Significance
Phosphorothioate (PT) modification of the DNA sugar-phosphate backbone is an important microbial epigenetic modification governed by DndABCDE, which together with DndFGH, constitutes a restriction-modification system. We show that up to 45% of 1,349 identified bacterial dnd systems exhibit the form of solitary dndABCDE without the restriction counterparts of dndFGH. The combination of epigenomics, transcriptome analysis, and metabolomics suggests that in addition to providing a genetic barrier against invasive DNA, PT modification is a versatile player involved in the epigenetic control of gene expression and the maintenance of cellular redox homeostasis. This finding provides evolutionary and functional insights into this unusual epigenetic modification. Our results imply that PT systems might evolve similar to other epigenetic modification systems with multiple cellular functions.
Keywords: DNA modification, DNA phosphorothioation, restriction modification
Abstract
The chemical diversity of physiological DNA modifications has expanded with the identification of phosphorothioate (PT) modification in which the nonbridging oxygen in the sugar-phosphate backbone of DNA is replaced by sulfur. Together with DndFGH as cognate restriction enzymes, DNA PT modification, which is catalyzed by the DndABCDE proteins, functions as a bacterial restriction-modification (R-M) system that protects cells against invading foreign DNA. However, the occurrence of dnd systems across a large number of bacterial genomes and their functions other than R-M are poorly understood. Here, a genomic survey revealed the prevalence of bacterial dnd systems: 1,349 bacterial dnd systems were observed to occur sporadically across diverse phylogenetic groups, and nearly half of these occur in the form of a solitary dndBCDE gene cluster that lacks the dndFGH restriction counterparts. A phylogenetic analysis of 734 complete PT R-M pairs revealed the coevolution of M and R components, despite the observation that several PT R-M pairs appeared to be assembled from M and R parts acquired from distantly related organisms. Concurrent epigenomic analysis, transcriptome analysis, and metabolome characterization showed that a solitary PT modification contributed to the overall cellular redox state, the loss of which perturbed the cellular redox balance and induced Pseudomonas fluorescens to reconfigure its metabolism to fend off oxidative stress. An in vitro transcriptional assay revealed altered transcriptional efficiency in the presence of PT DNA modification, implicating its function in epigenetic regulation. These data suggest the versatility of PT in addition to its involvement in R-M protection.
For bacteria, phages are arguably the most abundant and challenging antagonists present in their environment. Consequently, bacteria have evolved multiple defense mechanisms, such as restriction-modification (R-M) systems, abortive infection systems, and CRISPR-Cas proteins, which target nearly every step of the phage life cycle (1). The most extensively studied of these defense barriers are the methylation-based R-M systems, R-M systems typically aid resistance to phage infection by encoding a methyltransferase (MTase) that methylates a particular sequence of “self”-DNA and a cognate restriction endonuclease that discriminates and destroys nonmodified invasive DNA. Despite their tremendous diversity, all R-M systems are thought to perform DNA methylation on nucleobase moieties, generating N6-methyl-adenine, N4-methyl-cytosine, and C5-methyl-cytosine, for example, to distinguish self-DNA from non–self-DNA.
Phosphorothioate (PT) modification, in which the nonbridging oxygen in the phosphate moiety of the DNA sugar-phosphate backbone is replaced by sulfur, was originally developed as an artificial approach to protect oligonucleotides against nuclease degradation (2). However, we recently discovered that PT modification occurs naturally in bacteria and that its function extends our understanding of R-M systems (3–5). The DNA PT system is composed of two parts: gene products of the dndABCDE cluster function as the M component to confer the DNA PT modification in a stereo- and sequence-specific manner, whereas products of the dndFGH function together as the R component to recognize and hydrolyze non–PT-protected foreign DNA (6, 7). DndA possesses cysteine desulfurase activity and is functionally substituted by an IscS homolog in some bacterial strains (8). Thus, in a subset of strains, dndA is absent from the region surrounding the dndBCDE genomic locus. DndB binds to the promoter region of the dnd operon to regulate the transcription of dnd genes (9). DndC possesses ATP pyrophosphatase activity and exhibits high sequence homology with phosphoadenosine phosphosulfate reductase. The DndD protein has ATPase activity and is predicted to provide energy during the incorporation of sulfur (10), whereas DndE exhibits preferential binding to nicked double-strand DNA in vitro via positively charged lysine residues on its surface (11). Without PT protection, DNA is susceptible to double-strand damage caused by DndFGH in Salmonella enterica serovar Cerro 87, resulting in induction of the cellular SOS response (6, 7). The similarity of the DNA PT defensive mechanism to the methylation-based R-M system makes the DNA PT system a member of the prokaryotic innate immune systems.
Four major types of R-M systems (I, II, III, and IV) have been classified based on subunit composition, sequence recognition, cofactor requirements, and cleavage mechanism. Considering their genetic composition and requirement of ATP, dnd systems are more similar to type I R-M systems, which consist of three subunits of R (restriction), M (modification), and S (specificity) in the R2M2S1 complex (12). This finding agrees with the observation that DndC, DndD, DndE, and IscS form a large protein complex to confer DNA PT modification (13). Interestingly, our recent study revealed an interaction between DNA PT and methylation systems, demonstrating that DndABCDE and DNA adenine methylase (Dam) are capable of sharing the same recognition motif, 5′-GATC-3′, to generate hybrid 5′-GPS6mATC-3′ [PT internucleotide linkage (PS), N6-methyldeoxyadenosine (6mA)]. However, the PT modification 5′-GPSATC-3′ reduces Dam activity in vitro, whereas the methylation modification 5′-G6mATC-3′ can substitute for PT to confer resistance to DndFGH. This finding indicates the complex interactions between DNA defense systems and raises the possibility of the coevolution of these systems to avoid conflicts in functionality (14).
Due to their nuclease resistance, PT-linked dinucleotides are generated in addition to canonical mononucleotides upon enzymatic DNA hydrolysis, enabling the detection of PT modifications by liquid chromatography-coupled tandem quadrupole mass spectrometry (LC-MS/MS) (15). For example, S. enterica serovar Cerro 87 and Escherichia coli B7A harbor PT-modified d(GPSA) and d(GPST) dinucleotides at a ratio of 1:1, Hahella chejuensis KCTC2396 contains d(GPSA) modifications, and Pseudomonas fluorescens pf0-1 and Streptomyces lividans possess PT-linked d(GPSG) dinucleotides. Thus, we adopted single-molecule, real-time (SMRT) sequencing technology to map PT sites across bacterial genomes. The data confirmed that d(GPSA) and d(GPST) in E. coli B7A occur in a conserved 5′-GPSAAC-3′/5′-GPSTTC-3′ context without any apparent strict consensus beyond four nucleotides (16). Unexpectedly, only 12% of the genomic 5′-GAAC-3′/5′-GTTC-3′ sites were PT-protected, and the PT modifications were partial at any particular genomic 5′-GAAC-3′/5′-GTTC-3′, even in the presence of the active DndFGH restriction counterpart. These features differentiate DNA PT modification from known R-M mechanisms and indicate that Dnd proteins have an unusual target selection mechanism or that PT has alternative functions.
Since the original report in S. lividans, dnd genes and PT modifications have been identified in a subset of taxonomically distant bacterial genomes and environmental samples (15, 17). However, the development and evolution of dnd systems across a large number of bacterial genomes, as well as the alternative function(s) of PT modification, remain uncharacterized. In this study, we first conducted a comparative genomic survey and observed that ∼45% of 1,349 dnd systems harbored only dndBCDE M genes, without pairing with dndFGH cognate R genes. Interestingly, in some organisms, the complete PT R-M pairs appear to be assembled from distantly related R and M parts in the evolutionary process. Although the trans-omics analysis did not address the direct correlation between PT sites and gene regulation in P. fluorescens pf0-1, an in vitro transcriptional assay revealed that the saturated PT modification at a given site is able to influence the transcriptional efficiency, suggesting that PT modification can affect epigenetic control. It is noteworthy that the loss of PT in R−-M+ P. fluorescens pf0-1 did not cause impaired growth, as was observed in R+-M+ S. enterica serovar Cerro 87, but instead triggered significant changes in the overall cellular redox state and induced metabolic reprogramming to combat oxidative stress. These data suggest that PT modification maintains redox homeostasis and is involved in epigenetic gene regulation in addition to defense systems.
Results
Solitary PT R−-M+ Systems Are Widely Distributed in Bacterial Genomes.
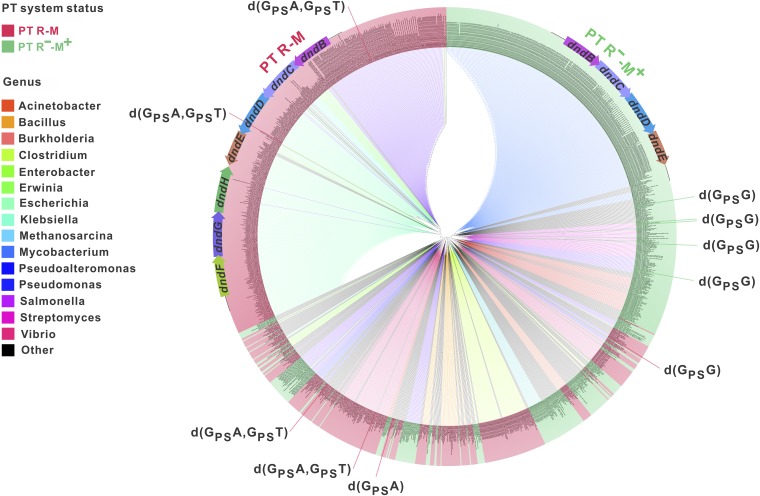
Although DNA PT modifications have been detected in ∼20 taxonomically diverse bacterial strains, there is little information regarding the occurrence of PT systems across a large number of bacterial genomes. We first set out to perform a comparative genomic survey to investigate the frequency and diversity of PT systems. A total of 1,349 positive hits were obtained from National Center for Biotechnology Information (NCBI) nucleotide databases (including the nonredundant nucleotide collection, Reference Sequence genome database, high-throughput genomic sequences, genomic survey sequences, and NCBI Genome) using the DndBCD protein sequences of S. enterica serovar Cerro 87 and H. chejuensis KCTC2396, which possess functionally identified PT R-M systems, as determined by querying with the M component (Dataset S1). DndE was not included in this search procedure because this protein is too small to be annotated in a number of genomes. A stringent e-value cutoff of 10−10 was used to reduce false-positive results and ensure the identification of true PT M genes. To exclude false-positive results arising from high local homology, we filtered out BLAST hits with aligned lengths shorter than 30% of the query proteins. The 1,349 identified bacterial strains, including S. enterica serovar Cerro 87 and H. chejuensis KCTC2396, represent 162 genera and 346 species and are preferentially distributed within the genera Mycobacterium (288, 21.3%), Escherichia (235, 17.4%), Salmonella (174, 12.9%), Vibrio (82, 6.1%), and Clostridium (60, 4.4%) (Dataset S1). We subsequently explored 30 ORFs upstream and downstream of dndBCD to identify dndFGH, which encodes the R component of the PT R-M system. Because DndF, DndG, and DndH are all required to fulfill the restriction function, the PT R component was considered “complete” only when all three genes encoding the proteins were present in a genome. Surprisingly, the coexistence of the complete PT R and M components was only identified in 734 strains, representing 54.4% of the total of 1,349 strains. A large number of PT systems (615, 45.6%) occur in the form of PT R−-M+, which possesses the PT M component but is not paired with a complete DndFGH (Fig. 1 and Dataset S1).
Fig. 1.
Phylogenetic distribution of DndBCD homologs. The phylogenetic tree of DndBCD homologs in 1,349 strains was created using MEGA5.1 (41) with the maximum likelihood method and 500 bootstrap replications and was visualized using Archaeopteryx (42). Different genera are colored accordingly. The strains shaded red and green indicate that DndBCD homologs are associated with complete and incomplete DndFGH, respectively (i.e., the PT R-M and R−-M+ systems). The PT sequence contexts identified in some strains are displayed as noted.
Phylogenetic Analysis of PT R-M and PT R−-M+ Systems.
To analyze the phylogenesis of DNA PT systems, a phylogenetic tree using the DndBCD sequences of 1,349 strains was constructed (Fig. 1). The topology of the tree was observed to largely follow the corresponding species tree, although several potential horizontal gene transfer events were apparent. For example, the majority of Salmonella strains clustered together in a large clade adjacent to Escherichia, consistent with their phylogenetic positions. In contrast, two Salmonella clades, were instead grouped with Aeromonas, Klebsiella, and Citrobacter strains, among others. This result suggests that the dndBCD genes in Salmonella might be acquired by horizontal gene transfer from bacteria of diverse origins.
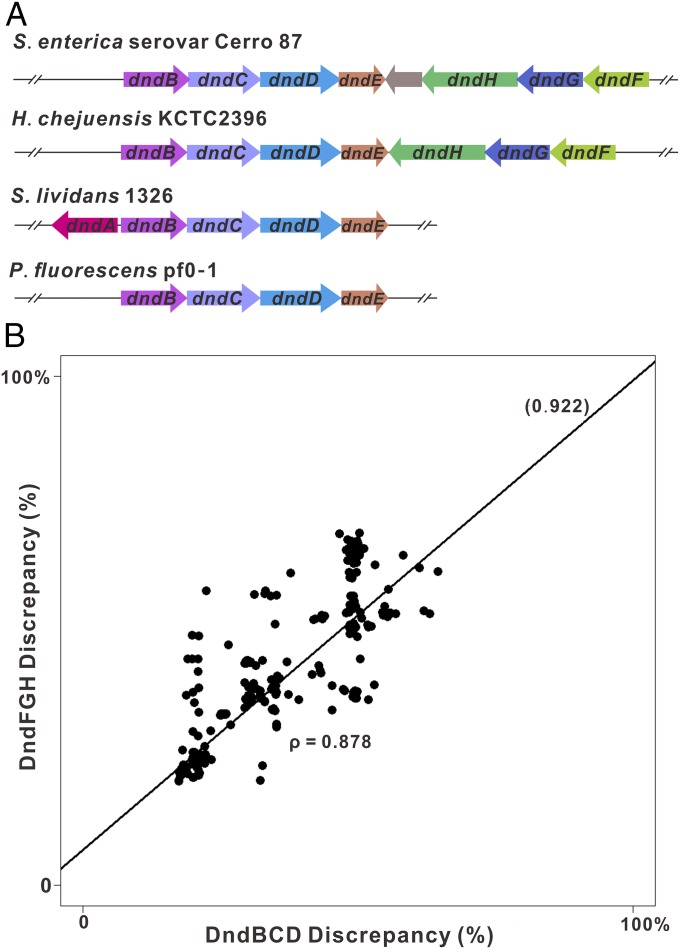
In contrast to the diverse recognition motifs of the methylation-based R-M systems, the consensus contexts of PT systems are limited; for example, DndABCDE confers the PT modifications 5′-GPSGCC-3′/5′-GPSGCC-3′ in S. lividans, Vibrio splendidus ZS-139, and Enterovibrio calviensis 1F-230, and the modifications 5′-GPSAAC-3′/5′-GPSTTC-3′ in E. coli B7A, S. enterica serovar Cerro 87, and Vibrio tasmaniensis 1F-267 (15). Thus, we examined whether the M and R components consistently evolved together or separately from each other. If the PT M and R components coevolved (i.e., they have similar rates of divergence from their corresponding ancestors), the sequence discrepancies of the M and R components between a given strain and the query strains should be proportional to each other. To examine this finding, the DndFGH distance to the queries was regressed on the DndBCD distance to the queries in 734 strains with both M and R components. The regression line slope, 0.922, indicated a nearly linear relationship of evolutionary variance from the reference between DndBCD and DndFGH. Accordingly, a correlation coefficient ρ of 0.878 was obtained for the divergences of the 734 M and R components (Fig. 2). Such a linear deviation in the protein sequences of the M and R components from the reference suggests a consistent coevolution of the two components in intact PT R-M systems.
Fig. 2.
Scatterplot of alignment distances of PT M and R genes in 734 complete PT R-M pairs in response to queries. (A) Four typical PT R-M and PT R−-M+ systems are displayed. The DndBCD and DndFGH sequences of S. enterica serovar Cerro 87 and H. chejuensis KCTC2396 were used as queries for BLAST searches. (B) Many strains have identical values for the two distances, and thus overlap. The high coefficient correlation, ρ = 0.878, suggests the coevolution of the M and R components in DNA PT systems.
To corroborate the coevolution of the PT M and R components, a tanglegram of juxtaposed trees formed by DndBCD and DndFGH from 734 PT R-M systems was generated (SI Appendix, Fig. S1). The topological difference between the DndBCD and DndFGH trees is significantly smaller, with a P value of 0.003, in comparison to that between tree pairs formed by randomly shuffled strains, suggesting the coevolution of DndBCD and DndFGH (Materials and Methods). Interestingly, the two trees were not identically shaped. In some strains, the PT R-M system appeared to be assembled from DndBCD and DndFGH components from different organisms. For example, DndFGH of Yersinia ruckeri strain YRB grouped with those of Yersinia mollaretii 64/02, Yersinia pseudotuberculosis MW109-2, Yersinia enterocolitica FE80217, and Yersinia sp. FDAARGOS 228. In contrast, DndBCD of Y. ruckeri strain YRB split from the other Yersinia strains and localized in a distant part of the tree with Erwinia strains (SI Appendix, Fig. S1). This result suggested that PT R and M components originating from distantly related species are likely compatible to constitute a functional PT defense system. The limited sequence selectivity of Dnd proteins might contribute to the generation of chimeric DndBCDE-DndFGH across bacterial species. This hypothesis is supported by the observation that DndFGH-induced growth inhibition of PT-deficient E. coli B7A is rescued by PT M genes from S. enterica serovar Cerro 87 due to the identical PT recognition motif, 5′-GPSAAC-3′/5′-GPSTTC-3′ (SI Appendix, Fig. S2).
Correlation Between the Genomic Distribution and Global Transcriptional Impact of PT Modification in P. fluorescens pf0-1.
The PT R−-M+ systems lacking DndFGH are reminiscent of orphan DNA MTases, which methylate nucleobases in select sequence contexts but are not associated with any restriction enzymes. The best-studied orphan MTases include Dam, DNA cytosine methylase (Dcm), and cell cycle-regulated methylase (CcrM), which play important roles beyond R-M systems, such as in chromosome replication, DNA mismatch repair, transcriptional regulation, cell cycle regulation, and pathogenesis (18–20). This similarity hints that PT modification is not merely a component of the R-M system and might have evolved additional functions, such as the epigenetic control of gene expression. To explore the global impact of PT modification on cellular physiology, genomic PT mapping and global transcriptome analysis were concurrently performed in P. fluorescens pf0-1, which harbors dndBCDE with d(GPSG) PT dinucleotides but lacks the cognate dndFGH.
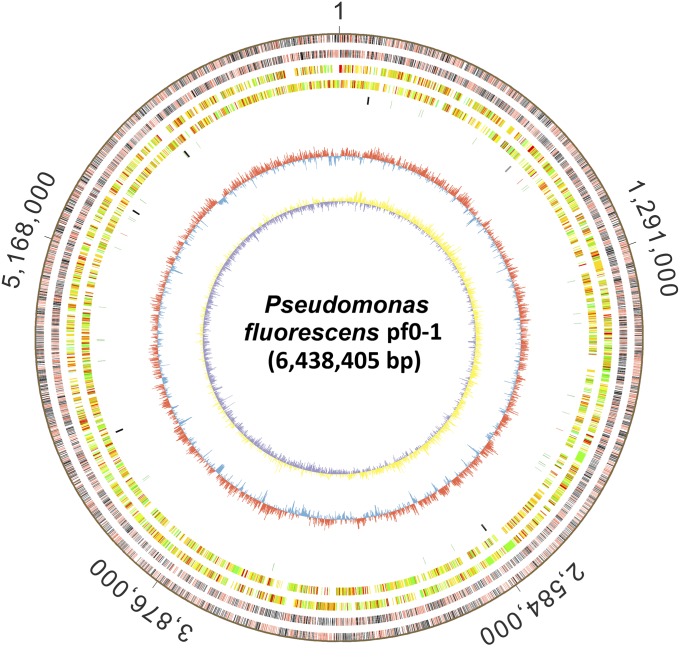
We subsequently performed SMRT sequencing to identify the distribution of genomic d(GPSG) sites based on variations in the interpulse duration of DNA polymerase (21). A fragmented SMRTbell library with an average insert size of 8.4 kb was constructed and sequenced using SMRT sequencing. A total of 5,202 PT signatures, PT-modified d(GPSG), were detected in the pf0-1 genome; of these, 3,704 were located in the complementary 5′-GPSGCC-3′/5′-GPSGCC-3′ context and 1,498 occurred in single-stranded 5′-GPSGCC-3′ (Fig. 3 and Dataset S2). Alignment of the 50-nt flanking sequences revealed no strict further sequence-context constraint beyond 5′-GPSGCC-3′, although the fifth base demonstrated a relative underrepresentation of C residues (SI Appendix, Table S1). To profile the distribution features of the 5,202 PT sites across the pf0-1 genome, we mapped 5′-GPSGCC-3′ using the pf0-1 genome sequence (GenBank accession no. CP000094) as a reference and determined that 2,168 of 5,722 ORFs, one of 73 tRNA genes, seven of 18 rRNA genes, and 2,045 noncoding regions contained at least one PT (Fig. 3 and Dataset S2).
Fig. 3.
PT annotation across the pf0-1 genome. From the outer to inner circles: 1 and 2 (forward, reverse strands), PT sites in ORFs (gray), RNA (blue) and nonencoding regions (red); 3 and 4, predicted protein-coding sequences colored according to COG functional categories; 5, tRNA/rRNA operons; 6, guanine–cytosine content; 7, guanine–cytosine skew.
In addition to ORFs containing one and two PT sites, two ORFs contained seven PT sites, three contained five PT sites, seven contained four PT sites, and 28 contained three PT sites. The following 12 genes had enriched PT sites (four or more): an ATP-dependent helicase (Pfl01_1420), a hybrid sensor histidine kinase/response regulator (Pfl01_1660), a putative nonribosomal peptide synthetase (Pfl01_2212), a hypothetical protein (Pfl01_2644), a DNA topoisomerase III (Pfl01_3364), a periplasmic sensor diguanylate cyclase/phosphodiesterase (Pfl01_5150), a glycine dehydrogenase alpha subunit (Pfl01_5426), a pyoverdine synthetase I (Pfl01_1845), a conserved hypothetical protein (Pfl01_3171), a putative toxin A (Pfl01_0609), a large adhesive protein (Pfl01_0133), and a pyoverdine synthetase (Pfl01_1846). Notably, the average gene length of these 12 ORFs was 8,494 bp, which is markedly longer than that of the 28 ORFs (3,563 bp) possessing three PT sites and the 318 ORFs (1,773 bp) with two PT sites, indicating that the enrichment of PT sites in these regions is attributable to the longer lengths of these genes. Among the 5,722 possible ORF promoters, 5.49% (314) were PT-modified. The promoters (200 bp upstream of the start codons) of four ORFs, namely, ORFs encoding a hypothetical protein (Pfl01_0873), a putative membrane protein (Pfl01_4826), a hypothetical ABC transporter ATP-binding protein (Pfl01_5442), and a putative phospholipase D (Pfl01_5532), contained two PT sites, whereas the other PT-modified promoters had only one PT site. Among RNA genes, 38.89% (seven of 18) of rRNA genes harbored PT-modified site(s) in their internal region and only one (Cys tRNA) of 73 tRNAs contained two PT sites, whereas no PT sites were detected in the promoter regions.
An analysis of PT occurrence frequencies by the Poisson exact test revealed that ORF and tRNA regions exhibited PT levels that were similar to the genomic mean level (0.4 PT per 103 nt), whereas ORF promoters, rRNA genes, and noncoding regions showed a lower PT density (SI Appendix, Table S2). To determine whether PT modifications are involved in the epigenetic control of the expression of specific genes, an RNA-sequencing (RNA-Seq) analysis was conducted to define the global transcriptional changes in early log-phase cultures of wild-type pf0-1 and the dndBCDE deletion mutant TT-5. Ten genes showed more than twofold differential expression [log2 ratio > 1 or < −1, false discovery rate (FDR) ≤ 0.001] in TT-5 compared with the wild-type pf0-1 strain, and these genes included six up-regulated genes and four down-regulated genes (SI Appendix, Table S3). Quantitative RT-PCR was performed to validate the gene expression levels observed by RNA-Seq (SI Appendix, Fig. S3). Due to the in-frame dndBCDE deletion in the TT-5 mutant, these genes were the most significantly down-regulated, unlike the pf0-1 samples. Two of the six up-regulated genes encoded phage-related proteins (Pfl01_1169 and Pfl01_3496), and the other genes encoded a putative membrane protein (Pfl01_0900), a xanthine dehydrogenase (Pfl01_1797), an N-acetylmuramoyl-l-alanine amidase (Pfl01_5657), and a hypothetical protein (Pfl01_2488). The transcription levels of the differentially expressed genes, which belonged to different clusters of orthologous groups (COGs) and had no apparent functional connection to each other, varied to different degrees. The combination of a genomic PT profile and transcriptome analysis revealed that none of the promoter regions of the 10 differentially transcribed genes contained PT sites, whereas the three genes that possessed one PT within their coding region were either up- or down-regulated (SI Appendix, Table S3).
PT Modifications Disturb Gene Transcription in Vitro.
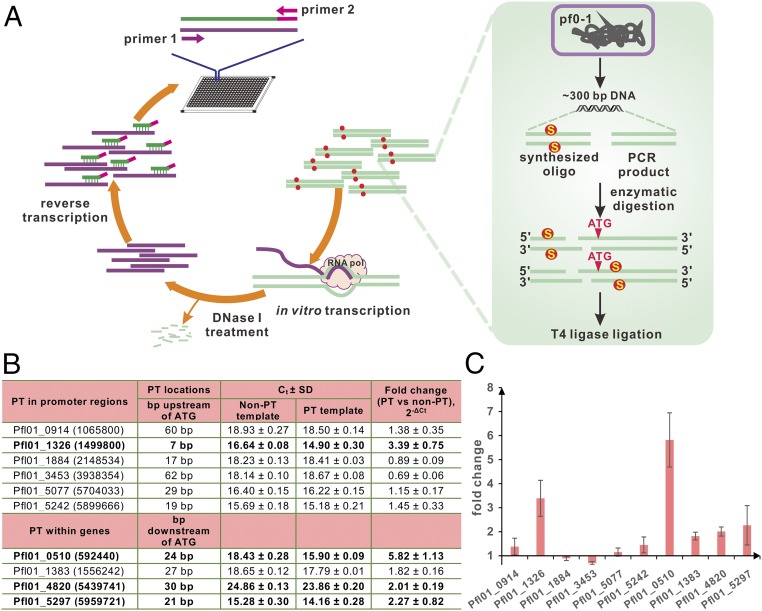
Considerable evidence has shown that DNA modifications (e.g., methylation) are important for regulating gene expression. For instance, the methylation of adenosine at the 5′-GATC-3′ site in the dnaA P2 promoter results in threefold to fivefold enhanced transcription, whereas methylation of 5′-GATC-3′ in the −35 region of the trpR gene, which encodes a tryptophan repressor, reduces its expression by threefold (22). Because only a subset of cells within a population of cells might have a PT modification at a given site, it raises the possibility of multiple phenotypically distinct pf0-1 cell types with heterologous PT patterns. The differentially expressed genes would not be identified by RNA-Seq analysis because the observed expression values represent the sum of transcriptional changes in a population of cells. To test this possibility, an in vitro transcription assay was performed to assess the effects of PT modification on gene transcription. To ensure complete PT modification at a given position, internucleotide PT linkages were chemically synthesized. According to the genomic mapping data, 10 PT-modified 5′-GPSGCC-3′ sites located in promoter regions or within a P. fluorescens pf0-1 gene were selected for the in vitro transcription assay (Fig. 4A and SI Appendix, Table S4). Four of the 10 genes were differentially expressed compared with the non–PT-modified counterparts, with a difference of at least twofold and an FDR ≤ 0.001. The use of promoters of Pfl01_0914, Pfl01_1884, Pfl01_3453, Pfl01_5077, and Pfl01_5242 for direct transcription resulted in the transcription of DNA templates with or without 5′-GPSGCC-3′ modification with no significant changes in efficiency. In contrast, 3.39 ± 0.75-fold up-regulation was observed for the promoter of Pfl01_1326 containing 5′-GPSGCC-3′, 7 bp upstream of the ATG codon. The occurrence of PT-modified 5′-GPSGCC-3′ sites within the Pfl01_5297, Pfl01_0510, and Pfl01_4820 genes (e.g., 15 bp, 18 bp, and 24 bp downstream of the ATG codon) resulted in 2.27-, 5.82-, and 2.01-fold increases in transcription efficiency, respectively. Conversely, the 5′-GPSGCC-3′ site located 21 bp downstream of the ATG codon in Pfl01_1383 had no pronounced effect on transcription (Fig. 4 B and C). The different influences of PT on a subset of genes imply that PT modification is capable of being involved in epigenetic regulation.
Fig. 4.
Effects of PT on in vitro transcription. (A) Schematic of the approach used to prepare DNA templates. In vitro transcription (B) and assessment (C) of the impact of PT on transcriptional efficiency using real-time PCR and the comparative Ct method. The Ct and fold change values represent the average values of Ct ± SD and gene expression ± SD from three independent experiments, respectively. The text in bold in B indicates genes with an expression ratio greater than twofold, with an FDR ≤ 0.001.
Metabolomic Alterations in Response to PT Deficiency.
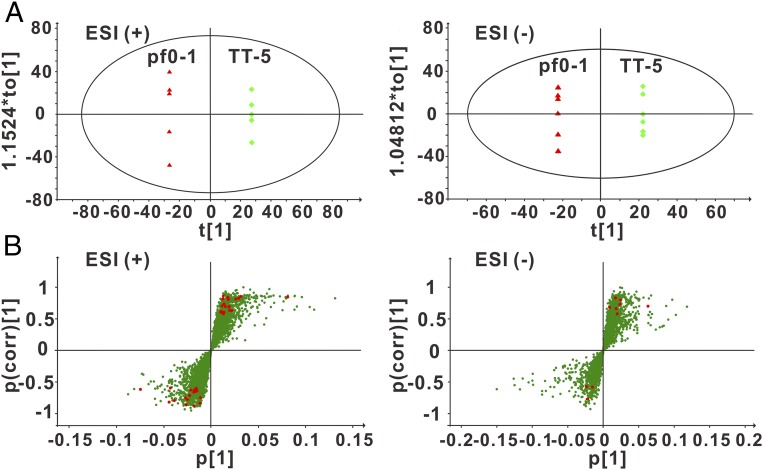
Unlike the growth defect of the PT-deficient S. enterica mutant, the dndBCDE-deleted P. fluorescens mutant TT-5 displayed a similar growth profile to the wild-type pf0-1 strain in minimal medium supplemented with different concentrations of a carbon source (glucose) and a sulfur source (sulfate), indicating the occurrence of physiological effects that are distinct from those of PT R-M systems (SI Appendix, Fig. S4). To further explore the impact of orphan PT systems on cell physiology, we subsequently assessed the metabolomic response of P. fluorescens to PT deficiency. An orthogonal projection to a latent structure-discriminant analysis (OPLS-DA) model was examined using the UPLC-ESI-Q-TOF-MS (ultraperformance liquid chromatography–electrospray ionization–quadrupole time-of-flight mass spectrometry) spectral data obtained from the wild-type pf0-1 strain and the dndBCDE-deleted mutant TT-5. The plot of OPLS-DA scores exhibited a clear separation, indicating that pf0-1 and TT-5 showed significantly different levels of cellular metabolites (Fig. 5A). Metabolites that contributed to these distinctions were identified based on the variable importance in projection (VIP) value (VIP > 1) and Student’s t test (P < 0.05). A total of 48 metabolites, highlighted in the S-plots, significantly changed due to PT loss, and these included 29 and 19 metabolites that showed increased and decreased levels, respectively, in TT-5 compared with wild-type pf0-1 (Fig. 5B and SI Appendix, Table S5).
Fig. 5.
Identification of discriminatory metabolites from pf0-1 and TT-5. (A) OPLS-DA score plot derived from the UPLC-ESI-Q-TOF-MS metabolomic analysis showed the differential clustering of pf0-1 and TT-5 in both the positive and negative modes. (B) S-plot represents the impact of the metabolites on the discriminative clustering of pf0-1 and TT-5. All 48 metabolites with VIP > 1 and P < 0.05 are marked with red circles. These metabolites lie in the upper right and lower left quadrants, showing high contribution and correlation to the discrimination of the metabolomes of pf0-1 and TT-5.
Notably, a 5.3-fold increase in proline content was observed as a consequence of PT deficiency. Proline is a proteinogenic amino acid that is essential for primary metabolism. It is also well-documented that proline accumulates as a physiological response against different stress conditions, such as oxidative stress, high salinity, intense light and UV irradiation, and heavy metals (23). The high proline concentration observed during stress responses is generally believed to result from the up-regulation of proline synthesis and a corresponding down-regulation of the proline degradation pathway (24). The observed proline levels agreed with our observations of up-regulated proline biosynthesis-related genes (albeit at marginal levels), such as proB, proA, and proC, which encode γ-glutamyl kinase, γ-glutamyl phosphate reductase, and ∆1-pyrroline-5-carboxylate reductase, respectively (SI Appendix, Table S6). The ability of proline to abate cell stress is not fully understood but has been proposed to involve a plethora of mechanisms, such as serving as an osmolyte, a scavenger of reactive oxygen species (ROS), and a chemical protein chaperone helping to balance intracellular redox homeostasis, among other functions (24). The increase in proline content indicated that the PT loss in P. fluorescens resulted in the generation of cellular stresses, which agrees with the profound changes observed in lipid metabolites, including phosphatidylcholine, phosphatidylserine, phosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, and ceramides. In contrast, a set of metabolites, including quinic acid, l-methionine, and glycine, showed more than twofold decreases and was correlated with ROS. Quinic acid, which is capable of improving the survival of Caenorhabditis elegans under oxidative stress, both directly by scavenging free radicals and indirectly by increasing the expression of stress-responsive genes (25), showed a significant decrease (by approximately sevenfold) in the TT-5 mutant. A group of caffeoyl quinic acid derivatives from the roots of Dipsacus asper Wall also exhibits antioxidant activity (26). The TT-5 metabolome showed approximately twofold significant reductions in the concentrations of glycine and methionine. Interestingly, both glycine and methionine are involved in protective effects against oxidative stress (27, 28).
Discussion
The discovery of the physiological importance of PT modification in bacteria has expanded our understanding of the composition and structure of DNA. DNA PT modification, catalyzed by the DndABCDE proteins, resembles methylation-based R-M defense systems using DndFGH as the cognate restriction enzyme. Despite the previous identification of PT modifications in ∼20 bacterial strains, there is little information regarding the occurrence of dnd systems across a large number of sequenced bacterial genomes. Our data showed that nearly half of 1,349 identified dnd systems take the form of PT R−-M+, which are in possession of only the PT M genes and not the DndFGH restriction enzyme-encoding counterparts. The frequency of PT R−-M+ systems explains the observation that PT-based R-M defense was only observed in a subset of strains. For example, based on the growth profiles and RNA-Seq results, the PT deficiency in P. fluorescens pf0-1 does not lead to DndFGH-mediated genome damage or cellular SOS responses.
With respect to intact PT R-M systems, a mathematical model and phylogenetic comparison provided evidence that the M and R components evolved together, but a number of M and R components might originate from different ancestors. This finding raises two possibilities: (i) The limited types of PT modification-catalyzing enzymes facilitate the generation of chimeric PT R-M systems, and (ii) this might allow PT-disturbed cells to effectively survive the toxic effects of DndFGH. In terms of the occurrence of PT R−-M+ systems, one possibility is that these systems might have evolved from an ancestor that is different from that of the PT R-M pairs. However, based on DndBCD phyologenesis, we did not observe a clear phylogenetic distinction between PT R−-M+ and R-M systems. The other possibility is that PT R−-M+ systems share a common ancestor with PT R-M systems and are derived from the degradation of PT R-M ancestors. If the PT M component or the entire PT R-M system is disturbed, PT-deficient DNA becomes vulnerable to lethal pressure from the cognate R components. To survive this pressure, selective degradation or mutation and subsequent loss of the R parts might occur, yielding PT R−-M+ systems. However, no significant DndFGH degradation event was observed in 615 PT R−-M+ systems, with the exception of Tolumonas auensis DSM 9187. T. auensis DSM 9187 harbors clustered dndBCDE genes (GenBank accession nos. TOLA_RS08085, TOLA_RS08090, TOLA_RS08095, and TOLA_RS08100) associated with a possible degradation intermediate of DndFGH. A gene, annotated as dndF (TOLA_RS08125), which is in the vicinity of dndBCDE and encodes a 242-aa protein, shares 57% and 56% sequence similarity to the N termini of the DndF proteins in S. enterica and H. chejuensis KCTC2396 (549 aa and 532 aa), respectively. No detectable DndG- or DndH-encoding genes were observed in the neighborhood of dndF, even at a relaxed e-value of 10−2. It is noteworthy that single dndF or dndH homologs were observed to be present in eight and one bacterial strains, respectively, but these were not accompanied by other dnd genes (Dataset S3). It is currently unclear whether these individual dndF and dndH genes are products of PT R-M degradation. However, the observation that the overwhelming majority of PT systems occur as either complete R-M pairs or R−-M+ with no trace of DndFGH indicates that this degradation, if it occurs, might not be a recent event.
A degradation scenario is observed in methylation-based R-M systems, and the occurrence of orphan MTases overwhelms the complete R-M systems (29). Given sufficient time for evolution, orphan MTases achieve one of two fates. First, in the absence of selective pressure for their maintenance, orphan MTases might undergo further degradation and loss. Second, orphan MTases might acquire other cellular functions, such as the control of gene expression by Dam and the regulation of cell cycle progression by CcrM. To determine whether orphan PT modification enzymes have developed alternative physiological functions other than R-M, we profiled the genomic PT sites, global transcriptional changes, and metabolomic alterations in response to PT loss in P. fluorescens pf0-1. In response to PT loss, the PT distribution exhibited no apparent influence on global gene expression. One plausible explanation for the partial PT modification at a given site within a population of cells involves an epigenetic mechanism similar to that observed in a number of phase-variable type I and III R-M systems, in which M components are expressed in a phase-variable manner, leading to epigenetic heterogeneity in the bacterial population (30). For example, the R-M locus retains its function as a typical type III R-M system in some strains of Haemophilus influenzae, whereas in others, it has evolved from a DNA restriction system to coordinate the expression of multiple genes via differential methylation of the genome (31). The fact that PT modification does not occur consistently at a given site in a population of cells raises the possibility of multiple phenotypically distinct pf0-1 cells with heterologous PT patterns. Each PT pattern might influence a different set of genes, although these differentially altered genes are not reflected in an RNA-Seq analysis, which reveals transcriptional changes in a population of cells. This hypothesis is supported by the in vitro transcriptional assay, in which PT sites induced changes in transcription efficiency.
The global metabolic profile indicated that the PT-deleted mutant suffered intracellular oxidative stress based on the observations of proline accumulation, lipid metabolite alterations, and significant decreases in ROS scavengers. The alteration in metabolic profiles is reminiscent of the reducing chemical property of the PT structure. Sulfur replacement enables an enhancement in the tolerance of DNA to ROS, as demonstrated through the following observations: (i) Upon H2O2 treatment, oxidative damage occurred at a 2.6-fold lower rate in plasmid DNA with PT modifications than in non-PT DNA, and (ii) S. lividans exhibited a two- to 10-fold higher survival rate than the PT-deficient mutant when treated with 20 mM H2O2 (32, 33). However, Dai et al. (32) noticed no significant transcriptional increases in antioxidant genes (e.g., catalase, alkyl hydroperoxide reductases, organic hydroperoxide resistance genes) following treatment with peroxides in S. lividans. Our metabolomics data indicated the contribution of PT to the maintenance of cellular redox homeostasis, the loss of which led to an increase in cellular oxidative stress. Consequently, the bacterial host reconfigures its metabolic networks to fend off excessive ROS rather than induce antioxidant genes.
Additionally, to test the possibility that PT modifications in PT R−-M+ systems have evolved to pair with other nucleases and thereby generate new PT R-M modules, we searched for nucleases in close proximity (1 bp–5 kb) to dndBCD or dndBCD-dndFGH. Sixty-three (10%) of 615 PT R−-M+ systems have encoded nucleases, including HNH family nucleases, uma2 family endonucleases, and unclassified restriction nucleases, within the defined neighborhood, whereas 168 (23%) of 734 complete PT R-M pairs have surrounding nucleases (Dataset S1). These data show no apparent enrichment of conserved nucleases in PT R−-M+ systems. It is not clear at this point whether the neighboring endonucleases in PT R−-M+ systems target PT sites or whether they have evolved to be PT-dependent endonucleases (e.g., ScoMcrA), while the M genes become defective (34). Interestingly, we also observed the occurrence of solitary dndFGH in 82 bacterial strains without an adjacent dndBCD or other potentially alternative DNA modification enzymes (Dataset S3). It is currently unclear whether solitary DndFGHs are still functional, and future investigations are needed to address this question.
In summary, we discovered that DNA PT systems are widely distributed in diverse bacterial genomes, and nearly half of them are present as PT R−-M+, and thus not associated with the cognate restriction enzyme DndFGH. In addition to being a component of defense systems, DNA PT modification has evolved and acquired alternative cellular functions, including the epigenetic control of gene transcription and the maintenance of cellular redox homeostasis, an indication of the versatility of PT.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains, plasmids and primers used in this study are listed in SI Appendix, Tables S7 and S8. The E. coli and P. fluorescens strains were grown in Luria-Bertani (LB) broth at 37 °C and 28 °C, respectively. When necessary, the antibiotics ampicillin and kanamycin were used at final concentrations of 100 μg/mL and 50 μg/mL, respectively. In addition, the medium used to grow the donor E. coli strain WM3064 was supplemented with 10 μg/mL diaminopimelic acid (DAP) (Sigma).
Construction of the dndBCDE In-Frame Deletion Mutant.
In-frame deletions of P. fluorescens pf0-1 genes were created using the pSR47s suicide plasmid and a two-step homologous recombination procedure. First, fragments upstream and downstream of the target gene were generated by PCR using the primer pairs spf01/spf02 and spf03/spf04 (SI Appendix, Table S8). Second, the recombinant fragment was amplified using a mixture of the upstream and downstream PCR products, which overlapped by ∼40 bp, as the templates and spf01/spf04 as the primers. The amplicon was inserted into the pEASY-Blunt Zero cloning vector, resulting in pWHU3345. After sequencing, the correct recombinant fragments were released from pWHU3345 by NotI digestion and cloned into pSR47s to generate pWHU3350, which was used to make in-frame deletions of dndBCDE. pWHU3350 was transformed into the DAP auxotroph E. coli WM3064 as a donor and was then conjugated to P. fluorescens pf0-1 as the recipient. Transconjugants were picked from plates and purified by serial streaking. The purified transconjugants were used for double-crossovers on an LB agar plate supplemented with 5% sucrose at 28 °C to obtain the dndBCDE in-frame deletion mutant of TT-5. The absence of dndBCDE in TT-5 was further confirmed by PCR with the primer pair spfup/spfdown.
Illumina RNA-Seq and Data Analysis.
Total RNA was isolated from Pseudomonas bacteria in the exponential growth phase (OD600 = 1.0) using RNAprotect Bacteria Reagent (Qiagen) and an RNeasy Mini Kit (Qiagen). The mRNA enrichment, RNA fragment interruption, adapter addition, size selection, PCR amplification, and RNA-Seq were performed at the Beijing Genome Institute (35). The general methods for RNA sequencing, information processing, and analysis were described previously (7).
The raw reads were filtered via the following three steps to generate clean reads: (i) removal of reads with adapters, (ii) removal of reads with more than 10% unknown bases, and (iii) removal of low-quality reads (more than 50% low-quality bases). In total, we obtained 14,508,066 clean reads from the TT-5 mutant, containing 1,305,725,940 bp, among which 14,398,801 (99.25%) reads were successfully mapped to the reference P. fluorescens pf0-1 genome using BWA (v0.7.10-r789) software. The numbers of clean reads with perfect homology and with unique matches are 12,135,231 (83.64%) and 14,190,690 (97.81%), respectively. From wild-type pf0-1, a total of 14,502,382 clean reads were generated, containing 1,305,214,380 bp and 14,395,562 (99.26%) reads mapped to the reference genome. The numbers of reads with perfect homology and with unique matches are 12,142,920 (83.73%) and 14,188,041 (97.83%), respectively. Reads per kilobase of gene per megabase of library size were calculated according to a protocol from Chepelev et al. (36). Genes with an FDR ≤ 0.001 and fold change ≥ 2 were identified as being differentially expressed. Gene ontology category and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were then performed on the differentially expressed genes.
In Vitro Transcription.
The PT-modified DNA templates (∼300 bp) used for the transcriptional assay consisted of two segments, the chemically synthesized oligonucleotides with PT modifications and unmodified regular PCR products (Fig. 4A). First, the oligonucleotides (55–86 bp) with PT were chemically synthesized at TSINGKE Biological Technology and annealed in buffer containing10 mM Tris (pH 8.0), 1 mM EDTA, and 50 mM NaCl. The rest of the DNA template segment without PT was then generated by PCR amplification. The PCR products were enzymatically digested to produce a cohesive end and ligated with annealed oligonucleotides possessing the complementary end, yielding the full-length DNA templates. Non–PT-modified DNA templates were constructed using the same procedure as that used for the control. Full-length DNA templates were gel-purified and quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific). To validate the quantitation, real-time PCR was concurrently conducted using SYBR Green Master Mix (Vazyme) with a 7900HT Fast Real-Time PCR System (Applied Biosystems) to determine the threshold cycle (Ct) of the same amount of PT- and non–PT-modified DNA template (ΔCt ≤ 0.3). One hundred nanograms of DNA template was transcribed in a 20-μL reaction mixture containing 0.5 units of E. coli RNA polymerase, holoenzyme (New England Biolabs), 4 μL of 5× E. coli RNA polymerase reaction buffer, 0.5 mM of each nucleotide (ATP, CTP, GTP, and UTP), and 20 units of RNase inhibitor (Yeasen). The in vitro transcription was performed at 37 °C for 10 min and then increased to 85 °C for 10 min to terminate the reaction in a Bio-Rad c1000 thermal cycler.
RT and Quantitative Real-Time PCR.
Two microliters of the in vitro transcriptional mRNA product was treated with DNase I for 1 h at 37 °C and then subjected to RT using a RevertAid first-strand cDNA synthesis kit (Thermo Scientific). The sequence 5′-AATTGGTGACACTCAGGCAC-3′ was fused in the 5′ terminus of the reverse primer to serve as a tag to allow differentiation from the template DNA (Fig. 4). RT was performed according to the manufacturer’s instructions. The resulting cDNA was then analyzed by real-time PCR to determine the Ct value. Real-time PCR was performed with a program consisting of 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The comparative 2−ΔCt method was used to determine the transcriptional changes affected by PT (37).
Tree Bootstrap and Topological Difference.
To quantitatively measure the topological similarity/difference between two phylogenetic trees, we calculated a series of metrics of a tree. Specifically, we defined the distance between two tips or leaves (bacterial strains in our study), A and B, of a tree x as
where n is the least number of nodes that have to be passed when going from A to B along the tree. The total internal “variation” V of a tree x was represented by the vectors, whose elements indicate the distance of a tip pair in that tree, namely,
In this equation, xi (i = 1, 2, …, m) is the distance between two tips of the tree, and m = C2l (choose 2 from l items), where l is the total number of tips in the tree. Under this setting, the topological structure of a tree can be represented uniquely by its total internal variation vector V. Thus, the similarity/difference of the topological structures of two trees, x and y, was measured by the Manhattan distance between their total internal variations, as determined by the following formula:
Trees that are more topologically similar have a shorter distance D, and vice versa. Theoretically, when two trees have the same topological structure, D = 0. However, calculating the maximum value of D of two trees that are completely topologically different is difficult, particularly if they have a high total number of tips. In our study, to overcome this problem and to measure how far the similarity between the BCD tree and the FGH tree is from randomness, we bootstrapped two trees 331 times by randomly reshuffling all of the tips of each tree while maintaining their topological structures. The total internal variations V of simulated trees and their distances D to the original BCD and FGH trees were calculated and compared with D(BCD)(FGH) to calculate the P value for the topological similarity with our original trees.
Bioinformatics Analysis.
Protein sequences and genomic location information were downloaded from NCBI databases using E-Utilities (https://www.ncbi.nlm.nih.gov/books/NBK25501/), and all analytical computations were performed using R-3.2.2 (38). A tanglegram was constructed using Dendroscope 4.4.4 (39), and Cluster 3.0 (40) was used to perform k-means clustering.
Metabolic Profiling and Data Analysis.
Metabolites were extracted from 50 mg of cell pellets with 1 mL of methanol. Ten microliters of 2.9 mg/mL DL-o-chlorophenylalanine was used as an internal standard. In general, the samples were vortexed for 30 s, homogenized for 2 min, ultrasonicated for 30 min, and centrifuged at 13,800 × g and 4 °C for 10 min. Six biological replicates of each bacterial strain were used. Analysis of metabolites was performed on the ACQUITYTM UPLC-ESI-Q-TOF-MS platform using a Waters ACQUITY UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm), and the column was maintained at 40 °C. Elution was performed at a flow rate of 0.3 mL⋅min−1 with a gradient of buffer A (water with 0.1% formic acid) and buffer B (acetonitrile with 0.1% formic acid) according to the following profile: 5% buffer B for 2 min, followed by a gradient to 95% buffer B over 10 min, maintenance at 95% buffer B for 3 min, and reequilibration in 5% buffer B for 3 min. The following parameters were optimized for maximal sensitivity in + and − ESI mode in separate runs: source temperature, 120 °C; desolvation temperature, 350 °C; cone gas flow rate, 50 L⋅h−1; desolvation gas flow rate, 600 L⋅h−1; collision energy, 10–40 V; ion energy, 1 V; scan time, 0.03 s; interscan time, 0.02 s; capillary voltage, 1.4 kV in the positive mode and 1.3 kV in the negative mode; and sampling cone, 40 V in the positive mode and 23 V in the negative mode.
The generated data were processed using a Waters MassLynx 4.1 MS workstation, which provides automated peak detection based on peak alignment and normalization to the total peak area. SIMCA-P (13.0; Umetrics) was used for multivariate statistical calculations and plotting. Accurate masses of features presenting significant differences were searched against the METLIN and LIPIDMAPS databases.
Supplementary Material
Acknowledgments
We thank Yizhou Zhang for preparing Fig. 4 and Yuquan Xu and Ping Xu for providing the vectors. This work was supported by grants from the National Science Foundation of China (Grants 31720103906 and 31520103902), the 973 program of the Ministry of Science and Technology (Grant 2013CB734003), and the Young One Thousand Talent program of China. The work of S.Y.L. was further supported by the Systems Metabolic Engineering for Biorefineries Program (Grants NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and Information and Communications Technology through the National Research Foundation of Korea.
Footnotes
The authors declare no conflict of interest.
Data deposition: The pf0-1 SMRT sequencing data obtained using the PacBio RSII instrument have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP127485). The RNA-sequencing data are available in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE110489).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721916115/-/DCSupplemental.
References
- 1.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014;24:374–387. doi: 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- 3.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 5.Zou X, et al. Genome engineering and modification toward synthetic biology for the production of antibiotics. Med Res Rev. 2018;38:229–260. doi: 10.1002/med.21439. [DOI] [PubMed] [Google Scholar]
- 6.Cao B, et al. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Mol Microbiol. 2014;93:776–785. doi: 10.1111/mmi.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gan R, et al. DNA phosphorothioate modifications influence the global transcriptional response and protect DNA from double-stranded breaks. Sci Rep. 2014;4:6642. doi: 10.1038/srep06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An X, et al. A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. PLoS One. 2012;7:e51265. doi: 10.1371/journal.pone.0051265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He W, et al. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Sci Rep. 2015;5:12368. doi: 10.1038/srep12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao F, Xu T, Zhou X, Deng Z, You D. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009;583:729–733. doi: 10.1016/j.febslet.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 11.Hu W, et al. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Res. 2012;22:1203–1206. doi: 10.1038/cr.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong W, Zhao G, Yu H, He X. Interactions of Dnd proteins involved in bacterial DNA phosphorothioate modification. Front Microbiol. 2015;6:1139. doi: 10.3389/fmicb.2015.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, et al. Convergence of DNA methylation and phosphorothioation epigenetics in bacterial genomes. Proc Natl Acad Sci USA. 2017;114:4501–4506. doi: 10.1073/pnas.1702450114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci USA. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao B, et al. Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat Commun. 2014;5:3951. doi: 10.1038/ncomms4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X, et al. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65:1034–1048. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 18.Heusipp G, Fälker S, Schmidt MA. DNA adenine methylation and bacterial pathogenesis. Int J Med Microbiol. 2007;297:1–7. doi: 10.1016/j.ijmm.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plumbridge J. The role of dam methylation in controlling gene expression. Biochimie. 1987;69:439–443. doi: 10.1016/0300-9084(87)90081-2. [DOI] [PubMed] [Google Scholar]
- 23.Szabados L, Savouré A. Proline: a multifunctional amino acid. Trends Plant Sci. 2010;15:89–97. doi: 10.1016/j.tplants.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Liang X, Zhang L, Natarajan SK, Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal. 2013;19:998–1011. doi: 10.1089/ars.2012.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Zhang J, Zhao B, Zhao-Wilson X. Quinic acid could be a potential rejuvenating natural compound by improving survival of Caenorhabditis elegans under deleterious conditions. Rejuvenation Res. 2012;15:573–583. doi: 10.1089/rej.2012.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung TM, et al. Antioxidant activity of caffeoyl quinic acid derivatives from the roots of Dipsacus asper Wall. J Ethnopharmacol. 2006;108:188–192. doi: 10.1016/j.jep.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 27.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alhasawi A, Castonguay Z, Appanna ND, Auger C, Appanna VD. Glycine metabolism and anti-oxidative defence mechanisms in Pseudomonas fluorescens. Microbiol Res. 2015;171:26–31. doi: 10.1016/j.micres.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Seshasayee AS, Singh P, Krishna S. Context-dependent conservation of DNA methyltransferases in bacteria. Nucleic Acids Res. 2012;40:7066–7073. doi: 10.1093/nar/gks390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 31.Fox KL, et al. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–5252. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai D, et al. DNA phosphorothioate modification plays a role in peroxides resistance in Streptomyces lividans. Front Microbiol. 2016;7:1380. doi: 10.3389/fmicb.2016.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, et al. DNA backbone sulfur-modification expands microbial growth range under multiple stresses by its anti-oxidation function. Sci Rep. 2017;7:3516. doi: 10.1038/s41598-017-02445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu G, et al. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin N, et al. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep. 2014;4:5467. doi: 10.1038/srep05467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.R Development Core Team 2015. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.2.2.
- 39.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.