Abstract
The evolution of mammalian olfaction is manifested in a remarkable diversity of gene repertoires, neuroanatomy and skull morphology across living species. Olfactory receptor genes (ORGs), which initiate the conversion of odorant molecules into odour perceptions and help an animal resolve the olfactory world, range in number from a mere handful to several thousand genes across species. Within the snout, each of these ORGs is exclusively expressed by a discrete population of olfactory sensory neurons (OSNs), suggesting that newly evolved ORGs may be coupled with new OSN populations in the nasal epithelium. Because OSN axon bundles leave high-fidelity perforations (foramina) in the bone as they traverse the cribriform plate (CP) to reach the brain, we predicted that taxa with larger ORG repertoires would have proportionately expanded footprints in the CP foramina. Previous work found a correlation between ORG number and absolute CP size that disappeared after accounting for body size. Using updated, digital measurement data from high-resolution CT scans and re-examining the relationship between CP and body size, we report a striking linear correlation between relative CP area and number of functional ORGs across species from all mammalian superorders. This correlation suggests strong developmental links in the olfactory pathway between genes, neurons and skull morphology. Furthermore, because ORG number is linked to olfactory discriminatory function, this correlation supports relative CP size as a viable metric for inferring olfactory capacity across modern and extinct species. By quantifying CP area from a fossil sabertooth cat (Smilodon fatalis), we predicted a likely ORG repertoire for this extinct felid.
Keywords: cribriform plate, olfactory receptor gene, olfactory sensory neuron, pseudogene, skull morphology, sabertooth cat
1. Introduction
Reliance on olfaction for existence varies widely across mammal species. Mammalian olfactory systems have acquired and lost olfactory traits in response to shifts in chemosensory demands over evolutionary time, resulting in extensive genomic and anatomical diversity. The keen sense of smell shared by most mammals is mediated by the olfactory receptor gene (ORG) superfamily [1]. Through repeated gene duplication, gene loss and pseudogenization, ORG repertoires have diverged [2] and the number of ORGs (functional as well as pseudogenes) now spans approximately two orders of magnitude across living taxa [3,4].
Inside the mammalian nose, or snout, ORGs are the primary agents of odorant detection. Each intact ORG encodes a specific odorant receptor that is expressed in the membrane of olfactory sensory neurons (OSNs) [1]. Each OSN expresses a single ORG [5], and each ORG is expressed by its own unique population of thousands of OSNs [6,7]. Multiple ORG-specific populations of neurons contribute to the olfactory epithelium, which is variable in size across species [8,9]. The olfactory pathway is completed via axonal projections from all OSN populations, which travel from the periphery through the cribriform plate's (CP) foramina, or perforations, to reach their first relay station in the brain, the olfactory bulb [10].
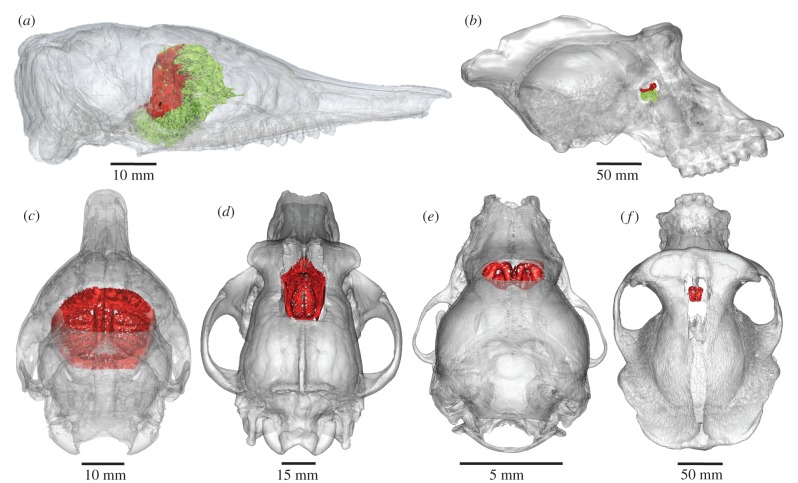
The CP (figure 1) is a part of the ethmoid bone that forms early in development to surround assembling olfactory nerves [11] and is the only passageway to the brain for OSN axons in adult mammals. Consequently, its foramina are a high-fidelity imprint of the relative innervation within an animal's snout [12] (electronic supplementary material, movie S1). Aside from small amounts of connective tissue, cerebrospinal fluid, terminal nerve fibres and occasional small vessels, the tissue passing through the CP is composed solely of OSN axon fascicles [13]. Therefore, the size of the CP and its foramina area may be reasonable indicators of the relative number of OSNs transmitting odour signals to the olfactory bulb. The CP is a signature trait of Mammalia [14] and is present in all crown species, with the exception of odontocetes (toothed whales), where it is thought to be lost [15]. CP morphology varies widely across mammal species (figure 1) likely reflecting the relative importance of olfaction [9,12,16]. Two metrics of CP morphology, overall surface area and cumulative cross-sectional area of foramina, covary across species [12]. Previous studies have established strong correlations between the surface area of CP and other olfactory structures, such as ethmoturbinal bones and the olfactory epithelium that lines them [9,12].
Figure 1.
CP size varies markedly across mammal species. (a,b) Lateral views: (a) nine-banded armadillo (Dasypus novemcinctus) skull and (b) gorilla (Gorilla gorilla). (c–f) Dorsoposterior views: (c) armadillo, (d) dog (Canis familiaris, saluki breed), (e) little brown bat (Myotis lucifugus), and (f) gorilla. Red: CP. Green: turbinal bones (ethmoid, frontal and nasal) and partial nasal septum are only clearly viewable in lateral views.
The expression of individual ORGs by distinct populations of OSNs and the early formation of CP bone around developing OSN axon bundles suggest that evolutionary gains or losses in ORG repertoires will be manifested in skull morphology as either an expansion or reduction in the CP. However, the relationship between CP morphology and ORG repertoire size is poorly resolved. The sole paper to explore this relationship found a significant but weak positive correlation between absolute CP surface area and total number of ORGs that disappeared when CP size was adjusted for body size [17]. One limitation of this pioneering study was its sole reliance on linear measurements to estimate CP surface area and its handling of the irregularly shaped CP as a generalizable ellipsoid. Here, we revisit the link between CP skull morphology and the number of ORGs, both functional and pseudogenized [2–4,18–20] with updated digital methods. We use high-resolution computer tomography (CT) scans of skulls and three-dimensional imaging software to carefully map only the perforate surface of the CP in 27 species with identified ORG repertoires (electronic supplementary material, S2 and S3). Furthermore, where the previous work adjusted for body size by dividing estimated CP area by skull area, we use residuals from the scaling relationship between CP surface area and body mass to define relative CP size. These methods allow us to more precisely test the hypothesis that the developmental relationship between ORGs and OSNs is witnessed in skull morphology. Specifically, CP size is expected to covary with the number of functional ORGs. Additionally, because a number of studies have used the percentage of OR pseudogenes within species' genomes to infer relative olfactory ability [21–23], we examined the relationship between CP size and pseudogenes (fraction as well as absolute number).
In our investigation of the relationship between CP morphology and ORG count, we rely on the assumption that ORG repertoire size is an informative metric of relative olfactory function. We acknowledge there is no single definition for olfactory function or ability. Olfactory ability is variously described in terms of detection threshold [24,25], odorant discrimination [26] and identification [27], and breadth of detectable odorants [28]. Given that, we point to studies in which the number of functional ORGs is correlated with a species’ ability to discriminate between structurally related odorants [25,28]. Additionally, because odour detection is shown to be mediated by a combinatorial process, whereby a single OR detects multiple odorants and a single odorant activates multiple ORs [28,29], we reason here that a larger ORG repertoire may be coupled with a larger palette of detectable odorants as well. Despite this, a recent review [30] argued that there is no clear link between ORG repertoire and olfactory ability. Notably, the review overlooked behavioural studies mentioned above, which contradict this assumption [26,31].
Finally, using the significant relationship we found between CP size and ORG number, we applied our new osteological metric of relative CP size to a fossil skull to produce an estimate of ORG repertoire for the extinct mammal species, sabertooth cat (Smilodon fatalis) and to infer Smilodon's reliance on olfaction compared with extant felids.
2. Material and methods
(a). Specimens sampled
We sampled skulls from 27 living species with annotated genomes and OR subgenomes (electronic supplementary material, S2), plus two species, one extinct (sabertooth cat, S. fatalis) and one extant (gray wolf, Canis lupus), with no molecular data. Species body masses ranged from less than 0.1 kg (little brown bat, Myotis lucifugus) to greater than 2900 kg (African elephant, Loxodonta africana) and covered 14 mammalian orders and all superorders [32,33]. Availability permitting, we selected two (preferably one male, one female) wild-caught, adult specimens per species from museum and university collections (electronic supplementary material, S3). We recognize that the per-species sample size is low, but limited availability of specimens, high scanning costs and labour-intensive data collection prohibited a deeper sampling. We are confident the low specimen count does not bias our results, as the median intraspecific range in CP surface area (log10) is 1.49% of the overall sample range (electronic supplementary material, S4).
(b). Morphological data collection
With the exception of the elephant, all skulls were scanned on high-resolution industrial CT scanners (Phoenix v|tome|x-s™, nanotom-s, nanotom-m™, North Star Imaging ACTIS™ and Nikon Metrology XT-H-225-ST™) (electronic supplementary material, S3). In all but three specimens, voxel size ranged from 0.008 to 0.045 mm. Owing to its size, the elephant skull could only be accommodated by a Definition-AS64™ medical scanner (Siemens Healthineers, Forchheim, Germany) and thus was captured at somewhat lower resolution (0.5 mm voxels).
Three-dimensional digital models were constructed for each skull. CT scans were imported into the visualization software package Mimics (v. 15.0–18.0, Materialise, Leuven, Belgium). From each skull scan, the CP was segmented into two-dimensional masks delineating bone from non-bone, then reconstructed into three-dimensional models. The first metric, CP surface area, includes only the portion of the CP perforated by foramina carrying olfactory nerves (electronic supplementary material, S5). To quantify CP surface area, a continuous surface was generated in the imaging program 3-matic (v. 7.0.1, Materialise) with a wrapping function that digitally fills the foramina. This surface was digitally cut at the perimeter of the perforated area and its area was calculated in 3-matic [12]. A second CP metric, the cumulative cross-sectional area of the CP foramina, is an estimate of the relative area occupied by olfactory nerve branches. Splines, or coordinate point rings, were applied to foramina perimeters in each Mimics CP model. All splines were imported into modelling software Rhinoceros-4 (McNeel and Associates). Non-planar surface areas for all foramina were calculated and tallied in Rhinoceros. Wherever multiple foramina fed into one larger foramen from rostral to caudal, we measured the caudal-most opening [12]. Quantifying CP foramina area is intensely time-consuming and prohibitive with damaged skulls or low-resolution CT scans. Therefore, the strong correlation between CP and CP foramina areas was used only to validate CP surface area as a stand-alone metric (r2 = 0.91, p < 0.0001; electronic supplementary material, S6).
Although we determined that the bottlenose dolphin (Tursiops truncatus) ethmoid bone has paired foramina connecting the nasal passage to the brain (electronic supplementary material, S7a), it is beyond the scope of this paper to validate the presence of OSNs or a vestigial CP by testing nasal tissue for known olfactory markers. Therefore, we omitted the dolphin from our quantitative analysis.
(c). Genomic data
We used published counts for functional and non-functional ORGs, which apply similar criteria for defining OR functionality [2–4,18–20] (electronic supplementary material, S8). Briefly, these publications defined pseudogenes as ORGs containing one or more of the following conditions: (i) a premature stop codon, (ii) frameshift mutations, (iii) in-frame deletions within a single transmembrane region and/or deletions of conserved amino acid sites [34], and in one case, (iv) truncated genes with fewer than 250 amino acids and lacking all seven transmembrane domains [4]. OR pseudogenes are considered here to be non-functional [35]. ORGs are identified from whole genome sequences [34]. Wherever possible, studies using high-coverage genome assemblies were chosen; however, in order to construct a sufficient sample, published OR gene repertoires from some low-coverage assemblies were included in our study, including eight species with less than six times draft genomes [4] (assembly statistics; electronic supplementary material, S9). Genomic metrics used for this study include: (i) total number of ORGs, (ii) number of functional ORGs, (iii) absolute number of OR pseudogenes, and (iv) percentage of OR pseudogenes.
(d). Statistical analysis
Species means were derived for each morphological and genomic variable. Values were log-transformed in the analysis of linear relationships because data covered several orders of magnitude. ORG counts do not correlate with body size, so were presented here without scaling to body mass. By contrast, CP surface area does correlate with body size. Therefore, to determine CP size relative to body size, CP surface area was regressed against body mass values taken from the literature (electronic supplementary material, S8) using generalized least squares regression. Resulting residuals were used as relative CP size (RelCP) values in all subsequent analyses. When investigating the relationship between RelCP and ORG repertoires, reduced major axis (RMA) regressions were used so we could invert predictor and response variables and establish mutual prediction lines. Correlation coefficients and confidence intervals were derived from bootstrapped resampling of the data (10 000 iterations).
When a nonlinear, dual trend was apparent in one plot, an inflection point was first established based on grouping species with the largest gene counts (independent variable) and secondarily was calculated in the R package ‘rpart’, a recursive data-partitioning program [36], and resulting data subsets were regressed separately.
To estimate a probable ORG repertoire for the sabertooth cat (Smilodon fatalis), we applied Smilodon RelCP as new data to the regression equation derived from the relationship between RelCP and functional ORGs in our larger sample. An estimated ORG range for Smilodon was calculated within the 95% CI around the regression line, derived by bootstrap resampling of the data. Finally, to account for phylogenetic relatedness and covariance in all relationships between variables, we performed phylogenetic generalized least squares (PGLS) analysis (R Caper package) [37] with a time-calibrated mammal phylogeny [38]. Summary statistics for PGLS regressions are given in electronic supplementary material, S10. All analyses were conducted in R v. 3.3.1 [39]. Custom R code is available upon request from the corresponding author.
3. Results
(a). Cribriform plate size as a function of olfactory receptor gene repertoire
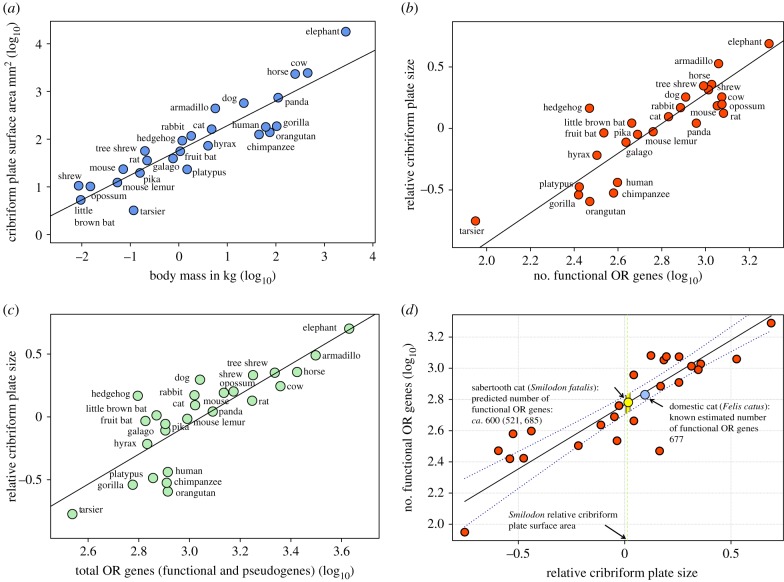
Because CP area is closely coupled to body mass (figure 2a, r2 = 0.82, p < 0.001), the relationship between CP and ORGs only comes into focus when relative CP surface areas, defined as the residuals from the regression of CP area on body mass, are considered. We found a strong positive linear correlation between relative CP size (RelCP) and the number of functional ORGs across our sample (figure 2b, r2 = 0.76, p < 0.001; electronic supplementary material, S10). After removing phylogenetic dependence with a phylogenetic generalized least squares (PGLS) regression [37,38], the correlation remained robust (PGLS-r2 = 0.75, p < 0.001). A similarly strong linear correlation exists between relative CP size and the total number of ORGs (functional and pseudogenes) (figure 2c, r2 = 0.68, p < 0.001; PGLS-r2 = 0.68, p < 0.001). Without correcting for body size, absolute CP surface area and number of functional ORGs are only weakly correlated (r2 = 0.22, p = 0.015) and this correlation disappears if data for the greatest absolute CP, corresponding to the largest species, the elephant, are removed (electronic supplementary material, S10).
Figure 2.
Linear correlation between relative CP size and ORG repertoire size was established and used to estimate likely ORG repertoire for the extinct sabertooth cat (S. fatalis). (a) Log–log regression plot of absolute CP surface area against body mass for 26 species (r2 = 0.82, p < 0.001). Residuals from regression constitute relative CP size (RelCP) for each species and are used in all subsequent analyses. (b) Strong positive linear correlation between RelCP and the number of functional ORG (log10) is shown in the RMA regression plot (r2 = 0.76, p < 0.001, n = 26). (c) Linear correlation between RelCP and total ORG number (functional, pseudogenes) (log10) (r2 = 0.68, p < 0.001). (d) Axes from figure 2b plot are inverted and the RelCP value (green vertical line) for the sabertooth cat is applied to the regression equation. This predicts Smilodon to have had approximately 600 (581 685) functional ORG. Dotted lines: 95% CI around regression line (see Material and methods for details; see electronic supplementary material S12d for species labels).
(b). Nonlinear relationship between cribriform plate size and olfactory receptor pseudogenes
Unlike the strong linear relationship between RelCP and functional ORGs, the association between RelCP and OR pseudogenes is complex. First, there is a weak linear correlation between RelCP and absolute number of pseudogenes (r2 = 0.36, p = 0.001, PGLS-r2 = 0.41, p < 0.001), but within this relationship, there appear to be two distinct trends (figure 3a). To further investigate this dichotomous pattern, we partitioned the sample data based on which species have particularly large pseudogene counts and ran linear regressions on resulting data subsets. In species with greater than or equal to approximately 750 pseudogenes, there is a strong positive correlation between RelCP and pseudogenes (r2 = 0.74, p = 0.027; PGLS-r2 = 0.72, p = 0.032; n = 6), but among species with lower pseudogene counts (less than approx. 500), there is no relationship. Cleaving the sample with the purely data-driven R program ‘rpart’ adds one more species to the group with the largest pseudogene counts but renders a similarly robust correlation within this group between RelCP and pseudogenes (electronic supplementary material, S10).
Figure 3.
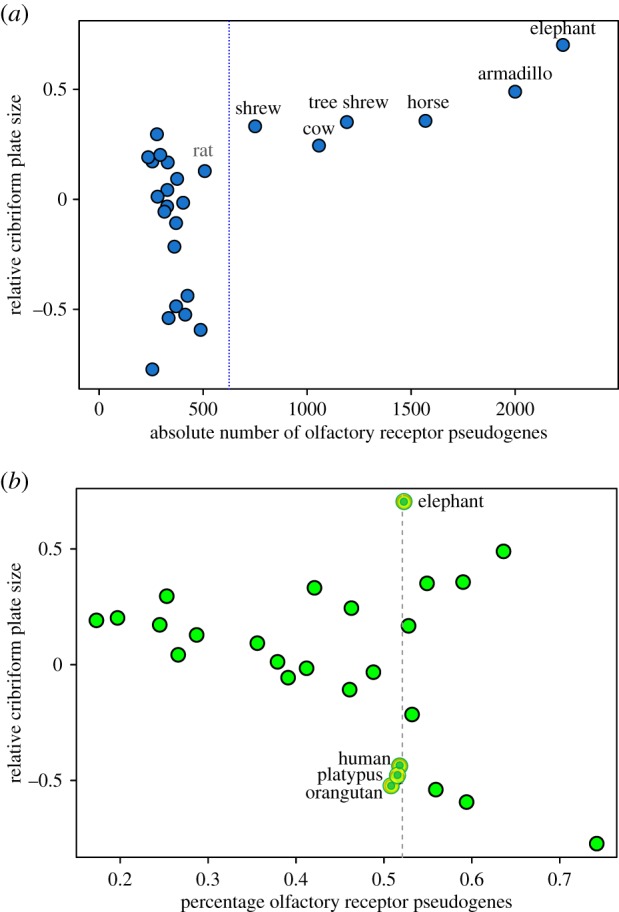
Nonlinear relationship between RelCP and OR pseudogene repertoires. (a) Dual trends are evident in the relationship between RelCP and the absolute number of OR pseudogenes, as delineated here by the vertical line: in species with the largest pseudogene counts (approx. greater than 750), the correlation is robust (r2 = 0.74, p = 0.026, PGLS-r2 = 0.72, p = 0.005); in the group of species with low pseudogene counts (approx. less than 500), there is no correlation. See electronic supplementary material, S11 for linear regressions on partitioned as well as non-partitioned data. (b) No linear relationship was found between RelCP and the percentage of pseudogenes present in ORG subgenomes. A high percentage of pseudogenes, a traditional measure of olfactory loss, is found here in taxa with the largest as well as smallest RelCPs (vertical line) and thus shown to be a poor indicator of relative olfactory ability. (Online version in colour.)
Second, we found no linear correlation between RelCP and the percentage of pseudogenes (r2 = 0.12, p = 0.09) (figure 3b). The pattern is somewhat V-shaped, as species with the largest (elephant) and smallest (platypus, among others) RelCPs have similarly high proportions of pseudogenes.
(c). Morphologic and genomic diversity
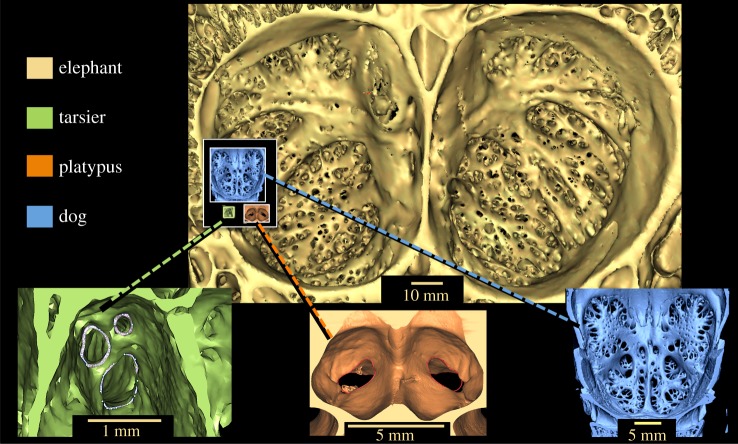
Our sample of species includes outstanding examples of olfactory disparity. The African elephant possesses the largest CP ever recorded, a deep concavity with approximately 1000 nerve passageways (figure 4). This is consistent with reports that elephants have undergone extreme expansion of their ORG repertoire [3]. The nine-banded armadillo (Dasypus novemcinctus) has the second largest RelCP, outranking even the domestic dog (C. familiaris) (figures 1a,c and 2b; electronic supplementary material, S8). All large apes share coupled reductions in ORGs and CP morphology (figures 1f and 2a–c). The smallest RelCP in our sample belongs to the Philippine tarsier (Tarsius syrichta), a species with a concordantly small functional ORG repertoire (approx. 90 compared to approx. 781 estimated for the most recent common ancestor of placental mammals) [3,4] (figure 4; electronic supplementary material, S7b). The notable outlier, the bottlenose dolphin (Tu. truncatus), possesses only approximately 12 functional ORGs [4] and has likewise lost all obvious CP landmarks. Closer inspection of digital models, however, revealed a pair of branched foramina in the ethmoid bone that connect nasal passages to the cranium but whose function is not certain (electronic supplementary material, S7a).
Figure 4.
Morphological disparity in CP seen in elephant, tarsier, platypus and dog skull models. Yellow top centre: CP of the African elephant (L. africana), viewed here from brain case, has the largest absolute and relative surface area and approximately 1000 foramina for passage of olfactory nerves to the brain. Extreme reduction in complexity is evident in CPs of the tarsier (T. syrichta) (green, lower left) and platypus (Ornithorhynchus anatinus) (orange, bottom centre) CPs. Dog (C. familiaris) CP (blue, lower right). The inset superimposed over elephant model shows tarsier, platypus and dog CPs to scale with the elephant's CP.
(d). Predicting olfactory function in an extinct sabertooth cat
The linear correlation between functional ORG number and relative CP size apparent in our sample of living species was used to estimate a likely repertoire of functional ORGs for an extinct mammal species. For this, we chose the Pleistocene sabertooth cat (S. fatalis) from the Rancho La Brea tar seeps, for which a CT scan was available [40]. With the aid of high-resolution CT scans and imaging software, we distinguished CP bone from its surrounding asphaltum matrix and quantified a fossil CP surface area for the first time. RelCP was estimated as the residual from the regression of CP surface area against body mass among all sample species (electronic supplementary material, S12a). Based on RelCP, Smilodon is predicted to have had approximately 600 (521, 685; 95% CI) functional ORGs (figure 2d), a number similar to that identified for the other felid in our sample, the domestic cat (Felis catus) [19]. When viewed in the context of existing morphological data from 10 living felids [12], CP surface area of Smilodon falls well within the range of variation found among these cats, but also has a relatively smaller RelCP size than all species but the cheetah (Acinonyx jubatus) and the fishing cat (Prionailurus viverrinus) (electronic supplementary material, S12b and S13)
4. Discussion
Our study places the CP at an intersection of olfactory genomics, development and evolution. The strong linear correlations between relative CP size and number of functional ORGs as well as total ORGs (figure 2b,c) support the hypothesis that skull morphology and ORGs are linked via the OSNs, whose axon fascicles are recorded in the CP bone. Previous studies have established that each individual ORG is expressed by thousands of OSNs, which together contribute to the size of the olfactory epithelium [6,7]. Additional research has shown that olfactory epithelium and CP surface areas are correlated [9]. Results from our study bridge these findings by establishing a direct, linear biological relationship between the size of the OR subgenomes and the relative size of the CP (RelCP).
Our results contradict previous research that found no correlation between ORG number and CP surface area adjusted to body mass [17], although the authors did find a weak correlation between ORG repertoire size and absolute CP surface area. In large part, the differences here can be explained by our contrasting approaches to the confounding factor of body size. As with most morphological features, CP area increases as a function of body size (r2 = 0.82), making comparisons across species that span several orders of magnitude in body mass difficult. For example, the absolute CP surface area is far smaller in the rat (Rattus norvegicus) than the human, yet the former has an ORG repertoire [3,4] and discriminatory ability [28] that exceed that of the latter. This suggests that a correlation between CP size and ORG number needs to be viewed relative to body size. Our ability to recover this relationship also benefited from the application of precise digital methods to estimate CP surface area from high-resolution CT scans. In a reanalysis of RelCP versus ORG number using CP surface area estimated from linear metrics in lieu of digitally derived data, no significant correlation was found.
Nevertheless, there is some scatter in the plot of RelCP size against ORG repertoire (figure 2b,c) that deserves attention. An important factor contributing to the scatter is likely the inclusion of a number of low-coverage draft genomes in our sample (electronic supplementary material, S9, S14a,b). Classification of ORGs is less reliable in low-coverage genomes owing to inherent sequencing gaps as well as over-collapse in regions with segmental duplications [4,41]. Because intact, functional ORGs are more likely to be misidentified as truncated and are generally undercounted [4], we expect their number to increase in low-coverage species as genome assemblies are improved. We note here four cases (elephant, rabbit, dog, cat) in which we can compare the number of functional ORGs identified from low-coverage draft genomes (1.5–2×) with those counted from current, higher coverage assemblies (greater than or equal to 6×). In these cases, the genomes rendered on average 39.7% more functional ORGs from higher coverage assemblies than from initial draft genomes. This measure of increase, applied to the 12 low-coverage species in our sample, strengthens the correlation between number of functional ORGs and RelCP (r2 = 0.84, PGLS-r2 = 0.81, p < 0.001; electronic supplementary material, S14b, compared with r2 = 0.76, figure 3b; electronic supplementary material, S14a). When we omit the 12 low-coverage species, the correlation between ORGs and CP tightens further (r2 = 0.89, PGLS-r2 = 0.82, p < 0.001; electronic supplementary material, S14c). If we make similarly derived adjustments to the total number of ORGs (functional and pseudogenes) in low-coverage species (10.68% increase), the correlation between CP and total ORGs tightens only slightly (r2 = 0.7 compared with 0.68, p < 0.001; PGLS-r2 = 0.68, p < 0.001).
OR pseudogenes are considered to be non-functional [2,3,22,35]. For this reason, it was surprising to find a distinct association between CP morphology and the absolute number of OR pseudogenes. While there is a weak linear correlation between RelCP and pseudogenes, there appear to be two separate trends in the data (figure 3a). Only among species with the largest pseudogene count is RelCP positively correlated with the absolute number of pseudogenes. This trend appears to be exponential, due to the strong influence of the armadillo and elephant. This distinct pattern raises the question why most species with high numbers of pseudogenes in their ORG repertoires have the largest RelCPs. Or, asked differently, when axes are inverted (electronic supplementary material, S15a), why have a number of species with particularly large RelCPs retained high numbers of pseudogenes in their OR repertoires? Here, we consider alternative evolutionary scenarios for this pattern. First, lineages leading to large-CP species may have undergone fairly recent ORG expansions generated by repeated gene duplication events. Although a fraction of new ORGs may have been conserved under adaptive pressure (e.g. to enhance detection of specific odorants), one would predict that many duplicates would degenerate by random mutations, due to relaxed selective pressure [35], and may have persisted in the genomes for short periods of time before degenerating to sequences no longer recognized as ORGs [42]. This is consistent with the observation that species with relatively high numbers of OR pseudogenes also have some of the largest repertoires of functional ORGs (electronic supplementary material, S15b). Alternatively, the pattern of high pseudogene concentration in large-CP species may signal constraints in either CP size or ORG repertoire size and a concomitant increase in pseudogenization of ORGs. A large ORG repertoire is costly, as every OSN in each OR-specific subpopulation is regenerated throughout an animal's lifetime [43]. Similarly, large CPs carry a structural risk, in that densely perforated bone is thinner and more easily fractured than surrounding cranial bone [44]. Notably, the two largest CPs occur in skulls with features that, while otherwise adaptive, may offer secondary reinforcement to the fragile CP, for example, dermal armour in the armadillo [45] and a thick periphery of highly pneumatized bone surrounding the brain in the elephant [46] (electronic supplementary material, S16). Finally, we consider the possibility that OR pseudogenes, traditionally assumed to be functionally inert and neutral [22,35], may be biologically active, transcribed into long non-coding RNAs (lncRNAs) and therefore conserved in species with expanded olfactory systems. As demonstrated for the tumour-suppressor PTEN gene, non-coding pseudogene transcripts can act as decoys for microRNAs (miRNA) that target the parent gene, thereby mitigating the effects of translational repression mediated by the miRNAs [47]. A potential role for OR-derived lncRNA transcripts as miRNA sponges might be investigated, particularly in the light of recent findings that coding ORG transcripts in mice have lost many miRNA binding sites, effectively reducing the modulatory impact of miRNAs on expression levels [48].
Numerous researchers have linked the evolutionary loss of olfactory function with the fraction of pseudogenes in ORG repertoires [4,21–23]. However, our study found no significant linear correlation between the percentage of pseudogenes and the RelCP (figure 3b). These results support the argument that the percentage of pseudogenes is a poor predictor of olfactory ability [3,18].
The strong link documented here between skull morphology and a wide dynamic range of ORG repertoires invites us to examine the olfactory ecology of individual sample taxa. Our sample offers outstanding examples of olfactory expansion and reduction that might be explained by differences in dependence on olfaction in life history. For example, the African elephant possesses by far the largest RelCP and ORG repertoire [3] of any mammal studied, suggesting that elephants are heavily reliant on olfaction for their survival. This is consistent with the elephant's numerous unparalleled olfactory specializations, among them, extremely dense ethmoid turbinals (electronic supplementary material, S17), compact multiple layers of glomeruli in the olfactory bulb [49] and its signature mobile proboscis. In accordance with these anatomical features, behavioural experiments reveal a striking complexity of odour recognition among elephants. Specifically, they recognize up to 30 individuals within their kin group from urine cues alone [50] and can differentiate between two human ethnic groups by sniffing odour cues from worn garments [51].
Interestingly, we found the nine-banded armadillo (D. novemcinctus) has one of the largest olfactory systems in our sample (figure 2b). It is reasonable to view the armadillo as reliant on a strong sense of smell, given its degenerate vision. As a rod monochromat, the armadillo has no cones, resulting in colour blindness and low visual acuity in dim-light and total blindness under phototopic, or bright light, conditions [52]. Given that the nine-banded armadillo shares rod monochromacy with at least two other xenarthrans, Hoffman's two-toed sloth (Choloepus hoffmanni) and the extinct ground sloth (Mylodon darwinii) [52], a broader examination of CP morphology among xenarthrans is warranted.
Domestic dogs (C. familiaris) are thought to have a well-developed sense of smell and are reliable drug sniffers and olfactory scouts [53]. However, dogs are surpassed by five other species in RelCP as well as ORG count. Conceivably, the breeds chosen for our sample may have variably influenced the estimated CP size for the dog. Of the four, only the dachshund is considered a scent hound, whereas two (borzoi and saluki) are considered sight hounds and one is brachycephalic, or short-snouted (French bulldog). It has not yet been established whether selection for keen sightedness or foreshortened snouts imposes constraints on CP morphology or ORG repertoires. A more focused examination of the CP/ORG relationship among dog breeds is a necessary next step. Additionally, we might consider that non-olfactory traits selected for in dogs, such as tameness and trainability [54], may play a role in dogs' success as odour detectives. In future comparative studies of the olfactory anatomy in domestic versus wild species, the dog will certainly be a target species. Interestingly, the domestic dog has a smaller CP for its size than does the gray wolf (C. lupus), whose ORG repertoire is presently unknown (electronic supplementary material, S12c) [12]. Based on its larger RelCP, the wolf is expected to have a larger ORG repertoire than the dog, but confirmation awaits the analysis of new genomic data [55].
Several of our sampled species with exceptionally small CPs and ORG repertoires rely on well-developed sensory modalities other than olfaction. For example, the tarsier, a nocturnal primate with pronounced visual specializations, such as enormous eyes and an enlarged primary visual cortex [56], has a negligible CP (figure 4; electronic supplementary material, S7b). Similarly, the simplified CP of the semi-aquatic platypus (figure 4) may in part have been shaped by preferential reliance on an elaborate sensory system of electroreception adapted to underwater foraging [57]. The most striking example of olfactory loss and sensory trade-off is the bottlenose dolphin, a fully aquatic odontocete that relies on echolocation to locate prey. Studies variously report that CP foramina, present in fetal odontocetes, fully close in adults or that a few remain open and carry fibres only from the terminal nerve, a cranial nerve thought to play a role in pheromone response [15,58]. Our digital models of adult dolphin skulls confirmed that there are indeed openings connecting the nasal passages to the brain in the ethmoid area (electronic supplementary material, S7a). To be certain, the dolphin shows profound degeneration in its olfactory system, including the pseudogenization of two genes involved in olfactory signal transduction CNGA2 and CNGA4 [59] and a nearly complete loss of a functioning OR subgenome. However, given that the dolphin has retained a small repertoire of approximately 12 intact ORGs and perforations in the ethmoid bone, nasal tissue surrounding these openings needs to be tested for possible expression of known OSN markers (e.g. OMP, Gng13, Stoml3) [59,60] before fully excluding a vestigial presence of sensory neurons and other olfactory anatomy in this odontocete.
An important goal of this study was to establish the CP as an osteological correlate of olfactory function, a metric applicable to extinct species. Here, we used the linear relationship between RelCP and ORGs to predict the likely number of functional ORGs for the extinct sabertooth cat (S. fatalis) from its CP morphology. Smilodon's CP and predicted ORG count positions the Pleistocene felid close to, but slightly below, the domestic cat in our olfactory scale (figure 2d). When compared with 10 living felids, Smilodon has, for its body size, a somewhat smaller CP than most felid species (electronic supplementary material, S12b), but this should be confirmed with a sample that includes multiple individuals of each species. If a relatively smaller CP for Smilodon is confirmed, then it may have been less reliant on olfaction than the similarly large extant felid, the African lion (Panthera leo). If so, it would be interesting to estimate Smilodon's relative reliance on alternative sensory modalities (e.g. hearing, vision) as possible evidence of compensatory adaptations.
5. Conclusion
The strong relationship we observed between CP morphology and ORGs contributes to our understanding of olfactory biology in several ways. First, the RelCP/ORG correlation illuminates an underlying developmental intersection of genes, neuroanatomy and skull morphology along the peripheral olfactory pathway from nose to brain and points to the need for future work that addresses the mechanisms linking ORGs and the generation of OSN populations in the nasal epithelium. Second, these results suggest that the coupling of genes and morphology provides a coherent molecular–morphological metric, with which we can make strong inferences about relative olfactory ability in individual mammal species. Finally, CP size is reinforced here as a stand-alone indicator of the relative importance of olfaction across mammals and a metric that can be applied to extinct species, from which molecular data can no longer be extracted.
Supplementary Material
Acknowledgements
We thank M. Faillace, R. Rudolph and J. Urbanski of General Electric Inspection Technologies for the training in and facilitation of high-res CT scanning, M. Colbert, R. Ketcham and J. Maisano of University of Texas HRCT Digital Morphology group, M. McNitt-Gray and J. Hoffman of UCLA for their skill and care in producing high-res CT scans, T. Smith, E. Seiffert, J. Kappelman, F. Wei, A. Curtis, for lending individual CT scans, curators and collection managers C. Conroy, K. Molina, J. Dines for providing skulls, and C. Griffis for his wizardry in R. Thanks to the Van Valkenburgh laboratory members and two anonymous reviewers for their comments.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material and are also available via the Dryad Digital Repository at: https://doi.org/10.5061/dryad.28sm8. CT scans created for this paper are available through DigiMorph at: http://digimorph.org/ or MorphoSource at: https://www.morphosource.org/.
Authors' contributions
D.J.B. conceived the project, designed digital methods, scanned specimens, collected and analysed data. W.J.M. supervised selection of genomic data and collaborated on interpretation of results. L.F.-R. and I.H. collected morphological data. I.H. created the animation. R.A.E. facilitated training in CT scanning through funding from Division of Physical Sciences UCLA; B.V.V. provided ideas and direction for every aspect of the project and facilitated acquisition of museum specimens and scans. D.J.B. and B.V.V. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NSF Graduate Research Fellowship Program grant DGE-1144087; NSF grants: IOS-1119768, IOS-1457106, BCS/IOS-0924592, BCS-1231350, BCS-1231717, BCS-0959438.
References
- 1.Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. ( 10.1016/0092-8674(91)90418-X) [DOI] [PubMed] [Google Scholar]
- 2.Niimura Y, Nei M. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708 ( 10.1371/journal.pone.0000708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niimura Y, Matsui A, Touhara K. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 24, 1485–1496. ( 10.1101/gr.169532.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20, 1–9. ( 10.1101/gr.099416.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chess A, Simon I, Cedar H, Axel R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834. ( 10.1016/S0092-8674(94)90562-2) [DOI] [PubMed] [Google Scholar]
- 6.Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. 1996. Visualizing an olfactory sensory map. Cell 87, 675–686. ( 10.1016/S0092-8674(00)81387-2) [DOI] [PubMed] [Google Scholar]
- 7.Bressel OC, Khan M, Mombaerts P. 2015. Linear correlation between the number of olfactory sensory neurons expressing a given mouse odorant receptor gene and the total volume of the corresponding glomeruli in the olfactory bulb. J. Comp. Neurol. 524, 199–209. ( 10.1002/cne.23835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyamichi K, Serizawa S, Kimura HM, Sakano H. 2005. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J. Neurosci. 25, 3586–3592. ( 10.1523/JNEUROSCI.0324-05.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pihlström H, Fortelius M, Hemilä S, Forsman R, Reuter T. 2005. Scaling of mammalian ethmoid bones can predict olfactory organ size and performance. Proc. R. Soc. Lond. B 272, 957–962. ( 10.1098/rspb.2004.2993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farbman AI. 1992. Cell biology of olfaction. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 11.Miller AM, Maurer LR, Zou D-J, Firestein S, Greer CA. 2010. Axon fasciculation in the developing olfactory nerve. Neural. Dev. 5, 20 ( 10.1186/1749-8104-5-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird DJ, Amirkhanian A, Pang B, Van Valkenburgh B. 2014. Quantifying the cribriform plate: influences of allometry, function, and phylogeny in Carnivora. Anat. Rec. (Hoboken) 297, 2080–2092. ( 10.1002/ar.23032) [DOI] [PubMed] [Google Scholar]
- 13.Vilensky JA. 2014. The neglected cranial nerve: nervus terminalis (cranial nerve N). Clin. Anat. 27, 46–53. ( 10.1002/ca.22130) [DOI] [PubMed] [Google Scholar]
- 14.Rowe T, Macrini TE, Luo Z-X. 2011. Fossil evidence on origin of the mammalian brain. Science 332, 955–957. ( 10.1126/science.1203117) [DOI] [PubMed] [Google Scholar]
- 15.Oelschläger HHA, Buhl EH. 1985. Development and rudimentation of the peripheral olfactory system in the harbor porpoise Phocoena phocoena (Mammalia: Cetacea). J. Morphol. 184, 351–360. ( 10.1002/jmor.1051840309) [DOI] [PubMed] [Google Scholar]
- 16.Bhatnagar KP, Kallen FC. 1974. Cribriform plate of ethmoid, olfactory bulb and olfactory acuity in forty species of bats. J. Morphol. 142, 71–90. ( 10.1002/jmor.1051420104) [DOI] [PubMed] [Google Scholar]
- 17.Garrett EC, Steiper ME. 2014. Strong links between genomic and anatomical diversity in both mammalian olfactory chemosensory systems. Proc. R. Soc. B 281, 20132828 ( 10.1098/rspb.2013.2828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsui A, Go Y, Niimura Y. 2010. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol. Biol. Evol. 27, 1192–1200. ( 10.1093/molbev/msq003) [DOI] [PubMed] [Google Scholar]
- 19.Montague MJ, et al. 2014. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc. Natl Acad. Sci. USA 111, 17 230–17 235. ( 10.1073/pnas.1410083111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes GM, Gang L, Murphy WJ, Higgins DG, Teeling EC. 2013. Using Illumina next generation sequencing technologies to sequence multigene families in de novo species. Mol. Ecol. Resour. 13, 510–521. ( 10.1111/1755-0998.12087) [DOI] [PubMed] [Google Scholar]
- 21.Rouquier S, Blancher A, Giorgi D. 2000. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl Acad. Sci. USA 97, 2870–2874. ( 10.1073/pnas.040580197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S. 2007. Correction: loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2, 50165 ( 10.1371/journal.pbio.0020005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kishida T, Kubota S, Shirayama Y, Fukami H. 2007. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol. Lett. 3, 428–430. ( 10.1098/rsbl.2007.0191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall DA, Blumer L, Moulton DG. 1981. Odor detection curves for n-pentanoic acid in dogs and humans. Chem. Senses 6, 445–453. ( 10.1093/chemse/6.4.445) [DOI] [Google Scholar]
- 25.Meisami E. 1989. A proposed relationship between increases in the number of olfactory receptor neurons, convergence ratio and sensitivity in the developing rat. Dev. Brain Res. 46, 9–19. ( 10.1016/0165-3806(89)90139-9) [DOI] [PubMed] [Google Scholar]
- 26.Laska M, Shepherd GM. 2007. Olfactory discrimination ability of CD-1 mice for a large array of enantiomers. Neuroscience 144, 295–301. ( 10.1016/j.neuroscience.2006.08.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson DA, Stevenson RJ. 2006. Learning to smell: olfactory perception from neurobiology to behavior. Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 28.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. 2009. Odor coding by a mammalian receptor repertoire. Sci. Signal. 2, ra9 ( 10.1126/scisignal.2000016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell 96, 713–723. ( 10.1016/S0092-8674(00)80581-4) [DOI] [PubMed] [Google Scholar]
- 30.McGann JP. 2017. Poor human olfaction is a 19th-century myth. Science 356, eaam7263 ( 10.1126/science.aam7263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizvanovic A, Amundin M, Laska M. 2013. Olfactory discrimination ability of Asian elephants (Elephas maximus) for structurally related odorants. Chem. Senses 38, 107–118. ( 10.1093/chemse/bjs097) [DOI] [PubMed] [Google Scholar]
- 32.Meredith RW, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 33.Foley NM, Springer MS, Teeling EC. 2016. Mammal madness: is the mammal tree of life not yet resolved? Phil. Trans. R. Soc. B 371, 3667–3679. ( 10.1098/rstb.2015.0140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niimura Y. 2013. Identification of olfactory receptor genes from mammalian genome sequences. Methods Mol. Biol. (Clifton, NJ) 1003, 39–49. ( 10.1007/978-1-62703-377-0_3) [DOI] [PubMed] [Google Scholar]
- 35.Nei M, Niimura Y, Nozawa M. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. ( 10.1038/nrg2480) [DOI] [PubMed] [Google Scholar]
- 36.Therneau TM, Atkinson B, Ripley B. 2010. rpart: Recursive partitioning. R Packag. version 3, 1–46.
- 37.Orme D. 2013. The caper package: comparative analysis of phylogenetics and evolution in R. R Packag. version 5.
- 38.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 39.Team RC. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 40.Stock C, Harris JM.. 1992. Rancho La Brea: a record of Pleistocene life in California. Los Angeles, CA: Natural History Museum of Los. [Google Scholar]
- 41.Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819. ( 10.1038/nature04338) [DOI] [PubMed] [Google Scholar]
- 42.Schrider DR, Costello JC, Hahn MW. 2009. All human-specific gene losses are present in the genome as pseudogenes. J. Comput. Biol. 16, 1419–1427. ( 10.1089/cmb.2009.0085) [DOI] [PubMed] [Google Scholar]
- 43.Ma L, Wu Y, Qiu Q, Scheerer H, Moran A, Yu CR. 2014. A developmental switch of axon targeting in the continuously regenerating mouse olfactory system. Science 344, 194–197. ( 10.1126/science.1248805) [DOI] [PubMed] [Google Scholar]
- 44.Pease M, Marquez Y, Tuchman A, Markarian A, Zada G. 2013. Diagnosis and surgical management of traumatic cerebrospinal fluid oculorrhea: case report and systematic review of the literature. J. Neurol. Surg. Rep. 74, 57–66. ( 10.1055/s-0033-1347902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen IH, Kiang JH, Correa V, Lopez MI, Chen PY, McKittrick J, Meyers MA. 2011. Armadillo armor: mechanical testing and micro-structural evaluation. J. Mech. Behav. Biomed. Mater. 4, 713–722. ( 10.1016/j.jmbbm.2010.12.013) [DOI] [PubMed] [Google Scholar]
- 46.Shoshani J, Kupsky WJ, Marchant GH. 2006. Elephant brain. Part I: gross morphology, functions, comparative anatomy, and evolution. Brain Res. Bull. 70, 124–157. ( 10.1016/j.brainresbull.2006.03.016) [DOI] [PubMed] [Google Scholar]
- 47.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038. ( 10.1038/nature09144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shum EY, Espinoza JL, Ramaiah M, Wilkinson MF. 2015. Identification of novel post-transcriptional features in olfactory receptor family mRNAs. Nucleic Acids Res. 43, 9314–9326. ( 10.1093/nar/gkv324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ngwenya A, Patzke N, Ihunwo A, Manger PR. 2011. Organisation and chemical neuroanatomy of the African elephant (Loxodonta africana) olfactory bulb. Brain Struct. Funct. 216, 403–416. ( 10.1007/s00429-011-0316-y) [DOI] [PubMed] [Google Scholar]
- 50.Bates LA, Sayialel KN, Njiraini NW, Poole JH, Moss CJ, Byrne RW. 2008. African elephants have expectations about the locations of out-of-sight family members. Biol. Lett. 4, 34–36. ( 10.1098/rsbl.2007.0529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bates LA, Sayialel KN, Njiraini NW, Moss CJ, Poole JH, Byrne RW. 2007. Elephants classify human ethnic groups by odor and garment color. Curr. Biol. 17, 1938–1942. ( 10.1016/j.cub.2007.09.060) [DOI] [PubMed] [Google Scholar]
- 52.Emerling CA, Springer MS. 2014. Genomic evidence for rod monochromacy in sloths and armadillos suggests early subterranean history for Xenarthra. Proc. R. Soc. B 282, 20142192 ( 10.1098/rspb.2014.2192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browne C, Stafford K, Fordham R. 2006. The use of scent-detection dogs. Irish Veterinary J. 59, 97. [Google Scholar]
- 54.Turcsán B, Kubinyi E, Miklósi Á. 2011. Trainability and boldness traits differ between dog breed clusters based on conventional breed categories and genetic relatedness. Appl. Anim. Behav. Sci. 132, 61–70. ( 10.1016/j.applanim.2011.03.006) [DOI] [Google Scholar]
- 55.Fan Z, et al. 2016. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 26, 163–173. ( 10.1101/gr.197517.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins CE, Hendrickson A, Kaas JH. 2005. Overview of the visual system of Tarsius. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 287A, 1013–1025. ( 10.1002/ar.a.20263) [DOI] [PubMed] [Google Scholar]
- 57.Pettigrew JD. 1999. Electroreception in monotremes. J. Exp. Biol. 202, 1447–1454. [DOI] [PubMed] [Google Scholar]
- 58.Ridgeway S. 1987. The terminal nerve in odontocete cetaceans. Ann. NY Acad. Sci. 519, 201–212. [DOI] [PubMed] [Google Scholar]
- 59.Springer MS, Gatesy J. 2017. Inactivation of the olfactory marker protein (OMP) gene in river dolphins and other odontocete cetaceans. Mol. Phylogenet. Evol. 109, 375–387. ( 10.1016/j.ympev.2017.01.020) [DOI] [PubMed] [Google Scholar]
- 60.Tan L, Li Q, Xie XS. 2015. Olfactory sensory neurons transiently express multiple olfactory receptors during development. Mol. Syst. Biol. 11, 1–9. ( 10.15252/msb.20156639) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material and are also available via the Dryad Digital Repository at: https://doi.org/10.5061/dryad.28sm8. CT scans created for this paper are available through DigiMorph at: http://digimorph.org/ or MorphoSource at: https://www.morphosource.org/.