Abstract
Chronic lymphocytic leukemia (CLL) is a heterogeneous B-cell cancer exhibiting a wide spectrum of disease courses and treatment responses. Molecular characterization of RNA and DNA from CLL cases has led to the identification of important driver mutations and disease subtypes, but the precise mechanisms of disease progression remain elusive. To further our understanding of CLL biology we performed isobaric labeling and mass spectrometry proteomics on 14 CLL samples, comparing them with B-cells from healthy donors (HDB). Of 8694 identified proteins, ∼6000 were relatively quantitated between all samples (q<0.01). A clear CLL signature, independent of subtype, of 544 significantly overexpressed proteins relative to HDB was identified, highlighting established hallmarks of CLL (e.g. CD5, BCL2, ROR1 and CD23 overexpression). Previously unrecognized surface markers demonstrated overexpression (e.g. CKAP4, PIGR, TMCC3 and CD75) and three of these (LAX1, CLEC17A and ATP2B4) were implicated in B-cell receptor signaling, which plays an important role in CLL pathogenesis. Several other proteins (e.g. Wee1, HMOX1/2, HDAC7 and INPP5F) were identified with significant overexpression that also represent potential targets. Western blotting confirmed overexpression of a selection of these proteins in an independent cohort. mRNA processing machinery were broadly upregulated across the CLL samples. Spliceosome components demonstrated consistent overexpression (p = 1.3 × 10−21) suggesting dysregulation in CLL, independent of SF3B1 mutations. This study highlights the potential of proteomics in the identification of putative CLL therapeutic targets and reveals a subtype-independent protein expression signature in CLL.
Chronic lymphocytic leukemia (CLL)1, the most common adult leukemia in the western world, is a CD5+ B-cell neoplasm with a heterogeneous clinical course (1, 2). B-cell receptor (BCR) signaling plays a role in the pathogenesis of CLL, and drugs targeting this pathway are revolutionizing CLL treatment (3). Indeed, the mutational status of the immunoglobulin heavy-chain variable-region gene (IGHV) within the BCR largely correlates with disease outcome. CLL cases with unmutated V-genes (U-CLL) typically have a progressive disease with patients often requiring early treatment, in contrast cases with mutated V-genes (M-CLL) are more indolent, and those presenting with a low tumor burden often never require treatment (4).
CLL transcriptomics analyses have identified minimal differences in gene expression between subtypes such as U- and M-CLL suggesting that common mechanisms elicit CLL pathogenesis (5, 6). More recently, the DNA methylation profile of CLL cases was shown to closely reflect that of the proposed cell of origin, namely memory B-cells (MBC) and naive B cells (NBC) for M-CLL and U-CLL, respectively. Interestingly, both studies identified a third epigenetic CLL subgroup with an intermediate methylation signature enriched within M-CLL with between 95 and 98% IGHV somatic mutations. These three CLL epitypes exhibit different clinico-biological features, with the MBC-like CLL cases exhibiting a more indolent clinical course (7–10).
Although no single aberration appears to drive disease development, many recurring gene mutations and chromosome abnormalities have been described in CLL, and several have prognostic and/or predictive significance. Deletion of 17p and 11q which results in the loss of TP53, baculoviral IAP repeat-containing 3 (BIRC3) or ataxia-telangiectasia mutated serine/threonine kinase (ATM) respectively, are frequently associated with TP53 and ATM mutations on the remaining allele and poor outcome following chemo-immunotherapy (11, 12). In contrast the most frequent cytogenetic abnormality in CLL, deletion of 13q, results in increased expression of the anti-apoptotic protein Bcl-2, largely because of loss of miRNA15 and miR16–1, and is associated with a good prognosis, particularly in M-CLL.
Other recurrent mutations also influence disease progression and treatment response. Next generation sequencing studies have confirmed that mutations of splicing factor 3B subunit 1 (SF3B1) (13) and neurogenic locus notch homolog protein 1 (NOTCH1), a transmembrane receptor and transcriptional regulator determining cell fate (14), are the most frequent recurring mutations in CLL, with an incidence of ∼18% and 12%, respectively, at the time of initial treatment. Mutations in either gene are associated with a poorer outcome following chemo or chemo-immunotherapy and NOTCH1 mutations are also predictive of a poor response to chemotherapy plus anti-CD20 antibody combinations (15, 16).
SF3B1 is a spliceosome component with a role in the regulation of pre-mRNA intron excision. Heterozygous missense mutations of the C-terminal HEAT domain are the most frequent alteration to SF3B1, impacting spliceosomal function (17). Indeed, SF3B1 mutation has been shown to induce large numbers of aberrantly spliced and altered gene products in CLL (18). The frequency of SF3B1 mutations and the resulting changes to spliceosomal activity suggest that dysregulation of the spliceosome plays a prominent role in CLL pathogenesis.
Liquid chromatography with mass spectrometry (LC-MS) proteomics provides an effective means of establishing global differential protein expression profiles. Several studies have previously employed MS proteomics in the analysis of CLL (recently reviewed (19, 20)); with the latest, a characterization of 9 U-CLL versus 9 M-CLL, identifying 3521 and relatively profiling 2024 proteins, of which ∼100 proteins were differentially expressed (±1.5-fold change, p < 0.05) (21). Several earlier studies provided limited proteome coverage but suggested a number of markers of poor prognosis, such as nucleophosmin, PDCD4 and TCL1 (22–27). Technological and methodological advances such as Orbitrap technology, isobaric labeling and two dimensional chromatography have greatly improved proteomics coverage, leading to the first comprehensive drafts of the human proteome (28–30).
Our current discovery-stage study has applied isobaric labels and LC-MS proteomics to the characterization of isolated B-cell material from 3 healthy donors and 14 CLL patients. CLL samples were selected to include a range of clinically relevant CLL subtypes associated with poor prognosis versus healthy donor B-cells with the aim of assessing CLL-specific differential protein expression. The resulting quantitative proteomes identified a strong signature common to CLL, highlighting several potential therapeutic targets and suggesting mechanisms, such as spliceosome overexpression, contributing to pathogenesis.
MATERIALS AND METHODS
Materials
SDS, HPLC and LC-MS grade ACN and formic acid (FA), methyl methanethiosulfonate (MMTS), tris(2-carboxyethyl)phosphine (TCEP), triethylammonium bicarbonate (TEAB), DMSO, hydroxylamine were purchased from Sigma, St. Louis, Missouri. EasySep human B-cell enrichment kits without CD43 depletion were purchased from Stem Cell Technologies, Cambridge, UK. Tandem mass tags (TMT) 10-plex isobaric labeling reagents and 100 μl C18 solid phase extraction tips were purchased from Thermo Scientific, Waltham, MA. Proteomics grade trypsin was purchased from Roche, Basel, Switzerland. Fetal calf serum (FCS) was purchased from Lonza, Basel, Switzerland, lymphoprep medium from Axis-Shield Diagnostics, Dundee, UK, and anti-human antibodies against CD3 (OKT3) and CD19 (HIB19) from Biolegend, San Diego, CA.
Isolation of Healthy Donor and CLL B-cells From Clinical Samples
Ethical approval for the use of human samples was granted under REC references 228/02/t (Southampton) and 06/Q2202/30 (Bournemouth). Peripheral blood mononuclear cells (PBMCs) were derived from healthy donors and CLL patients by Southampton Blood Services or Bournemouth Tissue Bank, respectively. PBMCs were isolated from whole blood or blood cones by density gradient centrifugation as previously described (3). PBMCs were frozen at 5 × 107 cells/ml in FCS containing 10% v/v DMSO. Healthy donor and CLL patient PBMCs were defrosted, washed and B-cells isolated by negative selection using the EasySep human B-cell enrichment kit without CD43 depletion, according to the manufacturer's instructions. The enriched B-cells were washed 3 times in excess PBS. B-cell purity was assessed by immunostaining with CD19 and CD3 by flow cytometry and yielded at least 90% purity in all samples (supplemental Table S1). Isolated cell pellets were snap frozen and stored in liquid nitrogen before lysate preparation.
Experimental Design and Statistical Rationale
CLL samples were selected to include a range of clinically relevant CLL subtypes associated with poor prognosis. Samples were assigned evenly across two batches of analyzed samples, to accommodate the capacity limits of the isobaric labels (supplemental Table S1). To simultaneously analyze and provide relative quantitative comparability of 14 CLL proteomes, samples were assigned to two tandem mass tag (TMT) 10-plex label sets. Three healthy donor B-cell (HDB) samples were used as both biological and inter-experimental bridging controls. HDB peptides were prepared and TMT-labeled before bifurcation and allocation to each 10-plex. The selected samples included (non-discretely) 5 Trisomy 12 cases, 3 CD38+ (>99%) cases, 8 unmutated IGHV cases, 5 NOTCH1-mutant cases and 5 SF3B1-mutant cases (supplemental Table S1).
To minimize batch effects, reduce technical noise and disregard inconsistent observations, a stringent approach considering only the smallest of the CLL to HDB fold changes was adopted; detailed in Quantitative and Statistical Analysis of MS Data below. CLL:HDB ratios were evaluated by an FDR-corrected 1 sample t test and compared alongside a value termed the regulation score (Rs) (31), a more robust measure than an average, also detailed below. With 14 samples, in those instances where variation was as low as 25%, a fold change of ∼1.25 could be concluded, based on the power analysis calculations presented by Levin (33). Use of standard deviation as the denominator in the regulation score adjusted for variation on a case by case basis. At a threshold of Rs>0.3, the lowest fold change defined as significantly regulated was 1.25, observed with an average variation of 18%.
Sample and Peptide Preparation and Labeling for LC-MS
Snap frozen cell pellets were lysed on ice by trituration with a 23 gauge needle in 0.5 m TEAB containing 0.05% w/v SDS. Disrupted cells were further sonicated on ice and lysates cleared by centrifugation at 16,000 g for 10 min at 4 °C. Protein concentration was determined by a Direct Detect Spectrometer (Millipore, Billerica, MA). One hundred μg of cell lysate (200 μg for each HDB sample, for use across two 10-plex experiments) was reduced with 50 mm TCEP and alkylated with 200 mm MMTS, before digestion overnight at RT with a 30:1 ratio of proteomics grade trypsin. Peptides were incubated with TMT 10-plex isobaric tags according to the manufacturer's instructions. CLL sample peptides were assigned to two TMT 10-plex label sets, A and B, respectively, as follows: 128N A,B: 3999, 2886; 128C A,B: 2497, 2483; 129N A,B: 568, 625; 129C A,B: 653, 4140; 130N A,B: 3405, 1194; 130C A,B: 3156, 1125; 131 A,B: 4621, 2459. For the healthy donors, all 200 μg of B-cell peptides were labeled in the same reaction with labels 126 A+B, 127N A+B and 127C A+B for donors 1, 2 and 3, respectively. Peptides were lyophilized at RT by vacuum concentration and reconstituted in 2% v/v ACN, 0.1% v/v FA and pooled to their respective 10-plex A and 10-plex B, with half the labeled material from each of the three HDBs being assigned to each 10-plex.
Peptide Prefractionation
Peptides were cleaned using three repetitions of solid phase extraction using 100 μl C18 zip tips according to manufacturer's instructions. Peptides were lyophilized and reconstituted in 2% v/v ACN, 0.1% v/v NH4OH, and resolved using high-pH RP C8 chromatography (150 mm × 3 mm ID x 3.5 μm particle, XBridge) (Waters, Milford, MA) at 300 μl/min with a LC-20AD HPLC system (Shimadzu, Kyoto, Japan) maintained at 30 °C, using the mobile phases (MP); A - 99.9% H2O, 0.1% NH4OH, B - 99.9% ACN, 0.1% NH4OH. The 120 min gradient was as follows; 0 min; 2% B, 10 min; 2% B, 75 min; 30% B, 105 min; 85% B, 120 min; 2% B. Fractions were collected in a peak-dependent manner and individually lyophilized. The top 25 high-abundance peptide fractions—reproducibly between both experiments—were selected for individual analysis and the remaining fractions were orthogonally concatenated.
Peptide Fraction Analysis by LC-MS/MS
Lyophilized peptide fractions were individually reconstituted in 2% ACN, 0.1% FA and ∼500 ng of peptides loaded by a Dionex Ultimate 3000 (Thermo Scientific) and analyzed by LC-MS/MS, described previously (31). In summary, peptides were trapped by C18 and eluted over a reverse phase gradient of 8 h (the richest fraction), 5 h (2nd–4th richest), 4 h (6th–13th richest), and 3 h (remaining and pooled). Gradients for each group (8, 5, 4 and 3 h, respectively) were as follows: 2% (0 mins), 22% (350, 200, 160, 110 mins), 45% (415, 250, 200, 145 mins), 85% (465, 285, 225, 165 mins) organic phase (94.9% ACN, 5% DMSO, 0.1% FA) in aqueous phase (2% ACN, 5% DMSO, 0.1% FA). The total MS time for each proteome was ∼210 h.
Peptides were analyzed with an LQT-Orbitrap Elite Velos Pro hybrid mass spectrometer (Thermo Scientific). MS analysis of eluting peptides was conducted between 350 and 1900 m/z at 120,000 mass resolution. The top 12 + 2 and +3 precursor ions per MS scan (minimum intensity 1000) were characterized by tandem MS with high-energy collisional dissociation (HCD) (30,000 mass resolution, 1.2 Da isolation window, 40 keV normalized collision energy) and CID (ion trap MS, 2 Da isolation window, 35 keV). Additionally, the DMSO ion at 401.922718 (MS1) (32) and the TMT-H+ ion at 230.170422 (MS2) were used as lockmasses and the MS was calibrated weekly.
MS Data Processing
Target-decoy searching of raw spectra data was performed with Proteome Discoverer software version 1.4.1.14 (Thermo Scientific). Spectra were subject to a two stage search, both using SequestHT (version 1.1.1.11), with Percolator used to estimate FDR with a threshold of q ≤ 0.01. In both searches, fragment ion mass tolerances of 0.02 Da and 0.5 Da were used for HCD and CID spectra, respectively. Fixed modifications of Methythio (C), TMT (K and N terminus) were used, searching for tryptic peptides. The first allowed only a single missed cleavage, minimum peptide length of 7, precursor mass tolerance of 5 ppm, no variable modifications and searched against the human UniProt Swissprot database (downloaded January 2015; 20,159 protein sequences). The second search used only spectra with q > 0.01 from the first search, allowed 2 missed cleavages, minimum peptide length of 6, searched against the human UniProt trembl database (downloaded 01/15, 67,812 protein sequences), precursor mass tolerance of 10 ppm and a maximum of 2 variable (1 equal) modifications of; TMT (Y), oxidation (M), deamidation (N,Q) or phospho (S,T,Y). PhosphoRS was used to predict the probability of specific phosphorylated residues. Reporter ion intensities were extracted from non-redundant PSMs with a tolerance of 20 ppm. Data from the two 10-plex experiments was searched separately and combined for protein grouping by Proteome Discoverer. The raw data and processed outputs have been deposited to the ProteomeXchange Consortium (33) via the PRIDE partner repository with the dataset identifier (PXD002004).
Quantitative and Statistical Analysis of MS Data
Log2 (ratios) were generated describing each CLL sample relative to the three HDB bridging controls (supplemental Table S2). To select for only the most robust and consistent findings, the ratio of CLL:HDB control was assessed for all 3 HDB controls and, rather than a potentially misrepresentative average value, the ratio with the lowest fold change was selected. Findings with both up and downregulation for a protein among the 3 CLL:HDB ratios were rejected. For example: for fold-changes of 1.4, 1.5 and 1.8, 1.4 was selected; for fold-changes of −2.2, −1.7, −1.9 -> −1.7 was selected; for fold-changes of 1.5, −1.9, 1.3 -> a value of 0 was taken.
To reduce ratio compression, peptide spectrum match data for proteins (q<0.01) were exported from Proteome Discoverer and submitted to Statistical Processing for Isobaric Quantitation Evaluation (SPIQuE) at spiquetool.com. This method weighted the contributions of each PSM quantitation to a protein's quantitation based on PSM features (manuscript in preparation). For example, high-intensity peptides with low isolation interference were given a greater weighting factor, as previously applied (31).
Because of the substantial differences in non-B-cells present in the HD and CLL PBMCs, contamination emerged in the HDB samples as a number of downregulated proteins. To accommodate for the contamination, the analyses primarily focused on proteins overexpressed, and therefore specifically attributable to CLL. As the most apparent source of contamination in the proteomics results were platelet-derived proteins, a filtering list consisting of the 1000 most abundant platelet proteins was generated from a prior proteomics analysis of platelets, defined by copy number (described previously (34)). Of these 1000 proteins, 194 were observed with apparent overexpression in the HDB samples which were subsequently removed from the dataset. Additionally, the WT CLL sample 4621 appeared as an outlier and was therefore excluded from further analyses.
For the minimally deviated log2 (ratios), differential expression in CLL, relative to HDB, was defined by an FDR-corrected one sample t test, where p < 0.05 and the regulation score (Rs), where Rs>0.3 or >–0.3. Rs provided a single, robust measure, representative of both the magnitude and consistency of differential expression (Rs = mean/(standard deviation +1)). For proteins with no, or inconsistent differential expression, the Rs tended toward 0.
Bioinformatics Analyses
Proteins reaching the thresholds outlined above were submitted to either Ingenuity Pathway Analysis (IPA) or Database for Annotation, Visualization and Integrated Discovery (DAVID). For DAVID analyses, the default settings were used for pathway and gene ontology (GO) term enrichment, with Benjamini-corrected p values of <0.05 considered significant. For IPA analyses, default settings were used. Annotations of biomarkers and drug targets were conducted by IPA. Hierarchical clustering was conducted using log2 (ratios) and Cluster 3.0 (University of Tokyo, Human Genome Centre) clustering based upon Euclidian distance and complete linkage. Java TreeView (version 1.1.6r2) was used to visualize the clustering. Chromosome enrichment analysis was performed in DAVID, using default setting and visualized using Ensembl geneome browser.
Flow Cytometry
Cells were stained with either the manufacturer's recommended concentration, or 10 μg/ml, of in-house antibody for 30 min in the dark at 4 °C, washed and analyzed by flow cytometry with a FACScalibur (BD Biosciences, Franklin Lakes, NJ) (35, 36).
Western Blotting
Independent cohorts of isolated B-cells and CLL cells were washed in cold PBS and lysed in RIPA buffer (150 mm NaCl, 1% Triton X-100, 0.5% Deoxycholate, 0.1% SDS, 50 mm Tris, pH 8). Protein content was quantified against a BSA standard curve using a standard Bradford assay (BioRad, Hemel Hempstead, UK). 40 μg protein was loaded into 10% Bolt Novex gels (Thermo Fisher) and transferred to a methylcellulose membrane (GE Lifesciences, UK) by standard techniques. Membranes were probed with anti-CKAP4 (G1/296, 1:500 dilution, Enzo Life Sciences, Farmingdale, NY), anti-INPP5F (H00022876-B01P, 1:500 dilution, Abnova, Taiwan), anti-ROR1 (4102, 1:500 dilution, CST, Danvers, MA), anti-Wee1 (D10D2, 1:1000 dilution, CST), anti-Bcl-2 (610539, 1:500 dilution, BD Biosciences), anti-CD79b (AT105–1, 2 μg/ml, Produced in house).
RESULTS
Quantitative Proteomics of CLL Relative to Healthy Donor B-cell Samples Identifies a Consistent, Reproducible CLL Phenotype
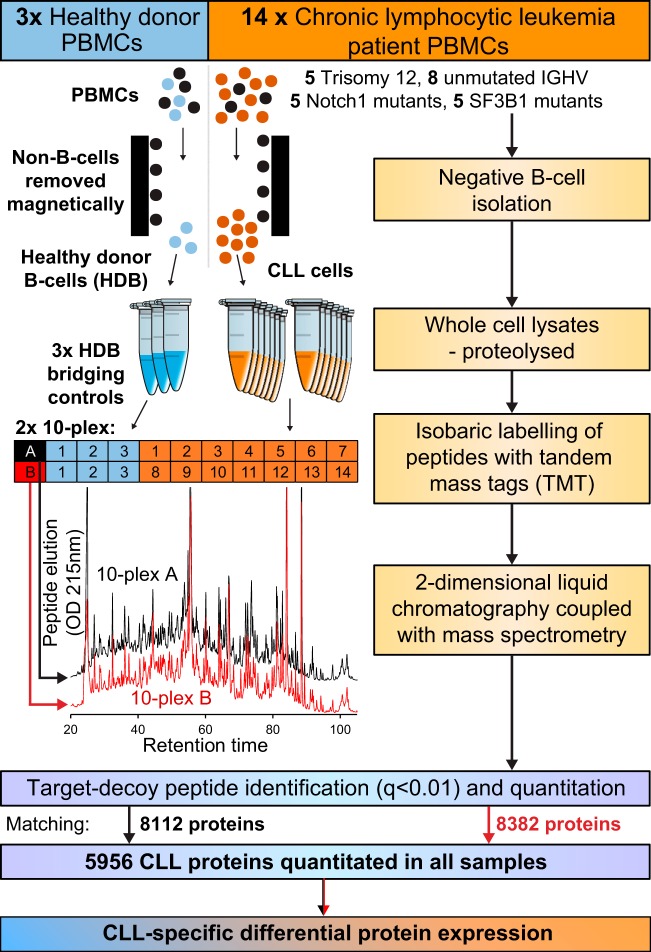
The MS and TMT workflow applied to measure relative differential expression in the proteomes of CLL cells versus HDB controls is summarized in Fig. 1. A total of 8694 proteins were identified (q<0.01) of which 5956 proteins were relatively quantified for all samples (supplemental Table S2).
Fig. 1.
CLL proteomics workflow. PBMCs from 14 CLL patients and 3 healthy donors were subjected to negative B-cell isolation followed by whole cell lysis, reduction, alkylation and trypsin digestion. 100 μg of peptides from each CLL sample was assigned to one of two tandem mass tag (TMT)-labeled 10-plex experiments. 200 μg of each healthy donor B-cell (HDB) protein lysate was labeled and bifurcated to provide bridging controls across the two 10-plex experiments. Each 10-plex was handled and analyzed separately using 2-dimensional liquid chromatography coupled with data-dependent mass spectrometry; in each case, 60 peak-dependent fractions were analyzed. Peptides were identified from mass spectra using target-decoy searching (false discovery rate of <1%). The identified proteins were quantitated from isobaric labels relative to HDB bridging controls and differential expression analyzed to identify CLL-specific differences in protein expression.
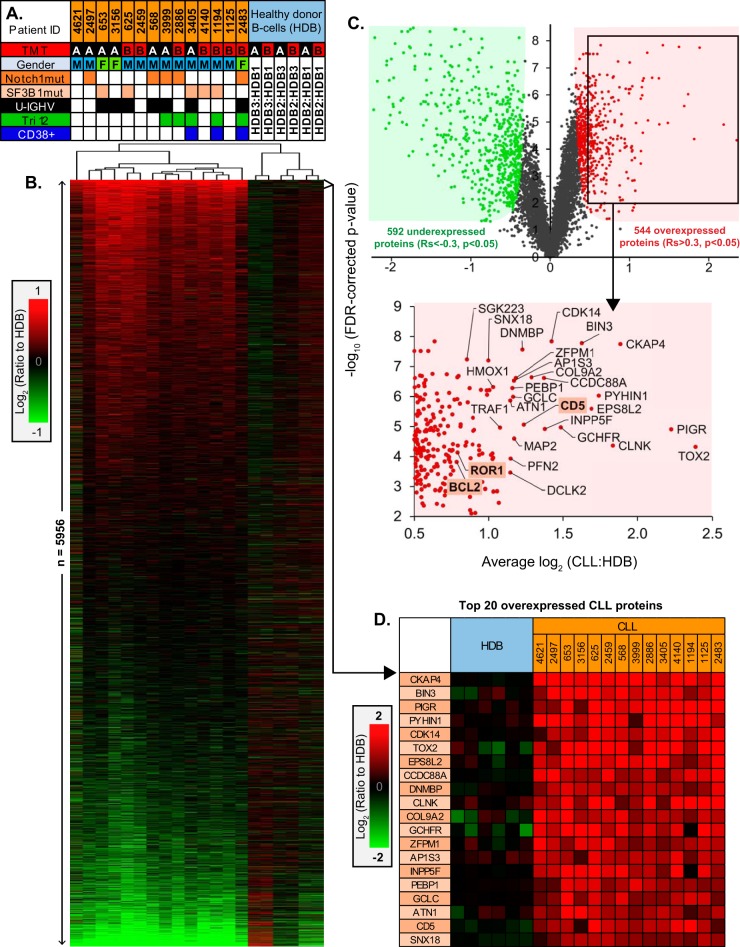
Hierarchical clustering of differential expression across the 14 CLL samples, relative to HDB, highlighted a broad CLL-specific signature with no distinctly clustered subtypes (Fig. 2A, 2B). The reproducibility of this signature was also apparent when comparing the average CLL expression determined in the two distinct 10-plex experiments (R2 = 0.799) and the technical reproducibility between the HDB bridging controls (R2 = 0.797) (supplemental Fig. S1).
Fig. 2.
Quantitative proteomics profiles and characteristics of CLL samples. A, Summary of the major characteristics of the 14 CLL samples, including; the assigned TMT 10-plex experiment (A or B), patient gender, presence of mutations to either NOTCH1 or SF3B1 genes, IGHV mutation status (U-IGHV, solid fill; unmutated IGHV, open), trisomy 12 status (Tri12), CD38+ (>99%) cases. B, Hierarchical clustering of 5956 protein log2 (ratios) for the 14 CLL samples relative to HDB, sorted by regulation score. C, Volcano plot demonstrating proteins observed with significant overexpression (Rs>0.3, p < 0.05) or underexpression (Rs<-0.3, p < 0.05) in CLL versus HDB. A selection of the most overexpressed proteins are annotated, identifying the accepted CLL-specific hallmarks BCL2, ROR1 and CD5. D, The top 20 most consistently overexpressed proteins in CLL identified by the proteomics.
544 significantly overexpressed and 592 significantly underexpressed CLL proteins, relative to HDB, were identified with a regulation score of >0.3 or <-0.3, respectively (Fig. 2C). Among the overexpressed proteins were examples of well-characterized hallmarks of CLL; CD5, BCL2, CD23 and ROR1. Eighteen proteins were identified with a greater overexpression than CD5, several of which were previously undescribed in CLL (Fig. 2D).
In addition to the overexpression of CD5, BCL2, CD23 and ROR1, several other previously reported CLL protein upregulations were present in the data; including ST14/matriptase, CD6, PTPN22, CDKN1B/p27, TRAF1, JUND, BMI1, IKZF3/Aiolos, TOSO, DIABLO/SMAC and SIT1 (Fig. 3A). Additionally, overexpression was also apparent, although more variably, for ZAP-70 and IL4Rα.
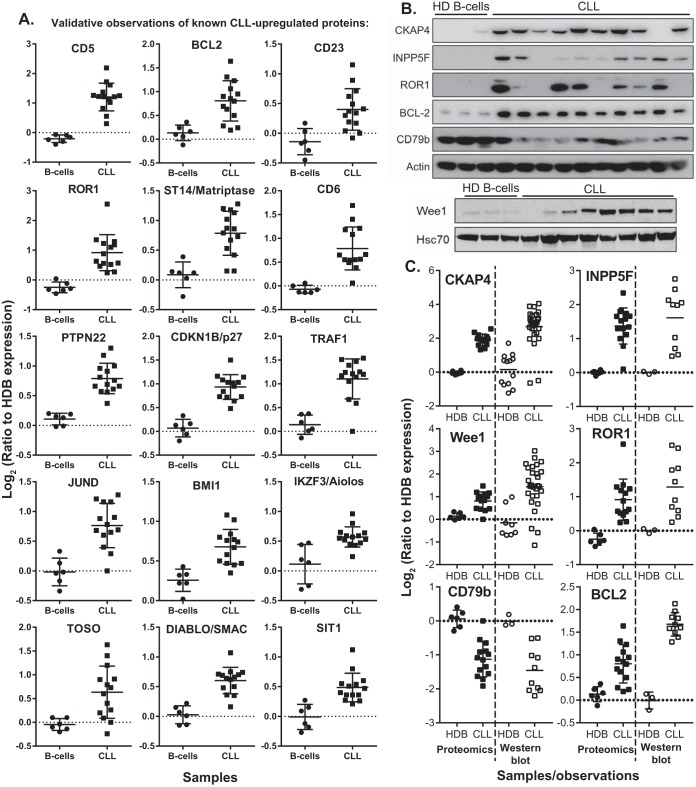
Fig. 3.
Validation of proteins deemed overexpressed by CLL proteomics. A, Proteomics-derived quantitations for proteins previously described with overexpression in CLL, relative to HDB controls. B, Western blot validation of differential protein expression observed in an independent cohort of CLL samples versus HDB controls. C, Comparison between the differential expressions observed for key proteins by Western blotting and proteomics for CLL and HDB controls.
Markers differentiating CLL subtypes were also observed by proteomics. Integrin alpha 4 (CD49d) expression was significantly higher in trisomy 12 cases as previously described (37), IgM expression associated with U-CLL cases (3), CD38 expression was correctly identified as significantly higher in CD38+ (>99%) cases, and the Y-chromosome-encoded proteins (EIF1AY and RPS4Y1) allowed the differentiation of patient genders (supplemental Fig. S1). Subtype specific differences emerging from the proteomics were considered (supplemental Fig. S3, Table S3), however lacked the statistical power for significant findings.
Putative Novel CLL Immunophenotypes and Drug Targets
In addition to confirmation of established characteristics, several novel observations emerged among the most upregulated proteins (Fig. 2C, 2D). Cytoskeleton-associated protein 4 (CKAP4), a cell surface receptor for antiproliferative factor (APF), was the most significantly and consistently upregulated CLL protein (250–590% HDB expression, p = 1.8 × 10−8) with 34 unique peptides identified from over 500 PSMs and has not previously been reported in CLL at the protein level. At the mRNA level, no overexpression was detected (supplemental Fig. S2) (38). To validate this finding, Western blotting was employed, evaluating CKAP4 expression in the proteomics-analyzed samples and an independent cohort of 10 additional CLL samples, relative to HDB controls (Fig. 3B, 3C). CKAP4 was observed to be substantially overexpressed in all but two CLL samples, with little CKAP4 expression observed in healthy B-cells.
INPP5F (SAC2), an inositol 4-phosphatase with a role in AKT signaling, was also found to be among the top 20 overexpressed CLL proteins (200–500% HDB expression in 12/14 samples, p = 1.2 × 10−5). Western blot validation (Fig. 3B, 3C) confirmed this overexpression, but appeared more variable in the independent cohort than observed by proteomics. The G2 checkpoint kinase, Wee1 was observed with significant overexpression in CLL (>175% HDB expression in 9/14 samples, p = 4.3 × 10−5). Annotation by IPA highlighted Wee1 as a potential therapeutic target of the inhibitor MK1775. Again, overexpression was confirmed by Western blotting with the majority of CLL samples having greater Wee1 expression than HDB samples. BCL2, ROR1 and CD79b expression were also evaluated to demonstrate the concordance between the proteomics- and Western blotting- derived ratios for proteins with known over- or under-expression in CLL.
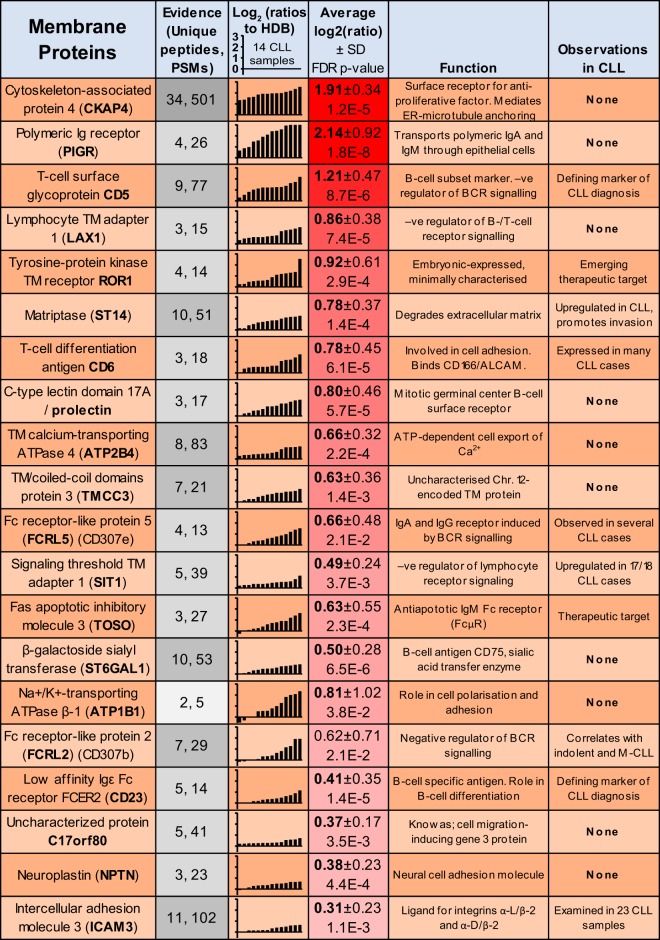
In addition to the proposed cell surface expression of CKAP4, CLL proteomics identified 20 consistently upregulated proteins annotated with surface localization (Fig. 4). This list highlighted 10 putative novel markers, CKAP4, PIGR, LAX1, CLEC17A, ATP2B4, TMCC3, ST6GAL1, ATP1B1, C17orf80, and NPTN (supplemental Fig. S4), with no previous descriptions of protein upregulation in CLL, providing them as targets for future evaluation for roles in CLL biology or as bio-markers.
Fig. 4.
Proteomics identification of CLL-overexpressed cell surface proteins. Proteomics-derived quantitations for the 20 most consistently upregulated cell surface-expressed proteins in CLL, relative to HDB. The data represent the number of unique peptides and PSMs, the Log2 (ratios) relative to HDB, average Log2 (ratios) and proposed function and evidence for prior observations in CLL.
Although for CKAP4, and to some extent PIGR, surface localization may be affected by internalisation, other candidates, such as lymphocyte transmembrane adapter 1 (LAX1), transmembrane calcium-transporting ATPase (ATP2B4) and prolectin (CLEC17A) are likely primarily localized to the cell periphery. LAX1, ATP2B4, and CLEC17A may additionally present potential effectors of BCR signaling (39–42).
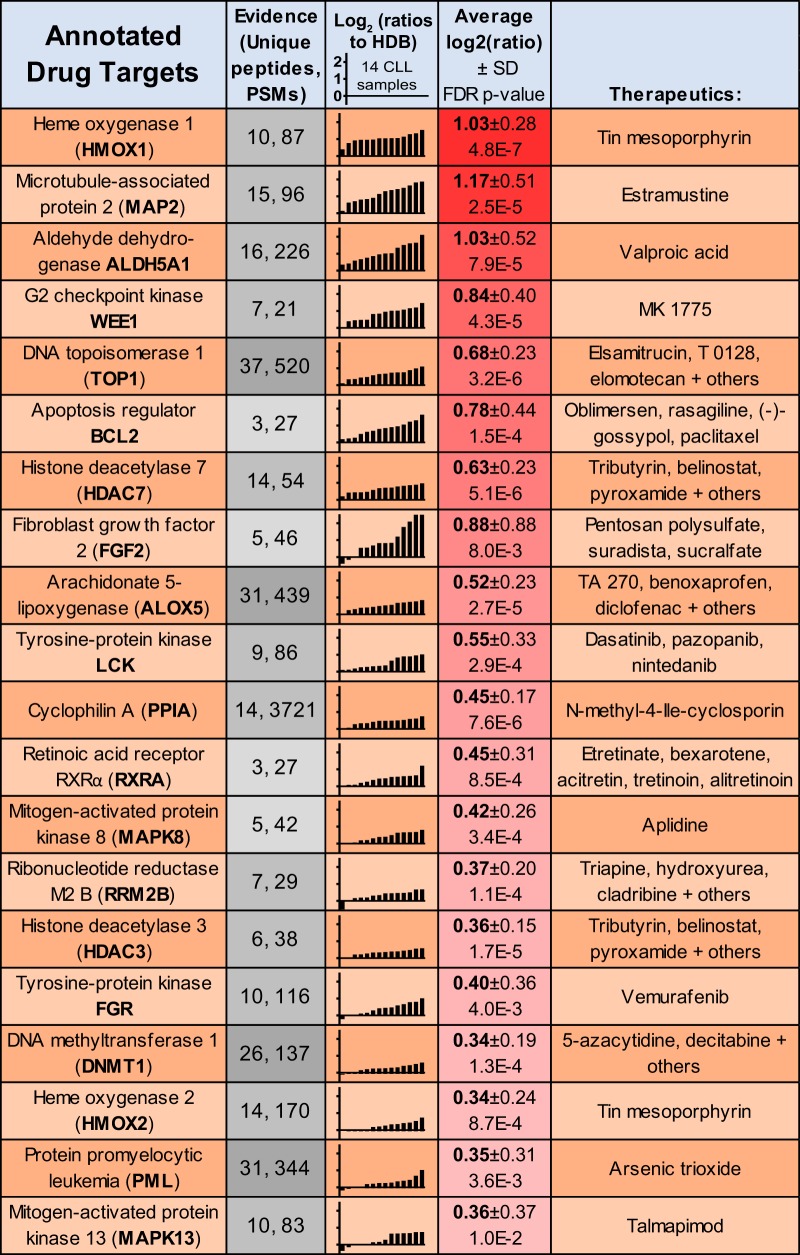
Next, overexpressed CLL proteins were annotated for their potential as therapeutic targets based on existing drug/inhibitor knowledge using IPA (Fig. 5). Both functional heme oxygenase isoforms (HMOX1 and HMOX2), which offer cytoprotective effects through free heme degradation, were observed upregulated and annotated as targets of tin mesoporphyrin. Histone deacetylases HDAC3 and HDAC7, and to a lesser extent HDAC1 and sirtuin 5, also presented putative targets of inhibition. This list additionally contained several kinases as therapeutic targets, such as two mitogen-activated protein kinases, MAPK8/JNK1 and MAPK13; two tyrosine-protein kinase proto-oncoproteins, FGR and LCK; and two cell cycle progression kinases, cyclin-dependent kinase 7 (CDK7) and the G2 checkpoint kinase WEE1.
Fig. 5.
Proteomics identification of CLL-overexpressed drug targets. Proteomics-derived quantitations for the 20 most consistently upregulated annotated targets of small molecular inhibitors in CLL, relative to HDB. Proteins were annotated using IPA. The data represent the number of unique peptides and PSMs, the Log2 (ratios) relative to HDB, average Log2 (ratios) and IPA-annotated drugs known to target each protein.
mRNA Processing Upregulation in CLL
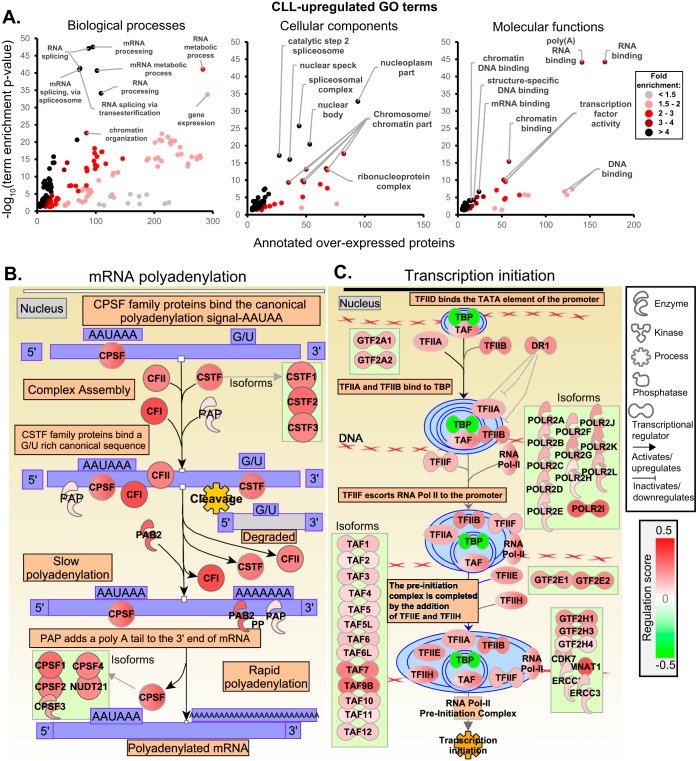
Gene ontology (GO) term enrichment was used to identify over-represented features of CLL biology (Fig. 6A). Transcription and transcript processing were dominant features among the 544 overexpressed proteins, with “mRNA processing” the most significantly enriched term (p = 2.9 × 10−48) with 95 annotated proteins. Most of the most significant biological process terms additionally described mRNA-related processes. Four terms describing RNA splicing, for instance, were highly enriched (n>72 proteins, p < 10−40). Another emerging trend was that of chromatin modification and organization (n>84, p < 2.3 × 10−23). Enrichment of cellular component and molecular function terms further described the dysregulation of mRNA-related processes, such as “nucleoplasm” (n = 258, p = 2.1 × 10−69), “spliceosomal complex” (n = 44, p = 2.0 × 10−26) and “RNA binding” (n = 167, p = 6.0 × 10−45). IPA canonical pathway enrichment identified broad upregulation of proteins and isoforms involved in “cleavage and polyadenylation of pre-mRNA” (p = 2.0 × 10−10) (Fig. 6B) and “preinitiation complex assembly” (p = 1.7 × 10−4) (Fig. 6C).
Fig. 6.
Bioinformatics analysis of the CLL-overexpressed proteome. A, GO term enrichment for the 544 overexpressed proteins (Rs>0.3, p < 0.05). The benjamini-corrected GO term enrichment p values were plotted against the number of CLL-upregulated proteins annotated with each term, additionally highlighting the observed fold-enrichment relative to the number expected by chance. B, IPA enriched canonical pathway “cleavage and polyadenylation of pre-mRNA” (p = 2.0 × 10−10) and C, “preinitiation complex assembly” (p = 1.7 × 10−4).
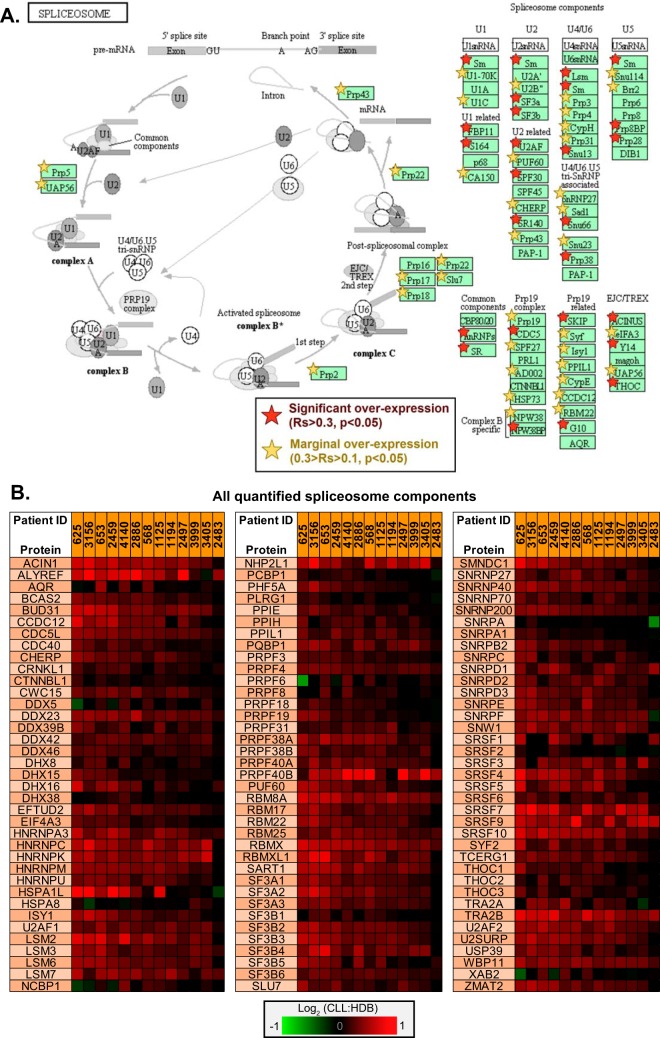
Upregulated KEGG pathways were also interrogated, identifying a 9-fold enrichment of spliceosome components (n = 36, p = 1.3 × 10−21) (Fig. 7A). A further 60 components were identified with marginal upregulation (0.1<Rs<0.3, p < 0.05). Fig. 7B presents all proteins within the quantified proteome which map to the KEGG spliceosome pathways, highlighting a near-consistent trend of some degree of over-expression across the CLL samples relative to HDB.
Fig. 7.
Significant enrichment of spliceosome components in CLL. A, Overexpressed proteins demonstrated significant enrichment of the components of the KEGG pathway “spliceosome” (n = 36, p = 1.3 × 10−21). This pathway is annotated with those proteins identified as significantly overexpressed (p < 0.05), with both substantial (Rs>0.3) and marginal (0.1<Rs<0.3) overexpression annotated using red and yellow stars, respectively. B, The differential expressions observed for all proteins and the individual log2 (ratios) for each CLL sample, mapping to the KEGG “spliceosome” pathway.
DISCUSSION
CLL has been the subject of numerous investigations applying genomics and transcriptomics that have contributed greatly to the clinical and biological understanding of the disease (6, 43–45). However, low correlations observed between mRNA and protein expression limits insight from these studies (46, 47). Indeed, a comparison between a previous transcriptomics analysis of CLL (38) and these proteomics results highlight minimally correlated differential expression (supplemental Fig. S2). Proteomics has been applied to CLL in several investigations providing insight into potential differences between subtypes and some CLL-specific signatures (21, 22, 25, 26, 48, 49). To date, however, CLL proteomics studies have lacked sufficient coverage to identify most expressed proteins and are yet to fully explore comparisons with healthy donor B-cell controls.
This study aimed to implement advances in quantitative LC-MS proteomics to provide a detailed characterization of a broad spectrum of CLL samples and evaluate changes in protein expression relative to B-cells derived from healthy donors. Overall, this investigation provided a substantial, reproducible (supplemental Fig. S1) and representative (Fig. 3) description of the CLL proteome to a depth of almost 6000 proteins. Additionally, the accuracy of the results for individual samples was highlighted by the expression of key subtype markers (supplemental Fig. S1) suggesting the potential of the presented methods for the dissection of subtype-specific differences in CLL and other cancers in the future.
The most striking finding was that of a consistent subtype-independent expression profile across the CLL samples (Fig. 2B). Given the heterogeneous clinical nature of CLL, some variation and clustering of subtypes was anticipated, although homogeneity among CLL subtypes has also been observed previously by transcriptomics (5, 6). This suggests that the phenotypic differences between CLL subtypes may either be a product of post-translational modifications, microenvironment interactions or CLL niche-specific characteristics. Further studies with greater sample numbers will be required to better understand these potentially subtle differences in protein expression.
Phenotypic differences exist between CLL cells in lymph nodes and peripheral blood (50), suggesting that evaluation of CLL cells from additional niches may be required to understand the differences in disease behaviors observed between subtypes. Furthermore, evaluation of fractionated B-cell subsets from several niches would serve as more informative controls. Indeed, given recent insights from methylation studies relating to the likely cell of origin for different CLL subtypes, B-cell subsets relevant to each CLL sample (i.e. MBC for M-CLL and NBC for U-CLL) should be evaluated, guided by their CpG methylation signatures (7–10). Currently, we are not aware of proteomics data describing these B-cell subsets.
The findings presented here provide several potential novel hypotheses for further investigation. CKAP4, for instance (Fig. 2D, 3), was robustly identified as a highly abundant, overexpressed, putative surface protein in CLL and validated in a separate cohort by Western blotting. This offers potential mechanistic insight into CLL and presents a prospective clinical tool. In addition to roles in the endoplasmic reticulum and as a transcription factor (51), CKAP4, also known as CLIMP-63, can act as a cell surface receptor for tissue plasminogen activator (tPA) (52), surfactant protein A (SP-A) (53) and anti-proliferative factor (APF) (54). APF treatment of a bladder cancer cell line resulted in reduced proliferation, attributable to substantially reduced phosphorylation of AKT and GSK3β and an increased expression of p53 (55). Interestingly, substantial CKAP4 overexpression was also observed in tumors of the Eμ-TCL1 mouse model of CLL suggesting a potential means of investigation (31). Targeting CKAP4 with either ligands or immunotherapy may offer means of treating CLL.
INPP5F mRNA overexpression in CLL has previously been linked with low progression-free survival for patients undergoing fludarabine-based therapy (56). The proteomics and Western blot validation presented here indicate that the INPP5F protein is commonly overexpressed in CLL. Together these observations suggest INPP5F may present a therapeutic target in CLL, especially in treatment-resistant cases and its mechanism in CLL warrants further investigation.
The identification of several consistently upregulated membrane proteins in CLL versus HDBs (Fig. 4, supplemental Fig. S4) highlighted the potential of proteomics approaches to identify novel immunotherapy targets for selective targeting with monoclonal antibodies. Additionally, identification of proteins linked with BCR signaling should enable a better understanding of this process in CLL which may enable improved therapeutic targeting of this currently incurable disease (39–42). LAX1 was shown to be phosphorylated upon BCR stimulation by Src and Syk (39), with B-cell hyper-responsiveness observed in LAX1−/− mice suggesting a regulatory role in BCR signaling (57). ATP2B4 has a role in BCR-induced calcium efflux (40) and Prolectin, also known as CLEC17A, is expressed in germinal center B-cells where expression correlates with proliferation (58). Prolectin also has a potential role in BCR signaling, through an association with BLNK (42).
The analysis of drug targets further highlighted the potential of proteomics in the identification of putative clinical tools (Fig. 5). Wee1 overexpression, for instance (Fig. 3B), suggests a potential target to inhibit the cell cycle, with the inhibitor MK1775 shown to have therapeutic benefit in other cancers (59). The upregulation of HMOX1 and 2 suggested an increased degradation of free heme in CLL, combined inhibition of which could induce apoptosis (60). A trend of HDACs upregulated in these results highlighted the potential of targeted HDAC inhibitors (HDACi). HDAC1 and HDAC3 were both observed upregulated and specifically targetable, for instance, with Entinostat; previously identified to induce pro-apoptotic effects in CLL cells (61). HDAC7 additionally exhibited consistent upregulation, observed previously at the mRNA level (62), suggesting the possibility of more targeted means of HDAC interference, with fewer off-target effects compared with those seen in previous pan-HDACi trials in CLL (62–64).
The strong signature of upregulated mRNA processes highlighted by the bioinformatics analyses (Fig. 6) suggests a general underlying defect in CLL, independent of subtype. SF3B1 mutations (65) and observations of major dysregulation of splicing patterns (44) have previously indicated aberrant spliceosome activity in CLL. Additionally, inhibition of SF3B1 by spliceostatin-A was toxic to CLL, independent of SF3B1 mutational status, suggesting a broader role for aberrant splicing in CLL biology (66). The observation of broadly consistent overexpression of spliceosomal proteins (Fig. 7) may therefore offer some explanation for these previous observations. This reinforces the notion that interference with aberrant splicing activity could offer a means of better understanding and potentially treating CLL.
A limitation to our study was the observation of non-B-cell contaminants in the HDB samples resulting from the substantial difference in percentage of non-B-cells and platelets between healthy donor and CLL patient PBMCs; varying from 95% in healthy PBMCs to below 10% in CLL patient PBMCs. Emphasis was therefore placed upon overexpressed proteins in CLL and conclusions based upon downregulated proteins - potentially attributable to contamination in the HDB samples - were avoided.
In summary, these results offer the first comprehensive insight into the molecular composition of CLL compared with healthy donor B-cells at the proteome level. They demonstrate the potential of proteomics to identify protein-specific differences between cancers and healthy tissues. The application of such approaches on a larger scale promises the elucidation of putative therapeutic targets and prognostic and diagnostic indicators, in addition to the dissection of the underlying cancer biology.
DATA AVAILABILITY
The raw data and processed outputs have been deposited in the PRIDE archive: https://www.ebi.ac.uk/pride/archive/projects/PXD002004.
Supplementary Material
Acknowledgments
We thank all patients who contributed to this study. We would like to thank Roger Allsopp and Derek Coates for raising funds the FT-MS proteomics platform at the University of Southampton. Thanks are due to Professor Paul Townsend for his role in securing this funding. Karen Kimpton and Dr Zadie Davis, PhD, are acknowledged for technical assistance and for collating the clinical and laboratory data. We would like to acknowledge the use of the IRIDIS High Performance Computing Facility and associated support services at the University of Southampton and the PRIDE team in the submission of our data to the PRIDE database. We would also like to thank Cory White for SPIQuE development, Dr. Xunli Zhang for HPLC access and Sofia Macari for assistance with HPLC. Also, thanks to Graham Packham and Chris Sutton for critical feedback on this work.
Footnotes
* This work was supported by an MRC iCASE studentship, a supplementary MRC fellowship award to H.J. and grants from Bloodwise (10012 and 12050; 11052, 12036), the Kay Kendall Leukaemia Fund (873), Cancer Research UK (C34999/A18087, ECMC C24563/A15581), and the Bournemouth Leukaemia Fund.
Conflict of Interest Disclosure: M.S.C. is a retained consultant for Bioinvent and has performed educational and advisory roles for Baxalta. He has received research funding from Roche, Gilead and GSK. A.J.S. is a consultant and has also received research funding and honoraria from Portola Pharmaceuticals (USA) and Gilead (UK).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CLL
- Chronic lymphocytic leukaemia
- HDB
- Healthy donor B-cells
- IGHV
- Immunoglobulin heavy-chain variable-region gene
- MBC
- Memory B-cells
- M-CLL
- IGHV-mutated CLL
- NBC
- Naive B cells
- NOTCH1
- Neurogenic locus notch homolog protein 1
- PBMCs
- Peripheral blood mononuclear cells
- Rs
- Regulation score
- SF3B1
- Splicing factor 3B subunit 1
- TMT
- Tandem mass tags
- Tri12
- Trisomy 12
- U-CLL
- IGHV-unmutated CLL.
REFERENCES
- 1. Hallek M., Cheson B. D., Catovsky D., Caligaris-Cappio F., Dighiero G., Dohner H., Hillmen P., Keating M. J., Montserrat E., Rai K. R., Kipps T. J., and International Workshop on Chronic Lymphocytic, L. (2008) Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 111, 5446–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marti G. E., Rawstron A. C., Ghia P., Hillmen P., Houlston R. S., Kay N., Schleinitz T. A., Caporaso N., International Familial, C. L. L. C. (2005) Diagnostic criteria for monoclonal B-cell lymphocytosis. Br. J. Haematol. 130, 325–332 [DOI] [PubMed] [Google Scholar]
- 3. Mockridge C. I., Potter K. N., Wheatley I., Neville L. A., Packham G., and Stevenson F. K. (2007) Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood 109, 4424–4431 [DOI] [PubMed] [Google Scholar]
- 4. Damle R. N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S. L., Buchbinder A., Budman D., Dittmar K., Kolitz J., Lichtman S. M., Schulman P., Vinciguerra V. P., Rai K. R., Ferrarini M., and Chiorazzi N. (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847 [PubMed] [Google Scholar]
- 5. Rosenwald A., Alizadeh A. A., Widhopf G., Simon R., Davis R. E., Yu X., Yang L., Pickeral O. K., Rassenti L. Z., Powell J., Botstein D., Byrd J. C., Grever M. R., Cheson B. D., Chiorazzi N., Wilson W. H., Kipps T. J., Brown P. O., and Staudt L. M. (2001) Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 194, 1639–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein U., Tu Y., Stolovitzky G. A., Mattioli M., Cattoretti G., Husson H., Freedman A., Inghirami G., Cro L., Baldini L., Neri A., Califano A., and Dalla-Favera R. (2001) Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194, 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oakes C. C., Seifert M., Assenov Y., Gu L., Przekopowitz M., Ruppert A. S., Wang Q., Imbusch C. D., Serva A., Koser S. D., Brocks D., Lipka D. B., Bogatyrova O., Weichenhan D., Brors B., Rassenti L., Kipps T. J., Mertens D., Zapatka M., Lichter P., Dohner H., Kuppers R., Zenz T., Stilgenbauer S., Byrd J. C., and Plass C. (2016) DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 48, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oakes C. C., Claus R., Gu L., Assenov Y., Hullein J., Zucknick M., Bieg M., Brocks D., Bogatyrova O., Schmidt C. R., Rassenti L., Kipps T. J., Mertens D., Lichter P., Dohner H., Stilgenbauer S., Byrd J. C., Zenz T., and Plass C. (2014) Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 4, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Queiros A. C., Villamor N., Clot G., Martinez-Trillos A., Kulis M., Navarro A., Penas E. M., Jayne S., Majid A., Richter J., Bergmann A. K., Kolarova J., Royo C., Russinol N., Castellano G., Pinyol M., Bea S., Salaverria I., Lopez-Guerra M., Colomer D., Aymerich M., Rozman M., Delgado J., Gine E., Gonzalez-Diaz M., Puente X. S., Siebert R., Dyer M. J., Lopez-Otin C., Rozman C., Campo E., Lopez-Guillermo A., and Martin-Subero J. I. (2015) A B-cell epigenetic signature defines three biologic subgroups of chronic lymphocytic leukemia with clinical impact. Leukemia 29, 598–605 [DOI] [PubMed] [Google Scholar]
- 10. Kulis M., Heath S., Bibikova M., Queiros A. C., Navarro A., Clot G., Martinez-Trillos A., Castellano G., Brun-Heath I., Pinyol M., Barberan-Soler S., Papasaikas P., Jares P., Bea S., Rico D., Ecker S., Rubio M., Royo R., Ho V., Klotzle B., Hernandez L., Conde L., Lopez-Guerra M., Colomer D., Villamor N., Aymerich M., Rozman M., Bayes M., Gut M., Gelpi J. L., Orozco M., Fan J. B., Quesada V., Puente X. S., Pisano D. G., Valencia A., Lopez-Guillermo A., Gut I., Lopez-Otin C., Campo E., and Martin-Subero J. I. (2012) Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 44, 1236–1242 [DOI] [PubMed] [Google Scholar]
- 11. Rossi D., Rasi S., Spina V., Bruscaggin A., Monti S., Ciardullo C., Deambrogi C., Khiabanian H., Serra R., Bertoni F., Forconi F., Laurenti L., Marasca R., Dal-Bo M., Rossi F. M., Bulian P., Nomdedeu J., Del Poeta G., Gattei V., Pasqualucci L., Rabadan R., Foa R., Dalla-Favera R., and Gaidano G. (2013) Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 121, 1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marasca R., Maffei R., Martinelli S., Fiorcari S., Bulgarelli J., Debbia G., Rossi D., Rossi F. M., Rigolin G. M., Martinelli S., Gattei V., Del Poeta G., Laurenti L., Forconi F., Montillo M., Gaidano G., and Luppi M. (2013) Clinical heterogeneity of de novo 11q deletion chronic lymphocytic leukaemia: prognostic relevance of extent of 11q deleted nuclei inside leukemic clone. Hematol. Oncol. 31, 88–95 [DOI] [PubMed] [Google Scholar]
- 13. Rossi D., Bruscaggin A., Spina V., Rasi S., Khiabanian H., Messina M., Fangazio M., Vaisitti T., Monti S., Chiaretti S., Guarini A., Del Giudice I., Cerri M., Cresta S., Deambrogi C., Gargiulo E., Gattei V., Forconi F., Bertoni F., Deaglio S., Rabadan R., Pasqualucci L., Foa R., Dalla-Favera R., and Gaidano G. (2011) Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 118, 6904–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabbri G., Rasi S., Rossi D., Trifonov V., Khiabanian H., Ma J., Grunn A., Fangazio M., Capello D., Monti S., Cresta S., Gargiulo E., Forconi F., Guarini A., Arcaini L., Paulli M., Laurenti L., Larocca L. M., Marasca R., Gattei V., Oscier D., Bertoni F., Mullighan C. G., Foa R., Pasqualucci L., Rabadan R., Dalla-Favera R., and Gaidano G. (2011) Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 208, 1389–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stilgenbauer S., Schnaiter A., Paschka P., Zenz T., Rossi M., Dohner K., Buhler A., Bottcher S., Ritgen M., Kneba M., Winkler D., Tausch E., Hoth P., Edelmann J., Mertens D., Bullinger L., Bergmann M., Kless S., Mack S., Jager U., Patten N., Wu L., Wenger M. K., Fingerle-Rowson G., Lichter P., Cazzola M., Wendtner C. M., Fink A. M., Fischer K., Busch R., Hallek M., and Dohner H. (2014) Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123, 3247–3254 [DOI] [PubMed] [Google Scholar]
- 16. Quesada V., Conde L., Villamor N., Ordonez G. R., Jares P., Bassaganyas L., Ramsay A. J., Bea S., Pinyol M., Martinez-Trillos A., Lopez-Guerra M., Colomer D., Navarro A., Baumann T., Aymerich M., Rozman M., Delgado J., Gine E., Hernandez J. M., Gonzalez-Diaz M., Puente D. A., Velasco G., Freije J. M., Tubio J. M., Royo R., Gelpi J. L., Orozco M., Pisano D. G., Zamora J., Vazquez M., Valencia A., Himmelbauer H., Bayes M., Heath S., Gut M., Gut I., Estivill X., Lopez-Guillermo A., Puente X. S., Campo E., and Lopez-Otin C. (2011) Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 44, 47–52 [DOI] [PubMed] [Google Scholar]
- 17. Wan Y., and Wu C. J. (2013) SF3B1 mutations in chronic lymphocytic leukemia. Blood 121, 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L., Brooks A. N., Fan J., Wan Y., Gambe R., Li S., Hergert S., Yin S., Freeman S. S., Levin J. Z., Fan L., Seiler M., Buonamici S., Smith P. G., Chau K. F., Cibulskis C. L., Zhang W., Rassenti L. Z., Ghia E. M., Kipps T. J., Fernandes S., Bloch D. B., Kotliar D., Landau D. A., Shukla S. A., Aster J. C., Reed R., DeLuca D. S., Brown J. R., Neuberg D., Getz G., Livak K. J., Meyerson M. M., Kharchenko P. V., and Wu C. J. (2016) Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell 30, 750–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thurgood L. A., Chataway T. K., Lower K. M., and Kuss B. J. (2017) From genome to proteome: Looking beyond DNA and RNA in chronic lymphocytic leukemia. J. Proteomics 155, 73–84 [DOI] [PubMed] [Google Scholar]
- 20. Diez P., Gongora R., Orfao A., and Fuentes M. (2017) Functional proteomic insights in B-cell chronic lymphocytic leukemia. Expert Rev. Proteomics 14, 137–146 [DOI] [PubMed] [Google Scholar]
- 21. Eagle G. L., Zhuang J., Jenkins R. E., Till K. J., Jithesh P. V., Lin K., Johnson G. G., Oates M., Park K., Kitteringham N. R., and Pettitt A. R. (2015) Total proteome analysis identifies migration defects as a major pathogenetic factor in immunoglobulin heavy chain variable region (IGHV)-unmutated chronic lymphocytic leukemia. Mol. Cell. Proteomics 14, 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alsagaby S. A., Khanna S., Hart K. W., Pratt G., Fegan C., Pepper C., Brewis I. A., and Brennan P. (2014) Proteomics-based strategies to identify proteins relevant to chronic lymphocytic leukemia. J. Proteome Res. 13, 5051–5062 [DOI] [PubMed] [Google Scholar]
- 23. Rees-Unwin K. S., Faragher R., Unwin R. D., Adams J., Brown P. J., Buckle A. M., Pettitt A., Hutchinson C. V., Johnson S. M., Pulford K., Banham A. H., Whetton A. D., Lucas G., Mason D. Y., and Burthem J. (2010) Ribosome-associated nucleophosmin 1: increased expression and shuttling activity distinguishes prognostic subtypes in chronic lymphocytic leukaemia. Br. J. Haematol. 148, 534–543 [DOI] [PubMed] [Google Scholar]
- 24. Cochran D. A., Evans C. A., Blinco D., Burthem J., Stevenson F. K., Gaskell S. J., and Whetton A. D. (2003) Proteomic analysis of chronic lymphocytic leukemia subtypes with mutated or unmutated Ig V(H) genes. Mol. Cell. Proteomics 2, 1331–1341 [DOI] [PubMed] [Google Scholar]
- 25. Barnidge D. R., Jelinek D. F., Muddiman D. C., and Kay N. E. (2005) Quantitative protein expression analysis of CLL B cells from mutated and unmutated IgV(H) subgroups using acid-cleavable isotope-coded affinity tag reagents. J. Proteome Res. 4, 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnidge D. R., Tschumper R. C., Jelinek D. F., Muddiman D. C., and Kay N. E. (2005) Protein expression profiling of CLL B cells using replicate off-line strong cation exchange chromatography and LC-MS/MS. J. Chromatogr. 819, 33–39 [DOI] [PubMed] [Google Scholar]
- 27. Perrot A., Pionneau C., Nadaud S., Davi F., Leblond V., Jacob F., Merle-Beral H., Herbrecht R., Bene M. C., Gribben J. G., Bahram S., and Vallat L. (2011) A unique proteomic profile on surface IgM ligation in unmutated chronic lymphocytic leukemia. Blood 118, e1–e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., and Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
- 29. Kim M. S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L. D., Patil A. H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S. K., Marimuthu A., Sathe G. J., Chavan S., Datta K. K., Subbannayya Y., Sahu A., Yelamanchi S. D., Jayaram S., Rajagopalan P., Sharma J., Murthy K. R., Syed N., Goel R., Khan A. A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T. C., Zhong J., Wu X., Shaw P. G., Freed D., Zahari M. S., Mukherjee K. K., Shankar S., Mahadevan A., Lam H., Mitchell C. J., Shankar S. K., Satishchandra P., Schroeder J. T., Sirdeshmukh R., Maitra A., Leach S. D., Drake C. G., Halushka M. K., Prasad T. S., Hruban R. H., Kerr C. L., Bader G. D., Iacobuzio-Donahue C. A., Gowda H., and Pandey A. (2014) A draft map of the human proteome. Nature 509, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P. H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., and Ponten F. (2015) Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- 31. Johnston H. E., Carter M. J., Cox K. L., Dunscombe M., Manousopoulou A., Townsend P. A., Garbis S. D., and Cragg M. S. (2017) Integrated Cellular and Plasma Proteomics of Contrasting B-cell Cancers Reveals Common, Unique and Systemic Signatures. Mol. Cell. Proteomics 16, 386–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hahne H., Pachl F., Ruprecht B., Maier S. K., Klaeger S., Helm D., Medard G., Wilm M., Lemeer S., and Kuster B. (2013) DMSO enhances electrospray response, boosting sensitivity of proteomic experiments. Nat. Methods 10, 989–991 [DOI] [PubMed] [Google Scholar]
- 33. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burkhart J. M., Vaudel M., Gambaryan S., Radau S., Walter U., Martens L., Geiger J., Sickmann A., and Zahedi R. P. (2012) The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood 120, e73–e82 [DOI] [PubMed] [Google Scholar]
- 35. Hussain K., Hargreaves C. E., Roghanian A., Oldham R. J., Chan H. T., Mockridge C. I., Chowdhury F., Frendeus B., Harper K. S., Strefford J. C., Cragg M. S., Glennie M. J., Williams A. P., and French R. R. (2015) Upregulation of FcgammaRIIb on monocytes is necessary to promote the superagonist activity of TGN1412. Blood 125, 102–110 [DOI] [PubMed] [Google Scholar]
- 36. Ivanov A., Cragg M. S., Erenpreisa J., Emzinsh D., Lukman H., and Illidge T. M. (2003) Endopolyploid cells produced after severe genotoxic damage have the potential to repair DNA double strand breaks. J. Cell Sci. 116, 4095–4106 [DOI] [PubMed] [Google Scholar]
- 37. Zucchetto A., Caldana C., Benedetti D., Tissino E., Rossi F. M., Hutterer E., Pozzo F., Bomben R., Dal Bo M., D'Arena G., Zaja F., Pozzato G., Di Raimondo F., Hartmann T. N., Rossi D., Gaidano G., Del Poeta G., and Gattei V. (2013) CD49d is overexpressed by trisomy 12 chronic lymphocytic leukemia cells: evidence for a methylation-dependent regulation mechanism. Blood 122, 3317–3321 [DOI] [PubMed] [Google Scholar]
- 38. Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G. T., Bene M. C., De Vos J., Hernandez J. M., Hofmann W. K., Mills K. I., Gilkes A., Chiaretti S., Shurtleff S. A., Kipps T. J., Rassenti L. Z., Yeoh A. E., Papenhausen P. R., Liu W. M., Williams P. M., and Foa R. (2010) Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 28, 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhu M., Janssen E., Leung K., and Zhang W. (2002) Molecular cloning of a novel gene encoding a membrane-associated adaptor protein (LAX) in lymphocyte signaling. J. Biol. Chem. 277, 46151–46158 [DOI] [PubMed] [Google Scholar]
- 40. Chen J., McLean P. A., Neel B. G., Okunade G., Shull G. E., and Wortis H. H. (2004) CD22 attenuates calcium signaling by potentiating plasma membrane calcium-ATPase activity. Nat. Immunol. 5, 651–657 [DOI] [PubMed] [Google Scholar]
- 41. Breiman A., Lopez Robles M. D., de Carne Trecesson S., Echasserieau K., Bernardeau K., Drickamer K., Imberty A., Barille-Nion S., Altare F., and Le Pendu J. (2016) Carcinoma-associated fucosylated antigens are markers of the epithelial state and can contribute to cell adhesion through CLEC17A (Prolectin). Oncotarget 7, 14064–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Satpathy S., Wagner S. A., Beli P., Gupta R., Kristiansen T. A., Malinova D., Francavilla C., Tolar P., Bishop G. A., Hostager B. S., and Choudhary C. (2015) Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 11, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strefford J. C. (2015) The genomic landscape of chronic lymphocytic leukaemia: biological and clinical implications. Br. J. Haematol. 169, 14–31 [DOI] [PubMed] [Google Scholar]
- 44. Ferreira P. G., Jares P., Rico D., Gomez-Lopez G., Martinez-Trillos A., Villamor N., Ecker S., Gonzalez-Perez A., Knowles D. G., Monlong J., Johnson R., Quesada V., Djebali S., Papasaikas P., Lopez-Guerra M., Colomer D., Royo C., Cazorla M., Pinyol M., Clot G., Aymerich M., Rozman M., Kulis M., Tamborero D., Gouin A., Blanc J., Gut M., Gut I., Puente X. S., Pisano D. G., Martin-Subero J. I., Lopez-Bigas N., Lopez-Guillermo A., Valencia A., Lopez-Otin C., Campo E., and Guigo R. (2014) Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 24, 212–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haslinger C., Schweifer N., Stilgenbauer S., Dohner H., Lichter P., Kraut N., Stratowa C., and Abseher R. (2004) Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J. Clin. Oncol. 22, 3937–3949 [DOI] [PubMed] [Google Scholar]
- 46. Gygi S. P., Rochon Y., Franza B. R., and Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vogel C., and Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Glibert P., Vossaert L., Van Steendam K., Lambrecht S., Van Nieuwerburgh F., Offner F., Kipps T., Dhaenens M., and Deforce D. (2014) Quantitative proteomics to characterize specific histone H2A proteolysis in chronic lymphocytic leukemia and the myeloid THP-1 cell line. Int. J. Mol. Sci. 15, 9407–9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miguet L., Bechade G., Fornecker L., Zink E., Felden C., Gervais C., Herbrecht R., Van Dorsselaer A., Mauvieux L., and Sanglier-Cianferani S. (2009) Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J. Proteome Res. 8, 3346–3354 [DOI] [PubMed] [Google Scholar]
- 50. Pasikowska M., Walsby E., Apollonio B., Cuthill K., Phillips E., Coulter E., Longhi M. S., Ma Y., Yallop D., Barber L. D., Patten P., Fegan C., Ramsay A. G., Pepper C., Devereux S., and Buggins A. G. (2016) Phenotype and immune function of lymph node and peripheral blood CLL cells are linked to transendothelial migration. Blood 128, 563–573 [DOI] [PubMed] [Google Scholar]
- 51. Sandoz P. A., and van der Goot F. G. (2015) How many lives does CLIMP-63 have? Biochem. Soc. Trans. 43, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Razzaq T. M., Bass R., Vines D. J., Werner F., Whawell S. A., and Ellis V. (2003) Functional regulation of tissue plasminogen activator on the surface of vascular smooth muscle cells by the type-II transmembrane protein p63 (CKAP4). J. Biol. Chem. 278, 42679–42685 [DOI] [PubMed] [Google Scholar]
- 53. Gupta N., Manevich Y., Kazi A. S., Tao J. Q., Fisher A. B., and Bates S. R. (2006) Identification and characterization of p63 (CKAP4/ERGIC-63/CLIMP-63), a surfactant protein A binding protein, on type II pneumocytes. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L436–L446 [DOI] [PubMed] [Google Scholar]
- 54. Conrads T. P., Tocci G. M., Hood B. L., Zhang C. O., Guo L., Koch K. R., Michejda C. J., Veenstra T. D., and Keay S. K. (2006) CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J. Biol. Chem. 281, 37836–37843 [DOI] [PubMed] [Google Scholar]
- 55. Shahjee H. M., Koch K. R., Guo L., Zhang C. O., and Keay S. K. (2010) Antiproliferative factor decreases Akt phosphorylation and alters gene expression via CKAP4 in T24 bladder carcinoma cells. J. Exp. Clin. Cancer Res. 29, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Palermo G., Maisel D., Barrett M., Smith H., Duchateau-Nguyen G., Nguyen T., Yeh R. F., Dufour A., Robak T., Dornan D., Weisser M., and investigators R. (2015) Gene expression of INPP5F as an independent prognostic marker in fludarabine-based therapy of chronic lymphocytic leukemia. Blood Cancer J. 5, e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu M., Granillo O., Wen R., Yang K., Dai X., Wang D., and Zhang W. (2005) Negative regulation of lymphocyte activation by the adaptor protein LAX. J. Immunol. 174, 5612–5619 [DOI] [PubMed] [Google Scholar]
- 58. Graham S. A., Jegouzo S. A., Yan S., Powlesland A. S., Brady J. P., Taylor M. E., and Drickamer K. (2009) Prolectin, a glycan-binding receptor on dividing B cells in germinal centers. J. Biol. Chem. 284, 18537–18544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang M., Dominguez D., Chen S., Fan J., Qin L., Long A., Li X., Zhang Y., Shi H., and Zhang B. (2017) WEE1 inhibition by MK1775 as a single-agent therapy inhibits ovarian cancer viability. Oncol. Lett. 14, 3580–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou Y., Hileman E. O., Plunkett W., Keating M. J., and Huang P. (2003) Free radical stress in chronic lymphocytic leukemia cells and its role in cellular sensitivity to ROS-generating anticancer agents. Blood 101, 4098–4104 [DOI] [PubMed] [Google Scholar]
- 61. Lucas D. M., Davis M. E., Parthun M. R., Mone A. P., Kitada S., Cunningham K. D., Flax E. L., Wickham J., Reed J. C., Byrd J. C., and Grever M. R. (2004) The histone deacetylase inhibitor MS-275 induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia cells. Leukemia 18, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 62. Van Damme M., Crompot E., Meuleman N., Mineur P., Bron D., Lagneaux L., and Stamatopoulos B. (2012) HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics 7, 1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Garcia-Manero G., Yang H., Bueso-Ramos C., Ferrajoli A., Cortes J., Wierda W. G., Faderl S., Koller C., Morris G., Rosner G., Loboda A., Fantin V. R., Randolph S. S., Hardwick J. S., Reilly J. F., Chen C., Ricker J. L., Secrist J. P., Richon V. M., Frankel S. R., and Kantarjian H. M. (2008) Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood 111, 1060–1066 [DOI] [PubMed] [Google Scholar]
- 64. Byrd J. C., Marcucci G., Parthun M. R., Xiao J. J., Klisovic R. B., Moran M., Lin T. S., Liu S., Sklenar A. R., Davis M. E., Lucas D. M., Fischer B., Shank R., Tejaswi S. L., Binkley P., Wright J., Chan K. K., and Grever M. R. (2005) A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 105, 959–967 [DOI] [PubMed] [Google Scholar]
- 65. Quesada V., Conde L., Villamor N., Ordonez G. R., Jares P., Bassaganyas L., Ramsay A. J., Bea S., Pinyol M., Martinez-Trillos A., Lopez-Guerra M., Colomer D., Navarro A., Baumann T., Aymerich M., Rozman M., Delgado J., Gine E., Hernandez J. M., Gonzalez-Diaz M., Puente D. A., Velasco G., Freije J. M., Tubio J. M., Royo R., Gelpi J. L., Orozco M., Pisano D. G., Zamora J., Vazquez M., Valencia A., Himmelbauer H., Bayes M., Heath S., Gut M., Gut I., Estivill X., Lopez-Guillermo A., Puente X. S., Campo E., and Lopez-Otin C. (2012) Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Gen. 44, 47–52 [DOI] [PubMed] [Google Scholar]
- 66. Larrayoz M., Blakemore S. J., Dobson R. C., Blunt M. D., Rose-Zerilli M. J., Walewska R., Duncombe A., Oscier D., Koide K., Forconi F., Packham G., Yoshida M., Cragg M. S., Strefford J. C., and Steele A. J. (2016) The SF3B1 inhibitor spliceostatin A (SSA) elicits apoptosis in chronic lymphocytic leukaemia cells through downregulation of Mcl-1. Leukemia 30, 351–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and processed outputs have been deposited in the PRIDE archive: https://www.ebi.ac.uk/pride/archive/projects/PXD002004.