Abstract
Background
Abundant aspartyl-asparaginyl-β-hydroxylase (ASPH) expression supports robust neuronal migration during development, and reduced ASPH expression and function, as occur in fetal alcohol spectrum disorder, impair cerebellar neuron migration. ASPH mediates its effects on cell migration via hydroxylation-dependent activation of Notch signaling networks. Insulin and Insulin-like growth factor (IGF-1) stimulate ASPH mRNA transcription and enhance ASPH protein expression by inhibiting Glycogen Synthase Kinase–3β (GSK-3β). This study examines the role of direct GSK-3β phosphorylation as a modulator of ASPH protein expression and function in human cerebellar-derived PNET2 cells.
Methods
Predicted phosphorylation sites encoded by human ASPH were ablated by S/T→A site-directed mutagenesis of an N-Myc-tagged wildtype (WT) cDNA regulated by a CMV promoter. Phenotypic and functional features were assessed in transiently transfected PNET2 cells.
Results
Cells transfected with WT ASPH had increased ASPH protein expression, directional motility, Notch-1 and Jagged-1 expression, and catalytic activity relative to control. Although most single- and multi-point ASPH mutants also had increased ASPH protein expression, their effects on Notch and Jagged expression, directional motility and adhesion, and catalytic activity varied such that only a few of the cDNA constructs conferred functional advantages over WT. Immunofluorescence studies showed that ASPH phosphorylation site deletions can alter the subcellular distribution of ASPH and therefore its potential interactions with Notch/Jagged at the cell surface.
Conclusions
Inhibition of ASPH phosphorylation enhances ASPH protein expression, but attendant alterations in intra-cellular trafficking may govern the functional consequences in relation to neuronal migration, adhesion and Notch activated signaling.
Keywords: Neuronal migration, Phosphorylation, Aspartyl-asparaginyl-β- hydroxylase, Hydroxylation, Notch, Cell migration
Introduction
Fetal alcohol spectrum disorder (FASD) is the most common preventable cause of neurodevelopmental birth defects [1,2] leading to sustained cognitive-motor dysfunction [3-5]. Alcohol mediated teratogenesis of the cerebellum causes sustained impairments in motor function. Prenatal alcohol exposures, whether chronic or binge, disrupt neuronal growth, survival, migration, and plasticity in various brain regions, including the cerebellum [6-8]. Previous studies linked impairments in neuronal migration to inhibition of insulin and insulin-like growth factor, type 1 (IGF-1) signaling and activation of phosphoinositol- 3-kinase (PI3K) and Akt pathways that regulate expression of aspartyl-asparaginyl-β- hydroxylase (ASPH; formally AAH) [8-10]. ASPH has a demonstrated role in cell motility and invasion [11-14]. High levels of ASPH, mediated by increased signaling through insulin/IGF-1 receptors, insulin receptor substrate (IRS) proteins, and PI3K-Akt (AKT signalling pathway), or overexpression of ASPH, increase cell motility, whereas low levels of ASPH induced by siRNA [10] small molecule inhibitors of ASPH’s catalytic activity [15] or impaired signaling through insulin/IGF-IRS-PI3K-Akt [8] reduce cell motility.
ASPH is an ~86 kD [16] type 2 trans-membrane protein located within the endoplasmic reticulum (ER) [17,18]. In immature cells and transformed malignant neoplastic cells, ASPH is also expressed along the plasma membrane [14,16]. ASPH is physiologically cleaved into a ~30-34 kD N-terminal fragment that is identical to Humbug, a truncated isoform that binds to calcium and promotes cell adhesion [19,20] and a ~52- 56 kD catalytically active C-terminal region that contains a 675-His residue that is essential for hydroxylation [21,22]. ASPH hydroxylates β-carbons of specific aspartate (Asp) and asparagine (Asn) residues within consensus sequences of epidermal growth factor (EGF)-like domains of target molecules [23] such as Notch and Jagged [24]. Furthermore, the findings that, A) the consensus sequences for ASPH hydroxylation are expressed in Notch-1 and Jagged-1, its natural ligand [24,25]. B. ASPH physically interacts with both Notch-1 and Jagged-1 [11]; and C) ASPH, Notch-1, and Jagged-1 are abundantly expressed in cells that are highly motile including placental trophoblasts, malignant neoplasms, and immature neuronal cells of central nervous system (CNS) origin [11,14,15,26] support the concept that ASPH functionally modulates notch signaling networks [10,11,27].
Notch signaling is activated by the binding of Jagged (or Delta-like proteins) to its extracellular domain, triggering metalloprotease- dependent cleavage of Notch’s membrane-associated extracellular domain, followed by rapid presenilin protease cleavage of its intracellular domain [28]. The free Notch intracellular domain (NID) translocates to the nucleus where it complexes with the CBF1 transcriptional activator (Centromere binding factor), suppressor of hairless or lag-1 (CSL), and transcriptional co-activators of the mastermind- like family of proteins. The NID-complex then binds to CSL recognition sequences on DNA leading to recruitment of additional factors, followed by increased transcription of downstream target genes, such as hairy and enhancer of split-1 (HES-1) and HES- related proteins [28,29].
ASPH gene expression is regulated by insulin/IGF signaling through ERK/MAPK [30-32] (extracellular signal-regulated kinases/Mitogen-activated protein kinase) and PI3K-Akt [26]. In addition, corresponding with the protein subsequence analysis that identified multiple potential glycogen synthase kinase-3β (GSK-3β), protein kinase A (PKA), protein kinase C (PKC), and casein kinase 2 (CK2) phosphorylation sites on ASPH, several studies demonstrated that ASPH protein expression can be regulated by each of these kinases [8-10,26]. Our focus has been on the role of GSK-3β as a modulator of ASPH expression and function because in FASD, impaired neuronal migration is associated with inhibition of insulin/IGF-1 signaling through PI3K-Akt and increased activation of GSK-3β [8-10]. The inhibitory effects of GSK-3β activity on ASPH protein expression have been characterized. For example, treatment with chemical inhibitors or siRNA targeting GSK-3β increases ASPH protein expression and cell motility, whereas increasing GSK-3β activity decreases ASPH protein expression and cell migration [8,10,33]. However, those studies do not inform as to whether the effects of GSK-3β are indirect or direct, yet studies showing that ASPH can be phosphorylated [8,26,33,34] support the concept that kinases can phosphorylate ASPH and thereby modulate its expression and function. Furthermore, the notion that ASPH undergoes post-translational modification is supported by its ~140 kD size on SDS-PAGE versus the predicted Mr ~86 kD.
The proposed dual mechanisms of ASPH modulation at the post-transcriptional level are summarized in Figure 1A. Previous studies documented that increasing GSK- 3β activity inhibits ASPH expression and cell motility, whereas inhibiting GSK-3β enhances ASPH protein expression and cell motility [8,26,34]. The additional proposed method of post-transcriptional regulation of ASPH is via direct phosphorylation of the protein by GSK-3β, CK2, PKA, or PKC as diagramed based on the predicted phosphorylation sites (Figure 1B) [8,33]. Importantly, post-transcriptional regulation of ASPH can be mediated by trophic factor stimulation such as with insulin or IGF-1, or modulating GSK-3β activity by other mechanisms such as oxidative stress. In a previous study using hepatocellular carcinoma cells, we demonstrated that increased ASPH protein expression and translocation to the plasma membrane enables its C-terminal catalytic domain to interact with and hydroxylate Notch [11,34]. Attendant cleavage of Notch liberates its intracellular domain which enters the nucleus to activate the CSL complex and initiate transcription of Notch signaling target genes HES and HEY and promotes cell motility [33,35]. In addition, the N-terminal fragment of ASPH could remain attached to the ER membrane where it may serve a similar function as Humbug, i.e. regulating Ca2+ flux needed for actin filament polymerization and de-polymerization [21,33,35,36].
Figure 1.
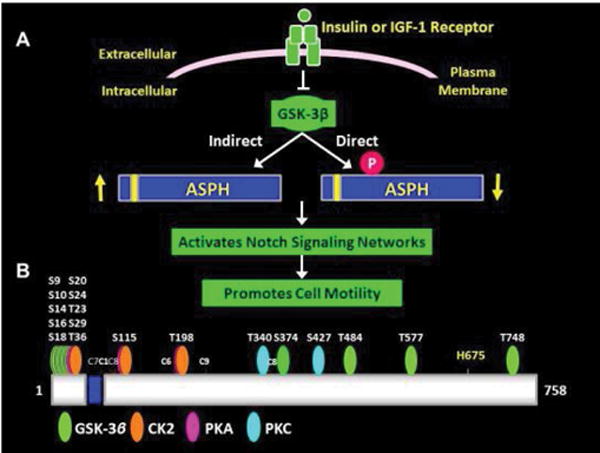
Proposed mechanisms of ASPH phosphorylation and strategy for generating site-directed mutants with the WT N-Myc-ASPH construct: (A) Diagram of insulin/IGF-1 regulation of ASPH. Insulin/IGF-1 stimulation of PI3K-Akt inhibits GSK-3β, whereas inhibition of insulin/IGF-1 signaling or increased oxidative stress increases GSK- 3β activity and phosphorylation of ASPH (direct). Increased GSK-3β activity can activate other kinases that phosphorylate ASPH (indirect). Depending on the positions and nature of ASPH phosphorylation, ASPH’s protein levels can either increase (enhanced stability) or decrease (enhanced cleavage and degradation). Increased ASPH protein expression and hydroxylation of Notch protein causes Notch to cleave, activating Notch networks. Notch’s intracellular domain translocates to the nucleus where it promotes transcription of genes that increase cell adhesion, motility, and invasion. (B) Diagram displaying the relative positions of predicted phosphorylation sites on ASPH. Most of sites correspond to GSK-3β and are distributed in the N-terminus of the protein. CK2, PKA, and PKC also can phosphorylate ASPH. The H675 residue is critical for hydroxylase activity. Ser/Thr residues within predicted phosphorylation sites were mutated to Ala using the Quick- Change Site-Directed mutagenesis kit and primer pairs listed in S-Table 1.
The present study examines the functional consequences of mutating GSK-3β, PKA, PKC, and CK2 phosphorylation sites on ASPH in human CNS (cerebellar)-derived primitive neuroectodermal tumor cells (PNET2). This work has relevance to FASD because ASPH expression is reduced vis-à-vis impaired insulin/IGF-IRS-PI3K-Akt signaling and increased GSK-3β activity in the brain [27,37,38]. Understanding how phosphorylation of ASPH regulates neuronal cell motility (migration) could help in the identification of novel targets to prevent teratogenic neurodevelopmental effects of prenatal alcohol exposure. Given the functional relationship between ASPH and Notch, these studies may also have general relevance to neuronal migration disorders in humans.
Reagents and Methods
Reagents
The QuickChange Site-directed Mutagenesis Kit was obtained from Stratagene (La Jolla, CA, USA). Escherichia coli DH5α cells, Dulbecco’s Modified Eagle Medium (DMEM), Lipofectamine 2000 Transfection Reagent, TRIzol, Amplex UltraRed, and 4-methylumbelliferyl phosphate (4-MUP) were obtained from Invitrogen (Carlsbad, CA, USA). The pcDNA 3 vector containing a 6x Myc-tag was generated as previously described [39]. The QIAquick Gel Extraction, QIAprep Spin Miniprep, and RNeasy Lipid Tissue Mini Kits were purchased from Qiagen (Valencia, CA, USA). All disposable assay plates and plasticware were obtained from Thermo Fisher Scientific Inc, (East Providence, RI, USA). ATPLite reagents were from Perkin-Elmer (Waltham, MA, USA). The Myc antibody was from Cell Signaling Technologies (Danvers, MA, USA). The bicinchoninic assay kit, enhanced chemiluminescence reagents, and Dylight 547 Conjugated to Streptavidin were purchased from Pierce Chemical Company (Rockford, IL, USA). Rabbit polyclonal antibody to large acidic ribosomal protein (RPLPO) was purchased from Proteintech (Chicago, IL, USA). Alkaline Phosphatase Conjugated to Streptavidin was purchased from Vector Laboratories (Burlingame, CA, USA). Positive- charge glass microscope slides were from Fisher Scientific (Pittsburgh, PA, USA). The Shandon Cytospin Centrifuge 3 was obtained from Thermo Shandon (Pittsburgh, PA, USA). The SpectraMax M5 Microplate Reader and Kodak PhosphorImager Screen S0230 with Cassette were from Molecular Dynamics (Sunnyvale, CA, USA). Histofix was purchased from Amresco (Solon, Ohio, USA). The AMV First Strand cDNA Synthesis Kit, probe-based primer pairs (Universal ProbeLibrary Assay Design Center), and LightCycler 480 System were from Roche (Indianapolis, IN, USA). MacVector 10 software was purchased from MacVector, Inc. (Cary, NC, USA). Re-usable Boyden chambers were obtained from Neuro Probe (Gaithersburg, MD, USA). Nucleofector 2b device and transfectction reagents were obtained from Lonza (Portsmouth, NH, USA). α-[14C]- Ketoglutaric acid was purchased from NEN Life Science (Boston, MA). Glass fiber filter paper (GF/C) was purchased from Packard Instruments (Meriden, CT). The GE Storm 820 Phosphor Imager was purchased from Molecular Devices (Sunnyvale, CA, USA). Other fine chemicals were purchased from CalBiochem (Carlsbad, CA, USA) or Sigma-Aldrich (St. Louis, MO, USA).
Recombinant ASPH plasmid constructs: The methods of generating recombinant ASPH plasmid construct were described previously [34]. In brief, the coding region of human ASPH was polymerase chain reaction (PCR) amplified from a 293T cDNA library (forward primer: 5′-CGGAATTCATGGCCCAGCGTAAGAATGCCA-3′; reverse primer: 5′- CCGCTCGAGCTAAATTGCTGGAAGGCTGC-3′; Pfu DNA polymerase). The EcoRI and XhoI digested PCR product was gel purified (QIAquick Gel Extraction Kit), and cloned into pcDNA 3 containing a 5′ 6x Myc-tag (pN-Myc-ASPH). Recombinant plasmid was purified (QIAprep Spin Miniprep Kit) from Ampicillin-resistant clones of DH5α transformed cells. Cloned inserts and their orientations were verified by DNA sequencing of both strands, and Western blot analysis of recombinant protein expressed in 293 cells. Point mutations were made by converting Serine and Threonine codons predicted to be phosphorylated by GSK-3β, PKC, PKA, or CK2 to Alanine (S/T→A) (Figure 1B) [8] using gene-specific primers (Supplementary Table 1; S-Table 1) and the QuickChange Site-directed Mutagenesis kit. The relative position of the point mutants in relation to the cDNA are depicted in Figure 1. Mutations were confirmed by direct sequencing of both cDNA strands. Expression of the correct size protein was demonstrated by Western blot analysis using Myc and ASPH antibodies.
Cell culture
Human central nervous system (CNS) derived cerebellar primitive neuroectodermal tumor 2 (PNET2) cells were maintained in DMEM supplemented with heat-inactivated 10% fetal bovine serum (FBS), 4 mM L-glutamine, 0.8 g/L glucose, and non-essential amino acids in 5% CO2 at 37°C. To examine the effects of ASPH over-expression and deletion of specific ASPH phosphorylation sites, CYZ cells were transiently transfected with wildtype or mutant pN-Myc-ASPH by electroporation using the Nucleofector 2b device and reagents (Lonza, Portsmouth, NH). Transfection efficiency was monitored by co-transfecting with recombinant plasmid containing the green fluorescent protein (GFP) cDNA. Recombinant ASPH and GFP expression were regulated by a cytomegalovirus (CMV) promoter.
Detection of ASPH immunoreactivity
For the ASPH measurements, we used two monoclonal antibodies. A85G6-ASPH binds to the C-terminal region of ASPH which contains a catalytic domain [40] required to promote cell motility and neuronal plasticity. A85E6 binds to the N-terminal Humbug-homologous region of ASPH; Humbug has a role in regulating calcium sequestration in the ER. ASPH is regulated by insulin/IGF-1 signaling through IRS-1 and Akt. Inhibition of ASPH perturbs cell motility and adhesion, and in the case of FASD, ethanol’s inhibitory effects on ASPH expression correlate with impairments in cerebellar neuronal migration and motor dysfunction [40].
Duplex Enzyme-linked Immunosorbent Assay (ELISA)
Duplex ELISAs sequentially measured immunoreactivity to target and loading control (RPLPO) proteins [40]. CYZ cells were homogenized in radio-immunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors [10]. Protein concentration was measured with the BCA assay. Protein samples (100 ng/50 μl) were adsorbed to the well bottoms of FluoroNunc MaxiSorp 96-well plates by overnight incubation. Non-specific binding was blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS) and primary antibody incubations were carried out overnight at 4°C. Immunoreactivity was detected with horseradish peroxidase (HRP)-conjugated secondary antibodies and Amplex UltraRed soluble fluorophore (Ex 530 nm/Em 590 nm). RPLPO expression, which corresponds with sample loading, was measured by incubating the proteins with biotinylated anti-RPLPO followed by streptavidin-conjugated alkaline phosphatase and 4-MUP (Ex 360 nm/Em 460 nm). Fluorescence was measured in a SpectraMax M5 microplate reader. Calculated ratios of target protein (Myc or ASPH) and RPLPO fluorescence were used for inter-group comparisons. Eight replicate culture samples were analyzed per group in each experiment.
Immunofluorescence
50,000 cells suspended in 10% FBS culture medium were cytocentrifuged onto glass microscope slides using a Shandon Cytospin Centrifuge 3 (Thermo Fisher Scientific Inc, East Providence, RI USA). After fixation in Histofix, the cells were permeabilized with 0.05% saponin in TBS and then non-specific sites were blocked with SuperBlock in TBS. Following overnight incubation with primary antibody (1:1000), immunoreactivity was detected with biotinylated secondary antibody and streptavidin-conjugated DyLight 547 (Ex 557 nm/Em 574 nm) [11,14]. Cells were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) and imaged by confocal microscopy.
14CO2 capture assay
The 14CO2 capture assay measures 14C-labeled CO2 released by the ASPH hydroxylase reaction [41]. To evaluate the effects of ASPH over- expression and mutation of specific phosphorylation sites, 400 ng of A85G6+A85E6+FB50 monoclonal ASPH antibodies were adsorbed to the well bottoms of MaxiSorp 96-well plates. After blocking with PBS containing 1% BSA and 0.05% Tween-20 (PBST), 25 μg of CYZ proteins were added to the wells to capture ASPH. Negative and positive control reactions included 200 ng BSA and 200 ng recombinant ASPH, respectively. For hydroxylase activity, 40 μL reaction cocktails containing 60 μM epidermal growth factor (EGF)-like peptide (60 μM), 40 μM [14C]-α-ketoglutarate acid in 50 mM PIPES pH 7.0, 100 mM NaCl, 100 μM FeCl2, and 0.2 mg/ml casein were added to each well. The plates were tightly covered with GF/C soaked in 30 mM Ca(OH)2 and incubated for 1 hour at 25°C. Radioactivity was detected and quantified on the dried filters using a GE Storm 820 Phosphor Imager (Molecular Devices, Sunnyvale, CA USA).
Quantitative reverse transcriptase-PCR (qRT-PCR)
The RNeasy Lipid Tissue Mini Kit was used to isolate total RNA from TRIzol-lysed PNET2 cells. cDNAs generated with the AMV First Strand cDNA synthesis kit and random oligodeoxynucleotide primers were used to measure gene expression via hydrolysis probe based duplex qRT-PCR with hypoxanthine-guanine phosphoribosyltransferase (HPRT) as reference gene and the Taqman Gene expression master mix. Reactions (20:l) contained 400 nM of primers for both HPRT and the target gene, and 200 nM of probes for HPRT (Y555 labeled) and the target gene (FAM labeled). Amplification reactions were conducted using the LightCycler 480 as described [34]. Primer sequences and matched probes were determined by using Roche Probe Finder Software.
Directional motility assay
Directional motility was measured with a Boyden Chamber type ATP luminescence-based motility invasion (ALMI) assay [42]. Briefly, culture media containing 1% FBS was placed in the lower chambers that were separated from the upper chambers with 12-μm pore polycarbonate membranes. 100,000 viable cells suspended in 0.1% FBS culture media were seeded into each upper chamber. After 30 minutes incubation in a 37°C CO2 incubator, cells harvested from the upper chamber (non-motile), undersurface of the membrane (migrated-adherent), and lower chamber (migrated, non-adherent) were quantified using the ATPlite luminescence assay kit. Six replicate assays per condition were used for statistical analysis.
Statistical analysis
Data depicted in the graphs represent the means ± standard deviation (S.D.) for each group. Inter-group comparisons were made using repeated measures of one-way analysis of variance (ANOVA) with the post hoc Fisher’s Least Significant Difference test. Statistical analyses were performed using GraphPad Prism 6 software.
Results
Initial studies characterized ASPH expression in transiently transfected PNET2 cells
Western blots were probed with Myc and A85G6-ASPH monoclonal antibodies to detect the Myc tag and ASPH. As a control for loading, the blots were stripped and re-probed with polyclonal anti-p85 (subunit of PI3 Kinase). Cells transfected with empty vector (EV) had relatively low levels of endogenously expressed ASPH (Figure 2). Myc immunoreactivity was not detected in EV transfected cells by Western blot analysis due to the small size of the peptide compared with Myc-ASPH fusion proteins. In contrast, cells transfected with Myc-tagged wildtype (WT) expressed the Myc-fusion protein and had higher than endogenous levels of ASPH. In addition, two bands were associated with A85G6-ASPH correspond to super-shifted endogenous (lower band) and Myc-tagged phosphorylated ASPH, whereas only the lower band was detected in EV-transfected cells. Among the 18 single S/T→A point-mutated ASPH constructs studied, 16 had higher levels of Myc expression than in WT-ASPH transfected cells (Figure 2). However, recombinant A85G6-ASPH immunoreactivity (upper band) varied such that 13-14 of the 16 mutant constructs produced higher levels of recombinant ASPH (upper band) relative to WT-ASPH and the remainder had either similar or lower levels of ASPH expression. Cells transfected with M10 or M12 had sharply lower levels of Myc-tag and ASPH than in cells transfected with WT or other point mutated ASPH constructs. The p85 control Western blot results confirmed that sample loading was approximately even. Since the results were comparable to PNET2 cells transfected with empty vector yet their mRNAs were detectable by qRT-PCR analysis, most likely the corresponding proteins generated with the M10 and M12 constructs were unstable and had short half-lives. Therefore, these cDNAs were not further studied.
Figure 2.
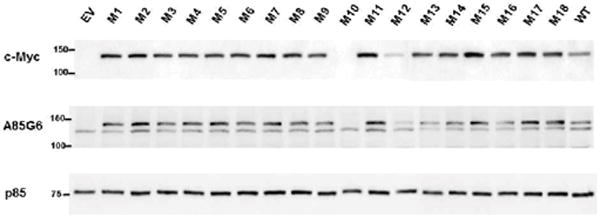
Effects of S/T→A mutations on ASPH protein expression: PNET2 cells were transfected with wildtype (WT) or point-mutated (M#:S/T→A) ASPH cDNAs in which a Myc tag was fused in-frame (N-Myc-ASPH); See Table 1 for mutations map. Gene expression was under the control of a CMV promoter. Myc-empty vector (EV) served as a negative control. 24 hours after transfection, the cells were harvested and subjected to Western blot analysis using antibodies to mouse monoclonal Myc or A85G6-ASPH. The blots were stripped and re-probed with rabbit polyclonal antibodies to p85-PI3K as a sample loading control.
Effects of ASPH GSK-30 phosphorylation site mutation on cell adhesion and directional motility
ASPH promotes cell adhesion and motility [8,10,11,13,26,34,43,44]. The ATPLyte-based directional motility and adhesion blind-well chamber assay [42] was used to examine the effects of WT and mutant-ASPH (M5, M7, M9, M14, M15, or M17) over-expression on PNET2 cell motility. The percentages of total motile, motile adherent, and motile-nonadherent cells were calculated and depicted graphically (Figure 3). Statistical comparisons with EV control and WT-ASPH transfected cells were made by one-way repeated-measures ANOVA and post hoc Fisher LSD tests (S-Table 2). Cells transfected with WT-ASPH had significantly higher percentages of total motile, motile adherent, and motile non-adherent cells relative to EV-transfected cells. All 7 ASPH mutant constructs produced significantly higher mean percentages of total motile (Figure 3A) and motile non-adherent (Figure 3C) cells, and all except for M7 significantly increased the percentages of motile adherent cells (Figure 3B). Furthermore, the M9, M14 and M17 mutants had significantly higher total motility and motile adherent indices compared with WT, and all except for M5 had significantly higher percentages of motile non-adherent cells relative to WT (S-Table 2; Figure 3C). Therefore, over-expression of ASPH cDNAs with selected point S/T→A point mutations in GSK-3β phosphorylation sites, increased recombinant ASPH expression and directional motility.
Figure 3.
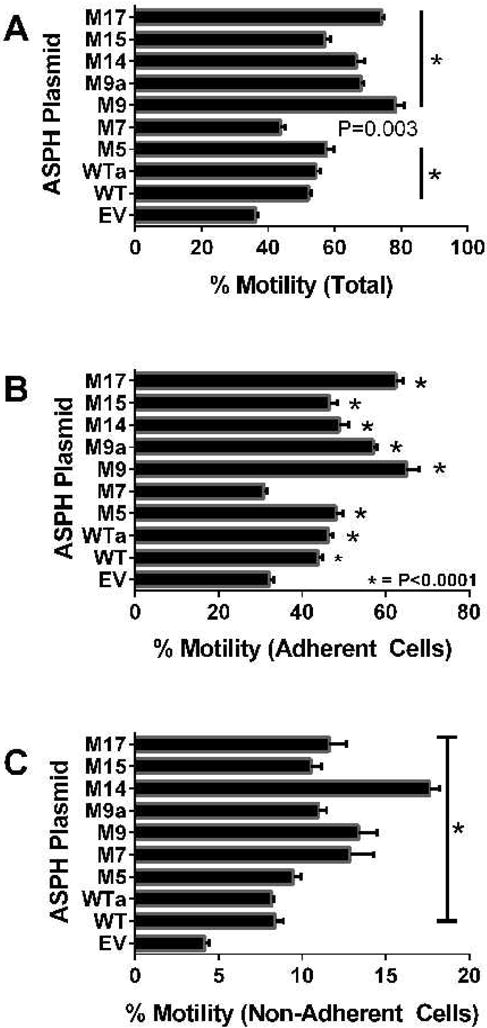
Directional motility and adhesion assays: PNET2 cells transfected with EV, WT N-Myc ASPH, or point-mutated N-Myc-ASPH were used in an ATP luminescence-based Boyden-chamber type directional motility and adhesion assay in which 1% FBS in culture medium was supplied as the trophic factor in the lower chamber (see Methods). Graphs depict mean (± S.D.) percentages of (A) all motile, (B) motileadherent, and (C) motile non-adherent cells (see Methods). Four replicate cultures from independent transfections were analyzed simultaneously. Inter-group comparisons were made by one-way ANOVA with post-hoc Fisher LSD tests. *P<0.0001 for comparisons with WT (S-Table 2).
ASPH point mutations alter cellular localization of ASPH
The effects of S/T→A mutations on ASPH protein expression and subcellular localization were examined by immunofluorescence staining and confocal microscopy (Figure 4). PNET2 cells transfected with WT N-Myc-ASPH cDNA exhibited abundant Myc-immunoreactivity in the perinuclear zone, central cytoplasm, surface membrane, and in podocytes (Figure 4A). In contrast, cells transfected with point-mutated (M#:S/T→A) N-Myc-ASPH cDNAs exhibited altered levels and cellular distributions of Myc immunoreactivity. For example, the M7-S24A mutant resulted in Myc immunoreactivity predominantly localized to the surface membrane and podocytes (Figure 4C), whereas the M18 mutant produced a particulate predominantly cytoplasmic patterns of immunoreactivity that most likely corresponded to microsomal or mitochondrial localizations (Figure 4D). Importantly, the M18 mutant altered the distributions of Myc immunoreactivity such that peripheral cytoplasmic and podocyte labeling were not detected. Cells transfected with M19-H675Q exhibited mainly nuclear and tight perinuclear distributions without plasma membrane or podocyte localizations (Figure 4E). Cells transfected with Myc-EV served as a negative control (Figure 4B).
Figure 4.
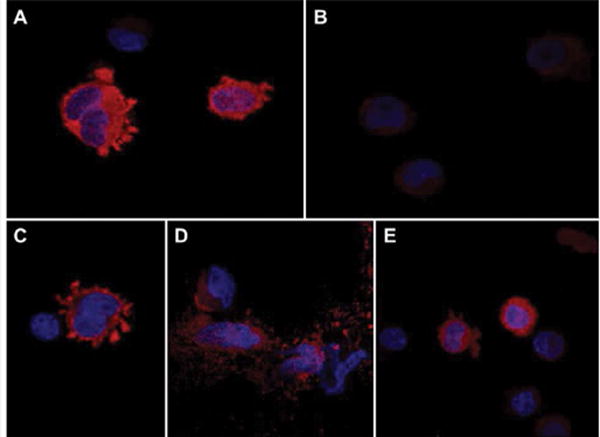
Effects of S/T→A mutations on ASPH protein expression and subcellular localization: PNET2 cells were transiently transfected with wildtype (WT) or a pointmutated (M#:S/T→A) N-Myc-ASPH cDNA. Myc-empty vector (EV) served as a negative control. The M19-H675Q mutant, disrupting ASPH’s catalytic activity, served as a positive control. Representative results obtained by immunofluorescence staining and confocal imaging of cells transfected with (A) WT, (B) M7-S24A, (C) M18-T748A, (D) M19-H675Q, or (E) EV and stained by immunofluorescence with anti-Myc. Immunoreactivity was detected with biotinylated secondary antibody and Streptavidin-conjugated Dylight 547 (red). Cells were counterstained with DAPI (blue). (Merged images: 600× magnification, 2× digital zoom).
ASPH over-expression enhances signaling through Notch pathways: Previous studies in hepatic cells demonstrated that ASPH-mediated enhancement of cell motility was linked to increased signaling through Notch networks [10,11,43,44]. To demonstrate similar responses in the present experiments, immunofluorescence staining and confocal microscopic imaging were used to qualitatively examine effects of ASPH over-expression on ASPH, Notch-1 and Jagged-1 immunoreactivity (Figure 5). As negative controls, cells were transfected with a recombinant plasmid expressing GFP or Humbug, a truncated homolog of ASPH that lack the critical catalytic domain needed to hydroxylate/activate Notch and Jagged [16]. ASPH was detected with two different monoclonal antibodies: A85G6-ASPH, which binds to the C-terminal region of ASPH and FB50-ASPH/Humbug, which binds to the N-terminal regions of ASPH and Humbug. Cells transfected with WT-ASPH had higher levels of both A85G6-ASPH and FB50-ASPH/Humbug immunoreactivity than in GFP or Humbug transfected cells (Figures 5A-5F). In addition, Notch-1 and Jagged-1 immunoreactivity were higher in the WT-ASPH-transfected compared with GFP- and Humbug-transfected cells (Figures 5G-5L). These findings suggest that ASPH over- expression enhances Notch-1 and Jagged-1 immunoreactivity in PNET2 CNS-derived neuronal cells.
Figure 5.
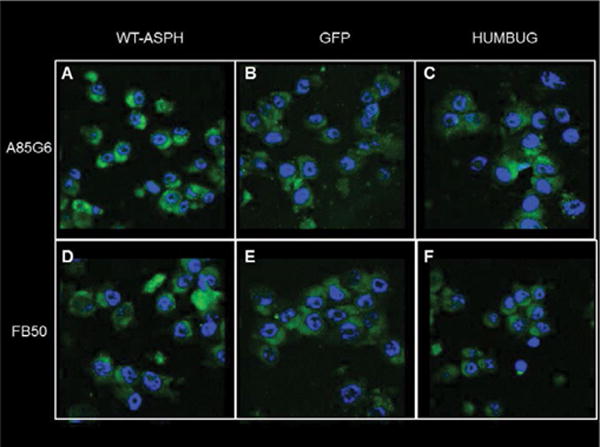
Effects of WT N-Myc-ASPH Over-expression on ASPH and Notch Pathway Protein Immunoreactivity: PNET2 cells transfected with recombinant plasmids carrying full-length cDNAs encoding (A, D, G, J) WT-ASPH, (B, E, H, K) GFP, or (C, F, I, L) Humbug (truncated form of ASPH) were harvested after 48 hours to generate cytospin preparations for immunofluorescence assays. Formalin-fixed, permeabilized cells were incubated with (A-C) A85G6-ASPH, (D-F) FB50-ASPH/Humbug, (G-I) Notch-1, or (J-L) Jagged-1 monoclonal antibodies. Immunoreactivity was detected with biotinylated secondary antibody and Streptavidin-conjugated Dylight 547 (green) or Dylight 647 (red). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue). Panels depict merged confocal microscopy images. (600× original magnification, 2× digitalzoom).
Effects of ASPH GSK-3β S/T→A single point mutations on Notch pathway genes
qRT-PCR assays examined effects of deleting GSK-3β sites on mRNA expression of Notch-1, Jagged-1, HES-1 and HEY-1. For comparison, the M19-H675Q mutant which lacks catalytic activity and inhibits cell motility [45] was studied in parallel. Corresponding with the immunofluorescence studies, cells transfected with WT N-Myc-ASPH had significantly higher levels of Notch-1 and Jagged-1 expression relative to EV, whereas HES-1 or HEY-1 expression were not significantly altered (Figure 6; S-Table 3). Despite higher levels of ASPH immunoreactivity, Notch 1 mRNA was significantly elevated in cells transfected with the M1 M3 and M7 mutants and significantly inhibited in cells transfected with M6, M11, M12, M13, M15, M18 or M19 (Figure 6A; S-Table 3A). Jagged-1 mRNA expression was increased in cells transfected with M3, M6 or M7, and either significantly inhibited (n=4) or unchanged (n=3) by the other 7 mutants (Figure 6B; S-Table 3B). HES-1 mRNA expression was largely unchanged by transfection with mutant ASPH (Figure 6C; S-Table 3C). The two exceptions were M1 and M11 which were modestly but significantly increased relative to EV HEY-1 expression was significantly modulated by 7 of the 10 mutants. However, only M3 significantly increased HEY-1 relative to EV. M6, M7, M13, M15, M18, and M19 significantly inhibited HEY-1, whereas M1, M11 and M12 had no significant effect (Figure 6D; S-Table 3D).
Figure 6.
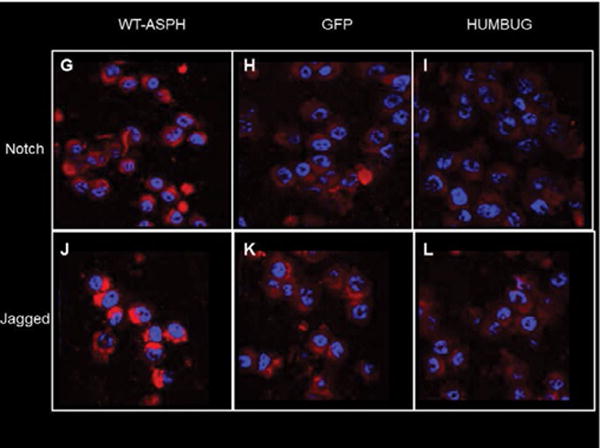
Effects of WT and point mutated ASPH on Notch signaling genes: PNET2 cells transfected with WT or point-mutated N-Myc-ASPH cDNAs were used in duplex qRT-PCR assays to measure (A) Notch-1, (B) Jagged-1, (C) HES-1, and (D) HEY-1 expression. Data were normalized to HPRT measured simultaneously in the same reactions. EV-transfected cells served as negative controls, whereas M19-H◊Q transfected cells served as positive controls for inhibiting Notch signaling due to loss of catalytic activity. Graph depict relative mean (± S.D.) levels of target gene (mRNA) expression. Inter-group comparisons were made by one-way ANOVA with the post-hoc Fisher LSD test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001 for comparisons with EV (S-Table 3).
Additive effects of ASPH S/T→A mutations on ASPH protein expression-ELISAs
To investigate additive effects of S/T→A mutations, PNET2 cells transfected with ASPH cDNAs carrying 2 to 5 point mutations were probed for Myc and ASPH immunoreactivity by Western blot analysis (Figure 7) and ELISA (Figure 8). The cells were analyzed 48 hours after transfection. Cells transfected with WT-ASPH, ASPH cDNAs with single point mutations (M2, M7, M9, M14, M15, and M17), or EV were studied in parallel. Among the single point mutant constructs, only M9 resulted in lower levels of Myc than observed in WT-ASPH transfected cells. In cells transfected with M2, M7, M14, M15, or M17, Myc- ASPH fusion protein expression was conspicuously higher than in the WT-ASPH transfected cells (Figure 7). Stepwise additions of each of these point mutations did not further increase Myc-ASPH fusion protein expression. Probing the blot with A85G6- ASPH monoclonal antibody demonstrated higher levels of ~140 kD ASPH with over-expression of each mutant cDNA (single-site and multi-site) relative to WT. The lower bands corresponding to native (~86 kD) and fragmented ASPH were detected in all cells, including EV- and GFP-transfected (Figure 7).
Figure 7.
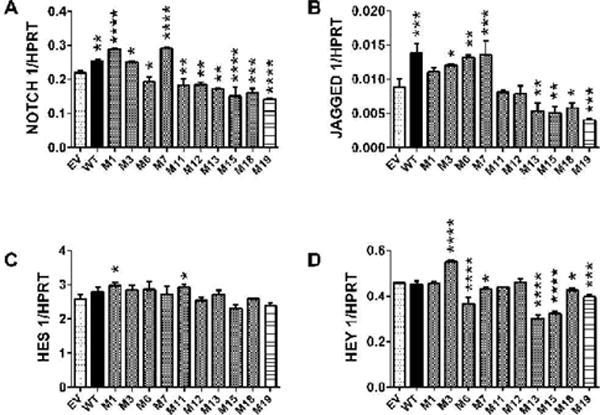
Additive ASPH point mutation effects in PNET2 cells on Myc and ASPH immunoreactivity. PNET2 cells were transfected with ASPH cDNAs carrying from 0 (WT) to 6 point mutations (M#:S/T◊A). The Myc tag was fused in-frame (N-Myc-ASPH) and gene expression was under the control of a CMV promoter. Myc-empty vector (EV) and GFP-transfected cells served as negative controls. 48 hours after transfection, the cells were harvested and subjected to Western blot analysis using antibodies to mouse monoclonal Myc or A85G6-ASPH. The blots were stripped and re-probed with rabbit polyclonal antibodies to p85-PI3K as a sample loading control.
Figure 8.
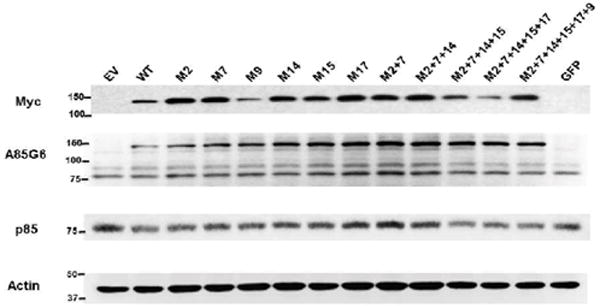
Effects of single versus multiple ASPH point mutations on Myc and ASPH immunoreactivity and catalytic activity. PNET2 cells were transfected with ASPH cDNAs carrying from 0 (WT) to 6 point mutations (M#:S/T◊A). Myc tag was fused in-frame (NMyc- ASPH). Gene expression was under the control of a CMV promoter. Myc-empty vector (EV) served as a negative control. 48 hours after transfection, the cells were harvested and subjected to duplex ELISA tests of (A) Myc and (B) A85G6-ASPH protein expression, with results normalized to RPLPO. Three independently transfected cultures were assayed with 4 technical replicates. (C) ASPH hydroxylase activity was measured with the 14CO2 capture assay using recombinant EGF peptide as the substrate. Released 14CO2 was measured with a Phosphor-Imager. The graph depicts the mean (± S.E.M.) levels of released 14CO2 (cpm)/μg protein. Graph depict mean (± S.D.) levels of immunoreactivity or catalytic activity. Inter-group comparisons were made by one-way repeated measures ANOVA with the post-hoc Fisher LSD test. A, B- *P<0.0001 for comparisons with EV; ξ P<0.0001 for comparisons with WT. C- *P<0.05; **P<0.01 relative to EV (S-Table 4).
Duplex ELISAs were used to measure Myc and A85G6-ASPH protein expression in transfected PNET2 cells and confirm results obtained by Western blot analysis. To distinguish recombinant from endogenous ASPH, immunoreactivity was detected with Myc antibody. Similar levels of Myc immunoreactivity were measured in EV, WT, M2, M17, and M2+M7+M14 transfected cells (Figure 8A; S-Table 4A). In contrast, Myc expression was significantly higher in M7, M9, M14, M15, M2+M7, M2+M7+M14+M15, M2+M7+M14+M15+M17, and M2+M7+M14+M15+M17+M9 transfected relative to EV and WT transfected cells (all P<0.0001). Corresponding with results obtained by Western blot analysis, A85G6-ASPH immunoreactivity was significantly elevated in WT transfected relative to EV controls (P<0.0001). In addition, the levels of A85G6-ASPH immunoreactivity were significantly elevated in PNET2 cells transfected with each of the single and multiple mutant constructs relative to EV and WT (all P<0.0001) (Figure 8B; S-Table 4B). Furthermore, although the degrees of enhanced expression varied somewhat among the mutants, the differences were unrelated to position or multiplicity of S/T phosphorylation site deletion.
Effects of S/T→A GSK-3β site-directed mutagenesis on ASPH hydroxylase activity: Since ASPH promotes cell migration and adhesion through hydroxylation of its target molecules, we investigated the effects of S/T→A mutations on ASPH’s catalytic activity. Our focus on the GSK-3β sites was based on inhibition of GSK-3β with LiCl or transfection with siRNA targeting GSK-3β enhances ASPH expression and cell motility, whereas increased GSK-3β expression reduces ASPH expression and cell motility [8,26]. ASPH catalytic activity was measured with a microplate scaled assay that quantifies 14C-labeled CO2 released as a by-product of hydroxylase activity [41]. PNET2 cells overexpressing WT N-Myc-ASPH released significantly higher levels of 14CO2 (P<0.0001) relative to EV control cells (Figure 8C; S-Table 4C). Among the ASPH constructs carrying a single point mutation, 4 of the 6 had significantly increased hydroxylase activity relative to EV, but 3 had reduced catalytic activity relative to WT, 2 had comparable levels and one had significantly higher levels of catalytic activity relative to WT (Figure 8C; S-Table 4C). In cells transfected with ASPH mutants carrying between 2 and 6 S/T→A point mutations, 4 of the 5 constructs produced significantly higher levels of ASPH activity relative to EV, and comparable levels of catalytic activity relative to WT.
Discussion
ASPH expression and function resulting in increased cell adhesion and motility are regulated by insulin and IGF-1 [8,11-13,26,44,46]. The finding that inhibition of GSK-3β [8,10] enhances ASPH protein expression without altering its mRNA led to further investigations that showed: 1) ASPH protein has intrinsic phosphorylation sites, many of which correspond to GSK-3β motifs; 2) ASPH protein can be phosphorylated by GSK-3β [3,8,10], and 3) inhibition of other kinases predicted to phosphorylate ASPH, i.e. PKC, PKA, and CK2, enhance insulin/IGF stimulated ASPH expression/immunoreactivity and motility [26]. However, those earlier studies did not evaluate direct effects of ASPH phosphorylation on its protein expression and function. One goal of this study was to examine functional consequences of deleting predicted ASPH phosphorylation sites. Responses measured were ASPH protein expression, subcellular localization, catalytic (hydroxylase) activity, and the effects on cell adhesion, directional motility, and Notch pathway gene expression.
The Myc-tagged ASPH construct enabled us to distinguish recombinant from endogenous ASPH protein, particularly since ASPH is physiologically cleaved and its half- life is approximately 30 minutes based on metabolic labeling studies [10]. Transient transfection studies demonstrated that the recombinant protein was highly expressed for 48 hours, and persisted for 96 hours (data not shown). Except for 2 constructs (M10, M12), S/T→A point mutation of specific phosphorylation sites mainly increased Myc- ASPH expression above the levels observed with the WT-ASPH cDNA. However, the effects were more varied when the blots were probed with the A85G6-ASPH antibody because the combined endogenous and transgene proteins and cleavage products were detected. The findings with respect to the Myc-tagged ASPH mutant cDNAs suggest that deletion of GSK-3β phosphorylation sites on ASPH increases ASPH protein levels, consistent with previous reports showing that LiCl inhibition of GSK-3β activity enhances ASPH expression [8, 26]. In contrast to observations made in Huh7 human hepatocellular carcinoma cells [34], Myc-ASPH protein expression was not differentially altered by N- terminus versus C-terminus positions of the point mutations.
In contrast to the relatively consistent effects of phosphorylation site deletion on ASPH expression, the functional consequences pertaining to cell motility and adhesion varied. Total motility was significantly higher in cells transfected with WT or any of the point-mutated ASPH cDNAs compared with EV control cells. However, relative to WT, point mutations in the N-terminus of ASPH produced no significant increases in total motility, whereas 4 of the 5 phosphorylation site mutations within the C-terminus significantly increased total motility compared with WT. Therefore, ASPH modulation of directional motility was governed by position of the deleted phosphorylation site. Analysis of the sub-populations of motile cells revealed that although the fractions of motile- adherent cells were significantly increased relative to WT for 3 of 6 the mutant constructs, the dominant and consistent effect of the point mutations was to increase the percentage of motile non-adherent cells.
Functional alterations induced by the point mutations could have been due to conformational changes, altering ASPH’s subcellular distribution and ability to interact with and hydroxylate Notch and Jagged. Immunofluorescence and confocal microscopy studies demonstrated abundant perinuclear, particulate (microsomal-type), cell surface, and podocyte immunoreactivity in PNET2 cells transfected with WT ASPH, and conspicuously altered patterns of immunoreactivity in cells transfected with point-mutated ASPH cDNAs. For example, immunoreactivity corresponding to the M7-S24A mutant was mainly localized in the perinuclear zone rather than at the cell periphery and in podocytes. This phenomenon may account for the reduced levels of total and migrated adherent cell motility indices and catalytic activity relative to WT, despite higher levels of ASPH protein expression.
The M18 mutant located in the C-terminus of ASPH was abundantly expressed and at levels above WT, yet its subcellular distribution was strikingly altered and largely particulate in nature, corresponding to a microsomal pattern. In contrast, the M19-H675Q mutant in which directional motility was impaired due to deletion of the critical His residue required for catalytic function, exhibited ASPH immunoreactivity localized to the perinuclear zone rather than along the surface membrane and within podocytes. This suggests that besides loss of hydroxylase activity, the inhibitory effects of the M19 mutant were mediated by abnormal cellular distributions of ASPH such that potential interactions with Notch or Jagged at the cell surface would have been compromised. Therefore, functional alterations in ASPH produced by point mutations were likely mediated in part by changes in intracellular trafficking. Mechanistically, these effects could impact ASPH’s activation of Notch signaling networks needed to promote cell motility and adhesion.
Immunofluorescence with confocal microscopy studies were used to demonstrate that over-expression of WT ASPH selectively increased Notch and Jagged immunoreactivity in PNET2 cells. Further qRT-PCR studies were used to examine effects of point-mutated ASPH on Notch-1, Jagged-1, HES-1, and HEY-1 mRNA levels. Although the responses were complex and somewhat varied, the main conclusions were that with few exceptions, the point-mutated ASPH cDNAs: 1) did not significantly increase Notch- 1, Jagged-1, HES-1, or HEY-1 expression above WT-ASPH; 2) mainly inhibited expression of Notch-1, Jagged-1, and HEY-1 relative to EV and WT-ASPH; and 3) had minimal effect on HES-1 expression. In light of the higher levels of ASPH expression and increased motility, these seemingly disparate results most likely indicate that in short-term studies (24-96 hours), the phenotypic consequences of over-expressing GSK-3β phosphorylation site mutant ASPH cDNAs are positive, but the long-term effects associated with suppression of Notch networks at the level of transcription could be detrimental to neuronal cellmotility.
Since it is possible that more than one ASPH phosphorylation site is functional, we examined the effect of cumulative point mutations on ASPH protein expression and catalytic activity. Western blot analysis and ELISA studies showed differential effects of adding GSK-3β phosphorylation site mutations on Myc-ASPH fusion protein and A85G6- ASPH expression. The results demonstrated that although most of the mutant constructs significantly increased Myc-ASPH and A85G6-ASPH above EV and WT-ASPH, the highest levels of ASPH achieved were associated with single point mutations. The greater variability in the levels of Myc-ASPH relative to A85G6-ASPH may have been due to physiological cleavage of the recombinant protein, as suggested by the increased levels of several bands corresponding to A85G6-ASPH by Western blotanalysis.
One of the key functions of ASPH is its catalytic/hydroxylase activity which activates Notch signaling [16,45]. Previous studies showed that large fold-increases in ASPH expression were not required to confer increased cell motility, and that catalytic activity was most important [45]. Therefore, it was important to interpret the effects of the phosphorylation site mutations in the context of ASPH’s catalytic activity. Using the 14CO2 capture assay to quantify 14C-labeled CO2 released with hydroxylase activity [41], we demonstrated significantly increased levels of catalytic activity in WT Myc-ASPH transfected cells relative to EV. The effects of single or multiple phosphorylation site mutations on catalytic activity varied. Six of the mutant cDNAs produced mean levels of catalytic activity that were similar to WT, two (M2+M7 and M2+M7+M124+M15) had higher than EV but lower than WT levels of catalytic activity, and three (M2, M17, M2+M7+M14) were similar to EV and reduced relative to WT. It is noteworthy that the 3 cDNAs in which catalytic activity was not increased above control also had similar levels of Myc-ASPH relative to EV. mutated cDNAs. Although the other discordances are not readily explained, the aggregate results could be interpreted to indicate that catalytic activity is saturable and therefore rate-limited. However, with sustained higher levels of ASPH protein expression, effects on cell motility may be longer sustained as suggested by the findings in directional motility assays. In conclusion, these studies support the hypothesis that phosphorylation of ASPH protein regulates expression and function. The findings suggest an important mechanistic consideration related to phosphorylation- dependent conformational changes in ASPH protein that may alter its expression, sub- cellular distribution, and catalytic activity. Further studies are needed to address this question.
Supplementary Material
Acknowledgments
Research reported in this publication as supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award # T32GM077995. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. Diana Borgas was a student in the Brown Graduate Program in Molecular Pharmacology and Physiology when she conducted research contained within this manuscript.
References
- 1.Roozen S, Peters GJ, Kok G, Townend D, Nijhuis J, et al. Worldwide Prevalence of Fetal Alcohol Spectrum Disorders: A Systematic Literature Review Including Meta-Analysis. Alcohol Clin Exp Res. 2016;40:18–32. doi: 10.1111/acer.12939. [DOI] [PubMed] [Google Scholar]
- 2.Del Campo M, Jones KL. A review of the physical features of the fetal alcohol spectrum disorders. Eur J Med Genet. 2017;60:55–64. doi: 10.1016/j.ejmg.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Marquardt K, Brigman JL. The impact of prenatal alcohol exposure on social, cognitive and affective behavioral domains: Insights from rodent models. Alcohol. 2016;51:1–15. doi: 10.1016/j.alcohol.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murawski NJ, Moore EM, Thomas JD, Riley EP. Advances in Diagnosis and Treatment of Fetal Alcohol Spectrum Disorders: From Animal Models to Human Studies. Alcohol Res. 2015;37:97–108. doi: 10.35946/arcr.v37.1.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid N, Dawe S, Shelton D, Harnett P, Warner J, et al. Systematic Review of Fetal Alcohol Spectrum Disorder Interventions Across the Life Span. Alcohol Clin Exp Res. 2015;39:2283–2295. doi: 10.1111/acer.12903. [DOI] [PubMed] [Google Scholar]
- 6.de la Monte SM, Kril JJ. Human alcohol-related neuropathology. Acta Neuropathol. 2014;127:71–90. doi: 10.1007/s00401-013-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontaine CJ, Patten AR, Sickmann HM, Helfer JL, Christie BR. Effects of pre- natal alcohol exposure on hippocampal synaptic plasticity: Sex, age and methodological considerations. Neurosci Biobehav Rev. 2016;64:12–34. doi: 10.1016/j.neubiorev.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Carter JJ, Tong M, Silbermann E, Lahousse SA, Ding FF, et al. Ethanol impaired neuronal migration is associated with reduced aspartyl-asparaginyl-beta-hydroxylase expression. Acta Neuropathol. 2008;116:303–315. doi: 10.1007/s00401-008-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Monte SM. Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol (Fayetteville, NY) 2009;43:225. doi: 10.1016/j.alcohol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silbermann E, Moskal P, Bowling N, Tong M, de la Monte SM. Role of aspartyl- (asparaginyl)-beta-hydroxylase mediated notch signaling in cerebellar development and function. Behav Brain Funct. 2010;6:68. doi: 10.1186/1744-9081-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantarini MC, de la Monte SM, Pang M, Tong M, D’Errico A, et al. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006;44:446–457. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- 12.de la Monte SM, Tamaki S, Cantarini MC, Ince N, Wiedmann M, et al. Aspartyl-(asparaginyl)-beta-hydroxylase regulates hepatocellular carcinoma invasiveness. J Hepatol. 2006;44:971–983. doi: 10.1016/j.jhep.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T, Sepe P, Lahousse S, Tamaki S, Enjoji M, et al. Antisense oligodeoxynucleotides directed against aspartyl (asparaginyl) beta-hydroxylase suppress migration of cholangiocarcinoma cells. J Hepatol. 2003;38:615–622. doi: 10.1016/s0168-8278(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 14.Sepe PS, Lahousse SA, Gemelli B, Chang H, Maeda T, et al. Role of the aspartyl-asparaginyl-beta-hydroxylase gene in neuroblastoma cell motility. Lab Invest. 2002;82:881–891. doi: 10.1097/01.lab.0000020406.91689.7f. [DOI] [PubMed] [Google Scholar]
- 15.Sturla LM, Tong M, Hebda N, Gao J, Thomas JM, et al. Aspartatebeta-hydroxylase (ASPH): A potential therapeutic target in human malignant gliomas. Heliyon. 2016;2:e00203. doi: 10.1016/j.heliyon.2016.e00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavaissiere L, Jia S, Nishiyama M, de la Monte S, Stern M, et al. Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J Clin Invest. 1996;98:1313–1323. doi: 10.1172/JCI118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia S, VanDusen WJ, Diehl RE, Kohl NE, Dixon RA, et al. cDNA cloning and expression of bovine aspartyl (asparaginyl) beta-hydroxylase. J Biol Chem. 1992;267:14322–14327. [PubMed] [Google Scholar]
- 18.Wang Q, Vandusen WJ, Petroski CJ, Garsky VM, Stern AM, et al. Bovine liver aspartyl B-hydroxylase purification and characterization. J Biol Chem. 1991;266:14004–14010. [PubMed] [Google Scholar]
- 19.Lee JH. Overexpression of humbug promotes malignant progression in human gastric cancer cells. Oncol Rep. 2008;19:795–800. [PubMed] [Google Scholar]
- 20.Treves S, Feriotto G, Moccagatta L, Gambari R, Zorzato F. Molecular cloning, expression, functional characterization, chromosomal localization, and gene structure of junctate, a novel integral calcium binding protein of sarco(endo)plasmic reticulum membrane. J Biol Chem. 2000;275:39555–39568. doi: 10.1074/jbc.M005473200. [DOI] [PubMed] [Google Scholar]
- 21.Dinchuk JE, Henderson NL, Burn TC, Huber R, Ho SP, et al. Aspartyl beta-hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J Biol Chem. 2000;275:39543–39554. doi: 10.1074/jbc.M006753200. [DOI] [PubMed] [Google Scholar]
- 22.Jia S, McGinnis K, VanDusen WJ, Burke CJ, Kuo A, et al. A fully active catalytic domain of bovine aspartyl (asparaginyl) beta-hydroxylase expressed in Escherichia coli: characterization and evidence for the identification of an active- site region in vertebrate alpha-ketoglutarate-dependent dioxygenases. Proceedings of the National Academy of Sciences. 1994;91:7227–7231. doi: 10.1073/pnas.91.15.7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinnis K, Ku GM, VanDusen WJ, Fu J, Garsky V, et al. Site-directed mutagenesis of residues in a conserved region of bovine aspartyl (asparaginyl) beta-hydroxylase: evidence that histidine 675 has a role in binding Fe2+ Biochemistry. 1996;35:3957–3962. doi: 10.1021/bi951520n. [DOI] [PubMed] [Google Scholar]
- 24.Dinchuk JE, Focht RJ, Kelley JA, Henderson NL, Zolotarjova NI, et al. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J Biol Chem. 2002;277:12970–12977. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 25.Gronke RS, VanDusen WJ, Garsky VM, Jacobs JW, Sardana MK, et al. Aspartyl beta-hydroxylase: in vitro hydroxylation of a synthetic peptide based on the structure of the first growth factor-like domain of human factor IX. Proc Natl Acad Sci U S A. 1989;86:3609–3613. doi: 10.1073/pnas.86.10.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahousse SA, Carter JJ, Xu XJ, Wands JR, de la Monte SM. Differential growth factor regulation of aspartyl-(asparaginyl)-beta-hydroxylase family genes in SH- Sy5y human neuroblastoma cells. BMC Cell Biol. 2006;7:41. doi: 10.1186/1471-2121-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gundogan F, Elwood G, Longato L, Tong M, Feijoo A, et al. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29:148–157. doi: 10.1016/j.placenta.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenwald I, Kovall R. Notch signaling: genetics and structure. WormBook. 2013 doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng AP, Aster JC. Multiple niches for Notch in cancer: context is everything. Curr Opin Genet Dev. 2004;14:48–54. doi: 10.1016/j.gde.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Boyault S, Rickman DS, de Reyniès A, Balabaud C, Rebouissou S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 31.Dürr R, Caselmann WH. Carcinogenesis of primary liver malignancies. Langenbecks Arch Surg. 2000;385:154–161. doi: 10.1007/s004230050259. [DOI] [PubMed] [Google Scholar]
- 32.Tovar V, Alsinet C, Villanueva A, Hoshid Y, Chiang DY. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. Journal of hepatology. 2010;52:550. doi: 10.1016/j.jhep.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borgas DL, Gao JS, Tong M, Roper N, de la Monte SM. Regulation of Aspartyl- (Asparaginyl)-beta-Hydroxylase Protein Expression and Function by Phosphorylation in Hepatocellular Carcinoma Cells. J Nat Sci. 2015;1 [PMC free article] [PubMed] [Google Scholar]
- 34.Borgas DL, Gao JS, Tong M, de la Monte SM. Potential Role of Phosphorylation as a Regulator of Aspartyl-(asparaginyl)-beta-hydroxylase: Relevance to Infiltrative Spread of Human Hepatocellular Carcinoma. Liver Cancer. 2015;4:139–153. doi: 10.1159/000367731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan X, Wu H, Xu H, Xiong H, Chu Q, et al. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer Lett. 2015;369:20–27. doi: 10.1016/j.canlet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 36.Feriotto G, Finotti A, Breveglieri G, Treves S, Zorzato F, et al. Transcriptional activity and Sp 1/3 transcription factor binding to the P1 promoter sequences of the human AßH-J-J locus. FEBS Journal. 2007;274:4476–4490. doi: 10.1111/j.1742-4658.2007.05976.x. [DOI] [PubMed] [Google Scholar]
- 37.de la Monte S, Derdak Z, Wands JR. Alcohol, insulin resistance and the liver-brain axis. J Gastroenterol Hepatol. 2012;27(Suppl 2):33–41. doi: 10.1111/j.1440-1746.2011.07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Monte SM, Yeon JE, Tong M, Longato L, Chaudhry R, et al. Insulin resistance in experimental alcohol-induced liver disease. J Gastroenterol Hepatol. 2008;23:e477–486. doi: 10.1111/j.1440-1746.2008.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 40.Lizarazo D, Zabala V, Tong M, Longato L, de la Monte SM. Ceramide inhibitor myriocin restores insulin/insulin growth factor signaling for liver remodeling in experimental alcohol-related steatohepatitis. J Gastroenterol Hepatol. 2013;28:1660–1668. doi: 10.1111/jgh.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang JH, Qi RC, Chen T, Chung TD, Stern AM, et al. Development of a carbon dioxide-capture assay in microtiter plate for aspartyl-beta-hydroxylase. Anal Biochem. 1999;271:137–142. doi: 10.1006/abio.1999.4135. [DOI] [PubMed] [Google Scholar]
- 42.de la Monte SM, Lahousse SA, Carter J, Wands JR. ATP luminescence-based motility-invasion assay. Biotechniques. 2002;33:98–100. doi: 10.2144/02331rr01. [DOI] [PubMed] [Google Scholar]
- 43.Gundogan F, Bedoya A, Gilligan J, Lau E, Mark P, et al. siRNA inhibition of aspartyl-asparaginyl ß-hydroxylase expression impairs cell motility, Notch signaling, and fetal growth. Pathol Res Pract. 2011;207:545–553. doi: 10.1016/j.prp.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawton M, Tong M, Gundogan F, Wands JR, de La Monte SM. Aspartyl- (asparaginyl) ß-hydroxylase, hypoxia-inducible factor-1a and notch cross-talk in regulating neuronal motility. Oxid Med Cell Longev. 2010;3:347–356. doi: 10.4161/oxim.3.5.13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ince N, de la Monte SM, Wands JR. Overexpression of human aspartyl (asparaginyl) beta-hydroxylase is associated with malignant transformation. Cancer Res. 2000;60:1261–1266. [PubMed] [Google Scholar]
- 46.Gundogan F, Elwood G, Greco D, Rubin LP, Pinar H, et al. Role of aspartyl-(asparaginyl) beta-hydroxylase in placental implantation: Relevance to early pregnancy loss. Hum Pathol. 2007;38:50–59. doi: 10.1016/j.humpath.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


