Significance
The gene pool of modern Europeans was shaped through prehistoric migrations that reached the Western Mediterranean last. Obtaining biomolecular data has been challenging due to poor preservation related to adverse climatic conditions in this region. Here, we study the impact of prehistoric (Neolithic–Bronze Age) migrations in Iberia by analyzing genomic and dietary data, demonstrating that farming practices were introduced by a population genetically distinct from the first farmers in central and northern Europe. After recovering from a founder bottleneck, these first farmers mixed with local hunter-gatherers. Finally, post-Neolithic migrations had a much smaller impact on the Iberian gene pool than they had in other parts of Europe. Stable isotope analysis reveals a homogenous terrestrial diet throughout this period.
Keywords: archaeogenomics, Iberia, migrations, diversity, palaeodiet
Abstract
Population genomic studies of ancient human remains have shown how modern-day European population structure has been shaped by a number of prehistoric migrations. The Neolithization of Europe has been associated with large-scale migrations from Anatolia, which was followed by migrations of herders from the Pontic steppe at the onset of the Bronze Age. Southwestern Europe was one of the last parts of the continent reached by these migrations, and modern-day populations from this region show intriguing similarities to the initial Neolithic migrants. Partly due to climatic conditions that are unfavorable for DNA preservation, regional studies on the Mediterranean remain challenging. Here, we present genome-wide sequence data from 13 individuals combined with stable isotope analysis from the north and south of Iberia covering a four-millennial temporal transect (7,500–3,500 BP). Early Iberian farmers and Early Central European farmers exhibit significant genetic differences, suggesting two independent fronts of the Neolithic expansion. The first Neolithic migrants that arrived in Iberia had low levels of genetic diversity, potentially reflecting a small number of individuals; this diversity gradually increased over time from mixing with local hunter-gatherers and potential population expansion. The impact of post-Neolithic migrations on Iberia was much smaller than for the rest of the continent, showing little external influence from the Neolithic to the Bronze Age. Paleodietary reconstruction shows that these populations have a remarkable degree of dietary homogeneity across space and time, suggesting a strong reliance on terrestrial food resources despite changing culture and genetic make-up.
The Mediterranean region has unquestionably played a central role in human history, partly deriving from the navigable nature of the sea that connects southern Europe, western Asia, and North Africa. This unique setting has led to its being one of the most important and dynamic areas throughout prehistory.
The period of the Mesolithic to the Bronze Age (in western Eurasia) covers two major cultural shifts that are arguably among the most important transitions in human prehistory, heralding the change from hunter-gatherer subsistence to food production and later the emergence of metallurgy, changes that fundamentally transformed human culture. Recent large-scale studies of ancient human genomic variation (e.g., refs. 1–8) have focused mainly on central and northern Europe and have revealed that changes during the Neolithic and later during the Bronze Age were driven by population movements into Europe from the southeast and east, first by early farmers from Anatolia and the Levant (1–3, 9, 10) and second by herders from the Pontic-Caspian steppe (4, 6). These migrations profoundly reshaped the genetic and cultural landscape of Europe. However, studying the genetic impacts of these cultural transitions in southernmost Europe, especially the Mediterranean, has usually focused on single time periods (10–16).
The full Neolithic package reached the Iberian Peninsula and northern (modern-day) Morocco ca. 7,500 Cal BP, with the Cardial pottery culture coming from the central Mediterranean (17). This was rapidly followed by a regional diversification of ceramics and lithics with the Cardial pottery type present in most of the Mediterranean fringe and the interior of the Iberian Peninsula represented by the Boquique type (e.g., El Portalón de Cueva Mayor) (18) potentially introduced through the north via the Pyrenees (19). In southern Iberia (Andalusia), however, the early Neolithic is characterized by a type of impressed non-Cardial ceramic decorated a la Almagra (20). This type of pottery culture reached central Andalusia by 7,300 Cal BP, soon replacing the Cardial pottery, and is found at the Murciélagos de Zuheros cave (21). It has been proposed that North Africa played a significant part in the origins of the Neolithic in southern Spain (22), although this has recently been challenged (23). The prehistory of the Iberian Peninsula as a whole is of particular interest, given its specific geographic location at the westernmost edge of the continent, naturally making it the furthest point from the documented prehistoric migrations originating from eastern Eurasia. This location holds potential for a complex demographic history with migrations from diverse sources, as it is connected with mainland Europe in the north, is surrounded by two potential maritime migration routes along the Mediterranean Sea and the Atlantic Ocean, and furthermore is in close geographic proximity to North Africa. Previous studies on early Iberian farmers have shown that these populations represent the descendants of migrants from Anatolia (6, 13) followed by admixture with local hunter-gatherers (5, 6, 9–11, 13, 24). Furthermore, modern-day southwestern Europeans are genetically closer to Early and Middle Neolithic Europeans than are modern-day central Europeans, who are more closely related to Late Neolithic and Bronze Age populations (1, 2, 4, 6, 11, 25–28), suggesting diverse and regionally distinctive demographic histories.
To investigate the demographic history of the westernmost edge of the prehistoric Eurasian migrations, we have sequenced the genomes of 13 individuals excavated from six prehistoric Iberian sites in the north and south of modern-day Spain (SI Appendix, Section S1 and Table S2.1). These sites cover the Neolithic, Late Neolithic/Copper Age (LNCA), and Bronze Age chronologies between 7,245 and 3,500 Cal BP (SI Appendix, Fig. S3.2 and Table S3.2), including the oldest sequenced genome in southern Iberia, from the Murciélagos de Zuheros cave. This individual is directly dated to 7,245–7,024 y Cal BP and represents the first genome of an individual from the Neolithic Almagra Pottery Culture, the early agriculturalists of the south of the Iberian Peninsula. For the El Portalón cave, we generated additional DNA sequence data for published individuals (11) as well as sequencing five additional individuals, enabling the genomic analysis of a population that spans a temporal sequence comprising the Neolithic, Copper Age, and Bronze Age periods (directly dated to between 7,165 and 3,500 y Cal BP). These genomic data, combined with published data (6, 9, 11, 13, 24), allow us to comprehensively study demographic changes through time in the Iberian Peninsula in general and in one single location in particular. We contrast these developments with contemporary populations in other parts of Europe where two major population turnovers took place during this time span (1, 4, 6, 25). Our genomic analysis was combined with stable isotope analysis to investigate the role of aquatic resources in the diets of the individuals tested here. Previous genomic studies have revealed increasing amounts of genetic hunter-gatherer admixture in the farmer population after the initial arrival of Neolithic people in Europe (1–3, 11, 13), implying the continued survival of hunter-gatherer populations or at least their lineages. In contrast, paleodietary stable isotope studies of the Iberian late Neolithic–Early Bronze Age have indicated that aquatic resources are not abundant in the diet, despite their likely availability at many sites (29–35). Detecting aquatic resources can be difficult using only bulk isotope analysis, and it has been posited that freshwater and terrestrial C3 diets can be more easily distinguished using stable carbon isotope values of amino acids (36, 37). Ten of the sequenced individuals were investigated for amino acid stable carbon isotopes of bone collagen to reconstruct paleodietary preferences (38, 39), in particular the presence of aquatic food intake (SI Appendix, Section S3) (36, 40).
Results and Discussion
We sequenced the genomes of 10 individuals from northern and southern Spain either contextually or directly radiocarbon dated to the Neolithic, LNCA, and Bronze Age (Fig. 1A and SI Appendix, Table S4.1), and we increased the sequencing depth of three individuals from a previous study (ATP16, ATP2, and ATP12) (11) using additional bone material (SI Appendix, Table S2.1). Altogether, our 13 sequenced genomes range from 0.01× to 12.9× (SI Appendix, Table S4.1) with six individuals having >2.0× genome coverage. Our sequence data show postmortem damage and fragmentation, as expected from endogenous ancient DNA (aDNA) molecules (41). Eleven individuals were genetically inferred to be males and two to be females. We obtained contamination estimates based on the X chromosome in male samples (42), which were all ≤5% or lower (SI Appendix, Table S4.1). Mitochondrial contamination estimates based on reads mapping to the mitochondrial genome (43) suggest >5% mitochondrial contamination for two samples (POR003 and ATP019) (SI Appendix, Table S4.1), and the sequence data from these individuals were subsequently filtered to retain fragments displaying postmortem damages to conservatively restrict the analysis to these sequences (44).
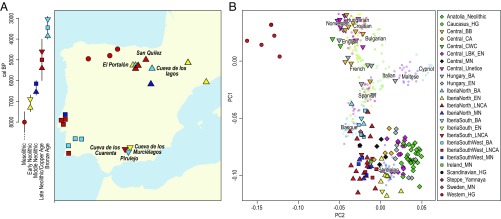
Fig. 1.
(A) Sampling locations of individuals included in this study. Sites with newly generated sequencing data (SI Appendix, Table S4.1) are labeled. (B) Enlarged section of the PCA plot (Dataset S2) showing the part of the PC1–PC2 space occupied by the ancient Iberians as well as other ancient groups. BA, Bronze Age; BB, Bell Beaker; CA, Copper Age; CWC, Corded Ware Culture; EN, Early Neolithic; HG, hunter-gatherer; LBK_EN, Linearbandkeramik_Early Neolithic; LNCA, Late Neolithic/Copper Age; MN, Middle Neolithic.
Nine of the 13 ancient Iberian individuals were found to carry mitochondrial haplogroups associated with European early farmers, namely K, J, N, and X (SI Appendix, Table S4.1), distributed throughout the Early Neolithic to the Bronze Age (6, 45). Two individuals have haplogroups HV0 and H, known in both European early farmers and hunter-gatherers (25, 45) and are present during the LNCA. Further, haplogroup U5, characteristic of hunter-gatherers (46, 47), is found in a Late Copper Age individual. Consistent with the mitochondrial haplogroup composition of the ancient Iberians, the Y chromosome composition (Dataset S1) displays a mix of haplogroups associated with both European farmers and hunter-gatherers. Among the Early Neolithic individuals, we find the European farmer-associated haplogroup G2a2 (9) and the European farmer-associated haplogroup H2 (1, 6), while in the LNCA we observe haplogroup I2, previously found in both hunter-gatherers and farmers (SI Appendix, Table S4.1) (1, 6). Both Bronze Age males carried haplogroup R1b-M269 (SI Appendix, Table S4.1), which is frequent among Late Neolithic and Bronze Age samples from other parts of Europe (4, 6). This uniparental marker composition is in agreement with the well-known admixture between resident hunter-gatherers and incoming farmers.
To obtain an overview of the genetic variation within prehistoric Iberia, we performed principal component analysis (PCA) on a reference panel of 26 modern-day populations from western Eurasia (1) on which we projected our 13 ancient individuals, together with relevant Mesolithic (n = 17), Neolithic/Chalcolithic (n = 98), and Late Neolithic/Bronze Age (n = 78) genomes from Europe and Anatolia (Fig. 1B and Dataset S2). This PCA replicates previous findings: (i) a clear genetic distinction between early farmers and resident hunter-gatherers, (ii) affinity of the former with the southwestern modern-day European variation, and (iii) an increased affinity of prehistoric farmers to western hunter-gatherers over time due to increased admixture between the two populations (1–3, 11, 25, 26). Despite the geographic proximity of southern Iberia to northern Africa, we do not see substantial affinities of any individual to modern-day African populations, but the lack of ancient North African genomes limits our abilities to test these connections (SI Appendix, Fig. S5.1).
Zooming into the genetic variation within prehistoric Iberia, we do not find a geographic stratification pattern between North and South Iberian populations; instead, we observe stratification directly associated with chronology. Three clusters are identified among the Iberian farmers on the PCA. The first cluster comprises the early Neolithic Iberians (Fig. 1B, yellow triangles) that falls within the modern-day Sardinian genomic variation showing the highest affinity to Sardinians among all early European farmers. The second group includes the Middle Neolithic (Fig. 1B, blue triangles and squares) and LNCA (Fig. 1B, red triangles and squares) populations falling within the modern-day southern European variation but differentiating from the early Neolithic Iberians, a pattern which can be explained by the subsequent admixture with local hunter-gatherers (6, 11, 24). These Iberian farmers were likely the group that later developed Bell Beaker pottery (widely found in Western Europe during the third millennium BC), which then spread without major migrations toward central and northwestern Europe (48). The third cluster encompasses the Bronze Age individuals (Fig. 1B, turquoise triangles and squares) and is differentiated from the other two groups, showing the highest affinity to modern-day Iberians. There is a strong genetic difference among Iberian populations dating to the Mesolithic, early and middle Neolithic, LNCA, and Bronze Age; this genetic pattern is likely the result of numerous migrations and admixture events over time.
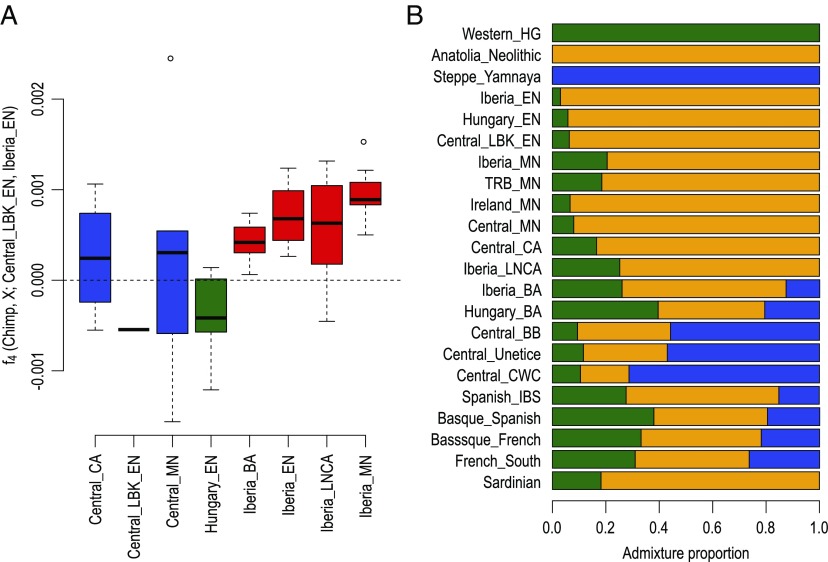
A striking feature of the PCA (Fig. 1B) is the genetic difference between Iberian and central European farmers (orange, purple, gray, and black diamonds in Fig. 1B). This division could represent slightly different gene pools of the migrating populations along the two different routes for early Neolithic farmers: one following the Danube river into central Europe and a second along the Mediterranean coast, which has been suggested based on the analysis of smaller datasets (5, 12, 24). To formally test this separation, we calculated f4 statistics to investigate if Iberian farmers form a clade to the exclusion of central European farmers. The statistic of the form f4(Chimp, X; Central_LBK_EN and Iberia_EN) measures whether an individual X shares more genetic drift with early Neolithic central Europeans (if the value is negative) or early Neolithic Iberians (if the value is positive). For this analysis, we only used SNP-captured individuals for the reference populations (Central LBK_EN and Iberia_EN) to avoid spurious affinities between references and X due to technological artifacts (49, 50). In contrast to prehistoric populations from central Europe, this statistic is consistently shifted toward positive values for prehistoric Iberian groups from different chronologies (Fig. 2A) and is qualitatively similar when using other individuals as reference (Dataset S4). This pattern does not seem to be driven by hunter-gatherer–related admixture into the farming populations (Dataset S5). This observation suggests that all Neolithic Iberians trace most of their ancestry to the first Neolithic migrants arriving in the peninsula and that later contributions from contemporary central Europeans were only minor. The overall pattern is consistent with two independent Neolithic migrations of genetically slightly different populations that spread farming practices across Europe. The Mediterranean route migrants show a strong connection with modern-day population isolates in southwestern Europe. Modern-day Sardinians have been suggested to be relatively direct descendants of the early Neolithic individuals (27, 51), and modern-day Basques also trace a high proportion of their ancestry to the first Mediterranean farmers with only minor additional admixture since the Neolithic (11).
Fig. 2.
(A) f4 statistics testing affinities of prehistoric European farmers to either early Neolithic Iberians or central Europeans, restricting these reference populations to SNP-captured individuals to avoid technical artifacts driving the affinities. The boxplots in A show the distributions of all individual f4 statistics belonging to the respective groups. The signal is not sensitive to the choice of reference populations and is not driven by hunter-gatherer–related admixture (Datasets S4 and S5). (B) Estimates of ancestry proportions in different prehistoric Europeans as well as modern southwestern Europeans. Individuals from regions of Iberia were grouped together for the analysis in A and B to increase sample sizes per group and reduce noise.
A large migration of Pontic-Caspian steppe herders (the Yamnaya culture) during the Late Neolithic/Early Bronze Age has been found to have a substantial impact on the gene pool of central and northern European populations (4, 6–8), but the impact of this migration on contemporary southern and western Europeans has been unclear. To estimate the genetic contributions of different prehistoric groups (hunter-gatherers, early Anatolian farmers, and Steppe herders) to other ancient populations, we inferred admixture fractions using both unsupervised (ADMIXTURE, ref. 52) (Dataset S3) and supervised approaches (qpAdm and ADMIXTURE, refs. 6 and 52). Neolithic European populations share different proportions of hunter-gatherer and Neolithic farmer genetic material, with a tendency toward more hunter-gatherer–related ancestry in later groups (Fig. 2B) (11). Late Neolithic and Bronze Age north-central Europeans display substantial fractions of Pontic Steppe ancestry (up to 71% estimated with qpAdm; 93% based on supervised ADMIXTURE) (SI Appendix, Fig. S5.3) at the onset of the Bronze Age. However, steppe ancestry in Bronze Age individuals from Iberia (13%; 18%) and Hungary (21%; 38%) is lower than in their north-central European counterparts (Fig. 2B), a pattern previously suggested but not directly quantified (24). The estimates for Bronze Age Iberians are close to the 15% steppe ancestry estimated for the modern Spanish population (Fig. 2B). Consistently, Bronze Age populations from Greece and Anatolia also show a limited increase in steppe ancestry compared with their Neolithic ancestors (15). This reduced impact of Steppe herders on these populations could reflect a decrease in the number of migrants or a dilution of Steppe ancestry during this process. In contrast to the events in north-central Europe, the arrival of most of the Yamnaya-related ancestry in Iberia postdates the onset of Bell Beaker pottery in Iberia, suggesting that the Bell Beaker culture spread culturally (48), while steppe ancestry was brought into Iberia through later migrations. Notably, both male Bronze Age Iberian individuals in this study as well as all three Iberian Bronze Age males in ref. 24 carried R1b-M269 Y chromosomes (SI Appendix, Table S4.1) also found with high frequency in individuals associated with the Yamnaya culture, the source population of steppe ancestry (4, 6), indicating a continuing male-driven migration from central Europe into southwestern Europe (8, 24, 53).
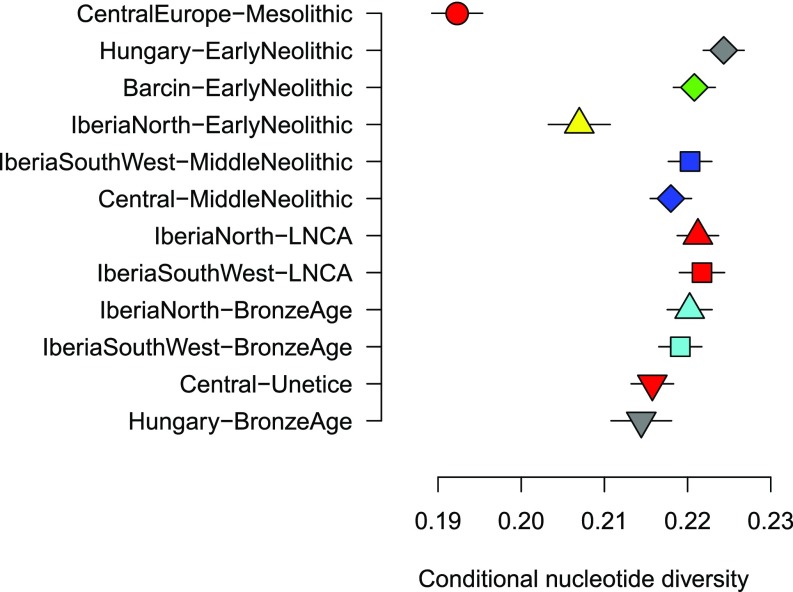
To obtain additional insights into the demographic development of prehistoric populations—their diversity as well as their effective population sizes—we estimated genetic diversity through time in all shotgun-sequenced Iberian, Anatolian, and central European farmers as well as in Mesolithic hunter-gatherer populations (Fig. 3). As previously observed (2), Mesolithic hunter-gatherers had the lowest diversity of the groups tested. Interestingly, the group with lowest genetic diversity among farmers was early Neolithic Iberians. There is a significant increase in diversity from the early Neolithic to the middle Neolithic in Iberia, a pattern not observed in central Europe. The low diversity levels in early Neolithic Iberian farmers could potentially reflect a bottleneck resulting from the initial migration along the Mediterranean coast. The subsequent increase in genetic diversity over time represents a recovery from this bottleneck and is likely due to an increase in population size and admixture with local hunter-gatherers.
Fig. 3.
Conditional nucleotide diversity (2) for different prehistoric populations. Each population is represented by the two shotgun-sequenced individuals with the highest sequencing coverage. Error bars show two SEs estimated using a block jackknife. The estimates for early Neolithic Hungarians are based on the shotgun data published in ref. 25.
Paleodietary analysis (SI Appendix, Section S3) of these same individuals shows that they had a remarkable degree of homogeneity in their diets, temporally and geographically. The analyses indicate that the individuals maintained a C3 terrestrial diet and that neither freshwater nor marine sources of protein significantly contributed to the diet, in agreement with previous studies (e.g., refs. 29 and 30). Even though the Early Neolithic individuals from northern and southern Iberia were found >600 km apart, there is a clear similarity in dietary preferences for terrestrial foods. In addition, although there is evidence of contact between groups that have different subsistence strategies (an increasing hunter-gatherer genetic component through time), dietary preferences remain constant from the Early Neolithic to the beginning of the Bronze Age.
Conclusions
We present a comprehensive biomolecular dataset spanning four millennia of prehistory across the whole Iberian Peninsula. Our results highlight the power of archaeogenomic studies focusing on specific regions and covering a temporal transect. The 4,000 y of prehistory in Iberia were shaped by major chronological changes but with little geographic substructure within the Peninsula. The subtle but clear genetic differences between early Neolithic Iberian farmers and early Neolithic central European farmers point toward two independent migrations, potentially originating from two slightly different source populations. These populations followed different routes, one along the Mediterranean coast, giving rise to early Neolithic Iberian farmers, and one via mainland Europe forming early Neolithic central European farmers. This directly links all Neolithic Iberians with the first migrants that arrived with the initial Mediterranean Neolithic wave of expansion. These Iberians mixed with local hunter-gatherers (but maintained farming/pastoral subsistence strategies, i.e., diet), leading to a recovery from the loss of genetic diversity emerging from the initial migration founder bottleneck. Only after the spread of Bell Beaker pottery did steppe-related ancestry arrive in Iberia, where it had smaller contributions to the population compared with the impact that it had in central Europe. This implies that the two prehistoric migrations causing major population turnovers in central Europe had differential effects at the southwestern edge of their distribution: The Neolithic migrations caused substantial changes in the Iberian gene pool (the introduction of agriculture by farmers) (6, 9, 11, 13, 24), whereas the impact of Bronze Age migrations (Yamnaya) was significantly smaller in Iberia than in north-central Europe (24). The post-Neolithic prehistory of Iberia is generally characterized by interactions between residents rather than by migrations from other parts of Europe, resulting in relative genetic continuity, while most other regions were subject to major genetic turnovers after the Neolithic (4, 6, 7, 9, 25, 48). Although Iberian populations represent the furthest wave of Neolithic expansion in the westernmost Mediterranean, the subsequent populations maintain a surprisingly high genetic legacy of the original pioneer farming migrants from the east compared with their central European counterparts. This counterintuitive result emphasizes the importance of in-depth diachronic studies in all parts of the continent.
Materials and Methods
Archaeological Samples.
Thirteen individuals from northern Spain (El Portalón, San Quílez, and Cueva de los Lagos) and Andalusia in the south (Murciélagos de Zuheros, Cueva de los Cuarenta, and El Pirulejo) were sampled for aDNA analyses. All six sites cover a chronological period from the Early Neolithic to the Bronze Age. Eleven samples were directly radiocarbon dated using accelerator mass spectrometry, and the remaining two were associated with archaeological contexts. See SI Appendix, Section S1.
Sequencing.
DNA was extracted from bones and teeth (54); DNA extracts were converted into blunt-end Illumina libraries (55). All samples were prepared in dedicated aDNA facilities at the Evolutionary Biology Center in Uppsala, Sweden. The libraries were sequenced on Illumina HiSeq platforms 2500 or X Ten at the SNP&SEQ Technology Platform at the Science for Life Laboratory Sequencing Centre in Uppsala. All 13 samples were screened for human DNA and yielded over 1% human DNA content; thus, all were included for downstream analysis. See SI Appendix, Section S2.
Stable Isotopes.
A subsample of the individuals studied for DNA underwent amino acid stable carbon isotope analysis of bone collagen to determine the long-term dietary preferences of these individuals. Bone collagen was isolated from bone samples following a modified Longin method (56, 57), and amino acids were prepared via hydrolysis of collagen. The δ13C values of the amino acid fractions were measured using a Thermo Fisher LC-isotope ratio MS (LC-IRMS) system following methods similar to those described in ref. 38. These data were supplemented by bulk δ13C and δ15N values provided with the radiocarbon dates of the bone collagen. See SI Appendix, Section S3.
Next-Generation Sequencing Data Processing and Authentication.
Overlapping paired-end reads were merged, the remaining adapters were trimmed (58), and the fragments were mapped to the human reference genome using bwa (59). Fragments with identical start and end positions were considered PCR duplicates, and all duplicates were collapsed into consensus sequences. Contamination was estimated based on heterozygous sites on the X chromosome in males (42) and in the mitochondrial genome (43). All samples show indications of characteristic aDNA damage, and for samples with high levels of mitochondrial contamination (>15%), the analysis was restricted to fragments indicating postmortem damage (44). See SI Appendix, Section S4.
Uniparental Haplogroups.
HAPLOFIND (60) was used to infer the most likely haplogroup for mitochondrial consensus sequences. Y chromosomal haplogroups were assigned by investigating up to 732 haplotype-informative single base substitutions obtained from the Phylotree version of March 9, 2016 (61). Sites presenting more than one allele were not taken into account for the classification. Derived sites for all samples are shown in Dataset S1. See SI Appendix, Section S4.
Reference Datasets.
Newly sequenced individuals were analyzed with a large set of published prehistoric European genomes as well as 203 modern populations from the Human Origins panel (1). At each SNP position, a single read (minimum mapping and base quality of 30) was randomly drawn to represent the ancient individual. Transitions were coded as missing data to exclude potential postmortem damage. See SI Appendix, Section S5.
Population Genetic Analysis.
For each ancient individual, a PCA was conducted together with modern Europeans from the Human Origins panel using smartpca (62). All ancient individuals were projected on one plot using Procrustes analysis. We used popstats (63) to calculate D and f statistics (64) to estimate shared drift between populations and to test tree topologies. To estimate genetic diversity (2) within prehistoric groups, we calculated conditional nucleotide diversity for shotgun sequence data and at transversion polymorphisms ascertained in Yorubans. See SI Appendix, Section S5.
The model-based clustering approach implemented in ADMIXTURE (52) was run with all modern individuals of the Human Origins dataset and all ancient individuals. Common modes among the different runs were identified, and clusters were aligned across different values of K using pong (65). The full unsupervised ADMIXTURE results are shown in Dataset S3. We used supervised ADMIXTURE and qpAdm (6) to estimate the ancestry proportions attributable to early Anatolian farmers, hunter-gatherers, and steppe herders in Neolithic and Bronze Age Europeans (SI Appendix, Section S5).
Supplementary Material
Acknowledgments
We thank everyone involved in the excavations for their contributions, Rafael Carmona at Museo de Priego de Córdoba for facilitating access to material, Arielle R. Munters and Robin Olsson for initial processing and curating of next-generation sequencing data, and Elena Santos for editorial assistance with text and figures. This project was supported by grants from the Transforming Human Societies Research Focus Area and Bridging Fellowship from La Trobe University (C.V.), the Wenner-Gren Foundation (T.G.), the Knut and Alice Wallenberg Foundation (M.J. and A.G.), the Swedish Research Council (M.J. and A.G.), the European Research Council (M.J.), Australian Research Council Grant FT0992258 (to C.I.S.), the Junta de Andalucía Research Project of Excellence Grant HUM-1510 (to R.M.M.-S.), and Spanish Ministerio de Economía y Competitividad Grant CGL2015-65387-C3-2-P (MINECO/FEDER) (to C.V., I.U., R.R.-V., E.I., L.R., J.-M.B.d.C., E.C., J.-M.C., and J.L.A.). Sequencing was performed at the National Genomics Infrastructure, Uppsala, and all computations were conducted through the Uppsala Multidisciplinary Centre for Advanced Computational Science under projects b2013203 and b2013240.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB23467).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717762115/-/DCSupplemental.
References
- 1.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 3.Skoglund P, et al. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science. 2012;336:466–469. doi: 10.1126/science.1216304. [DOI] [PubMed] [Google Scholar]
- 4.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 5.Cassidy LM, et al. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci USA. 2016;113:368–373. doi: 10.1073/pnas.1518445113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones ER, et al. The Neolithic transition in the Baltic was not driven by admixture with early European farmers. Curr Biol. 2017;27:576–582. doi: 10.1016/j.cub.2016.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saag L, et al. Extensive farming in Estonia started through a sex-biased migration from the steppe. Curr Biol. 2017;27:2185–2193.e6. doi: 10.1016/j.cub.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omrak A, et al. Genomic evidence establishes Anatolia as the source of the European Neolithic gene pool. Curr Biol. 2016;26:270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Günther T, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA. 2015;112:11917–11922. doi: 10.1073/pnas.1509851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmanová Z, et al. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci USA. 2016;113:6886–6891. doi: 10.1073/pnas.1523951113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olalde I, et al. A common genetic origin for early farmers from Mediterranean cardial and central European LBK cultures. Mol Biol Evol. 2015;32:3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kılınç GM, et al. The demographic development of the first farmers in Anatolia. Curr Biol. 2016;26:2659–2666. doi: 10.1016/j.cub.2016.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazaridis I, et al. Genetic origins of the Minoans and Mycenaeans. Nature. 2017;548:214–218. doi: 10.1038/nature23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Fortes G, et al. Paleogenomic evidence for multi-generational mixing between Neolithic farmers and Mesolithic hunter-gatherers in the lower Danube basin. Curr Biol. 2017;27:1801–1810.e10. doi: 10.1016/j.cub.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manen C, Marchand G, Calvalho AF. Le Néolithique ancien de la Péninsule Ibérique: Vers une nouvelle évaluation du mirage africain? In: Évin J, editor. Congrès du Centenaire: Un siècle de construction du discours scientifique en Préhistoire. Société Préhistorique Française. Actes du XXVIe Congrès Préhistorique de France; Paris: 2007. pp. 133–151. [Google Scholar]
- 18.Alday A, et al. La Cerámica Boquique: Caracteres, cronología y Contexto. EDAR Arqueologia y Patrimonio; Milan: 2009. Reflejos del Neolítico Ibérico; p. 179. [Google Scholar]
- 19.Utrilla P. Actes Del Congrés Internacional Xarxes Al Neolític. Rubricatum. Revista del Museu de Gavà; Barcelona: 2012. Caminos para el Neolítico Aragonés: La aportación del radiocarbono y del arte rupestre; pp. 555–564. [Google Scholar]
- 20.García Borja P, Aura Tortosa JE, Jordá Pardo JF, Salazar-García DC. La cerámica neolítica de la Cueva de Nerja (Málaga, España): Salas del Vestíbulo y la Mina. Archivo de Prehistoria Levantina. 2014;30:81–131. [Google Scholar]
- 21.Peña-Chocarro L, Pérez Jordà G, Morales J, Vera Rodríguez JC. Y llegaron los agricultores: Agricultura y recolección en el occidente Mediterráneo. Menga Revista de Prehistoria de Andalucía. 2013;4:15–34. [Google Scholar]
- 22.Cortés Sánchez M, et al. The Mesolithic–Neolithic transition in southern Iberia. Quat Res. 2012;77:221–234. [Google Scholar]
- 23.Martínez-Sánchez RM, Vera-Rodríguez JC, Pérez-Jordà G, Peña-Chocarro L, Bokbot Y. The beginning of the Neolithic in northwestern Morocco. Quat Int. 2017 doi: 10.1016/j.quaint.2017.05.052. [DOI] [Google Scholar]
- 24.Martiniano R, et al. The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLoS Genet. 2017;13:e1006852. doi: 10.1371/journal.pgen.1006852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 27.Sikora M, et al. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean Iceman and the genetic structure of Europe. PLoS Genet. 2014;10:e1004353. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipson M, et al. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontanals-Coll M, Díaz-Zorita Bonilla M, Subirà ME. A palaeodietary study of stable isotope analysis from a high-status burial in the Copper Age: The Montelirio megalithic structure at Valencina de la Concepción–Castilleja de Guzmán, Spain. Int J Osteoarchaeol. 2016;26:447–459. [Google Scholar]
- 30.Fontanals-Coll M, Eulàlia Subirà M, Díaz-Zorita Bonilla M, Gibaja JF. First insight into the Neolithic subsistence economy in the north-east Iberian Peninsula: Paleodietary reconstruction through stable isotopes. Am J Phys Anthropol. 2017;162:36–50. doi: 10.1002/ajpa.23083. [DOI] [PubMed] [Google Scholar]
- 31.Fontanals-Coll M, Subirà ME, Bonilla MD-Z, Duboscq S, Gibaja JF. Investigating palaeodietary and social differences between two differentiated sectors of a Neolithic community, La Bòbila Madurell-Can Gambús (north-east Iberian Peninsula) J Archaeol Sci Rep. 2015;3:160–170. [Google Scholar]
- 32.Lubell D, Jackes M, Schwarcz H, Knyf M, Meiklejohn C. The Mesolithic-Neolithic transition in Portugal: Isotopic and dental evidence of diet. J Archaeol Sci. 1994;21:201–216. [Google Scholar]
- 33.McClure SB, García O, Roca de Togores C, Culleton BJ, Kennett DJ. Osteological and paleodietary investigation of burials from Cova de la Pastora, Alicante, Spain. J Archaeol Sci. 2011;38:420–428. [Google Scholar]
- 34.Waterman AJ, Tykot RH, Silva AM. Stable isotope analysis of diet-based social differentiation at Late Prehistoric collective burials in south-western Portugal. Archaeometry. 2016;58:131–151. [Google Scholar]
- 35.Carvalho AF, Petchey F. Stable isotope evidence of Neolithic palaeodiets in the coastal regions of southern Portugal. J Island Coast Archaeol. 2013;8:361–383. [Google Scholar]
- 36.Webb EC, et al. Compound-specific amino acid isotopic proxies for detecting freshwater resource consumption. J Archaeol Sci. 2015;63:104–114. [Google Scholar]
- 37.Webb EC, et al. The influence of varying proportions of terrestrial and marine dietary protein on the stable carbon-isotope compositions of pig tissues from a controlled feeding experiment. Sci Technol Archaeol Res. 2017;3:36–52. [Google Scholar]
- 38.Mora A, et al. High-resolution palaeodietary reconstruction: Amino acid δ13C analysis of keratin from single hairs of mummified human individuals. Quat Int. 2017;436:96–113. [Google Scholar]
- 39.Smith CI, Fuller BT, Choy K, Richards MP. A three-phase liquid chromatographic method for δ13C analysis of amino acids from biological protein hydrolysates using liquid chromatography-isotope ratio mass spectrometry. Anal Biochem. 2009;390:165–172. doi: 10.1016/j.ab.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Honch NV, McCullagh JSO, Hedges REM. Variation of bone collagen amino acid δ13C values in archaeological humans and fauna with different dietary regimes: Developing frameworks of dietary discrimination. Am J Phys Anthropol. 2012;148:495–511. doi: 10.1002/ajpa.22065. [DOI] [PubMed] [Google Scholar]
- 41.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7:e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green RE, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skoglund P, et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc Natl Acad Sci USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandt G, et al. Genographic Consortium Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science. 2013;342:257–261. doi: 10.1126/science.1241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 47.Malmström H, et al. Ancient mitochondrial DNA from the northern fringe of the Neolithic farming expansion in Europe sheds light on the dispersion process. Philos Trans R Soc Lond B Biol Sci. 2015;370:20130373. doi: 10.1098/rstb.2013.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olalde I, et al. 2017. The Beaker phenomenon and the genomic transformation of northwest Europe. bioRxiv:10.1101/135962.
- 49.Monroy Kuhn JM, Jakobsson M, Günther T. 2017. Estimating genetic kin relationships in prehistoric populations. bioRxiv:10.1101/100297. [DOI] [PMC free article] [PubMed]
- 50.Günther T, et al. Genomics of Mesolithic Scandinavia reveal colonization routes and high-latitude adaptation. PLoS Biol. 2018;16:e200370. doi: 10.1371/journal.pbio.2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiang CWK, et al. 2016. Population history of the Sardinian people inferred from whole-genome sequencing. bioRxiv:10.1101/092148.
- 52.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg A, Günther T, Rosenberg NA, Jakobsson M. Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations. Proc Natl Acad Sci USA. 2017;114:2657–2662. doi: 10.1073/pnas.1616392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010:pdb.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 56.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 57.Richards MP, Hedges REM. Stable isotope evidence for similarities in the types of marine foods used by Late Mesolithic humans at sites along the Atlantic coast of Europe. J Archaeol Sci. 1999;26:717–722. [Google Scholar]
- 58.Kircher M. Analysis of high-throughput ancient DNA sequencing data. Methods Mol Biol. 2012;840:197–228. doi: 10.1007/978-1-61779-516-9_23. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vianello D, et al. HAPLOFIND: A new method for high-throughput mtDNA haplogroup assignment. Hum Mutat. 2013;34:1189–1194. doi: 10.1002/humu.22356. [DOI] [PubMed] [Google Scholar]
- 61.van Oven M, Van Geystelen A, Kayser M, Decorte R, Larmuseau MH. Seeing the wood for the trees: A minimal reference phylogeny for the human Y chromosome. Hum Mutat. 2014;35:187–191. doi: 10.1002/humu.22468. [DOI] [PubMed] [Google Scholar]
- 62.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skoglund P, et al. Genetic evidence for two founding populations of the Americas. Nature. 2015;525:104–108. doi: 10.1038/nature14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.