Abstract
Increasing evidence implicates lysosomal dysfunction in the pathogenesis of neurodegenerative diseases, including the rare inherited lysosomal storage disorders (LSDs) and the most common neurodegenerative diseases, such as Alzheimer's and Parkinson's disease (AD and PD). Although the triggers of the lysosomal impairment may involve the accumulated macromolecules or dysfunction of the lysosomal enzymes, the role of the lysosomal glycocalyx in the lysosomal (dys)function has not been studied. The goal of this work was to analyze whether there are changes in the lysosomal glycocalyx in a cellular model of a LSD Niemann-Pick type C disease (NPC). Using the ferrofluid nanoparticles we isolated lysosomal organelles from NPC1-null and CHOwt cells. The magnetically isolated lysosomal fractions were enriched with the lysosomal marker protein LAMP1 and showed the key features of NPC disease: 3-fold higher cholesterol content and 4–5 fold enlarged size of the particles compared with the lysosomal fractions of wt cells. These lysosomal fractions were further processed to isolate lysosomal membrane proteins using Triton X-114 and their N-glycome was analyzed by HILIC-UPLC. N-glycans presented in each chromatographic peak were elucidated using MALDI-TOF/TOF-MS. We detected changes in the N-glycosylation pattern of the lysosomal glycocalyx of NPC1-null versus wt cells which involved high-mannose and sialylated N-glycans. To the best of our knowledge this study is the first to report N-glycome profiling of the lysosomal glycocalyx in NPC disease cellular model and the first to report the specific changes in the lysosomal glycocalyx in NPC1-null cells. We speculate that changes in the lysosomal glycocalyx may contribute to lysosomal (dys)function. Further glycome profiling of the lysosomal glycocalyx in other LSDs as well as the most common neurodegenerative diseases, such as AD and PD, is necessary to better understand the role of the lysosomal glycocalyx and to reveal its potential contribution in lysosomal dysfunction leading to neurodegeneration.
Since their first description scientists have tried to fully characterize lysosomal composition and function. Today, many facts are known about lysosomal physiology. The acidic pH, ionic gradients and the membrane potential make lysosomes an ideal environment for activity of luminal lysosomal hydrolases and a cellular center for nutrient sensing and recycling (1). Lysosome's primary role is to digest a cargo from endocytic, phagocytic or autophagocytic pathways. More than 50 lysosomal hydrolases have been characterized and dysfunction in their activity/levels leads to accumulation of the lysosomal cargo which causes lysosomal storage disorders (LSDs)1 (2). Besides luminal hydrolases, integral lysosomal membrane proteins are also important for proper function of lysosomes. Mutations in genes encoding these proteins lead to defects in the transport of lysosomal cargo and/or ions across the lysosomal membrane also causing the LSDs (3). Individuals with lysosomal storage disorders often develop symptoms early in life and in majority of LSDs the brain, especially neurons, are affected (4).
Niemann-Pick Type C (NPC) disease is a rare, autosomal recessive, progressive and fatal disorder characterized by abnormal cholesterol trafficking and intracellular accumulation of cholesterol and glycosphingolipids in late endosomes and lysosomes. NPC is caused by loss of function of either NPC1, a multi-transmembrane lysosomal protein, or NPC2, a small luminal lysosomal protein that mediate intracellular cholesterol transport (5). Recently, NPC disease has shown to share several pathological features with the most common and complex Alzheimer's disease (AD) (6). Our previous work has suggested that dysfunction of the late endosomal/lysosomal compartments in NPC1-cellular model is, most likely, responsible for the AD-like features in NPC (7, 8), and that increased levels of free cholesterol in NPC play an important role in compartmentalization of the key AD-proteins (β-amyloid precursor protein - APP and β-secretase - BACE1) within lipid rafts (9) or in modulation of membrane stiffness lading to sequestration of both APP and BACE1 within the endolysosomal pathway (10). Besides cholesterol, other lipids, especially phospholipids and sphingolipids are also involved in these processes (11).
Glycosylation is one of the most common co-translational and post-translational modification which regulates the structure, stability, localization and function of various proteins, and N-glycosylation has been the most studied type of it (12). N-glycans are known to be versatile and responsive to environmental stimuli and undergo significant changes in numerous diseases including those of central nervous system (13–17). Moreover, it has been recently shown that modulation of glycosylation of APP, a key protein in the pathogenesis of AD, may represent a potential target for AD therapy (18). It has been previously shown that glycoproteins accumulate in NPC model (19), and that blocking the O-linked glycosylation lowers cholesterol levels and increases the number of lysosomes (20), thus rescuing the NPC cellular defects. In the present work, we studied N-glycosylation profile of the lysosomal membrane proteins in NPC1-null cells versus wt-cells. We tested the hypothesis that alteration of the lysosomal glycocalyx is an additional feature of the lysosomal dysfunction in NPC disease, as well as in other LSDs.
To the best of our knowledge, here we describe the first complete N-glycome of the lysosomal glycocalyx in NPC disease cellular model, which potentially could be useful for restoring lysosomal storage defects in NPC disease and other LSDs as well as for rescuing pathological processes occurring in AD.
EXPERIMENTAL PROCEDURES
Cell Culture
Chinese hamster ovary wild type cells (CHOwt), CHO cells lacking NPC1 protein (NPC1-null) and NPC1-null cells stably express human NPC1 protein (NPC1-null + NPC1) were kindly provided by Dr. Daniel Ory. The cells were grown in DMEM/F12 medium supplemented with 10% FBS, 2 mm l-glutamine and antibiotic/antimycotic mix, all from Sigma-Aldrich (MO).
Magnetic Separation of Lysosomes From Whole Cells
Lysosomes were purified according the Walker and Lloyde Evans protocol (21). Briefly, the cells were grown in T75 flasks, they were incubated with 10% ferrofluid solution (superparamagnetic iron oxide nanoparticles, 10 mg/ml of 40 kDa dextran-stabilized magnetite, Liquids Research Ltd, UK) and 10 mm HEPES pH 7.2 in growth medium for 24 h. After washing and the chase period in the regular medium for 24 h, cells were trypsinized, harvested and resuspended in 2 ml of hypotonic buffer (15 mm KCL, 1.5 mm MgAc, 1 mm DTT, 10 mm HEPES and proteinase inhibitor (Roche, Switzerland)). After homogenization in dounce homogenizer (30 times) and passing through 23G needle (10 times), 0.5 ml of hypertonic buffer was added (220 mm HEPES pH 7.2, 0.1 mm sucrose, 375 mm KCl, 22.5 mm MgAc, 1 mm DTT, 50 ml of DNase 1 (Roche Applied Science)). Following incubation for 5 min, the cellular homogenate was centrifuged and the same amount of proteins in the supernatant of CHOwt and NPC1-null cells was subjected to MS column (MACS Miltenyi Biotec, Germany) activated with 0.5% BSA in PBS and attached to QuadroMACS magnetic separator. Flow-through was collected, and column was washed with DNase solution and Phosphate Buffer Saline (PBS) supplemented with 0.1 mm sucrose. The column was removed from magnetic separator and lysosomes were eluted with 0.5 ml of PBS supplemented with 0.1 mm sucrose and proteinase inhibitor mixture (Roche Applied Science). All collected fractions were stored at −80 °C before analysis.
Western Blot
Isolated fractions were mixed with sample buffer (6 times concentrated: 60% glycerol, 12% SDS, 3% DTT, 1/8 v/v 0.5 m Tris pH 6.8, bromphenol blue) and 20 μl of each fraction including the cell lysate (the input) was subjected to SDS-PAGE on 8% Tris-glycine gel. Proteins were transferred to PVDF membrane (Roche Applied Science), and blocked with I-block (Tropix, Thermo Fisher Scientific, MA). LAMP1 (rabbit polyclonal, Sigma-Aldrich, MO) was used as a lysosomal marker and EEA1 (mouse monoclonal, BD Transduction Laboratories, CA) was used as an early endosomal marker. Actin (rabbit polyclonal, Sigma-Aldrich) was used as a loading control. For detection, HRP conjugated secondary antibodies mouse/rabbit (Bio-Rad, CA) were used. Proteins were visualized by chemiluminescence using POD chemiluminescence blotting substrate (Roche Applied Science) on UviTec (UVItec ltd. Cambridge, UK). Western blots were quantified using ImageJ software (National Institutes of Health). Statistical validation of the data was achieved by Student t test.
Protein and Cholesterol Levels
Total protein concentration was measured using commercially available Pierce BCA Protein Assay Kit (Thermo Scientific) according to manufacturer's protocol on Multiskan EX (Thermo Scientific). Total cholesterol concentration was measured using commercially available AmplexRed Cholesterol Assay (Molecular Probes) according to manufacturer's protocol on Fluoroskan Ascent FL (Thermo Scientific).
Enzymatic Activity Measurement
N-Acetyl-β-d-glucosaminidase (NAG) activity was measured using 4-Nitrophenyl N-acetyl-β-d-glucosaminide substrate (Alfa Aesar). Ten μl of lysate fraction and lysosomal fraction were mixed with 90 μl of substrate (1 mg/ml in 0.09 m Citrate buffer solution, pH 4.8). After 30 min of incubation at 37 °C the reaction was stopped my adding 200 μl of 0.4 m Na2CO3. The absorbance of p-nitrophenylate ion was measured at 405 nm.
Particle Size Measurement
The hydrodynamic size of isolated lysosomes was measured using a Zetasizer Nano ZS instrument (Malvern, UK). The hydrodynamic size was measured from a dilute (1:20) suspension of the sample in PBS pH 7.4 in a disposable plastic cuvette at 25 °C. The results were analyzed using the Zetasizer software v.6.32 provided by the manufacturer.
Dextran Uptake Assay
Cells were grown on 12 mm glass covered slips and treated with 40 kDa dextran labeled with Fluorescein (Molecular Probes, Invitrogen, CA) in growth medium for 24 h. Cells were washed with PBS and further grown in the fresh growth medium for additional 0, 2, 4, 6, 8, and 24 h. After that cells were washed again in PBS and fixed in 4% paraformaldehyde (Sigma-Aldrich) and mounted with Polyvinyl alcohol mounting medium with DABCO antifading (Fluka). All samples were analyzed using inverted fluorescent confocal microscope Leica SP8 X FLIM.
For measuring cellular and secreted fluorescein labeled dextran, cells were grown in 6-well plates, they were incubated with 40 kDa fluorescein-labeled dextran for 24 h, following washing and 24 h chase in the regular medium. As a control the cells which were not incubated with dextran were used. Cells were lysed (50 mm Tris pH 7.6, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, proteinase inhibitor) and fluorescein levels in the cell lysate and in the medium were measured on Fluoroskan Ascent FL (Thermo Scientific).
Mass Spectrometric Proteomics Analysis
Lysosomal fractions were prepared in triplicate and processed using filter aided sample preparation (FASP) protocol (22) with some modifications. For the analysis, 8 μg of total protein from lysosomal fraction were mixed with the FASP-urea buffer (8 m urea in 0.1 m Tris-HCl pH 8.5) to final volume of 150 μl. SDS (Sigma-Aldrich) was added to final concentration of 1% (v/v). Before FASP, samples were sonicated for 10 min using ultrasonic bath, reduced (10 mm DTT, 30 min, 55 °C) and mixed with 100 μl of FASP-urea buffer. After transferring to the 10-kDa membrane filter units (Microcon YM-10, Merck Millipore), samples were centrifuged (13 000 × g, 45 min, 22 °C) and washed subsequently with 200 μl of FASP-urea buffer followed by centrifugation. Before digestion, proteins were alkylated (50 mm IAA, 20 min at room temperature in the dark), washed twice with FASP-urea buffer and then twice with ammonium bicarbonate (50 mm NH4HCO3 pH 7.6) followed by centrifugation (13 000 × g, 30 min). Protein digestion was achieved by adding trypsin (Promega) (enzyme-to-protein ratio 1:30, v/v) and by incubation at 37 °C overnight. Tryptic peptides were collected from filter units by centrifugation (13,000 × g, 30 min), washing with 50 μl of ammonium bicarbonate (50 mm NH4HCO3 pH 7.6) and subsequent centrifugation, followed by vacuum drying.
Peptides were dissolved in loading solvent (1% ACN, 0.1% formic acid) and separated using Ultimate 3000 RSLCnano system (Dionex, CA) before on-line ESI-MS/MS analysis by Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). A total of amount of 1.5 μg was injected onto the trapping column (C18 PepMap100, 5 μm, 100A, 300 μm × 5 mm). After washing for 15 min with loading solvent at a flow rate of 15 μl/min, peptides were eluted onto the analytical column (PepMap™ RSLC C18, 50 cm × 75 μm) using linear gradient 5–45% mobile phase B (0.1% formic acid in 80% ACN) over 150 min, 45% to 90% for 2 min, held at 80% for 2 min and re-equilibrated at 5% B for 20 min at a flow rate of 300 nL/min. Mobile phase A consisted of 0.1% formic acid in water. Eluate from the column was introduced into the mass spectrometer via Nanospray Flex ion source and SilicaTip emitter (New Objective). The ionization voltage was set at 1.9 kV and the ion transfer capillary temperature at 250 °C. MS was operating in positive ion mode using HCD MS2 in data dependent acquisition mode. Full scan FTMS spectra were acquired in range from m/z 350.0 to 1800.0 with a resolution of 70,000. The maximum injection time for FTMS full scan was set at 100 ms reaching an automatic gain control (AGC) target value of 1 × 106. Top 15 most intense precursor ions were chosen for further HCD fragmentation with a resolution of 17500 using injection time 60 ms and MS2 AGC target of 1 × 105. The collision energy was set as 28% NCE. A ± 1.7 Da isolation window was applied to isolate precursor ions with dynamic exclusion of 15 s. MS raw files were processed by Proteome Discoverer software (version 2.0.0.802., Thermo Fisher Scientific) and SEQUEST search against Homo sapiens FASTA files (42116 sequences, downloaded November 5, 2016 from SwissProt database, TaxID = 9609 and subtaxonomies). Static peptide modification included carbamidomethylation (C), and dynamic oxidation (M) and deamidation (N,Q). Maximum two trypsin missed cleavage sites were allowed. Precursor tolerance and ion fragment tolerance were set at 10 ppm and 0.05 Da, respectively. Percolator confidence levels were set at 1% false discovery rate (FDR) (high) and 5% FDR (middle), for both peptide and protein levels FDR is determined automatically by Percolator node based on targeted-decoy strategy (23). For peptide confidence, validation was based on q-value (minimal FDR at which the identification is considered correct (24)) which was set at 0.01 (high). At least two unique peptides and 5% FDR were required for reporting confidently identified proteins.
Gene ontology analysis of identified proteins was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (version 6.8) (25). The significance of enrichment (p value) and enrichment rate (fold enrichment) was calculated by DAVID tool.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (26) partner repository with the dataset identifier PXD008438.
Membrane Protein Isolation Using Triton-X114, Protein Extraction, Glycan Release, and Fluorescent Labeling
The whole procedure was performed as previously reported (27). Briefly, the most hydrophilic molecules from commercial Triton X-114 were eliminated by adding of 490 ml of 10 mm Tris-HCl pH 7.4, 150 mm NaCl and keeping the solution at 4 °C for Triton X-114 dissolution and at 37 °C for its condensation. The whole procedure was repeated three times. Isolated lysosomes were homogenized in Triton lysis buffer (10 mm Tris-HCl pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% (v/v) Triton X-114 in PBS and protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany)), using an ultrasonic processor (UP100H Hielscher, Teltow, Germany) (four cycles, 15–20 s). Samples were then incubated overnight at 4 °C and the lysosome lysate was clarified by centrifugation (30 min, 10,000 × g, 4 °C). The clear supernatant was overlaid on 200 μl of sucrose cushion (6% (w/v) sucrose, 10 mm Tris-HCl pH 7.4, 150 mm NaCl and 0.06% Triton X-114) and incubated at 37 °C for 20 min. Clouding of the solution occurred. Samples were centrifuged for 3 min, 400 × g, 37 °C and two phases (detergent-rich and detergent-poor) formed. The detergent-poor phase was transferred to a fresh tube and kept on ice. The detergent-rich phase was resuspended in 500 μl of cold PBS and the phase separation was repeated. This aqueous phase was pooled with initial one and re-extracted by adding 50 μl of Triton stock solution and the phase separation was performed as described previously.
Proteins from each phase were isolated by adding four times the sample volume of methanol and twice the initial sample volume of chloroform (Merck, Darmstadt, Germany) and vortexed well. At the end, three times of the initial sample volume of water was added, samples were vortexed vigorously and centrifuged for 1 min, 9 000 × g, 4 °C. After centrifugation the proteins were in the liquid interphase. The aqueous top layer was removed, an additional three volumes of methanol were added after which the samples were vortexed and centrifuged again for 2 min, 9,000 × g, 4 °C to pellet the proteins. Supernatant was removed, as much as possible without disturbing the precipitate, and samples were left to air-dry.
The isolated proteins were denatured with the addition of SDS (Invitrogen) and by incubation at 65 °C. The excess of SDS was neutralized with Igepal-CA630 (Sigma-Aldrich) and N-glycans were released following the addition of PNGase F (Promega, WI) in PBS. The released N-glycans were labeled with 2-AB. Free label and reducing agent were removed from the samples using hydrophilic interaction liquid chromatography solid-phase extraction (HILIC-SPE). Glycans were eluted with ultrapure water and stored at −20 °C until use.
The cell lysate proteins were prepared using the same protocol except there was not membrane protein isolation.
Ultraperformance Liquid Chromatography (UPLC)
Fluorescently labeled N-glycans were separated by HILIC on an Acquity UPLC instrument (Waters) consisting of a quaternary solvent manager, sample manager, and an FLR fluorescence detector set with excitation and emission wavelengths of 250 and 428 nm, respectively. The instrument was under the control of Empower 3 software, build 3471 (Waters Corp., MA). Labeled N-glycans were separated on a Waters BEH Glycan chromatography column, 150 × 2.1 mm i.d., 1.7 μm BEH particles, with 100 mm ammonium formate, pH 4.4, as solvent A and ACN as solvent B. The separation method used a linear gradient of 30–47% solvent A at flow rate of 0.56 ml/min in a 23 min analytical run. Samples were maintained at 10 °C before injection, and the separation temperature was 25 °C. Data processing was performed using an automatic processing method with a traditional integration algorithm, after which each chromatogram was manually corrected to maintain the same intervals of integration for all the samples. The chromatograms were all separated in the same manner into 37 and 25 chromatography peaks for the lysosomal and cell lysate N-glycome, respectively. HILIC-UPLC chromatograms were used for quantification, and abundance of each glycan was expressed as percentage of total integrated area.
Glycan Ethyl Esterification and Determined by Matrix-assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF/TOF-MS) Analysis
The identity of N-glycans separated by HILIC-UPLC was determined by MALDI-TOF/TOF-MS. Prior to MS analysis, fractions of each N-glycan chromatography peak were collected, dried down in a vacuum concentrator and resuspended in 10 μl of ultrapure water. To stabilize and distinguish α2,3-linked from α2,6-linked sialic acids ethyl esterification was performed as previously described (28). Aliquots of 2 μl were spotted onto a MTP AnchorChip 384 BC MALDI target (Bruker Daltronics, Bremen, Germany), mixed on plate with 1 μl of matrix solution (5 mg/ml 2,5-DHB, 1 mm NaOH in 50% acetonitrile) and left to dry by air. Recrystallization was performed by adding 0.2 μl of ethanol to each spot. Analyses were performed in positive-ion reflectron mode on an UltrafleXtreme MALDI-TOF/TOF-MS equipped with a Smartbeam-II laser and FlexControl 3.4 software Build 119 (Bruker Daltonics). The instrument was calibrated using a plasma N-glycome standard. A 25-kV acceleration voltage was applied after a 140 ns extraction delay. A mass window of m/z 1000 to 5 000 with suppression up to m/z 900 was used for N-glycan samples. For each spectrum, 10,000 laser shots were accumulated at a laser frequency of 2000 Hz, using a complete sample random walk with 200 shots per raster spot. N-glycan structures were assigned using GlycoMod (29) (http://web.expasy.org/glycomod/) for finding oligosaccharide composition from obtained mass using following settings: entering monoisotopic mass which in case of sialic acid was subtracted for 28 Da for each ester and increased for 18 Da for each lacton, mass tolerance of 0.5 Da and mass of 143 Da which included mass of 2-AB and Na+ as an adduct. NIBRT Glycobase (30) (http://glycobase.nibrt.ie/glycobase/browse_glycans.action) was used to confirm that a suggested N-glycan elutes in a distinct chromatographic peak using glucose units as well as knowledge of the biosynthetic pathway of N-glycans with an emphasis on specificity of N-glycosylation in CHO cell line (31, 32). Finding of informative fragments after MS/MS analysis to confirm a structure was performed using GlycoWorkbench (33).
Statistical Analysis
In order to remove experimental variation from measurements, normalization was performed on UPLC glycan data. To make measurements across samples comparable, normalization by total area was performed in which the area of each glycan peak was divided by the total area of the corresponding chromatogram. Differences in lysosomal (and whole-cell-lysate) N-glycomes between NPC1-null and CHOwt cells were determined using a general linear model. Before the analyses, normalized glycan measurements were log-transformed because of right skewness of their distributions. False discovery rate was controlled using Benjamini- Hochberg procedure. Data was analyzed and visualized using R programming language (version 3.0.1).
RESULTS
Lysosomal Isolation from NPC1-null and CHOwt Cells Using Magnetic Beads
Traditionally, lysosomes have been isolated using different gradient centrifugation techniques. In this work we adopted recently published method for lysosomal isolation using ferrofluid particles (21). Using this method we isolated lysosomal fractions which were positive for LAMP1 (lysosomal marker) and negative for EEA1 (early endosome marker) (Fig. 1A). It should be noted that the isolated lysosomal fractions contained 10-fold lower protein content than the cell lysate and that we were able to load only 3–5 μg of protein per each lane. Interestingly, we observed lower LAMP1-signal in the lysosomal fraction in NPC1-null cells compared with CHOwt cells (Fig. 1A). To further characterize the isolated lysosomal fractions we measured the activity of the lysosomal enzyme NAG (Fig. 1B) and the total cholesterol levels (Fig. 1C). We detected significantly increased activity of the NAG in the lysosomal fraction compared with the cell lysate both in CHOwt and in NPC1-null cells, suggesting an enrichment of lysosomes in the isolated fractions. In addition, a 3-fold increase of cholesterol was detected in the isolated lysosomal fractions from NPC1-null compared with CHOwt cells (p < 0.01) (Fig. 1C). This result agrees with our previous work using this model (7–9). We next performed MS-based proteomic analysis of the isolated lysosomal fractions. We identified 1577 and 2051 proteins in the lysosomal fractions (found in at least two out of three replicates) of CHOwt and NPC1-null cells, respectively. DAVID Gene Ontology (GO) tool was used for the classification and the detection of the most relevant annotation terms associated with protein set of each lysosomal fraction to get the insight into their subcellular localization. GO functional annotation tool was used to list top terms with the minimum of 2-fold enrichment and p < 0.05. In the lysosomal fractions of CHOwt and NPC1-null cells the identified proteins were mainly represented as “membrane-bounded vesicle” and as “membrane-enclosed lumen” (Fig. 1D).
Fig. 1.
Characterization of the lysosomal fractions isolated using the magnetic beads from NPC1-null and CHOwt cells. A, Western blot analysis of the isolated fractions: LAMP1 - lysosomal marker, EEA1 - early endosomal marker, actin - loading control. Lysosomal fractions are positive for LAMP1 and are negative for EAA1 early endosomal markers; B, N-Acetyl-β-d-glucosaminidase (NAG) activity shows a significant increase in the isolated lysosomal fractions versus the cells lysate, * p < 0.05; C, cholesterol levels in the isolated lysosomal fractions from NPC1-null cells are significantly increased compared with CHOwt cells, ** p < 0.01; (D) GO analysis of proteins identified in lysosomal fractions (2-fold enrichment, p < 0.05).
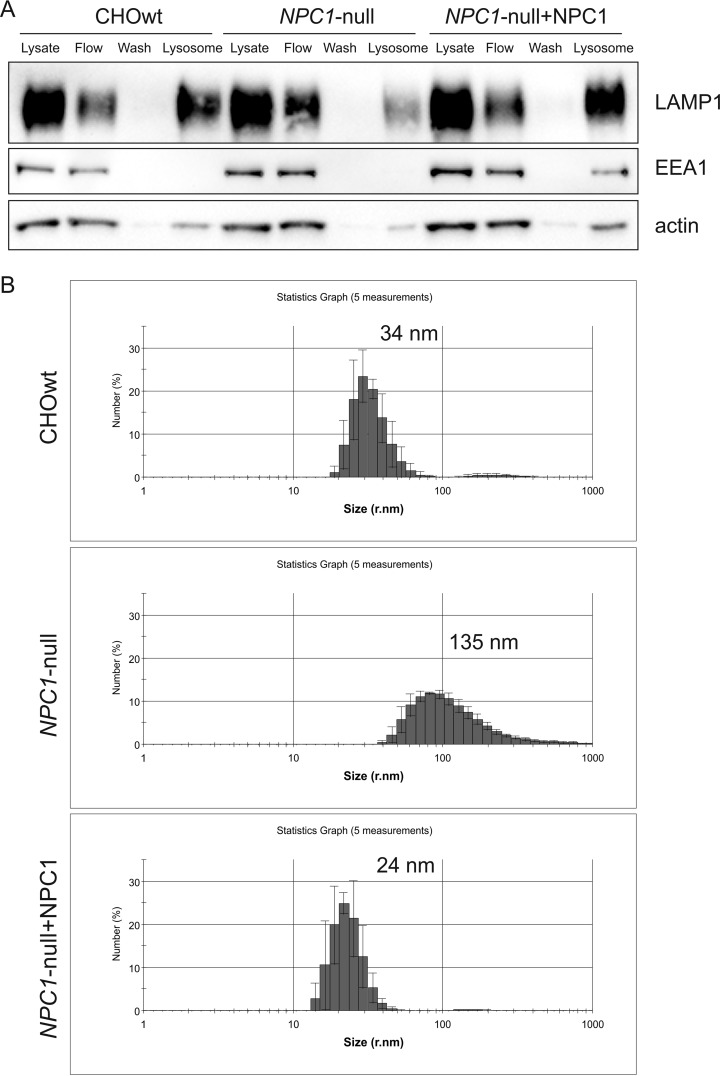
Because we noticed decreased LAMP1 signal in the isolated lysosomal fraction in NPC1-null versus wt cells (Fig. 1A), we further investigated this. Using the same markers as in Fig. 1A we additionally analyzed all fractions collected from the isolation procedure (including flow and wash through fractions) by Western blotting (Fig. 2A). Because of the no or very low LAMP1 signal detected in the cell lysate (Fig. 1A) we loaded the same volume of each fraction and the input (cell lysate, which was normalized per protein amount prior the lysosomal isolation). We observed increased amount of LAMP1 in the flow fraction in the NPC1-null cells compared with CHOwt cells (a fraction containing the rest of the cell lysate which did not capture ferrofluid particles and thus was passed through the magnetic field). Interestingly, this feature was fully recovered by back expression of NPC1 protein in NPC1-null cells, suggesting that the observed effect is NPC1-specific (Fig. 2A). Furthermore, we characterized the particle size in the isolated lysosomal fractions. Although in the lysosomal fraction from CHOwt cells the size of the particles was in the narrow range with average radius of 34 nm, the particles in the lysosomal fraction from NPC1-null cells were in a broader size range with average radius of 135 nm (Fig. 2B). NPC1 overexpression in NPC1-null cells reverted particle size distribution and average radius to that as in CHOwt cells (Fig. 2B).
Fig. 2.
NPC1 overexpression in NPC1-null cells reverts lysosomal isolation efficiency and the particle size distribution to that as in wild type cells. A, Western blot analysis of the isolated fractions using the “ferrofluid particles” method: LAPM1 - lysosomal marker, EEA1 - early endosomal marker, actin - loading control. Upon NPC1 overexpression in NPC1-null cells lysosomal isolation efficiency was comparable to that as in CHOwt cells; B, Particle size measurement revealed broader distribution and enlarged lysosomes in NPC1-null cells compared with CHOwt and NPC1-transfected NPC1-null cells.
To further characterize lower LAMP1 signal in the isolated lysosomal fraction in NPC1-null versus CHOwt cells using the “ferrofluid particles” method, we monitored the uptake, trafficking and degradation of dextran in these cells. For that we used fluorescein labeled dextran that had similar molecular weight as the one used in the ferrofluid particle suspension for lysosomal isolation. First, we monitored its trafficking after initial 24 h of intake (pulse period). At the time zero (chase period), we observed no significant difference in the uptake and distribution of fluorescein labeled dextran between CHOwt and NPC1-null cells (Fig. 3A). During several hours of chase we observed that small dots were accumulating to larger dots (which was especially visible after 6 h of chase) in both CHOwt and NPC1-null cells, suggesting dextran delivery into the lysosomes. However, soon after that (at 8 h and at 24 h time points), in CHOwt cells the signal continued to accumulate in larger aggregates, whereas in NPC1-null cells the signal was noted to fade (Fig. 3A). We reasoned the two possible explanations for this observation: (1) that NPC1-null cells somehow excrete dextran in the medium and (2) that dextran leaks from lysosomes into the cytosol, where it is diluted and, thus, is not visible any more, which could also explain decreased lysosomal isolation efficiency in NPC1-null versus wt-cells. To test these two possibilities, we measured the levels of fluorescein signal in the cell lysate and in the medium 24 h after initial uptake. We did not observe significant differences of the intracellular and secreted fluorescein signal between the NPC1-null and CHOwt cells (Fig. 3B), suggesting that dextran is not being secreted out of the NPC1-null cells and that it is present within the cells. This observation indirect supports our second hypothesis of the lysosomal leakage of dextran in NPC1-null versus CHOwt cells.
Fig. 3.
Decreased lysosomal isolation efficiency in NPC1-null cells versus CHOwt is not because of different cellular uptake and distribution of ferrofluid dextran labeled particles, but rather is because of a lysosomal leakage in NPC1-null cells. A, Cells were incubated with fluorescein labeled dextran for 24 h and chased in regular medium for 0–24 h. In CHOwt cells labeled dextran accumulates in enlarged vesicles, i.e. lysosomes, whereas in NPC1-null cells signal is fading; B, After 24 h incubation with dextran (+) or without dextran (−) and 24 h chase, fluorescein signal was measured in cell lysates and in the medium. There was a non-significant trend of cellular accumulation and decreased excretion of fluorescein labeled dextran in NPC1-null cells compared with CHOwt cells.
N-glycosylation Analysis of the Lysosomal Glycocalyx of NPC1-null and CHOwt Cells
The lysosomal glycocalyx consists of the oligosaccharides, from the lysosomal membrane glycoproteins (mainly LAMP1 and LAMP2), that are thought to protect the phospholipid bilayer with embedded proteins from the hydrolytic destruction by hydrolases in the lysosomal lumen.
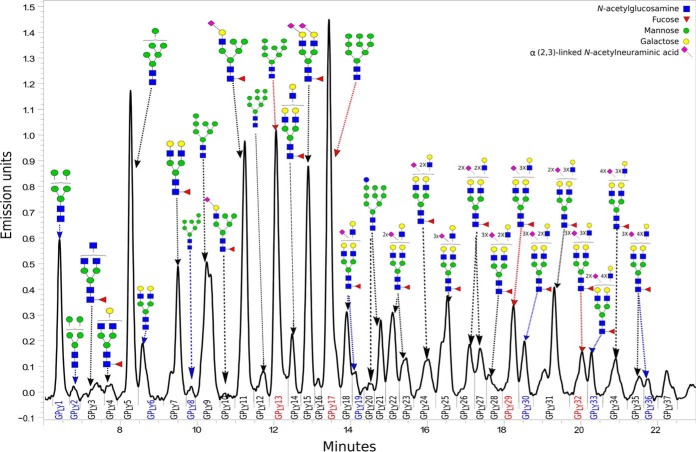
We assumed that altered glycocalyx composition may represent an additional feature of lysosomal (dys)function that may contribute to the potential lysosomal leakage effect thought to occur in lysosomal storage diseases as well as in the most common neurodegenerative disorders, such as Alzheimer's and Parkinson's disease (34, 35). To analyze N-glycome of the lysosomal glycocalyx between NPC1-null and CHOwt cells from the isolated lysosomal fractions, we first separated the lysosomal membrane proteins from the lysosomal luminal proteins using Triton X-114 (Fig. 4). N-glycomic profiles of lysosomal membrane proteins were obtained after PNG-ase F treatment using HILIC-UPLC analysis. The obtained chromatograms were separated into 37 (Fig. 5) distinct chromatographic peaks. Identity of N-glycan structures present in each peak was elucidated by MALDI-TOF MS approach. The most abundant structures (representing > 10% of individual peak area) present in each peak are given in Table I and Fig. 5. Relative abundance of the N-glycan structures was quantitatively assessed as areas under the peaks; the most abundant N-glycan (including isomers) among the lysosomal membrane proteins of both NPC1-null and CHOwt cells was a high-mannose type glycan -H9N2 (Fig. 5). Even though the high-mannose glycans comprise the same amount of the lysosomal membrane proteins' N-glycome (∼ 38%) in NPC1-null and CHOwt cells; the statistically significant decrease of H9N2 and H8N2 glycans, and the increase of H7N2 and H5N2 glycans was detected in NPC1-null cells compared with the CHOwt. In addition, we observed the statistically significant increase of sialylated complex glycans (H6N5F1L1, H9N8F1L2 and H9N8F1L3 showed the most significant increase) in the N-glycome of lysosomal membrane proteins of NPC1-null versus CHOwt cells (Table II, supplemental Fig. S1). However, the same changes in the high-mannose glycans of the lysosomal membrane proteins were also observed in N-glycome of the whole-cell-lysate of NPC1-null versus CHOwt cells (supplemental Fig. S2, supplemental Table S1), indicating that the observed high-mannose N-glycosylation alterations upon NPC1-loss of function are not specific to the lysosomal glycocalyx. On the contrary, the changes of the sialylated complex glycans in the whole-cell-lysate N-glycome compared with the changes in the lysosomal membrane proteins' N-glycome of NPC1-null versus CHOwt cells were opposite. Namely, there was a significant decrease of more complex and sialylated glycans and the increase of small high-mannose and neutral complex/hybrid glycans in the whole-cell-lysate N-glycome of NPC1-null versus CHOwt cells (supplemental Fig. S2, supplemental Table S1).
Fig. 4.
Overlaid chromatograms of HILIC-UPLC N-glycan profiles of intraluminal, soluble fraction (dark gray) and hydrophobic membrane fraction (light gray).
Fig. 5.
Representative chromatogram of 2-AB labeled N-linked glycans released from lysosomal membrane proteins isolated from CHOwt cells and separated by HILIC-UPLC. The integration areas, together with a major structure (except for GPLY16 and GPLY37 because of the limited evidence) presented in each glycan peak are given as shown in Table I and using glucose units as well as knowledge of the biosynthetic pathway of N-glycans in the CHO cell line (26, 27). Glycan peaks (glycan peaks of lysosomal glycocalyx - GPLY) are numbered from GPLY1-GPLY37, as used in the manuscript. Glycan peaks which percentage of area increased and decreased in lysosomal glycocalyx of NPC1-null cells compared with CHOwt cells (adjusted p value < 0.05) are shown blue and red, respectively.
Table I. Assumed composition of N-glycome of the lysosomal membrane proteins. The lysosomal N-glycome was separated into 37 chromatographic peaks (glycan peaks of lysosomal glycocalyx - GPLY) by HILIC-UPLC and masses of individual glycan structures were detected by MALDI-TOF/TOF-MS. Abbreviations used are hexose (H), N-acetylhexosamine (N), fucose (F) and sialic acid with α2,3-linkage as indicated by lactonization (NA–N-acetylneuraminic; NG–N-glycolylneuraminic acid). I–Intensity, GU–Glucose unites after HILIC-UPLC analysis.
| Peak | m/z | Δa/Da | I | GU | Composition | Peak | m/z | Δa/Da | I | GU | Composition |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GPLY1 | 1377.391 | -0.142 | 10137 | 6.54 | H5N2 | GPLY22 | 2841.682 | -0.390 | 78 | 10.75 | H6N5F1NA2 |
| 1564.487 | −0.130 | 335 | H4N3F1 | 2660.404 | 0.389 | 15 | H7N6F1 | ||||
| GPLY2 | 1377.715 | 0.181 | 133 | 6.71 | H5N2 | GPLY23 | 2841.386 | 0.333 | 1079 | 10.99 | H6N5F1NA2 |
| GPLY3 | 1808.807 | 0.083 | 563 | 6.84 | H3N5F1 | 2787.365 | 0.324 | 365 | H7N6NA1 | ||
| 1580.686 | 0.073 | 578 | H5N3 | GPLY24 | 2968.413 | 0.334 | 311 | 11.38 | H6N5NA3 | ||
| 1523.653 | 0.061 | 414 | H5N2F1 | 2933.450 | 0.351 | 1838 | H7N6F1NA1 | ||||
| 1767.788 | 0.090 | 301 | H4N4F1 | 2857.404 | 0.357 | 440 | H6N5F1NA1NG 1 | ||||
| GPLY4 | 2011.857 | 0.053 | 91 | 7.10 | H3N6F1 | GPLY25 | 3114.627 | 0.490 | 61 | 11.73 | H6N5F1NA3 |
| 1767.694 | −0.003 | 434 | H4N4F1 | GPLY26 | 3206.493 | 0.308 | 379 | 12.12 | H7N6F1NA2 | ||
| GPLY5 | 1539.531 | −0.055 | 35791 | 7.29 | H6N2 | GPLY27 | 3206.676 | 0.491 | 2467 | 12.31 | H7N6F1NA2 |
| GPLY6 | 1971.084 | 0.307 | 60 | 7.40 | H4N5F1 | 3130.594 | 0.462 | 289 | H6N5F1NA2NG 1 | ||
| 1783.875 | 0.182 | 414 | H5N4 | GPLY28 | 3333.496 | 0.285 | 89 | 12.45 | H7N6NA3 | ||
| GPLY7 | 1929.660 | −0.090 | 2973 | 7.74 | H5N4F1 | 3206.412 | 0.227 | 98 | H7N6F1NA2 | ||
| GPLY8 | 1701.786 | 0.147 | 1511 | 7.88 | H7N2 | GPLY29 | 3298.545 | 0.313 | 422 | 12.85 | H8N7F1NA1 |
| GPLY9 | 1701.780 | 0.141 | 577 | 8.03 | H7N2 | 3206.918 | 0.412 | 112 | H7N6F1NA2 | ||
| GPLY10 | 2133.056 | 0.226 | 243 | 8.24 | H5N5F1 | GPLY30 | 3479.530 | 0.261 | 131 | 13.03 | H7N6F1NA3 |
| 2056.986 | 0.209 | 244 | H5N4NA1 | GPLY31 | 3752.761 | 0.406 | 560 | 13.47 | H7N6F1NA4 | ||
| 2016.027 | 0.277 | 418 | H6N3NA1 | 3571.660 | 0.343 | 876 | H8N7F1NA2 | ||||
| GPLY11 | 2202.993 | 0.158 | 524 | 8.46 | H5N4F1NA1 | 3495.576 | 0.312 | 193 | H8N6NA3 | ||
| GPLY12 | 1863.564 | −0.127 | 1392 | 8.70 | H8N2 | 3390.579 | 0.300 | 1224 | H9N8F1 | ||
| GPL13 | 2294.743 | −0.139 | 248 | 8.87 | H6N5F1 | GPLY32 | 3844.792 | 0.391 | 942 | 13.88 | H8N7F1NA3 |
| 1863.608 | −0.083 | 9140 | H8N2 | 3768.654 | 0.305 | 142 | H7N6F1NA3NG 1 | ||||
| GPLY14 | 2294.953 | −0.083 | 11 | 9.10 | H6N5F1 | GPLY33 | 3860.665 | 0.269 | 76 | 14.00 | H8N7F1NA2NG 1 |
| GPLY15 | 2476.283 | 0.363 | 209 | 9.34 | H5N4F1NA2 | 3936.708 | 0.259 | 162 | H9N8F1NA2 | ||
| 2422.185 | 0.276 | 17 | H6N5NA1 | GPLY34 | 4117.825 | 0.338 | 72 | 14.33 | H8N7F1NA4 | ||
| GPLY17 | 2025.916 | 0.171 | 5989 | 9.65 | H9N2 | GPLY35 | 4210.126 | 0.481 | 131 | 14.68 | H9N8F1NA3 |
| GPLY18 | 2568.174 | 0.207 | 2619 | 9.95 | H6N5F1NA1 | GPLY36 | 4210.116 | 0.480 | 437 | 14.83 | H9N8F1NA3 |
| GPLY19 | 2568.344 | 0.377 | 733 | 10.09 | H6N5F1NA1 | GPLY37 | 4483.312 | 0.351 | 68 | 15.16 | H9N8F1NA4 |
| GPLY20 | 2188.050 | 0.252 | 2023 | 10.36 | H10N2 | 3816.288 | −0.087 | 85 | H7N6F1NG 4 | ||
| GPLY21 | 2695.314 | 0.319 | 262 | 10.52 | H6N5NA2 | ||||||
| 2188.100 | 0.302 | 2081 | H10N2 | ||||||||
aDeviation between experimental and theoretical mass using GlycoMod.
Table II. Differences in the lysosomal membrane proteins' N-glycomes (glycan peaks of lysosomal glycocalyx - GPLY) between CHOwt and NPC1-null cells. Abbreviations used are hexose (H), N-acetylhexosamine (N), fucose (F) and sialic acid with α2,3-linkage as indicated by lactonization (NA–N-acetylneuraminic; NG–N-glycolylneuraminic acid).
| Glycan | Composition | Effect | p value | Adjusted p value | Glycan | Composition | Effect | p value | Adjusted p value |
|---|---|---|---|---|---|---|---|---|---|
| GPLY1 | H5N2 | 0.209 | 0.0002 | 0.0079 | GPLY26 | H7N6F1NA2 | 2.394 | 0.0889 | 0.1644 |
| GPLY19 | H6N5F1NA1 | 0.224 | 0.0006 | 0.0113 | GPLY3 | H5N3 | 0.427 | 0.0978 | 0.1723 |
| GPLY2 | H5N2 | 0.252 | 0.0025 | 0.0262 | GPLY15 | H5N4F1NA2 | 2.303 | 0.1054 | 0.1773 |
| GPLY13 | H8N2 | 3.937 | 0.0028 | 0.0262 | GPLY12 | H8N2 | 0.45 | 0.1238 | 0.1991 |
| GPLY36 | H9N8F1NA3 | 0.266 | 0.0043 | 0.032 | GPLY23 | H6N5F1NA2 | 0.463 | 0.1388 | 0.2139 |
| GPLY33 | H9N8F1NA2 | 0.276 | 0.0062 | 0.0383 | GPLY22 | H6N5F1NA2 | 1.896 | 0.2251 | 0.3332 |
| GPLY6 | H5N4 | 0.284 | 0.0081 | 0.0427 | GPLY5 | H6N2 | 0.55 | 0.2587 | 0.3681 |
| GPLY29 | H8N7F1NA1 | 3.453 | 0.0092 | 0.0427 | GPLY9 | H7N2 | 0.578 | 0.3029 | 0.4151 |
| GPLY17 | H9N2 | 3.372 | 0.0113 | 0.0463 | GPLY35 | H9N8F1NA3 | 0.59 | 0.319 | 0.4216 |
| GPLY8 | H7N2 | 0.302 | 0.013 | 0.0467 | GPLY10 | H6N3NA1 | 0.611 | 0.3465 | 0.4421 |
| GPLY30 | H7N6F1NA3 | 0.309 | 0.0151 | 0.0467 | GPLY21 | H10N2 | 0.712 | 0.526 | 0.6487 |
| GPLY32 | H8N7F1NA3 | 3.244 | 0.0151 | 0.0467 | GPLY27 | H7N6F1NA2 | 0.74 | 0.575 | 0.6863 |
| GPLY14 | H6N5F1 | 0.342 | 0.0302 | 0.0798 | GPLY18 | H6N5F1NA1 | 1.3 | 0.6486 | 0.7499 |
| GPLY16 | / | 0.34 | 0.0292 | 0.0798 | GPLY7 | H5N4F1 | 1.234 | 0.698 | 0.7596 |
| GPLY31 | H8N7F1NA2 | 2.791 | 0.04 | 0.0925 | GPLY25 | H6N5F1NA3 | 1.242 | 0.6888 | 0.7596 |
| GPLY37 | H7N6F1NG4 | 0.358 | 0.0394 | 0.0925 | GPLY34 | H8N7F1NA4 | 1.162 | 0.7944 | 0.8398 |
| GPLY4 | H4N4F1 | 0.379 | 0.0535 | 0.11 | GPLY20 | H10N2 | 0.897 | 0.8408 | 0.8642 |
| GPLY11 | H5N4F1NA1 | 2.651 | 0.0535 | 0.11 | GPLY24 | H7N6F1NA1 | 1.015 | 0.9775 | 0.9775 |
| GPLY28 | H7N6F1NA2 | 0.385 | 0.0586 | 0.114 | |||||
DISCUSSION
In this work we describe changes in the lysosomal glycocalyx in a cellular model of a lysosomal storage disease Niemann-Pick type C (NPC). Lysosomal impairment is considered to play an important role in the pathogenesis of neurodegenerative diseases including the rare inherited lysosomal storage disorders, like Niemann-Pick type C disease (NPC), as well as the most common and complex Alzheimer's and Parkinson's disease (AD and PD). Here, we have characterized the N-glycan profile of the lysosomal glycocalyx of NPC1-null versus CHOwt cells. Notably, in our previous studies we have substantially used this NPC disease cellular model to elucidate the molecular and cellular details of an AD-like phenotype in NPC (7–9, 36). Based on our results we speculate that changes in the N-glycome of the lysosomal membrane proteins may contribute to lysosomal (dys)function in NPC disease.
Lysosomes were isolated using the magnetic ferrofluid nanoparticles instead of the traditionally used ultracentrifugation fractionation procedures because the isolation using the magnetic beads reveals more pure lysosomal fractions. Indeed, our Western blot analysis of endosomal and lysosomal marker proteins in the magnetically isolated lysosomal fractions confirmed that they were mainly LAMP1-positive, indicating that we have successfully isolated lysosomal organelles. However, we noticed that the yield of the isolated lysosomes was substantially lower in NPC1-null versus CHOwt cells. This contrasts with the recently reported lysosomal isolation from HeLa NPC1-KO cells using similar approach (37). In contrast to our dextran-coated ferrofluid nanoparticles, Tharkeshwar et al. used much smaller size coated ferrofluid nanoparticles which may be the reason of the different cellular behavior of the NPC1-lacking lysosomes. Our further analysis of the dextran uptake and its lysosomal accumulation between CHOwt and NPC1-null cells revealed that leakage of lysosomes may likely explain decreased lysosomal isolation efficiency in NPC1-null versus wt-cells by the “dextran-coated ferrofluid nanoparticles” method. Indeed, lysosomal leakage has been proposed to occur in the most common neurodegenerative disorders, such as AD (38), as well as the rare LSDs, such as NPC (39). This defect was fully recovered upon NPC1-back expression in NPC1-null cells, suggesting that the observed feature of decreased lysosomal isolation in the NPC1-null cells is dependent on NPC1-function. Moreover, particles in the isolated lysosomal fractions from NPC1-null cells presented with substantially increased radius (4–5 fold) and contained 3-fold more cholesterol than the lysosomal fraction of CHOwt cells. Altogether, these results (enlarged lysosomal organelles together with cholesterol accumulation) reveal that the magnetically isolated lysosomal fractions contain the key features of lysosomal impairment in NPC disease, indicating that these lysosomes could be further used for the organelle profiling.
Next, we sought to analyze the N-glycome of the lysosomal membrane proteins from the isolated lysosomal fractions in order to elucidate if the N-glycosylation pattern of the lysosomal glycocalyx is altered in NPC1-null versus CHOwt cells and, thus, may contribute to the lysosomal dysfunction. Indeed, our findings support this as we observed both changes in the high-mannose and sialylated N-glycome patterns of the lysosomal membrane proteins between the two cell lines. To the best of our knowledge this study is the first to report N-glycome profiling of the lysosomal glycocalyx. All of the detected structures have the characteristics of N-glycosylation in CHO cell line; myriad of LacNAc structures, core fucosylation, α2,3 linked sialic acid and higher abundance of N-acetylneuraminic than of N- glycolylneuraminic acid (31, 32, 40). Compared with CHO cellular glycocalyx where the main N-glycan structures were a series of core-fucosylated asialoglycans (40), we detected that the main structures of the lysosomal N-glycome belong to the high-mannose glycans with H9N2 as the most abundant structure, regardless of the cell line, whereas core-fucosylated sialylated N-glycans were the second most abundant group of the glycans. Although the function of the lysosomal glycocalyx remains to be elucidated, it is known that sialic acids can protect molecules from attack by glycosidases and proteases (34) which may indicate that higher level of sialylation plays a role in protecting the limiting lysosomal membrane from the action of degradative lysosomal enzymes in the lumen. Yet, this is the first study where the specific changes of N-glycosylation of the lysosomal glycocalyx were observed between the NPC1-null (NPC disease cellular model) and CHOwt cell lines. More precisely, we identified higher abundance of more complex sialylated and smaller high-mannose N-glycans in the lysosomes derived from NPC1-null versus wt-cells. Higher glycosylation of the lysosomal proteins in NPC disease was previously observed by two independent studies (41, 42). Chen et al. suggested that only NPC2 protein is more glycosylated whereas others remain unchanged. However, Dixit et al. proved that higher level of glycosylation is not limited to NPC2 but rather widely present in the lysosomal proteome. Indeed, in this study we detected that more complex and sialylated N-glycans are present in the lysosomal glycocalyx of the NPC1-null versus wt-cells whereas the same cannot be said for the whole-cell-lysate N-glycome. Even though, the changes were solely related to the lysosomal N-glycome, the changes of the high-mannose N-glycans were equally distributed in N-glycans of the NPC1-null cells regardless of their origin, i.e. cellular versus lysosomal. It was noticed in previous studies that expression of α-mannosidase is higher in NPC disease which can explain higher abundance of smaller high-mannose glycans that we observed in NPC1-null cells. This is further supported by findings which showed that lysosomal α-mannosidase is capable of mannose digestion of the native proteins (42, 43). Moreover, higher abundance of H5N2 can be explain by lower expression of dolichol-phosphate-mannose (DPM) biosynthesis regulatory protein in NPC disease (44) as this protein is needed to stabilize expression and localization of DPM1, a catalytic subunit of DPM synthase (45). Indeed, an accumulation of H5N2 was observed in the cells with a mutation in DPM synthase (46). Interestingly, it has been recently reported that decreasing the lysosomal glycocalyx density causes cells to become less dependent on NPC1 protein function to transfer cholesterol across the lysosomal membrane. Namely, inhibition of O-glycosylation reduced accumulation of cholesterol in NPC1-null cells which is the primary feature of NPC disease (20). Because in this study we observed higher level of glycosylation in the NPC1-null versus wt-cells this may indicate that glycosylation could be actively involved in cholesterol accumulation and NPC disease progression.
Thus, modulation of the lysosomal glycocalyx may not only cause impairment of the lysosomal function but may also be used as a tool to bypass certain lysosomal defects such as cholesterol accumulation in NPC disease. Further glycome profiling of the lysosomal glycocalyx in other lysosomal storage disorders as well as the most common neurodegenerative diseases, such as AD and PD, is necessary to better understand the function of the lysosomal glycocalyx and to reveal its potential contribution in lysosomal dysfunction leading to neurodegeneration.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (26) partner repository with the dataset identifier PXD008438.
Supplementary Material
Acknowledgments
We would like to thank Lucija Horvat for the assistance with confocal microscopy and Dr. Maja Dutour Sikiric for assistance with Zetasizer instrument.
Footnotes
* This work was funded by the Unity Through Knowledge Fund (S.H.), by the European Commission FP7 grants MIMOmics (contract #305280, G.L.), HTP-GlycoMet (contract #324400, G.L.), VetMedZg (contract #621394, A.H.), H2020 grants GlySign (contract #722095, G.L.), SYSCID (contract #733100, G.L.) and IMforFuture (contract #721815, G.L.) as well as by funding for the Croatian National Centre of Research Excellence in Personalized Healthcare and IRI project Nova generacija visokoprotočnih glikoservisa.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- LSD
- Lysosomal storage disorder
- AD
- Alzheimer's disease
- APP
- β-amyloid precursor protein
- BACE1
- β-secretase
- CHO
- Chinese hamster ovary
- HCD
- Higher energy collisional dissociation
- HILIC
- Hydrophilic interaction liquid chromatography
- EEA1
- Early Endosome Antigen 1 - early endosomal marker
- ESI
- Electrospray ionization
- FASP
- Filter aided sample preparation
- GO
- Gene ontology
- LAMP1
- Lysosomal Associated Membrane Protein 1 - lysosomal marker
- MALDI
- Matrix-assisted laser desorption/ionization
- NAG
- N-Acetyl-β-D-glucosaminidase
- NPC
- Niemann-Pick Type C
- PD
- Parkinson's disease
- Rab7
- RAS-related GTP-binding protein 7- late endosomal marker
- SPE
- Solid-phase extraction
- TfR
- Transferrin receptor – a marker of recycling endosomes
- TOF
- Time-of-flight
- UPLC
- Ultra-performance liquid chromatography.
REFERENCES
- 1. Xu H., and Ren D. (2015) Lysosomal Physiology. Annu. Rev. Physiol. 77, 57–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Futerman A. H., and van Meer G. (2004) The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 5, 554–565 [DOI] [PubMed] [Google Scholar]
- 3. Saftig P., and Klumperman J. (2009) Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635 [DOI] [PubMed] [Google Scholar]
- 4. Schultz M. L., Tecedor L., Chang M., and Davidson B. L. (2011) Clarifying lysosomal storage diseases. Trends Neurosci. 34, 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pacheco C. D., and Lieberman A. P. (2008) The pathogenesis of Niemann-Pick type C disease: a role for autophagy? Expert. Rev. Mol. Med. 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malnar M., Hecimovic S., Mattsson N., and Zetterberg H. (2014) Bidirectional links between Alzheimer's disease and Niemann-Pick type C disease. Neurobiol. Dis. 72 Pt A, 37–47 [DOI] [PubMed] [Google Scholar]
- 7. Malnar M., Kosicek M., Lisica A., Posavec M., Krolo A., Njavro J., Omerbasic D., Tahirovic S., and Hecimovic S. (2012) Cholesterol-depletion corrects APP and BACE1 misstrafficking in NPC1-deficient cells. Biochim. Biophys. Acta - Mol. Basis Dis. 1822, 1270–1283 [DOI] [PubMed] [Google Scholar]
- 8. Malnar M., Kosicek M., Mitterreiter S., Omerbasic D., Lichtenthaler S. F., Goate A., and Hecimovic S. (2010) Niemann-Pick type C cells show cholesterol dependent decrease of APP expression at the cell surface and its increased processing through the β-secretase pathway. Biochim. Biophys. Acta - Mol. Basis Dis. 1802, 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kosicek M., Malnar M., Goate A., and Hecimovic S. (2010) Cholesterol accumulation in Niemann Pick type C (NPC) model cells causes a shift in APP localization to lipid rafts. Biochem. Biophys. Res. Commun. 393, 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. von Einem B., Weber P., Wagner M., Malnar M., Kosicek M., Hecimovic S., von Arnim C. A. F., and Schneckenburger H. (2012) Cholesterol-dependent energy transfer between fluorescent proteins-insights into protein proximity of APP and BACE1 in different membranes in Niemann-pick type C disease cells. Int. J. Mol. Sci. 13, 15801–15812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kosicek M., and Hecimovic S. (2013) Phospholipids and Alzheimer's disease: Alterations, mechanisms and potential biomarkers. Int. J. Mol. Sci. 14, 1310–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stanley P., Schachter H., and Taniguchi N. (2009) Chapter 8. N-Glycans, Essentials of Glycobiology, 2nd Edition [Google Scholar]
- 13. Vučković F., Krištić J., Gudelj I., Teruel M., Keser T., Pezer M., Pučić-Baković M., Štambuk J., Trbojević-Akmačić I., Barrios C., Pavić T., Menni C., Wang Y., Zhou Y., Cui L., Song H., Zeng Q., Guo X., Pons-Estel B. A., McKeigue P., Leslie Patrick A., Gornik O., Spector T. D., Harjaček M., Alarcon-Riquelme M., Molokhia M., Wang W., and Lauc G. (2015) Association of systemic lupus erythematosus with decreased immunosuppressive potential of the IgG glycome. Arthritis Rheumatol. 67, 2978–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gudelj I., Baciarello M., Ugrina I., De Gregori M., Napolioni V., Ingelmo P. M., Bugada D., De Gregori S., Ðerek L., Pučić-Baković M., Novokmet M., Gornik O., Saccani Jotti G., Meschi T., Lauc G., and Allegri M. (2016) Changes in total plasma and serum N-glycome composition and patient-controlled analgesia after major abdominal surgery. Sci. Rep. 6, 31234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrios C., Zierer J., Gudelj I., Štambuk J., Ugrina I., Rodríguez E., Soler M. J., Pavić T., Šimurina M., Keser T., Pučić-Baković M., Mangino M., Pascual J., Spector T. D., Lauc G., and Menni C. (2015) Glycosylation Profile of IgG in Moderate Kidney Dysfunction. J. Am. Soc. Nephrol. 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freidin M. B., Keser T., Gudelj I., Štambuk J., Vučenović D., Allegri M., Pavić T., Šimurina M., Fabiane S. M., Lauc G., and Williams F. M. K. (2016) The Association Between Low Back Pain and Composition of IgG Glycome. Sci. Rep. 5, 26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bieberich E. (2014) Synthesis, Processing, and Function of N-glycans in N-glycoproteins. Adv. Neurobiol. 9, 47–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jacobsen K. T., and Iverfeldt K. (2011) O-GlcNAcylation increases non-amyloidogenic processing of the amyloid-β precursor protein (APP). Biochem. Biophys. Res. Commun. 404, 882–886 [DOI] [PubMed] [Google Scholar]
- 19. Mbua N. E., Flanagan-Steet H., Johnson S., Wolfert M. A., Boons G.-J., and Steet R. (2013) Abnormal accumulation and recycling of glycoproteins visualized in Niemann-Pick type C cells using the chemical reporter strategy. Proc. Natl. Acad. Sci. 110, 10207–10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li J., Deffieu M. S., Lee P. L., Saha P., and Pfeffer S. R. (2015) Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc. Natl. Acad. Sci. 112, 14876–14881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walker M. W., and Lloyd-Evans E. (2015) A rapid method for the preparation of ultrapure, functional lysosomes using functionalized superparamagnetic iron oxide nanoparticles. Methods Cell Biol. 126, 21–43 [DOI] [PubMed] [Google Scholar]
- 22. Wisniewski J. R., Zougman A., Nagaraj N., Mann M., and Wi J. R. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 377–362 [DOI] [PubMed] [Google Scholar]
- 23. Spivak M., Weston J., Bottou L., Kall L., and Noble W. S. (2009) Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J. Proteome Res. 8, 3737–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Käll L., Storey J. D., MacCoss M. J., and Noble W. S. (2008) Posterior error probabilities and false discovery rates: Two sides of the same coin. J. Proteome Res. 7, 40–44 [DOI] [PubMed] [Google Scholar]
- 25. Dennis G., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H., and Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, R60. [PubMed] [Google Scholar]
- 26. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pavić T., Gudelj I., Keser T., Pučić-Baković M., and Gornik O. (2016) Enrichment of hydrophobic membrane proteins using Triton X-114 and subsequent analysis of their N-glycosylation. Biochim. Biophys. Acta - Gen. Subj. 1860, 1710–1715 [DOI] [PubMed] [Google Scholar]
- 28. Reiding K. R., Blank D., Kuijper D. M., Deelder A. M., and Wuhrer M. (2014) High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 86, 5784–5793 [DOI] [PubMed] [Google Scholar]
- 29. Cooper C. A., Gasteiger E., and Packer N. H. (2001) GlycoMod - A software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1, 340–349 [DOI] [PubMed] [Google Scholar]
- 30. Campbell M. P., Royle L., and Rudd P. M. (2015) Glycobase and autoGU: Resources for interpreting HPLC-glycan data. Methods Mol. Biol. 1273, 17–28 [DOI] [PubMed] [Google Scholar]
- 31. Xu X., Nagarajan H., Lewis N. E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., Andersen M. R., Neff N., Passarelli B., Koh W., Fan H. C., Wang J., Gui Y., Lee K. H., Betenbaugh M. J., Quake S. R., Famili I., Palsson B. O., and Wang J. (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 29, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin N., Mascarenhas J., Sealover N. R., George H. J., Brooks J., Kayser K. J., Gau B., Yasa I., Azadi P., and Archer-Hartmann S. (2015) Chinese hamster ovary (CHO) host cell engineering to increase sialylation of recombinant therapeutic proteins by modulating sialyltransferase expression. Biotechnol. Prog. 31, 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: A tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 34. Umeda T., Tomiyama T., Sakama N., Tanaka S., Lambert M. P., Klein W. L., and Mori H. (2011) Intraneuronal amyloid β oligomers cause cell death via endoplasmic reticulum stress, endosomal/lysosomal leakage, and mitochondrial dysfunction in vivo. J. Neurosci. Res. 89, 1031–1042 [DOI] [PubMed] [Google Scholar]
- 35. Burbulla L. F., Song P., Mazzulli J. R., Zampese E., Wong Y. C., Jeon S., Santos D. P., Blanz J., Obermaier C. D., Strojny C., Savas J. N., Kiskinis E., Zhuang X., Krüger R., Surmeier D. J., and Krainc D. (2017) Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science 357, 1255–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kosicek M., Wunderlich P., Walter J., and Hecimovic S. (2014) GGA1 overexpression attenuates amyloidogenic processing of the amyloid precursor protein in Niemann-Pick type C cells. Biochem. Biophys. Res. Commun. 450, 160–165 [DOI] [PubMed] [Google Scholar]
- 37. Tharkeshwar A. K., Trekker J., Vermeire W., Pauwels J., Sannerud R., Priestman D. A., te Vruchte D., Vints K., Baatsen P., Decuypere J.-P., Lu H., Martin S., Vangheluwe P., Swinnen J. V., Lagae L., Impens F., Platt F. M., Gevaert K., and Annaert W. (2017) A novel approach to analyze lysosomal dysfunctions through subcellular proteomics and lipidomics: the case of NPC1 deficiency. Sci. Rep. 7, 41408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cataldo A. M., Barnett J. L., Berman S. a., Li J., Quarless S., Bursztajn S., Lippa C., and Nixon R. a. (1995) Gene Expression and Cellular Content of Cathepsin D in Alzheimer's Disease Brain: Evidence for Early Up-Regulation of the EndosomaI-Lysosomal System. Neuron 14, 671–680 [DOI] [PubMed] [Google Scholar]
- 39. Amritraj A., Peake K., Kodam A., Salio C., Merighi A., Vance J. E., and Kar S. (2009) Increased Activity and Altered Subcellular Distribution of Lysosomal Enzymes Determine Neuronal Vulnerability in Niemann-Pick Type C1-Deficient Mice. Am. J. Pathol. 175, 2540–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. North S. J., Huang H. H., Sundaram S., Jang-Lee J., Etienne A. T., Trollope A., Chalabi S., Dell A., Stanley P., and Haslam S. M. (2010) Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 285, 5759–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen F. W., Gordon R. E., and Ioannou Y. a. (2005) NPC1 late endosomes contain elevated levels of non-esterified ('free') fatty acids and an abnormally glycosylated form of the NPC2 protein. Biochem. J. 390, 549–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dixit S. S., Jadot M., Sohar I., Sleat D. E., Stock A. M., and Lobel P. (2011) Loss of niemann-pick C1 or C2 protein results in similar biochemical changes suggesting that these proteins function in a common lysosomal pathway. PLoS ONE 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Damme M., Morelle W., Schmidt B., Andersson C., Fogh J., Michalski J.-C., and Lubke T. (2010) Impaired Lysosomal Trimming of N-Linked Oligosaccharides Leads to Hyperglycosylation of Native Lysosomal Proteins in Mice with -Mannosidosis. Mol. Cell Biol. 30, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vázquez M. C., del Pozo T., Robledo F. A., Carrasco G., Pavez L., Olivares F., González M., and Zanlungo S. (2011) Alteration of gene expression profile in niemann-pick type C mice correlates with tissue damage and oxidative stress. PLoS ONE 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maeda Y., Tomita S., Watanabe R., Ohishi K., and Kinoshita T. (1998) DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: Correct subcellular localization and stabilization of DPM1, and binding of dolichol phosphate. EMBO J. 17, 4920–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stoll J., Robbins A. R., and Krag S. S. (1982) Mutant of Chinese hamster ovary cells with altered mannose 6-phosphate receptor activity is unable to synthesize mannosylphosphoryldolichol. Proc. Natl. Acad. Sci. U.S.A. 79, 2296–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (26) partner repository with the dataset identifier PXD008438.