Abstract
Rationale: Cystic fibrosis, like primary ciliary dyskinesia, is an autosomal recessive disorder characterized by abnormal mucociliary clearance and obstructive lung disease. We hypothesized that genes underlying the development or function of cilia may modify lung disease severity in persons with cystic fibrosis.
Objectives: To test this hypothesis, we compared variants in 93 candidate genes in both upper and lower tertiles of lung function in a large cohort of children and adults with cystic fibrosis with those of a population control dataset.
Methods: Variants within candidate genes were tested for association using the SKAT-O test, comparing cystic fibrosis cases defined by poor (n = 127) or preserved (n = 127) lung function with population controls (n = 3,269 or 3,148, respectively). Associated variants were then tested for association with related phenotypes in independent datasets.
Results: Variants in DNAH14 and DNAAF3 were associated with poor lung function in cystic fibrosis, whereas variants in DNAH14 and DNAH6 were associated with preserved lung function in cystic fibrosis. Associations between DNAH14 and lung function were replicated in disease-related phenotypes characterized by obstructive lung disease in adults.
Conclusions: Genetic variants within DNAH6, DNAH14, and DNAAF3 are associated with variation in lung function among persons with cystic fibrosis.
Keywords: respiratory function tests, ciliary motility disorders, cystic fibrosis, genetic association studies
Identification of genes and pathways that modify the clinical characteristics of Mendelian conditions can facilitate the development of new pharmaceuticals and treatment strategies (1, 2). Cystic fibrosis (CF; [MIM:219700]) is an autosomal recessive condition for which the genetic basis has been known for 25 years. The observation that there is wide interindividual variation in severity of lung disease in persons with the same cystic fibrosis transmembrane conductance regulator gene (CFTR) genotype (i.e., p.Phe508del homozygotes) has prompted an aggressive search for genetic and environmental modifiers that might point to novel treatment strategies. To this end, candidate gene analyses and genome-wide linkage and association studies have identified modifiers for severity of airway obstruction (e.g., TGFβ-1, APIP) and infection with Pseudomonas aeruginosa (e.g., MBL2, SLC6A14, DCTN4, CAV2, TMC6) (3–7), both of which contribute to disease progression and early death. Nevertheless, it is suspected that many additional modifiers of CF-related lung disease remain to be discovered.
CF results from disrupted epithelial cell chloride and bicarbonate transport that alters the characteristics of exocrine gland secretions in multiple tissues (8). This leads to abnormal lung host defenses and reduced mucociliary clearance (8–10) that cause chronic airway infection, inflammation, and injury (e.g., bronchiectasis [9, 10]). Bronchiectasis and chronic airway inflammation/infection are hallmarks of several other conditions, including primary ciliary dyskinesia (PCD; [MIM:244400]), chronic bronchiectasis ([MIM:PS211400]), and chronic obstructive pulmonary disease (COPD; [MIM:606963]). Chronic bronchiectasis and COPD are complex traits for which the genetic basis remains largely unknown. In contrast, PCD is a rare autosomal recessive disorder (11, 12) caused by mutations in any one of several genes that encode proteins involved in ciliary structure and function (13). We hypothesized that genetic variants within cilia genes are associated with severity of lung disease in individuals with CF. To test this hypothesis, we used a phenotypic extreme versus population control study design (6) to test for associations between candidate genes encoding cilia proteins and both higher- and lower-than-expected lung function in persons with CF. Some of these results were reported previously in the form of an abstract (14).
Methods
Discovery Cohort
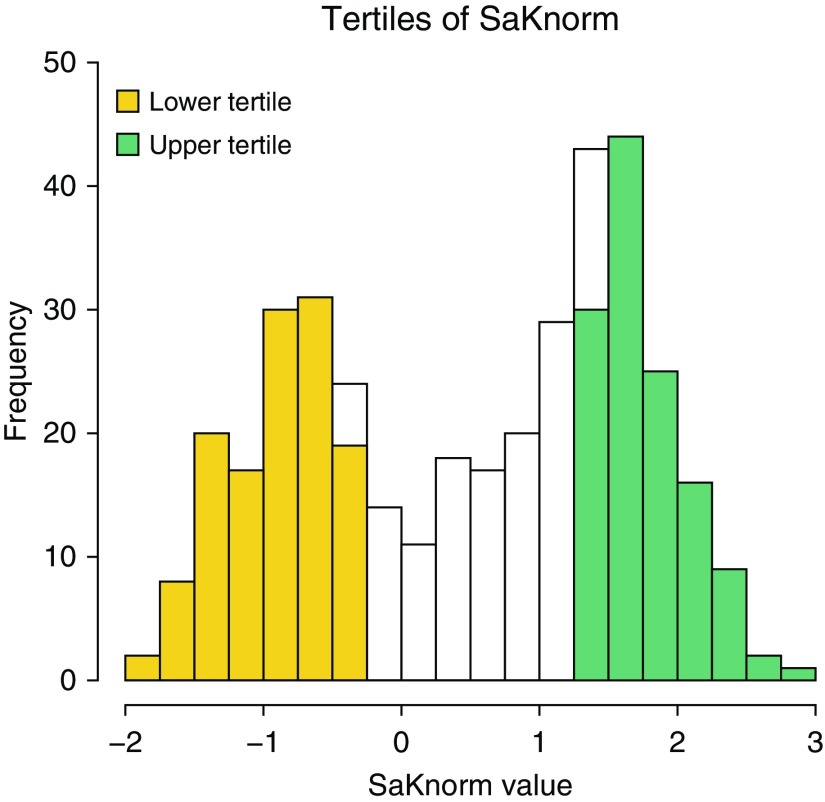
Cases were drawn from the EPIC (Early Pseudomonas Infection Control [15]) Observational Study and the Genetic Modifier Study (16), a member of the North American CF Gene Modifier Consortium. The phenotype assessed was SaKnorm, a measure of lung function based on forced expiratory volume in 1 second (FEV1) adjusted for age, height, and sex among individuals with CF (17) and then further adjusted for mortality (18). Subsets of these data were reported in a prior publication (7). Principal components (PCs) were used to identify outliers and to adjust for population stratification and/or batch effects (see Figures E1 and E2 in the online supplement). From a total of 404 participants with non-Hispanic European ancestry for whom exome sequencing data were available, we excluded all but one sibling, PC outliers, and those without SaKnorm values, leaving a sample of 381 persons. These 381 persons include 190 females and 191 males, and predominantly belong to CFTR risk group 1, defined as having two minimal function CFTR mutations (n = 352), with 308 p.Phe508del homozygotes and 10 p.Phe508 heterozygotes. The mean patient age in 2017 was 30.34 ± 0.57 years, with a range of 14–68 years. Phenotypic extremes were drawn from the observed tails of the SaKnorm distribution (Figure 1). The 127 persons with the lowest SaKnorm values were defined as the lower tertile (SaKnorm range, −1.912 to −0.341), and the 127 persons with the highest SaKnorm values were defined as the upper tertile (SaKnorm range, 1.34–2.861). Controls were ascertained from the National Heart, Lung, and Blood Institute “Grand Opportunity” Exome Sequencing Project (ESP) Lung and Heart cohorts (19), excluding individuals with lung disease. This research was approved by the Seattle Children’s Hospital Institutional Review Board (approval numbers 12974 and 11686). Written informed consent or assent was collected from all study participants for the use of their DNA and sequence data to be used in studies for the genetic risk of CF and for broad data sharing. The phenotype and exome sequence data have been described previously (6) and are available through dbGaP (database of Genotypes and Phenotypes), with the accession numbers summarized on the ESP website.
Figure 1.
Distribution of SaKnorm (lung function based on forced expiratory volume in 1 s adjusted for age, height, and sex, then mortality among individuals affected by cystic fibrosis) values in the discovery cohort. The lower tertile is highlighted in yellow, and the upper tertile is highlighted in green.
Positions are given for the hg19/GRCh37 reference build of the human genome. Variants were annotated using SeattleSeq build 8.00 to define gene sets, as well as the Variant Effect Predictor tool [18] and the ENCODE (Encyclopedia of DNA Elements) data in the University of California Santa Cruz Genome Browser for potential regulatory effects. These annotations included reference allele frequencies, predicted variant function, Genomic Evolutionary Rate Profiling (GERP) scores indicating variant conservation (21), and combined annotation–dependent depletion (CADD [22]), PolyPhen-2 (Polymorphism Phenotyping software tool [21]), and SIFT (Sorting Tolerant from Intolerant algorithm [24]) scores predicting variant deleteriousness. Variants with GERP scores greater than 3 were considered conserved, and variants with CADD phred-scaled scores greater than 15 were considered deleterious. Default qualitative variant classification was used for both PolyPhen-2 and SIFT. Additional details on the discovery cohort, inclusion criteria, and analyses performed are provided in the online supplement.
By-variant association testing was performed using adjusted SKAT-O (v0.81), including PC1 and PC2 as covariates. The candidate gene list consisted of 93 genes that either encode cilia proteins and/or were known to underlie PCD (see Table E1 in the online supplement). We used two approaches to correct for multiple testing: a strict Bonferroni correction for the number of variants tested and a correction for the number of independent tests (25). Linkage disequilibrium (LD) was measured using the r2 statistic, calculated by PLINK v1.90 (26) using the entire ESP cohort. Odds ratios (ORs) for the additive effects of associated single-nucleotide polymorphism (SNP) alleles were estimated by logistic regression in PLINK v1.90, adjusting for PC1 and PC2.
Extension Cohorts
Adjusted SKAT-O was used to test for association between candidate variants and the extreme tertiles of FEV1 percent predicted values after adjustment for PC1 and PC2 in the 338 persons with COPD in the Lung Health Study (LHS) with self-reported European ancestry and phenotype data, excluding those with reactive airways. We also tested for association between candidate variants and subphenotypes often observed in persons with CF (27, 28) and PCD (29–33): chronic sinusitis, chronic bronchitis, and bronchiectasis. Illumina Human Core Exome Chip genotypes from 7,418 individuals from the Marshfield dataset (34) were available for testing using both the chi-square test and Fisher’s exact test (35, 36). One variant, rs17522489, was not observed and was instead represented by rs12032942, whose genotypes were highly correlated with rs17522489 in a reference dataset with European ancestry (r2 = 0.94). Additional details on the extension cohorts, inclusion criteria, and analyses performed are provided in the online supplement.
Results
Discovery Analysis
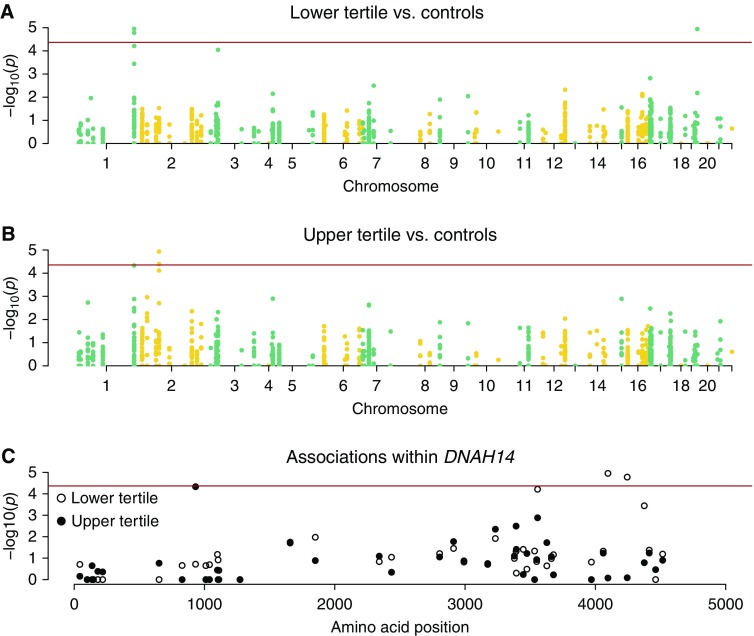
Association testing was performed to compare the lower SaKnorm tertile and population controls for a total of 1,165 single-nucleotide variants (SNVs) passing inclusion criteria within 88 of 93 candidate genes. No variants passed all inclusion criteria in five candidate genes. Significant association was observed between two variants in DNAH14 (dynein axonemal heavy chain 14, rs3856154 and rs950210) and rs58824375 in DNAAF3 (dynein axonemal assembly factor 3) and the lower SaKnorm tertile after Bonferroni correction (Table 1 and Figure 2A). A third variant in DNAH14 reached significance after correction for the number of independent tests (rs17522489) (Table 1). The majority of test statistics followed expectation (Figure E3), indicating well-controlled type I error. An excess of −log10 P values near zero was a consequence of comparing a small number of cases with a large number of controls and is expected in this situation.
Table 1.
Single variants significantly associated with the lower SaKnorm tertile in adjusted SKAT-O analysis
| Variants Associated with the Lower SaKnorm Tertile | ||||
|---|---|---|---|---|
| Chromosome | 1 | 19 | 1 | 1 |
| hg19 position | 225565015 | 55673145 | 225569241 | 225533931 |
| Allele | T>C | C>T | T>G | A>G |
| Gene | DNAH14 | DNAAF3 | DNAH14 | DNAH14 |
| P valueall | 1.11E-05* | 1.14E-05* | 1.66E-05* | 6.13E-05 |
| P valueCFTRhomozygotes | 9.50E-08* | 7.06E-07* | 1.72E-07* | 8.48E-05 |
| Odds ratio (allele) | 1.43 (T) | 2.85 (T) | 1.42 (T) | 1.42 (G) |
| rsID | rs3856154 | rs58824375 | rs950210 | rs17522489 |
| AAFcases | 0.43 | 0.07 | 0.43 | 0.21 |
| AAFcontrols | 0.33 | 0.03 | 0.33 | 0.15 |
| Change | p.Leu4096Pro | p.Gly245Ser | p.Phe4244Cys | p.Gln3556Arg |
| GERP | 4.32 | 2.52 | 1.16 | 5.45 |
| CADD (phred) | 8.00 | 8.90 | 8.54 | 7.09 |
| PolyPhen-2 | Benign | Benign | Benign | Benign |
| SIFT | Tolerated | Tolerated | Deleterious | Tolerated |
Definition of abbreviations: AAF = alternate allele frequency; CADD = combined annotation–dependent depletion; CFTR = cystic fibrosis transmembrane conductance regulator; GERP = Genomic Evolutionary Rate Profiling; PolyPhen-2 = Polymorphism Phenotyping software tool; rsID = single-nucleotide polymorphism identifier; SaKnorm = lung function based on forced expiratory volume in 1 second adjusted for age, height, and sex, then mortality among individuals affected by cystic fibrosis; SIFT = Sorting Tolerant from Intolerant algorithm.
Test results are presented when all cases within the lower SaKnorm tertile were included (P valueall) and when cases were restricted to CFTR p.Phe508del homozygotes (P valueCFTRhomozygotes). GERP scores (21) greater than 3 suggest that a variant occurs at a conserved sequence position; CADD (22) phred–scaled scores greater than 15 suggest that a variant is likely to be deleterious; and both PolyPhen-2 (23) and SIFT (24) scores predict variant deleteriousness.
Significant after strict Bonferroni correction.
Figure 2.
Single-variant adjusted SKAT-O association test results. (A) Results for the lower SaKnorm (lung function based on forced expiratory volume in 1 s adjusted for age, height, and sex, then mortality among individuals affected by cystic fibrosis) tertile. (B) Results for the upper SaKnorm tertile. (C) Results within a single gene, DNAH14. The red line indicates the Bonferroni significance level. A minor excess of P values near zero for both the lower tertile and the upper tertile versus controls resulted from the comparison of a very small number of cases with a large number of controls.
rs58824375 in DNAAF3 has a minor allele frequency (MAF) near 2% in Europeans (EUR) in the 1000 Genomes Project (1KG) (37) and was at least twice as common among persons with CF within the lower tertile of SaKnorm than among the ESP control subjects (Table 1). All three associated missense variants in DNAH14 are common, and both rs3856154 and rs17522489 are highly conserved. rs3856154 and rs950210 are in strong LD (r2 = 0.88) in the discovery dataset, suggesting that their associations with the lower tertile of SaKnorm are not independent. None of the variants associated with the lower tertile of SaKnorm were flagged as deleterious or damaging by PolyPhen-2, SIFT, or CADD (Table 1).
Association testing was performed to compare the upper SaKnorm tertile and population controls for 1,142 SNVs within 88 of 93 candidate genes. No variants passed all inclusion criteria in five candidate genes. A significant association was observed between two missense variants, rs1192269 and rs28375417, in DNAH6 (dynein axonemal heavy chain 6) and the upper tertile of SaKnorm after Bonferroni correction (Table 2 and Figure 2B). A third missense variant in DNAH14, rs115366080, was significantly associated with the upper tertile after correcting for the number of independent tests (Table 2). The distribution of test statistics fit well with the expectation (Figure E4).
Table 2.
Single variants significantly associated with the upper SaKnorm tertile in adjusted SKAT-O analysis
| Variants Associated with Upper SaKnorm Tertile | |||
|---|---|---|---|
| Chromosome | 2 | 2 | 1 |
| hg19 position | 84932836 | 84880445 | 225268106 |
| Allele | G>A | G>C | C>A |
| Gene | DNAH6 | DNAH6 | DNAH14 |
| P valueall | 1.17E-05* | 4.03E-05* | 4.65E-05 |
| P valueCFTRhomozygotes | 2.50E-05* | 1.23E-05* | 1.94E-05* |
| Odds ratio (allele) | 2.15 (A) | 1.70 (C) | 2.03 (A) |
| rsID | rs1192269 | rs28375417 | rs115366080 |
| AAFcases | 0.09 | 0.18 | 0.08 |
| AAFcontrols | 0.04 | 0.13 | 0.04 |
| Change | p.Val2898Ile | p.Gly1694Ala | p.Ala931Asp |
| GERP | 6.05 | 5.03 | −1.3 |
| CADD (phred) | 20.20 | 8.71 | 16.55 |
| PolyPhen-2 | Probably damaging | Benign | Benign |
| SIFT | Deleterious | Tolerated | Tolerated |
Definition of abbreviations: AAF = alternate allele frequency; CADD = combined annotation–dependent depletion; CFTR = cystic fibrosis transmembrane conductance regulator; GERP = Genomic Evolutionary Rate Profiling; PolyPhen-2 = Polymorphism Phenotyping software tool; rsID = single-nucleotide polymorphism identifier; SaKnorm = lung function based on forced expiratory volume in 1 second adjusted for age, height, and sex, then mortality among individuals affected by cystic fibrosis; SIFT = Sorting Tolerant from Intolerant algorithm
Test results are presented when all cases within the upper SaKnorm tertile were included (P valueall) and when cases were restricted to CFTR p.Phe508del homozygotes (P valueCFTRhomozygotes). GERP scores (21) greater than 3 suggest that a variant occurs at a conserved sequence position; CADD (22) phred–scaled scores greater than 15 suggest that a variant is likely to be deleterious; and both PolyPhen-2 (23) and SIFT (24) scores predict variant deleteriousness.
Significant after strict Bonferroni correction.
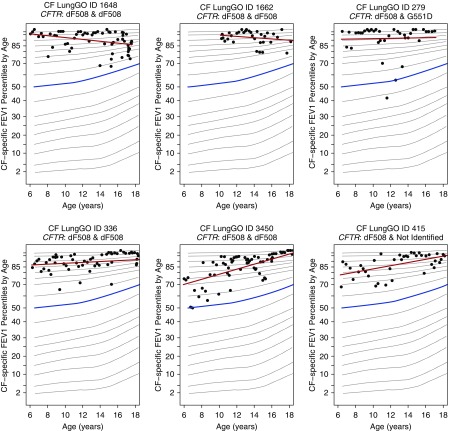
rs1192269 and rs28375417 are not in strong LD (r2 = 0.23) in the discovery dataset, suggesting that they represent independent association signals. rs1192269 is a highly conserved missense variant in DNAH6 with MAF of 4% in the ESP control subjects, predicted to be probably damaging by PolyPhen-2, deleterious by SIFT, and deleterious by CADD (Table 2). rs28375417 is a common missense variant in DNAH6 that is not highly conserved or predicted to be deleterious (Table 2). The missense variant, rs115366080, in DNAH14 is twice as common in the upper SaKnorm tertile (MAF = 8%) as in the ESP control subjects or the 1KG EUR subjects (MAF = 4%) (Table 2). rs115366080 is not highly conserved but is predicted to be deleterious by CADD alone (Table 2). Persons with CF who were heterozygous for rs1192269 showed improved lung function relative to their peers (Figure 3).
Figure 3.
Cystic fibrosis (CF)-specific percentiles of forced expiratory volume in 1 second (FEV1) over time for six patients heterozygous for rs1192269. Variant rs1192269 is the DNAH6 variant most strongly associated with the upper tertile of SaKnorm (lung function based on forced expiratory volume in 1 s adjusted for age, height, and sex, then mortality among individuals affected by cystic fibrosis). The six patients described were selected to represent variations in SaKnorm among members of the upper tertile across cystic fibrosis transmembrane conductance regulator (CFTR) genotype backgrounds. The four subjects on the left are homozygous for the p.Phe508del CFTR allele, whereas the other two carry one p.Phe508del allele and a second CFTR variant with minimal CFTR function. All six subjects were classified as having CFTR risk group 1 genotypes, indicating defects in the synthesis of the full-length protein. The median FEV1 percentile from reference data is highlighted in blue, and the individual-specific best-fit line is highlighted in red. Over the course of a decade, these six subjects tracked near the 85th percentile of lung function relative to their peers with CF (17).
Discoveries made by an extreme phenotype versus control approach can be unduly affected by outliers or sampling bias (38). Variation in causal CFTR genotype, if correlated with the variant being tested, could be the source of such a bias because CFTR genotype is also correlated with severity of lung disease (33). To assess whether our tests were affected by this source of bias, we restricted cases to p.Phe508del homozygotes and reanalyzed each variant significantly associated with either the upper or lower SaKnorm tertile. Each association remained statistically significant (Tables 1 and 2). Moreover, the P values for several of the associations were smaller than with use of the entire discovery cohort.
Vetting Study Design
We have previously used two different strategies to discover genetic modifiers of CF: extreme phenotypes (5) and extreme phenotype versus a population control (6). The extreme phenotype approach has greater power with increasing sample size and effect size, whereas the extreme phenotype versus population control design has more power when the enrichment/depletion of a variant or variants in a given gene is observed in a single extreme of the phenotype (6). We chose to use an extreme phenotype versus population control design based on an analysis that indicated greater power to detect a significant association than if we had used compared phenotypic extremes. Nevertheless, to determine whether we could replicate our findings using an extreme phenotype design, we performed a post hoc analysis comparing the lower versus upper SaKnorm tertiles. The directions of the effects of each SNV significantly associated with SaKnorm in the discovery phase were identical using an extreme phenotype analysis (Table 3). The association between lower (“case”) and upper (“control”) tertiles was nominally significant for rs115366080, whereas most other variants trended toward significance with P values less than 0.1. These results are consistent with the power simulations. The association between tertiles of SaKnorm and DNAH14 and DNAH6 could be observed using an extreme phenotypes design, but the statistical tests were underpowered to find that signal statistically significant.
Table 3.
Extreme vs. extreme results for variants associated with the lower or upper tertile of SaKnorm
| Chromosome | hg19 Position | rsID | Lower Tertile |
Upper Tertile |
P Value | ||
|---|---|---|---|---|---|---|---|
| No. of Samples | No. of Samples with Variant | No. of Samples | No. of Samples with Variant | ||||
| 1 | 225565015 | rs3856154 | 114 | 77 | 115 | 68 | 0.0577 |
| 19 | 55673145 | rs58824375 | 87 | 13 | 94 | 5 | 0.0401 |
| 1 | 225569241 | rs950210 | 113 | 76 | 114 | 67 | 0.0578 |
| 1 | 225533931 | rs17522489 | 113 | 44 | 115 | 38 | 0.6006 |
| 2 | 84932836 | rs1192269 | 114 | 10 | 115 | 18 | 0.0546 |
| 2 | 84880445 | rs28375417 | 112 | 21 | 113 | 36 | 0.0729 |
| 1 | 225268106 | rs115366080 | 114 | 7 | 115 | 18 | 0.0122 |
Definition of abbreviations: rsID = single-nucleotide polymorphism identifier; SaKnorm = lung function based on forced expiratory volume in 1 second adjusted for age, height, and sex among individuals affected by cystic fibrosis.
Extension to Related Airway Diseases
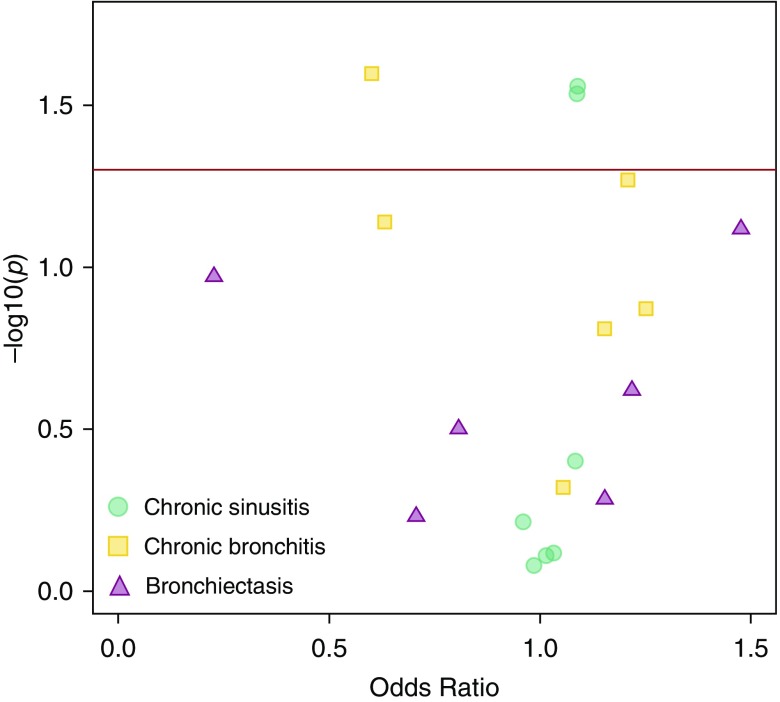
Four of the seven SNVs associated with lung function in the discovery analyses were associated with similar effects on a related airway disease in the extension datasets. In the LHS COPD cohort, rs17522489 in DNAH14 was significantly associated with the lower tertile of lung function after correction for multiple testing (P = 0.0083) (Table 4). rs58824375 could not be tested, because it failed to meet inclusion criteria. A restricted phenome-wide association study analysis in the Marshfield dataset replicated the associations between variants in DNAH14 and lung function (Figure 4, Table 5). Specifically, rs115366080, the variant in DNAH14 associated with the upper SaKnorm tertile in CF, was nominally associated with protection from chronic bronchitis (OR, 0.60; chi-square P = 0.0294). rs3586154 and rs950210, both variants associated with the lower SaKnorm tertile in CF, were both nominally associated with increased risk of chronic sinusitis (OR, 1.09; chi-square P = 0.0276 and 0.0285, respectively), and association between increased risk of chronic bronchitis and a tagging variant for rs17522489 nearly reached nominal significance (OR, 1.2; P = 0.0537). These results suggest that the variants, genes, or pathways that influence lung function in CF may influence other airway phenotypes characterized by abnormal mucociliary clearance.
Table 4.
Lung Health Study adjusted SKAT-O results comparing forced expiratory volume in 1 second lower or upper tertile with population controls
| Chromosome | hg19 Position | Ncases | Ncases with variant | Ncontrols | Ncontrols with variant | P Value |
|---|---|---|---|---|---|---|
| LHS lower tertile vs. ESP controls | ||||||
| 1 | 225565015 | 73 | 44 | 763 | 427 | 0.84636 |
| 1 | 225569241 | 73 | 44 | 755 | 423 | 0.82658 |
| 1 | 225533931 | 73 | 26 | 763 | 204 | 0.00828 |
| LHS upper tertile vs. ESP controls | ||||||
| 2 | 84932836 | 78 | 5 | 642 | 53 | 0.20146 |
| 2 | 84880445 | 73 | 14 | 618 | 148 | 0.31755 |
| 1 | 225268106 | 78 | 6 | 642 | 53 | 1 |
Definition of abbreviations: ESP = National Heart, Lung, and Blood Institute “Grand Opportunity” Exome Sequencing Project; LHS = National Heart, Lung, and Blood Institute “Grand Opportunity” Lung Health Study; N = number of samples.
Figure 4.
Association between lung phenotypes in the Marshfield dataset and the variants identified in the discovery cohorts. The data are listed in Table 5. The x-axis is the odds ratio, and the y-axis is the −log10 P value derived by Fisher’s exact test. The red horizontal line represents a P value of 0.05, or nominal significance. Points are color coded by phenotype.
Table 5.
Phenotype association results for candidate variants in the Marshfield dataset
| Lower-Tertile SNVs | Upper-Tertile SNVs | ||||||
|---|---|---|---|---|---|---|---|
| Chromosome | 1 | 19 | 1 | 1 | 2 | 2 | 1 |
| hg19 position | 225565015 | 55673145 | 225569241 | 223578706 | 84932836 | 84880445 | 225268106 |
| rsID | rs3856154 | rs58824375 | rs950210 | rs12032942 | rs1192269 | rs28375417 | rs115366080 |
| A1 | T | T | T | G | A | C | A |
| Chronic sinusitis DX_473 (2,462 cases, 3,587 controls) | |||||||
| Odds ratio | 1.089 | 1.032 | 1.088 | 0.9867 | 0.9604 | 1.015 | 1.084 |
| P value* | 0.0276 | 0.772 | 0.0285 | 0.8374 | 0.6199 | 0.7786 | 0.4049 |
| P value† | 0.0276 | 0.7638 | 0.0291 | 0.8317 | 0.6123 | 0.7765 | 0.3965 |
| Chronic bronchitis DX_491 (397 cases, 6,759 controls) | |||||||
| Odds ratio | 1.055 | 0.6314 | 1.055 | 1.209 | 1.251 | 1.153 | 0.6008 |
| P value* | 0.4796 | 0.0807 | 0.4796 | 0.0529 | 0.1308 | 0.166 | 0.0294 |
| P value† | 0.4793 | 0.0725 | 0.4793 | 0.0537 | 0.1341 | 0.1552 | 0.0252 |
| Bronchiectasis DX_494 (73 cases, 7,345 controls) | |||||||
| Odds ratio | 1.219 | 0.2265 | 1.218 | 0.8077 | 1.154 | 1.477 | 0.7068 |
| P value* | 0.2427 | 0.1057 | 0.2444 | 0.2957 | 0.6784 | 0.0676 | 0.4931 |
| P value† | 0.2403 | 0.1068 | 0.2403 | 0.3154 | 0.5203 | 0.0762 | 0.5875 |
Definition of abbreviations: A1 = allele 1; DX = International Classification of Diseases, Ninth Revision, Clinical Modification diagnosis code; rsID = single-nucleotide polymorphism identifier; SNV = single-nucleotide variant.
Chi-square statistic P value.
Fisher’s exact test P value.
Discussion
We identified significant associations between variants in both DNAH14 and DNAAF3 and poor lung function in persons with CF, as well as variants in both DNAH6 and DNAH14 and preserved lung function in persons with CF. Risk variants in DNAH14 associated with preserved versus poor lung function in DNAH14 localize to regions of the gene that encode different domains of the dynein heavy chain (Figure 2C). DNAH14 variants associated with poor lung function are clustered within the highly conserved region D6 of the dynein motor that has been directly implicated in force generation (39). The DNAH14 variant associated with preserved lung function falls within the N-terminal region 2 that is part of the cargo-binding “tail” of dynein (40–42). These observations suggest that variants associated with poor lung function may modify the force produced by respiratory cilia, whereas the variant associated with preserved lung function interferes with dynein subunits or cargo selection. Although rare variants in DNAAF3, DNAH6, and DNAH14 have been reported in patients with PCD, none of the variants associated with lung function in persons with CF cause PCD (20, 43, 44).
Cystic fibrosis is associated with abnormalities in lung defense that contribute to progressive obstructive lung disease and bronchiectasis (9). Abnormalities in lung defense associated with CFTR dysfunction include inactivation of airway antimicrobial peptides by a more acidic airway surface liquid and alterations in mucociliary clearance (41, 45, 46). We speculate that variants in motile cilia genes may further impair or augment mucociliary transport in patients with CF to affect lung disease severity. These findings further highlight the efforts to develop mucociliary clearance outcome measures and therapeutics to improve mucociliary transport in patients with CF (46).
Our results have prioritized specific proteins for functional studies exploring how abnormalities in axonemal heavy chains or axonemal assembly proteins affect lung function in CF. Both DNAH14 and DNAH6 encode axonemal force-generating proteins of respiratory cilia. Variation in DNAH6 is associated with multiple lung phenotypes, because it is required for normal airway ciliary motility (47–49). Mutations in other dynein axonemal heavy chains (e.g., DNAH11, DNAH5) are associated with alterations in ciliary motility and power stroke (41). DNAAF3 is required for the assembly of inner and outer dynein arms into complexes before they are transported into cilia, and variants in DNAAF3 are associated with ciliary defects in patients with PCD (41). Accordingly, DNAAF3, DNAH6, and DNAH14 (43, 48) are all compelling biological candidates for influencing the severity of lung function in persons with CF and other conditions characterized by abnormal mucociliary clearance and obstructive lung disease. The concordant direction of effect of the DNAH14 risk variants in CF and other chronic airway disease phenotypes in this study provides validation and suggests a shared pathogenesis of altered ciliary function. Together with the overrepresentation of variation in CFTR and cilia genes among patients with pulmonary nontuberculous mycobacterial infection (50), these results continue to build the accumulation of evidence that variation in cilia genes contribute to numerous pulmonary and respiratory phenotypes.
These discoveries are the direct result of selecting cases from phenotypic extremes while using a candidate gene approach to reduce the multiple testing burden. Six of the seven variants associated with lung function in this study exhibited the same direction of effect in a recent large-scale genome-wide association study analyzing SaKnorm as a quantitative trait (7); two of our SNPs associated with preserved lung function, rs1192269 in DNAH6 and rs115366080 in DNAH14, were also nominally significant (P < 0.05 [7]). Our focus on extreme phenotypes strengthened the association signals for these SNPs, whereas the candidate gene approach reduced the significance threshold, permitting their discovery.
Identifying genetic modifiers of a monogenic condition can facilitate identification of novel biological pathways influencing disease pathogenesis and new therapeutic targets. We identified three genes associated with variation in lung function in persons with CF by interrogating variation in candidate genes in phenotypic extremes. Studies designed to identify additional genetic modifiers of CF using extreme phenotypes sampled from considerably larger sample sizes (e.g., phenotypes harmonized across different CF cohorts) are underway and should provide additional statistical power to detect variants influencing CF severity.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the families for their participation, and specifically the EPIC (Early Pseudomonas Infection Control) study site investigators and research coordinators for their work. These investigators and research coordinators are named in the online supplement.
Footnotes
Supported by grants from the Cystic Fibrosis Foundation (GIBSON07K0, KNOWLE00A0, and OBSERV04K0); the National Heart, Lung, and Blood Institute (NHLBI) and numerous institutions, investigators, staff, and study participants who made this research possible. The “Grand Opportunity” Exome Sequencing Project (GO-ESP) was funded by NHLBI grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO [M.J.B., K.C.B.]), and RC2 HL-102924 (Women’s Health Initiative Sequencing Project [WHISP]). Additional support was provided by National Institutes of Health grants National Library of Medicine (NLM) K22LM011938 (S.J.H.), NIGMS R01GM114128 (S.J.H.), and NCATS UL1TR000427. Exome sequencing was funded by NHLBI grants RC2 HL-102925 (BroadGO), and RC2 HL-102926 (SeattleGO).
Author Contributions: E.B., J.X.C., R.L.G., M.J.B., and M.J.E.: participated in overall study design; E.B., T.L.L., S.J.H., K.C.B., N.M.R., and M.J.E.: performed analyses; and R.L.G., S.J.H., K.C.B., M.R.K., NHLBI GO ESP, and LungGO: contributed samples and sequence data. All authors revised and approved the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: U.S. National Heart, Lung, and Blood Institute “Grand Opportunity” Exome Sequencing Project (LungGO)
References
- 1.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 2.Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 3.Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, et al. Gene Modifier Study Group. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 4.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR, et al. National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project; Lung GO. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emond MJ, Louie T, Emerson J, Chong JX, Mathias RA, Knowles MR, et al. NHLBI GO Exome Sequencing Project. Exome sequencing of phenotypic extremes identifies CAV2 and TMC6 as interacting modifiers of chronic Pseudomonas aeruginosa infection in cystic fibrosis. PLoS Genet. 2015;11:e1005273. doi: 10.1371/journal.pgen.1005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corvol H, Blackman SM, Boëlle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haq IJ, Gray MA, Garnett JP, Ward C, Brodlie M. Airway surface liquid homeostasis in cystic fibrosis: pathophysiology and therapeutic targets. Thorax. 2016;71:284–287. doi: 10.1136/thoraxjnl-2015-207588. [DOI] [PubMed] [Google Scholar]
- 9.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 10.Stoltz DA, Meyerholz DK, Welsh MJ. Origins of cystic fibrosis lung disease. N Engl J Med. 2015;372:1574–1575. doi: 10.1056/NEJMc1502191. [DOI] [PubMed] [Google Scholar]
- 11.Morgan LC, Birman CS. The impact of primary ciliary dyskinesia on the upper respiratory tract. Paediatr Respir Rev. 2016;18:33–38. doi: 10.1016/j.prrv.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Fretzayas A, Moustaki M. Clinical spectrum of primary ciliary dyskinesia in childhood. World J Clin Pediatr. 2016;5:57–62. doi: 10.5409/wjcp.v5.i1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horani A, Brody SL, Ferkol TW. Picking up speed: advances in the genetics of primary ciliary dyskinesia. Pediatr Res. 2014;75:158–164. doi: 10.1038/pr.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blue EE, Emond MJ, Louie TL, Chong JX, Gibson RL, Bamshad MJ. Alternative study designs identify genes associated with variation in lung function among patients with cystic fibrosis [abstract] Genet Epidemiol. 2015;39:534. [Google Scholar]
- 15.Treggiari MM, Rosenfeld M, Mayer-Hamblett N, Retsch-Bogart G, Gibson RL, Williams J, et al. Early anti-pseudomonal acquisition in young patients with cystic fibrosis: rationale and design of the EPIC clinical trial and observational study. Contemp Clin Trials. 2009;30:256–268. doi: 10.1016/j.cct.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulich M, Rosenfeld M, Campbell J, Kronmal R, Gibson RL, Goss CH, et al. Disease-specific reference equations for lung function in patients with cystic fibrosis. Am J Respir Crit Care Med. 2005;172:885–891. doi: 10.1164/rccm.200410-1335OC. [DOI] [PubMed] [Google Scholar]
- 18.Taylor C, Commander CW, Collaco JM, Strug LJ, Li W, Wright FA, et al. A novel lung disease phenotype adjusted for mortality attrition for cystic fibrosis genetic modifier studies. Pediatr Pulmonol. 2011;46:857–869. doi: 10.1002/ppul.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auer PL, Reiner AP, Wang G, Kang HM, Abecasis GR, Altshuler D, et al. NHLBI GO Exome Sequencing Project. Guidelines for large-scale sequence-based complex trait association studies: lessons learned from the NHLBI Exome Sequencing Project. Am J Hum Genet. 2016;99:791–801. doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper GM, Stone EA, Asimenos G, NISC Comparative Sequencing Program, Green ED, Batzoglou S, Sidow A.Distribution and intensity of constraint in mammalian genomic sequence Genome Res 200515901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis SD, Ferkol TW, Rosenfeld M, Lee HS, Dell SD, Sagel SD, et al. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am J Respir Crit Care Med. 2015;191:316–324. doi: 10.1164/rccm.201409-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia: recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med. 2013;188:913–922. doi: 10.1164/rccm.201301-0059CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Moylan B, Leopold DA, Kim J, Rubenstein RC, Togias A, et al. Mutation in the gene responsible for cystic fibrosis and predisposition to chronic rhinosinusitis in the general population. JAMA. 2000;284:1814–1819. doi: 10.1001/jama.284.14.1814. [DOI] [PubMed] [Google Scholar]
- 30.Mackay IS, Djazaeri B. Chronic sinusitis in cystic fibrosis. J R Soc Med. 1994;87:17–19. [PMC free article] [PubMed] [Google Scholar]
- 31.Raju SV, Solomon GM, Dransfield MT, Rowe SM. Acquired cystic fibrosis transmembrane conductance regulator dysfunction in chronic bronchitis and other diseases of mucus clearance. Clin Chest Med. 2016;37:147–158. doi: 10.1016/j.ccm.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dransfield MT, Wilhelm AM, Flanagan B, Courville C, Tidwell SL, Raju SV, et al. Acquired cystic fibrosis transmembrane conductance regulator dysfunction in the lower airways in COPD. Chest. 2013;144:498–506. doi: 10.1378/chest.13-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiler CA, Drumm ML. Genetic influences on cystic fibrosis lung disease severity. Front Pharmacol. 2013;4:40. doi: 10.3389/fphar.2013.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebbring SJ, Rastegar-Mojarad M, Ye Z, Mayer J, Jacobson C, Lin S. Application of clinical text data for phenome-wide association studies (PheWASs) Bioinformatics. 2015;31:1981–1987. doi: 10.1093/bioinformatics/btv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Ye Z, Mayer JG, Hoch BA, Green C, Rolak L, et al. Phenome-wide association study maps new diseases to the human major histocompatibility complex region. J Med Genet. 2016;53:681–689. doi: 10.1136/jmedgenet-2016-103867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Z, Mayer J, Ivacic L, Zhou Z, He M, Schrodi SJ, et al. Phenome-wide association studies (PheWASs) for functional variants. Eur J Hum Genet. 2015;23:523–529. doi: 10.1038/ejhg.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas MP, Höök P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun. 2015;6:6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, et al. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horani A, Ferkol TW, Dutcher SK, Brody SL. Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oiwa K, Sakakibara H. Recent progress in dynein structure and mechanism. Curr Opin Cell Biol. 2005;17:98–103. doi: 10.1016/j.ceb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Berg JS, Evans JP, Leigh MW, Omran H, Bizon C, Mane K, et al. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: implications for application to clinical testing. Genet Med. 2011;13:218–229. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, et al. DNAH6 and its interactions with PCD genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet. 2016;12:e1005821. doi: 10.1371/journal.pgen.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, et al. Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science. 2014;345:818–822. doi: 10.1126/science.1255825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donaldson SH, Corcoran TE, Laube BL, Bennett WD. Mucociliary clearance as an outcome measure for cystic fibrosis clinical research. Proc Am Thorac Soc. 2007;4:399–405. doi: 10.1513/pats.200703-042BR. [DOI] [PubMed] [Google Scholar]
- 47.Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res. 2013;14:38. doi: 10.1186/1465-9921-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pazour GJ, Agrin N, Walker BL, Witman GB. Identification of predicted human outer dynein arm genes: candidates for primary ciliary dyskinesia genes. J Med Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang IV, Coldren CD, Leach SM, Seibold MA, Murphy E, Lin J, et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax. 2013;68:1114–1121. doi: 10.1136/thoraxjnl-2012-202943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szymanski EP, Leung JM, Fowler CJ, Haney C, Hsu AP, Chen F, et al. Pulmonary nontuberculous mycobacterial infection: a multisystem, multigenic disease. Am J Respir Crit Care Med. 2015;192:618–628. doi: 10.1164/rccm.201502-0387OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.