Significance
Protein sequencing remains a challenge for small samples. A sensitive sequencing technology will create the opportunity for single-cell proteomics and real-time screening for on-site medical diagnostics. To resolve protein identity, we previously developed a computational algorithm that analyzes the ordered sequence of only two types of amino acids within a protein species. Through modification of a biological nanomachine, here, we developed single-molecule fluorescence technology to linearize protein molecules and to read signals from labeled amino acids in an ordered manner. This proof of concept of single-molecule fingerprinting will open the door to single-molecule protein sequencing and pave the road toward the development of a new, fast, and reliable diagnostic tool.
Keywords: single-molecule FRET, ClpXP, protein analysis, peptides, single-molecule protein sequencing
Abstract
Proteomic analyses provide essential information on molecular pathways of cellular systems and the state of a living organism. Mass spectrometry is currently the first choice for proteomic analysis. However, the requirement for a large amount of sample renders a small-scale proteomics study challenging. Here, we demonstrate a proof of concept of single-molecule FRET-based protein fingerprinting. We harnessed the AAA+ protease ClpXP to scan peptides. By using donor fluorophore-labeled ClpP, we sequentially read out FRET signals from acceptor-labeled amino acids of peptides. The repurposed ClpXP exhibits unidirectional processing with high processivity and has the potential to detect low-abundance proteins. Our technique is a promising approach for sequencing protein substrates using a small amount of sample.
Proteomic analyses provide essential information on molecular pathways of cellular systems and the state of a living organism (1). Therefore, for understanding of biological processes and their (dys)regulation, including disease, it is critical to monitor the protein composition of cells by sequencing (i.e., determination of the amino acid sequence). Mass spectrometry is currently the first choice for protein sequencing. However, mass spectrometry analysis often fails to recognize minor species embedded among other dominant species since sequence prediction is made through analysis of complex spectral peaks (2). As many cellular proteins exist in low abundance (3), it is difficult to obtain large-scale proteomic information. DNA sequencing presents similar challenges, but they are overcome by amplifying DNA samples until a high signal-to-noise ratio is achieved. This solution cannot be applied to protein analysis since there is no natural machinery that can amplify proteins.
Single-molecule techniques have the potential to provide radically new protein-sequencing tools that can quantify cellular proteins with accuracy as high as for mass spectrometry while requiring sample amounts as small as a single cell. However, despite several recent explorations (4–8), bona fide single-molecule protein sequencing has not yet been achieved due to the complexity that arises from primary protein sequences. Whereas DNA consists of only four building blocks (A, G, C, T), proteins are built from 20 distinct amino acids. Independent of the readout method of choice, full protein sequencing would require the detection of 20 distinguishable signals, which has so far not been demonstrated in single-molecule detection. Recently, our team and another have computationally demonstrated that readout of only a subset of the 20 building blocks is sufficient to identify proteins at the single-molecule level (9, 10). In brief, the number of protein species in an organism is finite and predictable. Through bioinformatics-based comparison with proteomics databases, ordered detection of only two types of amino acids can still allow for protein identification. For example, ordered detection of cysteine and lysine residues, which can be modified using orthogonal chemistries, is sufficient to sequence the human proteome (10). We named this approach “single-molecule protein fingerprinting” to distinguish it from full protein sequencing. Here, we demonstrate a proof of concept of a single-molecule fingerprinting technology that reads out fluorescently labeled amino acids of synthetic peptides and a model cellular protein.
To obtain ordered determination of fluorescently labeled amino acids, we needed a molecular probe that can scan a peptide in a processive manner. We adopted a naturally existing molecular machinery, the AAA+ protease ClpXP from Escherichia coli. The ClpXP protein complex is an enzymatic motor that unfolds and degrades protein substrates. ClpX monomers form a homohexameric ring (ClpX6) that can exercise a large mechanical force to unfold proteins using ATP hydrolysis (11, 12). Through iterative rounds of force-generating power strokes, ClpX6 translocates substrates through the center of its ring in a processive manner (13, 14), with extensive promiscuity toward unnatural substrate modifications, including fluorescent labels (15–17). Protein substrates are recognized by ClpX6 when they display specific disordered sequences, such as the 11-aa C-terminal ssrA tag (18). ClpX6 targets substrates for degradation by feeding them into ClpP14, a homotetradecameric protease that contains 14 cleavage sites and self-assembles into a barrel-shaped complex that encloses a central chamber (19).
Results
Single-Molecule Fingerprinting Platform.
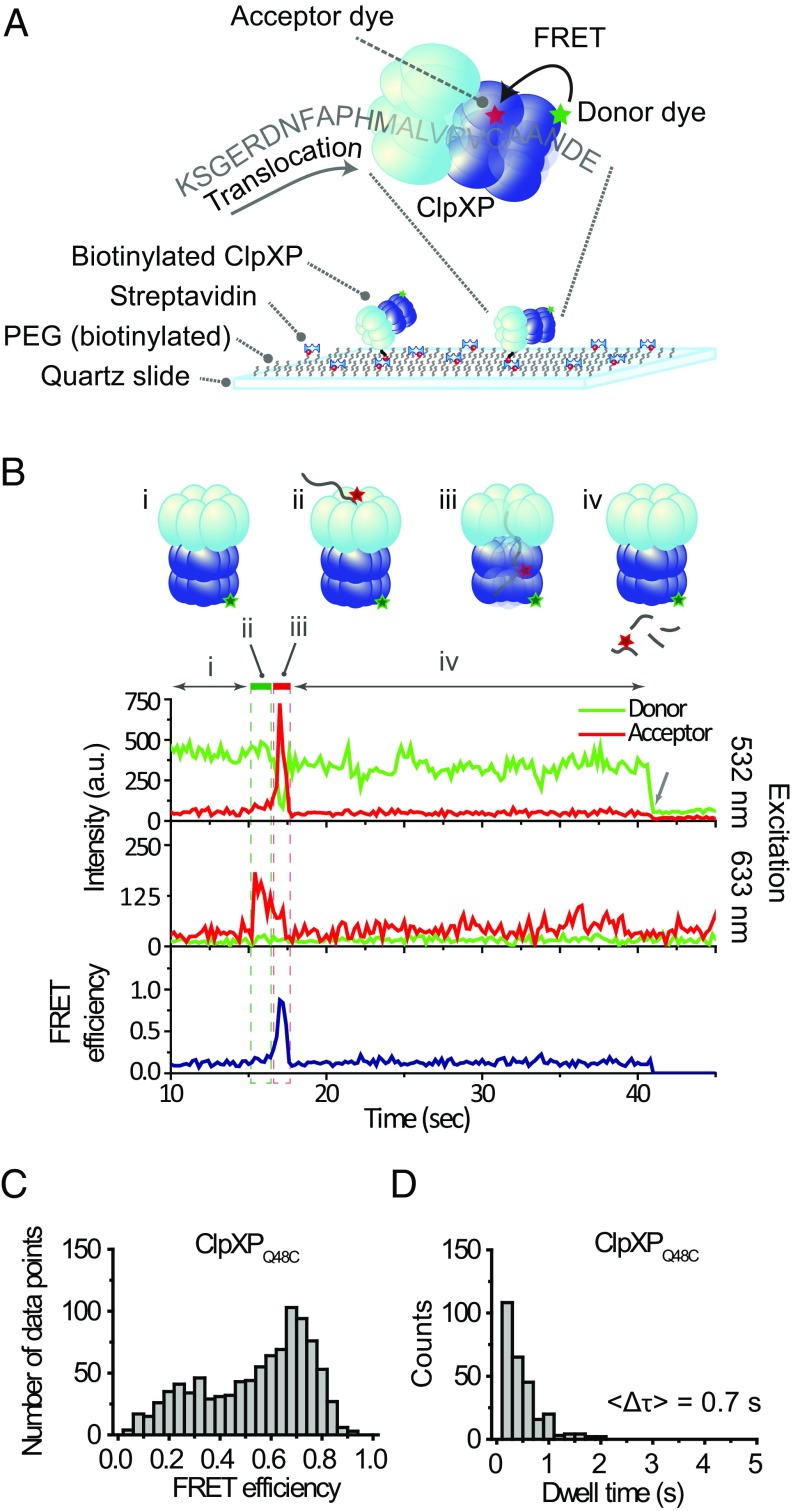
To immobilize ClpXP (ClpX6P14) for single-molecule imaging, we biotinylated ClpX6 and bound ClpX6P14 to a PEG-coated quartz surface through biotin–streptavidin conjugation (Fig. 1A). A combination of total internal reflection fluorescence microscopy (TIRF) and alternating laser excitation (ALEX) imaging (20, 21) was used to monitor individual ClpXP complexes bind, translocate, and degrade dye-labeled substrates in real time.
Fig. 1.
Single-molecule observation of ClpXP translocation. (A) Schematics of the single-molecule fingerprinting platform. Donor-labeled ClpXP is immobilized on a PEG-coated slide via biotin–streptavidin conjugation. ClpX6 recognizes an acceptor-labeled substrate (K-38-CCy5-ssrA, Table S1) and translocates it into the ClpP14 chamber, upon which FRET occurs. (B) A typical fluorescence time trace. (i) The donor signal is from Cy3-labeled ClpXP (Top trace) upon green excitation (532 nm). (ii, green box) The sudden appearance of acceptor signal (time ∼16 s) during acceptor-direct excitation with red (633 nm) reports on binding of acceptor-labeled substrate to ClpXP (Middle trace). (iii, red box) The high FRET (time ∼17 s) reports on the presence of the substrate in ClpP14 (Top and Bottom traces). (iv) Loss of fluorescence signal indicates the release of the substrate. The arrow at time ∼40 s indicates the photobleaching of Cy3. (C) FRET distribution of stage iii (n = 239). (D) Dwell time distribution of stage iii (n = 239).
To detect the progression of fluorescently labeled amino acids through the ClpX6 pore with nanometer-scale accuracy, we employed FRET (Förster resonance energy transfer) (22–24). We used two different types of model substrates for fingerprinting— short synthetic peptides and a small protein (the titin I27 domain). These substrates were labeled with acceptor fluorophores and were also appended with the ssrA tag. We constructed a FRET scanner by adding a fluorophore (donor) to the ClpP14 chamber (Fig. 1A). We introduced cysteines to the Q48 residue of ClpP (ClpPQ48C), A139 (ClpPA139C), or F31 (ClpPF31C), labeled them with maleimide-functionalized fluorophores (Fig. S1A), and evaluated the suitability for FRET-based substrate detection. ClpPQ48C and ClpPA139C showed higher FRET than ClpPF31C (Fig. S1B). Among the first two, ClpPQ48C was chosen for our final scanner due to its higher efficiency of fluorophore labeling (Methods).
The donor fluorophores on ClpPQ48C are located near the center of the ClpP14 chamber, which is ∼12 nm away from the substrate entry portal of ClpX6 (Fig. S1A) (25, 26). This distance is longer than the Förster radius of a standard single-molecule FRET pair (∼5 nm). This physical separation enabled us to selectively detect signals from only the fluorophores (acceptors) on a protein substrate that had been translocated through a ClpX6 central channel. We obtained FRET time traces reporting on translocation, as shown in Fig. 1B, by presenting a labeled peptide substrate to immobilized ClpXP complexes. The sudden appearance of acceptor signal during direct acceptor excitation indicated binding of acceptor-labeled peptide to ClpXP (Fig. 1B, Middle trace, stage ii). The subsequent appearance of a high FRET state indicated translocation of the substrate by ClpX6 into the ClpP14 chamber (Fig. 1B, stage iii). When a slowly hydrolyzable ATP analog (ATPγS) was used, the probability of high-FRET appearance decreased by one order of magnitude (Fig. S2D). Loss of FRET signal occurred upon the release of the dye-labeled peptide fragment (Fig. 1B, stage iv). When a cleavage inhibitor [diisopropyl fluorophosphate (DFP)] was used (27) (Fig. S2 A and B), the dwell time of high FRET increased 3.5-fold (Fig. S2C).
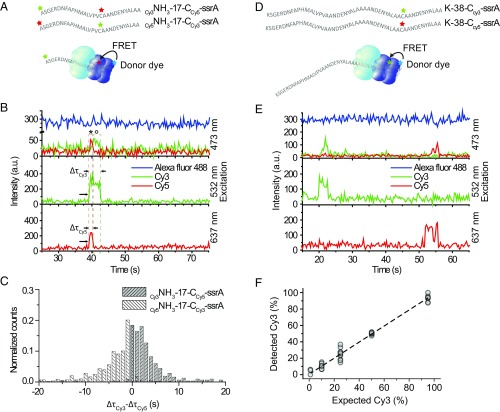
Our single-molecule fingerprinting concept requires detection of the order of fluorophores on a single substrate. To demonstrate fingerprinting, we functionalized a peptide with one type of fluorophore (Cy3) at the N-terminal site and a second type of fluorophore (Cy5) on an internal cysteine residue. We monitored the order in which the two fluorophores passed through Alexa488-labeled ClpP14 (Fig. 2A). The positions of the Cy3 and Cy5 fluorophores relative to the ssrA tag on the substrate should dictate the order of Alexa488-Cy3 FRET and Alexa488-Cy5 FRET signals since an ssrA-tagged substrate was translocated through ClpX6, starting from its C terminus. Fig. 2B depicts a representative time trace obtained from a substrate (Cy3NH3-17-CCy5-ssrA). The simultaneous appearance of Cy3 and Cy5 signals upon direct excitation with 532 nm and 637 nm (Fig. 2B, Middle and Bottom, t ∼ 40 s, indicated with arrows) indicates binding of a substrate containing both labels. In the FRET trace (Fig. 2B, Top), Alexa488-Cy5 FRET (marked * in the time trace) was observed before Alexa488-Cy3 FRET (marked °). This observation confirms that the ClpXP fingerprinter reads an ssrA-tagged substrate from the C-terminal to the N-terminal site.
Fig. 2.
Single-molecule fingerprinting. (A) Substrates with two acceptor dyes were labeled at the N-terminal end and on cysteine residues (Table S1). The green star stands for Cy3, red for Cy5, and blue for Alexa488. (B) A typical time trace for Cy3-NH3-17-CCy5-ssrA from three-color ALEX (Top) showed FRET between Alexa488 and Cy3; and Alexa488 and Cy5 upon excitation with blue laser light (473 nm). Concurrent signals from Cy3 (Middle) and Cy5 (Bottom) upon direct excitation, respectively, with green (532 nm) and red (637 nm). For clarity, an arbitrary offset of 200 a.u. was applied to the Alexa488 trace, and the sum of the Cy3 and Cy5 signals was plotted (Middle). For the original trace, see Fig. S1E. The symbol * indicate Alexa488-Cy5 FRET. The symbol ° indicates Alexa488-Cy3 FRET. The arrows indicate the start and end of each FRET. (C) Comparison of acceptor dwell times. The dwell time of Cy5 (ΔτCy5) was subtracted from that of Cy3 (ΔτCy3) for each event from Cy3-NH3-17-CCy5-ssrA (gray) (n = 318). The mean of the distribution is 3.5 ± 4.90 (s). In white is the same analysis for Cy5-NH3-17-CCy3-ssrA (n = 610). The mean of the distribution is −3.9 ± 5.67 (s). (D) Substrates labeled with one acceptor dye. Cysteine residues of the substrates (K-38-C-ssrA) were labeled with either Cy3 or Cy5. The green star stands for Cy3, red for Cy5, and blue for Alexa488. (E) A time trace from substrates in D. At time ∼20 s, a Cy3-labeled substrate binds (Top, Alexa488-Cy3 FRET) and 532-nm direct excitation (Middle). At time ∼50 s, a Cy5-labeled substrate binds (Top, Alexa488-Cy5 FRET) and 637-nm direct excitation (Bottom). An offset of 200 a.u. applied to Alexa488. (F) The percentage of processed Cy3-labeled substrates was plotted against the expected percentage of Cy3-labeled substrates. The line is a linear fit (slope of 0.98 ± 0.02, intercept 0.58 ± 0.79, R2 = 0.99). Data points are from 100-s recordings, repeated 10 times per condition (each n = 24.4 ± 1.50, in total n = 1,194).
We applied this fingerprinting scheme to the titin I27 domain. We labeled two Cys residues of titin (Cys64 and Cys80) with acceptor fluorophores. Because we did not have control over which dyes were attached to which Cys residues, we tagged both residues with the same dye, Cy5. Using Cy3 as a donor, we observed two separate FRET peaks within the time trajectories (Fig. S3A). The time interval between the two peaks was elongated when ATPγS was mixed with ATP (Fig. S3B), indicating that the two peaks represented the sequential probing of Cys80 and Cys64 residues.
We extracted the length of time that Cy3 and Cy5 acceptor fluorophores were engaged with ClpXP (ΔτCy3, ΔτCy5). We observed positive differences in dwell time (Δτ Cy3-Cy5 = ΔτCy3 − ΔτCy5, 〈Δτ Cy3-Cy5〉 = 3.5 s) for a substrate with N-terminal Cy3 labeling and internal Cy5 labeling (Fig. 2C, gray, Cy3NH3-17-CCy5-ssrA). For a substrate with exchanged dye positions (Cy5NH3-17-CCy3-ssrA), we observed negative differences (Fig. 2C, white, 〈Δτ Cy3-Cy5〉 = −3.9 s). Thus, dye-labeled amino acids located closer to the C-terminal ssrA tag were retained in the ClpXP complex for shorter amounts of time than labeled amino acids located more closely to the N terminus. We can conclude that our fingerprinter can detect dyes in an order matching the amino acid sequence. The ordered disappearance of the Cy3 and Cy5 signals further implies that uncleaved or partially cleaved substrate does not accumulate within the ClpP14 chamber, which would otherwise hamper accurate fingerprinting.
Performance of the Single-Molecule Fingerprinter.
A single-molecule fingerprinter should perform without any bias to fluorophores and with high dynamic range. To determine the sensitivity of our fingerprinter, we performed a population study in which ClpP14 was labeled with donor fluorophore (Alexa488) and substrate peptides were singly labeled with either Cy3 or Cy5 as an acceptor fluorophore (Fig. 2 D and E). We mixed Cy3- and Cy5-labeled substrates in varying proportions (1:99, 10:90, 25:75, 50:50, 95:5) and quantified the number of translocation events. We observed a linear relationship between the percentage of Cy3-labeled substrates we detected versus the expectation, with an offset of 0.58 ± 0.79% and a slope of 0.98 ± 0.02 (adjusted R2 = 0.99) (Fig. 2F). We conclude that both FRET pairs are detected with equal sensitivity and that our FRET scanner has the potential to detect low-abundance proteins.
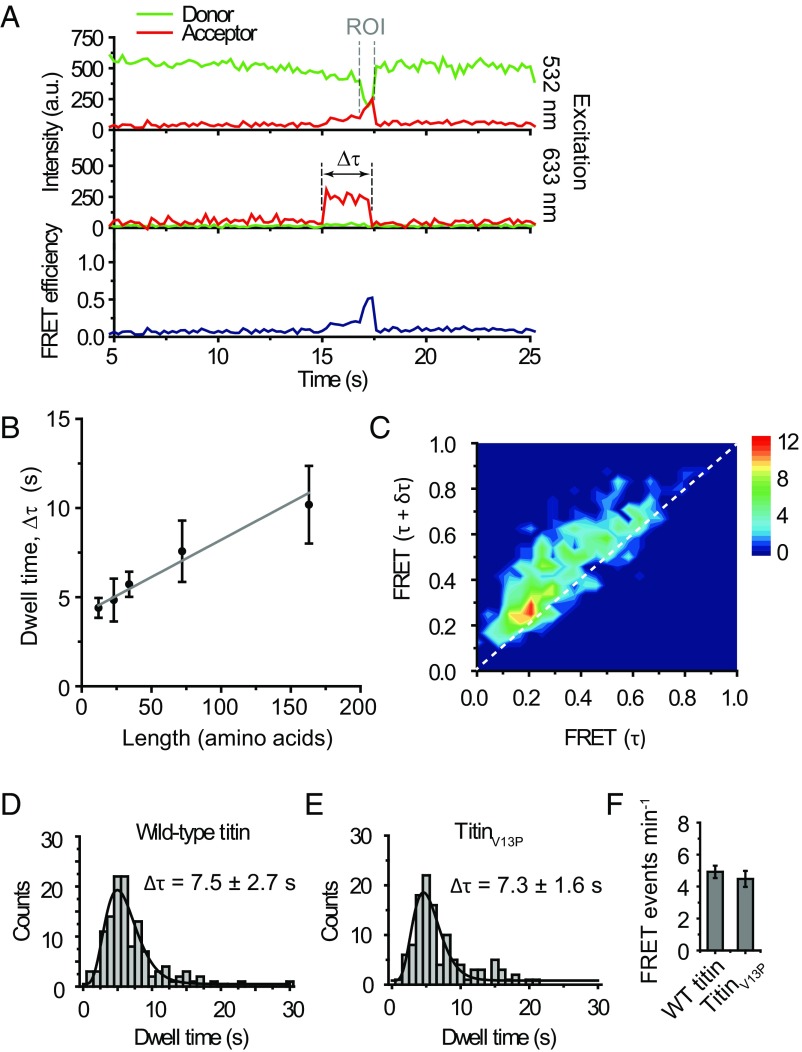
Our previous computational analysis indicated that the precision of our fingerprinting method would be enhanced if the distance between labeled cysteine and lysine residues could additionally be determined, as well as their order (10). A uniform speed of the scanner, represented by ClpX6, is crucial to extract distance information. To determine whether the processing time of ClpXP is proportional to the length of protein substrates, we determined the processing times (the dwell time of fluorescence signals emitted by Cy5 labels on substrates, upon direct excitation) for three peptides [29, 40, and 51 amino acids (aa) in length] (Table S1) and monomeric (119-aa) and dimeric (210-aa) versions of titin (all labeled at Cys) (Table S1). Plotting the total time (∆τ) (Fig. 3A) that a substrate was bound and processed by ClpXP versus the length of the substrates showed a linear increase, with an average processing speed of 23.9 aa per second (Fig. 3B and Fig. S4), which agrees with previous results obtained from both bulk (28) and single-molecule assays (11, 12, 29). We obtained a similar processing speed of 14.5 aa per second (translocation of 16 aa from labeled Cys64 to labeled Cys80 for 1.1 s) from the doubly labeled titin (Fig. S3B). In Fig. 3B, the y axis offset of 4.2 s reports on the initial docking phase and the eventual retention within ClpP14. Our data indicate that the ClpXP fingerprinter has the potential to determine both the order and spacing distance of labeled residues.
Fig. 3.
ClpXP performs unidirectional scanning with a constant speed. (A) Representative time trace. ROI (region of Interest) is where the FRET efficiency gradually increases. ∆τ: the total docking time. (B) Total dwell time vs. substrate length. The average time, 〈∆τ〉, was obtained by fitting data in Fig. S4 with a gamma distribution. Five different substrates were used: K-16-C-ssrA (n = 227), K-16-C-11-ssrA (n = 131), K-16-C-22-ssrA (n = 290), titin monomer (n = 85), and titin dimer (n = 81). The substrate length is the number of amino acids between the C terminus and a dye the most proximal to the N terminus. Error bars obtained by bootstrapping with 1,000 resamples. A linear fit results in an offset of 4.0 ± 0.20 s and a speed of 23.9 ± 2.86 aa per second. (C) Transition density plot. FRET change was analyzed by measuring FRETt = τ and FRETt = τ + δτ, with δτ = 0.4 s, for every point in ROI. The dotted line represents FRETδτ = FRETτ + δτ. K-38-C-ssrA was used (n = 44). (D and E) Total dwell times (∆τ) for WT titin (n = 124) and titinV13P (n = 114). Δτ = 7.5 ± 2.7 s and 7.3 ± 1.6 s were obtained, respectively, by fitting with a gamma distribution. Errors obtained by bootstrapping with 1,000 resamples. WT titin, n = 123. TitinV13P, n = 112. (F) The number of traces showing FRET events for WT titin and titinV13P. Error bars are SDs from 15 measurements.
Unidirectional translocation is also of utmost importance for our technology. Backtracking of ClpXP would result in insertion errors in the observed fingerprint and thus reduce the detection precision. To evaluate the occurrence of backtracking, we determined the change in FRET over time during processing of peptide substrates. We created a 2D heat map by plotting the change of FRET over a given time interval δτ. In Fig. 3C, FRET (t = τ + δτ) versus FRET (t = τ) is deposited for every time point along a time trace reporting on translocation [Fig. 3A, region of interest (ROI)]. We set δτ = 0.4 s, a time scale longer than our time resolution (0.2 s) but shorter than the average translocation time (0.7 s, Fig. 1D), to visualize the gradual increase of FRET. Any backtracking of ClpX6 along the substrate would result in momentary FRET decrease during translocation, which would appear as FRET (t = τ + δτ) values lower than FRET (t = τ) (population below the diagonal). We observed FRET (t = τ + δτ) higher than FRET (t = τ) (upper diagonal population) for a major fraction (92.5%) of the data points. The remaining fraction is likely due to the backtracking of ClpX, the statistical noise of the fluorescence signals, and the photoblinking of acceptor dyes. This degree of experimental error is predicted not to interfere with the ability to extract length information according to our computational simulation (10).
A single-molecule protein fingerprinter should be able to process any structural element of a protein. Single-molecule force spectroscopy studies of ClpXP have shown that ClpX6 stalls on substrates with rigid secondary structures (30, 31), which would inhibit the extraction of sequence information. We therefore explored the possibility of disrupting such tightly folded structures to enable fingerprinting. Perturbation of cysteine residues in the titin protein has been shown to interfere with the secondary structure of the protein, making it behave as an unstructured polypeptide chain (32, 33). We purified the I27 domain of both WT titin, known to make ClpX6 stall (30), and titinV13P, a variant that is still folded but is degraded at a rate close to denatured titin (33). By fluorophore labeling the cysteine residues of WT titin and titinV13P, we sought to determine the degree of structural influence of the cysteine-dye conjugation on ClpXP processing. We obtained equivalent total dwell times for processing stable WT titin (Δτ = 7.5 ± 2.7 s, Fig. 3D) and titinV13P (Δτ = 7.3 ± 1.6 s, Fig. 3E). A similar number of both substrates were processed by ClpXP within our time interval of observation (Fig. 3F), indicating that ClpX6 can process labeled WT and V13P substrates with the same efficiency. These results suggest that preparing substrates for sequencing by labeling cysteine residues (and likely lysine residues as well) might sufficiently destabilize their protein structures. This will allow for fingerprinting of any protein regardless of structural stability.
Discussion
We have demonstrated a FRET-based detection platform utilizing an AAA+ protease as a scanner of peptides and proteins. In our approach, we conjugate fluorophores to thiol groups of cysteine residues and amine groups of the N-terminal site (which can be extended to lysine residues) because these chemical groups can be labeled with high efficiency and specificity. Our platform, however, is not limited to these two modifications. With appropriate chemistry, one could target other residues or even posttranslational modifications. Detection of these moieties could be implemented by extending our current three-color FRET scheme to four-color FRET (21).
Several outstanding perspectives remain in order for our method to be directly applied to a protein sequencing technology. First, for proteomics analysis, our sequencing technique has to work for all cellular proteins without sequence bias. ClpX, a core of our platform, only recognizes substrates displaying specific sequence tags including ssrA. The substrate selectivity and specificity of ClpX would need to be broadened equally for every substrate, perhaps through targeted mutations in the substrate-recognition loops of the ClpX channel, or the use of engineered adaptor proteins (e.g., modified SspB) that nonspecifically deliver substrates to ClpX. Second, a challenge of cellular protein analysis is to detect low-abundance proteins within a complex sample, such as a clinical tissue sample. The depth of the sequencing coverage might be increased by removing housekeeping proteins chromatographically (34). Third, to cover the whole proteome in a reasonable amount of time, the throughput should be enhanced. Under the standard conditions used in this work (10 nM substrate, 512 × 512 pixel camera, ClpX6), we obtained ∼10 productive reactions per minute per imaging area. By using a complementary metal-oxide semiconductor (CMOS) camera that has a larger number of pixels (e.g., 2,084 × 2,084 pixels) (35), as well as a zero-mode waveguide platform that allows for single-molecule imaging of a higher concentration of substrate (e.g., 1 μM) (36), the throughput would be improved by a factor of ∼1,000. We also observed that productive reactions (a trace ending with high FRET) make up only ∼10% of the total population (Fig. S2D). We suspect that this low yield is due to the lack of adaptor proteins that facilitate binding, such as sspB, in our experiments. By adding adaptor proteins and replacing the hexameric linked ClpXΔN with WT, monomeric ClpX, which contains the N-terminal domain essential for binding adaptor proteins, we expect that the percentage of the productive population will reach near 100%. When our sequencer is improved with these changes, we expect to cover 1× of a single human-cell proteome (∼108 proteins) in nearly 10 h (10 events per minute × 16 × 100 × 10 ≈ 107 events per hour). In practice, sequencing errors will occur, and 1× coverage might not be sufficient to accurately determine a protein fingerprint. However, cellular protein copy numbers typically range from 100 to 107, resulting in an effective coverage of 100–107× per protein species. Reliable coverage of the lowest abundance protein species would require increased multiplexing beyond 1×, which could be achieved through further equipment improvements.
Our method has the potential to scan full-length proteins from end to end without the need for fragmentation. Sequencing substrates are processed at a constant speed, allowing for accurate protein identification (10). In this proof-of-concept study, we show our capability to detect low-frequency subpopulations of differentially labeled substrates, as well as our capacity to detect distinct acceptor fluorophores on a single substrate in a sequential manner. The platform we present here has the capability to transform proteomics from a basic research tool into an invaluable asset to clinical diagnostics.
Methods
Constructs used in this study are listed in Table S1. Additional methods may be found in SI Methods.
Single-Molecule Sample Preparation.
To reduce the nonspecific binding of proteins, acidic piranha-etched quartz slides (G. Finkenbeiner) were passivated with two rounds of polyethylene glycol [mPEG-Succinimidyl Valerate, MW 5000, Laysan, followed by MS(PEG)4, Piercenet] as described previously (37). After assembly of a microfluidic flow chamber, slides were incubated with 5% Tween 20 for 10 min (38), and excess Tween 20 was washed with T50 buffer (10 mM Tris⋅HCl, pH 8.0, 50 mM NaCl), followed by 1-min incubation with streptavidin (0.1 mg/mL; Sigma). Unbound streptavidin was washed with 100 μL of T50 buffer, followed by 100 μL of PD buffer (25 mM Hepes, pH 8.0, 5 mM MgCl2, 40 mM KCl, 0.148% Nonidet P-40, 10% glycerol). A ClpX6:ClpP14 = 1:3 molar ratio was used to ensure ClpXP complex formation with a 1:1 molar ratio (39). Then, 30 nM ClpX6 and 90 nM ClpP14 (either WT or mutant) were preincubated for 2 min at room temperature in the presence of 10 mM ATP in PD buffer. After preincubation, the sample was diluted 10 times in PD buffer to reach an expected final ClpXP complex concentration of 3 nM. The diluted sample was applied to the flow chamber and incubated for 1 min. Unbound ClpXP complexes were washed with 100 μL of PD buffer containing 1 mM ATP. Then, 10 to 20 nM acceptor-labeled substrate was introduced to the flow chamber in the presence of an imaging buffer (0.8% dextrose [Sigma], 1 mg/mL glucose oxidase [Sigma], 170 mg/mL catalase [Merck], and 1 mM Trolox [(±)-6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, 238813] [Sigma]). Donor-labeled ClpP14 added into a chamber without ClpX6 led to very few nonspecifically immobilized ClpP protein complexes, ruling out any nonspecific adsorption of ClpP14 to the surface (Fig. S1B). All experiments were performed at room temperature (23 ± 2 °C).
Single-Molecule Fluorescence.
Single-molecule fluorescence measurements were performed with a prism-type total internal reflection fluorescence microscope. For two-color measurements, Cy3 molecules were excited using a 532-nm laser (Compass 215M-50; Coherent), and Cy5 molecules were excited using a 633-nm laser (25 LHP 928; CVI Melles Griot). Fluorescence signals from single molecules were collected through a 60× water immersion objective (UplanSApo; Olympus) with an inverted microscope (IX71; Olympus). Scattered light from the 532-nm and 633-nm laser beams was blocked by a triple-notch filter (NF01-488/532/635; Semrock). The Cy3 and Cy5 signals were separated with a dichroic mirror (635 dcxr; Chroma) and imaged using an EM-CCD camera (Andor iXon 897 Classic; Andor Technology).
For three-color measurements, Alexa488 molecules were excited using a 473-nm laser (OBIS 473 nm LX, 75 mW; Coherent), Cy3 molecules were excited using a 532-nm laser (Sapphire 532 nm, 100 mW; Coherent), and Cy5 molecules were excited using a 637-nm laser (OBIS 637 nm LX, 140 mW; Coherent). Fluorescence signals from single molecules were collected through a 60× water immersion objective (UplanSApo; Olympus) with an inverted microscope (IX73; Olympus). The 473-nm laser beam was blocked by a 473-nm long pass filter (BLP01-473R-25; Semrock), the 532 nm laser beam was blocked by a 532-nm notch filter (NF03-532E-25; Semrock), and the 637-nm laser beam was blocked by a 633-nm notch filter (NF03-633E-25; Semrock). The Alexa488, Cy3, and Cy5 signals were separated by dichroic mirrors (540dcxr and 635 dcxr; Chroma) and imaged using an EM-CCD camera (Andor iXon 897 Classic; Andor Technology).
Data Acquisition.
Samples were excited alternatingly with different colors and using a custom-made program written in Visual C++ (Microsoft). A series of CCD images with an exposure time of 0.1 s were recorded. The time traces were extracted from the CCD image series using an IDL (ITT Visual Information Solution) algorithm that identifies fluorescence spots with a defined Gaussian profile and with signals above the average of the background signals. Colocalization between Alexa488, Cy3, and Cy5 signals was carried out with a custom-made mapping algorithm written in IDL. The extracted time traces were processed using Matlab (MathWorks) and Origin (Origin Lab).
Supplementary Material
Acknowledgments
We thank Marek Noga, Ivo Severins, and Anna Haagsma for technical support; Stanley Chandradoss and Margreet Docter for building the multicolor optical setup; Luuk Loeff and Misha Klein for assisting with data analysis; and Tim Blosser, Laura Restrepo, Margreet Docter, Stephanie Heerema, Noortje de Haan, and Viktorija Globyte for critical reading of the manuscript. ClpX6 expression plasmids were a generous gift from Andreas Martin. ClpP expression plasmids were a generous gift from Tania Baker and Robert Sauer. Titin expression plasmids were a generous gift from Victor Muñoz and Jörg Schönfelder. C.J. and A.S.M. were funded by the Foundation for Fundamental Research on Matter (Grants Projectruimte 12PR3029 and Vrije Programma SMPS).
Footnotes
Conflict of interest statement: J.v.G., C.J., and A.S.M. hold a patent (“Single molecule protein sequencing”; WO2014014347).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707207115/-/DCSupplemental.
References
- 1.Harper JW, Bennett EJ. Proteome complexity and the forces that drive proteome imbalance. Nature. 2016;537:328–338. doi: 10.1038/nature19947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zubarev RA. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics. 2013;13:723–726. doi: 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- 3.Talapatra A, Rouse R, Hardiman G. Protein microarrays: Challenges and promises. Pharmacogenomics. 2002;3:527–536. doi: 10.1517/14622416.3.4.527. [DOI] [PubMed] [Google Scholar]
- 4.Nivala J, Marks DB, Akeson M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat Biotechnol. 2013;31:247–250. doi: 10.1038/nbt.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen CB, Rodriguez-Larrea D, Bayley H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat Biotechnol. 2014;32:179–181. doi: 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat Nanotechnol. 2014;9:466–473. doi: 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohshiro T, et al. Detection of post-translational modifications in single peptides using electron tunnelling currents. Nat Nanotechnol. 2014;9:835–840. doi: 10.1038/nnano.2014.193. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy E, Dong Z, Tennant C, Timp G. Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat Nanotechnol. 2016;11:968–976. doi: 10.1038/nnano.2016.120. [DOI] [PubMed] [Google Scholar]
- 9.Swaminathan J, Boulgakov AA, Marcotte EM. A theoretical justification for single molecule peptide sequencing. PLoS Comput Biol. 2015;11:e1004080. doi: 10.1371/journal.pcbi.1004080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Y, Docter M, van Ginkel J, de Ridder D, Joo C. Single-molecule protein sequencing through fingerprinting: Computational assessment. Phys Biol. 2015;12:055003. doi: 10.1088/1478-3975/12/5/055003. [DOI] [PubMed] [Google Scholar]
- 11.Aubin-Tam M-E, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson MW, Singh SK, Maurizi MR. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J Biol Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 15.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolygo K, et al. Studying chaperone-proteases using a real-time approach based on FRET. J Struct Biol. 2009;168:267–277. doi: 10.1016/j.jsb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 19.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, et al. Single-molecule four-color FRET. Angew Chem Int Ed Engl. 2010;49:9922–9925. doi: 10.1002/anie.201005402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Oijen AM. Single-molecule approaches to characterizing kinetics of biomolecular interactions. Curr Opin Biotechnol. 2011;22:75–80. doi: 10.1016/j.copbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Lamichhane R, Solem A, Black W, Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: Surface passivation and immobilization. Methods. 2010;52:192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae W, Choi MG, Hyeon C, Shin YK, Yoon TY. Real-time observation of multiple-protein complex formation with single-molecule FRET. J Am Chem Soc. 2013;135:10254–10257. doi: 10.1021/ja404276g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flanagan JM, Wall JS, Capel MS, Schneider DK, Shanklin J. Scanning transmission electron microscopy and small-angle scattering provide evidence that native Escherichia coli ClpP is a tetradecamer with an axial pore. Biochemistry. 1995;34:10910–10917. doi: 10.1021/bi00034a025. [DOI] [PubMed] [Google Scholar]
- 26.Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- 27.Maurizi MR, Clark WP, Kim SH, Gottesman S. Clp P represents a unique family of serine proteases. J Biol Chem. 1990;265:12546–12552. [PubMed] [Google Scholar]
- 28.Martin A, Baker TA, Sauer RT. Protein unfolding by a AAA+ protease is dependent on ATP-hydrolysis rates and substrate energy landscapes. Nat Struct Mol Biol. 2008;15:139–145. doi: 10.1038/nsmb.1380. [DOI] [PubMed] [Google Scholar]
- 29.Shin Y, et al. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc Natl Acad Sci USA. 2009;106:19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordova JC, et al. Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell. 2014;158:647–658. doi: 10.1016/j.cell.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nivala J, Mulroney L, Li G, Schreiber J, Akeson M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano. 2014;8:12365–12375. doi: 10.1021/nn5049987. [DOI] [PubMed] [Google Scholar]
- 32.Iosefson O, Nager AR, Baker TA, Sauer RT. Coordinated gripping of substrate by subunits of a AAA+ proteolytic machine. Nat Chem Biol. 2015;11:201–206. doi: 10.1038/nchembio.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 34.Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juette MF, et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 37.Chandradoss SD, et al. Surface passivation for single-molecule protein studies. J Vis Exp. 2014:e50549. doi: 10.3791/50549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan H, Xia Y, Qin M, Cao Y, Wang W. A simple procedure to improve the surface passivation for single molecule fluorescence studies. Phys Biol. 2015;12:045006. doi: 10.1088/1478-3975/12/4/045006. [DOI] [PubMed] [Google Scholar]
- 39.Singh SK, et al. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.