Abstract
Flagellated, Gram-negative, anaerobic, crescent-shaped Selenomonas species are colonizers of the digestive system, where they act at the interface between health and disease. Selenomonas sputigena is also considered a potential human periodontal pathogen, but information on its virulence factors and underlying pathogenicity mechanisms is scarce. Here we provide the first report of a Selenomonas glycoprotein, showing that S. sputigena produces a diversely and heavily O-glycosylated flagellin C9LY14 as a major cellular protein, which carries various hitherto undescribed rhamnose- and N-acetylglucosamine linked O-glycans in the range from mono- to hexasaccharides. A comprehensive glycomic and glycoproteomic assessment revealed extensive glycan macro- and microheterogeneity identified from 22 unique glycopeptide species. From the multiple sites of glycosylation, five were unambiguously identified on the 437-amino acid C9LY14 protein (Thr149, Ser182, Thr199, Thr259, and Ser334), the only flagellin protein identified. The O-glycans additionally showed modifications by methylation and putative acetylation. Some O-glycans carried hitherto undescribed residues/modifications as determined by their respective m/z values, reflecting the high diversity of native S. sputigena flagellin. We also found that monosaccharide rearrangement occurred during collision-induced dissociation (CID) of protonated glycopeptide ions. This effect resulted in pseudo Y1-glycopeptide fragment ions that indicated the presence of additional glycosylation sites on a single glycopeptide. CID oxonium ions and electron transfer dissociation, however, confirmed that just a single site was glycosylated, showing that glycan-to-peptide rearrangement can occur on glycopeptides and that this effect is influenced by the molecular nature of the glycan moiety. This effect was most pronounced with disaccharides. This study is the first report on O-linked flagellin glycosylation in a Selenomonas species, revealing that C9LY14 is one of the most heavily glycosylated flagellins described to date. This study contributes to our understanding of the largely under-investigated surface properties of oral bacteria. The data have been deposited to the ProteomeXchange with identifier PXD005859.
Flagellar motility is one of the most extensively studied processes in prokaryotic microbiology. The bacterial flagellum, a complex multiprotein assembly, is best known as the locomotive organelle that allows microbes to actively move toward favorable environments by chemotaxis (1). In many pathogenic bacteria such as Escherichia coli, Vibrio cholerae, Helicobacter spp., Campylobacter spp., Pseudomonas aeruginosa, Borrelia burgdorferi, Treponema spp., and Salmonella spp. the flagellum represents an essential structure that is crucial for full pathogenesis (2). Although it was initially believed that flagella are involved in bacterial pathogenicity solely by conferring motility, it has now become evident that this organelle can play a role in every step of the infection cycle (3).
The typical bacterial flagellum consists of six components—a basal body, a motor, a switch, a hook, a filament, and an export apparatus (4). Escherichia coli and Salmonella enterica sv. typhimurium have long served as the model organisms for studying flagellar assembly (4), with more than 50 genes involved in flagellar biosynthesis and function. Approximately half of these genes encode the structural components of the flagellum, and the rest is responsible for either the regulation of flagellar assembly or the detection and processing of environmental signals to which flagella respond (5). However, there is extensive diversity among bacteria in the contents and organization of the gene complexes that specify flagella as well as structural variation in the flagellum itself (6, 7).
The flagella filament is built from ∼20,000–30,000 copies of flagellin monomers (8). The major structural flagella proteins and their specific glycosylation have become a focus of recent research. Following the first description about 30 years ago, an increasing number of reports on flagella post-translational modifications from archaea and bacteria has been published and reviewed (9–11). In most bacterial flagellins described to date, the glycans are attached to the protein via an O-glycosidic linkage. Genes for glycosyltransferases and the genetic information governing carbohydrate biosynthetic pathways that are required for flagellin modification are, in many cases, located near the genes encoding the flagellar apparatus. Interestingly, flagellin glycosylation is mainly found in Gram-negative species and seems to be more commonplace in, but not limited to, organisms producing polar flagella (9). Especially in pathogenic bacteria such as Campylobacter spp., Helicobacter pylori and Pseudomonas aeruginosa, the important role of glycosylation for both flagellar assembly as well as a means for exerting biological interactions has clearly been established (12–15).
Selenomonas sputigena is a flagellated, motile, anaerobic, crescent-shaped, Gram-negative bacterium that was originally isolated from the periodontal pockets of patients suffering from periodontitis (16). Oral members of the bacterial genus Selenomonas have repeatedly been associated with periodontal disease and recent studies support the association of Selenomonas with the etiology of periodontitis, based on the detection of Selenomonas species at significant levels in subgingival biofilm samples of subjects with both chronic (17) and aggressive (18) periodontitis. It appears that Selenomonas species contribute considerably to the structural organization of multispecies oral biofilms (“dental plaque”), which is the condition under which periodontitis develops (19). Periodontitis continues to be the most frequently occurring inflammatory disease world-wide; in its chronic form, it is the major cause of tooth loss and can also impact systemic health (20), underlining the urgent need for therapeutic interference. However, although a group of distinct, nonmotile Gram-negative anaerobes (the “red complex”) has been identified as keystone periodontal pathogens, the pathogenesis of periodontitis is still not fully understood (20). Flagellation as present on S. sputigena cells might represent a hitherto unknown/unrecognized strategy of the bacterium to colonize its niche in the multispecies oral biofilm, thereby contributing to the establishment of the disease.
To date there is essentially no knowledge available on flagellin glycosylation of oral Selenomonas spp. Unlike in mammalian glycoprotein synthesis, glycosylation pathways in bacterial species are highly diverse (21), making different orthogonal approaches a necessity to collect information on monosaccharide identity, glycan composition, and protein attachment. Mass spectrometry has always been a core technology for determining the primary structure of glycoprotein glycans. Glycopeptide product ion spectra provide a highly sensitive and selective opportunity to investigate glycosylation levels and sites of glycan attachment. Collision-induced dissociation (CID)1 of glycopeptides usually results in a prominent Y1-ion that facilitates identification of the peptide-linked monosaccharide (22). Such Y1-assigments are, however, increasingly difficult if more than one site of glycosylation is present on a single peptide or if gas phase monosaccharide rearrangements occur (23). Halim and Zauner independently reported the detection of small product ion signals in some O-glycopeptide product ion spectra acquired from human urinary and human fibrinogen glycopeptides that they assigned to a hexose rearrangement (24, 25). Because these pseudo Y1-glycopeptide signals were, if detected at all, just of very low intensity, they did not interfere with accurate assignment. Most of the monosaccharide rearrangements are reported for the analysis of protonated glycans, in particular when one or more fucose residues are present on the glycan (26). In the analysis of N-glycopeptides, deoxyhexose or hexose rearrangements have only been found to occur between N-glycan antennae or toward the innermost N-acetylglucosamine (GlcNAc) of N-glycans, as comprehensively reviewed by Wuhrer et al. (26). However, besides these few reports, monosaccharide rearrangement and its possible effects on glycoproteomic data analysis has hardly been further investigated.
In this study, we employed a variety of glycomic and glycoproteomic approaches to investigate the glycosylation of the S. sputigena flagellin protein (UniProt Accession C9LY14), demonstrating that C9LY14 is a heavily glycosylated protein exhibiting a surprising glyco-heterogeneity at multiple sites of O-glycosylation and carrying hitherto not described O-glycan structures. We also show that CID of O-glycan carrying glycopeptides induced a glycan structure dependent gas phase monosaccharide rearrangement resulting in the formation of strong pseudo Y1-ions. These indicated the presence of two sites of glycosylation on a single glycopeptide, whereas electron transfer dissociation (ETD) confirmed that a disaccharide was present on a single site of glycosylation only.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Selenomonas sputigena ATCC 35185 was obtained from the Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). Bacteria were grown anaerobically at 37 °C in 37 g/L Brain-Heart-Infusion (BHI) broth (Oxoid, Hampshire, UK), supplemented with 5 g/L yeast extract (Oxoid, Hampshire, UK), 0.5 g/L l-cysteine (Sigma), 5 μg/ml hemin (Sigma) and 2.0 μg/ml menadione (Sigma). For the preparation of BHI-agar plates (swarming plates), 0.5% w/v agar was added. Cells were harvested by centrifugation (5000 × g, 20 min, 4 °C), washed twice with phosphate-buffered saline (PBS; pH 7.5), and either used instantly or stored at −20 °C.
Escherichia coli DH5α cells and E. coli BL21 (DE3) cells (both Life Technologies, Carlsbad, CA) were grown at 37 °C with shaking at 200 rpm in Luria-Bertani (LB) broth or on LB agar plates supplemented with 50 μg/ml kanamycin.
Flagellin Enrichment from Bacterial Cultures
Enrichment of native S. sputigena flagellin protein was achieved by differential centrifugation as described previously (27), with minor modifications. Briefly, bacterial cells from BHI-swarming plates were inoculated into 10 ml of BHI medium and incubated overnight at 37 °C. Five ml of this pre-culture were then transferred to 500 ml of half-strength BHI medium (diluted 1:1 with PBS) and incubated again overnight at 37 °C. Cells were harvested by centrifugation (5000 × g, 20 min) and resuspended in PBS (20 ml per 1 g of wet weight of cells). The suspension was blended for 2 min using a commercial blender to shear off flagella, followed by centrifugation (5000 × g, 30 min). The collected supernatant was centrifuged at 16,000 × g for 15 min and further processed by centrifugation at 40,000 × g for 3 h. All centrifugation steps were carried out at 4 °C. The remaining pellet was resuspended in 0.5 ml of Milli-Q (MQ) water and freeze-dried. The purity of the sample was analyzed by SDS-PAGE.
SDS-PAGE and Western-Immunoblotting
SDS-PAGE was carried out on 12% slab gels according to Laemmli (28). Protein bands were visualized using Coomassie Brilliant Blue G-250 (CBB; Serva, Heidelberg, Germany) and carbohydrates were stained with periodic acid-Schiff (PAS) reagent (29). For Western-immunoblotting, proteins were transferred onto a nitrocellulose membrane (Peqlab, Erlangen, Germany) and detection was performed at 800 nm on an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE). For visualization of native flagellin C9LY14, a flagellin-specific antiserum was used in combination with IR Dye 800CW goat anti-mouse antibody (LI-COR). Detection of His6-tag on recombinant C9LY14 (C9LY14R) was done with a mouse anti-His6 antibody (Life Technologies) in combination with the IR Dye 800CW goat anti-mouse antibody (LI-COR).
General Molecular Biology Methods
Restriction enzymes and ligase were purchased from Thermo Scientific (Vienna, Austria). Genomic DNA of S. sputigena ATCC 35185 was isolated as described previously (30). Plasmid DNA was purified from transformed cells using the GeneJETTM Plasmid MiniPrep Kit (Thermo Scientific). PCR fragments were amplified by Phusion® High-Fidelity DNA Polymerase (Thermo Scientific) according to the manufacturer′s protocol. PCR fragments and digested plasmids were purified from agarose gels using the GeneJETTM Gel Extraction Kit (Thermo Scientific). Chemically competent E. coli DH5α cells and E. coli BL21 (DE3) cells were transformed according to the protocol provided by the manufacturer. Recombinant cells were analyzed by restriction mapping and confirmed by sequencing (Microsynth, Austria).
Expression and Purification of Recombinant Flagellin
Recombinant S. sputigena ATCC 35185 flagellin (C9LY14R) was expressed as C-terminally hexahistidine (His6)-tagged protein in E. coli BL21 (DE3). The coding sequence for C9LY14 was amplified from S. sputigena ATCC 35185 genomic DNA by PCR using the primers (Thermo Scientific) 002-fw-fla-NcoI (5′-CCGACACCATGGCATTGGTAGTTAAGAACAAC-3′) and 001-rev-fla-XhoI (5′-GACGACTCGAGCAGGCTGAGAACGCCGGAG-3′). The PCR product was digested with NcoI/XhoI and ligated into the pET28a(+) expression vector (Novagen, Wisconsin). Subsequently, pET28a_C9LY14R_His6 was transformed into E. coli BL21 (DE3) and freshly transformed cells were grown in LB medium at 37 °C with shaking at 200 rpm to an OD600 of ∼0.6. Protein expression was induced with a final concentration of 1 mm isopropyl β-d-1-thiogalactopyranoside (IPTG; Thermo Scientific) and cultures were grown for additional 3 h.
His6-tagged C9LY14R for detection purposes was purified under denaturing conditions using Ni-NTA Agarose (Qiagen, Hilden, Germany) according to the manufacturer′s protocol. The purified protein was dialyzed against MQ water overnight at 4 °C.
Purification of Recombinant Flagellin for Immunization of Mice
Escherichia coli BL21 (DE3) cells harboring the expression plasmid were harvested by centrifugation (5000 × g, 30 min, 4 °C) and resuspended in 50 mm sodium citrate buffer (pH 6.2) containing 0.1% Triton X-100. After addition of lysozyme (800 μg/ml; Sigma-Aldrich) and benzonase (50 U/ml; Sigma-Aldrich), cells were incubated for 30 min at 37 °C. Ultrasonication (Branson sonifier, duty cycle 50%; output 6) was used to further break open bacteria applying ten cycles of 10 pulses with 30-s breaks, each, and soluble protein was separated from the insoluble material by centrifugation (25,000 × g, 30 min, 4 °C), with both fractions containing C9LY14R. To increase the yield of C9LY14R, the remaining protein was extracted from the pellet with 50 mm sodium citrate buffer (pH 5.5) containing 5 m GdHCl, 20 mm imidazole and 0.5 m NaCl for 1.5 h at 4 °C and shaking at 200 rpm. The extract was centrifuged (45,000 × g, 1 h, 4 °C) and the supernatant subjected to membrane-filtering (0.45 μm pore size). The resulting protein extracts were combined and applied to a 1-ml HisTrap HP column (GE Healthcare, Little Chalfont, UK). The recombinant protein was recovered in elution buffer (50 mm sodium citrate buffer [pH 5.5], 5 m GdHCl, 1 m imidazole, 0.5 m NaCl) followed by dialysis against 50 mm sodium citrate buffer (pH 5.5). Immunization of mice and preparation of polyclonal antiserum against purified C9LY14R was done at EF-BIO s.r.o. (Bratislava, Slovakia).
Protein Digestion and Glycan Release
For glycoproteomic analyses, the preparation enriched in native S. sputigena flagellin was separated by SDS-PAGE and protein bands of interest were excised and destained (31). Any cysteine residues were reduced with 10 mm dithiothreitol (D0632; Sigma-Aldrich) for 1 h at 56 °C and carbamidomethylated by incubation in 55 mm iodoacetamide (I6125; Sigma-Aldrich) for 1 h at room temperature in the dark. Subsequently, the protein was in-gel digested at 37 °C with trypsin (11047841001; Roche, Basel, Switzerland) or chymotrypsin (11418467001; Roche), respectively, in 25 mm ammonium bicarbonate buffer using an enzyme-to-protein ratio of 1:50 (w/w). Tryptic digests were carried out overnight, whereas chymotrypsin digests were terminated after 4 h. Resulting (glyco)peptides were extracted from the gel pieces and dried in a centrifugal evaporator (in vacuo). For reversed-phase nanoLC-ESI-MSMS analyses, the samples were resolubilized in 40 μl of 0.1% formic acid (94318; Sigma-Aldrich) from which 1-μl aliquots were injected per analysis. Glycoproteomic data has been acquired for three flagellin preparations that were performed on three independently grown S. sputigena cultures. A technical triplicate analysis was performed from a single batch for the in E. coli-expressed recombinant flagellin protein.
O-glycans were released from tryptic glycopeptides by reductive β-elimination. Dried glycopeptides were incubated with 50 μl of 0.5 m sodium borohydride (71321; Sigma-Aldrich) in 50 mm potassium hydroxide (221473; Sigma-Aldrich) for 16 h at 50 °C. The released O-glycans were desalted over an AG 50W-X8 resin (142–1451; Bio-Rad Laboratories, Hercules, CA) and purified by solid-phase extraction with porous graphitized carbon (PGC; 210101, Grace Bio-Labs, Oregon) (32). For glycomic analyses by PGC nanoLC-ESI-MSMS, the dried O-glycans were resolubilized in 15 μl of 10 mm ammonium bicarbonate of which 5 μl were injected per analysis. Because of limitations in sample material, glycomic data has been acquired for a single S. sputigena flagellin preparation.
Protein and Glycan Analysis
Peptides and glycopeptides were analyzed by reversed-phase nanoLC-ESI-MSMS on a Dionex Ultimate 3000 UHPLC online coupled to a Bruker amaZon Speed ETD ion trap mass spectrometer in positive ion mode. Reversed phase chromatography was performed on PepMap C18 columns (precolumn: Thermo Fisher, PepMap100, 100 μm [ID] × 2 cm, C18, 5 μm particle size, 100 Å pore size, P/N 164564; analytical column: Thermo Fisher, Acclaim PepMap RSLC, 75 μm [ID] × 15 cm, C18, 2 μm particle size, 100 Å pore size, P/N 164534) using a linear gradient of 0.1% formic acid (solvent A) and 90% acetonitrile (ACN) containing 0.1% formic acid (solvent B) at a constant flow rate of 400 nl/min at 45 °C. The samples were loaded onto the trap column in solvent A. The analytical gradient started at 5% of solvent B and gradually increased to 45% over 30 min. The columns were flushed with 90% solvent B for 10 min after each sample before re-equilibrating to starting conditions.
The ion trap was set to scan from m/z 400 to 1600 with an SPS Target Mass of m/z 1350 using the instrument's Enhanced Resolution mode. The ICC target was set to 200,000 with a maximum accumulation time of 50 ms. The three most intense signals of each MS scan were selected for CID and the resulting fragments were recorded from m/z 100 to 2000 in the UltraScan mode. Besides CID, electron transfer dissociation (ETD) was employed for (glyco)peptide fragmentation in separate analyses. The ETD reagent (flouranthene) ICC Target was set to 500,000 with a maximum accumulation time of 10 ms. The ETD reaction time was set to 100 ms. The glycopeptide signal intensities were further enhanced using a Bruker CaptiveSpray NanoBooster™ ionization source using nitrogen as dry gas (3 L/min at 150 °C) and ACN as dopant solvent. An Active Exclusion setting (exclude after three spectra, release after 0.5 min) was employed in the data-dependent acquisition (DDA) method.
The acquired MSMS data was analyzed using Bruker DataAnalysis 4.2 and Bruker ProteinScape 3.1 with Matrix Science MASCOT Server 2.3. A custom database of 3,308 S. sputigena proteins was retrieved from UniProt (http://www.uniprot.org/uniprot/?query = selenomonas+sputigena) on June 10, 2014. This database was employed for the MASCOT protein search (the search parameters are listed in supplemental Table S2). Glycopeptides were identified manually from the CID and ETD fragment spectra. The ExPASy tool “PepSweetener” (http://glycoproteome.expasy.org/pepsweetener/app/) assisted in the initial identification of the peptide and glycan moieties of the glycopeptides based on precursor masses (33). The online tool Fragment Ion Calculator (Institute for Systems Biology, Seattle, WA; http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html) was further used to verify the detected fragment spectra. A technical duplicate analysis of each proteolytically digested sample was performed.
The O-glycans released by reductive β-elimination (32) were analyzed by PGC nano-flow chromatography online coupled to a Bruker amaZon Speed ETD ion trap mass spectrometer in negative ion mode. A Dionex Ultimate 3000 UHPLC was equipped with Hypercarb columns (precolumn: KappaGuard 0.3 mm [ID] × 30 mm, 5 μm particle size, 250 Å pore size, P/N 35005–030315; analytical column: 75 μm [ID] × 100 mm, 3 μm particle size, 250 Å pore size, P/N 35003–100065, both Thermo Scientific). Solvent A was 10 mm ammonium bicarbonate and solvent B was 60% ACN in 10 mm ammonium bicarbonate. The flow rate was held constant at 0.9 μl/min at 40 °C. The samples were loaded onto the trap column in solvent A. The analytical gradient increased from 2 to 35% solvent B over a period of 33 min. The ion trap was operated in the UltraScan mode in the m/z range from 200 to 1000 with an SPS Target Mass of m/z 600. The three most intense signals were selected for CID and the MSMS scan range was set to m/z 100 to 2500 in the Enhanced Resolution mode. The acquired mass spectra (MS and MSMS) were analyzed manually using Bruker DataAnalysis 4.2. O-glycans were analyzed as a technical duplicate.
Monosaccharide Analysis by Reversed Phase (RP)-HPLC
To determine the identity of the flagellin O-glycan monosaccharide constituents, the monosaccharides obtained after acid hydrolysis of released flagellin O-glycans and of glycopeptides were fluorescence-tagged with anthranilic acid (AA) and analyzed by RP-HPLC (34). Further investigation on the nature and methylation status of the sugars was performed by hypermethylation of the AA-labeled sample (35). All monosaccharide standard compounds (d-Glc, d-Gal, d-Man, d-Xyl, l-Ara, l-Fuc, l-Rha, 6-deoxy-d-glucose, d-GlcN, d-GalN, d-GlcUA, d-GalUA) as well as AA and methyl-d3 iodide (CD3I) were obtained from Sigma-Aldrich, 6-deoxy-talose was prepared in-house as described previously (35). Sequencing-grade trypsin was purchased from Promega (Madison, WI).
SDS-PAGE-separated flagellin glycoprotein bands were excised, destained, carbamidomethylated, and incubated with trypsin overnight at 37 °C. The extracted material was loaded onto RP cartridges (Phenomenex; Strata C-18E [55 μm, 70 Å] 1 ml [50 mg]) equilibrated with 80 mm formic acid (buffered to pH 3.0 with ammonia). After washing of the sample three times with 0.5 ml of the above-mentioned buffer, the retained (glyco)peptides were eluted with 0.4 ml of 65% ACN in buffer and dried in vacuo. In parallel, O-glycans were released from excised gel pieces by in-gel reductive β-elimination. For this purpose, excised, destained and dried gel pieces were covered with 1 ml of 50 mm NaOH containing 1 m sodium borohydride and incubated overnight at 50 °C. The alkaline solution containing the released O-glycans was loaded onto PGC cartridges (Supelco; Supelclean ENVI Carb SPE Tubes 3 ml [0.25 g]) previously equilibrated with 80 mm formic acid (buffered to pH 9.0 with ammonia). After washing the sample three times with 1 ml of buffer, glycans were eluted with 1.2 ml of 50% ACN in buffer and dried in vacuo. Dried glycopeptides and glycans were dissolved in 0.3 ml of 4 m trifluoroacetic acid and hydrolyzed at 100 °C for 4 h followed by drying in vacuo. Hydrolyzed samples and the monosaccharide standards (1 nmol each) were derivatized with AA following the microvial procedure (34). In short, dry samples were resuspended in 10 μl of MQ water, mixed with 70 μl of labeling solution and incubated at 80 °C for 1 h. Labeled sugars were then analyzed by RP-HPLC using a volatile buffer system (35). Separation was performed with a core-shell Kinetex C18 column (250 × 4.6 mm, 5 μm particle size, 100 Å pore size; Phenomenex, Torrance, CA) with a Security Guard Ultra precolumn (Phenomenex) on a Nexera X2 HPLC system with a RF-20Axs Fluorescence Detector, equipped with a semimicro flow cell (Shimadzu, Korneuburg, Austria). Solvent A consisted of 80 mm formic acid, buffered to pH 3.4 with ammonia, solvent B was 80% ACN in solvent A. The applied gradient started at 9.5% solvent B, raised to 21.4% over 27 min and further to 65% in 4 min at a flow rate of 0.85 ml/min; the column oven and flow cell thermostat were set to 33 °C. Fluorescence was measured with wavelengths Ex/Em 360 nm and 425 nm. Data was analyzed in Postrun Analysis with LabSolutions 5.73 (Shimadzu, Germany). For larger injection volumes, e.g. for fractionation of peaks of interest for inspection by MS, labeled samples were diluted with MQ water ad 200 μl. Up to 2 μl were directly injected without dilution. The remainder of the solution was directly used for hypermethylation of AA-labeled sugars.
For the fractionation of peaks followed by MS analysis, dried fractions after HPLC were redissolved in MQ water and separated on a Biobasic column (C18, 320 μm × 150 mm, 5 μm particle size; Thermo Scientific) run at a flow rate of 6 μl/min at 35 °C with an Ultimate 3000 system coupled to an amaZon ion trap mass spectrometer (Bruker, Bremen, Germany). Solvent A consisted of 80 mm formic acid (buffered to pH 3.0 with ammonia) and solvent B of 80% ACN in solvent A. A gradient was applied from 1.3% to 15% solvent B in 5 min, to 56% B over 6 min and 95% B in 2.5 min. After maintaining a solvent concentration of 95% B for 0.4 min, the column was flushed back to starting conditions over a period of 0.6 min and equilibrated in starting conditions. The ion trap was operated in the positive ion mode with DDA. Specific parameters were optimized for a low mass range; target mass m/z 300 and a m/z range from 150 to 600 in the Enhanced Resolution mode with 100,000 ICC target and 200 ms maximum accumulation time.
For the hypermethylation of AA-labeled sugars excess reagent was removed via a self-made microcrystalline cellulose cartridge in HILIC mode (36). The eluate was transferred into a glass vessel, dried over a stream of nitrogen, redissolved in 100 μl of dimethyl sulfoxide and mixed with a few crumbs of triturated NaOH. Subsequently, 25 μl of CD3I were added followed by incubation for 30 min with slight shaking. The reaction was stopped by addition of 0.8 ml of 30% acetic acid and products were extracted with trichloromethane. After washing the organic phase once with 30% acetic acid and twice with MQ water, it was dried under a stream of nitrogen and redissolved in 25 μl of MQ water for capillary HPLC with ESI-MS detection. Five μl of hypermethylated AA-monosaccharides were injected with a 20 μl sample loop in Microliter PickUp mode and separated on a Dionex Acclaim Pepmap (C18, 300 μm × 150 mm, 2 μm particle size; Thermo Scientific) at a flow rate of 6 μl/min at 35 °C with an Ultimate 3000 system coupled to an amaZon ion trap mass spectrometer (Bruker, Bremen, Germany). Solvent A was 80 mm formic acid (pH 3.0) and solvent B was 80% ACN in solvent A. After 5 min of flushing with 1.3% solvent B it was increased to 38.8% in 2 min and subsequently to 55% in minute 32. The ion trap was operated in the positive ion mode with DDA. Specific parameters were optimized for a low mass range - target mass m/z 400 and m/z range from 150 to 600 in the Enhanced Resolution mode with 100,000 ICC target and 200 ms maximal accumulation time. MSMS parameters used were cut-off selection default, fragmentation time 40 ms, activated FxD, UltraScan mode and m/z 80–800 scan range. Selected ion chromatograms and average MSMS spectra were generated using Bruker Data Analysis 4.0. Monosaccharide analyses were performed in duplicate from two biological replicates.
Experimental Design and Statistical Rationale
A detailed glycomics and glycoproteomics evaluation of the major flagellin glycoprotein of S. sputigena was performed using a variety of orthogonal LC-MS based techniques. Glycopeptides were generated from the SDS-PAGE separated flagellin by proteolytic in-gel digestion using two proteases with orthogonal cleavage specificities. Reversed phase LC-ESI MSMS analyses using CID and ETD fragmentation were used to identify and characterize the (glyco)peptides generated by the proteolytic enzymes. O-glycans were released by reductive β-elimination from flagellin tryptic glycopeptides extracted from gel pieces obtained after SDS-PAGE separation. The released O-glycans were identified by PGC-nanoLC ESI MSMS. Monosaccharides were produced by acid hydrolysis of flagellin glycopeptides and released O-glycans using anthranilic acid labeling and reversed phase HPLC and MS. The same hydrolyzed monosaccharide samples have been used for hypermethylation analyses. Sample preparation, followed by MS analysis was performed in triplicates (glycopeptides/peptides) and duplicates (O-glycans/monosaccharides/hypermethylation).
Data Availability
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (37) with the data set identifier PXD005859.
RESULTS
Indications of Flagellin Glycosylation in S. sputigena ATCC 35185
The separation of a crude S. sputigena cell extract by SDS-PAGE and subsequent staining with PAS reagent revealed a single prominent carbohydrate-positive protein band with an apparent size of ∼60 kDa (Fig. 1A). The identity of this band was determined based on tryptic peptides as S. sputigena flagellin UniProt Accession C9LY14 (supplemental Table S1).
Fig. 1.
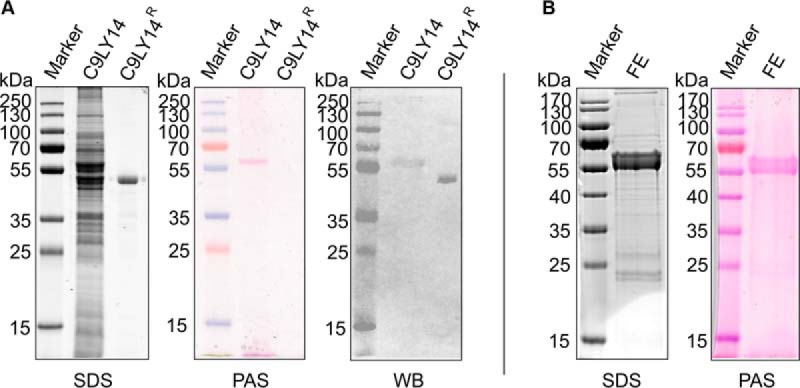
Comparison of native flagellin C9LY14 and E. coli-produced C9LY14R. A, SDS-PAGE (SDS), periodic acid-Schiff (PAS) staining and Western-immunoblotting (WB) reveal a clear shift in molecular size between native flagellin C9LY14 from a crude S. sputigena cell extract and C9LY14R indicating post-translational modification of C9LY14 by glycosylation. PageRuler Prestained Protein Ladder (Thermo Scientific) was used as marker. B, Flagellin enrichment (FE) from a S. sputigena culture followed by separation by SDS-PAGE (SDS) and PAS staining (PAS).
Although native C9LY14 protein obtained from a crude S. sputigena cell extract traveled at 55–60 kDa, a recombinant His6-tagged flagellin (C9LY14R) produced in E. coli was downshifted on the SDS-PAGE gel migrating as a PAS-negative band around the size calculated based on amino acid composition (46.1 kDa; Fig. 1A). Western-immunoblotting using a flagellin specific antibody revealed a ∼10–15 kDa difference in molecular size between C9LY14 and C9LY14R, providing a first indication of native C9LY14 being post-translationally modified.
C9LY14 is the Only Detectable Flagellin in S. sputigena ATCC 35185
Enriched native S. sputigena flagellin was separated by SDS-PAGE and the CBB-stained protein band was first analyzed using traditional bottom-up proteomics. The protein was proteolytically in-gel digested and the (glyco)peptides were extracted from the gel. Reversed-phase nanoLC-ESI-MSMS confirmed the identity of the protein being S. sputigena flagellin C9LY14 (supplemental Fig. S3). The initial MASCOT search allowed annotation of 63.2% of the protein sequence. Manual analysis of the fragment spectra showed the presence of several O-glycopeptides. Taking all the identified glycopeptides into account, this data resulted in a total sequence coverage of 80.8% (supplemental Fig. S3; for further details please see section Glycoproteomics).
Native C9LY14 exhibited a broad protein band in SDS-PAGE analysis (Fig. 1B), which is likely to be caused by its protein glycosylation. Heavy post-translational modification of C9LY14 was also indirectly supported by the fact that a plain tryptic digest performed on equal amounts of nonglycosylated C9LY14R resulted in 30% higher sequence coverage (94.5% versus 63.2% for the native protein, supplemental Fig. S3).
Glycomics on C9LY14 Flagellin Identifies Novel O-glycan Structures
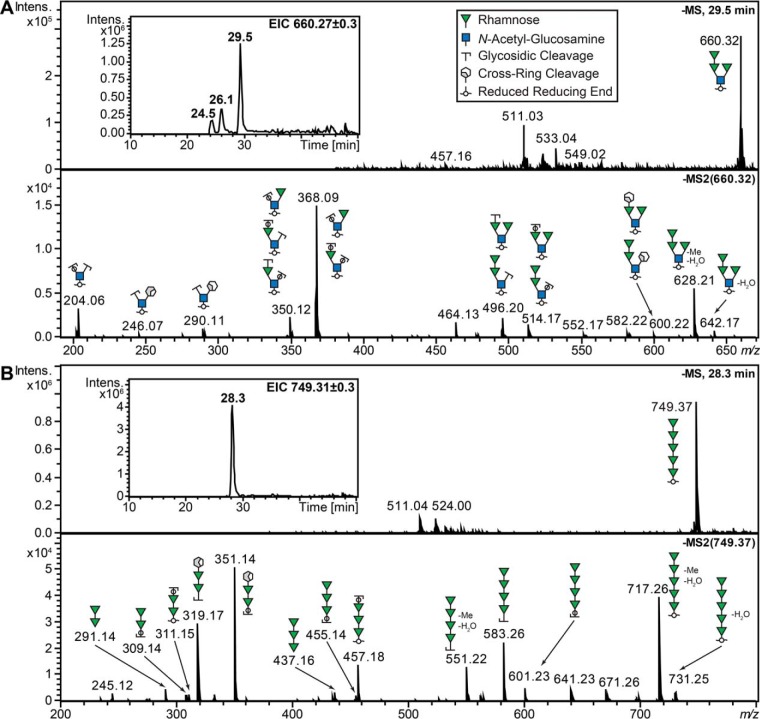
O-glycans were released from tryptic glycopeptides of S. sputigena flagellin using reductive β-elimination and analyzed using negative mode PGC nanoLC-ESI-MSMS. We identified six different O-glycans in the range from tri- to pentasaccharides that consisted mostly of deoxyhexose (dHex) and N-acetylhexosamine (HexNAc) residues (Table I; supplemental Table S5). Separate monosaccharide analyses identified these sugars to be rhamnose (Rha) and GlcNAc, respectively (vide infra). The GlcNAc residue in the Rha3GlcNAc tetrasaccharide was identified in its reduced state, indicating that the glycan is attached via the GlcNAc to the protein (Fig. 3). Specific fragments at m/z 246 and 290 also indicated the presence of a branched glycan structure as these ions were not predicted for a linear glycan of the same composition by in silico fragmentation using Glycoworkbench 2 (38). In the case of the Rha5 pentasaccharide, MSMS data confirmed the calculated composition. Taken together, these data provided evidence that at least two different forms of O-glycosylation are present on S. sputigena flagellin (Fig. 3). In addition, three modified oligosaccharides were detected exhibiting +14-Da or +42-Da modifications. The exact molecular nature of these oligosaccharide modifications could not be clarified based on this analysis alone (Table I).
Table I. O-glycans released from tryptic peptides of S. sputigena C9LY14 flagellin using reductive β-elimination. Released O-glycans were detected using PGC nanoLC-ESI MSMS. RT, retention time in minutes; all mass values are in Dalton; calc. m/z, theoretical m/z value. Structures of O-glycans in bold letters were determined based on MSMS spectra in negative ion mode. For all others, no product ion spectrum was generated, assignment based on MS1 level only.
| RT | m/z | z | calc. m/z | Δm/z | Composition | Modification | Relative abundance [%] |
|---|---|---|---|---|---|---|---|
| 24.7 | 457.16 | 1 | 457.19 | -0.03 | Rha3 | 7.6 | |
| 28.3 | 749.34 | 1 | 749.31 | 0.03 | Rha5 | 59.0 | |
| 39.9 | 763.34 | 1 | 763.32 | 0.02 | Rha5 | +14 Da | 0.9 |
| 38.6 | 791.36 | 1 | 791.32 | 0.04 | Rha5 | +42 Da | 0.5 |
| 24.5 | 660.30 | 1 | 660.27 | 0.03 | Rha3 GlcNAc | 3.8 | |
| 26.1 | 660.30 | 1 | 660.27 | 0.03 | Rha3 GlcNAc | 8.4 | |
| 29.5 | 660.30 | 1 | 660.27 | 0.03 | Rha3 GlcNAc | 19.3 | |
| 37.9 | 702.31 | 1 | 702.28 | 0.03 | Rha3 GlcNAc | +42 Da | 0.5 |
Fig. 3.
Negative mode CID product ion spectra of PGC-nanoLC separated Rha3GlcNAc and Rha5O-glycans obtained using reductive β-elimination from tryptic peptides of S. sputigena C9LY14 flagellin. The monosaccharide identities are assigned based on separate monosaccharide analyses (Fig. 4). The putative glycan structures are represented using SNFG nomenclature (63). A, Three isoforms of the Rha3GlcNAc tetrasaccharide were detected (extracted ion chromatogram (EIC) inset), but product ion spectra were only obtained for the tetrasaccharide eluting at 29.5 min. The m/z 204 oxonium ion indicated that the glycan was attached to the protein via the GlcNAc residue, as this value corresponds to the reduced GlcNAc oxonium ion when analyzed as deprotonated molecules in negative ion mode. Fragments m/z 246 and 290 indicated a branched glycan structure as these ions were not predicted for a linear structure of the same composition (in silico fragmentation using Glycoworkbench 2 (38)). B, The Rha5 pentasaccharide was detected as a single structure (see EIC inset). Based on the product ion spectra a linear structure provides the best explanation for this oligosaccharide. The fragment ions m/z 717 and 551 corresponded to the loss of water and a methyl group (Me).
C9LY14 Flagellin O-glycans Consist of Rhamnose and N-Acetylglucosamine
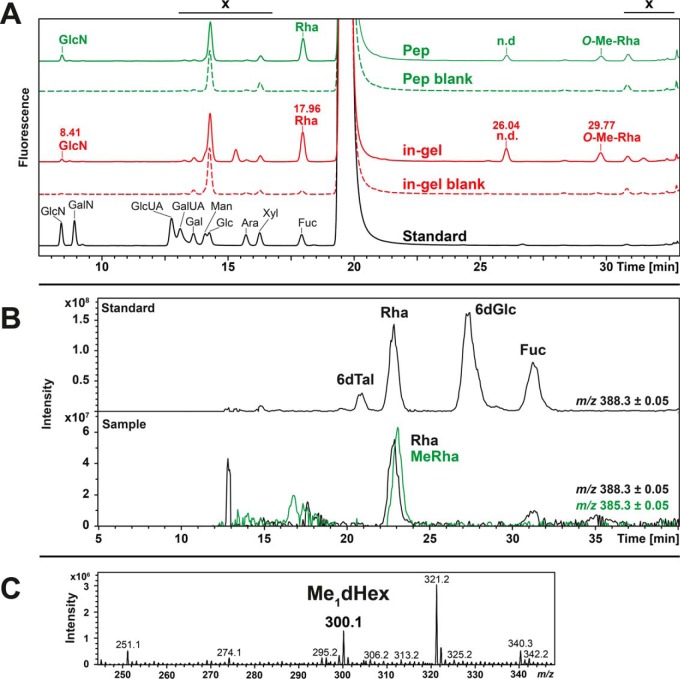
Without any supporting information on S. sputigena's biosynthetic protein glycosylation machinery it was impossible to assign monosaccharide identities directly from the glycomics data. Therefore, monosaccharide analyses were performed on the released O-glycans as well as on glycopeptides obtained after tryptic digestion. Following acid hydrolysis and AA-labeling, the monosaccharide constituents were identified by RP-HPLC. The main monosaccharides present on S. sputigena flagellin C9LY14 were assigned based on the comparison with defined monosaccharide standards. Blank SDS-PAGE gel pieces from the same run were used to estimate background signals.
From the trypsin-digested glycopeptides and the β-eliminated O-glycan samples, Fuc/Rha were identified as the major monosaccharide constituents (Fig. 4A). However, Fuc and Rha share the same retention time in the classical reversed phase HPLC separation with FLD and could, thus, at first not be distinguished. Hypermethylation of the alkaline-released AA-labeled samples, however, allowed differentiation of these two deoxysugars, revealing that Rha was the deoxysugar incorporated into C9LY14 O-glycans. Minor amounts of Fuc were detected that were derived from the sample matrix as they were also present in HPLC-FLD of the gel blank (Fig. 4A).
Fig. 4.
Identification of monosaccharide constituents of the S. sputigena C9LY14 flagellin O-glycan. A, HPLC with fluorescence detection of anthranilic acid (AA)-labeled monosaccharides revealed four major residues. The C9LY14 flagellin O-glycan monosaccharide constituents were derived from tryptic peptides (Pep, green traces) and in-gel alkaline glycan release (in-gel, red traces). Empty SDS-PAGE gel pieces (blank) were used as controls. The identity of Rha (17.96 min) and O-Me-Rha (29.77 min) was determined by hypermethylation (see parts b and c). X refers to nonglycan related matrix peaks. B, Capillary RP-LC-ESI-MS of CD3I hypermethylated AA-labeled monosaccharides. Comparison with appropriate standards (top base peak chromatogram) allowed identification of Rha and methylated Rha as the deoxyhexoses present on S. sputigena C9LY14 flagellin (bottom base peak chromatogram). Retention time shifts were because of the deuterium effect (64). C, MS spectrum of a fraction collected after HPLC of AA-labeled monosaccharides. The m/z of the detected signal corresponded to the singly methylated, AA-labeled version of Rha.
Selenomonas sputigena flagellin C9LY14 contained two additional sugars for which the retention times did not agree with any of the available standards (retention times 26 min and 29.8 min; Fig. 4A). Their late elution during reversed-phase chromatography (after 25 min) indicated that these compounds exhibited a higher hydrophobicity, potentially introduced by modifications such as methylation. Peaks of interest were collected for further investigations by LC-ESI-MS. For the peak eluting at 26 min, so far, no solid conclusions could be drawn; for the fraction containing the peak with retention time 29.8 min, a signal was detected at m/z 300.1, which corresponded to a protonated, singly methylated dHex residue carrying the AA fluorescence tag (Fig. 4C). Hypermethylation of the AA-labeled sample, performed with deuterated methyliodide, identified this dHex sugar as methylated Rha (Fig. 4B).
Because of the acid hydrolysis applied during sample processing, GlcNAc residues were detected as GlcN (Fig. 4A). The reducing end sugar should in principal no longer be available for labeling via reductive amination after reductive β-elimination. Given the fact that the peak area of the GlcN peak was clearly increased in the tryptic glycopeptide sample and no further peaks could be assigned either to ManN (eluting between GlcN and GalN; (35)) or GalN, the GlcN signal detected in the β-eliminated sample was a matrix background effect rather than a nonreducing end GlcNAc of O-glycans. These results agreed with the nanoLC-ESI-MSMS O-glycomics results. In summary, they confirm that the GlcNAc-containing O-glycans are linked to the peptide via the GlcNAc.
Glycoproteomics Uncovers C9LY14 Flagellin to be a Highly Heterogeneous Glycoprotein
In total we identified 22 unique glycopeptide species derived from S. sputigena flagellin C9LY14 (Table II). This list of unique glycopeptides was distilled from 64 detected compounds exhibiting glycopeptide-specific features (supplemental Table S3). For 27 of these compounds, the glycopeptide identity was confirmed from the CID and ETD data (supplemental Table S4). The remaining 37 compounds were classified as likely glycopeptides based on their precursor masses, oxonium ion peaks, and/or peaks corresponding to the sequential loss of monosaccharides from glycopeptides, but no peptide specific fragment ions were detected in these spectra (supplemental Fig. S1). A precursor mass (deconvoluted from measured m/z and z) was taken as an indicator for a glycopeptide if it was equal (max. mass error ± 0.3 Da) to the sum of a peptide (from in silico digests) and a plausible glycan mass (composition range GlcNAc0–2Rha0–5 plus possible modifications). In addition, oxonium ions and/or specific fragmentation patterns were required for confident glycopeptide assignment (supplemental Fig. S1).
Table II. Summary of all 22 unique glycopeptide species identified from S. sputigena flagellin C9LY14. Identified glycosylation sites are indicated in bold, red letters. RT, retention time in minutes; all mass values are in Dalton. * Indicates glycopeptides where the site of glycosylation could not be unambiguously identified.
| RT | m/z | z | calc. m/z | Δm/z | Peptide | Glycan | Modification |
|---|---|---|---|---|---|---|---|
| 23.2 | 785.42 | 2 | 785.36 | -0.06 | 191VNQGKTETTSF201 | Rha2 | +14 Da +52.1 Da |
| 23.4 | 842.48 | 2 | 842.39 | -0.09 | 179NIHSQDTITVSY190 | Rha2 | +14 Da |
| 25.8 | 956.02 | 2 | 956.04 | 0.02 | 179NIHSQDTITVSY190* | Rha2 | +14 Da +227.3 Da |
| 25.1 | 1164.09 | 2 | 1164.08 | -0.01 | 326AQALGLQGSDGSTLNISTR344 | Rha3 | |
| 25.4 | 1171.08 | 2 | 1171.07 | -0.01 | 326AQALGLQGSDGSTLNISTR344 | Rha3 | +14 Da |
| 25.3 | 1310.12 | 2 | 1310.13 | 0.01 | 326AQALGLQGSDGSTLNISTR344* | Rha5 | |
| 16.1 | 581.79 | 2 | 581.77 | -0.02 | 196TETTSFK202 | RhaGlcNAc | |
| 17.0 | 561.28 | 3 | 561.23 | -0.05 | 191VNQGKTETTSF201 | GlcNAc | +267.1 Da |
| 17.4 | 780.84 | 2 | 780.87 | 0.03 | 191VNQGKTETTSF201 | RhaGlcNAc | |
| 17.7 | 787.84 | 2 | 787.87 | 0.03 | 191VNQGKTETTSF201* | RhaGlcNAc | +14 Da |
| 18.8 | 801.91 | 2 | 801.92 | 0.01 | 191VNQGKTETTSF201 | RhaGlcNAc | +42 Da |
| 18.9 | 947.92 | 2 | 947.93 | 0.01 | 191VNQGKTETTSF201 | Rha3GlcNAc | +42 Da |
| 19.2 | 780.88 | 2 | 780.87 | -0.01 | 142NQKVTATTTTF152* | RhaGlcNAc | |
| 20.1 | 801.86 | 2 | 801.94 | 0.08 | 142NQKVTATTTTF152 | RhaGlcNAc | +42 Da |
| 20.5 | 947.92 | 2 | 947.95 | 0.03 | 142NQKVTATTTTF152 | Rha3GlcNAc | +42 Da |
| 20.9 | 742.43 | 3 | 742.40 | -0.03 | 136LVDGTHNQKVTATTTTF152* | RhaGlcNAc | +42 Da |
| 24.3 | 842.94 | 2 | 842.93 | -0.01 | 256AAGTGVGKSVGGITF270 | RhaGlcNAc | +14 Da |
| 24.5 | 982.05 | 2 | 981.99 | -0.06 | 256AAGTGVGKSVGGITF270 | Rha3GlcNAc | |
| 24.7 | 835.97 | 2 | 835.93 | -0.04 | 256AAGTGVGKSVGGITF270 | RhaGlcNAc | |
| 24.9 | 856.89 | 2 | 856.98 | 0.09 | 256AAGTGVGKSVGGITF270 | RhaGlcNAc | +42 Da |
| 24.9 | 1003.08 | 2 | 1002.99 | -0.09 | 256AAGTGVGKSVGGITF270 | Rha3GlcNAc | +42 Da |
| 25.3 | 724.35 | 2 | 724.34 | -0.01 | 245TASGESAISLY255* | RhaGlcNAc |
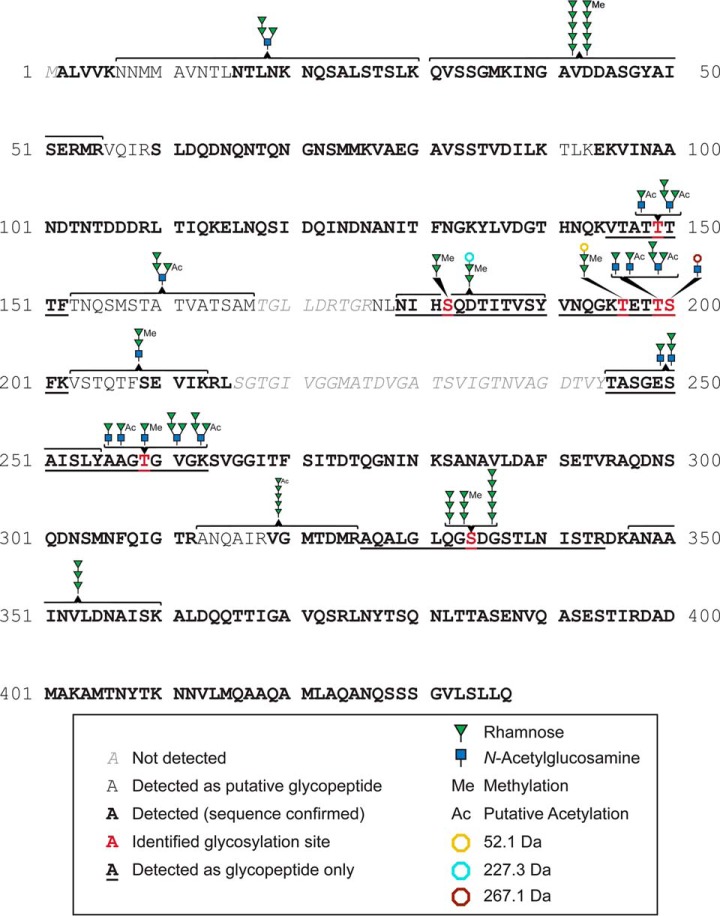
From all detected glycopeptides, six were exclusively detected as glycopeptides, whereas six additional ones were also found in their nonglycosylated form, which lead us to conclude that C9LY14 flagellin also shows a considerable degree of glyco-macroheterogeneity (Fig. 2). The site of glycosylation could be unambiguously identified for five glycopeptides at amino acid positions Thr149, Ser182, Thr199, Thr259, and Ser334. In addition to these five sites, Thr196 and Ser200 were also identified as being glycosylated carrying di- and monosaccharides, respectively, with further not yet defined substitutions of +52.1 Da and +267.1 Da (Fig. 2, Table II, supplemental Table S4). Substantial glyco-microheterogeneity was present on most detected glycopeptides (Fig. 5, supplemental Table S4, supplemental Fig. S5). In agreement with the glycomics data and the monosaccharide analyses, the attached O-glycans were mainly comprised of Rha and GlcNAc residues. Interestingly, the O-glycan compositions detected on the glycopeptides exhibited even larger composition diversity than found by the glycomics analyses. Many glycopeptides were detected having one, two, three or five Rha residues attached to the peptide backbone, others were found to carry GlcNAc extended by up to five additional Rha residues. Glycans consisting of an odd number of Rha residues appeared more frequent compared with glycans with two or four Rha residues (supplemental Table S4).
Fig. 2.
Schematic overview of C9LY14 flagellin primary structure, combining data obtained from tryptic and chymotryptic (glyco)peptides (Table II, supplemental Table S4). Bold amino acids indicate sequence confirmation on MSMS level, amino acid stretches containing nonbold letters indicate assignment as glycopeptide by characteristic glycosylation fragments, but insufficient peptide fragment coverage. Five sites of glycosylation (Thr149, Ser182, Thr199, Thr259, and Ser334) were unambiguously identified using CID and ETD fragmentation (red labeled Ser/Thr residues). In addition, Thr196 and Ser200 were found to carry di- and monosaccharides, respectively, with further not yet defined substitutions of +52.1 Da and +267.1 Da. The putative glycan structures detected at those sites are described using the SNFG nomenclature (63). If more than a single structure was found attached to a glycopeptide (microheterogeneity), the structures indicated on top of a glycopeptide are sorted according to their mass. The identity of the respective monosaccharides was determined by separate monosaccharide analyses (Fig. 4). Six glycopeptides were detected exclusively in their glycosylated form (underlined sequence stretches), whereas the others were also found in a nonglycosylated form (macroheterogeneity). Six additional glycopeptides, for which no specific glycosylation site could be assigned, are indicated by the wide brackets pointing toward the glycan. These glycopeptides were assigned based on their precursor mass, oxonium ions, and ions representing neutral loss of monosaccharide residues from the glycopeptide.
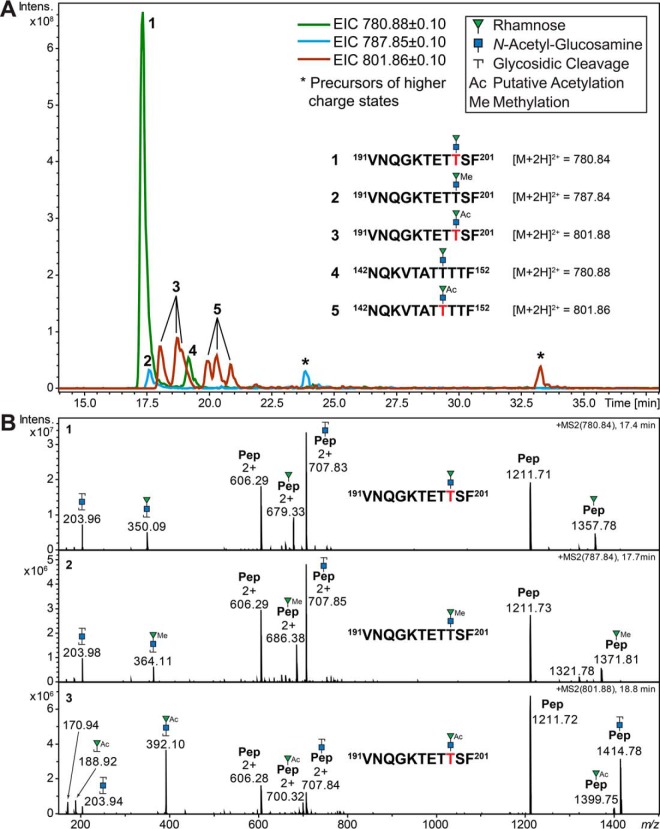
Fig. 5.
Example of glyco-microheterogeneity shown for five selected glycopeptides with a peptide backbone mass difference of just 0. 036 Da (191VNQGKTETTSF201, M = 1210.583 Da and 142NQKVTATTTTF152, M = 1210.619 Da). A, Extracted ion chromatograms (EIC) showing three m/z values reflecting the RhaGlcNAc disaccharide and its modifications (methylation and putative acetylation) attached to the two peptide backbones. Although the nonmodified disaccharide is the dominant form on glycopeptide 191–201, the later eluting glycopeptide 142–152 shows higher levels of putative acetylation. A single peak was detected for the methylated version of glycopeptide 191–201, but none for glycopeptide 142–152. Both glycopeptides exhibited three peaks for the putative acetylated glycoform, possibly indicating that different hydroxyl groups are modified on the rhamnose (see also (B)) in a site-specific manner. B, Representative CID product ion spectra for the three glycoforms of glycopeptide 191VNQGKTETTSF201. The oxonium ions clearly indicate that methylation (364.1 Da) and putative acetylation (392.1 Da) are exclusively occurring on the nonreducing end rhamnose. The native disaccharide and its methylated form exhibited a strong pseudo Y1-glycopeptide signal reflecting the peptide plus one Rha that is best explained by a Rha rearrangement (signals at 679.332+/1357.78+ and 686.382+/1371.81+, respectively). This is almost entirely abolished when the Rha residue is modified by putative acetylation (bottom spectrum). Red letters indicate sites of glycosylation unambiguously identified in separate ETD product ion spectra (see also Fig. 6 and supplemental Fig. S5).
Additionally, either of two distinct modifications was detected on 31 out of all 64 detected glycopeptides contributing to high glyco-microheterogeneity. A +14-Da mass shift was present on several precursor ions indicating that part of the glycans experienced some form of methylation during biosynthesis. Furthermore, several glycopeptide molecules exhibited a +42 Da shift possibly corresponding to putative acetylation (Table II, Fig. 5, supplemental Fig. S5). The glycomics data and glycopeptide fragment spectra clearly showed that these modifications were present on the glycan moieties and not on any of the amino acids (Fig. 5, supplemental Fig. S5). Besides methylation and putative acetylation, further modifications were detected on some glycopeptides that could not yet be unambiguously annotated. On Ser200 of peptide 191VNQGKTETTSF201, one GlcNAc residue was detected that showed an additional mass increment of 267.1 Da, which yet could not be associated with any known molecule (Table II, supplemental Table S4, supplemental Fig. S2). The same peptide sequence was also detected with another modified glycan corresponding to two Rha residues, one methylation and an additional +52.1-Da mass increment, all forming one molecule attached to Thr196 (Table II, supplemental Fig. S5J). Peptide 179NIHSQDTITVSY190 was found having an additional mass of 533.4 Da attached to it (Table II, supplemental Fig. S5V). CID fragmentation showed that two Rha residues and one methylation are part of that modification, but to date the remaining 227.3 Da could not be explained by any of the known glycosylation modifications.
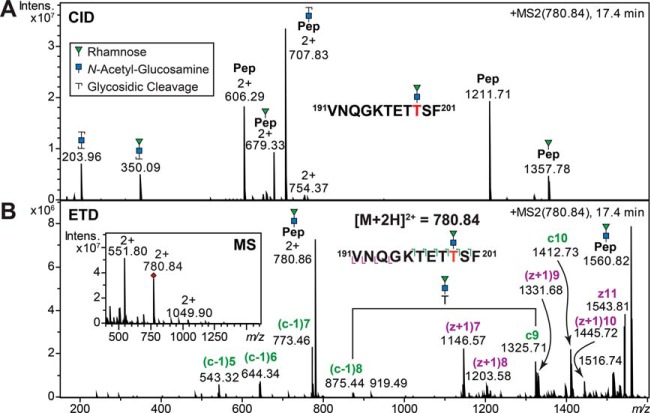
Nonreducing End Rhamnose Rearrangement Produces Pseudo Y1-Ion Fragments
Manual data evaluation revealed a series of glycopeptides carrying a RhaGlcNAc disaccharide including their methylated and putatively acetylated species. Interestingly, in numerous glycopeptide CID product ion spectra, intense Y1-glycopeptide ions were detected in the same product ion spectrum that indicated the attachment of one GlcNAc and one Rha residue at independent sites within a single peptide backbone (Figs. 5 and 6). Rha-linked O-glycans were also identified on S. sputigena flagellin C9LY14 before (Table I, Fig. 3), thus the occurrence of a glycopeptide with two sites of glycosylation was entirely plausible. However, the detected oxonium ions contradicted this interpretation as these hinted toward the attachment of a single disaccharide. Subjecting the respective glycopeptides to ETD analyses showed that a disaccharide moiety was attached to a single amino acid (Fig. 6). These results led us to conclude that the observed Rha-Y1 glycopeptide signals were more likely the product of a Rha rearrangement resulting from the migration of the Rha residue toward the peptide (Fig. 6B). This interpretation was further substantiated by the observation that the level of Rha migration was influenced by the molecular nature of the glycan moiety. Similar high levels of Rha rearrangement were observed for the native RhaGlcNAc disaccharide and its singly methylated form. However, if Rha was modified by putative acetylation, hardly any pseudo Rha-Y1 signals were detected in the product ion spectra but a large singly charged oxonium ion at m/z 392.1 (Fig. 5). In the case of these RhaGlcNAc glycopeptides, this behavior appeared to be independent of the peptide backbone, as all glycopeptides carrying these disaccharides exhibited similar rearrangement fragmentation patterns.
Fig. 6.
Example of Rha rearrangement occurring on glycopeptides. A, Under low energy CID conditions this rearrangement results in the formation of a pseudo Y1-glycopeptide signal simulating two independent sites of glycosylation. B, ETD analysis of the very same precursor ion verified that just a single disaccharide was attached on one site rather than two monosaccharides on two independent sites.
DISCUSSION
In this study, we provide the first clear evidence that S. sputigena ATCC 35185, a Gram-negative bacterium that has emerged as a potential periodontal pathogen, glycosylates its flagella. We show that the S. sputigena flagellin C9LY14 is modified with a heterogeneous set of O-linked glycans at multiple serine and threonine residues and report hitherto not described prokaryotic O-glycans that S. sputigena uses to heavily modify its flagellar glycoprotein. From the multiple sites of glycosylation, five (i.e. Thr149, Ser182, Thr199, Thr259, and Ser334) could be unambiguously identified and from our data it is evident that the 437-amino acid long flagellin protein exhibits extensive glycan macro- and microheterogeneity. This glyco-heterogeneity explains why the PAS-stained flagellin-enriched sample migrated as a rather broad glycoprotein band on the SDS-PAGE gel (Fig. 1B). It needs to be mentioned that next to C9LY14, a second protein (UniProt Accession F4EYJ4) has been annotated as flagellar protein FliS in the S. sputigena ATCC 35185 genome sequence (Nucleotide Accession NC_015437.1). C9LY14 and F4EYJ4 show 58% amino acid identity and both proteins share similarities with flagellins from Selenomonas ruminantium and other oral Selenomonas species, as well as with Anerovibrio lipolyticus and species belonging to the genus Mitsokuella, all of them members of the Acidaminococcaceae family. F4EYJ4, despite its mass difference of only ∼2 kDa to C9LY14, was not detectable in any of the analyzed SDS-PAGE separated bands, confirming that all presented data on flagellin glycosylation are attributable to C9LY14.
A considerably heterogeneous pool of O-glycans ranging from mono- to hexasaccharides was identified on C9LY14, consisting mainly of Rha and GlcNAc residues (Table I, Fig. 2). Our data on these hitherto not described O-glycans supported evidence for the presence of branched and linear structures, indicating that the S. sputigena glycosylation machinery can produce and attach a large variety of different glycans onto different sites of glycosylation (Fig. 3). Glycopeptides were detected carrying glycans composed of either one, two, three or five Rha residues or one GlcNAc residue extended by one to five Rha residues. Future work will show whether S. sputigena O-glycosylation is accomplished by stepwise addition of single monosaccharides or whether preformed glycan precursors are transferred onto the flagellin protein, and which glycosyltransferases are responsible for C9LY14 glycosylation. The CAZy database for Carbohydrate Active EnZymes (http://www.cazy.org/b1644.html) indicates 45 glycosyltransferases affiliated to various glycosyltransferase families to be present in the S. sputigena ATCC 35185 genome. Four of them (i.e. C9LY10–13) are encoded by genes that are in immediate vicinity of the flagellin structural protein C9LY14, and, thus, constitute promising candidates for involvement in flagellin glycosylation. Further, F4EWP7 possesses a Wzy_C motif typical of O-antigen polymerases, ligases and oligosaccharyltransferases. However, because S. sputigena ATCC 35185 has been recalcitrant to genetic manipulation so far, it was impossible to prove the involvement of any of these enzymes in S. sputigena flagellin glycosylation. The observed glycan microheterogeneity together with the occurrence of both threonine and serine residues as acceptor amino acids rather hints toward stepwise addition and elongation, as bacteria with preformed precursors usually exhibit a more homogenous glycosylation pattern (39). Nevertheless, relaxed specificities of bacterial O-oligosaccharyltransferases have also been described previously (40).
The O-glycan monosaccharide constituents were unambiguously identified after AA-labeling using RP-HPLC as Rha, O-Me-Rha and GlcNAc (Fig. 4). The deoxyhexose Rha is widely distributed among bacteria and plants. Although d-Rha is mainly found as a constituent of EPS or LPS of Gram-negative bacteria, the l-Rha isomer is more common (41). In flagellar glycans, however, Rha is rather unusual. So far Rha has been described as a component of flagellin O-glycosylation in the opportunistic human pathogen Pseudomonas aeruginosa, and in the plant-growth promoting rhizobacterium Azospirillum brasilense. In P. aeruginosa strains, l-Rha represents the reducing end sugar, either as a single modification or further substituted through sequential extension with a heterogeneous glycan (11). In Azospirillum brasilense, both l-Rha and l-Fuc have been reported in a repetitive glycan structure linked to the bacterial flagellin (42). Furthermore, Rha is part of a common unique trisaccharide on the flagellins of the phytopathogenic bacteria Pseudomonas syringae pv. glycinea race 4 and P. syringae pv. tabaci 6605 (43). The genomic information available for S. sputigena ATCC 35185 supports the glycoproteomics data of l-Rha modification of flagellin as presented in this study, because the bacterium possesses homologs of the four enzymes RmlA-D required for the biosynthesis of dTDP-l-rhamnose (41). Specifically, these are C9LUR1 (glucose-1-phosphate thymidilyltransferase, RmlA), C9LUQ9 (dTDP-d-glucose 4,6-dehydratase, RmlB), C9LUR0 (dTDP-4-dehydrorhamnose 3,5-epimerase, RmlC), and C9LUI6 (dTDP-4-dehydrorhamnose reductase, RmlD). However, a dedicated rhamnosyltransferase for the transfer of Rha from its nucleotide-activated form as the reducing end sugar of a glycan to a polypeptide backbone remains to be found.
N-Acetylglucosamine is a common precursor in many carbohydrate biosynthetic pathways in bacteria, including that of peptidoglycan and, thus, is synthesized during the bacterium's housekeeping functions. There is, however, the necessity of a dedicated O-GlcNAc transferase that catalyzes the transfer of the GlcNAc residue (or a preassembled glycan with a reducing end GlcNAc) to the protein. Twine et al. (44) identified an O-linked HexNAc modification - later to be recognized as O-GlcNAc (45) - on the flagellin of Clostridium difficile and described a glycosyltransferase (UniProt Accession CD0240) from the flagellar biosynthesis gene cluster to be involved in the glycosylation process. In the S. sputigena genome, two genes encoding glycosyltransferase family 2 proteins (UniProt Accessions C9LY12 and C9LY13) are present adjacent to the flagellin protein C9LY14 encoding gene that exhibit similarity to the C. difficile transferase (36 and 32% sequence identity with C9LY12 and C9LY13, respectively). Other genes on the S. sputigena ATCC 35185 genome annotated to encode glycosyltransferases are C9LY10 and C9LY11. Future work will show what role these candidate enzymes play in flagellin glycosylation and whether the gene locus encompassing the genes C9LY10 through C9LY14 represents a real flagellin glycosylation gene locus of S. sputigena ATCC 35185. Probing a flagellin-enriched preparation or a S. sputigena cell lysate with a β-O-linked GlcNAc specific monoclonal antibody (OGT Monoclonal Antibody RL2, Thermo Scientific) did not give any signal (Cornelia B. Rath, Christina Schäffer, unpublished observation). However, as most GlcNAc residues on C9LY14 are further substituted by Rha residues and/or other modifications (Fig. 3) it is likely that antibody recognition is impaired by these modifications.
Mass spectrometry is one of the most frequently applied methods to determine the structure of glycoconjugates. In the analysis of released glycans, monosaccharide rearrangement occurs commonly when protonated glycans are analyzed by CID. However, this has rarely been reported to occur on glycopeptides. In these reports, however, it did not significantly influence correct glycopeptide assignment (24–26). Our data clearly shows that monosaccharide rearrangement observed in CID analyses of protonated glycopeptides can also significantly interfere with correct data interpretation as pseudo Y1-ions are generated that pretend the presence of more than a single site of glycosylation (Figs. 5 and 6). The extent of this pseudo Y1-ion formation was glycan structure- and size-dependent, whereas the peptide moiety did not seem to influence the observed behavior. In agreement with earlier studies performed on methylated and peracetylated glycans, the rearrangement was essentially abolished in the presence of a putative acetylation (Fig. 5) (46, 47). Nevertheless, the question remains if and how a specific glycan structure influences the extent of Rha migration and whether the observed fragmentation patterns possibly offer additional opportunities to extract more details on the glycan structure. It would be highly interesting if information such as Rha linkage and/or stereochemistry could be gathered from these observed fragmentation patterns, however earlier studies performed on protonated oligosaccharides indicated that this phenomenon occurred for all glycosidic linkage types and seemed to be independent of the type of residue lost (48). In any case our data support that in addition to conventional CID a combination of orthogonal fragmentation techniques such as ETD should be used in the primary structure evaluation of glycoconjugates from poorly understood organisms.
Besides glycosylation, additional modifications were observed on both the released flagellin C9LY14 glycans and the glycoprotein. Observed mass increments of +14 Da or +42 Da were annotated here as methylation, or are indicative of putative acetylation, respectively. The presence of Rha methylation was confirmed by hypermethylation of AA-labeled monosaccharide samples. Modification by acetylation, however, needs further verification by orthogonal methods such as NMR. As only a limited amount of purified flagellin material was available, future work will clarify this matter. Besides methylation and putative acetylation other modifications were also detected on some glycopeptides. These could, however, not be annotated based on the detected mass increments. Peptide 191VNQGKTETTSF201 was identified carrying a single GlcNAc residue attached to Ser200 that further exhibited a mass increment of +267.1 Da, which molecular nature needs yet to be determined (supplemental Fig. S2). A methylated di-Rha glycosylated glycopeptide was detected with a +52.1-Da increment (Table II). This modified disaccharide was attached to Thr196 on the very same peptide (Supplemental Fig. 5J). Glycopeptide 179NIHSQDTITVSY190 was found to carry an additional mass of +533.4 Da, which could be partially characterized to consist of a singly methylated Rha disaccharide carrying an additional modification of +227.3 Da (supplemental Fig. S5V). This mass increment, too, requires future investigations to uncover its detailed molecular nature. Our data clearly indicate that the modifications present on the S. sputigena flagellin glycoprotein require a yet to be identified subset of different enzymes that introduce the multitude of identified modifications. The fact that even scrupulous manual data investigations did not allow annotation of distinct peptides present in the native flagellin (e.g. amino acid residues 153–178 and 216–244), indicates that the native flagellin is subject to other forms of modification that we were not able to decipher, yet. Both were clearly identified in the recombinant form of the protein (supplemental Fig. S3).
The glycosylation sites of most flagellin glycoproteins described to date appear to be focused around the central variable region of the protein (49, 50), which forms the outside surface-exposed domains (D2 and D3) in the assembled filament (11). In contrast, multiple amino acid sequence alignment indicates that the N- and C-terminal regions, corresponding to the D0 and D1 domains, are well conserved in flagellin proteins (11). Several amino acids in the D0/D1 region have, in many cases, proven to be crucial for the formation of the flagellar filament and, therefore, motility (51). Taking into consideration that we identified potential glycopeptides located at the N-terminal part of the S. sputigena flagellin in addition to C-terminally located ones (Fig. 2), it could well be that glycosylation in these parts of the protein influences flagellin monomer interaction and, therefore, filament stability.
The innate immune receptor Toll-like receptor 5 (TLR5), of which flagellin is the natural ligand, recognizes a site of the protein that lies predominantly in the N-terminal D1 domain, where it is centered on a stretch of eight amino acids (89–96 in Salmonella flagellin), with contribution from the C-terminal D1 domain additionally required (51–53). Recently, a β-hairpin structure adjacent to the N-terminal D1 domain in S. enterica ssp. enteritidis 90–13-706 has been identified as a third region fundamental for TLR5 activation in mammals (54). In the S. sputigena flagellin C9LY14, the glycosylated residues T149, S182 and T199 are very close to the N-terminal D1/β-hairpin region (Fig. 2). Glycosylation of flagellin so close toward the N terminus of the protein is unusual and has so far only been described for C. jejuni FlaA (55) and for the periplasmic flagella of the spirochete Treponema denticola, for which modification with a novel O-glycan within the conserved D1 domain has been reported very recently (56). It is tempting to speculate that glycans in the proteins' N-terminal region could potentially affect recognition of flagellin monomers by TLR5 and it will be an interesting future task to establish the role post-translational modifications of these residues play in a larger systems biology context.
Still, the intriguing question remains what function S. sputigena flagellin glycosylation fulfills. Being exposed on the bacterial cell surface and especially being part of an important motility structure, an essential function for flagella assembly, motility or bacterium-host/bacterium-environment interactions can be assumed. Only a relatively small subset of oral bacteria produce flagella and, in most cases (with the exception of Treponema denticola (57)) flagella and motility have not been strongly correlated with the colonization or virulence of bacteria identified from dental plaque. It is, however, likely that oral Selenomonas—constituting a relevant portion of the biomass (58, 59)—do in some way contribute to the pathogenic processes involved in periodontal disease. Being motile, S. sputigena could locate a favorable environment within dental plaque and develop a strong association with distinct bacteria on selective recognition of the partner cell surface (60). As unfortunately no genetic tools are currently available that allow manipulation of the S. sputigena genome, it will remain difficult to elucidate the functional role of S. sputigena flagellin glycosylation.
Considering the diversity of unique glycopeptide species and their distribution over a large portion of the protein sequence, the S. sputigena flagellin currently represents one of the most heavily glycosylated flagellins. This study represents the first step toward understanding how these Rha and GlcNAc/Rha containing O-glycans with their various modifications influence or control flagellum function and, eventually, pave the way for S. sputigena's establishment within dental plaque.
This study increases our understanding of the surface properties of oral bacteria, covering a critical gap not only in the biology of the oral cavity, but of flagellated Selenomonas species in general. These are associated with different parts of the digestive system—mouth, throat, gut (61, 62)—where they can be beneficial or involved in the degradation of local site health, either by facilitating the co-organization of other bacteria or by stimulating the host negatively. Digestive tract health is a vital component of systemic health. Various metagenomics approaches are currently undertaken to understand the biology of the bacterial cell surface and bacterial assembly in the context of systemic health. These, however, are frequently limited by the lack of experimental data for mechanistic validation. Bacterial cell surface glycoproteins are known regulators of bacterial cell-cell interactions and modulators of host cells' functions. Detailed glycoproteomics insights into their individual glycoprotein signatures thus are ideally suited to complement metagenomics data sets to decipher the biological basis of digestive tract systemic health.
DATA AVAILABILITY
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (37) with the data set identifier PXD005859.
Supplementary Material
Footnotes
* This work was supported by the Austrian Science Fund FWF, projects P26836-B22 (to C.S.) and the PhD Programme “Biomolecular Technology of Proteins” W1224. F.S, P.H.S. and D.K. gratefully acknowledge generous funding by the Max Planck Society. D.K. acknowledges support by the European Union (Seventh Framework Programme “Glycoproteomics”, grant number PCIG09-GA-2011-293847 and IBD-BIOM project, grant number 305479). DK is the recipient of an Australian Research Council Future Fellowship (project number FT160100344) funded by the Australian Government.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CID
- collision-induced dissociation
- AA
- anthranilic acid
- ACN
- acetonitrile
- BHI
- Brain-Heart Infusion
- CBB
- Coomassie Brilliant Blue G-250
- d-Gal
- d-galactose
- d-GalN
- d-galactosamine
- d-GalUA
- d-galacturonic acid
- d-Glc
- d-glucose
- d-GlcN
- d-glucosamine
- d-GlcUA
- d-glucuronic acid
- dHex
- deoxyhexose
- d-Man
- d-mannose
- d-Xyl
- d-xylose
- ETD
- electron transfer dissociation
- FLD
- fluorescence detection
- GdHCl
- guanidinium hydrochloride
- GlcNAc
- N-acetylglucosamine
- HexNAc
- N-acetylhexosamine
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- l-Ara
- l-arabinose
- LB
- Luria-Bertani
- l-Fuc
- l-fucose
- l-Rha
- l-rhamnose
- O-Me-Rha
- O-methyl-rhamnose
- PAS
- periodic acid-Schiff
- PBS
- phosphate-buffered saline
- PGC
- porous graphitized carbon.
REFERENCES
- 1. Adler J. (1966) Chemotaxis in bacteria. Science 153, 708–716 [DOI] [PubMed] [Google Scholar]
- 2. Josenhans C., and Suerbaum S. (2002) The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614 [DOI] [PubMed] [Google Scholar]
- 3. Duan Q., Zhou M., Zhu L., and Zhu G. (2013) Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8 [DOI] [PubMed] [Google Scholar]
- 4. Macnab R. M. (2003) How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100 [DOI] [PubMed] [Google Scholar]
- 5. Macnab R. M. (2004) Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694, 207–217 [DOI] [PubMed] [Google Scholar]
- 6. Penn C. W., and Luke C. J. (1992) Bacterial flagellar diversity and significance in pathogenesis. FEMS Microbiol. Lett. 100, 331–336 [DOI] [PubMed] [Google Scholar]
- 7. Bardy S. L., Ng S. Y., and Jarrell K. F. (2003) Prokaryotic motility structures. Microbiology 149, 295–304 [DOI] [PubMed] [Google Scholar]
- 8. Namba K., and Vonderviszt F. (1997) Molecular architecture of bacterial flagellum. Q. Rev. Biophys. 30, 1–65 [DOI] [PubMed] [Google Scholar]
- 9. Logan S. M. (2006) Flagellar glycosylation - a new component of the motility repertoire? Microbiology 152, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 10. Hayakawa J., and Ishizuka M. (2012) Flagellar glycosylation: current advances. Glycosylation, 127–152 [Google Scholar]
- 11. Merino S., and Tomas J. M. (2014) Gram-negative flagella glycosylation. Int. J. Mol. Sci. 15, 2840–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goon S., Kelly J. F., Logan S. M., Ewing C. P., and Guerry P. (2003) Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50, 659–671 [DOI] [PubMed] [Google Scholar]
- 13. Guerry P., Ewing C. P., Schirm M., Lorenzo M., Kelly J., Pattarini D., Majam G., Thibault P., and Logan S. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schirm M., Soo E. C., Aubry A. J., Austin J., Thibault P., and Logan S. M. (2003) Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48, 1579–1592 [DOI] [PubMed] [Google Scholar]
- 15. Arora S. K., Neely A. N., Blair B., Lory S., and Ramphal R. (2005) Role of motility and flagellin glycosylation in the pathogenesis of Pseudomonas aeruginosa burn wound infections. Infect. Immun. 73, 4395–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanner A. C., Haffer C., Bratthall G. T., Visconti R. A., and Socransky S. S. (1979) A study of the bacteria associated with advancing periodontitis in man. J. Clin. Periodontol. 6, 278–307 [DOI] [PubMed] [Google Scholar]
- 17. Paster B. J., Boches S. K., Galvin J. L., Ericson R. E., Lau C. N., Levanos V. A., Sahasrabudhe A., and Dewhirst F. E. (2001) Bacterial diversity in human subgingival plaque. J. Bacteriol. 183, 3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faveri M., Mayer M. P., Feres M., de Figueiredo L. C., Dewhirst F. E., and Paster B. J. (2008) Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 23, 112–118 [DOI] [PubMed] [Google Scholar]
- 19. Drescher J., Schlafer S., Schaudinn C., Riep B., Neumann K., Friedmann A., Petrich A., Gobel U. B., and Moter A. (2010) Molecular epidemiology and spatial distribution of Selenomonas spp. in subgingival biofilms. Eur. J. Oral Sci. 118, 466–474 [DOI] [PubMed] [Google Scholar]
- 20. Hajishengallis G. (2015) Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schaffer C., and Messner P. (2017) Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41, 49–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ritchie M. A., Gill A. C., Deery M. J., and Lilley K. (2002) Precursor ion scanning for detection and structural characterization of heterogeneous glycopeptide mixtures. J. Am. Soc. Mass Spectrom. 13, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 23. Hinneburg H., Stavenhagen K., Schweiger-Hufnagel U., Pengelley S., Jabs W., Seeberger P. H., Silva D. V., Wuhrer M., and Kolarich D. (2016) The art of destruction: optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J. Am. Soc. Mass Spectrom. 27, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halim A., Nilsson J., Ruetschi U., Hesse C., and Larson G. (2012) Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell. Proteomics 11, M111 013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zauner G., Hoffmann M., Rapp E., Koeleman C. A., Dragan I., Deelder A. M., Wuhrer M., and Hensbergen P. J. (2012) Glycoproteomic analysis of human fibrinogen reveals novel regions of O-glycosylation. J. Proteome Res. 11, 5804–5814 [DOI] [PubMed] [Google Scholar]
- 26. Wuhrer M., Deelder A. M., and van der Burgt Y. E. (2011) Mass spectrometric glycan rearrangements. Mass Spectrom. Rev. 30, 664–680 [DOI] [PubMed] [Google Scholar]
- 27. Montie T. C., Craven R. C., and Holder I. A. (1982) Flagellar preparations from Pseudomonas aeruginosa: isolation and characterization. Infect. Immun. 35, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Doerner K. C., and White B. A. (1990) Detection of glycoproteins separated by nondenaturing polyacrylamide gel electrophoresis using the periodic acid-Schiff stain. Anal. Biochem. 187, 147–150 [DOI] [PubMed] [Google Scholar]
- 30. Cheng H. R., and Jiang N. (2006) Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28, 55–59 [DOI] [PubMed] [Google Scholar]
- 31. Kolarich D., Jensen P. H., Altmann F., and Packer N. H. (2012) Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 32. Jensen P. H., Karlsson N. G., Kolarich D., and Packer N. H. (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 33. Domagalski M. J., Alocci D., Almeida A., Kolarich D., and Lisacek F. (2017) PepSweetener: a web-based tool to support manual annotation of intact glycopeptide MS spectra. Proteomics Clin. Appl. 10.1002/prca.201700069 [DOI] [PubMed] [Google Scholar]
- 34. Anumula K. R. (1994) Quantitative determination of monosaccharides in glycoproteins by high-performance liquid chromatography with highly sensitive fluorescence detection. Anal. Biochem. 220, 275–283 [DOI] [PubMed] [Google Scholar]
- 35. Windwarder M., Figl R., Svehla E., Mocsai R. T., Farcet J. B., Staudacher E., Kosma P., and Altmann F. (2016) “Hypermethylation” of anthranilic acid-labeled sugars confers the selectivity required for liquid chromatography-mass spectrometry. Anal. Biochem. 514, 24–31 [DOI] [PubMed] [Google Scholar]
- 36. Hitchcock A. M., Costello C. E., and Zaia J. (2006) Glycoform quantification of chondroitin/dermatan sulfate using a liquid chromatography-tandem mass spectrometry platform. Biochemistry 45, 2350–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 39. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Hojrup P., Packer N. H., Larsen M. R., and Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031–MCP000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwashkiw J. A., Vozza N. F., Kinsella R. L., and Feldman M. F. (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89, 14–28 [DOI] [PubMed] [Google Scholar]
- 41. Maki M., and Renkonen R. (2004) Biosynthesis of 6-deoxyhexose glycans in bacteria. Glycobiology 14:1R–15R, [DOI] [PubMed] [Google Scholar]
- 42. Belyakov A. Y., Burygin G. L., Arbatsky N. P., Shashkov A. S., Selivanov N. Y., Matora L. Y., Knirel Y. A., and Shchyogolev S. Y. (2012) Identification of an O-linked repetitive glycan chain of the polar flagellum flagellin of Azospirillum brasilense Sp7. Carbohydr. Res. 361, 127–132 [DOI] [PubMed] [Google Scholar]
- 43. Takeuchi K., Ono H., Yoshida M., Ishii T., Katoh E., Taguchi F., Miki R., Murata K., Kaku H., and Ichinose Y. (2007) Flagellin glycans from two pathovars of Pseudomonas syringae contain rhamnose in D and L configurations in different ratios and modified 4-amino-4,6-dideoxyglucose. J. Bacteriol. 189, 6945–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Twine S. M., Reid C. W., Aubry A., McMullin D. R., Fulton K. M., Austin J., and Logan S. M. (2009) Motility and flagellar glycosylation in Clostridium difficile. J. Bacteriol. 191, 7050–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Faulds-Pain A., Twine S. M., Vinogradov E., Strong P. C., Dell A., Buckley A. M., Douce G. R., Valiente E., Logan S. M., and Wren B. W. (2014) The post-translational modification of the Clostridium difficile flagellin affects motility, cell surface properties and virulence. Mol. Microbiol. 94, 272–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brüll L. P., Heerma W., Thomas-Oates J., Haverkamp J., Kovácik V., and Kovác P. (1997) Loss of internal 1 → 6 substituted monosaccharide residues from underivatized and per-O-methylated trisaccharides. J. Am. Soc. Mass Spectrom. 8, 43–49 [Google Scholar]
- 47. Zaia J. (2004) Mass spectrometry of oligosaccharides. Mass Spectrom. Rev. 23, 161–227 [DOI] [PubMed] [Google Scholar]
- 48. Brull L. P., Kovacik V., Thomas-Oates J. E., Heerma W., and Haverkamp J. (1998) Sodium-cationized oligosaccharides do not appear to undergo ‘internal residue loss’ rearrangement processes on tandem mass spectrometry. Rapid Commun. Mass Spectrom. 12, 1520–1532 [DOI] [PubMed] [Google Scholar]
- 49. Samatey F. A., Imada K., Vonderviszt F., Shirakihara Y., and Namba K. (2000) Crystallization of the F41 fragment of flagellin and data collection from extremely thin crystals. J. Struct. Biol. 132, 106–111 [DOI] [PubMed] [Google Scholar]
- 50. Beatson S. A., Minamino T., and Pallen M. J. (2006) Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14, 151–155 [DOI] [PubMed] [Google Scholar]
- 51. Andersen-Nissen E., Smith K. D., Strobe K. L., Barrett S. L., Cookson B. T., Logan S. M., and Aderem A. (2005) Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. U.S.A. 102, 9247–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., and Aderem A. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 53. Smith K. D., Andersen-Nissen E., Hayashi F., Strobe K., Bergman M. A., Barrett S. L., Cookson B. T., and Aderem A. (2003) Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 54. de Zoete M. R., Keestra A. M., Wagenaar J. A., and van Putten J. P. (2010) Reconstitution of a functional Toll-like receptor 5 binding site in Campylobacter jejuni flagellin. J. Biol. Chem. 285, 12149–12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zampronio C. G., Blackwell G., Penn C. W., and Cooper H. J. (2011) Novel glycosylation sites localized in Campylobacter jejuni flagellin FlaA by liquid chromatography electron capture dissociation tandem mass spectrometry. J. Proteome Res. 10, 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurniyati K., Kelly J. F., Vinogradov E., Robotham A., Tu Y., Wang J., Liu J., Logan S. M., and Li C. (2017) A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol. Microbiol. 103, 67–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lux R., Miller J. N., Park N. H., and Shi W. (2001) Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 69, 6276–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kumar P. S., Griffen A. L., Barton J. A., Paster B. J., Moeschberger M. L., and Leys E. J. (2003) New bacterial species associated with chronic periodontitis. J. Dent. Res. 82, 338–344 [DOI] [PubMed] [Google Scholar]
- 59. Ebersole J. L., Holt S. C., Hansard R., and Novak M. J. (2008) Microbiologic and immunologic characteristics of periodontal disease in Hispanic americans with type 2 diabetes. J. Periodontol. 79, 637–646 [DOI] [PubMed] [Google Scholar]
- 60. Kolenbrander P. E., Andersen R. N., and Moore L. V. (1989) Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57, 3194–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kingsley V. V., and Hoeniger J. F. (1973) Growth, structure, and classification of Selenomonas. Bacteriol. Rev. 37, 479–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ricke S. C., Martin S. A., and Nisbet D. J. (1996) Ecology, metabolism, and genetics of ruminal selenomonads. Crit. Rev. Microbiol. 22, 27–56 [DOI] [PubMed] [Google Scholar]
- 63. Varki A., Cummings R. D., Aebi M., Packer N. H., Seeberger P. H., Esko J. D., Stanley P., Hart G., Darvill A., Kinoshita T., Prestegard J. J., Schnaar R. L., Freeze H. H., Marth J. D., Bertozzi C. R., Etzler M. E., Frank M., Vliegenthart J. F., Lutteke T., Perez S., Bolton E., Rudd P., Paulson J., Kanehisa M., Toukach P., Aoki-Kinoshita K. F., Dell A., Narimatsu H., York W., Taniguchi N., and Kornfeld S. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology 25, 1323–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang R., Sioma C. S., Wang S., and Regnier F. E. (2001) Fractionation of isotopically labeled peptides in quantitative proteomics. Anal. Chem. 73, 5142–5149 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (37) with the data set identifier PXD005859.
The mass spectrometry glycoproteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (37) with the data set identifier PXD005859.