Significance
Although there has been tremendous progress in understanding hormone action and its relationship to human physiology and disease, there has been no comprehensive approach to search the viral genome for the presence of human-like hormones. Here, using a bioinformatics approach, we have identified 16 different human peptide hormones/growth factors, including four insulin/insulin growth factor (IGF)1-like peptides (VILPs) that have homologous sequences in viruses. When these VILPs were chemically synthesized, the resulting peptides could bind to human and murine insulin and IGF1 receptors, stimulate postreceptor signaling, increase glucose uptake, and activate proliferation of cells. Injection of VILPs into mice can significantly lower the blood glucose. Thus, VILPs are members of the insulin superfamily and first characterized viral hormones.
Keywords: insulin, insulin-like growth factor, viral pathogenesis, diabetes, viral hormones
Abstract
Viruses are the most abundant biological entities and carry a wide variety of genetic material, including the ability to encode host-like proteins. Here we show that viruses carry sequences with significant homology to several human peptide hormones including insulin, insulin-like growth factors (IGF)-1 and -2, FGF-19 and -21, endothelin-1, inhibin, adiponectin, and resistin. Among the strongest homologies were those for four viral insulin/IGF-1–like peptides (VILPs), each encoded by a different member of the family Iridoviridae. VILPs show up to 50% homology to human insulin/IGF-1, contain all critical cysteine residues, and are predicted to form similar 3D structures. Chemically synthesized VILPs can bind to human and murine IGF-1/insulin receptors and stimulate receptor autophosphorylation and downstream signaling. VILPs can also increase glucose uptake in adipocytes and stimulate the proliferation of fibroblasts, and injection of VILPs into mice significantly lowers blood glucose. Transfection of mouse hepatocytes with DNA encoding a VILP also stimulates insulin/IGF-1 signaling and DNA synthesis. Human microbiome studies reveal the presence of these Iridoviridae in blood and fecal samples. Thus, VILPs are members of the insulin/IGF superfamily with the ability to be active on human and rodent cells, raising the possibility for a potential role of VILPs in human disease. Furthermore, since only 2% of viruses have been sequenced, this study raises the potential for discovery of other viral hormones which, along with known virally encoded growth factors, may modify human health and disease.
Viruses are among the most abundant biological entities (1). Humans and other animals are continuously exposed to viruses through inhalation, injection, or skin/mucosal contact. While most interactions are nonconsequential, in some cases viruses penetrate host defense barriers, and this may lead to disease through direct tissue damage or as a result of inflammatory or secondary immunological responses (2). Viruses may also manipulate their hosts by expressing host-like proteins (3–5). These can have immunomodulatory actions (6) or serve as transforming growth factors, as in the case of Simian sarcoma virus-derived platelet-derived growth factor (PDGF, v-sis) (7) and the epidermal and transforming growth factor-like molecules of vaccinia virus (8, 9). It is now clear that viruses are also present as part of a larger microbiome carried by the host in the gut as well as on skin, mucosal, and other surfaces (10).
Peptide hormones and growth factors have a central role in regulating metabolism, growth, and development. This occurs by binding to and activating specific receptors on target cells throughout the body. Since viruses have been shown to encode growth factor-like molecules, we hypothesized that, given their abundance and diversity, viruses might also produce peptide hormone-like molecules and that these molecules might have the potential to affect host pathophysiology by binding to these hormone receptors and mimicking the actions of these peptides and/or stimulating an autoimmune response.
Results
Viruses Carry Human Hormone-Like Sequences.
To begin to explore this hypothesis, we performed a comprehensive bioinformatics search for the presence of peptide sequences with homology to 62 human peptide hormones, metabolism-related cytokines, or growth factor precursors and their processed products in the viral/viroid genome database at the National Center for Biotechnology Information (NCBI), which includes the International Nucleotide Sequence Database Collaboration databases [DNA Data Bank of Japan (DDBJ) and European Molecular Biology Laboratory (EMBL) database] and GenBank (Dataset S1). Using this approach, we identified viral sequences that showed significant (e-value ≤0.06) alignment with either major domains or the full-length sequences of 16 different human peptide hormones including insulin, insulin-like growth factor-1 and -2 (IGF1 and IGF2), tumor necrosis factor, endothelin-1 and -2, transforming growth factor β-1 and -2 (TGF-β 1/2), FGF-19 and FGF-21, interleukin 6, inhibin, adiponectin, resistin, adipsin, and irisin (Dataset S2). We did not find any significant analogs in viral genomes for the other 46 human regulatory proteins searched (Dataset S3). Although the presence of TGF- and IL-6–like sequences have been reported (8, 11), our analysis identified multiple previously unidentified viral sequences that might encode peptides with hormone-like domains. In some cases, such as adiponectin, the homology involved either limited domains or repetitive sequence structure, while others, including insulin/IGF, inhibin, FGF-19 and -21, adipsin, and endothelin-1, had major domains of high sequence similarity. Most of these hormone-like sequences were identified in dsDNA viruses including Poxviruses, Herpesviruses, and Iridoviridae, which are also known for their ability to encode growth-factor and other host-like sequences (12, 13).
Members of the Iridoviridae Family Carry Genes Potentially Encoding Insulins.
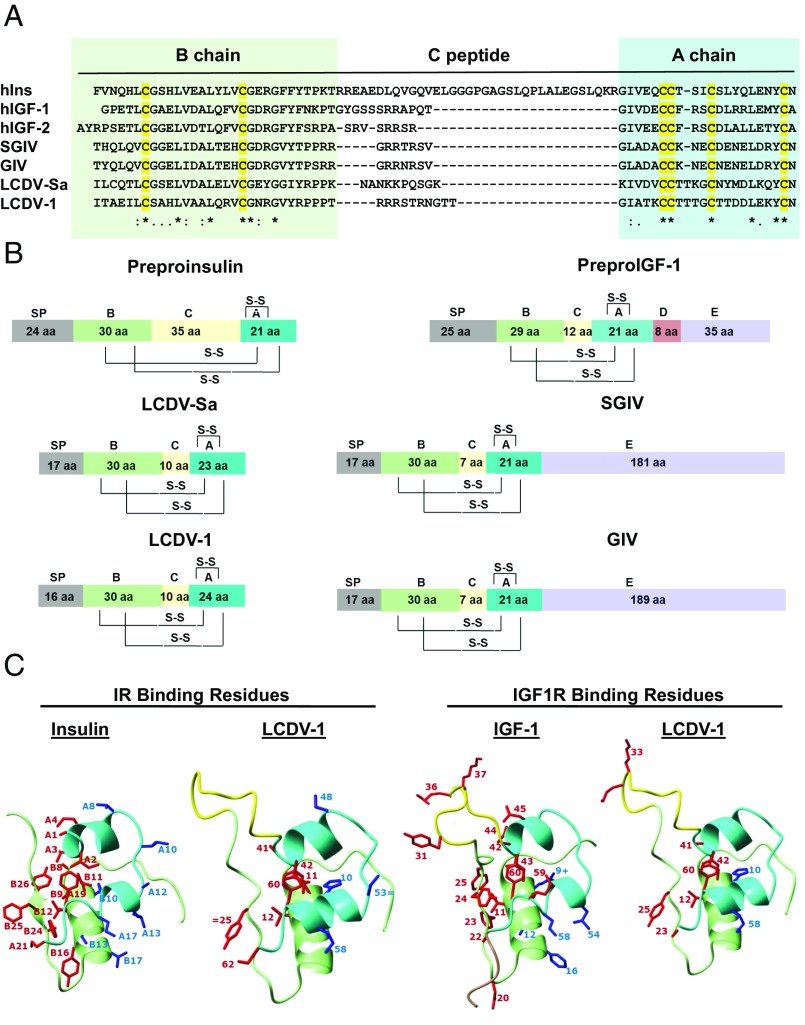
Among the sequences showing homology, one striking family was a group of four viral insulin/IGF1-like peptides (known as “VILPs” for short). These had not only a high sequence alignment across the full-length of the peptide but also conservation of all cysteine residues essential for 3D folding (Fig. 1A and Fig. S1A). VILPs were identified in lymphocystis disease virus-1 (LCDV-1), lymphocystis disease virus-Sa (LCDV-Sa), grouper iridovirus (GIV), and Singapore grouper iridovirus (SGIV) (14–17). Comparative alignment analysis showed that the A- and B-chain regions of LCDV-1 and LCDV-Sa VILPs are 52–56% and 46–47% identical to human insulin A- and B-chains, while those of GIV and SGIV VILPs are 38% and 30–33% identical, respectively. All six critical cysteine residues that form intrachain and interchain disulfide bonds in the insulin tertiary structure are conserved among the four VILPs (Fig. 1A). Each of the VILPs has a potential signal peptide, suggesting that they are likely to be secreted (Table S1). The VILPs also contain 7- to 10-aa connecting peptides, similar to IGF1 and -2. While the two LCDV VILPs are only 80 aa long with a stop codon at the end of the A-chain–like insulin, both the SGIV and GIV VILPs have potential IGF1-like E-terminal extensions (Fig. 1B); however, these extensions are not homologous with the E-domains of IGF1 and IGF2 but are caused by a variable number of tandem repeats at the C-terminal end of the proposed peptide.
Fig. 1.
Viral insulin/IGF-like peptides are structurally a part of the insulin superfamily. (A) Sequence alignment of the B-, C-, and A-chains of insulin, IGF1, IGF2, and four VILPs. Cysteines are highlighted in yellow. Identical residues are denoted by asterisks; low and high degrees of similarity are represented by a period and a colon, respectively. (B) Domain structure of human insulin, IGF1, and four VILPs. The domains are indicated as follows: A, A-chain; B, B-chain; C, C-peptide; S-S, disulfide bonds; and SP, signal peptide. (C) Predicted 3D structure of LCDV-1VILP and comparison with insulin and IGF-I. The A-chain is cyan; the B-chain is light green; the C-peptide is yellow; and the D-domain is pale brown. The conserved or conservatively substituted side chains of residues of the ligands involved in binding to site 1 of the IR/IGF1R are shown in red, and binding site 2 residues are shown in blue. Conservative substitutions are indicated by an equal sign. One substitution that increases affinity in IGF-I (B10 His to Glu) is indicated by a plus sign.
Structural modeling by I-TASSER (18) showed that the sequence of all four VILPs could be easily threaded on the canonical structure of insulin-like peptides, including the two A-chain α-helices, the central B-chain α-helix, and the positioning of the three disulfide bridges (Fig. 1C and Fig. S1 B and C). In all four VILPs, at least 50% of the receptor-binding residues are conserved or are conservatively substituted, including, for example, the equivalent to insulin’s Tyr A19 critical for insulin receptor (IR) binding and the pair of arginines in the C-domains of all VILPs (Tables S2–S5) important for IGF1 receptor (IGF1R) binding (19–21). However, some important residues are missing, such as the valine equivalent to insulin position A3 and IGF1 residue 44, suggesting these VILPs may have reduced receptor affinities. Although these viruses were isolated from fish, assessment of the VILPs in a phylogenetic context against the sequences of insulins and IGFs from 30 different species, including insects, fish, birds, and mammals, showed that VILPs are equally well-related to humans and other species as to fish insulins/IGFs (Fig. S2 and Table S6).
VILPs Can Bind to Human Insulin and IGF1 Receptors.
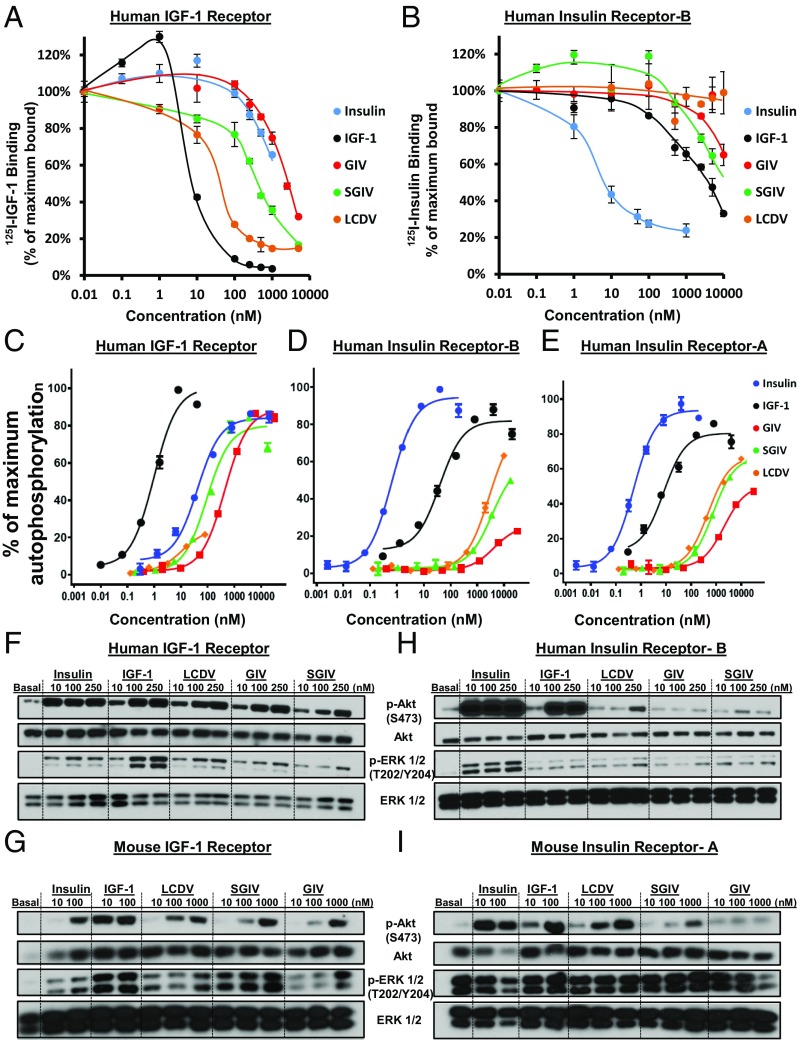
Although there is a report transfecting SGIV-VILP to fish cells showing its mitogenic effects, there is no study that characterized the all VILPs with their insulin/IGF1-like functions using mammalian/human cells and determining their insulin/IGF1 receptor binding and postreceptor signaling stimulation capacity (22). To functionally characterize these VILPs and determine if they could be active in a mammalian system, LCDV-1, SGIV, and GIV VILPs were chemically synthesized as single-chain peptides (Fig. S3) (23) and were assessed for their ability to compete with 125I-insulin and 125I-IGF1 for binding to murine brown preadipocyte cell lines in which the endogenous IR and IGF1R had been inactivated and the cells reconstituted by stable transfection with either human IR B-isoform (hIR-B) or human IGF1R (hIGF1R) (24, 25). As expected, human IGF1 (hIGF1) had highest affinity for the hIGF1R with half-maximal inhibition at ∼5 nM. LCDV-1 VILP competed with about 10-fold lower affinity (ED50 ∼50 nM), while SGIV VILP was less active (ED50 ∼500 nM), followed by GIV VILP and human insulin (Fig. 2A). Both SGIV and GIV also showed a similar ability to compete for binding to the hIR, which was slightly less than that of human IGF1 (Fig. 2B). More importantly, all VILPs were able to stimulate tyrosine autophosphorylation of human IR-A, IR-B, and IGF1R in vitro as assessed by ELISA using HEK293 cells overexpressing each receptor isoform. For IGF1R autophosphorylation, the VILPs showed activity similar to that of human insulin (Fig. 2C), whereas on human IR-A and IR-B, VILPs were about an order of magnitude less active than IGF1 and were about 200- to 500-fold less potent than insulin (Fig. 2 D and E and Table S7).
Fig. 2.
VILPs bind to hIR-B and hIGF1R and stimulate downstream insulin/IGF1-signaling pathways. (A and B) Binding competition dose–response curves showing the competing effect of VILPs for the hIGF1R (A) and hIR-B (B). The data are plotted as the percentage of the maximal binding of 125I-IGF1 alone or 125I-Insulin alone and are expressed as mean ± SEM (n = 3). (C–E) Stimulation of IR and IGF1R autophosphorylation using HEK293 cells overexpressing hIGF1R (C), hIR-B (D), or hIR-A (E) and a phosphotyrosine-specific ELISA. (F–I) Western blot analyses of the phosphorylation of Akt and ERK1/2 in lysates of murine DKO brown preadipocytes overexpressing hIGF1R cells (F), mIGF1R (G), hIR-B (H), and mIR-A (I) stimulated with the indicated concentrations of insulin, IGF1, or VILPs for 15 min.
VILPs Stimulate Downstream Insulin/IGF1-Signaling Pathways.
The two major pathways of IR and IGF1R signaling are the PI3K/Akt and the Ras-MAPK pathways. The former is responsible for most metabolic effects of these hormones; the latter is more important for cell growth and differentiation (26, 27). To determine if the VILPs could stimulate these pathways, we assessed Erk1/2 and Akt phosphorylation using double-knockout (DKO) preadipocytes overexpressing hIR-B and hIGF1R. As shown in Fig. 2F, all VILPs acting via hIGF1R strongly stimulated Akt phosphorylation at 15 min. They also stimulated Erk1/2 phosphorylation, albeit with very low potency, like insulin acting via the IGF1R. On cells expressing mouse IGF1R (mIGF1R), all VILPs produced dose-dependent stimulation of Akt phosphorylation and in this case also produced dose-dependent Erk1/2 phosphorylation, in some cases greater than that produced by human insulin (Fig. 2G). In general, the VILPS were less active on the hIR-B isoform compared with the A isoform, with LCDV-1 VILP stimulating Akt phosphorylation only at the highest concentration tested (250 nM); similar to IGF1, none produced significant stimulation of Erk1/2 phosphorylation (Fig. 2H). However, VILPs were able to stimulate postreceptor signaling on cells expressing the mouse IR-A isoform (mIR-A) with strong stimulation of Akt phosphorylation by both LCDV-1 and SGIV VILPs (Fig. 2I). While limiting material prevented studying full-time courses, it is worth noting that SGIV and GIV stimulated both Erk1/2 and Akt phosphorylation through human IR-B more robustly at 60 min than at 15 min, suggesting possible delayed kinetics of action for these ligands (Fig. S4). Thus, VILPs can activate both murine and human insulin and IGF1 receptors, and, as is consistent with binding and autophosphorylation results, VILPs stimulate postreceptor signaling better through IGF1R than through IR. It also appears that VILPs may act as biased ligands, preferentially activating the Akt pathway more than the Erk1/2 pathway, and have delayed kinetics.
VILPs Stimulate Proliferation and Glucose Uptake.
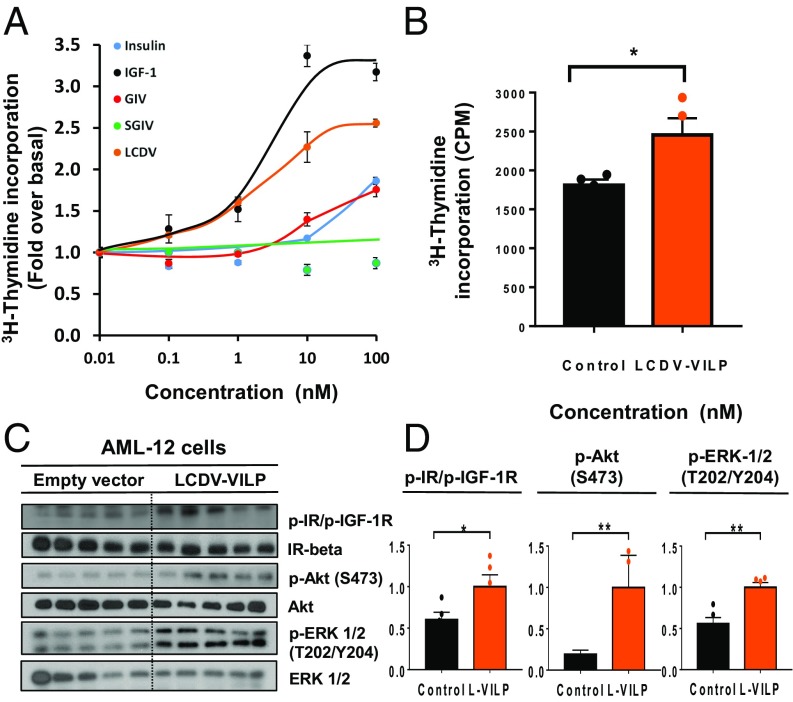
A classic effect of IGF1 is stimulation of thymidine incorporation into DNA and cell proliferation (28). LCDV-1 VILP was almost as potent as IGF1 in the stimulation of 3H-thymidine incorporation into DNA in human fibroblasts (Fig. 3A). GIV VILP also stimulated DNA synthesis, but, similar to insulin, required higher ligand concentrations. To mimic the expression of VILPs as might occur in viral infection, we transfected mouse AML-12 hepatocytes with LCDV-1 VILP cDNA with an N-terminal flag tag and assessed its effects on cellular signaling. Overexpression of LCDV-1 VILP caused a significant increase of 3H-thymidine incorporation into DNA (Fig. 3B). The LCDV-1 VILP-transfected cells also exhibited increased stimulation of IR/IGF1R autophosphorylation and phosphorylation of Akt and Erk1/2, indicating the biological potential of these peptides following viral infection of cells (Fig. 3 C and D).
Fig. 3.
Endogenous and exogenous mitogenic potency of VILPs illustrated by [3H]-thymidine incorporation and endogenous stimulation of postreceptor signaling. (A) Human fibroblasts (GM00409) were treated with increasing concentrations of the ligands, and [3H]-thymidine incorporation into DNA was assessed. Results are illustrated as the fold-increase over the basal level and are plotted as mean ± SEM (n = 4). (B) AML-12 cells were transiently transfected with either mock vector or plasmid encoding the LCDV-1 VILP gene. Results are shown as raw cpm values (*P < 0.05, t test, n = 4). (C) Immunoblotting of phosphorylation of IR/IGF1R beta subunits, Akt, and ERK1/2 in lysates from AML-12 cells transfected with mock vector or LCDV-1 VILP cDNA (n = 5). (D) Densitometric analysis of phosphorylated IR/IGF1R, Akt, and ERK1/2. Data are shown as mean ± SEM, normalized to each total protein level (*P < 0.05; **P < 0.01; t test; n = 5).
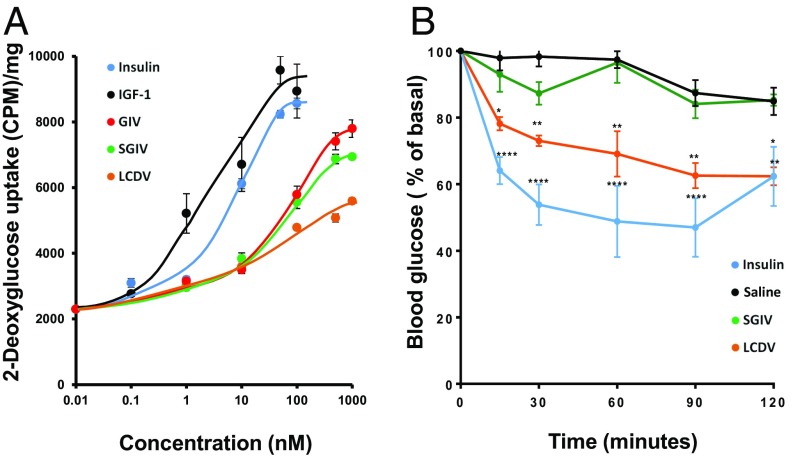
The VILPs also produced insulin-like effects on glucose metabolism in vitro and in vivo. Thus, all VILPs produced a strong dose response for stimulation of 2-deoxyglucose uptake in differentiated 3T3-L1 adipocytes (Fig. 4A). More importantly, following i.p. administration of LCDV-1 VILP, there was a significant lowering of blood glucose in mice over a 120-min time course (Fig. 4B). Interestingly, the onset of action of LCDV-1 VILP appeared to be slower but to persist longer than that of human insulin, suggesting potential differences in the in vivo kinetics of VILP action. This might be related to intrinsic differences in signaling, differences in internalization, or differences in the kinetics of clearance of these ligands. SGIV VILP also had a trend to lower blood glucose at 30 min but showed no persistent effect at the dose tested.
Fig. 4.
In vitro and in vivo effects of VILPs on glucose metabolism. (A) Differentiated 3T3-L1 cells were stimulated for 30 min with various concentrations of ligands, and uptake of 2-deoxyglucose was determined. Data are expressed as cpm/mg of protein ± SEM (n = 4). (B) Mice were injected i.p. with LCDV-1 VILP (1 µmol/kg; n = 4), SGIV VILP (1 µmol/kg; n = 4), insulin Humulin R (Eli Lilly), 6 nmol/kg; n = 6], or saline (n = 6). Blood glucose was measured at 0–120 min. Data are shown as mean ± SEM [*P < 0.05; **P < 0.01, ****P < 0.0001; two-way repeated-measures ANOVA (grouped by time) followed by Tukey 6 correction; n = 4 or 6].
Humans Are Exposed to VILP-Carrying Viruses.
Classically, Iridoviridae are known to infect fish, amphibians, and insects. The four Iridoviridae carrying VILPs had all been originally isolated from fish, where they were known to cause systemic infections with a broad range of tissue tropism, including liver, pancreas, brain, kidney, and skin, and in the last cause tumor-like growths (29). As noted above, however, the sequences of VILPs are related to human insulin/IGF1 as closely or more closely than they are to fish or other species (Fig. S2). More importantly, two recent analyses of the human fecal virome have identified DNA sequences belonging to LCDV-1 and SGIV (30, 31), and LCDV-1 has also been identified in the sequences of circulating DNAs in human blood (32, 33).
We have also reanalyzed the shotgun sequencing data from three different gut microbiome studies to search for evidence of human exposure. In a study comparing the enteric viromes from healthy children and children with islet cell autoantibodies, a marker of high risk for the development of type 1 diabetes, we were able to identify sequences of LCDV-Sa in samples of two healthy children (Dataset S4) (34). In an enteric virome study of HIV patients from Uganda, Iridoviridae had been reported as one of the most abundant 20 viral families in the gut (35), and on reanalysis we identified GIV viral sequences (Dataset S5). Last, in a study investigating the evolution of human gut virome following the same individuals for 2.5 y (36), we were also able to identify the sequences of LCDV-1. (Dataset S6). In each case, the specificity of the identified sequences to the viruses was confirmed with an additional Blastn analysis, and only unique sequences are reported in the datasets. Taken together with published data, these data support the hypothesis that humans are exposed to or can carry these Iridoviruses and that VILPs produced by these viruses could be involved in either triggering or protection from disease. Current studies of the human virome may also underestimate the presence of these viruses, since many studies use 0.2- to 0.45-µm–filtered samples, which might trap larger viruses like Iridoviridae (10).
Discussion
Insulin and IGFs have been shown to play important roles as hormonal regulators in species from Caenorhabditis elegans to Homo sapiens. Mammals have separate IRs and IGF1Rs, and, in most species, one insulin and two IGFs that interact with them. Drosophila and C. elegans, on the other hand, express multiple insulin-like peptides that interact with a single receptor tyrosine kinase (37, 38). In mammals, the IR primarily controls metabolism, and the IGF1R controls growth (24), whereas in lower organisms, the single receptor predominately regulates longevity and stress resistance (39). Across the whole phylogenetic tree, these ligands and receptors have a high degree of structural similarity, although there is considerable evolutionary diversity even among mammals. As a result, insulins and IGFs are able to bind to each other’s receptors, as do insulins and IGFs of different species, albeit with different affinities (40, 41). In this study, we demonstrate the presence of insulin/IGF1-like peptides in viruses that are active on mammalian IRs and IGF1s. These VILPs also mimic the postreceptor actions of insulin and IGF1, including stimulation of cell growth, stimulation of glucose uptake in vitro, and stimulation of glucose lowering in vivo.
Although the VILPs have higher affinity for the IGF1R than for the IR, they have conservation of structure and sequence which allows binding to both sites 1 and 2 of the IR. At the postreceptor levels, VILPs exhibit a bias to stimulation of Akt. How the VILPs might interact with IR/IGF1R hybrids, which occur in different cells to different extents, remains to be determined, but understanding how these molecules initiate insulin/IGF1 action may be useful in designing new, unique insulin analogs. Furthermore, our data suggest that VILPs, especially SGIV- and GIV-VILP, can stimulate autophosphorylation of the IR-A isoform slightly better than the IR-B isoform (compare Fig. 2 D and E). This difference was also observed in postreceptor signaling experiments in which LCDV-VILP acting on mIR-A–overexpressing preadipocytes was able to stimulate AKT phosphorylation even at the lowest concentrations (compare Fig. 2 H and I). IR-A is known to have a higher affinity for IGF1 and IGF2 than IR-B (42), suggesting that the VILPs behave more like IGF on the two IR isoforms. This could predict differences in potential effects in vivo, since IR-B is the major isoform expressed in liver and fat, while IR-A is the dominant isoform in muscle (43). In addition, IR-A tends to be enriched in various cancer cells (44). Further characterization of the IR isoform-specificity of VILPs will be needed to understand if this leads to tissue-preferential actions of VILPs and increases their potential role in cancer.
Although Iridoviridae are generally regarded as pathogens primarily in cold-blooded species (45), both our analysis and published data (30–33) indicate that DNA from the members of Iridoviridae may be found in the human fecal and blood samples. To the extent that humans might carry or be exposed to these viruses, the structural and activity similarities of VILPs to human insulin and IGF1 raise possibilities for the potential role of VILPs in hypoglycemia, insulin resistance, tumor formation, and production of/protection from type 1 diabetes. While we have not yet been able to test whether cells infected with these viruses can secrete VILPs into the bloodstream of the host, all VILPs possess an N-terminal sequence which has features of a signal peptide, and in fish which harbor these viruses the most common phenotype is the presence of tumor-like growths on the skin (45). Additionally, a previous study using grouper embryonic cells showed that SGIV VILP is transcribed at very early stages of the SGIV infection (6 h) with increasing enrichment of the transcripts at later time points, suggesting a role of VILPs in the infection-related disease process (22).
While viruses are known to encode growth factors such as PDGF(v-sis) (7), EGF (9), and TGF (8), and transfection of SGIV-VILP into fish cells has been shown to stimulate cell proliferation (22), the present study directly demonstrates that microbes—in this case four different viruses—can produce insulin-like molecules with structural similarities, receptor binding, and postreceptor actions of mammalian insulins/IGFs. Although there have been earlier suggestions for the presence of insulin-like peptides in bacteria based on activity assays (46), at least at the level of significant DNA sequence homology, our current search did not identify any insulin-like peptides in bacteria, archaea, or fungi. There are 7,455 complete viral genomes in the NCBI database as of January 11, 2018, including genomes of 21 Iridoviridae. Among these, we were able to identify VILPs in only four viruses. Thus, not all members of the Iridoviridae family carry insulin-like sequences, including some isolates of LCDV. On the other hand, it is important to keep in mind that the number of available viral genomes is still less than 2% of the predicted number of mammalian viruses (47). Thus, the viral hormones identified here may be the tip of the iceberg, with many additional viral species carrying VILPs and other hormone-like molecules.
There are also other reasons for believing the current study may underestimate the number of viral hormone-like molecules. Searching viral or microbial DNA/RNA sequences for hormone analogs is impossible for molecules other than peptides and also can fail to detect potentially active small peptide hormones, where search programs may miss homologs because of the statistical filters used when searching large viral/microbial databases. In addition, we focused only on primary sequence homology between viral sequences and human hormones, and it is well known that hormones with different primary sequences but with similar tertiary structure may also be active in receptor binding and function. Along these lines, for example, using an ACTH immunoassay, Qiang et al. (48) have identified a bacterial peptide fragment of Escherichia coli elongation factor-G that can mimic the antiinflammatory effects of α-melanocyte–stimulating hormone through the melanocortin-1 receptor. Likewise, by analysis of bacterial metagenomes, Cohen et al. (49) have shown that gastrointestinal bacteria can produce N-acyl amides that bear structural similarities to ligand of G protein-coupled receptors and can mimic their effects on GLP-1 secretion and glucose homeostasis, although such ligands would not be found searching the primary sequences of the microbial genomes.
Taken together with our studies, these findings make clear that both the viral and bacterial genomes may produce peptides and other small molecules with hormone-like sequences and activities. Further studies will be needed to determine the bioactivities of the multiple viral hormone-like sequences reported in this study. However, the identification and characterization of VILPs, as well as other viral hormone/growth factor-like molecules, not only expands our view of the insulin-like family of hormones but also indicates a potential mechanism of viral pathogenesis in which viruses encode biologically active hormone mimetics that can modify health and disease.
Methods
All animal studies were conducted in compliance with regulations and ethics guidelines of the NIH and were approved by the Institutional Animal Care and Use Committees of the Joslin Diabetes Center (no. 97-05) and Harvard Medical School (no. 05131).
Detailed materials and methods regarding bioinformatics, peptide synthesis and folding, receptor-binding and phosphorylation assays, insulin signaling, proliferation, glucose uptake experiments, plasmid transfections and insulin tolerance tests, and data availability are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Tine Glendorf (Novo Nordisk) for providing 125I-IGF-1 and 125I-insulin for binding-competition experiments; Prof. Jesse Roth, Prof. Donald Coen, and Prof. Max L. Nibert for useful discussions; Jonathan Dreyfuss (Joslin Bioinformatics Core) for comments on the database search; and Marie Solheim for help with the initial structural modeling of VILPs. This work was supported by NIH Grants R01DK031026 and R01DK033201 (to C.R.K.) and Joslin Diabetes and Endocrinology Research Center Grant P30 DK036836. E.A. was also supported by an Iacocca Family Foundation Senior Research Fellowship.
Footnotes
Conflict of interest statement: R.D. is currently an employee of Novo-Nordisk, but the work described in this paper was done in collaboration with his academic laboratory at Indiana University and has no commercial support or connection.
Data deposition: All the data used in this study, including PDB files and all the original codes that support the bioinformatics analysis in this study have been deposited with GitHub, https://github.com/jdreyf/viral-insulin-peptides. The viral genomes used in this study are available at https://www.ncbi.nlm.nih.gov/genome/viruses.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721117115/-/DCSupplemental.
References
- 1.Paez-Espino D, et al. Uncovering earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 2.Dimitrov DS. Virus entry: Molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enard D, Cai L, Gwennap C, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. eLife. 2016;5:e12469. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz F, et al. Giant viruses with an expanded complement of translation system components. Science. 2017;356:82–85. doi: 10.1126/science.aal4657. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert C, et al. Continuous influx of genetic material from host to virus populations. PLoS Genet. 2016;12:e1005838. doi: 10.1371/journal.pgen.1005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle RF, et al. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983;221:275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- 8.Brown JP, Twardzik DR, Marquardt H, Todaro GJ. Vaccinia virus encodes a polypeptide homologous to epidermal growth factor and transforming growth factor. Nature. 1985;313:491–492. doi: 10.1038/313491a0. [DOI] [PubMed] [Google Scholar]
- 9.Savory LJ, Stacker SA, Fleming SB, Niven BE, Mercer AA. Viral vascular endothelial growth factor plays a critical role in orf virus infection. J Virol. 2000;74:10699–10706. doi: 10.1128/jvi.74.22.10699-10706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki Y, et al. Angiogenesis and hematopoiesis induced by Kaposi’s sarcoma-associated herpesvirus-encoded interleukin-6. Blood. 1999;93:4034–4043. [PubMed] [Google Scholar]
- 12.Holzerlandt R, Orengo C, Kellam P, Albà MM. Identification of new herpesvirus gene homologs in the human genome. Genome Res. 2002;12:1739–1748. doi: 10.1101/gr.334302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odom MR, Hendrickson RC, Lefkowitz EJ. Poxvirus protein evolution: Family wide assessment of possible horizontal gene transfer events. Virus Res. 2009;144:233–249. doi: 10.1016/j.virusres.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tidona CA, Darai G. The complete DNA sequence of lymphocystis disease virus. Virology. 1997;230:207–216. doi: 10.1006/viro.1997.8456. [DOI] [PubMed] [Google Scholar]
- 15.Song WJ, et al. Functional genomics analysis of Singapore grouper iridovirus: Complete sequence determination and proteomic analysis. J Virol. 2004;78:12576–12590. doi: 10.1128/JVI.78.22.12576-12590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai CT, et al. Complete genome sequence of the grouper iridovirus and comparison of genomic organization with those of other iridoviruses. J Virol. 2005;79:2010–2023. doi: 10.1128/JVI.79.4.2010-2023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Bueno A, et al. Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead sea bream. J Virol. 2016;90:8768–8779. doi: 10.1128/JVI.01369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Zhang Y. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics. 2015;52:5.8.1–5.8.15. doi: 10.1002/0471250953.bi0508s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Meyts P. Insulin/receptor binding: The last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. BioEssays. 2015;37:389–397. doi: 10.1002/bies.201400190. [DOI] [PubMed] [Google Scholar]
- 20.Gauguin L, et al. Alanine scanning of a putative receptor binding surface of insulin-like growth factor-I. J Biol Chem. 2008;283:20821–20829. doi: 10.1074/jbc.M802620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menting JG, et al. Structural congruency of ligand binding to the insulin and insulin/type 1 insulin-like growth factor hybrid receptors. Structure. 2015;23:1271–1282. doi: 10.1016/j.str.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Yan Y, et al. An insulin-like growth factor homologue of Singapore grouper iridovirus modulates cell proliferation, apoptosis and enhances viral replication. J Gen Virol. 2013;94:2759–2770. doi: 10.1099/vir.0.056135-0. [DOI] [PubMed] [Google Scholar]
- 23.Zaykov AN, Mayer JP, Gelfanov VM, DiMarchi RD. Chemical synthesis of insulin analogs through a novel precursor. ACS Chem Biol. 2014;9:683–691. doi: 10.1021/cb400792s. [DOI] [PubMed] [Google Scholar]
- 24.Cai W, et al. Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression. Nat Commun. 2017;8:14892. doi: 10.1038/ncomms14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King GL, Goodman AD, Buzney S, Moses A, Kahn CR. Receptors and growth-promoting effects of insulin and insulinlike growth factors on cells from bovine retinal capillaries and aorta. J Clin Invest. 1985;75:1028–1036. doi: 10.1172/JCI111764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014;6:a009191. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucher J, Tseng YH, Kahn CR. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J Biol Chem. 2010;285:17235–17245. doi: 10.1074/jbc.M110.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King GL, Kahn CR, Rechler MM, Nissley SP. Direct demonstration of separate receptors for growth and metabolic activities of insulin and multiplication-stimulating activity (an insulinlike growth factor) using antibodies to the insulin receptor. J Clin Invest. 1980;66:130–140. doi: 10.1172/JCI109826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valverde EJ, Borrego JJ, Sarasquete MC, Ortiz-Delgado JB, Castro D. Target organs for lymphocystis disease virus replication in gilthead seabream (Sparus aurata) Vet Res (Faisalabad) 2017;48:21. doi: 10.1186/s13567-017-0428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Dinakaran V, et al. Elevated levels of circulating DNA in cardiovascular disease patients: Metagenomic profiling of microbiome in the circulation. PLoS One. 2014;9:e105221. doi: 10.1371/journal.pone.0105221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngoi CN, et al. The plasma virome of febrile adult Kenyans shows frequent parvovirus B19 infections and a novel arbovirus (Kadipiro virus) J Gen Virol. 2016;97:3359–3367. doi: 10.1099/jgv.0.000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramná L, et al. Gut virome sequencing in children with early islet autoimmunity. Diabetes Care. 2015;38:930–933. doi: 10.2337/dc14-2490. [DOI] [PubMed] [Google Scholar]
- 35.Monaco CL, et al. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minot S, et al. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duret L, Guex N, Peitsch MC, Bairoch A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. 1998;8:348–353. doi: 10.1101/gr.8.4.348. [DOI] [PubMed] [Google Scholar]
- 38.Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 40.Muggeo M, et al. The insulin receptor in vertebrates is functionally more conserved during evolution than insulin itself. Endocrinology. 1979;104:1393–1402. doi: 10.1210/endo-104-5-1393. [DOI] [PubMed] [Google Scholar]
- 41.De Meyts P, Van Obberghen E, Roth J. Mapping of the residues responsible for the negative cooperativity of the receptor-binding region of insulin. Nature. 1978;273:504–509. doi: 10.1038/273504a0. [DOI] [PubMed] [Google Scholar]
- 42.Frasca F, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–3288. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vienberg SG, et al. Receptor-isoform-selective insulin analogues give tissue-preferential effects. Biochem J. 2011;440:301–308. doi: 10.1042/BJ20110880. [DOI] [PubMed] [Google Scholar]
- 44.Heni M, et al. Insulin receptor isoforms A and B as well as insulin receptor substrates-1 and -2 are differentially expressed in prostate cancer. PLoS One. 2012;7:e50953. doi: 10.1371/journal.pone.0050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinchar VG, Waltzek TB, Subramaniam K. Ranaviruses and other members of the family Iridoviridae: Their place in the virosphere. Virology. 2017;511:259–271. doi: 10.1016/j.virol.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 46.LeRoith D, et al. Insulin-related material in microbes: Similarities and differences from mammalian insulins. Can J Biochem Cell Biol. 1985;63:839–849. doi: 10.1139/o85-106. [DOI] [PubMed] [Google Scholar]
- 47.Anthony SJ, et al. A strategy to estimate unknown viral diversity in mammals. MBio. 2013;4:e00598-13. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiang X, et al. New melanocortin-like peptide of E. coli can suppress inflammation via the mammalian melanocortin-1 receptor (MC1R): Possible endocrine-like function for microbes of the gut. NPJ Biofilms Microbiomes. 2017;3:31. doi: 10.1038/s41522-017-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen LJ, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 51.Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol. 2004;24:1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.