Significance
Deciphering which networks of genes allow cells to rapidly respond to environmental cues is key to understanding processes such as proliferation, differentiation, and death. Here, we describe a technique to quantitatively identify nascent protein synthesis using O-propargyl-puromycin (OPP), a specific covalent label of growing polypeptides, which are then isolated and measured by mass spectrometry. This technique, named OPP-mediated identification (OPP-ID), is applicable across a wide array of cell types for identification of proteins in situ. We illustrate the capability of OPP-ID by identifying a constellation of mammalian target of rapamycin (mTOR)-dependent gene networks and uncovering an array of genes involved in early erythroid progenitor expansion. OPP-ID is a powerful technique to study how cells modulate nascent protein expression to respond to stresses and stimuli.
Keywords: proteomics, translation, mTOR, erythropoiesis, puromycin
Abstract
Regulation of gene expression at the level of protein synthesis is a crucial element in driving how the genetic landscape is expressed. However, we are still limited in technologies that can quantitatively capture the immediate proteomic changes that allow cells to respond to specific stimuli. Here, we present a method to capture and identify nascent proteomes in situ across different cell types without disturbing normal growth conditions, using O-propargyl-puromycin (OPP). Cell-permeable OPP rapidly labels nascent elongating polypeptides, which are subsequently conjugated to biotin-azide, using click chemistry, and captured with streptavidin beads, followed by digestion and analysis, using liquid chromatography–tandem mass spectrometry. Our technique of OPP-mediated identification (OPP-ID) allows detection of widespread proteomic changes within a short 2-hour pulse of OPP. We illustrate our technique by recapitulating alterations of proteomic networks induced by a potent mammalian target of rapamycin inhibitor, MLN128. In addition, by employing OPP-ID, we identify more than 2,100 proteins and uncover distinct protein networks underlying early erythroid progenitor and differentiation states not amenable to alternative approaches such as amino acid analog labeling. We present OPP-ID as a method to quantitatively identify nascent proteomes across an array of biological contexts while preserving the subtleties directing signaling in the native cellular environment.
Understanding how cells swiftly direct gene expression in response to a myriad of environmental signals remains a poorly understood question. Despite advances in transcriptome analysis, transcript composition is an inaccurate surrogate for protein abundance (1) and argues for a need to comprehensively analyze gene expression at the proteomic level (2). Recent studies have uncovered translational control as a key determinant of protein abundance (3). However, quantitatively capturing de novo proteomic programs in primary cells undergoing differentiation in situ remains challenging.
Methods have evolved to quantify nascent proteomic changes in response to environmental cues. Stable isotope-labeled amino acids (4–7), bioorthogonal noncanonical amino acid tagging (BONCAT) [l-azidohomoalanine or homopropargylglycine (HPG) labeling] (8–13) or their combination (13) have been used to quantitatively identify nascent proteomes and visualize global protein synthesis in situ (14, 15). However, these methods are dependent on alteration of amino acids from cellular media, which imposes challenges on analysis of primary cell models of differentiation with fastidious, multistep media requirements. Puromycin-associated nascent chain proteomics (PUNCH-P) obviates the need for using specific growth conditions by labeling harvested ribosomes from sucrose-gradient polysome fractions with biotinylated puromycin (16). However, this technique may miss subtleties in dynamics of protein synthesis occurring uniquely within the actual cellular context.
To overcome these barriers, we set out to design a protocol that would allow identification and quantitative assessment of nascent proteomes in a short incubation time, be applicable irrespective of cellular background or culture conditions, and efficiently label proteomes to permit identification of proteomic alterations in situ. We took advantage of an analog of puromycin containing a terminal alkyne group, known as O-propargyl-puromycin (OPP) (17). As a tyrosine-tRNA mimetic, OPP can be catalytically incorporated by the ribosome into the C terminus of nascent polypeptide chains during elongation, which results in premature termination of translation and release of C-terminally truncated polypeptides bearing OPP. The alkyne group thus installed on nascent polypeptides can be conjugated to an array of biosensors, using copper(I)-catalyzed azide-alkyne cycloaddition (click chemistry), and has been used extensively to study dynamic changes in global translation both in vitro and in vivo (18–23). The combination of high cell permeability and signal-to-noise ratio makes OPP an ideal candidate compound to study nascent proteomes across a wide array of cellular types and conditions.
We wish to report a method for identifying and quantifying newly synthesized proteins, which we have termed OPP-ID (Fig. 1). In this approach, nascent polypeptides tagged with OPP are conjugated with biotin-azide and enriched using streptavidin beads. Immobilized samples are then subjected to on-bead digestion with trypsin, and the digests are analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS) to both identify and enable label-free quantification of nascent proteins. We show that OPP is robustly incorporated into a wide array of protein sizes and provides comparable coverage to BONCAT-based HPG methods. OPP-ID allows comprehensive and quantitative identification of nascent proteins in native cellular contexts, thus expanding our capability to study rapid changes in translation employed by cells to respond to stimuli such as pharmacologic perturbation or cellular differentiation.
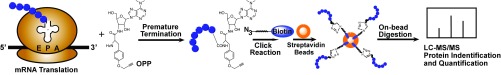
Fig. 1.
OPP-ID as a tool to identify nascent protein synthesis: By mimicking tyrosine-tRNA, OPP binds to the acceptor (A) site of the ribosome during elongation and leads to subsequent polypeptide chain termination instead of extension of the polypeptide through the peptidyl (P) site and release of tRNA at the exit (E) site. The alkyne group on OPP is conjugated to an azide group on biotin using cycloaddition. Biotin–polypeptide complexes are bound to streptavidin beads followed by on-bead digestion and analysis by LC-MS/MS for identification and quantification.
Results
Development of a method to quantify nascent protein abundance using OPP necessitated initial optimization of dosage and labeling time interval. Our goal was to identify a concentration and time pulse of OPP, which would allow observation of a maximal molecular weight diversity and fluorescence intensity of labeled proteins. K562 cells were pulsed with increasing concentrations of OPP over the course of 1 or 2 h. OPP-labeled polypeptides from cell lysates were conjugated to tetramethylrhodamine (TAMRA)-azide by cycloaddition, and fluorescence intensity was measured on equivalent total protein loading of SDS/PAGE-resolved gels. We observed a dose-dependent increase in fluorescence with a maximal intensity and protein size distribution when OPP was pulsed over the course of 2 h (Fig. S1A). The maximal duration of 2 h was chosen because prior results have shown decreased OPP labeling at 3 h of treatment (17). This label is specific to protein synthesis, as the fluorescent signal could be reduced to nonlabeled background levels with the inhibitor cycloheximide (Fig. S1A). The highest concentration of OPP used was 30 μM, which was well below published labeling doses (17) and had no effect on cell viability or markers of cellular stress, including PARP or caspase 3 cleavage or reactive oxygen species (ROS) generation (Fig. S2). We next asked whether our labeling strategy detected nascent protein synthesis at comparable levels as previously described methods employing the amino acid analog HPG. HPG-labeled proteins showed overall higher fluorescence than OPP-labeled proteins, especially for large molecular weight proteins (Fig. 2A and Fig. S1A).
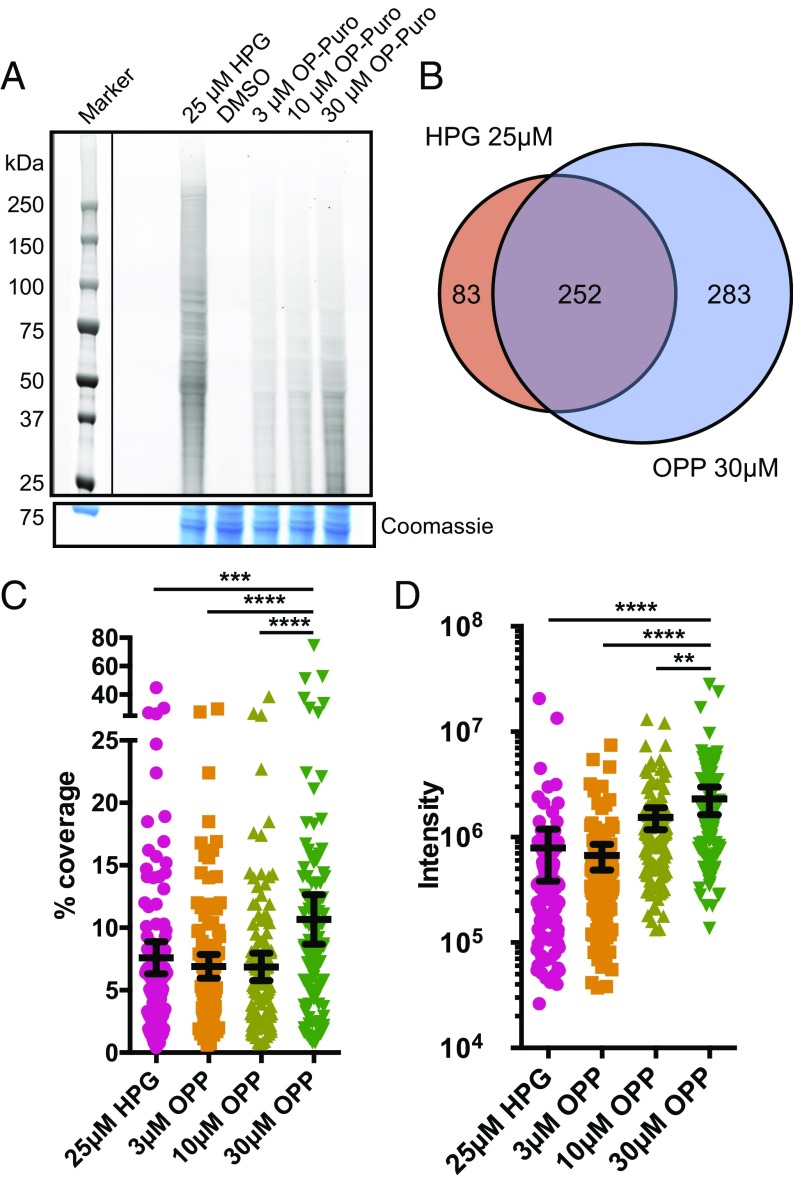
Fig. 2.
OPP labels nascent proteomes with comparable coverage, peptide intensity, and identification to HPG. (A) TAMRA-conjugated proteins labeled by specified concentrations of HPG or OPP in comparison with DMSO vehicle were isolated by SDS/PAGE and analyzed by fluorescence scan at 532 nm. Denoted box indicates overlaid image of molecular weight ladder captured at 647 nm. Representative section of Coomassie-stained gel for loading comparison. (B) Venn diagram of identified proteins in HPG 25 μM or OPP 30 μM after subtraction of overlapping proteins identified in DMSO vehicle. (C) Proteins identified in all labeling schemes were aligned, and mean percentage peptide coverage with 95% confidence interval plotted. ***P < 0.001; ****P < 0.0001. (D) Proteins identified in all labeling schemes were aligned and mean intensity (summed extracted ion chromatograms represented as arbitrary units) 95% confidence interval displayed. **P < 0.01; ****P < 0.0001.
To determine whether OPP labeling could isolate polypeptides of comparable length, protein coverage, and relative abundance to HPG-mediated labeling, we analyzed tryptic peptides obtained from HPG- or OPP-labeled proteins by LC-MS/MS (Dataset S1). Interestingly, comparison of the percentage of identified proteins across a distribution of polypeptide lengths did not show a pronounced difference between HPG and our maximal OPP dose of 30 μM (Fig. S1B). This result implies that despite the polypeptide chain terminator property of OPP, we are still able to identify proteins of a wide array of polypeptide sizes. We identified the highest number of proteins in the OPP-labeled cells, representing a 1.6× increase over HPG (535 in OPP vs. 335 in HPG) in unique proteins not found in the DMSO background dataset (Fig. 2B and Dataset S1). In addition, peptide sequence coverage of proteins identified was also augmented in an OPP dose-dependent manner to above that of HPG (Fig. 2C). Finally, to determine whether OPP labeling could be used to ascertain quantitative abundance of identified proteins, the relative abundance of identified proteins was calculated from the sum of the individual unique peptide intensities extracted ion chromatograms. We found that abundance per protein is also increased in an OPP dose-dependent manner, achieving levels above that of HPG (Fig. 2D). In summary, our method using 30 μM OPP can identify nascent polypeptides in cultured cells with comparable distribution of molecular weight, along with higher protein sequence coverage and abundance compared with that of HPG at 25 μM, without the necessity of employing specialized, amino acid-depleted media.
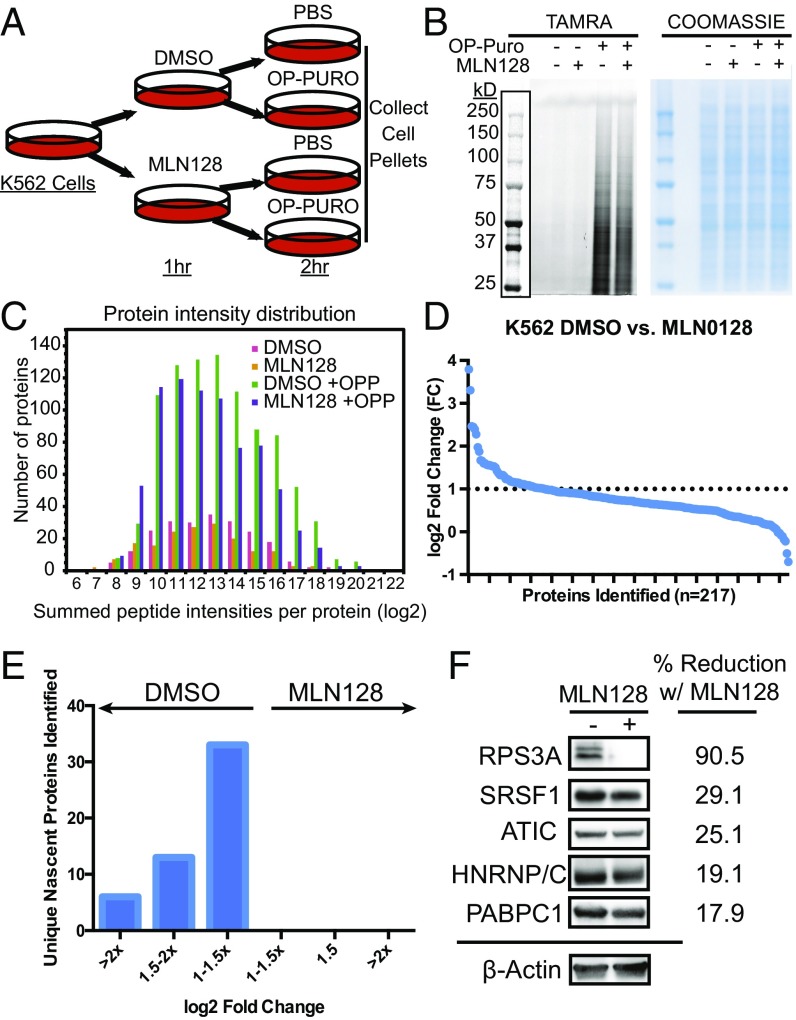
To demonstrate the utility of our technique, we asked whether OPP-ID could identify nascent proteomic changes on pharmacologic inhibition of the mammalian target of rapamycin (mTOR). mTOR is a key kinase downstream of several cues that acts as a hub to shape the cellular proteome to direct distinct molecular and cellular processes, including protein synthesis and cell growth (24, 25). Potent, specific ATP-binding site inhibitors of mTOR have been developed and extensively studied for their effect on gene expression (26–28), making these drugs ideal candidates to validate the specificity of OPP-ID in identifying nascent proteomes. To study proteomic output in the context of mTOR inhibition, we first pretreated K562 cells for 1 h with either vehicle or MLN128, an ATP-binding site inhibitor with 10-fold more potency than first-generation ATP-site inhibitors such as PP242 (28, 29). We then pulse-labeled with either vehicle or OPP for 2 h and collected cell pellets for lysis and analysis (Fig. 3A). Analysis of lysates showed a sensitive and specific OPP incorporation across an array of protein molecular weights with minimal decrease in global signal from the MLN128 treatment (Fig. 3B and Fig. S2D). Labeled protein was reacted with biotin-azide, conjugated to streptavidin beads, digested with trypsin, and analyzed using LC-MS/MS with CID on an LTQ Orbitrap Velos mass spectrometer. The experiment was performed in triplicate, and 1,001 proteins were quantified, with the majority seen in the OPP-labeled conditions (Fig. S3A). Proteins isolated from conditions without OPP were stringently removed, and nascent proteins from OPP-labeled conditions were further filtered to a total of 217 OPP-specific labeled nascent proteins (Fig. S3A). We validated efficacy of our MLN128-mediated mTOR repression by measuring reduction in phosphorylation of known downstream mTOR effectors 4EBP1, S6K, and rpS6 (Fig. S3B). To further evaluate whether our filtering strategy removed isolated proteins that were not specifically OPP-labeled, we used datasets contained in the Contaminant Repository for Affinity Purification (CRAPome) (30). We aligned our identified proteins with background contaminants from human cell experiments employing a similar streptavidin-based pull-down strategy (referred to here as the SA-CRAPome; Dataset S2). We considered proteins occurring in greater than 30% of the pull-down experiments in the SA-CRAPome as background in our experiment. Our filtering strategy effectively removed 84.4% of such background proteins (Dataset S2), confirming that our exclusion and inclusion criteria could specifically isolate OPP-labeled nascent proteomes. OPP-labeled cells showed a robust difference in the number of identified proteins across a broad distribution of peptide intensities over background conditions (Fig. 3C). Comparison of fold change of protein intensity demonstrated an array of de novo proteins inhibited by MLN128 at varying degrees (Fig. 3D). We identified 52 proteins with more than twofold repression of nascent proteins in MLN128-treated cells (Fig. 3E and Dataset S2). Proteins were further analyzed for statistical significance in up-regulation or repression in the presence of MLN128 (Fig. S3C and Dataset S2). DAVID Gene ontology analysis of proteins repressed by MLN128 strongly enriches for GO biological processes including nonsense-mediated decay, SRP protein targeting, translation initiation, and rRNA processing (Fig. S3D). Highly enriched categories of nascent proteomes inhibited by mTOR ATP site inhibitors showed strong conservation between OPP-ID and ribosome profiling-mediated experiments (28) (Fig. S3E). Western blot validation of MLN128-repressed targets of varying P values from the OPP-ID dataset showed differential levels of protein reduction with MLN128 (Fig. 3F) (31). Interestingly, qPCR of mRNA from MLN128-treated K562 cells does not implicate transcription as a mechanism for repression of specific nascent protein synthesis in these targets (Fig. S3F). This highlights an important concept that viewing gene expression strictly at the level of transcription neglects the contribution of de novo protein synthesis.
Fig. 3.
OPP-ID identifies a specific nascent proteome repressed by an active-site mTOR inhibitor. (A) Schematic for K562 cell-based analysis of the MLN128-repressed nascent proteome. (B) Cell lysates from culture conditions were analyzed for TAMRA-azide conjugated OPP proteins by SDS/PAGE with fluorescence detection at 532 nm. Denoted box indicates overlaid image of molecular weight ladder captured at 647 nm. Coomassie staining of gel for total protein loading shown adjacent to fluorescence image. (C) Plot of number of identified proteins segregated by binned average protein intensities (log2) in each of the four treatment conditions. (D) Fold change (log2) of nascent protein abundance in DMSO vehicle vs. MLN128-treated cells per individual protein. Dotted line denotes cutoff of log2 FC > 1. (E) Fold change analysis between DMSO vs. MLN128 subdivided into groups of log2 FC. (F) Western blot verification of OPP-ID identified nascent proteins with densitometric reduction normalized to beta-actin.
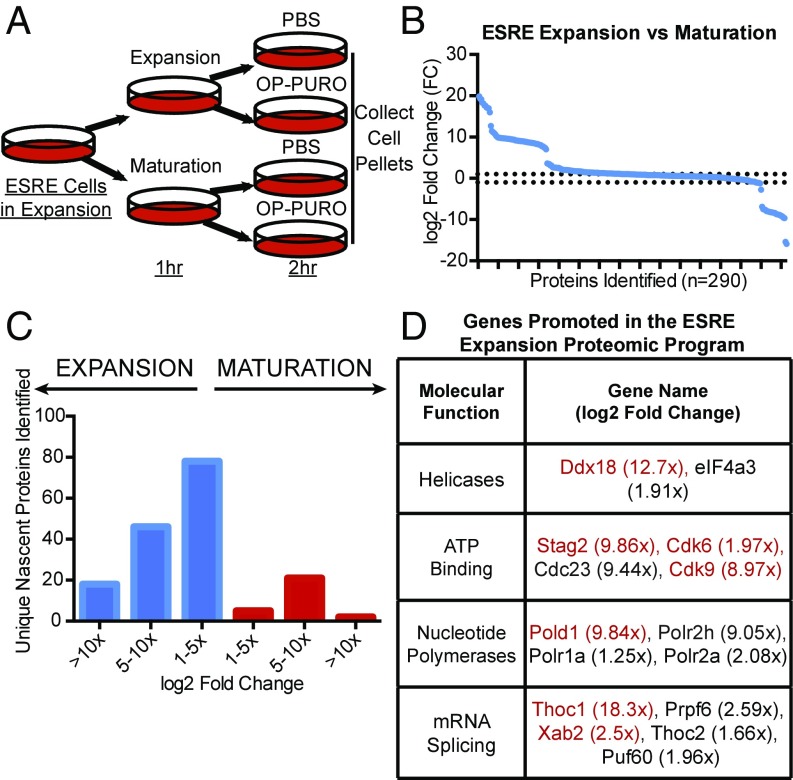
We next asked whether OPP-ID could be applied to decipher nascent proteomic changes in a model of physiologic cellular differentiation. Extensively self-renewing erythroblast (ESRE) cells are a murine yolk sac-derived cell system capable of recapitulating all phases of erythropoiesis (32–34). ESRE cells can expand in a progenitor state (predominantly proerythroblasts and basophilic erythroblasts), using a complex serum-free media containing an array of cytokines and growth factors known as expansion media. However, switching to maturation media induces terminal differentiation of erythroblasts over the course of 3 d to mature erythrocytes. Complex media requirements of this system present challenges to the use of strategies such as stable isotope-labeled amino acids or amino acid analogs. We used OPP-ID to interrogate the earliest nascent proteomic changes unique to growth conditions that maintain early erythroid precursors in comparison with conditions promoting maturation. ESRE cells were grown in expansion media, washed, and replated in either expansion or maturation media, followed by pulse labeling with either OPP or PBS vehicle (Fig. 4A). Protein lysates were processed as in prior experiments, but samples were run on a Q Exactive Plus hybrid quadrupole-orbitrap mass spectrometer with increased sensitivity, enabling quantification of 2,194 individual proteins (Fig. S4A) with minimal variance between replicates in MS1 intensity (Fig. S5). After background filtering, 290 OPP-labeled, nascently synthesized proteins were obtained (Fig. S4A and Dataset S3). After filtering proteins nonspecific to OPP-labeling, our filtering strategy removed 97.3% of mouse orthologs of our SA-CRAPome background proteins (Dataset S3). Within the first 3 h of switching growth conditions, expansion media promotes synthesis of 142 unique proteins, whereas maturation media directs synthesis of 28 unique proteins (Fig. 4 B and C). OPP-ID labeling enables LC-MS/MS-identified peptides spanning deeply across protein sequences, as shown for Ddx18, a protein promoted by expansion media (Fig. S6). Closer analysis of proteins uniquely synthesized in expansion media clusters processes of mRNA splicing, processing, export, and unwinding of secondary structure (Fig. S4B). Within these categories, known mediators of hematopoietic maintenance, expansion, and aging including Ddx18 (35), Cdk9 (36), Cdk6 (37), Stag2 (38), Thoc1 (39), Xab2 (40), and Pold1 (41) were observed (Fig. 4D). Identification of known regulators of early hematopoiesis lends credence to OPP-ID as a methodology to probe nascent proteomic programs and a platform to discover novel targets of complex biological processes.
Fig. 4.
OPP-ID identifies a specific nascent proteome inherent in maintaining expansion of ESRE erythroid precursors. (A) Schematic for ESRE-based analysis of the nascent proteome associated with either expansion or maturation media. (B) Fold change (log2) of nascent protein abundance in expansion vs. maturation media treated ESRE cells per individual protein. Dotted line denotes log2 FC > 1. (C) Fold change analysis between expansion vs. maturation subdivided into groups of log2 FC. (D) Subclasses of identified genes up-regulated in ESRE expansion media. Genes listed in red have been previously implicated in hematopoiesis.
Discussion
In this work, we describe a robust proteomic strategy to analyze newly synthesized proteins in situ. Our method is based on use of an alkyne derivative of puromycin to capture nascent proteins and mass spectrometry to establish their identity and relative amounts. Having the ability to interrogate rapid changes in gene expression networks at the protein level provides an extremely powerful tool to characterize the early cell decisions downstream of specific stimuli.
Interestingly, we observed higher fluorescence signal from HPG-labeled than from OPP-labeled proteins (Fig. 2A). This may be a result of a combination of favorable HPG incorporation in methionine-depleted media and the fact that HPG can potentially incorporate at multiple sites of a newly synthesized protein. On the contrary, OPP, as a chain terminator, can only incorporate a single label per polypeptide, which leads to lower in-gel fluorescence signal. Despite this, our OPP results show robust proteome labeling coverage with minimal perturbation of normal growth conditions in which amino acid analog labeling would not be feasible.
Strikingly, OPP-ID identified nascent proteins encoded by distinct categories of mRNA downstream of mTOR inhibition previously observed by prior ribosome profiling-mediated approaches (27, 28). However, the number of genes identified in each category was wider in ribosomal profiling. This is not surprising because of differences in cell types, slight difference in inhibitors, and the stringency in filtering protein identifications performed here. Overall, conservation of functional categories is a testament to the power of OPP-ID as an invaluable tool to directly measure differences between ribosomal occupancy and proteomic output.
Another strong advantage of techniques such as OPP-ID is that proteins with high stability may not have marked changes in abundance in response to stimuli when visualized by Western blot. Important effects on de novo synthesis of proteins with longer half-lives may be missed without techniques such as OPP-ID to detect subtle, yet important, proteomic changes (42).
The ability to interrogate rapid changes in gene expression at the level of protein production opens a window into how the cellular proteome responds to cues directing cell fates such as proliferation and differentiation. A paradigm example is erythropoiesis, which is dependent on an intricate balance of maintaining erythroid precursors and rapid differentiation to accommodate hematopoietic stresses. In this study, we employed ESRE cells to define the earliest proteomic programs unique to the decision between expanding erythroid precursors and terminal maturation. A major alteration in ESRE culture media between expansion and maturation media is the withdrawal of dexamethasone. How dexamethasone, a synthetic glucocorticoid crucial in in vitro expansion of erythroid precursors (32, 33, 43) and rescue of failed erythropoiesis in red cell aplasia disorders such as Diamond Blackfan Anemia (44–47), directs erythropoiesis at the proteomic level is still largely unknown. Using OPP-ID, we discovered a network of genes involved in splicing, nucleic acid secondary structure unwinding, and cell cycle regulation that are rapidly repressed in the absence of dexamethasone. Although we observed genes previously known to be associated with early hematopoietic precursor states (such as Ddx18, Stag2, and Pold1), we also identified multiple genes such as Prpf6, Sart1, Psen2, and Mycbp2, which have not been previously implicated in erythropoiesis. In future studies, it will be intriguing to decipher how these factors maintain early erythroid precursors and how dexamethasone supports their expression.
Given the ability of OPP to rapidly incorporate into multiple tissues in vivo after i.p. injection (17), OPP-ID has the potential to ask innumerable questions on dynamic proteomic networks at the organismal level. Combining the universal applicability of OPP-ID with other next-generation techniques such as RNAseq or ribosome profiling will create a powerful toolbox to study early cellular events in signal transduction, differentiation, and oncogenesis.
Materials and Methods
Cell Culture.
K562 cells were cultured in RPMI media 1640-GlutaMax (Gibco) supplemented with 10% FBS (Axenia BioLogix), 100 U/mL penicillin, and 100 μg/mL streptomycin. HPG experiments were performed using methionine-free RPMI (Invitrogen) with the supplementation listed here and 25 μM HPG (Invitrogen). ESRE cells were cultured as previously described (33). Briefly, ESREs were expanded in expansion media consisting of StemSpan SFE (Stem Cell Technologies), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 U/mL erythropoietin (Peprotech), 100 ng/mL stem cell factor (SCF) (Peprotech), 1 × 10−6 M dexamethasone (Sigma), 40 ng/mL IGF1 (Stem Cell Technologies), and 0.4% ExCyte (Millipore) grown on gelatin-coated tissue culture dishes. Maturation of ESREs was driven by washing cells and switching media to maturation media consisting of 1× Iscove’s modified Dulbecco’s medium (IMDM), 2 U/mL erythropoietin, 100 ng/mL SCF, 10% serum replacement (Invitrogen), 5% plasma-derived serum (PDS) (Animal Technologies), 1× glutamine, 10% protein-free hybridoma medium (PFHM)-II (Gibco) and 12.7 μL/100 mL of 1:10 MTG (Sigma). Culture density was kept <2 × 106/mL. All cells were maintained in a humidified 37 °C incubator with 5% CO2.
Treatment of Cells with Probes and Preparation of Cell Lysates.
For labeling experiments, K562 or ESRE cells were cultured to a density of 1.2 × 106/mL in a total of 20 mL media. In the OPP dose titration experiment, cells were treated directly with 3–30 μM OPP (MedChem) for 1 or 2 h in the full medium at 37 °C. For labeling of mTOR-dependent nascent proteomes, K562 cells were cultured with either DMSO vehicle or MLN128 (300 nM) for 1 h. For labeling of nascent maturation proteomes, ESRE cells were cultured with either expansion or maturation media (see earlier) for 1 h. Cultures were then treated with either PBS or OPP (30 μM) for 2 h with intermittent gentle agitation. After treatment, cells were collected by centrifugation at 300 × g at 4 °C and then resuspended in lysis buffer containing 100 mM Hepes at pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 2 mM PMSF, and 1× complete EDTA-free protease inhibitor mixture (Roche). Cellular debris was removed by centrifugation at 16,000 × g for 30 min at 4 °C. Protein concentration was determined by the BCA assay (Bio-Rad). Cell lysates were diluted with lysis buffer to give a final protein concentration of 1.8 mg/mL (1.2 mL total volume) or 1.6 mg/mL (0.4 mL total volume) for the K562 or ESRE cells, respectively, for the subsequent Cu(I)-catalyzed click reactions. Portions of the samples to be reacted with biotin-azide for affinity purification were precleared by overnight incubation with high-capacity streptavidin agarose resin (Thermo Scientific) at 4 °C with slow rotation.
Click Conjugation with TAMRA-Azide or Biotin-Azide.
For click chemistry, a reagent mix was freshly prepared by sequential addition of stock solutions to achieve 6.25% SDS, 0.625 mM TAMRA-azide, or biotin-azide (Thermo Scientific), 6.25 mM TCEP (Sigma), 0.625 mM TBTA (Sigma), and 6.25 mM CuSO4. Appropriate volumes of reagent mix were then delivered to lysates (1:5.25, vol/vol) to achieve 1% SDS, 100 μM TAMRA-azide or biotin-azide, 1 mM TCEP, 100 μM TBTA, and 1 mM CuSO4. Typically, 42 μL lysate was reacted for fluorescence visualization and 0.4–1.0 mL for affinity purification. After 1.5 h at room temperature, reactions were quenched as described here.
In-Gel Fluorescence Visualization.
After click conjugation with TAMRA-azide, 15 μL samples were treated with 4× Laemmli sample buffer and resolved by SDS/PAGE. Gels were scanned for fluorescently labeled proteins using excitation wavelength 532 nm, and also for the molecular weight marker, using excitation wavelength 647 nm (Typhoon Imaging System; Molecular Dynamics), then stained with Coomassie blue reagent.
Streptavidin Affinity Enrichment of Biotinylated Proteins.
After click conjugation with biotin-azide, samples were mixed with 5 volumes cold acetone and stored overnight (−20 °C). Precipitated proteins were pelleted by centrifugation at 3,500 × g at 4 °C for 5 min and washed twice with 2 mL cold methanol. Protein pellets were resuspended in 120 µL PBS containing 1% SDS and desalted by passing through Zeba spin 0.5 mL 7K cutoff desalting columns (Thermo Scientific) equilibrated with 1% Nonidet P-40, 0.1% SDS in PBS. Protein concentrations were measured (BCA assay) and normalized to the lowest concentration by dilution with the elution buffer. Labeled proteins (typically in 80 µL total volume) were then immobilized with magnetic streptavidin beads (typically from 80 μL slurry; Pierce) at 4 °C overnight with slow rotation. Beads were washed twice with 0.5 mL 1% Nonidet P-40, 0.1% SDS in PBS for 10 min, three times with 0.5 mL ice-cold 6 M urea in PBS for 15 min, and three times with 0.5 mL ice-cold PBS for 10 min, all at 4 °C with slow rotation. Finally, beads were rinsed with buffer containing 20 mM Tris⋅HCl at pH 8.0 and 2 mM CaCl2.
On-Bead Trypsin Digestion of Pull-Down Products.
Proteins immobilized on magnetic streptavidin beads were suspended in trypsinization buffer (20 mM Tris⋅HCl at pH 8.0 and 2 mM CaCl2). Samples were treated with 5 mM DTT at 56 °C for 30 min, followed by alkylation with 20 mM iodoacetamide at room temperature for 30 min in the dark. Samples were treated once more with 5 mM DTT (5 min), followed by incubation with 500 ng sequencing grade trypsin (Promega) overnight at 37 °C on a shaking incubator. Digestion was stopped by adding 1% formic acid. The resulting peptides were enriched with C18 Omix Tips (Agilent Technologies) and eluted with 50% acetonitrile containing 0.1% formic acid. The samples were dried down by SpeedVac and then resuspended in 0.1% formic acid for analysis by LC-MS/MS.
LC-MS/MS Analysis.
Peptides resulting from trypsinization were analyzed on an LTQ-Orbitrap Velos or a Q Exactive Plus hybrid quadrupole-orbitrap mass spectrometer (both from Thermo Fisher Scientific) connected to a NanoAcquity Ultra Performance UPLC system (Waters). An EASY-Spray PepMap RSLC C18, 3 μm, 75 μm × 15 cm column (Thermo Scientific) was used to resolve peptides using a binary solvent system consisting of 0.1% formic acid in water as mobile phase A and 0.1% formic acid in acetonitrile as mobile phase B. With the LTQ-Orbitrap Velos, typical 90-min gradients were as follows: load at 2% B at 600 nL/min for 18 min, reduce flow rate to 300 nL/min over 2 min, gradient from 2 to 30% B over 57 min, increase to 50% B over 2 min, decrease to 2% B over 2 min, and equilibrate at 2% B for 9 min. Slightly different elution conditions were used on the Q Exactive Plus: load at 2% B at 600 nL/min for 15 min, reduce flow to 400 nL/min, gradient from 2 to 25% B over 48 min, increase to 37% B over 6 min, increase to 40% B over 3 min, elute with 40–80% B over 3 min, decrease to 2% B over 3 min, and equilibrate to 2% B over 13 min. Both mass spectrometers were operated in the data-dependent mode to automatically switch between MS and MS/MS. On the LTQ-Orbitrap Velos (full MS resolution, 30,000), the top six precursor ions with a charge state of 2+ or higher were selected with an isolation width of 3.0 m/z and fragmented by CID, using a dynamic exclusion time of 60.0 s. On the Q Exactive Plus (full MS resolution, 70,000), the top 10 precursor ions with a charge state of 2+ or higher were selected with an isolation width of 4.0 m/z and fragmented by HCD (resolution, 17,500), with a 15.0 s dynamic exclusion time. Using Protein Prospector (48) (v 5.19.1), all peak lists from LC-MS/MS runs in a given experiment were searched in combination against the appropriate SwissProt database (2015.12.1), either human (20,194/549,832 entries searched) or mouse (16,740/549,832 entries searched), concatenated with a corresponding random decoy database to allow for FDR determinations. The database search was performed with the following parameters: a mass tolerance of 20 ppm for precursor masses, ±0.8 Da for MS/MS fragment ions on the LTQ-Orbitrap Velos or 30 ppm for MS/MS fragment ions on the Q Exactive Plus, cysteine carbamidomethylation as a fixed modification, and several common variable modifications (including acetylation of protein N termini, oxidation of methionine, and cyclization of N-terminal glutamine). Up to two modifications per peptide were allowed, and the enzyme was specified as trypsin, with one missed cleavage allowed. After homology filtering, we identified a total of 1,063 and 2,264 proteins in our K562 (LTQ-Orbitrap Velos) and ESRE (Q Exactive Plus) experiments, respectively, both at a protein FDR of <0.4%. For details on methods using label-free quantification and filtering of MS results see SI Materials and Methods.
Western Blot.
Western blot analysis was performed as previously described (31) with antibodies specific for 4EBP1, p-4EBP1Thr37/46, rpS6, p-rpS6Ser235/236, S6K, p-S6KThr389, PABPC1, PARP, Caspase 3 (cell signaling), β-actin (Sigma), HNRNP/C, ATIC, RPS3A (Abcam), and SRSF1 (Santa Cruz). Densitometry analysis was completed using ImageLab (Bio-Rad).
ROS Analysis.
ROS was analyzed using the DFCDA Cellular ROS Detection kit (Abcam) per provided protocol. K562 cells were treated with either H2O2 (250 µM) for a 24-h pretreatment or a 2-h pulse of PBS, OP-Puro (30 µM). Mock sample was treated with PBS, but without DFCDA reagent. Samples were analyzed by flow cytometry using BD LSRII.
qPCR Analysis.
RNA was isolated using the manufacturer’s protocol for RNA extraction with TRIzol reagent (Invitrogen) and RNA was DNase-treated with TURBO DNase (Thermo Fisher). DNase-treated RNA was transcribed to cDNA (cDNA) with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), and 1 µL cDNA was used to run each SYBR Green detection qPCR assay (SYBR Green Supermix and MyiQ2; Bio-Rad). Primers were used at 200 nM. Oligomer sequences were designed to span exon-exon junctions using Primer Blast and are listed in Fig. S7.
Supplementary Material
Acknowledgments
C.M.F. is funded by the Campini Foundation, The Leukemia and Lymphoma Foundation Career Development Grant, and University of California, San Francisco, Department of Pediatrics K12 (Award 5K12HDO72222-05). A.L.B., N.J.P., A.U., R.J.C., and J.A.O.-P. are funded by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, Howard Hughes Medical Institute, and NIH Grants 8P41GM103481 and 1S10OD16229. D.R. is funded by NIH Grants R01CA140456, R01CA184624, and R01CA154916.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.T.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707514115/-/DCSupplemental.
References
- 1.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowling H, et al. Antipsychotics activate mTORC1-dependent translation to enhance neuronal morphological complexity. Sci Signal. 2014;7:ra4. doi: 10.1126/scisignal.2004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jovanovic M, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: Application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol Cell Proteomics. 2008;7:1067–1076. doi: 10.1074/mcp.M700387-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong S-E, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beatty KE, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 10.Hatzenpichler R, et al. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol. 2014;16:2568–2590. doi: 10.1111/1462-2920.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballikaya S, et al. De Novo proteome analysis of genetically modified tumor cells by a metabolic labeling/azide-alkyne cycloaddition approach. Mol Cell Proteomics. 2014;13:3446–3456. doi: 10.1074/mcp.M113.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howden AJM, et al. QuaNCAT: Quantitating proteome dynamics in primary cells. Nat Methods. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.tom Dieck S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Methods. 2015;12:411–414. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ullrich M, et al. Bio-orthogonal labeling as a tool to visualize and identify newly synthesized proteins in Caenorhabditis elegans. Nat Protoc. 2014;9:2237–2255. doi: 10.1038/nprot.2014.150. [DOI] [PubMed] [Google Scholar]
- 16.Aviner R, Geiger T, Elroy-Stein O. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 2013;27:1834–1844. doi: 10.1101/gad.219105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider RK, et al. Rps14 haploinsufficiency causes a block in erythroid differentiation mediated by S100A8 and S100A9. Nat Med. 2016;22:288–297. doi: 10.1038/nm.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Signer RAJ, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Signer RAJ, et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 2016;30:1698–1703. doi: 10.1101/gad.282756.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco S, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–340. doi: 10.1038/nature18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hettmer S, et al. Functional genomic screening reveals asparagine dependence as a metabolic vulnerability in sarcoma. eLife. 2015;4:e09436. doi: 10.7554/eLife.09436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mateju D, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer. 2011;105:329–336. doi: 10.1038/bjc.2011.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126:1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoreen CC, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janes MR, et al. Efficacy of the investigational mTOR kinase inhibitor MLN0128/INK128 in models of B-cell acute lymphoblastic leukemia. Leukemia. 2013;27:586–594. doi: 10.1038/leu.2012.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellacheruvu D, et al. The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim AR, et al. Bmi-1 regulates extensive erythroid self-renewal. Stem Cell Reports. 2015;4:995–1003. doi: 10.1016/j.stemcr.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.England SJ, McGrath KE, Frame JM, Palis J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood. 2011;117:2708–2717. doi: 10.1182/blood-2010-07-299743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17:343–352. doi: 10.1016/j.cmet.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne EM, et al. Ddx18 is essential for cell-cycle progression in zebrafish hematopoietic cells and is mutated in human AML. Blood. 2011;118:903–915. doi: 10.1182/blood-2010-11-318022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottardi S, et al. The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. PLoS Genet. 2014;10:e1004827. doi: 10.1371/journal.pgen.1004827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurenti E, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015;16:302–313. doi: 10.1016/j.stem.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullenders J, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pitzonka L, et al. The Thoc1 encoded ribonucleoprotein is required for myeloid progenitor cell homeostasis in the adult mouse. PLoS One. 2014;9:e97628. doi: 10.1371/journal.pone.0097628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers SM, et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sincennes M-C, et al. The LMO2 oncogene regulates DNA replication in hematopoietic cells. Proc Natl Acad Sci USA. 2016;113:1393–1398. doi: 10.1073/pnas.1515071113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aebersold R, Burlingame AL, Bradshaw RA. Western blots versus selected reaction monitoring assays: Time to turn the tables? Mol Cell Proteomics. 2013;12:2381–2382. doi: 10.1074/mcp.E113.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Lindern M, et al. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-Kit to enhance and sustain proliferation of erythroid progenitors in vitro. Blood. 1999;94:550–559. [PubMed] [Google Scholar]
- 44.Narla A, et al. Dexamethasone and lenalidomide have distinct functional effects on erythropoiesis. Blood. 2011;118:2296–2304. doi: 10.1182/blood-2010-11-318543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narla A, Vlachos A, Nathan DG. Diamond Blackfan anemia treatment: Past, present, and future. Semin Hematol. 2011;48:117–123. doi: 10.1053/j.seminhematol.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebert BL, et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: Identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flygare J, Karlsson S. Diamond-Blackfan anemia: Erythropoiesis lost in translation. Blood. 2007;109:3152–3154. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Q, et al. Broad-spectrum kinase profiling in live cells with lysine-targeted sulfonyl fluoride probes. J Am Chem Soc. 2017;139:680–685. doi: 10.1021/jacs.6b08536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.