBI website (Supplementary Tables 1 & 2). The Tomato leaf curl New Delhi virus (ToLCNDV) was used as an out-group member.
Percent nucleotide identity and phylogenetic analysis
Pairwise percent nucleotide identity of both DNA A and DNA B and different ORFs were obtained using the software package SDTv1.2 [14]. The DNA A of Mg-mungbean-1was compared with 64 reported DNA A sequences: MYMIV (56), MYMV (5), HgYMV (1), DoYMV (1) and ToLCNDV (1) (Supplementary Table 1). Whereas, the DNA B of Mg-mungbean-1 was compared with 59 DNA B sequences: MYMIV (38), MYMV (18), HgYMV (1), DoYMV (1) and ToLCNDV (1) (Supplementary Table 2). DNA sequences were aligned using ClustalW algorithm of MEGA6 [25]. The Phylogenetic tree for DNA A and DNA B was constructed on the matrices of aligned sequences with1000 bootstrap replicates following neighbour-joining phylogeny of MEGA6.
Recombination analysis
Recombination analysis of DNA B of Mg-mungbean-1 was carried out using seven different methods (RDP, GENCOV, BOOTSCAN, MAXCHI, CHIMERA, SISCAN and 3SEQ) implemented in RDP4 (V4.22) [13]. On the basis of phylogenetic grouping of DNA B, 23 reference isolates identified including MYMIV (9 isolates) and MYMV (14 isolates) along with the Mg-mungbean-1 to perform the intra-component recombination (Supplementary Table 2). Recombination signals detected by at least three recombination detection methods, coupled with phylogenetic evidence of recombination, were considered as genuine recombination events.
Results
Genomic features of Mg-mungbean-1
The genomic features of Mg-mungbean-1 from Meghalaya has been presented in Supplementary Table 3. The nucleotide length was determined to be 2741 bp for DNA A and 2656 bp for DNA B. Both DNA A and DNA B encoded predicted ORFs typical of old world begomoviruses. The DNA A encodes seven ORFs, two (AV1 and AV2) in the virion-sense strand (5′–3′) and five (AC1-5) ORFs in the complementary sense strand (3′–5′). The second component DNA B encoded two predicted ORFs (BV1 and BC1), one in each orientation.
The pairwise alignment of non-coding regions between ORF AC1/AV2 in DNA A and BC1/BV1 in DNA B identified a CR in both DNA A and DNA B (Supplementary Fig. 1). The length of CR was 124 bp in DNA A (nucleotide 2644-26) and 121 bp in DNA B (nucleotide 2562-26) and they shared 90.4% nucleotide similarity to each other. The CR region of both DNA A and DNA B possessed the stem-loop structure with the loop containing the invariant nonanucleotide motif (TAATATTAC) that indicated the origin of virion-strand DNA replication. Moreover, the reported replication initiation protein binding iteron sequence (ATCGGTGT) was identified in CR of both DNA A and DNA B (Supplementary Fig. 1). However, instead of three iterons, commonly found, only two iterons has been identified in both DNA A and DNA B due to the nucleotide deletion (DNA A) and substitution (DNA B). All the iterons has been identified upstream of TATA box (TATATAT).
Percent identity and phylogeny
The full genomic component, as well as, different ORFs and CR of Mg-mungbean-1 was compared with the reference sequences. The DNA A component of Mg-mungbean-1 showed maximum (95.62–97.59%) nucleotide sequence similarity with isolate of MYMIV reported from India, Pakistan, Nepal, Bangladesh and Indonesia (Supplementary Table 4). The highest level of identity (97.59%) was recorded with an isolate of MYMIV originating from soybean from West Bengal, India (MYMIV: India: Bengal, HF922628). The percent identity with other species as follows: MYMV (82.64–86.89%) > HgYMV (82.87%) > DoYMV (64.80%) > ToLCNDV (64.08%). Comparison of different genes also showed higher identity with MYMIV isolates (> 93%) and lower levels of identity with MYMV (< 88%), HgYMV (< 86%), DoYMV (< 74%) and ToLCNDV (72%). All the genes showed highest identity with MYMIV:India:Bengal in the range of 96.60–99.26%. Among the genes, transcription activator (AC2) and replication enhancer (AC3) were found to be highly similar (> 99%) with MYMIV:India:Bengal. Interestingly, AV1 and AC5 of Mg-mungbean-1 showed 94.14 and 94.84% nucleotide identity, respectively with MYMV isolate originating from urdbean (MYMV:India:Urdbean:clone MF2, JQ398669). Rest of the genes maintained lower levels of identity with MYMV isolates (< 88%). Like the genes, the CR of DNA A of Mg-mungbean-1 showed maximum identity (88.78–98.98%) with MYMIV isolates. However, the highest identity of 98.98% was observed with most of the MYMIV isolates reported from Pakistan, Nepal and few isolates reported from Varanasi, India (Supplementary Table 4). Among the MYMIV, it showed the least identity (88.78%) with isolate reported from Bangladesh (MYMIV:Bangladesh, AF314145). On the other hand, the CR of present isolate showed highly variable identity to MYMV (47.97–71.13%), and lower identity with HgYMV (76.04%), DoYMV (44.26%) and ToLCNDV (52.85%).
A phylogenetic analysis of the complete DNA A sequence of Mg-mungbean-1 and reference isolates showed that the legume infecting YMV species (MYMIV, MYMV, HgYMV and DoYMV) clustered separately to that of begomoviruses infecting non-leguminous host (ToLCNDV) (Supplementary Fig. 2). The YMV cluster was further divided into species-specific clade, where the basal cluster was formed by DoYMV indicating it to be distinct from all other legume infecting YMV species. The DNA A of Mg-mungbean-1 clustered within MYMIV clade together with isolates originating from India (Bengal: HF922628, clone PPst 1: KP313758, Gujarat: AY937195 and AY618902), Bangladesh (AF314145) and Indonesia (JN368432-JN368439) and formed a distinct cluster basal to the remaining MYMIV DNA A sequences (Supplementary Fig. 2). These findings indicated that the YMV isolate from Meghalaya (Mg-mungbean-1) is an isolate of MYMIV, for which we proposed the isolate descriptor Mungbean yellow mosaic India virus, Meghalaya isolate (MYMIV:India: Mg-mungbean-1).
Comparison of DNA B sequence of Mg-mungbean-1 to the reference sequences indicated that it is most similar (~ 96% nucleotide sequence identity) to the sequences of DNA B components associated with MYMV isolates (LjKu01:KP319016; LjKu02:KP319017; LBG623:KF928962 and LBG623-EcoRI) from India reported to have recombinant DNA B (Supplementary Table 5). It was also noted that DNA B of Mg-mungbean-1 showed 90–94% nucleotide identity with both MYMIV and MYMV. The current isolate (Mg-mungbean-1) shared lower level of genetic identity (90.19–91.96%) with isolates reported from India, Pakistan and Nepal. Whereas, it showed higher genetic identity (94.00–94.57%) with isolates reported from Indonesia and one isolate originating from cowpea from Gujarat, India (AY937196 reported to have DNA B variant). Nucleotide identity of 92.20–94.09% was shared with MYMV isolates reported from India (Supplementary Table 5). The DNA B of Mg-mungbean-1 showed lower level of genetic identity with HgYMV (71.40%), DoYMV (61.49%) and ToLCNDV (48.78%). Both the genes in DNA B (BV1 and BC1) showed 91.83–95.43 and 95.07–97.15% nucleotide identity with MYMIV and MYMV isolates, respectively (Supplementary Table 5). Like the DNA B component, both BV1 and BC1 genes also showed lower level of genetic identity with HgYMV (73.80 and 79.15%, respectively), DoYMV (60.16 and 69.80%, respectively) and ToLCNDV (47.42 and 52.02%, respectively) (Supplementary Table 5). Interestingly, the CR of DNA B of Mg-mungbean-1 shared only 85.26–92.37% nucleotide identity with MYMIV reported from India, Pakistan and Nepal, but it had comparatively higher level of identity (96.34–96.77%) with MYMIV isolates reported from Indonesia and the DNA B variant reported from Gujarat, India (AY937196). Although the CR region of DNA B of Mg-mungbean-1 showed the highest identity (> 98%) with the recombinant MYMV DNA B, but very lower level of nucleotide identity (47.06–72.63%) was shared with remaining MYMV isolates reported from India. Thus, the DNA B of Mg-mungbean-1 showed higher level of similarity with MYMV isolates for full genome and genes, but for the CR region it showed higher similarity with MYMIV isolates.
The phylogenetic analysis based on DNA B sequences also showed species-specific clustering similar to that of DNA A (Supplementary Fig. 3). The MYMIV and MYMV isolates formed four groups viz., MYMIV(i), MYMIV(ii), MYMV(i) and MYMV(ii). The first group MYMIV(i) was composed of isolates reported from India, Pakistan and Nepal, whereas MYMIV(ii) was composed of isolates reported from Indonesia and the variant isolate reported from Gujarat, India (AY937196). In case of MYMV, the MYMV(i) was group of isolates reported from India and MYMV(ii) was of MYMV isolates having recombinant DNA B (Supplementary Fig. 3). Three groups [MYMIV(ii), MYMV(i) and MYMV(ii)] showed their position within a same sub-cluster where MYMV(ii) positioned between MYMIV(ii) and MYMV(i). The DNA B of Mg-mungbean-1 grouped with MYMV(ii) indicating variant or recombinant nature, which was also noticed during comparison of genomic components. Moreover, within MYMV(ii), Mg-mungbean-1 was the most distinct and basal to this group of sequences. Interestingly, the close association of Mg-mungbean-1 with an isolate originating from Tamil Nadu (LjKu01) indicated close relation between two isolates; those have geographical distance of 5000 km.
Intra-component recombination pattern
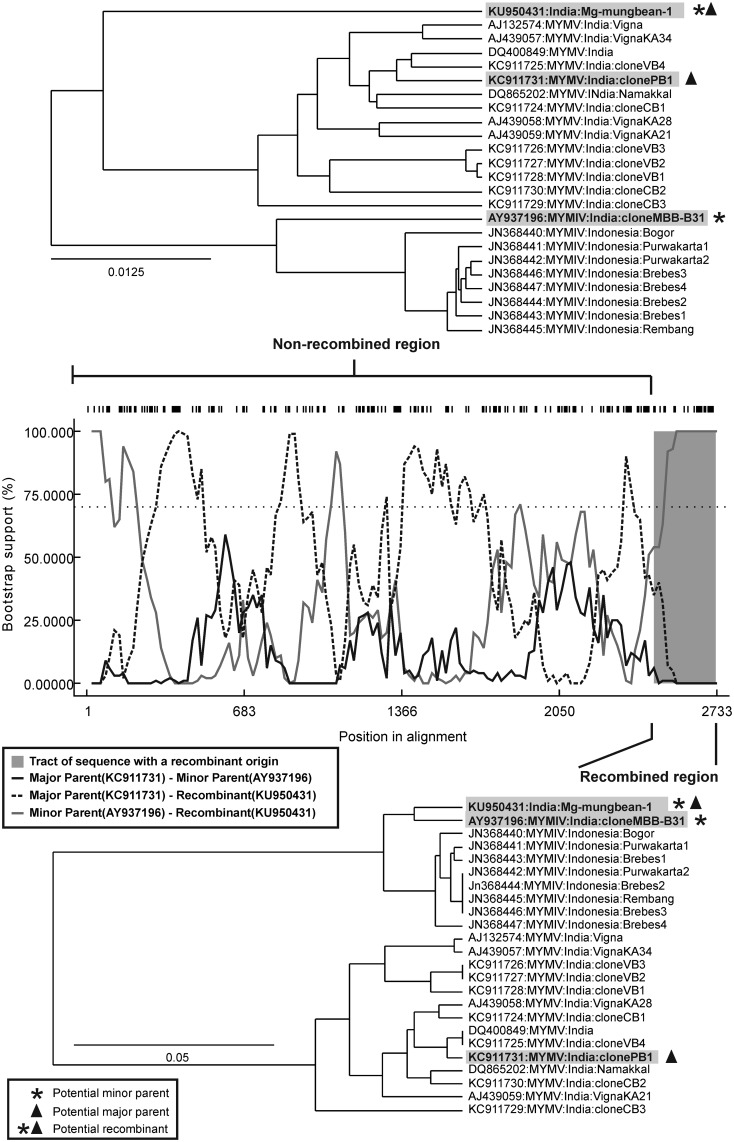
Intra-component recombination analysis was performed for DNA B of Mg-mungbean-1 including the isolates from MYMIV(ii) and MYMV(i) for better understanding on the evolution of DNA B of current isolate. In case of Mg-mungbean-1, a single intra-component recombination event was identified at nucleotide 2462-2729 (Fig. 1) corresponding to 5′ end region of CR (nucleotide position 2526-26). In this recombination event, one MYMV isolate from MYMV(i) (KC911731;MYMV:India:clonePB1) was the major parent and MYMIV:India:cloneMBB-B31 (AY937196) from MYMIV (ii) was the minor parent to Mg-mungbean-1. The P-values of different recombination methods has been presented in Supplementary Table 6. Thus, in this recombination event, a region encompassing the CR sequence of MYMV:India:clonePB1 was recombined with the corresponding region of MYMIV:India:cloneMBB-B31, resulting the recombinant DNA B of Mg-mungbean-1. Phylogenetic analysis of recombined and non-recombined regions of DNA B revealed significant differences in tree topologies (Fig. 1). The Mg-mungbean-1 clustered with major parent (MYMV:India:clonePB1) and all other isolates of MYMV(i) when only non-recombined region was considered for constructing the tree. Whereas, it clustered with minor parent (MYMIV:India:cloneMBB-B31) and other isolates of MYMIV(ii) based on the recombined region. Thus, a major genomic region of DNA B of Mg-mungbean-1 was derived from MYMV, but the CR region was replaced by MYMIV.
Fig. 1.
Recombination analysis of DNA B of Mg-mungbean-1 (KU950431). Bootstrap support plot showing a graphical overview of the recombination event in Mg-mungbean-1 at nucleotide position 2462–2729 spanning the common region (CR). Phylogenetic tree of the nucleotide sequence of the non-recombined region showing clustering of Mg-mungbean-1 with major parent (KC911731:MYMV:India:clonePB1). Phylogenetic tree of the nucleotide sequence of the recombined region showing clustering of Mg-mungbean-1 with minor parent (AY937196:MYMIV:India:cloneMBB-B31)
Discussion
In the present study, a new YMV isolate from mungbean grown under mid-hill condition of Meghalaya, India was characterized based on RCA followed by sequencing. RCA is a simple method based on the amplification of the circular DNA (Begomovirus) using bacteriophage ϕ29 DNA polymerase under an isothermal condition resulting exponential amplification of circular DNA [7]. RCA provides the opportunity to generate full genome size DNA fragments of the viruses for characterization [7, 8]. RCA of one symptomatic samples of mungbean from Meghalaya resulted in identification of two DNA molecules of ~ 2.7 Kb. Full sequence of one of them (digested with EcoRV) showed ~ 97% identity with MYMIV DNA A (HF922628), whereas the ORFs and full-length sequence analysis of other DNA molecule (digested with BglI) indicated it to be DNA B, which showed maximum nucleotide identity of ~ 96% with DNA B variants (KP319016 and KP319017). Earlier reports also showed specificity of restriction enzymes EcoRV and BglI for MYMIV DNA A and DNA B, respectively [15].
The sequence analysis showed that both DNA A and DNA B of the present isolate (Mg-mungbean-1) had all the characteristic features of begomoviruses viz., ORFs typical of Old World begomoviruses, the conserved replication initiation protein binding iterons, TATA box and stem-loop structure [4, 5, 8, 12]. The iterons located in the CR upstream of the nonanucleotide motifs are important for trans-replication. Although the sequence of the iterons of Meghalaya isolate is the same as that of earlier reported MYMV and MYMIV isolates (ATCGGTGT), but only two iterons instead of three have been identified in both DNA A and DNA B of Mg-mungbean-1. Whether these differences would prevent trans-replication remains to be determined. Further, nucleotide similarity between CR of DNA A and DNA B of Meghalaya isolate was 90.4%, which is much higher than the threshold (> 85%) for considering a DNA B molecule as cognate of DNA A [2]. Therefore, the YMV isolate from mungbean in mid-hills of Meghalaya was identified as a member of MYMIV (DNA A:KU95030 and DNA B:KU95031). Further the phylogenetic analysis of DNA A of Mg-mungbean-1 confirmed it to be a MYMIV showing clustering within MYMIV cluster. However, the DNA B of Mg-mungbean-1 grouped with variant MYMV isolates from India.
Genetic recombination is a significant contributor to the development of novel genetic variations in geminiviruses [24, 29]. In case of begomoviruses, putative recombination events happen throughout the virus genome, however, the intergenic region and AC1 are considered to be recombination hot spots [26]. In the present study, one recombination event was identified in the CR of DNA B for which MYMV:India:clonePB1 [from MYMV(i) group] and MYMIV:India:cloneMBB-B31 [from MYMIV(ii) group] were identified as major and minor parents, respectively. This clearly indicated that DNA B of Mg-mungbean-1 is a recombinant, with most of the sequence derived from MYMV but with the CR derived from MYMIV isolate reported to have variant DNA B [8]. This phenomenon of component exchange, called as pseudo-recombination for begomoviruses, is well documented [8, 22, 23]. As the interaction between replication initiation protein and iterons is highly specific, in most cases it prevents any functional interaction between components of members of distinct begomovirus species [4, 5, 18]. However, for the MYMIV:India: Mg-mungbean-1 DNA B this incompatibility might have been overcome by exchange of intergenic region/CR sequences as of earlier reported for MYMIV:India:cloneMBB-B31 (the minor parent).
Overall, the present study provided strong molecular evidence on occurrence of a MYMIV isolate associated with a recombinant DNA B in mid-hill condition of Meghalaya, India. However, to prove unequivocally that both the components; DNA A and DNA B of Mg-mungbean-1 are responsible for causing yellow mosaic symptoms in various leguminous crops, infectious clones should be produced and inoculated into test plants. To the best of our knowledge, this is the first molecular confirmation and characterization of legume infecting YMV from north-east India.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Department of Biotechnology, Government of India [grant number BT/324/NE/TBP/2012].
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13337-018-0429-5) contains supplementary material, which is available to authorized users.
References
- 1.Barnabas AD, Radhakrishnan GK, Usha R. Characterization of a begomovirus causing horsegram yellow mosaic disease in India. Eur. J. Plant Pathol. 2010;127:41–51. doi: 10.1007/s10658-009-9569-1. [DOI] [Google Scholar]
- 2.Briddon RW, Patil BL, Basavaraj B, Nawaz-ul-Rehman MS, Fauquet CM. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010;10:97. doi: 10.1186/1471-2148-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briddon RW, Stanley J. Subviral agents associated with plant single-stranded DNA viruses. Virology. 2006;344:198–210. doi: 10.1016/j.virol.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 4.Fontes EP, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 1994;269:8459–8465. [PubMed] [Google Scholar]
- 5.Fontes EP, Gladfelter HJ, Schaffer RL, Petty IT, Hanley-Bowdoin L. Geminivirus replication origins have a modular organization. Plant Cell. 1994;6:405–416. doi: 10.1105/tpc.6.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girish KR, Usha R. Molecular characterization of two soybean infecting begomoviruses from India and evidence for recombination among legume-infecting begomoviruses from South-East Asia. Virus Res. 2005;108:167–176. doi: 10.1016/j.virusres.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Inoue-Nagata AK, Albuquerque LC, Rocha WB, Nagata T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J. Virol. Methods. 2004;116:209–211. doi: 10.1016/j.jviromet.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 8.John P, Sivalingam PN, Haq QMI, Kumar N, Mishra A, Briddon RW, Malathi VG. Cowpea golden mosaic disease in Gujarat is caused by a Mungbean yellow mosaic India virus isolate with a DNA B variant. Arch. Virol. 2008;153:1359–1365. doi: 10.1007/s00705-008-0116-8. [DOI] [PubMed] [Google Scholar]
- 9.Karthikeyan A, Shobhana VG, Sudha M, Raveendran M, Senthil N, Pandiyan M, Nagarajan P. Mungbean yellow mosaic virus (MYMV): a threat to green gram (Vigna radiata) production in Asia. Int. J. Pest Manag. 2014;60:314–324. doi: 10.1080/09670874.2014.982230. [DOI] [Google Scholar]
- 10.Karthikeyan AS, Vanitharani R, Balaji V, Anuradha S, Thillaichidambaram P, Shivaprasad PV, Parameswari C, Balamani V, Saminathan M, Veluthambi K. Analysis of an isolate of Mungbean yellow mosaic virus (MYMV) with a highly variable DNA-B component. Arch. Virol. 2004;149:1643–1652. doi: 10.1007/s00705-004-0313-z. [DOI] [PubMed] [Google Scholar]
- 11.King AM, Adams MJ, Lefkowitz EJ, Carstens EB, Ball LA. Virus Taxonomy: IXth Report of the International Committee on Taxonomy of Viruses. London: Elsevier Academic Press; 2011. [Google Scholar]
- 12.Lazarowitz SG, Shepherd RJ. Geminiviruses: genome structure and gene function. Crit. Rev. Plant Sci. 1992;11:327–349. doi: 10.1080/07352689209382350. [DOI] [Google Scholar]
- 13.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer programme for analyzing recombination. Bioinfromatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naimuddin AM, Pratap A. First report of natural infection of Mungbean yellow mosaic India virus in two wild species of Vigna. New Dis. Rep. 2011;23:21–2. doi: 10.5197/j.2044-0588.2011.023.021. [DOI] [Google Scholar]
- 16.Naimuddin Akram M. Detection of mixed infection of begomoviruses in cowpea and their molecular characterization based on CP gene sequences. J. Food Legumes. 2010;23:191–195. [Google Scholar]
- 17.Nath PD, Saikia AK. Effects of time sowing on the incidence of mungbean yellow mosaic virus disease and whitefly (Bemisia tabaci Genn.) population in greengram. Ann. Agric. Res. 1995;16(4):483–484. [Google Scholar]
- 18.Orozeo BM, Gladfelter HJ, Settlage SB, Eagle PA, Gentry RN, Hanley-Bowdoin L. Multiple cis elements contribute to geminivirus origin function. Virology. 1998;242:346–356. doi: 10.1006/viro.1997.9013. [DOI] [PubMed] [Google Scholar]
- 19.Pant V, Gupta D, Choudhury NR, Malathi VG, Varma A, Mukherjee SK. Molecular characterization of the Rep protein of the blackgram isolate of Indian mungbean yellow mosaic virus. J. Gen. Virol. 2001;82:2559–2567. doi: 10.1099/0022-1317-82-10-2559. [DOI] [PubMed] [Google Scholar]
- 20.Qazi J, Ilyas M, Mansoor S, Briddon RW. Legume yellow mosaic viruses: genetically isolated begomoviruses. Mol. Plant Pathol. 2007;8:343–348. doi: 10.1111/j.1364-3703.2007.00402.x. [DOI] [PubMed] [Google Scholar]
- 21.Reddy BVB, Obaiah S, Prasanthi L, Sivaprasad Y, Sujitha A, Krishna TG. Mungbean yellow mosaic India virus is associated with yellow mosaic disease of blackgram (Vigna mungo L.) in Andhra Pradesh. India. Arch. Phytopathol. Plant Protect. 2015;48:345–353. doi: 10.1080/03235408.2014.888874. [DOI] [Google Scholar]
- 22.Roberts S, Stanley J. Lethal mutations within the conserved stem-loop of African cassava mosaic virus DNA are rapidly corrected by genomic recombination. J. Gen. Virol. 1994;75:3203–3209. doi: 10.1099/0022-1317-75-11-3203. [DOI] [PubMed] [Google Scholar]
- 23.Saunders K, Salim N, Mali VR, Malathi VG, Briddon RW, Markham PG, Stanley J. Characterization of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology. 2002;293:63–74. doi: 10.1006/viro.2001.1251. [DOI] [PubMed] [Google Scholar]
- 24.Seal SE, Van den Bosch F, Jeger MJ. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 2006;25:23–46. doi: 10.1080/07352680500365257. [DOI] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai WS, Shih SL, Venkatesan SG, Aquino MU, Green SK, Kenyon L, Jan FJ. Distribution and genetic diversity of begomoviruses infecting tomato and pepper plants in the Philippines. Ann. Appl. Biol. 2011;158:275–287. doi: 10.1111/j.1744-7348.2011.00462.x. [DOI] [Google Scholar]
- 27.Usharani KS, Surendranath B, Haq QMR, Malathi VG. Yellow mosaic virus infecting soybean in northern India is distinct from the species infecting soybean in southern and western India. Curr. Sci. 2004;86:845–850. [Google Scholar]
- 28.Varma A, Malathi VG. Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 2003;142:145–164. doi: 10.1111/j.1744-7348.2003.tb00240.x. [DOI] [Google Scholar]
- 29.Zhou X, Liu Y, Calvert L, Munoz C, Otim-Nape GW, Robinson DJ, Harrison BD. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 1997;78:2101–2111. doi: 10.1099/0022-1317-78-8-2101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.