Significance
Plant postembryonic development relies on a small pool of stem cells at the shoot and root tip. The question of how the cell cycle regulatory activities are integrated into the specific stem cell context is not well understood. This study identifies a previously unknown regulatory module in the flowering plant consisting of two regulatory genes, TSO1 and MYB3R1. TSO1 negatively regulates MYB3R1 to control cell division activity, maintain proper stem cell pool size, and balance cell proliferation with differentiation in shoot and root. Significantly, animal homologs of TSO1 and MYB3R1 are members of a cell cycle regulatory complex, suggesting that this conserved module operates in both plants and animals.
Keywords: MYB3R1, DREAM complex, cell cycle, meristem, TSO1
Abstract
Fundamental to plant and animal development is the regulated balance between cell proliferation and differentiation, a process intimately tied to cell cycle regulation. In Arabidopsis, mutations in TSO1, whose animal homolog is LIN54, resulted in severe developmental abnormalities both in shoot and root, including shoot meristem fasciation and reduced root meristematic zone. The molecular mechanism that could explain the tso1 mutant phenotype is absent. Through a genetic screen, we identified 32 suppressors that map to the MYB3R1 gene, encoding a conserved cell cycle regulator. Further analysis indicates that TSO1 transcriptionally represses MYB3R1, and the ectopic MYB3R1 activity mediates the tso1 mutant phenotype. Since animal homologs of TSO1 and MYB3R1 are components of a cell cycle regulatory complex, the DREAM complex, we tested and showed that TSO1 and MYB3R1 coimmunoprecipitated in tobacco leaf cells. Our work reveals a conserved cell cycle regulatory module, consisting of TSO1 and MYB3R1, for proper plant development.
An important distinction between animal and plant development is that the animal cell lineage is already determined during embryogenesis, while plant cells continuously develop and commit to new organs during postembryonic development, owing to the self-renewing stem cells at the two growing tips, the shoot apical meristem (SAM), and the root apical meristem (RAM). In addition, plant cells are highly plastic, capable of acquiring new cell identity in response to positional cues or environmental conditions. Despite the distinctions, plants and animals share conserved cell cycle regulatory machineries such as the cyclin–cyclin-dependent kinases (CDKs) and the DREAM complex (1–4). While tremendous insights into the cell cycle regulation were obtained from single cell studies such as yeast and mammalian cell culture (5, 6), the understanding of how the basic cell cycle machinery is integrated into the specific developmental context in animals and plants is lacking.
In Arabidopsis, the SAM consists of three periclinal cell layers (L1–L3) and is divided into the central zone (CZ), the peripheral zone (PZ) flanking CZ, and the rib zone (RZ) beneath the CZ. Slowly dividing stem cells are situated in the CZ and are promoted by the homeodomain transcription factor WUCHEL (WUS). Cells in the PZs give rise to leaf or flower primordia through rapid cell division (7). At the opposite pole, the RAM is organized distinctly from the SAM. The slow-dividing cells at the center of the root tip forms the quiescent center (QC), which is surrounded by initials that give rise to different root cell types. Arranged radially from the center to the peripheral of the root is the stele (vasculature and pericycle tissue), endodermis, cortex, epidermis, lateral root cap, and columella (8). The WUSCHEL-Related Homeobox 5 (WOX5) is expressed in the QC and maintains stem cell identity of the root initials (9). In the longitudinal axis of the root, cells near the root tip divide rapidly in the meristematic zone (MZ), then exit the mitotic cycle and enter the endocycle in the transition zone (TZ), and subsequently differentiate in the elongation zone (EZ).
Proper meristem organization and function relies heavily on proper cell cycle regulation. While slowly dividing cells such as stem cells must maintain themselves at the G1 or S phase for a long period of time, rapidly dividing cells like cells in emerging organ primordia must overcome cell cycle checkpoints in rapid successive cycles. Further, differentiating cells need to exit cell cycle permanently. Failure to maintain proper cell cycle regulation has devastating consequences. Retinoblastoma (RB), the first tumor suppressor gene found in animals, is a cell cycle regulator (10). Loss of the RB function in animals led to embryonic lethality and tumor (11, 12). Loss of the RB-Related (RBR) gene in Arabidopsis is gametophyte lethal (13–15). Conditional RNAi knockdown of RBR disrupts the stem cell maintenance and organ primordia initiation in the SAM (16). Investigations into how cell cycle regulation interacts with development will not only bring novel insights into the distinct developmental programs such as the SAM and RAM in higher plant development but will also uncover the conserved principles that balance cell proliferation with differentiation. This proper balance is critical to successful organogenesis and the prevention of undesirable growth such as tumor.
Central to this study is the Arabidopsis tso1 mutant with defects in the SAM and RAM (17). The tso1 mutant SAM has disorganized cell layers, enlarged cells with incompletely formed cell walls, and enlarged (fasciated) SAM. This is accompanied by failure to form floral organs, leading to complete sterility. These phenotypes suggest that loss of TSO1 activity leads to overproliferation of cells residing in the shoot apical meristem and failure in floral organ differentiation, indicating a role of TSO1 in balancing cell proliferation with differentiation. The TSO1 protein possesses two cysteine-rich (CXC) domains connected by a conserved hinge domain (18, 19). It is homologous to the animal LIN54, a core subunit of an evolutionarily conserved cell cycle complex found in Caenorhabditis elegans, Drosophila, and mammals (20–26). This multisubunit complex functions as a master coordinator of cell cycle transcription (3). Since the sequence similarity between TSO1 and LIN54 is limited only to the DNA-binding CXC-hinge-CXC domain, whether TSO1 plays a similar function in plants is not known.
Interestingly, two types of tso1 mutant alleles gave two distinct developmental phenotypes. The tso1-1 and tso1-2 mutations (type I), which alter one of the conserved cysteines in the CXC domain, caused strong and multiple mutant phenotypes ranging from meristem fasciation to complete sterility (17). In contrast, type II alleles such as tso1-3 to tso1-6 were caused by nonsense mutation or T-DNA insertion and only showed reduced fertility (19, 27). The strong tso1-1 mutation was shown to be recessive antimorphic as artificial miRNA targeting the mutant tso1-1 mRNA significantly reduced the phenotype severity. Further, the type II tso1-3 allele, when combined with a mutation in SOL2, TSO1’s closest paralog in Arabidopsis, showed a phenotype resembling tso1-1. Thus, tso1-1 is equivalent to the combined loss of both TSO1 and its paralog SOL2 (28).
In Arabidopsis, there are seven paralogs of TSO1, which are classified into two subtypes based on distinct motifs found outside the CXC-hinge-CXC domain. Type I consists of TSO1/At3G22780 and SOL1/At3g22760 (TCX3), SOL2/At4g14700 (TCX2), and At3g04850 (TCX4). Type II consists of four other members, At4g29000 (TCX5), At2g20110 (TCX6), At5g25790 (TCX7), and At3g16160 (27). With the exception of TSO1 and SOL2, there was no report of mutant phenotype associated with other family members. Interestingly, Kobayashi et al. (2) recently reported the isolation of two distinct plant DREAM complexes from Arabidopsis seedlings (4). The repressor complex acts in nonactively dividing cells to repress the cell cycle gene expression; it consists of ALY2/3, TCX5, RBR1, and repressors, E2FC and MYB3R3. The activator complex acts at the G2/M phase and consists of ALY3, TCX5, RBR1, DPA/B, and activators E2FB and MYB3R4 (2, 4). Both plant complexes contain TCX5, one of the TSO1 family members. The isolation of the DREAM complexes in Arabidopsis paved the way for further studies of how this conserved cell cycle complex acts in the context of plant development, for which little is understood.
To understand the mechanism of TSO1 function in the SAM and RAM, we conducted a genetic screen to isolate genetic modifiers of tso1-1. We hypothesized that if TSO1 acts in a plant DREAM complex, genes coding for the DREAM complex components or genes normally regulated by the complex could be identified as suppressors or enhancers of tso1-1. The identification of these genetic interactors should provide mechanistic insights into how TSO1 regulates the development of SAM and RAM. Among the 45 suppressors of tso1-1 identified in this study, 32 are mutations in the MYB3R1 gene, coding for an R1R2R3-MYB transcription factor (TF) homologous to the mammalian B-MYB (also called MYBL2). B-MYB is a known DREAM complex cofactor in animals (6, 20, 21). We showed that MYB3R1 is both a target of TSO1 and possibly a partner of TSO1. Loss-of-function mutations in the MYB3R1 gene suppressed all aspects of tso1-1 phenotype, strongly suggesting the existence of a TSO1-MYB3R1 regulatory module that operates in both the SAM and RAM to regulate proper shoot and root development. The study identifies a TSO1-MYB3R1 regulatory module, uncovers the module’s specific function during the development of the SAM and RAM, and indicates the evolutionary conservation of DREAM complex components in balancing cell proliferation with differentiation in higher eukaryotes.
Results
Identification of tso1-1 Suppressors.
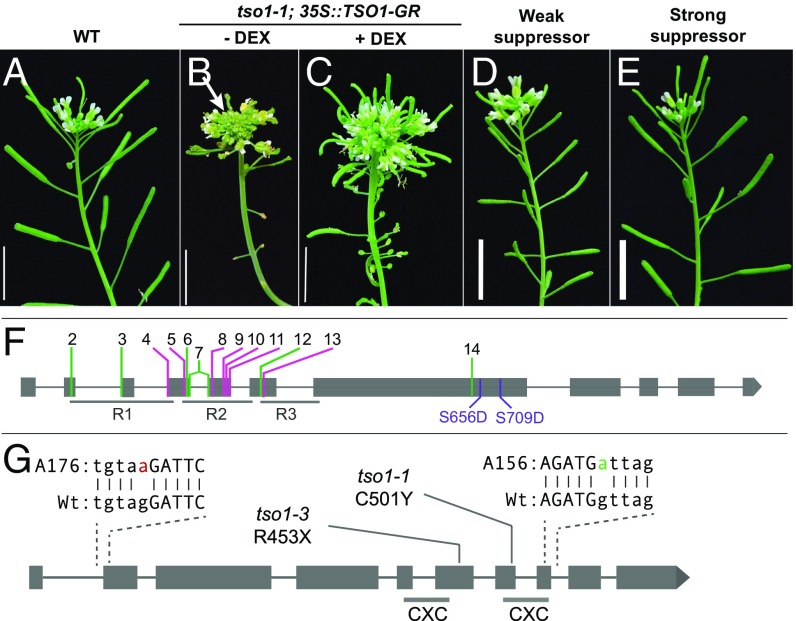
To determine the mechanism of TSO1 function, we conducted a mutagenesis screen to identify critical coregulators or downstream targets of TSO1. The genetic interactors may help place TSO1 into existing or novel regulatory pathways. tso1-1 allele was chosen for the mutagenesis as tso1-1 is 100% sterile, making fertility rescue easily identifiable.
Since tso1-1 plants produce no seed, we developed an inducible system to conditionally rescue tso1-1 to obtain tso1-1 homozygous seeds for mutagenesis. TSO1 full-length cDNA fused with the rat glucocorticoid receptor (GR) hormone binding domain was driven by the constitutive 35S promoter. The transgenic tso1-1 plants (tso1-1; 35S::TSO1-GR) produced seeds upon treatment with dexamethasone (DEX) (Fig. 1 B and C). These seeds were then mutagenized with EMS. By screening the M2 progeny of about 3,000 M1 tso1-1; 35S::TSO1-GR plants, 45 suppressors were identified based on their ability to develop elongated siliques in the absence of DEX treatment (SI Appendix, Fig. S1A). The degree of suppression ranges from weak to strong based on the length of the siliques (Fig. 1 D and E) and the number of seeds in each silique (SI Appendix, Fig. S2).
Fig. 1.
tso1-1 mutant phenotype and tso1-1 suppressors. (A) Wild-type (WT/Ler) inflorescence showing elongated siliques and flowers at the apex. (B) The inflorescence shoot of a tso1-1 mutant containing the 35S::TSO1-GR construct. No dexamethasone (DEX) was applied and a lack of siliques is evident. An enlarged shoot apex (arrow) indicates meristem fasciation. (C) DEX treatment (+DEX) of tso1-1 plants harboring the 35S::TSO1-GR transgene resulted in the rescue of the fertility defect as shown by the formation of long siliques. The inflorescence fasciation was not rescued due to the late stage of DEX application. (D) The inflorescence shoot of a weak suppressor of tso1-1, showing short siliques. Although the suppressor plant contains the 35S::TSO1-GR, no DEX was applied. (E) The inflorescence of a strong suppressor of tso1-1, showing normal size siliques. No DEX was applied. (F) Summary of myb3r1 mutations and their locations within the MYB3R1 gene. R1, R2, and R3 are the three DNA-binding domains. New myb3r1 alleles identified by the suppressors are respectively named myb3r1-2–myb3r1-14 (SI Appendix, Table S1). Nonsense or splicing mutations are labeled in green and missense mutations are labeled in pink. Serine 656 and serine 709 are changed into aspartic acid in the phosphomimic study. (G) Location of tso1-1, tso1-3, and the two intragenic suppressors tso1-7 (A156) and tso1-8 (A176). The exon sequence is in uppercase and the intron sequence is in lowercase. Specific changes in tso1-7 and tso1-8 are in magenta and green, respectively. The two CXC domains are underlined.
The Majority of Suppressors Map to the MYB3R1 Gene.
To determine the molecular identity of these suppressors, we used the bulk-segregant mapping-by-sequencing approach. Five suppressors were crossed with the tso1-1; 35S::TSO1-GR parental line to yield F2 mapping populations (SI Appendix, Fig. S1B). For each suppressor line, genomic DNA samples from 35–50 F2 plants showing the suppressed phenotype were pooled and sequenced. An analysis pipeline (Materials and Methods) mapped three suppressors (A317, B636, and B763) to the same region of chromosome 4 (SI Appendix, Fig. S3). Closer examination of the region revealed that the three suppressors caused either nonsense mutations (A317 and B763) or missense mutation (B636) in the same gene MYB3R1/At4G32730, and they were subsequently named myb3r1-14, -12, and -9, respectively (Fig. 1F and SI Appendix, Table S1).
To determine whether any of the remaining suppressors also reside in MYB3R1, we performed complementation tests through pairwise crosses among the suppressors. The results showed that 29 of the 40 suppressors failed to complement myb3r1 and hence defined additional alleles of myb3r1 (Fig. 1F and SI Appendix, Table S1). Sequence analysis of 22 such new myb3r1 suppressors identified specific mutational changes (SI Appendix, Table S1). Three alleles, myb3r1-3, myb3r1-4, and myb3r1-14, were isolated independently multiple times (SI Appendix, Table S1), suggesting saturation of the screen. In total, 13 independent new alleles (myb3r1-2–myb3r1-14) were confirmed (Fig. 1F and SI Appendix, Table S1). Seven are missense mutations, four are nonsense mutations, and two are splicing mutations. Twelve of the 13 alleles caused changes in the R1, R2, and R3 DNA-binding domains, indicating the essential function of the DNA-binding domains. Our data uncover myb3r1 as the major suppressor of tso1-1.
The second locus identified by the tso1-1 suppressors was TSO1 itself. Two suppressors (A156/tso1-7 and A176/tso1-8) were mapped to the same region of chromosome 3 (SI Appendix, Fig. S3) and each caused a mutation that affects the splicing of TSO1 mRNA (Fig. 1G and SI Appendix, Table S1). A third tso1 allele (B378/tso1-9) was identified among the remaining suppressors by complementation tests. The identification of intragenic tso1-1 suppressors confirms our previous studies, indicating that tso1-1 is a recessive antimorphic allele (28).
CRISPR-Induced myb3r1 Mutations Suppress tso1-1.
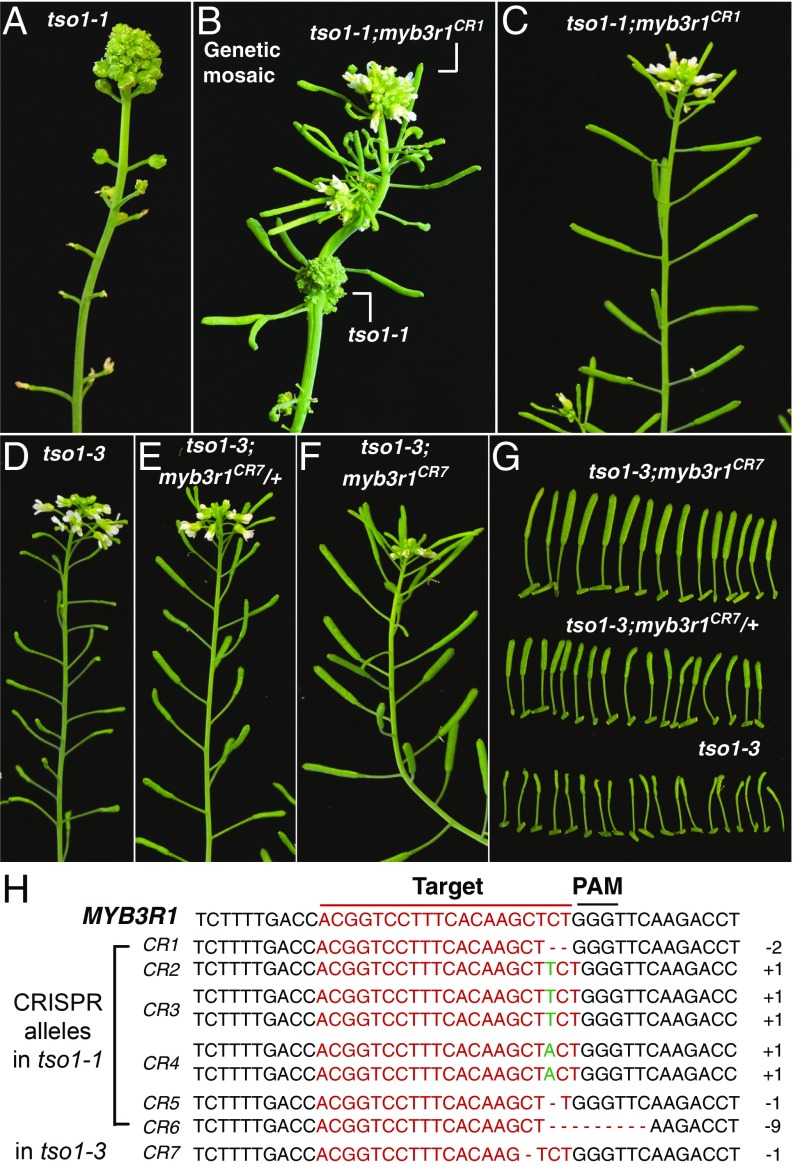
The EMS mutagen induced background mutations in the genome. Further, myb3r1 mutations could potentially suppress tso1-1 indirectly by facilitating TSO1-GR’s entry into the nucleus in the absence of DEX treatment. To eliminate these possibilities, we used CRISPR/Cas9 to knockout MYB3R1 directly in the tso1-1 background (without the TSO1-GR transgene). A gRNA targeting the fourth exon of MYB3R1 together with the Cas9 construct was transformed into plants heterozygous for tso1-1. In T2 generation, several tso1-1 mutant plants showed wild-type (WT) inflorescence branches and fertile siliques (Fig. 2B). These genetic mosaic plants contained CRISPR-induced biallelic mutations in the wild-type branches (Fig. 2H), indicating that the suppression of tso1-1 by myb3r1 is cell autonomous. Seeds collected from the wild-type branches gave rise to next generation plants that were indistinguishable from wild type (Fig. 2C). Therefore, germline-transmitted myb3r1CR completely suppressed tso1-1. This experiment firmly established that myb3r1 loss-of-function mutations suppress tso1-1 and eliminated any potential complication due to the TSO1-GR transgene.
Fig. 2.
CRISPR-edited myb3r1 alleles suppress both tso1-1 and tso1-3. (A) Inflorescence of tso1-1 mutant. (B) A genetic mosaic tso1-1 plant (T2 generation), which gives rise to wild-type branches with long siliques due to suppression of tso1-1 by the CRISPR-edited myb3r1CR allele. (C) T3 generation tso1-1; myb3r1CR plant derived from seeds collected from the wild-type branches of the mosaic plant shown in B. (D) tso1-3 plant showing severely reduced fertility; most seeds homozygous for tso1-3 are defective, leading to very small siliques. (E) tso1-3 plant heterozygous for a CRISPR-edited myb3r1CR7 allele. A total of 75% of seeds derived from the heterozygous mother should be normal, leading to increased seed yield and larger siliques. (F) tso1-3 plant homozygous for the CRISPR-edited myb3r1CR7 allele. The tso1-3; myb3r1CR7 double mutant developed normal siliques. Plants in D–F are siblings. (G) Closeup of the siliques made by plants in F (Upper), E (Middle), and D (Lower). (H) Sequence of MYB3R1 targeted by the gRNA and CRISPR-edited changes in the tso1-1 or tso1-3 background.
myb3r1 Mutations Suppress Different tso1 Alleles.
To determine whether myb3r1 also suppresses the other type of tso1 alleles, the same CRISPR/Cas9 construct was transformed into tso1-3, which harbors a nonsense mutation between the two CXC domains and shows reduced fertility (18, 28, 29) (Fig. 2D). In the tso1-3 background, plants heterozygous for the myb3r1CR mutation showed slightly elongated siliques compared with tso1-3 single mutant plants (Fig. 2 E and G) due to the formation of about a quarter homozygous myb3r1CR seeds. Further, tso1-3 plants homozygous for myb3r1CR are wild-type-like (Fig. 2 F and G). Therefore, myb3r1 mutations suppress both tso1-1 and tso1-3, and the effect of suppression is gene specific, not allele specific. The observation that myb3r1 mutations suppressed all defects of tso1 alleles indicates that MYB3R1 participates in all developmental processes regulated by TSO1.
Mutations in MYB3R4 Do Not Suppress tso1.
MYB3R1 is one of the five genes in Arabidopsis that encode homologs of the conserved cell cycle regulator B-MYB (MYBL2) in mammals (30–32). MYB3R1 and MYB3R4 were previously considered redundant MYB3Rs. While single myb3r1 and myb3r4 mutants had no phenotype, double my3r1; myb3r4 mutants showed a defect in mitosis and a reduction of G2/M cell cycle gene expression in the leaf stomatal cells (33). We thus asked why myb3r4 mutation was absent among the tso1-1 suppressors. Is the MYB3R4 region recalcitrant to the EMS mutagen? Or is MYB3R4 not involved in TSO1-regulated processes? The CRISPR/Cas9 system was used to target the second exon of MYB3R4 (Fig. 3E). tso1-1; myb3r4CR1 double homozygous mutants are indistinguishable from tso1-1 single mutants (Fig. 3 A and B). Hence, the my3r4 loss-of-function mutation did not suppress tso1-1.
Fig. 3.
MYB3R4 is not part of the TSO1-MYB3R1 module. (A) tso1-1 plant. (B) tso1-1; myb3r4CR1 plant exhibiting the same phenotype as tso1-1 single mutant. The specific mutations in MYB3R4CR1 are shown in E. (C) tso1-1; myb3r1-5 plant with a mild phenotype (reduced fertility). (D) tso1-1; myb3r1-5; myb3r4CR2 plant showing a similar fertility defect (small siliques) as the plant in C. The specific biallelic sequence changes in MYB3R4CR2 are shown in E. (E) MYB3R4 gene structure and CRISPR-edited mutations. R1R2R3 coding regions are underlined. The red sequence in the second exon of MYB3R4 was targeted by the gRNA. (Scale bars in A–D, 1 cm.)
To test whether MYB3R4 functions redundantly with MYB3R1 in interacting with tso1, we tested whether myb3r4CR could enhance the suppressor effect of myb3r1-5. myb3r1-5 is a hypomorphic allele and a weak suppressor of tso1-1 (Fig. 3 A and C). Compared with tso1-1; myb3r1-5, the triple mutant tso1-1; myb3r1-5; myb3r4CR2 did not appear to show any suppression of the fertility defect (Fig. 3 C and D). The smaller stature of the triple mutant was reminiscent of the myb3r1-1; myb3r4-1 double mutant (33). Taken together, we concluded that MYB3R1 and MYB3R4 do not act redundantly with respect to the TSO1-regulated processes.
MYB3R1 Is Misexpressed in tso1-1 SAM.
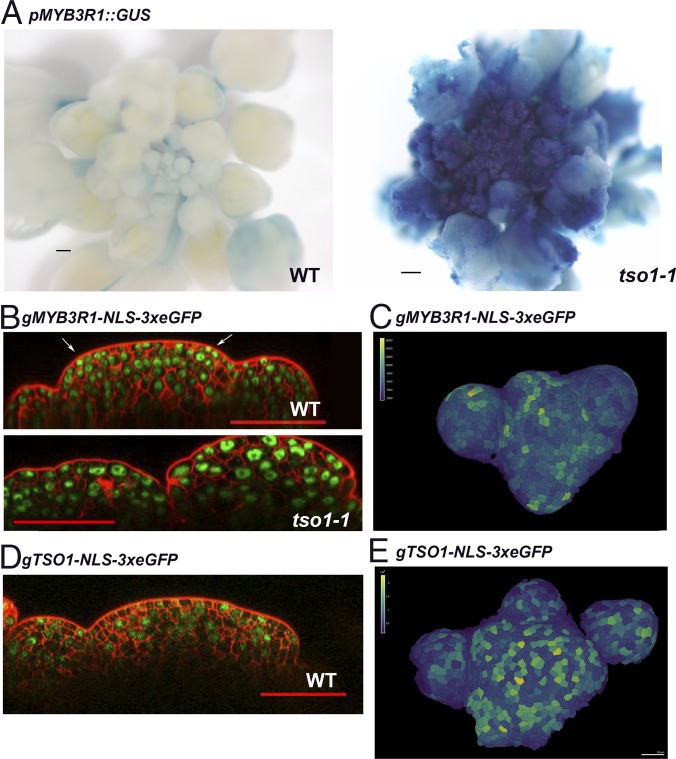
To investigate the molecular mechanism underlying the observed genetic interaction between tso1 and myb3r1, we examined MYB3R1 gene expression in tso1-1 mutants. A promoter pMYB3R1::GUS reporter construct published previously (31) was transformed into plants heterozygous for tso1-1. T1 transgenic plants heterozygous for tso1-1 gave rise to both tso1-1 and wild-type progeny, which were stained for 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-Gluc). The GUS expression pattern between tso1-1 and its wild-type siblings was compared. Consistently across multiple transgenic lines, increased intensity as well as ectopic expression of GUS were observed in tso1-1 inflorescences compared with their wild-type siblings (Fig. 4A). This reveals that the MYB3R1 promoter activity is affected by tso1-1, and the ectopic expression and overexpression of MYB3R1 likely mediate the tso1-1 phenotype. The data suggest that TSO1 normally represses the transcription of MYB3R1 in the SAM.
Fig. 4.
MYB3R1 is misexpressed in the tso1-1 SAM. (A) pMYB3R1::GUS expression in wild-type (Ler) and in tso1-1 inflorescence. The GUS signal is both ectopically expressed throughout the inflorescence and at a significantly higher level in tso1-1. The WT and tso1-1 plants are siblings harboring the same pMYB3R1::GUS transgene and stained in X-Gluc for the same amount of time. (B) gMYB3R1-NLS-3xeGFP expression in wild-type (Ler) and tso1-1 SAM. The WT and tso1-1 plants are siblings harboring the same gMYB3R1-NLS-3xeGFP insertion. (C) Top-down view of a WT inflorescence meristem containing the gMYB3R1-NLS-3xeGFP transgene. GFP fluorescent signals inside 3D segmented cells were visualized as heat map. Fluorescent quantification of segmented cells provided quantitative signal. (D) gTSO1-NLS-3xeGFP expression in WT SAM. (E) Top-down view of a WT inflorescence meristem containing the gTSO1-NLS-3xeGFP transgene. GFP fluorescent signals inside the 3D segmented cells were visualized as heat map. [Scale bars, 200 μm (A), 50 μm (B and D), and 20 μm (C and E).]
A translational fusion of MYB3R1 to 3xeGFP using its native promoter (gMYB3R1-NLS-3xeGFP) was made to visualize MYB3R1 protein accumulation. In the wild-type SAM, nuclear GFP signal is observed in most cells with varying intensity (Fig. 4B). A small number of cells with brighter GFP signal are at the L1 and L2 layers of the emerging primordia (Fig. 4B, arrows). At the surface view of the inflorescence apex (Fig. 4C), the cells in emerging floral meristems and organ primordia showed brighter GFP signals and different GFP intensity in different cells, which suggests cell cycle regulated expression (34). In the tso1-1 plant derived from the same transgenic line, the SAM longitudinal sections revealed the presence of larger cells with variable nuclei size and shape, and that the three-cell layer SAM organization was disrupted (17) (Fig. 4B). The gMYB3R1-NSL-3xeGFP signal appeared much brighter in tso1-1 SAM, reflecting an increased MYB3R1 protein level in tso1-1 (Fig. 4B).
A translational reporter of TSO1 (fused to NLS-3xeGFP) was also made, utilizing a 4.1-kb TSO1 genomic sequence including a 1-kb promoter and the TSO1 coding region. Wild-type plants containing the gTSO1-NLS-3xeGFP construct showed nuclear GFP signal in the inflorescence meristem and floral meristem cells (Fig. 4 D and E). The sporadic expression pattern is reminiscent of genes under cell cycle regulation (34). This study indicates that TSO1 and MYB3R1 are both expressed in the SAM; they may cooperate with each other to regulate cell cycle genes, which may include themselves.
myb3r1 Mutations also Suppress tso1-1 Root Phenotype.
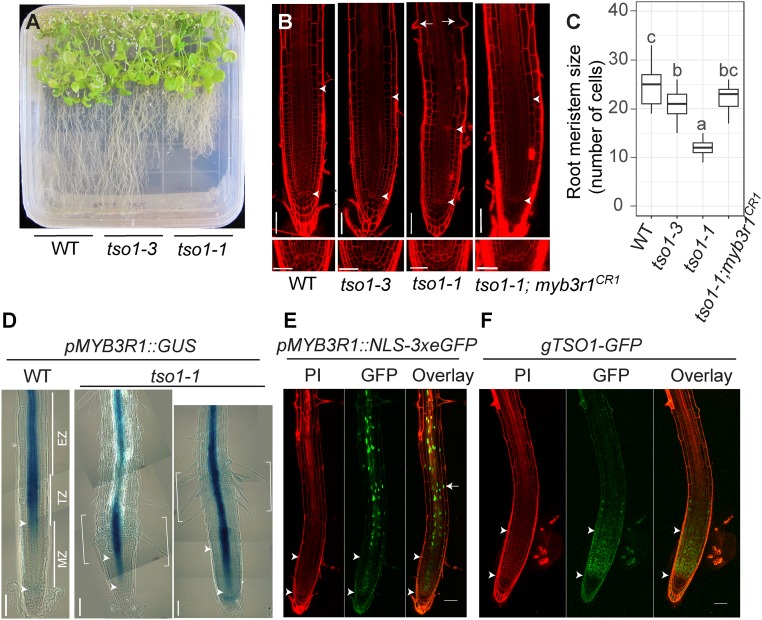
To determine whether tso1-1 affects root development, we took advantage of the TSO1-GR inducible system to obtain tso1-1 homozygous seeds. tso1-1 seeds germinated on MS plates showed significantly reduced root length (Fig. 5A). In contrast, tso1-3 did not exhibit this root defect (Fig. 5A). To investigate the underlying reason for the short root phenotype of tso1-1, we examined root cell organization using confocal microscopy. Roots were stained with propidium iodide (PI), and the root meristem size was measured by counting the number of cells in the cortex cell file in the MZ (Fig. 5B, the region between two arrowheads). The MZ was defined from the quiescent center to the first elongated cortex cell (35, 36). tso1-1 roots had significantly reduced root meristematic zone (Fig. 5 B and C); precocious differentiation of the root epidermal cells was indicated by the precocious root hair formation (Fig. 5B). The tso1-1 mutant root had all correct cell types with stereotypical division of the cortex/epidermal initials, and its QC did not terminate prematurely (Fig. 5B, Lower). Based on these results, we concluded that the short root phenotype of tso1-1 was due to precocious transition from proliferation to differentiation.
Fig. 5.
The tso1-1 short root phenotype is suppressed by myb3r1 mutations. (A) WT (Ler), tso1-3, and tso1-1 seedlings on a half strength MS media plate at 42 d postgermination (dpg). (B) Confocal images of root apical meristem of WT (Ler), tso1-3, tso1-1, and tso1-1; myb3r1CR1 at 7 dpg. The Upper arrowhead marks the upper boundary of the MZ as it marks the first cortex cell that is twice the size of the previous cell (cortex cell layer is the second from the epidermis). The Lower arrowhead marks the lower boundary of the MZ as well as the position of quiescent center (QC). Early differentiation of tso1-1 root is indicated by precocious formation of root hairs (arrows). Lower shows the stem cell niche in the root of respective genotype. [Scale bars, 50 μm (Upper) and 25 μm (Lower).] (C) Quantification of root MZ size by the cell number in cortex file in MZ (n = 10–15 roots). Different letters indicate statistically significant difference (P < 0.01, one-way ANOVA and Tukey test). (D) pMYB3R1::GUS expression in wild-type and tso1-1 root at 7 dpg. The MZ near the root tip is followed by the TZ, and then the EZ, which is shown by several images joined together with each image capturing a segment of the root. GUS signal, which reflects the promoter activity of MYB3R1, is restricted to the stele in the TZ and EZ. In tso1-1 mutant, the bracketed segment showing strong GUS signal is shifted downward. The region of ectopic MYB3R1 expression correlated with dense root hair formation. (E) In pMYB3R1::NLS-3xeGFP WT plant, nuclear GFP signals are found in the stele of the root in the EZ and the TZ, consistent with the GUS signal in D. (F) In WT plants containing the gTSO1-GFP construct, nuclear TSO1-GFP is largely restricted in the MZ and appears complementary to the MYB3R1 expression zone. Propidium iodide (PI) staining gives the red outline of the root cells. [Scale bars, 50 μm (B, E, and F) and 100 μm (D).]
Interestingly, the tso1-1 root phenotype appears opposite that of the triple myb3r1/3/5 mutant, whose root MZ is longer than wild type (2). This suggests that TSO1 and MYB3R1 may play opposite roles in regulating RAM, and that TSO1 likely represses MYB3R1 transcription in the root. To test this hypothesis, we examined the expression of the pMYB3R1::GUS transcriptional reporter in wild-type and tso1-1 plants. In wild type, pMYB3R1::GUS showed a strong and specific expression in the vascular tissue spanning the EZ and TZ (Fig. 5D). The expression decreases precipitously upon entering the MZ (Fig. 5D). In tso1-1 mutant root, the pMYB3R1::GUS expression extended further into the MZ. Furthermore, strong and precocious pMYB3R1::GUS expression (bracketed in Fig. 5D) often correlated with dense root hair formation. This pMYB3R1::GUS expression suggests that TSO1 normally represses MYB3R1 transcription in the MZ and that a loss of TSO1 in tso1-1 leads to ectopic MYB3R1 transcripts in the MZ. Precocious root differentiation occurs as a result, which is indicated by the formation of root hairs and a reduced size of the MZ. In support of this, the tso1-1; myb3r1CR1 double mutation, which removes the ectopic MYB3R1 in the tso1-1 background, suppressed the short root phenotype of tso1-1 (Fig. 5 B and C). Thus, ectopic/precocious MYB3R1 activity in tso1-1 mediates the short root phenotype.
The role of TSO1 in repressing MYB3R1 transcription in the root is consistent with the expression of gTSO1-GFP in the MZ (Fig. 5F) and the absence of (or low) pMYB3R1-3xeGFP transcription reporter signal in the MZ (Fig. 5E). In the TZ and EZ of the root, the reverse pattern was observed: the pMYB3R1-3xeGFP transcriptional reporter showed a strong GFP signal in the vascular tissue of TZ and EZ (Fig. 5E), where the gTSO1-GFP translational reporter had no signal (Fig. 5F). The MZ-specific expression of TSO1 was consistent with the previous “root map” data (37), where TSO1 transcripts were found in clusters 4 and 10 with the highest expression in the root MZ (SI Appendix, Fig. S4). Taken together, the complementary expression domains between TSO1-GFP protein and MYB3R1 promoter reporter, the opposite root mutant phenotypes, and the genetic suppression of tso1-1 by myb3r1CR provided strong support for a negative regulatory relationship between these two genes. TSO1 normally represses MYB3R1 transcription in the root MZ to prevent MYB3R1 expression and maintain meristematic activity. The data also suggest the existence of a TSO1-MYB3R1 regulatory module in the RAM as well as SAM.
Phosphomimics at S656 of MYB3R1 Enhances tso1-3 Fertility Defect.
To investigate whether the observed tso1-1 phenotype is mediated solely by misexpression of MYB3R1 at the transcriptional level, we tested whether overexpression of MYB3R1 could also cause the tso1-1 mutant phenotype. MYB3R1 cDNA was fused to the 35S promoter and transformed into wild-type plants, but the resulting transgenic plants were wild type in phenotype (SI Appendix, Table S2 and Fig. S5B). The same 35S::MYB3R1 transgene also failed to enhance the weak tso1-3 allele (SI Appendix, Table S2). Subsequently, we showed that the 35S::MYB3R1 construct failed to complement myb3r1-9 when introduced into tso1-1; myb3r1-9 plants (SI Appendix, Table S2 and Fig. S5 C and D). Hence, the 35S promoter could not be used to overexpress MYB3R1.
We then tested whether a hyperactive MYB3R1 could mediate a tso1-like phenotype. During late G1/S, mammalian B-MYB is phosphorylated by cyclin A/E-CDK2, which enhances its transactivation activity (38, 39). Therefore, we tested whether phosphomimics of MYB3R1 could lead to a tso1-like phenotype. By searching the Arabidopsis Protein Phosphorylation Site Database (PhosPhAt 4.0) (40, 41), serines at position 656 and 709 of MYB3R1 were found to be phosphorylated in previous large-scale phosphoproteomic studies (42, 43). Accordingly, we made a genomic construct of MYB3R1 (gMYB3R1) containing the MYB3R1 native promoter and coding sequence fused to GFP. The construct was shown to complement the myb3r1-9 when introduced into tso1-1; myb3r1-9 plants (SI Appendix, Table S2 and Fig. S5E). When introduced into WT or tso1-3 background, the gMYB3R1 construct had no phenotypic effect on the resulting plants (Fig. 6 B and F) even when the transgene is expressed at a high level (SI Appendix, Fig. S6C).
Fig. 6.
MYB3R1 phosphomimic S656D enhanced tso1-3 mutant. (A) The main inflorescence of WT (Ler). Lower shows the apex of the inflorescence. (B–D) Different versions of gMYB3R1 transgene constructs transformed into WT plants (genotype tso1-1/+). The specific version of transgene is indicated in parentheses. The main inflorescence shoot of gMYB3R1 (WT) (B), gMYB3R1 (S656D) (C), and gMYB3R1 (S709D) (D) all showed wild-type phenotype. (E) The inflorescence of tso1-3. Note the small silique due to reduced fertility and the minor phyllotaxy defect. (F–H) Different versions of gMYB3R1 transgene constructs transformed into tso1-3 plants. (F) The main inflorescence shoot of tso1-3; gMYB3R1 (WT) plant showing a phenotype that is identical to tso1-3 single mutant. (G) The inflorescence shoots of two transgenic lines of tso1-3; gMYB3R1 (S656D) plants showing a stronger phenotype than tso1-3 single mutants. The siliques did not form properly due to abnormal gynoecium and severely reduced fertility. (H) The inflorescence of a tso1-3; gMYB3R1 (S709D) plant. The phenotype is identical to that of tso1-3. [Scale bars, 1 cm (Upper) and 1 mm (Lower).]
Subsequently, we modified this gMYB3R1 by changing serine 656 and serine 709, respectively, into aspartic acid. These gMYB3R1 (S656D) and gMYB3R1 (S709D) “phosphomimic” constructs were respectively transformed into wild type and tso1-3. The gMYB3R1 (S709D) phosphomimic had no phenotypic impact on the resulting transgenic plants (Fig. 6 D and H). In contrast, the gMYB3R1 (S656D) transgene, while having no effect in the wild-type background (Fig. 6C), enhanced the tso1-3 fertility defect (Fig. 6G). Three tso1-3; gMYB3R1 (S656D) transgenic lines were analyzed further by quantifying their enhanced phenotype as well as the level of transgene expression (SI Appendix, Fig. S6). Line 8 had the lowest transgene expression and also the least enhancement of tso1-3 among the three transgenic lines. However, none of these lines showed a defect in the SAM fasciation (Fig. 6G and SI Appendix, Fig. S6A, Lower). Together, these results suggest that serine 656 is an important regulatory site of MYB3R1 and that phosphorylation at serine 656 (but not serine 709) may promote MYB3R1 activity. Our data also indicate that increased/ectopic MYB3R1 expression in tso1-1 mutants or hyperactive MYB3R1 via posttranscriptional modification (such as S656D) are both able to cause tso1-like phenotypes.
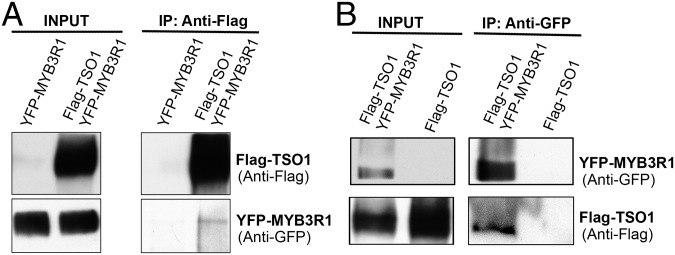
MYB3R1 Coimmunoprecipitates with TSO1 in Tobacco Leaves.
TSO1 and MYB3R1 both encode homologs of proteins that are components and cofactors of the animal and plant DREAM complex, thus we tested whether TSO1 and MYB3R1 associate with each other in a similar complex in plants. Since TSO1 is expressed at a low level in Arabidopsis, we used a tobacco transient expression system, where 35S::YFP-MYB3R1 and 35S::Flag-TSO1 were transiently coexpressed in the tobacco leaves. Antibody against the Flag-epitope was used to pull down extracted plant proteins that were subsequently subjected to Western blots. Both Flag-tagged TSO1 and YFP-tagged MYB3R1 were detected in the Western blots (Fig. 7A), indicating that TSO1 physically interacts with MYB3R1. The reciprocal pulldown was also performed using anti-GFP antibody in the pulldown; both Flag-TSO1 and YFP-MYB3R1 were detected in the immunoprecipitated fraction by Western blots (Fig. 7B). This coimmunoprecipitation suggests the possibility of a DREAM-like complex that includes TSO1 and MYB3R1. Our work in previous sections demonstrated that TSO1 represses MYB3R1 transcription; these co-IP data suggest that MYB3R1 may form a complex with TSO1 to participate in the regulation of its own transcription. The overlapping expression domains of the MYB3R1-GFP fusion protein and the TSO1-GFP fusion protein revealed by the gMYB3R1-NLS-3xeGFP and gTSO1-NLS-3xeGFP translational reporters in the SAM (Fig. 4 B and D) are consistent with this possibility.
Fig. 7.
TSO1 physically interacts with MYB3R1. Immunoprecipitation (IP) using protein extracts from tobacco leaves transiently expressing 35S::YFP-MYB3R1 and 35S::Flag-TSO1. (A) Coimmunoprecipitation using anti-Flag antibody. YFP-MYB3R1 was detected in the pulldown by Western blots using the anti-GFP antibody. The tobacco leaves transiently expressing only 35S::YFP-MYB3R1 were used as a negative control. The experiments were performed twice with the same result. (B) Coimmunoprecipitation using anti-GFP antibody. Flag-TSO1 was detected in the pulldown by Western blots using the anti-Flag antibody. Tobacco leaves transiently expressing only 35S::Flag-TSO1 were used as a negative control.
Discussion
The Animal DREAM Complex and Its Likely Conserved Function in Plants.
In the past 10–15 y, a new multiprotein complex, the DREAM complex, has gained increasing recognition as a critical cell cycle regulator. The DREAM complex not only coordinates G1/S with G2/M phases but also maintains cells at the G0 quiescent phase. Found in Drosophila, C. elegans, and mammals (22, 25, 26), the core complex of DREAM consists of five conserved members: LIN9, LIN37, LIN52, LIN54, and LIN53 (or RBBP4 in mammals). Depending on the organism, these components are also called MYB-interacting protein (MIP) or multivulva class B proteins (MuvB) (4). The core complex associates with different cofactors at different cell cycle phases. In G0 phase, the core complex binds to RB-like pocket proteins (p130 or p107) and repressor E2F/DP to repress all cell cycle-dependent gene expression. At the G1-to-S checkpoint, cyclin– CDKs phosphorylate RB-like pocket proteins to release E2F, which activates G1/S phase genes and promotes cell cycle progression. At the S phase, the DREAM core complex recruits MYB (B-MYB) and then recruits FOXM1 to the promoters of G2/M phase-expressed genes to promote their expression. Hence the core complex components support both repression and activation of gene expression and coordinate cell cycle phase-specific gene expression (3).
LIN54 (or mip120), the animal homolog of TSO1, is an essential core subunit of DREAM and confers DNA-binding activity to the DREAM complex (44). A predicted helix-coil-helix domain in LIN54 was required for its interaction with p130 and B-MYB. A reduction of LIN54 via RNAi disrupted the DREAM complex core, compromised the G1/S checkpoint, and down-regulated G2/M cell cycle genes in human cell lines (44–46). In Drosophila, mutation in mip120 caused complex phenotypes, including shortened longevity and severe eye defect (21). Interestingly, the Drosophila myb mutant adult lethality can be suppressed by mutations in the DREAM core component mip130 (47), demonstrating that mutating the interdependent DREAM complex components can lead to almost normal phenotype, which is similarly demonstrated in our study.
Arabidopsis genome encodes homologs of almost all DREAM complex components (2), suggesting the possibility of a DREAM-like complex in plants. Indeed, at least two distinct DREAM complexes were recently isolated from seedling and leaves that consist of RBR1, E2Fs, MYB3R3, ALYs, and TCX5 (2). MYB3R1 and TSO1 are respective paralogs of MYB3R3 and TCX5 in Arabidopsis, suggesting the likely existence of analogous DREAM complexes consisting of TSO1 and MYB3R1 in shoot and root tissues. This hypothesis is supported by our co-IP data (Fig. 7) showing an association between TSO1 and MYB3R1. Interestingly, TSO1 does not possess the helix-coil-helix domain present in LIN54 and may interact with MYB3R1 via distinct domains or other binding partners. The striking mutant phenotypes shown by tso1-1 and the complete suppression by myb3r1 provide an unusual opportunity to study the plant DREAM-like complex function in higher plant development.
Arabidopsis MYB3R1 Function and Regulation Bears Similarity to Mammalian B-MYB.
The mammalian B-MYB (MYBL2) resembles MYB3R1 in its cell cycle function and regulation (48). In G0/G1 cell cycle phase, B-MYB is repressed by the DREAM complex. In late G1/S, B-MYB is gradually released from the inhibition by the DREAM complex and instead associates with the core DREAM complex to cooperatively activate the G2/M cell cycle genes. Deregulation of B-MYB expression is involved in cancer initiation and progression; high B-MYB expression is correlated with poor patient outcome in numerous cancers (48, 49). The ectopic and extremely high level of MYB3R1 expression in tso1-1 mutant may cause what would be plant cancers: extreme meristem fasciation with many more meristem cells and failure of floral organ differentiation as shown in tso1-1 mutants.
We showed that the CRISPR knockout of MYB3R1 in tso1-1 provided a highly effective and comprehensive suppression of abnormal tso1-1 phenotypes. The tso1-1 suppressor screen provided unprecedented information on the critical domains and residues of MYB3R1, which is highly relevant to the understanding of the domains and functions of B-MYB and its role in human cell cycle regulation and cancer initiation, prognosis, and treatment. Among the 13 new MYB3R1 alleles, 12 reside in the DNA-binding domain, demonstrating the significance of this conserved domain in the MYB protein function. myb3r1-11 (C125Y), a strong suppressor of tso1-1, altered the conserved Cys125 residue in the R2 domain; Cys-125 was thought of as a redox sensor for the MYB protein (50) and an equivalent mutation in human MYB (Cys130) significantly impaired the activity of c-Myb and v-Myb (51, 52). Among the weak suppressors, myb3r1-5 (G88R) resides in the loop region connecting R1 and R2 (Fig. 1F). myb3r1-14 (R652X) causes a premature stop codon that removes the C-terminal region containing the serine 656. Alteration of this residue as demonstrated in the S656D phosphomimics has functional consequences. Future work will be necessary to test whether S656D might modulate TSO1-MYB3R1 protein–protein interaction, or constitutively activate downstream events, or both.
MYB3R1 Is both an Activator and Repressor.
The Arabidopsis genome has about 130 MYB genes, the majority of which are in the R2R3-MYBs (MYB2Rs) family. The MYB2Rs, with only two DNA-binding repeats, are not found in animals. In plants, they regulate diverse biological processes from development, metabolism, to response to environmental challenges (32). A second family of Arabidopsis MYB genes belong to the R1R2R3-MYBs (MYB3Rs) family. Only five MYB3Rs exist in Arabidopsis, which encode conserved cell cycle regulators found in animals (30–32). These five Arabidopsis MYB3Rs were shown to be either activators, MYB3R1 and MYB3R4, or repressors, MYB3R1, MYB3R3, and MYB3R5. The identification of only MYB3R1 and no other MYBs as the suppressor of tso1-1 strongly suggests a unique role of MYB3R1 in cell cycle regulation and plant development.
The repressor MYB3R3 and activator MYB3R4 were found to associate with the repressor and activator DREAM complexes, respectively, in Arabidopsis seedlings (2, 4). While the repressor MYB3Rs were shown to promote cell cycle exit and organ differentiation, the activator MYB3Rs were responsible for activating the G2/M-specific gene expression, including the B1-type cyclin CYCB1 and the plant-specific syntaxin KNOLLE (KN) for plant cell cytokinesis (2, 31, 33). Because of functional redundancy, none of the single mutants in the MYB3Rs showed any phenotypes (2, 31). Double mutants of activators my3r1/4 showed a defect in mitosis and reduced expression of G2/M cell cycle genes in leaf stomata (33). In contrast, triple mutants of the repressor MYB3Rs, myb3r1/3/5, showed a constitutive expression of G2/M genes even outside cell cycle phase and ectopic cell division and overproliferation; the triple repressor mutants also showed significantly increased root meristem size, suggesting a delayed exit from cell proliferation (2). Kobayashi et al. (2) proposed that the activator MYB3Rs and repressor MYB3Rs may work in different tissues and cell cycle phases, and they act in coordination rather than in competition. It is possible that MYB3R1 is critical to this coordination with its unique role both as an activator and a repressor in Arabidopsis.
TSO1 and MYB3R1 Form a Unique Partnership Regulating both the Shoot and Root.
In this study, 32 of the 45 suppressors of tso1-1 cause loss-of-function mutations in the MYB3R1 gene and none maps to the other four MYB3Rs, suggesting that MYB3R1, but not MYB3R2, -3, -4, and -5, is specifically regulated by TSO1. The ectopic and overexpression of pMYB3R1::GUS in the tso1-1 mutant SAM and RAM suggests that wild-type TSO1 is required to repress MYB3R1 expression in the shoot and root. Since myb3r1 loss-of-function mutations can suppress tso1-1 phenotype in the SAM and RAM, the ectopic MYB3R1 expression in tso1-1 is necessary to mediate all aspects of the tso1-1 mutant phenotypes. Our study functionally links TSO1 to MYB3R1 in plants and uncovers this critical TSO1-MYB3R1 module for proper shoot and root development.
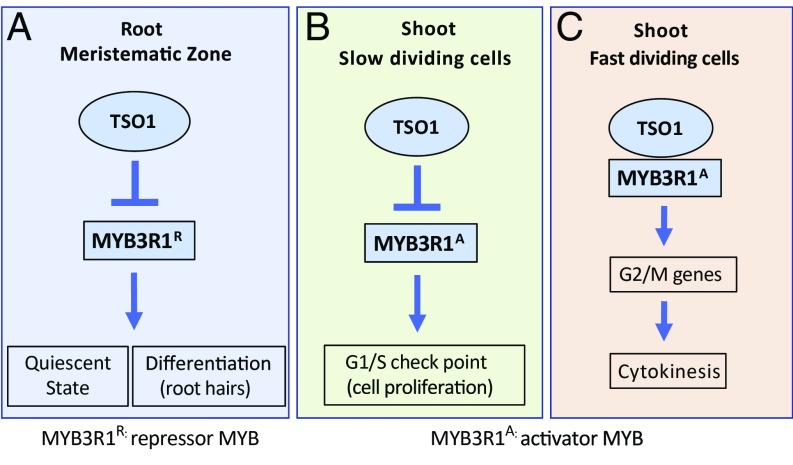
Fig. 8 is a model that proposes how the TSO1-MYB3R1 regulatory module controls the cell cycle in the RAM and SAM. Among the five MYB3Rs, TSO1 likely represses MYB3R1 only; the other four MYB3Rs are unlikely to be a target of TSO1 or are minimally regulated by TSO1, as none of tso1-1 suppressors maps to the other MYB3Rs. The effect of MYB3R1 as an activator (MYB3R1A) in the shoot and a repressor (MYB3R1R) in the root is also proposed. In the wild type, TSO1 represses the transcription of MYB3R1R so that cells can continuously proliferate in the root MZ (Fig. 8A). In tso1-1 mutants, the ectopic expression of MYB3R1 in the root MZ causes permanent cell cycle exit and premature differentiation, indicated by the root hair formation. However, in the SAM (Fig. 8B), TSO1 represses the MYB3R1A to prevent stem cells from entering the cell cycle. The repression of MYB3R1A by TSO1 mirrors the conserved function of the DREAM complex in repressing the B-MYB and other G1/S cell cycle genes in mammals (53, 54). In rapidly dividing cells such as those in floral primordia (Fig. 8C), TSO1 associates with the MYB3R1A to activate the G2/M cell cycle genes, just as their mammalian counterparts (LIN54 and B-MYB) do. This cooperation between TSO1 and the activator MYB3R1A provides an explanation for the incomplete cytokinesis phenotype found in the tso1-1 SAM and myb3r1/4 leaf stomata (17, 31) and is further supported by the co-IP data (Fig. 7).
Fig. 8.
A model of how the TSO1-MYB3R1 module balances cell proliferation with differentiation. TSO1 represses MYB3R1 expression in both the root and the shoot. Arrows and bars indicate positive and negative regulations, respectively, by which they can be either direct or indirect. (A) In the root MZ, TSO1 represses the expression of the repressor MYB3R1R so that cells will not enter differentiation. In tso1-1 mutants, ectopic expression of MYB3R1R in the root MZ causes cells to exit cell cycle and enter differentiation prematurely. (B) In the stem cells at the shoot apex, TSO1 represses the activator MYB3R1A to maintain a small pool of stem cells blocked at the G1/S checkpoint. (C) In rapidly proliferating floral primordia, TSO1 associates with the activator MYB3R1A to activate G2/M cell cycle genes, allowing smooth cell cycle progression and the completion of cytokinesis.
Our model provides a compelling explanation of why opposite effects were observed in the root and shoot of the tso1-1 mutant, which had the ectopic expression and overexpression of MYB3R1 in both the SAM and RAM. The overexpressed MYB3R1 in the SAM is an activator MYB, while the overexpressed MYB3R1 in the RAM acts as a repressor MYB. The suppression of the tso1-1 phenotype suggests that the devastating effect of misregulated MYB3R1 can be reversed by removing the MYB3R1 through CRISPR. As the overexpression of B-MYB correlated with poor prognosis in numerous cancers (48, 49), removing the overexpressed B-MYB in cancer cells by RNAi or by CRISPR could reverse cancer progression, pointing to the B-MYB as a promising cancer treatment target. Furthermore, the genetic mosaic plant (Fig. 2B) reveals the cell-autonomous nature of the misexpressed MYB3R1 and informs about the somatic suppression vs. germline suppression, providing unprecedented insights into how the cell cycle abnormality is manifested in the context of a whole organism. Our work demonstrates the existence of a highly conserved TSO1/LIN54 to MYB3R1/B-MYB regulatory module in plants and animals, which plays a critical role in coordinating the cell cycle with the cell fate commitment.
Materials and Methods
Plant Materials and Growth Conditions.
Plants were grown on Metromix soil (Griffin) under a 16-h light/8-h dark cycle at 20 °C. All mutants are in the Landsberg erecta (Ler) background and were described previously: tso1-1 (17), tso1-1+/+sup-5 (28), and tso1-3 (29). The tso1-3+/+sup-5 heterozygous line was created by pollinating the tso1-3 stigma with the sup-5 (55) pollen. Genotyping derived cleaved amplified polymorphic sequence (dCAPs) markers for the tso1 mutant alleles were described previously (28).
Vector Construction and Plant Transformation.
All transformations were conducted using floral dip and Agrobacterium strain GV3101. Because of the tso1-1 sterility, all constructs were transformed into tso1-1+/+sup-5 or tso1-3+/+sup-5 heterozygous plants. To construct 35S::TSO1-GR, full-length TSO1 cDNA was amplified from floral RNA with primers TSO1-GR.F and TSO1-GR.R (SI Appendix, Table S3) containing BamHI sites. The PCR fragment was inserted into pBI121 (Clontech)-based pBI-ΔGRBX vector (56) at the BamHI site. One transgenic line homozygous for the 35S::TSO1-GR transgene and tso1-1 was selected for mutagenesis. Starting at the stage of bolting, DEX was sprayed daily to plant shoots for 10 d at 50-μM concentration (in water containing 0.015% Silwet l-77).
To generate gTSO1-GFP and translational fusion lines, a 4.1-kb TSO1 genomic sequence containing a 1-kb promoter and full-length coding region (without the stop codon) was PCR amplified using Phusion (NEB) with primers TSO1.geno.F and TSO1.geno.R (SI Appendix, Table S3), and cloned into pCR8/GW/TOPO. After sequence confirmation, the genomic fragment was recombined into pMDC107 (57) to create the gTSO1-GFP. T2 plants homozygous for tso1-1 and the transgene were identified through PCR genotyping. Multiple transgenic lines showed the rescue of the mutant phenotype.
To generate gTSO1-NLS-3xeGFP with a stronger GFP signal, the 4.1-kb genomic fragment of TSO1 was PCR amplified with primers pTSO1.F and TSO10.3xGFP.n.R (SI Appendix, Table S3) to correct the reading frame and then recombined into pGreenII-NLS-3xEGFP (58).
The pMYB3R1::GUS reporter construct was generously provided by M. Ito, Nagoya University, Chikusa, Nagoya, Japan (31). Because of antibiotic selection incompatibility, the promoters from the above vector were cloned into pMDC162 (57). The promoter was first PCR amplified from the above vector using Q5 (NEB) and cloned into pCR8/GW/TOPO, which was then LR recombined into the pMDC162 vector. Five T1 transgenic lines were characterized. T2 progeny from each of these five T1 plants were stained with X-Gluc based on a published protocol (59). The root tips were then incubated with clearing solution (8 g chloral hydrate: 1 mL 100% glycerol: 1 mL water) for 1 h and mounted on slides in clearing solution. To generate fluorescent transcriptional reporter, the MYB3R1 promoter in the pCR8/GW/TOPO entry vector was recombined into pGreenII-NLS-3xEGFP (58).
To generate translational reporters for MYB3R1, a 6.4-kb genomic fragment containing the promoter and the gene body was PCR amplified using Q5 (NEB) with primers pMYB3R1.F and MYB3R1.non.stop.R (SI Appendix, Table S3) from Ler wild-type DNA and cloned into pCR8/GW/TOPO. After sequence confirmation, the genomic fragment of MYB3R1 was recombined into pMDC107 (57) to create gMYB3R1-GFP. To generate a fluorescent translational reporter line with a stronger GFP signal, the 6.4-kb genomic fragment of MYB3R1 was PCR amplified with primers pMYB3R1.F and MYB3R10.3xGFP.n.R (SI Appendix, Table S3) to correct the reading frame and recombined into pGreenII-NLS-3xEGFP (58).
To generate phosphomimic gMYB3R1-GFP constructs, the pCR8/GW/TOPO vector harboring the 6.4-kb MYB3R1 genomic fragment was modified using the Q5 Site-Directed Mutagenesis KIT (NEB) following the manufacturer’s instruction. Briefly, primers S656D.F/S656D.R (SI Appendix, Table S3) were used to amplify the MYB3R1 6.4-kb gDNA TOPO construct. The changes in the coding sequence resulted in the change of 656 serine to aspartic acid (S656D). Similarly, primers S709D.F/S709D.R (SI Appendix, Table S3) were used to generate the S709D. The modified MYB3R1 gDNA pCR8/GW/TOPO constructs were recombined into pMDC107 (57) to generate gMYB3R1 (S656D) and gMYB3R1 (S709D), respectively.
To generate constructs for co-IP assay, the TSO1 cDNA was cloned into PHB-FLAG-X vector (60) to generate 35S::Flag-TSO1. The 35S::MYB3R1(cDNA) construct was generously provided by M. Ito (30). The cDNA was PCR amplified using MYB3R1.F and MYB3R1.R (SI Appendix, Table S3) and then subcloned into pEarleyGate104 (61) to generate 35S::YFP-MYB3R1. The cDNA was also recombined into pEarleyGate100 (61) to generate 35S::MYB3R1.
Suppressor Screen and Complementation Tests.
The genetic screen scheme is shown in SI Appendix, Fig. S1. 35S::TSO1-GR; tso1-1 homozygous seeds were treated with 0.2% EMS solution for 12 h. After repeatedly washing off the mutagen with water, the seeds were germinated in soil and M1 plant fertility was restored by DEX spray. For the first 2,000 M1 plants, seeds were collected from individual M1 plants. For the next 1,000 M1 plants, seeds were pooled into one pool. M2 plants were screened for the ability to form elongated siliques. Forty-five suppressors were identified from screening the progeny of ∼3,000 M1 plants. These M2 suppressor plants were backcrossed to the parental line 35S::TSO1-GR; tso1-1 to generate the mapping population. For each suppressor, the F1 progeny at bolting was supplied with DEX to allow for seed production. The resulting F2 progeny showed suppressed (∼25%) and unsuppressed (∼75%) phenotype; these two populations served as the mapping populations (SI Appendix, Fig. S1).
For complementation test, M2 suppressor lines were crossed with one another. Since the suppressors are recessive, F1 progeny should show unsuppressed phenotype unless the suppressor mutations are allelic to each other, which would show suppressed phenotype in F1.
Mapping by Sequencing.
For each suppressor F2 mapping population, leaf tissues were collected and pooled from 35–50 plants, and genomic DNA was extracted from the pooled leaf tissue using the NucleoSpin Plant II Midi Kit (Macherey-Nagel). Each DNA pool was sequenced at 15–20× coverage with 51-bp single end reads (SI Appendix, Table S4). Reads were aligned to the TAIR10 Arabidopsis genome with Bowtie2 (62) and variants were called with SAMtools (63). Six independent suppressors (A144, A156, A176, A317, B636, and B763) were mapped and sequenced. A 35S::TSO1-GR; tso1-1 parental line was also sequenced at about 8× coverage (SI Appendix, Table S4). Variants shared among all six samples (556,864 total variants) are likely carried over from prior EMS mutagenesis that generated tso1-1 in the Ler background. They were removed from further analysis. SNPs unique to each suppressor pool were identified using the genotype calling feature of SAMtools (64). G-to-A or C-to-T changes were further selected as candidate SNPs as they are most likely caused by EMS. The enrichment of candidate SNPs in 100,000-bp window was plotted using R (SI Appendix, Fig. S3). Annotation of variants was done using the VariantAnnotation package (65) from Bioconductor to identify nonsynonymous SNPs within exons as well as SNPs affecting splicing.
CRISPR/Cas9 Gene Editing and Genotyping of the Resulting Mutants.
gRNA was designed using crispr.dbcls.jp/. The 20-bp seed RNA targets the fourth exon of MYB3R1 coding for a region between R1 and R2 of MYB3R1. Blast search against the Arabidopsis genome did not identify any other target homology. The target RNA sequence (ACGGTCCTTTCACAAGCTCT) was inserted between the AtU6 promoter and scaffold followed by the AtU6 terminator using overlapping PCR (SI Appendix, Table S3) with pCAMBIA-Cas9+gRNA (66) as the template. The above PCR fragment was cloned into pCR8/GW/TOPO, excised by SalI and KpnI, and ligated into SalI and KpnI sites in pCAMBIA-Cas9+sgRNA (66). T1 seeds were screened on 1/2 MS medium containing hygromycin. T2 plants showing chimeric wild-type branches in tso1-1 homozygous plants were further analyzed. DNA was extracted from the individual suppressed branches, and PCR primers (MYB3R1.CRISPR.con.F/R; SI Appendix, Table S3) were used to amplify DNA fragments spanning the gRNA target site within MYB3R1, which were then sequenced. For tso1-3+/+sup-5 plants transformed with the CRISPR constructs, DNA was extracted from six T2 plants showing slightly improved fertility and sequenced to reveal heterozygous mutations in MYB3R1. T3 progeny of these six T2 plants segregated 25% of completely rescued plants.
To construct CRISPR vector for MYB3R4 gene editing, gRNA was designed to target the second exon. Primers MYB3R4.CRIPSR.F/R (SI Appendix, Table S3) were annealed and ligated into the BsaI cutting site of the pHEE401E vector (67). The construct was introduced into tso1-1+/+sup-5 plants. CRISPR-generated mutations likely destroy a BbvCI restriction site at the gRNA target, allowing rapid screening of CRISPR-induced mutations. PCR primers MYB3R4.seq.pro.F and MYB3R4.fourth.exon.R (SI Appendix, Table S3) amplified the DNA fragment spanning the gRNA target site for sequencing.
Confocal Microscopy and Data Processing.
For RAM imaging, 7-d-old seedlings were immersed in 10 μg/mL PI in water for 2 min. The roots were then mounted on slides in water and observed using the 63× (1.2 HCXPLAPO CS) water objective of a Leica SPX5 confocal microscope. A fluorescent signal, excited by a white light laser at 536 nm, was detected with detection channel set at 550 nm–660 nm. Tile scans were taken and processed using Leica Application Suite 2.0.0 software.
For SAM imaging, inflorescences from 4-wk-old plants were dissected to remove flowers older than stage 5 (68). The inflorescence apexes were stained with 10 μg/mL PI for 3–5 min and mounted in a Nunc Lab-Tek chamber slide. Z stacks of PI (600–650 nm) and GFP (500–545 nm) channels were acquired with 63× (1.2 HCXPLAPO CS) water objective of a Leica SPX5 inverted confocal microscope. Scanning speed was set at 400 Hz to achieve 0.48 × 0.48 × 0.46 µm voxel size. Fiji (fiji.sc/) was used to extract .tiff files. The 3D segmentation of cells and flourescent signal heatmap were generated following instructions from LithoGraphX website section 4.3, Segmenting Cells in 3D (lithographx.org) (69).
Fertility Quantification and qRT-PCR Analysis of Transgenic Plants.
For fertility quantification (SI Appendix, Fig. S6B), siliques in the 5th–15th positions from the base were collected. Seeds per silique were quantified. The main shoots of 5–10 plants per transgenic line were quantified. For qRT-PCR (SI Appendix, Fig. S6C), inflorescence containing unopened flowers was collected for RNA extraction using TRIzol reagent (Ambion no. 66316). A total of 1 μg RNA from each sample was treated with RNase-Free DNase I (Thermo Scientific, no. EN0521) followed by first strand cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, no. K1621). Real-time quantitative PCR was performed using SsoFast EvaGreen supermix (Bio-Rad). Since the gMYB3R1 transgene is tagged by GFP, a reverse PCR primer against GFP (SI Appendix, Table S3) was used to distinguish the transgene from the endogenous MYB3R1 gene. Three biological replicates, each with three technical replicates, were performed for each transgenic line.
Coimmunoprecipitation Assays.
The Agrobacterium harboring 35S::Flag-TSO1 and 35S::YFP-MYB3R1 were used to cotransfect tobacco leaves by infiltration. Samples were collected after 48 h and ground in liquid nitrogen and homogenized in the lysis buffer (50 mM Tris⋅HCl pH7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% Triton-X-100, 1 mM Pefabloc, mixture, 50 μM MG132). After centrifugation, supernatant was incubated with 1 μL anti-Flag (Sigma) antibody [or with 5 μL anti-GFP (cat. no. A01388-100, rabbit; GenScript)] bound to 10 μL protein G (Invitrogen) beads for 2 h at 4 °C. Then the beads were washed three times with 1 mL of the lysis buffer and eluted by boiling with 20 μL 2× SDS sample buffer for 5 min and then separated on 10% SDS/PAGE gel. Anti-Flag (Sigma; 1:5,000) and anti-GFP (TransGen Biotech; 1:2,000) antibodies were used to detect Flag-TSO1 and YFP-MYB3R1, respectively.
Supplementary Material
Acknowledgments
We thank Drs. Masaki Ito, Qi-Jun Chen, and Donald Weeks for providing MYB3R1::GUS, 35S::MYB3R1, and CRISPR constructs; the undergraduate students who contributed to this project; Michael Wellen and Rao Fu for helping with seed collection at the early stage of the genetic screen; Dae Ik Yi, Xixi Zhou, and Song Yun for making constructs and assisting in transformation; Dr. Chunying Kang for comments on the manuscript; and Amy Beaven and the Department of Cell Biology and Molecular Genetics Imaging Core for assistance with confocal fluorescence microscopy. The work was supported by National Science Foundation Grant MCB0951460 (to Z.L.), and China Scholarship Council Postgraduate Scholarship (to W.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715903115/-/DCSupplemental.
References
- 1.Inzé D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi K, et al. Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 2015;34:1992–2007. doi: 10.15252/embj.201490899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadasivam S, DeCaprio JA. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13:585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischer M, DeCaprio JA. Does Arabidopsis thaliana DREAM of cell cycle control? EMBO J. 2015;34:1987–1989. doi: 10.15252/embj.201592196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 6.Litovchick L, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer A, Wenzl C, Lohmann JU. Beyond flexibility: Controlling stem cells in an ever changing environment. Curr Opin Plant Biol. 2017;35:117–123. doi: 10.1016/j.pbi.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annu Rev Plant Biol. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 10.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 11.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 12.Jacks T, et al. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 13.Ebel C, Mariconti L, Gruissem W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature. 2004;429:776–780. doi: 10.1038/nature02637. [DOI] [PubMed] [Google Scholar]
- 14.Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W. A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol. 2008;18:1680–1686. doi: 10.1016/j.cub.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 15.Johnston AJ, Gruissem W. Gametophyte differentiation and imprinting control in plants: Crosstalk between RBR and chromatin. Commun Integr Biol. 2009;2:144–146. doi: 10.4161/cib.8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borghi L, et al. Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell. 2010;22:1792–1811. doi: 10.1105/tpc.110.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Running MP, Meyerowitz EM. TSO1 functions in cell division during Arabidopsis flower development. Development. 1997;124:665–672. doi: 10.1242/dev.124.3.665. [DOI] [PubMed] [Google Scholar]
- 18.Song JY, Leung T, Ehler LK, Wang C, Liu Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development. 2000;127:2207–2217. doi: 10.1242/dev.127.10.2207. [DOI] [PubMed] [Google Scholar]
- 19.Hauser BA, He JQ, Park SO, Gasser CS. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development. 2000;127:2219–2226. doi: 10.1242/dev.127.10.2219. [DOI] [PubMed] [Google Scholar]
- 20.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 21.Beall EL, et al. Discovery of tMAC: A Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 2007;21:904–919. doi: 10.1101/gad.1516607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang J, Benson E, Bausek N, Doggett K, White-Cooper H. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with always early. Development. 2007;134:1549–1559. doi: 10.1242/dev.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmit F, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–1913. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 25.Korenjak M, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 26.Lewis PW, et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen SU, et al. The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J Exp Bot. 2007;58:3657–3670. doi: 10.1093/jxb/erm215. [DOI] [PubMed] [Google Scholar]
- 28.Sijacic P, Wang W, Liu Z. Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 2011;7:e1002352. doi: 10.1371/journal.pgen.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauser BA, Villanueva JM, Gasser CS. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics. 1998;150:411–423. doi: 10.1093/genetics/150.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito M. Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res. 2005;118:61–69. doi: 10.1007/s10265-005-0192-8. [DOI] [PubMed] [Google Scholar]
- 31.Haga N, et al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134:1101–1110. doi: 10.1242/dev.02801. [DOI] [PubMed] [Google Scholar]
- 32.Dubos C, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Haga N, et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Liu Z. Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell. 2006;18:350–365. doi: 10.1105/tpc.105.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dello Ioio R, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 36.Perilli S, Sabatini S. Analysis of root meristem size development. Methods Mol Biol. 2010;655:177–187. doi: 10.1007/978-1-60761-765-5_12. [DOI] [PubMed] [Google Scholar]
- 37.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 38.Sala A, et al. Activation of human B-MYB by cyclins. Proc Natl Acad Sci USA. 1997;94:532–536. doi: 10.1073/pnas.94.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson TK, Schweppe RE, Septer J, Lewis RE. Phosphorylation of B-Myb regulates its transactivation potential and DNA binding. J Biol Chem. 1999;274:36741–36749. doi: 10.1074/jbc.274.51.36741. [DOI] [PubMed] [Google Scholar]
- 40.Heazlewood JL, et al. PhosPhAt: A database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res. 2008;36:D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durek P, et al. PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umezawa T, et al. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal. 2013;6:rs8. doi: 10.1126/scisignal.2003509. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, et al. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J Proteomics. 2013;78:486–498. doi: 10.1016/j.jprot.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 44.Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–5716. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 45.Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 46.Matsuo T, Kuramoto H, Kumazaki T, Mitsui Y, Takahashi T. LIN54 harboring a mutation in CHC domain is localized to the cytoplasm and inhibits cell cycle progression. Cell Cycle. 2012;11:3227–3236. doi: 10.4161/cc.21569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beall EL, Bell M, Georlette D, Botchan MR. Dm-myb mutant lethality in Drosophila is dependent upon mip130: Positive and negative regulation of DNA replication. Genes Dev. 2004;18:1667–1680. doi: 10.1101/gad.1206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Musa J, Aynaud M-M, Mirabeau O, Delattre O, Grünewald TG. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8:e2895. doi: 10.1038/cddis.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amatschek S, et al. Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res. 2004;64:844–856. doi: 10.1158/0008-5472.can-03-2361. [DOI] [PubMed] [Google Scholar]
- 50.Heine GF, Hernandez JM, Grotewold E. Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J Biol Chem. 2004;279:37878–37885. doi: 10.1074/jbc.M405166200. [DOI] [PubMed] [Google Scholar]
- 51.Grässer FA, LaMontagne K, Whittaker L, Stohr S, Lipsick JS. A highly conserved cysteine in the v-Myb DNA-binding domain is essential for transformation and transcriptional trans-activation. Oncogene. 1992;7:1005–1009. [PubMed] [Google Scholar]
- 52.Guehmann S, Vorbrueggen G, Kalkbrenner F, Moelling K. Reduction of a conserved Cys is essential for Myb DNA-binding. Nucleic Acids Res. 1992;20:2279–2286. doi: 10.1093/nar/20.9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res. 2016;44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer M, Müller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol. 2017;52:638–662. doi: 10.1080/10409238.2017.1360836. [DOI] [PubMed] [Google Scholar]
- 55.Gaiser JC, Robinson-Beers K, Gasser CS. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell. 1995;7:333–345. doi: 10.1105/tpc.7.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- 57.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takada S, Jürgens G. Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development. 2007;134:1141–1150. doi: 10.1242/dev.02803. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi N, et al. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Curr Biol. 2013;23:1812–1817. doi: 10.1016/j.cub.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 60.Luo Q, et al. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell. 2014;26:2441–2456. doi: 10.1105/tpc.113.121657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Obenchain V, et al. VariantAnnotation: A bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014;30:2076–2078. doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang W, Yang B, Weeks DP. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One. 2014;9:e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z-P, et al. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barbier de Reuille P, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.