Abstract
The view that memory is encoded by variations in the strength of synapses implies that long-term biochemical changes take place within subcellular microdomains of neurons. These changes are thought ultimately to be an effect of transcriptional regulation of specific genes. Localized changes, however, cannot be fully explained by a purely transcriptional control of gene expression. The neuron-specific ELAV-like HuB, HuC, and HuD RNA-binding proteins act posttranscriptionally by binding to adenine- and uridine-rich elements (AREs) in the 3′ untranslated region of a set of target mRNAs, and by increasing mRNA cytoplasmic stability and/or rate of translation. Here we show that neuronal ELAV-like genes undergo a sustained up-regulation in hippocampal pyramidal cells only of mice and rats that have learned a spatial discrimination paradigm. This learning-specific increase of ELAV-like proteins was localized within cytoplasmic compartments of the somata and proximal dendrites and was associated with the cytoskeleton. This increase was also accompanied by enhanced expression of the GAP-43 gene, known to be regulated mainly posttranscriptionally and whose mRNA is demonstrated here to be an in vivo ELAV-like target. Antisense-mediated knockdown of HuC impaired spatial learning performance in mice and induced a concomitant down-regulation of GAP-43 expression. Neuronal ELAV-like proteins could exert learning-induced posttranscriptional control of an array of target genes uniquely suited to subserve substrates of memory storage.
Memory is thought to be stored in neurons by changes in the gene expression profile as a result of signal transduction events triggered by synaptic coincidence detectors. High-throughput analyses seem to confirm a memory-induced complex reprogramming of gene expression, implying both up-regulation and down-regulation of a number of genes (1, 2). The ultimate molecular determinants of these changes are thought to be transcription factors, some of which have been more or less conclusively related to memory (for a review see ref. 3). However, a purely transcriptional control of gene expression is insufficient to justify the occurrence of the “local” subcellular events that are thought to take place at the specific postsynaptic sites where learning-induced integration occurs. The participation of posttranscriptional control mechanisms acting on mRNA stability and translatability, posttranslational modifications, and protein turnover would provide a better opportunity for a local regulation of gene expression in “activated” neuronal microdomains. To date, no posttranscriptional mechanism of gene expression has been convincingly linked to memory formation.
ELAV-like proteins, also called Hu antigens, are mammalian orthologues of the elav (embryonic lethal abnormal vision) gene of Drosophila (4). As elav is necessary for the development and maintenance of the fly nervous system (5), the ELAV-like HuB, HuC, and HuD genes (expressed only in neurons, whereas HuA, also called HuR, is ubiquitous) are early markers of neuronal differentiation (6) and appear to be necessary to accomplish this cellular program (7, 8). All four members of this family code for RNA-binding proteins endowed with three RNA-interacting domains of the RRM type (4). The first two domains recognize and specifically associate with a target motif, the A+U-rich element (ARE), located in the 3′ untranslated region (UTR) of a subset of target mRNAs, whereas the third domain seems to bind the mRNA poly(A) tail (9). Recently, ELAV-like proteins have been shown both in vivo and in cultured cells to be coupled to translational enhancement of the bound mRNAs (4, 10–14).
The neuron-specific expression of HuB, HuC, and HuD; their demonstrated nucleocytoplasmic shuttling capability (15); their clear neurite-inducing activity (8); and their positive regulation of an array of target genes all make them very attractive candidates for a role in memory. In this work we clearly demonstrate such a role for two spatial learning paradigms in rodents, at the same time providing evidence for the involvement of posttranscriptional events in the learning-induced reprogramming of gene expression.
Materials and Methods
Behavioral Testing.
Male mice of the inbred strain C57BL/6, 14–15 weeks old, were used for the radial arm maze experiments. Mice were tested under computer control in an elevated eight-arm radial maze, as described (16), in which only three adjacent arms were used. During training, animals were exposed to 16 successive pairs of radial arms in a pseudorandom order. The baited center arm was in all pairs. Training was ended after 3 to 7 days, when an accuracy of less than 6 errors in 16 trials (per day) was achieved. Animals that did not reach the criterion within seven training sessions were not included in the trained (TR) group. One day after the final training day, each selected animal was tested in a probe session by eight exposures to all three arms opened simultaneously. Each TR animal was yoked to an active control (AC) animal that received exactly the same number of training trials. For this yoked AC group, food reinforcement was pseudorandomly located among the three arms for each trial. Thus, each active control animal received the same reinforcement and underwent the same locomotor activity as its yoked TR counterpart. Passive control (PC) animals remained in their home cages, were food deprived as above, and received food reinforcement each day.
Eight-week-old male Wistar rats were subjected to a water maze task in a swimming pool 1.5 m in diameter and 0.6 m high, filled with milky water (24 ± 1°C) and located in a well-lit room with distinct extra-maze cues. A 10-cm2 transparent square platform was hidden in a constant location (quadrant center) within the pool with its top surface submerged 1.5 cm below the water level. On day 0, rats were subjected to 2.5 min of swimming in the pool in the absence of the platform, for adaptation to the environment. For the next 5 days, rats were trained to locate the hidden island in four trials per day. Rats were then allowed to stay on the platform for 40 s before starting the next trial from another quadrant. Rats from the swimming control group swam for 2 min a day in the pool without the island for the next 5 days to equalize nonlearning behavior, such as locomotor activity and stress responses. Spatial memory of rats was assessed by the time required to find the platform (escape latency). Transfer tests performed on a separate group of animals demonstrated a clear spatial bias in rats' swim pathways after the same training. Naive control rats spent approximately the same number of days after delivery in their home cages. Five minutes after the probe session for mice and 24 h after the last session for rats, animals were killed, the brains were quickly removed, and the hippocampi were dissected and kept at −80°C until the biochemical assays.
Subcellular Fractionation and Western Blotting.
Subcellular fractions were prepared from mouse hippocampi as published (17) with minor modifications. Sample proteins were diluted in SDS protein gel loading solution, boiled for 5 min, separated by SDS/12% PAGE, and processed by standard procedures, with the use of the anti-Hu 16A11 mAb, the anti-BRUNO 3B1 mAb, and the anti-α-tubulin Ab.
Real-Time Quantitative Reverse Transcription–PCR (RT-PCR).
Total RNA from dissected mouse and rat hippocampi was extracted, treated with DNase, and subjected to reverse transcription by following standard procedures. PCR amplifications were carried out with the Lightcycler instrument (Roche Molecular Biochemicals). Aliquots of cDNA (0.1 μg) and known amounts of external standards (purified PCR products diluted in RT mix, 102 to 108 molecules) were amplified in parallel reactions (final volume 20 μl), with primers designed on the 3′ UTRs of the murine HuB, HuC, HuD, GAP-43, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and cycA mRNA sequences, and with the use of the FastStart SYBR Green master mix (Roche Molecular Biochemicals). The optimal cycle programs were determined for each single gene in preliminary PCR runs to obtain a single, specific PCR product, verified by melting curve analysis followed by gel electrophoresis and DNA sequencing. The temperature transition rate was 20°C/s and the fluorimeter gain value was 7. Quantification was performed by real-time detection of fluorimetric emission of SYBR green I measured at the end of each elongation phase (which indicates the amount of PCR product formed) and by comparison of the fluorescence of PCR products of unknown concentration with the fluorescence of the external standards. Only fluorescence values measured in the log-linear phase of amplification were considered, with the use of the second derivative maximum method of the Lightcycler data analysis software. For each single determination the PCRs were run in quadruplicate and gave an average variability of less than 10%.
Immunohistochemistry.
Twelve-micrometer-thick cryostat sections of the hippocampi from the three groups of mice were mounted on precoated glass slides and dried at room temperature. Sections were fixed for 30 min in freshly prepared 4% formaldehyde in PBS (pH 7.4), boiled for 30 min in 50 mM Tris (pH 8.0), and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Biotin nonspecific sites were blocked, and then the slices were exposed to the biotin-conjugated 16A11 anti-Hu mAb at 4°C overnight, followed by incubation with an avidin-peroxidase complex for 30 min at 37°C. Slides were rinsed twice with PBS, and bound peroxidase was visualized with the use of 0.05% 3,3-diaminobenzidine tetrahydrochloride (DAB) together with a 3% aqueous solution of H2O2 as chromogenic substrate. Color development was stopped by rinsing the slides in distilled water. Slides were finally dehydrated in graded ethanol and mounted for microscopic analysis.
In Situ Hybridization.
cDNA segments of about 300 bp were obtained by PCR on brain cDNA with the use of primers designed in the 3′ UTR of the murine GAP-43, HuC, and BRUNOL3 genes and of the rat HuC and BRUNOL3 genes. These cDNA segments were inserted into the pPCR-ScriptAmp vector (Stratagene) and sequenced. In situ hybridization on hippocampal slices was performed as described (18).
In Vivo RNA–Protein Binding.
One microgram of paramagnetic polystyrene microspheres covalently linked to streptavidin (Dynabeads M-280 Streptavidin; Dynal, Great Neck, NY) was washed twice in diethyl pyrocarbonate-treated solution A (0.1 M NaOH/0.05 M NaCl) and once in diethyl pyrocarbonate-treated solution B (0.1 M NaOH) by placing the tube in a magnetic particle concentrator for 3 min and then removing the free fraction, to ensure an RNase-free environment. For immunopurification of ELAV-like proteins together with the bound mRNAs, 10 μg of biotinylated 16A11 mAb was added to the beads in PBS + 1% BSA and incubated for 30 min at room temperature. The 16A11 mAb–beads complex was washed five times in PBS + 0.1% BSA and, after removal from the concentrator, incubated for 2 h with different aliquots of mouse total hippocampal tissue lysate, with continuous gentle mixing. After removal of the free fraction in the magnetic concentrator, beads were washed three times in PBS + 0.1% BSA, and total RNA was extracted from both fractions and subjected to qualitative RT-PCR. For purification of ELAV-like proteins by the GAP-43 mRNA binding site, RNase-free streptavidin-coated beads were incubated for 30 min at room temperature with 250 ng of 5′-biotinylated RNA oligonucleotides in binding buffer (2.5 μg of tRNA/1 mM DTT/40 units of RNasin/100 mM NaCl/10 mM Tris⋅HCl, pH 6.9) and washed five times with washing buffer 1 (100 mM NaCl/10 mM Tris⋅HCl, pH 6.9/1% BSA/0.5% Tween 20) and three times with washing buffer 2 (100 mM NaCl/10 mM Tris⋅HCl, pH 6.9). Bound proteins were eluted by the addition of 0.1 M triethylamine for 5 min, neutralized with Tris⋅HCl (1 M, pH 8), and then subjected to Western blotting.
Oligodeoxynucleotide (ODN) Design, Preparation, and Injection.
The sequence of the anti-HuC antisense (AS) ODN was chosen by cell culture prescreening with a panel of eight AS ODNs targeted to different regions of the murine HuC mRNA. All of the sequences were checked for the absence of significant homology with other mouse expressed genes and for minimization of potential hairpin and duplex formation. Primary hippocampal cell cultures were established from embryonic day 18 mouse embryos by described procedures (19) and treated with the different AS ODNs in triplicate at 1 μM final concentration for 3 days. The total RNA was extracted and subjected to semiquantitative RT-PCR for the HuC and GAPDH genes. The antisense with the sequence 5′-CCCAGTATCTGAGTGACCATTCTT-3′ was selected for the in vivo experiments because it induced the highest inhibition of HuC expression (72%). It is complementary to bases 401–424 of the mouse HuC gene (GenBank accession no. U29148). The AS ODN and the control, fully degenerate ODN (DN ODN) (5′-NNNNNNNNNNNNNNNNNNNNNNNN-3′, where N = A, T, G, or C) were phosphorothioate-protected by a 5′ and 3′ end double substitution, synthesized on a 10-μmol scale, and purified by HPLC. They were solubilized (4 mM) in N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate cationic lipid (13 μM) and sterile water. Intracerebroventricular injections were performed in freely moving mice via injection cannulae (8.5 mm long) attached to 1-μl Hamilton syringes with polyethylene catheter tubing. The syringes were fixed in a constant-rate infusion pump, and fluids (0.75 μl) were injected over a 3-min period. In all cases, injection flow rates were checked visually. Treatment began 3 days before the beginning of behavioral experiments, and injections were performed 2 h after the behavioral sessions. Surgery was performed on the mice while they were under general anesthesia (sodium thiopental, 70 mg/kg, and atropine sulfate, 0.01 ml, i.p.). A stainless steel guide cannula (30 gauge, 8 mm long) was stereotaxically implanted into the lateral ventricle (randomly in the left or right ventricle). Stereotaxic coordinates were 1.0 mm posterior to bregma, ±1.0 mm lateral to the midline, and 1.50 mm below the skull. After surgery the animals were given 10 days to recover before the experiments were started.
Data Analysis.
All experiments were repeated at least three times for each hippocampal tissue sample. Signals from in situ hybridization and Western blotting experiments were quantified by measuring the optic mean density of the relevant fields with the use of the National Institutes of Health image program. For statistical analysis, values were subjected to one-way ANOVA and a post hoc Tukey's test unless otherwise stated.
Results and Discussion
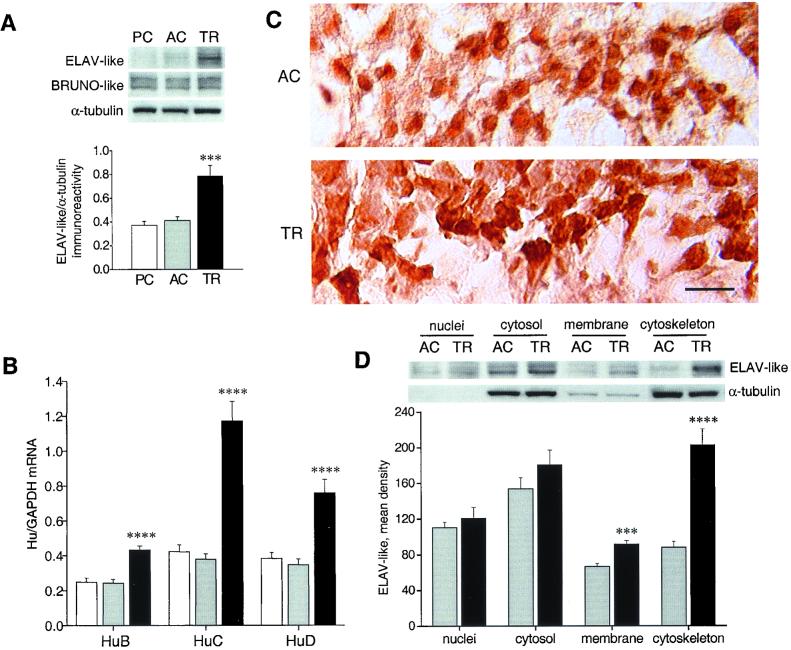
Western blot analysis of hippocampal lysates of mice obtained 24 h after the last training session in the radial maze (Fig. 1A) showed that ELAV-like immunoreactivity, tested by a monoclonal antibody able to recognize the three neuronal members of the family (6) and normalized to α-tubulin immunoreactivity, is significantly different among the three groups [one-way ANOVA, F(2,12) = 15.38, P < 0.001]. Further post hoc analysis with Tukey's test revealed an increase in the TR group with respect to the PC (P < 0.001) and, more importantly, to the AC animals (P < 0.005) but not between the two control groups, demonstrating a learning-specific effect. Immunoreactivity of the BRUNO-like proteins, the group of RNA-binding proteins phylogenetically closest to the Hu antigens, was unchanged among the groups. We then assessed at the mRNA level the contribution of individual Hu proteins to this increased expression, employing a quantitative, external standard-based, real-time PCR approach. A substantial enhancement in the mRNA steady-state levels, relative to the housekeeping GAPDH mRNA (and cycA mRNA, not shown), was confirmed for all of the three neuronal ELAV-like genes (post hoc pairwise comparisons between AC and TR mice, P < 0.001), with HuC showing the largest increase (about 3-fold; Fig. 1B). Immunohistochemical analysis of the AC and TR mice groups revealed a learning-specific dramatic increase in ELAV-like immunostaining, with a clear enhancement of cytoplasmic reactivity to the Hu antigens in proximal dendritic regions of the pyramidal neurons, in both the CA3 (Fig. 1C) and CA1 hippocampal (not shown) subfields. This pattern of distribution is reminiscent of that observed in PC12 cells, in which a strong neurite-inducing activity is strictly dependent on cytoplasmic localization of overexpressed HuD (8). To better identify the cell compartments of maximal ELAV-like protein increase, we examined different cellular fractions obtained from AC and TR mice hippocampi. The membrane and, particularly, the cytoskeletal fractions of TR animals showed a significantly increased ELAV-like immunoreactivity (Student's t test, P < 0.005 and P < 0.001, respectively), whereas the increase in quantity of nuclear and cytosolic ELAV-like protein was not statistically significant (Fig. 1D). The cytoskeletal accumulation of the learning-induced ELAV-like proteins is particularly meaningful, because HuB and HuD have been shown in cell cultures to bind to microtubules and to the associated ribosomes (20, 21), suggesting that assembly of this supramolecular complex may be critical for activity.
Figure 1.
Up-regulation of neuronal ELAV-like gene expression in mouse hippocampi after radial arm maze training. (A) (Upper) Representative Western blots showing whole-cell lysate immunoreactivity of the ELAV-like and BRUNO-like proteins (using the 16A11 and 3B1 mAbs, respectively) in the passive control (PC), active control (AC), and trained (TR) mouse groups. (Lower) Average results (means ± SEM) for ELAV-like proteins normalized to α-tubulin (n = 5 for each group; ***, P < 0.005, post hoc analysis between AC and TR mice). (B) Determination of the steady-state levels of HuB, HuC, and HuD mRNA by external standard-based, real-time quantitative RT-PCR. The values obtained from hippocampal RNA preparations of the three groups of mice (white bar, PC; light gray bar, AC; black bar, TR) have been normalized to the level of GAPDH mRNA and expressed as means ± SEM (n = 6 for each group; ****, P < 0.001, post hoc analysis between AC and TR mice). (C) Representative images showing the distribution of ELAV-like immunostaining for AC and TR animals in the pyramidal layer of the CA3 hippocampal subregion. (Scale bar, 25 μm.) (D) Representative Western blots (Upper) and average ELAV-like protein levels (Lower) after tissue fractionation. α-Tubulin levels are shown as a control of the nucleocytoplasmic separation, and ELAV-like protein levels are reported as means ± SEM (n = 6 for each group, ***, P < 0.005; ****, P < 0.001, Student's t test). All experiments were repeated at least three times for each hippocampal tissue sample. For statistical analysis, values were subjected to one-way ANOVA and a post hoc Tukey's test unless stated otherwise.
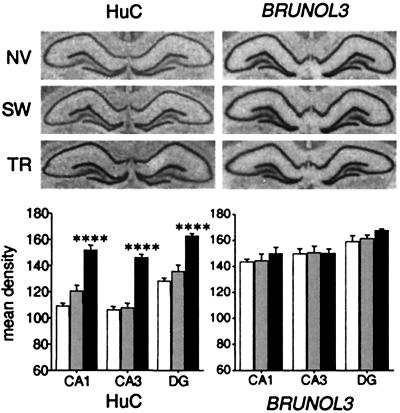
We then investigated the generality of the learning-induced ELAV-like gene up-regulation with the use of a different, well-established model of spatial memory, the Morris water maze. A group of rats was trained to locate an invisible submerged platform in a swimming pool, matched by an active control of rats swimming without any platform (SW) and a passive control of naive caged animals (NV). Hippocampal lysates obtained after the transfer test from these three groups were subjected to Western blotting for the neuronal ELAV-like proteins and to real-time RT-PCR for the HuC gene. The results showed a clear training-dependent increase in both ELAV-like immunoreactivity and HuC mRNA steady-state levels, similar in magnitude to that observed in the radial maze-trained mice (data not shown). In situ hybridization on the same hippocampi (Fig. 2A) showed that the enhancement of HuC expression was common to all of the three major hippocampal subfields (Fig. 2B, post hoc comparisons between TR and either NV or SW rats after one-way ANOVA, P < 0.001 for CA1, CA3, and dentate gyrus), whereas the levels of the rat BRUNOL3 always remained unchanged. Independent experiments performed by another group indicate up-regulation of HuD gene expression in a model of contextual fear conditioning in the rat, further extending the significance of this finding to nonspatial memory paradigms (N. I. Perrone-Bizzozzero, personal communication).
Figure 2.
Up-regulation of HuC gene expression in rat hippocampi after Morris water maze training. Representative in situ hybridization results are reported above for both the HuC and the BRUNOL3 genes in naïve (NV), swimming control (SW), and trained (TR) animals. Below, means ± SEM of the densitometric determinations of the mRNA signal in the CA1, CA3, and dentate gyrus (DG) subfields for six animals in each group (white bar, NV; light gray bar, SW; black bar, TR; ****, P < 0.001, post hoc analysis between SW and TR rats).
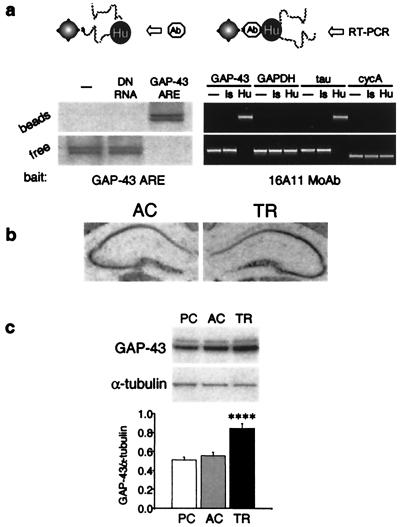
ELAV-like proteins are endowed with three RNA-binding domains of the RNP consensus sequence that predict an RNA-binding activity clearly demonstrated to be selective for AREs of the 3′ UTRs of various different mRNAs (22, 23). These AREs are known to be key determinants of mRNA turnover (24). Furthermore, a substantial body of evidence indicates that ELAV-like proteins exert a positive regulatory function on gene expression through posttranscriptional stabilization (10, 11) and possibly enhanced translation (12) of ARE-endowed mRNAs. One target of ELAV-like proteins is the mRNA of the GAP-43 gene (25), known to be a determinant of neural development, regeneration of neural connections, and synaptic plasticity in mature synapses (26). Whereas GAP-43 neuron-specific expression is controlled by elements in the promoter of this gene, its induction during neuritogenesis has been ascribed essentially to posttranscriptional events involving mRNA stabilization (27). We therefore hypothesized that the observed sustained activation of ELAV-like proteins by spatial learning could result in a downstream up-regulation of GAP-43 expression. To test this idea, we first looked at the in vivo binding of the neuronal ELAV-like proteins to GAP-43 mRNA. We set up a magnetic separation procedure with streptavidin-coated beads, with the use of, as bait, a biotinylated synthetic RNA oligonucleotide reproducing the Hu binding of GAP-43 mRNA or, alternatively, the biotinylated anti-Hu monoclonal antibody. Western blotting and RT-PCR on the bead-associated and free fractions showed (Fig. 3a) that the GAP-43 mRNA present in mouse hippocampal lysates is quantitatively complexed with the ELAV-like proteins. A degenerate RNA probe instead of the GAP-43 mRNA binding site was unable to associate with the Hu antigens, and, on the other hand, the ARE-negative GAPDH and cycA mRNAs were not recruited together with the antibody in the bead-associated fraction, as occurred with the ARE-positive and ELAV-like-bound (28) tau mRNA. Together with negative controls, these binding properties ensured the selectivity of the interaction. We then examined GAP-43 gene expression at the mRNA level [by in situ hybridization (Fig. 3b) and by real-time RT-PCR, not shown] and at the protein level (by Western blotting) in the group of mice subjected to radial arm maze training and in the AC and PC groups. We found a consistent up-regulation [one-way ANOVA of the protein determination: F(2,15) = 20.24, P < 0.001] for which a post hoc analysis revealed learning specificity (TR vs. either control, P < 0.001). We found that this key gene of neuronal plasticity is overexpressed as a consequence of behavioral training in mammals, a result consistent with the recent description of an enhancement of learning performance ability in a strain of transgenic mice that overexpress GAP-43 (29).
Figure 3.
In vivo binding of GAP-43 mRNA by neuronal ELAV-like proteins and enhancement of GAP-43 gene expression by learning. (a) (Lower) Magnetic bead-mediated coseparation of ELAV-like proteins and GAP-43 mRNA with the use of an RNA oligonucleotide reproducing the GAP-43 ELAV-like binding ARE (Left) and the 16A11 mAb (Right). (Upper) Schematic representation of the strategy used in either case. The presence of the protein and of different mRNAs in the bead-associated and the free fractions is assessed respectively by Western blotting with the 16A11 mAb and by qualitative RT-PCR with specific primer pairs. Negative controls for both experiments are the absence of the bead-linked ligand (RNA oligonucleotide or 16A11 mAb) and a nonligand (fully degenerate RNA oligonucleotide, DN RNA, or a mAb of the same 16A11 isotype, Is). (b) Representative in situ hybridization histochemistry obtained with a GAP-43 mRNA probe in brain slices of AC and TR mice. Note the absence of GAP-43 expression in the dentate gyrus. (c) Representative Western blot (Upper) and related average results normalized to α-tubulin immunoreactivity (Lower; means ± SEM) of GAP-43 protein levels in hippocampi of PC, AC, and TR mice (n = 6 for each group; ****, P < 0.001, post hoc analysis between AC and TR mice).
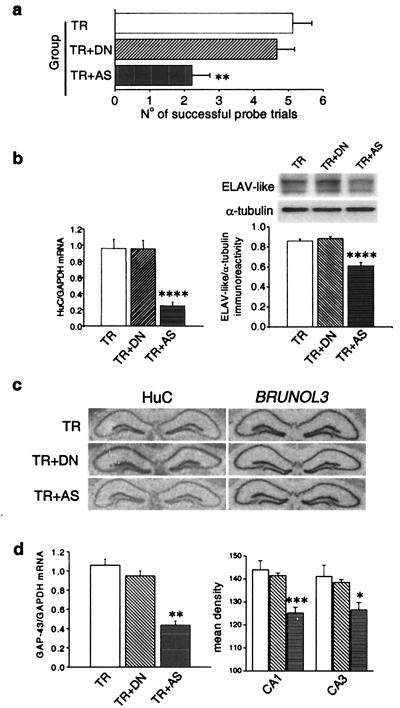
To more directly address the role of ELAV-like genes in spatial learning we adopted an antisense-mediated “knockdown” strategy targeted to the mRNA of HuC, the ELAV-like gene that was particularly up-regulated (cf. Fig. 1B) by radial maze training. We administered daily intracerebroventricular injections of 3 nmol of a phosphorothioate-capped AS ODN in the mice, from 3 days before and throughout the whole behavioral training period. As a control for the AS ODN, a sequence-independent DN ODN was administered to a group of mice undergoing the same radial arm maze training, and a third group served as the sham control. Fig. 4a shows the discrimination performance for the three groups of mice in a probe session conducted 24 h after the last training session. A one-way ANOVA indicates significant differences [F(2,23) = 8.99; P = 0.001]. Pairwise comparisons show that the anti-HuC AS ODN significantly impairs mice while performing in the probe task (AS ODN vs. sham controls: P = 0.002; AS ODN vs. DN ODN: P < 0.01), whereas sham controls do not differ from mice treated with the DN ODN. We then verified the AS ODN-induced down-regulation of the HuC gene at the mRNA level with real-time quantitative PCR, finding a steady-state level about four times less than in the sham controls. This antisense-induced down-regulation was reflected by a clear decrease in the overall ELAV-like immunoreactivity assessed by Western blotting. A post hoc pairwise comparison between AS ODN- and DN ODN-treated mice gave a P < 0.001 for both mRNA and protein level determinations, whereas the DN ODN showed no significant effect on ELAV-like immunoreactivity or HuC mRNA. The results are summarized in Fig. 4b. Specificity of the antisense treatment was assessed by in situ hybridization on the three groups of mice for the HuC and the BRUNOL3 gene (other names, Etr-3, NAPOR), belonging to the ELAV-related BRUNO-like subfamily. Representative in situ images are shown in Fig. 4c. Comparison between DN ODN-treated and AS ODN-treated mice showed an average hippocampal decrease for HuC (mean densities ± SEM, 137.9 ± 2.5 and 101.4 ± 2.4, respectively; n = 6 for each group, Student's t test: P < 0.001), confirming the RT-PCR results, whereas there was no effect for BRUNOL3. We then reasoned that if the observed coincident up-regulation of the neuronal ELAV-like proteins with GAP-43 was a cause–effect relationship, the specific antisense-mediated down-regulation of HuC should be reflected in a decrease in GAP-43 expression levels. Fig. 4d shows that there was indeed such a decrease in GAP-43 expression levels; a significant down-regulation of GAP-43 mRNA, revealed by one-way ANOVA and post hoc analysis, was observed for the anti-HuC antisense treatment by quantitative RT-PCR and confirmed by in situ hybridization in both the CA1 and CA3 hippocampal subfields (in all cases after pairwise comparisons AS ODN- and DN ODN-treated mice were different at P < 0.001). Similar to this finding, in PC12 cells antisense-mediated down-regulation of another ELAV-like gene, HuD, resulted in a correlated decrease in GAP-43 mRNA steady-state levels and stability and in defective neurite outgrowth (30).
Figure 4.
Behavioral and biochemical effects of antisense-mediated knockdown of HuC gene expression in mouse brain. (a) Performance in the probe session of the radial maze task in sham controls (TR) and in mice daily infused during the training with fully sequence-degenerate (TR + DN) or anti-HuC antisense (TR + AS) oligonucleotides (n = 9 for TR animals and n = 8 for TR + DN and TR + AS animals). Data are expressed as means ± SEM (**, P < 0.01, post hoc analysis between DN-treated and AS-treated mice). (b) Effect of the anti-HuC antisense treatment on HuC gene expression. (Left) Real-time quantitative RT-PCR determination of HuC mRNA steady-state levels normalized to GAPDH levels. (Right) (Upper) Representative Western blot of the overall ELAV-like immunoreactivity in the three groups of mice compared with α-tubulin. (Lower) Means ± SEM, n = 6 each group for both mRNA and protein determinations. ****, P < 0.001, post hoc analysis between DN-treated and AS-treated mice. (c) Sequence specificity of the antisense-mediated HuC gene expression down-regulation. Representative in situ hybridizations showing hippocampal mRNA levels for the HuC and for the phylogenetically related BRUNOL3 genes in the AS ODN-treated and the two control mice groups. (d) Effect of the anti-HuC antisense treatment on GAP-43 gene expression. (Left) Real-time quantitative RT-PCR determination of GAP-43 mRNA levels normalized to GAPDH levels in the hippocampi of the three groups of mice (means ± SEM, n = 6 for each group; **, P < 0.01, post hoc comparison between DN-treated and AS-treated mice). (Right) Densitometric analysis of GAP-43 mRNA levels in the CA1 and CA3 hippocampal subfields from in situ hybridization performed on brain slices of the same mice (means ± SEM, data collected from five to seven mice from each group; *, P < 0.05; ***, P < 0.005).
The 3′ UTRs of c-fos (31), egr-1 (23), Nf1 (32), and HuB (33) mRNAs, in addition to GAP-43 mRNA, have been shown to be specifically bound by members of the ELAV-like protein family. c-fos and egr-1 are well-known immediate early genes demonstrated to be up-regulated in the hippocampus and in other brain structures in a number of classical and instrumental conditioning paradigms in mammals, and the Nf1 gene has been proven to be required for spatial learning in mice (34) and for olfactory associative learning in flies (35). The HuB and GAP-43 genes have been implicated in spatial learning here. Moreover, ARE elements predicted to be potential ELAV-like binding sites are present in the mRNA 3′ UTR of other genes that have been related to neuronal plasticity, such as bcl-2 and neuritin.
The learning-induced increase in ELAV-like mRNA stabilizing proteins associated particularly with the cytoskeleton of hippocampal pyramidal cells, together with the interfering effect on learning of anti-HuC AS ODNs, suggests a role for these proteins in the establishment of memory traces. The learning-induced up-regulation of GAP-43 appears to be at least in part a direct effect of the increased levels of ELAV-like proteins, given that anti-HuC antisense treatment specifically also depresses GAP-43 expression. The established notion that ELAV-like proteins positively regulate gene expression by binding to AREs and by prolonging half-life of an array of target mRNAs provides us with a plausible molecular mechanism by which a single learning-elicited signal can be translated into the selective multiple enhancement of de novo protein synthesis of an array of memory effector proteins. These effector proteins could cooperatively orchestrate long-lasting biochemical changes and morphological rearrangements.
This report links posttranscriptional regulation of gene expression to memory formation. The ELAV-like proteins could be key components of a basic molecular machinery able to confer increased translatability, and therefore increased protein availability, at only those synaptic sites relevant to a specific learning event. This model has the advantage of suggesting a basis for learning-induced localized biochemical events, which cannot be fully provided for by reprogramming of the gene expression profile at the transcriptional level.
Abbreviations
- ARE
adenine- and uridine-rich element
- UTR
untranslated region
- AC
active control
- PC
passive control
- TR
trained
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- ODN
oligodeoxynucleotide: AS ODN, antisense ODN
- DN ODN
fully degenerate ODN
References
- 1.Cavallaro S, Meiri N, Yi C L, Musco S, Ma W, Goldberg J, Alkon D L. Proc Natl Acad Sci USA. 1997;94:9669–9673. doi: 10.1073/pnas.94.18.9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavallaro S, Schreurs B G, Zhao W, D'Agata V, Alkon D L. Eur J Neurosci. 2001;13:1809–1815. doi: 10.1046/j.0953-816x.2001.01543.x. [DOI] [PubMed] [Google Scholar]
- 3.Tischmeyer W, Grimm R. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antic D, Keene J D. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao K M, Samson M L, Reeves R, White K. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- 6.Marusich M F, Furneaux H M, Henion P D, Weston J A. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 7.Akamatsu W, Okano H J, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell R B, Okano H. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasashima K, Terashima K, Yamamoto K, Sakashita E, Sakamoto H. Genes Cells. 1999;4:667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- 9.Ma W J, Chung S, Furneaux H. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford L P, Watson J, Keene J D, Wilusz J. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antic D, Lu N, Keene J D. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng S S, Chen C Y, Xu N, Shyu A B. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan X C, Steitz J A. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascale A, Nogues X, Marighetto A, Micheau J, Battaini F, Govoni S, Jaffard R. NeuroReport. 1998;9:725–729. doi: 10.1097/00001756-199803090-00030. [DOI] [PubMed] [Google Scholar]
- 17.Leach K L, Ruff V A, Jarpe M B, Adams L D, Fabbro D, Raben D M. J Biol Chem. 1992;267:21816–21822. [PubMed] [Google Scholar]
- 18.Zhao W, Chen H, Xu H, Moore E, Meiri N, Quon M J, Alkon D L. J Biol Chem. 1999;274:34893–34902. doi: 10.1074/jbc.274.49.34893. [DOI] [PubMed] [Google Scholar]
- 19.Guo Q, Sebastian L, Sopher B L, Miller M W, Ware C B, Martin G M, Mattson M P. J Neurochem. 1999;72:1019–1029. doi: 10.1046/j.1471-4159.1999.0721019.x. [DOI] [PubMed] [Google Scholar]
- 20.Antic D, Keene J D. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 21.Gao F-B, Keene J D. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 22.Gao F-B, Carson C C, Levine T, Keene J D. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenenbaum S A, Carson C C, Lager P J, Keene J D. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C Y, Shyu A-B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 25.Chung S, Eckrich M, Perrone-Bizzozero N I, Kohn D T, Furneaux H. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 26.Aarts L H, Schotman P, Verhaagen J, Schrama L H, Gispen W H. Adv Exp Med Biol. 1998;446:85–106. doi: 10.1007/978-1-4615-4869-0_6. [DOI] [PubMed] [Google Scholar]
- 27.Perrone-Bizzozero N I, Cansino V V, Kohn D T. J Cell Biol. 1993;120:1263–1270. doi: 10.1083/jcb.120.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aranda-Abreu G E, Behar L, Chung S, Furneaux H, Ginzburg I. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Routtenberg A, Cantallops I, Zaffuto S, Serrano P, Namgung U. Proc Natl Acad Sci USA. 2000;97:7657–7662. doi: 10.1073/pnas.97.13.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobarak C D, Anderson K D, Morin M, Beckel-Mitchener A, Rogers S L, Furneaux H, King P, Perrone-Bizzozero N I. Mol Biol Cell. 2000;11:3191–3203. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung S, Jiang L, Cheng S, Furneaux H. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 32.Haeussler J, Haeusler J, Striebel A M, Assum G, Vogel W, Furneaux H, Krone W. Biochem Biophys Res Commun. 2000;267:726–732. doi: 10.1006/bbrc.1999.2019. [DOI] [PubMed] [Google Scholar]
- 33.Abe R, Yamamoto K, Sakamoto H. Nucleic Acids Res. 1996;24:2011–2016. doi: 10.1093/nar/24.11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva A J, Frankland P W, Marowitz Z, Friedman E, Lazlo G, Cioffi D, Jacks T, Bourtchuladze R. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- 35.Guo H F, Tong J, Hannan F, Luo L, Zhong Y. Nature (London) 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]