Abstract
Epigenetic pathways, including DNA methylation, are crucial for telomere maintenance. Deficient in DNA Methylation 1 (DDM1) encodes a nucleosome remodeling protein, required to maintain DNA methylation in Arabidopsis thaliana. Plants lacking DDM1 can be self-propagated, but in the sixth generation (G6) hypomethylation leads to rampant transposon activation and infertility. Here we examine the role of DDM1 in telomere length homeostasis through a longitudinal study of successive generations of ddm1-2 mutants. We report that bulk telomere length remains within the wild-type range for the first five generations (G1–G5), and then precipitously drops in G6. While telomerase activity becomes more variable in later generation ddm1-2 mutants, there is no correlation between enzyme activity and telomere length. Plants lacking DDM1 also exhibit no dysregulation of several known telomere-associated transcripts, including TERRA. Instead, telomere shortening coincides with increased G-overhangs and extra-chromosomal circles, consistent with deletional recombination. Telomere shortening also correlates with transcriptional activation of retrotransposons, and a hypersensitive DNA damage response in root apical meristems. Since abiotic stresses, including DNA damage, stimulate homologous recombination, we hypothesize that telomere deletion in G6 ddm1-2 mutants is a by-product of elevated genome-wide recombination in response to trans-poson mobilization. Further, we speculate that telomere truncation may be beneficial in adverse environmental conditions by accelerating the elimination of stem cells with aberrant genomes.
Keywords: Telomerase, Recombination, DNA damage, Stem cell, DNA methylation
Introduction
Telomeres serve two vital functions: they promote faithful replication of the chromosome terminus and they prevent chromosome ends from being recognized as double-strand breaks (DSBs). Maintaining proper telomere length is required for cell proliferation. Critically short telomeres trigger a powerful DNA damage response (DDR) that leads to end-to-end chromosome fusion, senescence or cell death (Sandell and Zakian 1993; Longhese 2008 Lazzerini-Denchi and Sfeir 2016). Likewise, ultra-long telomeres limit cell growth and promote tumorigenesis (McEachern and Blackburn 1995; Fairlie and Harrington 2015). Telomere length homeostasis is achieved through the establishment of a species-specific set point (i.e. 6–9 kb at birth in humans) and 2–9 kb in the model plant Arabidopsis thaliana (Slagboom et al. 1994; Richards and Ausubel 1988), and the maintenance of that set point through successive organismal generations.
To maintain telomere length, there must be a balance of forces that extend the telomere tract and those that shorten it. The primary mechanism for extending telomeres is through the action of the telomerase reverse transcriptase, which employs a catalytic subunit (TERT) to reiteratively copy an integral long noncoding RNA subunit (TER) for the continual addition of telomere repeats onto chromosome ends (Blackburn and Collins 2011). Telomere shortening occurs when telomerase is absent, due to the failure to fully copy nucleotides at the extreme 3′ end of the chromosome during conventional DNA replication (Watson 1972). In addition, because telomeres consist of long G-rich repeat arrays, a variety of accessory factors are necessary to promote replication fork progression through the telomere tract. Such factors include structure-specific proteins (primarily helicases) that help to resolve replication barriers including replication fork stalling (Martinez and Blasco 2015). If not resolved, these replication blocks can lead to telomere truncation by recombinational repair (Jain and Cooper 2010).
Telomere shortening can also occur if telomere binding proteins fail to protect chromosome ends from nucleolytic attack or deletional recombination of t-loops. T-loops are higher-order structures at chromosome ends that arise when the 3′ single-strand overhang on the telomere folds back and invades the duplex region of the telomere tract (Stansel et al. 2001; de Lange 2004). Because t-loops resemble a Holliday junction intermediate, they can be resolved through homologous recombination, resulting in extrusion of extra-chromosomal telomeric circles (ECTCs) and concomitant truncation of the telomere (Wang et al. 2004). This process, termed telomere rapid deletion (TRD), serves as a re-sizing mechanism to trim overly long telomeres back into the normal size range (Li and Lustig 1996). TRD must be tightly regulated to avoid catastrophic loss of telomeric DNA.
Arabidopsis thaliana is a useful model for telomere studies as a result of its extraordinary tolerance to telomere dysfunction (Watson and Riha 2010; Nelson and Shippen 2012). Plants lacking the telomerase catalytic subunit TERT undergo progressive telomere shortening (Fitzgerald et al. 1999; Riha et al. 2001). In the sixth generation of self-pollinated homozygous mutants (G6), telomeres reach a critical length threshold of 1 kb (Heacock et al. 2004), below which they become destabilized and susceptible to end-to-end chromosome fusion. Remarkably, telomerase-deficient plants can survive for up to four more generations, but they exhibit worsening developmental defects, including asymmetric leaf growth, decreased germination efficiency, and eventually vegetative growth arrest. These phenotypes are attributed to stem cell deficiency from telomere dysfunction (Riha et al. 2001).
As in mammals, plant telomeres are subjected to epigenetic modification. Two groups reported that A. thaliana telomeres are methylated by asymmetrical DNA methylation pathways directed by RNA-dependent DNA methylation (RdDM) (Cokus et al. 2008; Vrbsky et al. 2010). More recent studies suggest that the extent of DNA methylation at telomeres may be quite low or confined to subtelomeric regions (Vaquero-Sedas et al. 2011; Ogrocka et al. 2014; Vega-Vaquero et al. 2016). While both telomeric and subtelomeric regions of yeast and mammal chromosomes are exclusively heterochromatic (Blasco 2007; Ottaviani et al. 2008), subtelomeric regions in A. thaliana are reported to contain euchromatic (H3K4Me2 and H3K9Ac) as well as heterochromatic (H3K9Me2 and H3K27Me) features (Vaquero-Sedas et al. 2011, 2012; Vrbsky et al. 2010). In mammals histone methylation and DNA methylation are independently required for telomere maintenance (Blasco 2007). Mouse cell lines lacking histone methyltransferases Suv39h1 and Suv39h2 have abnormally long telomeres that are devoid of heterochromatin proteins (Garcia-Cao et al. 2004). Similarly, rodents lacking DNA methyltransferases have dramatically elongated telomeres (Gonzalo et al. 2006).
Two recent studies indicate that DNA methylation is required for telomere length homeostasis in Arabidopsis (Ogrocka et al. 2014; Vaquero-Sedas and Vega-Palas 2014). Plants bearing the ddm1-2 allele of Deficient in DNA Methylation 1 (DDM1) exhibit telomeres shorter than wild-type (Vaquero-Sedas and Vega-Palas 2014). Telomere shortening is also reported with a second DDM1 allele, ddm1-8 (Ogrocka et al. 2014).
DDM1 is a master regulator of DNA methylation. It functions as a SWI2/SNF2 family chromatin remodeler (Brzeski and Jerzmanowski 2003) that facilitates heterochromatin formation by promoting DNA methyltransferase access to the heterochromatin (Zemach et al. 2013). Plants lacking DDM1 experience a gradual genome-wide demethylation of up to 70% of all cytosines through successive generations (Vongs et al. 1993; Ronemus et al. 1996; Jeddeloh et al. 1999). Histone H3 methylation patterns are also altered in ddm1 mutants, contributing to genome-wide mobilization of transposable elements (TEs) and altered gene expression (Gendrel et al. 2002; Miura et al. 2001; Cokus et al. 2008). The cumulative change in DNA methylation status eventually leads to developmental pleiotropy: loss of apical dominance, shorter internode lengths, later flowering, increased cauline leaf number, and dramatically reduced fertility in the sixth generation of the homozygous ddm1 mutant (G6 ddm1) (Kakutani et al. 1996; Ronemus et al. 1996; Finnegan et al. 1996).
The mechanism by which DDM1 contributes to telomere maintenance in Arabidopsis is unknown. Here we investigate the molecular basis for telomere shortening through a longitudinal analysis across successive generations of ddm1-2 mutants. Specifically, we correlate changes in telomere length homeostasis with telomerase activity, telomere-related transcription, telomere recombination, chromosome end protection and stem cell viability. We report that telomere length is maintained through five consecutive generations of the mutants, but then drops precipitously in the sixth generation. Telomere shortening correlates with rampant transposon activation, telomere recombination and stem cell death. These findings reveal an unanticipated link between genome instability caused by DNA hypomethylation, telomere truncation and stem cell death.
Results
DDM1 is required for telomere length maintenance in A. thaliana
To investigate how DNA hypomethylation contributes to telomere length regulation in A. thaliana, we measured telomere length across multiple generations in ddm1-2 mutants, which bear a point mutation that causes aberrant splicing and results in a loss-of-function allele (Jeddeloh et al. 1999). Terminal restriction fragment (TRF) analysis was employed to assess bulk telomere length in the second generation (G2) and the terminal sixth generation (G6) of self-pollinated homozygous ddm1-2. Wild-type plants (Col-0) harbor telomeres within the range of 2–5 kb (Fig. 1a, lane 1) (Shakirov and Shippen 2004). Telomeres of G2 ddm1 mutants resembled those of wild-type (Fig. 1a, lanes 2–3). However, telomeres were markedly shorter in G6 mutants, with a dramatic loss of higher molecular weight telomere tracts, and ranged from only 1.5–2.5 kb (Fig. 1a, lanes 4–5).
Fig. 1.
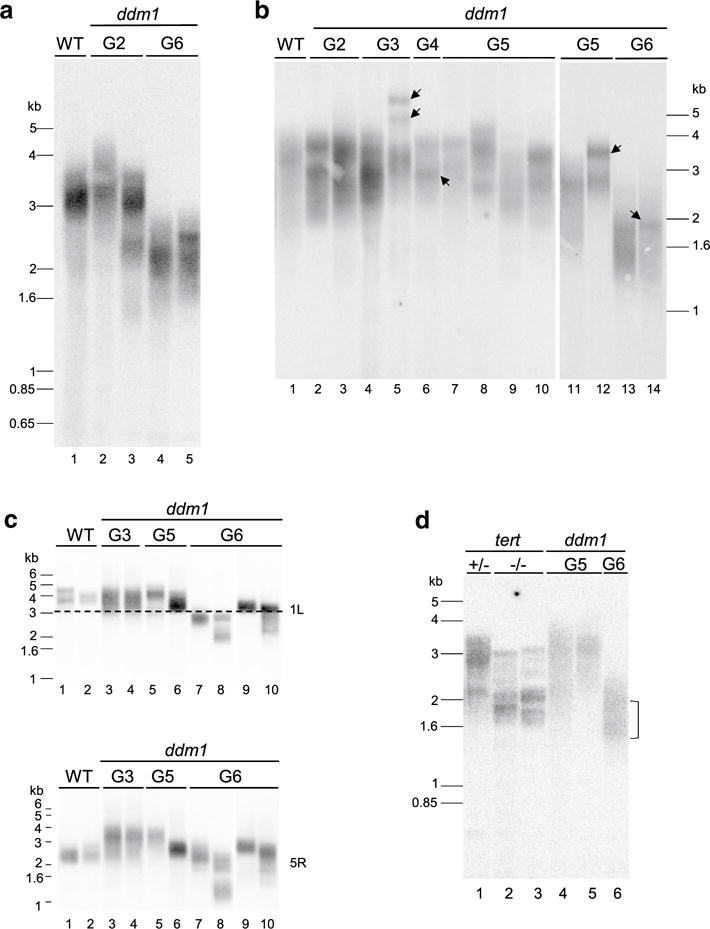
Plants lacking DDM1 display precipitous telomere shortening in the sixth generation. a TRF analysis of bulk telomere length of wild-type (WT), G2 and G6 ddm1-2 mutants is shown. DNA from an individual plant is analyzed in each lane. b TRF analysis of bulk telomere length in different generations of ddm1-2 mutants is shown. Some variation of telomere length is observed among individuals. Arrows indicate more homogeneous TRF bands within a given individual and may represent telomere recombination events. c Telomere length was assessed on individual chromosome arms by primer extension telomere repeat amplification (PETRA). 1L and 5R indicate PETRA analysis using subtelomeric primer for the left arm of chromosome 1 or right arm of chromosome 5, respectively. PETRA results for the two chromosome arms (1L and 5R) of an individual plant are shown in the top and bottom panel. d TRF analysis of G1 tert and G5 and G6 ddm1 mutants. Results for offspring of a tert heterozygous plant are analyzed (lanes 1–3). The bracket indicates telomeres below the 2 kb threshold
We examined the kinetics of telomere shortening by studying TRF profiles in successive generations of ddm1-2 plants (Figs. 1b, 2). Mean telomere length (mean TRF) was measured using the TeloTool (Gohring et al. 2014). The average bulk telomere length for at least five individual plants (biological replicates) from each generation was calculated. Telomeres in G2–G4 ddm1 generations were similar to wild-type, spanning 2–5 kb (Figs. 1b, 2). In the fifth generation of the mutants, the average telomere length (2.8 ± 0.2 kb) remained the same as wild-type (average = 2.9 ± 0.2 kb) (Fig. 2b) (p value > 0.05, t test), although in some individual plants the lower boundary of telomere tracts dipped below 2 kb (Figs. 1a, 2a). Strikingly, telomere length dropped dramatically in G6 ddm1 mutants for the majority of individuals tested (Figs. 1b, 2a). In G6 mutants the average telomere length was 2.1 ± 0.3 kb (Fig. 2b), a value that differed significantly from wild-type or ddm1 mutants in prior generations (p value < 0.01, t test). Notably, the amount of telomere shortening was variable: some G6 ddm1-2 individuals showed an average telomere length well-below 2 kb, while for others the average length remained within the wild-type range (Fig. 2a). Telomeres shortened by an average of ~ 700 bp from G5 to G6 (Fig. 2b). Loss of telomeric DNA was especially remarkable for long telomere tracts (Fig. 1a, b), which were depleted by up to 2 kb in a single generation. Due to reduced fertility, it was difficult to recover G7 ddm1-2 plants. Nevertheless, we were able to obtain sufficient DNA from two G7 individuals for analysis and found no additional telomere shortening (Fig. 2a), suggesting that telomeres in these plants may have reached a new stable set point.
Fig. 2.
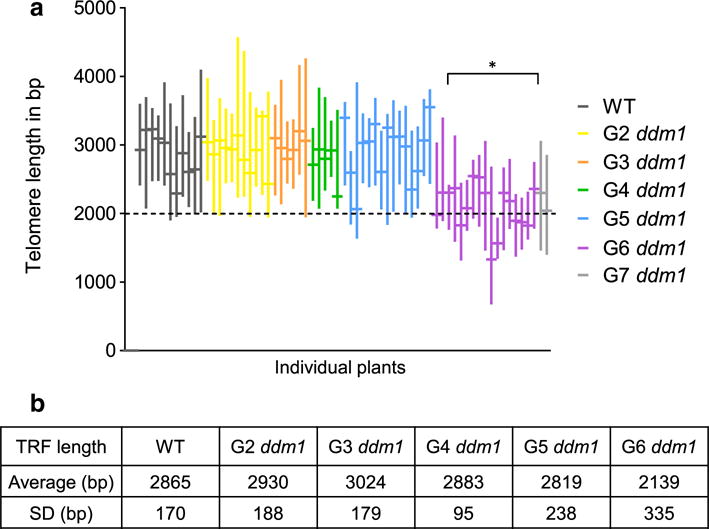
Telomere length quantification of different generations of ddm1-2 mutants using Telo-Tool. a Graphic representation of the telomere length range (vertical lines) and mean TRF (horizontal bars) of individual WT and ddm1 mutants. Dashes indicate the 2 kb threshold for minimal telomere length in WT. The asterisk indicates that mean telomere length of G6 ddm1 plants is significantly different from G5 ddm1 (p value < 0.01, t test). b Compiled data for telomere length analysis of each generation. SD standard deviation
For further insight into telomere length dynamics in ddm1-2 mutants, we performed primer extension telomere repeat amplification (PETRA) (Heacock et al. 2004). This method allows us to assess telomere length on specific chromosome arms from individual plants. As previously reported (Shakirov and Shippen 2004), there was variation in telomere length on different chromosome arms within the same plant (Fig. 1c, compare lanes 1 and 2 top and bottom panels; Fig. S1 top and bottom panels). In addition, in the same G6 ddm1 plant, not all telomeres were appreciably shorter than wild-type (Fig. 1c, compare lane 8 top and bottom panels, Fig. S1, lane 10, top and bottom). However, in agreement with bulk telomere length analysis, we observed obvious shortening of telomere tracts in several G6 ddm1 individuals. Compared with wild-type, G3 and G5 ddm1 mutants, three out of four G6 ddm1 plants showed marked telomere shortening on the left arm of chromosome 1 (1L), the left arm of chromosome 3 (3L) and the right arm of chromosome 4 (4R) (Fig. 1c, top panel; Fig. S1, top and bottom panels). Conversely, telomere tracts on the right arm on chromosome 5 (5R) remained in the wild-type range for two out of four G6 ddm1 individuals (Fig. 1c, lanes 7 and 9, bottom panel), while for two other individuals telomeres were substantially reduced (Fig. 1c, lanes 8 and 10, bottom panel). Taken together, these data indicate that a subset of chromosome arms experience a sudden loss of telomeric DNA in G6 ddm1-2 mutants.
Telomere shortening in ddm1‑2 mutants does not correlate with telomerase inactivation or changes in telomere‑related transcripts
The onset of pleiotropic developmental defects in late generation ddm1 mutants is remarkably similar to late generation A. thaliana tert mutants that experience telomere dysfunction (Riha et al. 2001). Therefore, we asked if telomerase activity is impaired in plants lacking DDM1 using the quantitative telomeric repeat amplification protocol (qTRAP) to monitor telomerase activity in flowers. In G2 ddm1-2 mutants, telomerase activity decreased to approximately 72% of the wild-type level (Fig. 3a). Telomerase activity declined further in G5 and G6 mutants (approximately 62 and 75% of wild-type) (Fig. 3a). Overall, the level of telomerase activity was much more variable in G6 ddm1-2 compared to earlier generations of the mutant or wild-type plants (Fig. 3a, b). Several G6 ddm1 individuals had less than 50% of the wild-type level of telomerase, with one G6 plant exhibiting only 23% of wild-type activity (Fig. 3b). These findings suggest that reduced telomerase activity may contribute to telomere shortening in a subset of G6 plants.
Fig. 3.
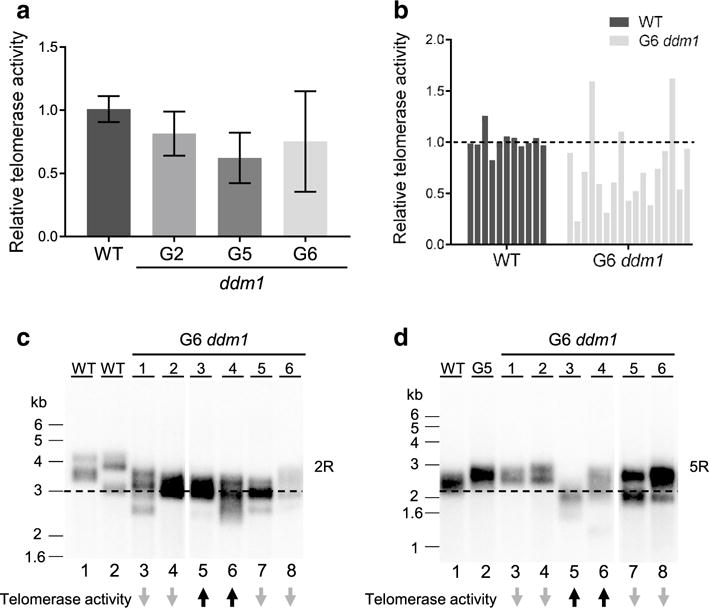
Telomerase activity in ddm1 mutants. a qTRAP results showing relative telomerase activity from flowers of ddm1 mutants. The average WT level of telomerase activity was set as 1. Each data point represents analysis of at least three biological replicates with two technical replicates. The standard deviation between biological replicates is represented by error bars. b Relative telomerase activity in individual WT and G6 ddm1-2 mutants is plotted. c, d Telomere length analysis for individual WT and G6 ddm1-2 mutants on specific chromosome arms using the PETRA assay. Representative data for the right arm of chromosome 2 (2R) are presented in c and for the right arm of chromosome 5 (5R) in d. The relative change in telomerase activity for each G6 ddm1 mutants is shown on the bottom. Upward black arrows indicate an increase in telomerase activity compared with WT; downward grey arrows indicate a decrease in telomerase activity for these specific samples. Dashes indicate the minimal telomere length in WT samples
To test whether there was a direct correlation between telomerase activity and telomere length in G6 ddm1-2 mutants, we compared the results of qTRAP with PETRA analysis performed on DNA samples from the same plants. Telomere length on several different chromosome arms was assessed (Fig. 3c, d; Fig. S2). Telomere shortening was observed in individuals with reduced telomerase activity (Fig. 3c, d, lanes 3, 4, 7 and 8) as well as in individuals with increased telomerase activity (Fig. 3c, d, lanes 5 and 6), revealing no direct correlation between the level of telomerase activity and telomere length in individual G6 ddm1-2 mutants.
Next we compared the TRF profile of plants completely null for telomerase activity (tert−/− mutants) (G1 tert) with that of G6 ddm1-2 mutants (Fig. 1d). As reported previously, telomeres in tert mutants shortened on average by ~ 250–500 bp per plant generation and gave rise to a discrete set of TRF bands that correspond to individual chromosome ends lacking telomerase extension (Riha et al. 2001) (Fig. 1d, lanes 2 and 3). In marked contrast, telomere tracts in G5 and G6 ddm1-2 mutants largely retained their size heterogeneity (Fig. 1d, lanes 4–6), consistent with sustained telomerase action on chromosome ends. Moreover, while a substantial fraction of telomeres in G1 tert mutants remained within the 2–5 kb wild-type size range, the majority of telomeres in G6 ddm1-2 fell below the bottom threshold of the wild-type range [see Fig. 1b, d (bracket)]. These data indicate that decreased telomerase activity is unlikely to be the main cause of telomere shortening in ddm1-2 mutants.
Loss of DNA methylation in ddm1 mutants causes de-repression of transposons and global transcriptional dysregulation (Zhang et al. 2006). We asked whether telomere shortening in G6 ddm1-2 mutants correlates with a change in the steady-state level of several known telomere-associated transcripts by employing quantitative RT-PCR (qRT-PCR). As a positive control, we monitored transcript levels for a mutator-like element (AtMu1) and a gypsy family retrotransposon (ATGP3). Consistent with previous studies (Hirochika et al. 2000; Tsukahara et al. 2009), we observed strong induction of the two TEs in G6 ddm1-2 mutants, but not in prior generations (Fig. S3).
We found no correlation between telomere shortening and the level of telomere-related transcripts in G6 ddm1-2 (p value ≥ 0.5, Table S1) for subunits of the telomeric G-overhang binding complex CST: CTC1 (Surovtseva et al. 2009), STN1 (Song et al. 2008) and TEN1 (Leehy et al. 2013), as well as Ku70, a core component of the non-homologous end-joining DNA repair pathway that is involved in telomere length regulation (Riha et al. 2002) and chromosome end stability (Kazda et al. 2012). Similarly, there was no change in telomerase subunits including the processivity factor POT1a (Surovtseva et al. 2007; Renfrew et al. 2014), the canonical telomerase RNA TER1 (Cifuentes-Rojas et al. 2011) and the telomerase regulatory lncRNA TER2 (Cifuentes-Rojas et al. 2012).
Telomeric repeat-containing RNA (TERRA) is a population of long non-coding RNAs transcribed from subtelomeric and telomeric DNA, which is implicated in telomere length regulation (Pfeiffer and Lingner 2012; Arora et al. 2014). Since epigenetic changes in subtelomeric chromatin impact TERRA production in human cells, (Arnoult et al. 2012), we monitored TERRA levels across multiple generations in ddm1-2 mutants by northern blotting. TERRA was detected as a heterogeneous smear sensitive to NaOH treatment (Figs. S4a, b) (Vrbsky et al. 2010). The overall TERRA hybridization signal in ddm1-2 mutants was similar to wild-type (Fig. S4c). The size distribution of TERRA transcripts of ddm1 mutants was also similar to wild-type except in G6 ddm1, where higher molecular weight TERRA transcripts were depleted (Fig. S4a). This change in the TERRA profile is consistent with shorter telomere tracts in G6 ddm1-2 plants. Taken together, these results indicate that telomere shortening in G6 ddm1-2 plants is not associated with a substantial change in telomere-related RNA transcripts.
The genome of late generation ddm1‑2 mutants is not grossly compromised
Another explanation for telomere shortening in G6 ddm1-2 mutants is loss of chromosome end protection. Therefore, we looked for evidence of end-to-end chromosome fusions using telomere fusion PCR (TF-PCR) (Fig. S5). In this assay, subtelomere-specific primers are used to amplify covalently linked telomeres (Heacock et al. 2004). A robust TF-PCR signal was detected with DNA from the stn1-1 positive control (Song et al. 2008), while no fusion products were obtained with wild-type or ddm1-2 mutants using two subtelomeric primer combinations (Fig. S5). To confirm these results, cytology of mitotic chromosomes spreads was conducted. The dicentric chromosomes formed from telomere fusion do not segregate properly during mitosis, and can be observed during anaphase as chromatin bridges. Analysis of over 50 anaphases in G5 and G6 mutants revealed no bridged chromosomes. The absence of chromatin bridges in ddm1-2 mutants was anticipated since, even in G6, the average length of telomeres does not fall below the critical end-protection threshold of 1 kb (Heacock et al. 2004). The absence of chromatin bridges also supports the conclusion that the genome of G6 ddm1-2 mutants is not severely destabilized.
To further assess genome stability, we used RT-PCR analysis to test whether G6 ddm1-2 mutants mount a global DNA damage response by monitoring transcripts that are elevated in plants experiencing telomere dysfunction (Boltz et al. 2012; Cifuentes-Rojas et al. 2012; Hashimura and Ueguchi 2011). We found no significant induction of GR1 (Gamma response 1), BRCA1 (Breast cancer susceptibility 1) or RAD51, although PARP1 (poly [ADP-ribose] polymerase 1) was somewhat increased (1.5-fold, p value > 0.05, t test) (Fig. S6). These findings indicate that in G6 ddm1-2 mutants, telomere integrity is largely preserved and plants do not suffer massive genome instability.
Increased G‑overhangs and ECTCs in late generation ddm1‑2 mutants
A more subtle indicator of telomere architecture is the status of the single-stranded G-overhang on the chromosomal terminus. Increased G-overhang signals are associated with defective telomere replication (e.g. incomplete C-strand fill-in after telomerase has extended the G-strand and uncoupling of telomeric G- and C-strand synthesis) as well as deprotection of the chromosome end leading to nucleolytic attack (O’Sullivan and Karlseder 2010). We assessed G-overhangs in ddm1-2 mutants using the in-gel hybridization method (Heacock et al. 2007). As expected (Riha and Shippen 2003), G-overhangs in our positive control (ku70 mutants) were significantly elevated relative to wild-type (Fig. 4a). We found essentially no difference in the G-overhang signal in G2 ddm1-2 (1.4 ± 0.6) compared to wild-type. However, the G-overhang signal was elevated in G5 (3.4 ± 1.7), and was even higher in G6 mutants (4.4 ± 1.5) (Figs. 4a, S7; p value < 0.01, t test). These findings indicate that although telomeres remain protected from end-to-end fusion in late generation ddm1-2 mutants, the structure of the chromosome terminus is perturbed.
Fig. 4.
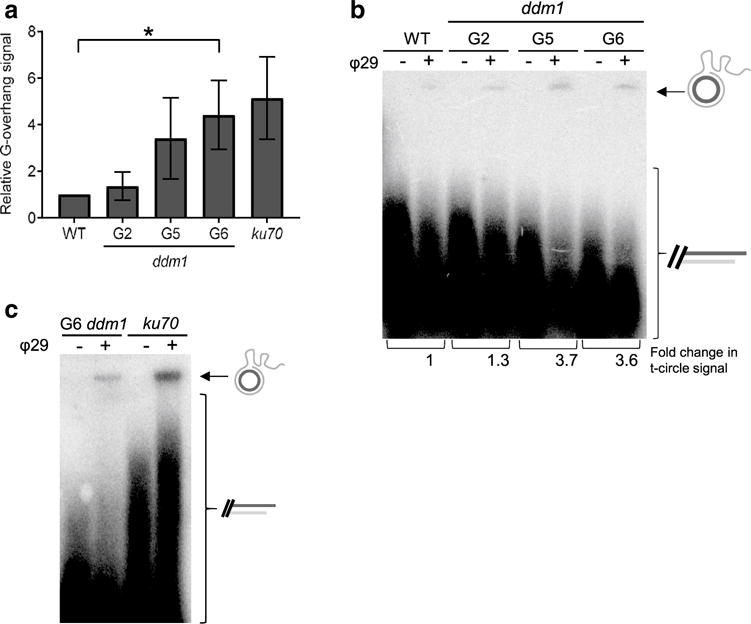
Evidence for telomere recombination in ddm1-2 mutants. a Quantification of the G-overhang signal from WT, ddm1 mutants and ku70 mutants as a positive control. Data are from at least three independent experiments and represented as mean ± SD. The G-overhang signal in WT was set to 1. The asterisk shows a statistically significant difference (p value < 0.01, t test) between WT and G6 ddm1. b, c Telomeric circle amplification (TCA) was carried out with DNA from individual WT, ddm1 and ku70 mutants in the presence (+) or absence (−) of phi 29 polymerase to amplify ECTCs. Circular and linear telomere repeats are indicated by the arrow and bracket. b TCA analysis of G2, G5 and G6 ddm1 mutants compared with WT is shown. Change in the ECTC signal compared with WT is indicated under corresponding lanes. c TCA analysis of G6 ddm1 and ku70 mutants
Finally, we considered the possibility that telomere shortening in G6 ddm1-2 mutants might be caused by deletional recombination. In wild-type A. thaliana, telomeres are typically heterogeneous in length (Fitzgerald et al. 1996; Riha et al. 2001), but in ddm1-2 mutants we occasionally observed a few homogeneous TRF bands, representing discrete populations of telomere tracts (Fig. 1b, lanes 5, 6, 12 and 14). A similar telomere profile has been reported for plants deficient in STN1 and TEN1 and has been attributed to an elevated frequency of recombination events due to compromised telomere tracts (Song et al. 2008; Leehy et al. 2013). A molecular signature of deletional recombination at telomeres is accumulation of extra-chromosomal telomeric circles (ECTCs) (Wang et al. 2004; Li et al. 2008). ECTCs can be detected using a telomeric circle amplification (TCA) assay (Zellinger et al. 2007). In this approach high molecular weight single-strand DNA produced by rolling circle amplification of ECTCs can be observed on an alkaline agarose gel by Southern blotting with a telomeric probe. We observed a slight increase in ECTC production in G2 ddm1-2 mutants (1.3-fold) relative to wild-type (Fig. 4b). However, the signal was markedly increased in G5 and G6 ddm1 mutants (3.6-fold). While the extent of ECTC production in G6 ddm1 mutants varied, in essentially all of our experiments the level of ECTCs was elevated compared to wild-type (Figs. 4b, S8). As an additional control, we compared t-circle formation in ddm1-2 mutants to plants deficient in KU70 (Fig. 4c). Telomeres are substantially elongated in ku70 plants, and both the TCA assay and 2D-gel analysis reveal evidence of telomeric recombination (Zellinger et al. 2007). We found a more dramatic increase in ECTC production in ku70 mutants than G6 ddm1-2 (Fig. 4c). We also compared ECTC formation in G6 ddm1-2 to stn1 mutants (Fig. S8), which display significantly shortened and de-protected telomere tracts (Song et al. 2008). Consistent with previous studies (Song et al. 2008), there was an increase in ECTC for homozygous stn1 mutants (twofold) compared to heterozygotes and wild-type plants. Notably, the fold increase in ECTC was approximately the same in two G6 ddm1-2 samples as in stn1 mutants (Fig. S8). These findings support the conclusion that telomeres in late generation ddm1-2 mutants are subject to deletional recombination.
Root apical meristems in G6 ddm1‑2 mutants are hypersensitive to DSBs
Genes associated with TE repression, including DDM1, DNA methyltransferases, and RdDM factors, are all upregulated in meristematic tissues to reinforce TE silencing and stable epigenetic inheritance (Baubec et al. 2014). Because plants deficient in DDM1 are hypersensitive to UV light and gamma irradiation and accumulate more DNA damage under genotoxic stress than wild-type (Shaked et al. 2006; Questa et al. 2013), we considered the possibility that the abrupt telomere shortening in G6 ddm1-2 mutants may be part of a targeted response to TE activation within the stem cell niche.
Since Arabidopsis stem cells are hypersensitive to DNA damage and prone to programmed cell death (PCD) (Fulcher and Sablowski 2009), our approach was to assess the frequency of PCD in the root apical meristem (RAM) of 5-day-old ddm1-2 mutant seedlings using propidium iodide (PI) staining (Fulcher and Sablowski 2009) (Fig. 5a). In the absence of genotoxic stress, only 2/47 (~ 4%) wild-type seedlings displayed positive PI staining in the RAM. Similarly, few if any of the RAM from G2 (0/37, 0%), G5 (1/51, ~ 2%) and G6 (2/46, ~ 4%) ddm1-2 mutants were PI-positive (Fig. 5b), indicating that the intrinsic genome instability associated with the loss of DDM1 does not trigger PCD in the RAM.
Fig. 5.
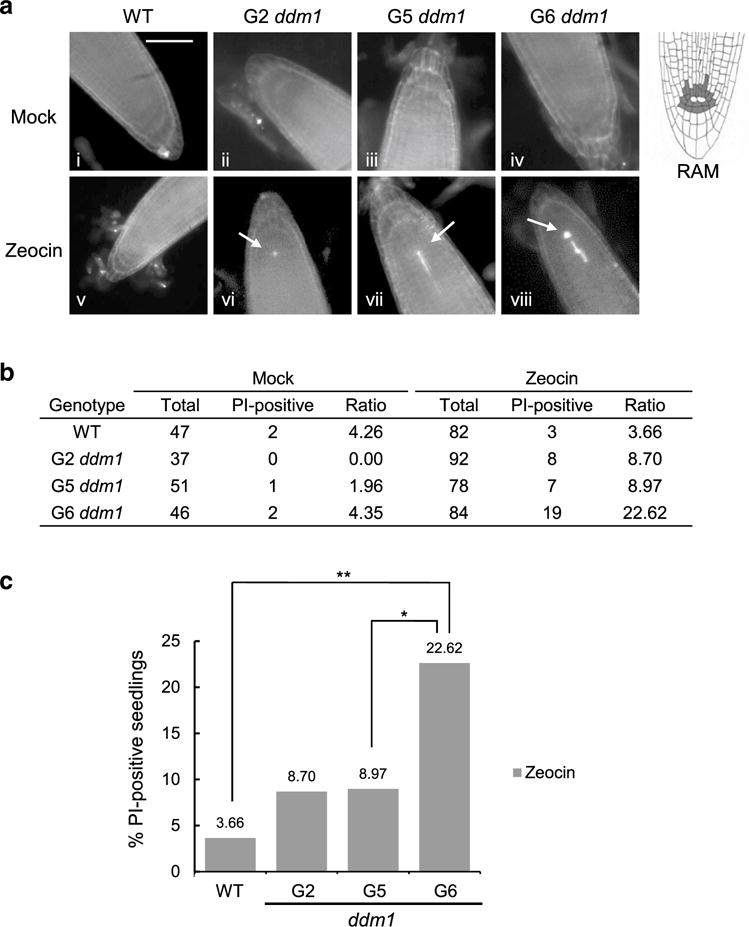
G6 ddm1-2 mutants are hypersensitive to DNA damage. a Representative images of root tips of 5-day-old WT and ddm1-2 mutant seedlings stained with propidium iodide (PI) to detect programmed cell death in the absence (i–iv) or presence (v–viii) of zeocin. Scale bar in a denotes 50 μm. Arrows denote PI-positive stem cells. For reference, a schematic diagram of the root tip is shown on the right. Dark gray cells in the root apical meristem (RAM) denote the stem and progenitor cells surrounding the quiescent center (in white). b Quantification of the percentage PI-positive RAM in WT and ddm1 mutants. c Graphic demonstration of the percentage of PI-positive RAM in the presence of zeocin. Single asterisk denotes p value < 0.05 (Fisher exact test) and double asterisks denote a p value < 0.005
To assess whether stem cells in ddm1 mutants might be hypersensitive to DNA damage, we treated ddm1-2 mutant seedlings with zeocin, a radiomimetic drug that induces DSBs (Fulcher and Sablowski 2009). The RAM of wild-type plants showed no substantial difference in PI staining in the absence (~ 4%) or presence of 20 μM zeocin (3/82, 3.66%), presumably due to the active DNA damage repair pathways and the low dose of the drug (Fulcher and Sablowski 2009). However, the percentage of PI-positive seedlings was elevated in G2 (8/92, ~ 9%) and G5 (7/78, ~ 9%) ddm1-2 mutants, consistent with a hypersensitive DNA damage response. Notably, a significantly higher fraction of the G6 ddm1-2 seedlings contained PI-positive RAMs (19/84, ~ 23%; p value < 0.005, Fisher exact test) upon zeocin treatment compared to wild-type (Fig. 5b, c). These findings indicate that stem cells in G6 ddm1-2 mutants are more sensitive to DNA damage than earlier generation mutants or wild-type. These results are consistent with some degree of chromosome instability in the RAM of G6 ddm1-2 mutants, and imply a mechanistic link between hypersensitivity to DNA damage and abrupt telomere shortening.
Discussion
Epigenetic regulation, particularly DNA methylation, is critical for plant growth and development. Here we performed a longitudinal study of ddm1-2 mutants and identified a new link between the loss of genome-wide DNA methylation, telomere length homeostasis and stem cell viability. A major function of DDM1 is to facilitate DNA methylation and thus repress TEs (Zemach et al. 2013). Gradual loss of DNA methylation in ddm1 mutants directs redistribution of H3K9 and H3K4 methylation at heterochromatin regions as early as the first generation, and ultimately results in loss of the heterochromatin state and increased transposition of TEs after multiple generation of self-pollination (Tsukahara et al. 2009). TE mobilization, in turn, leads to profound developmental defects and sterility (Finnegan et al. 1996). Consistent with the delayed onset of aberrant phenotypes in ddm1-2 mutants, we found that telomere length is maintained at the wild-type level throughout the first five generations of the mutants, but then it is abruptly disrupted in G6 when telomeres dramatically shorten. Thus, in addition to its role in maintaining heterochromatin and DNA methylation across the A. thaliana genome, our results support the conclusion of two previous studies (Ogrocka et al. 2014; Vaquero-Sedas and Vega-Palas 2014), indicating that DDM1 promotes telomere length homeostasis. Our data further argue that dysregulation of telomere length is correlated with a metastable genome caused by transposon mobilization.
We considered a number of different explanations for the molecular basis of telomere shortening in plants lacking DDM1. One notable finding was the delayed response of telomeres to DDM1 depletion. The ability of ddm1-2 mutants to faithfully maintain telomere length homeostasis through the first five generations implies that a threshold level of genome-wide DNA hypomethylation must be achieved to alter the integrity of telomere length control. DDM1 plays a minor role in gene methylation (Lippman et al. 2004), however the prolonged absence of DDM1 has been shown to profoundly impact chromatin structure leading to changes in gene expression (Zhang et al. 2006). Therefore, we asked if altered transcription of an essential telomere maintenance component correlated with telomere shortening in ddm1-2 mutants. We found no significant change in the most well-studied A. thaliana telomere-related genes, as well as the telomere-derived transcript, TERRA. These results are consistent with findings from Ogrocka et al. (2014) and with microarray data using inflorescences from wild-type and G6 ddm1 mutants (K. Slotkin, personal communication).
We also found that telomere shortening in G6 ddm1-2 mutants did not coincide with compromised chromosome end protection as measured by end-to-end chromosome fusion. In addition, since telomere shortening in G6 ddm1-2 is associated with only a subset of chromosome arms (Fig. 1c), the disruption of telomere length homeostasis is more likely to reflect stochastic telomere truncation rather than massive telomere erosion. Consistent with this conclusion, our transcription analysis did not reveal a significant activation of DDR in G6 ddm1-2 mutants, even though TEs have become globally de-repressed.
We considered the possibility that telomerase enzyme activity was down-regulated in response to loss of DDM1. We found that telomerase activity declined in ddm1 mutants over successive generations and by G6 was below 50% of the wild activity level in some plants. However, we also found numerous examples of G6 plants with wild-type levels of telomerase that nevertheless experienced telomere shortening. In addition, not only was telomere shortening greater in G6 ddm1-2 mutants than in plants null for telomerase, but also telomeres in late generation ddm1 mutants retained the length heterogeneity characteristic of telomerase extension. While we cannot rule out the possibility that deficient telomerase action contributes to telomere shortening in some G6 ddm1-2 individuals, it cannot explain the abrupt and dramatic loss of telomeric DNA in this setting.
There has been considerable debate concerning the presence of DNA methylation at A. thaliana telomeres and its impact on telomere length (Cokus et al. 2008; Vrbsky et al. 2010; Vega-Vaquero et al. 2016). One group reported that plants lacking RDR2, a core component of the DNA methylation pathway at asymmetric cytosines (such as telomeric repeats), do not display any telomere length dysregulation (Vrbsky et al. 2010), while other studies, including this one, find telomere shortening in plants lacking DDM1 (Ogrocka et al. 2014; Vaquero-Sedas and Vega-Palas 2014). Recent data indicate that subtelomeric sequences flanking telomere repeats, not telomeric DNA itself, are subject to methylation (Vega-Vaquero et al. 2016). Thus, it has been proposed that changes in telomeric chromatin are responsible for telomere shortening in ddm1 mutants (Vaquero-Sedas et al. 2011).
The extent to which loss of DDM1 impacts chromatin architecture at Arabidopsis telomeres remains an open question. Previous cytological analysis of ddm1 plants revealed dramatic de-condensation of centromeres, due to DNA hypomethylation and redistribution of histone marks, but no gross alteration of telomeres (Probst et al. 2003). However, we found that the G-overhang signal is significantly elevated in G5 and G6 ddm1-2 mutants. Telomere recombination is associated with invasion of G-overhangs into the duplex telomere region and is accompanied by the extrusion of ECTCs generated from t-loop resolution (Lustig 2003; Wang et al. 2004). Importantly, we observed a substantial increase in ECTCs in G5 and G6 ddm1 mutants, consistent with deletional recombination at telomeres. We note that telomere recombination has been associated with mutations in DNA methyltransferase in mammalian cells (Garcia-Cao et al. 2004; Gonzalo et al. 2006).
We hypothesize that hypomethylation of the A. thaliana genome leads to a global increase in homologous recombination (HR) that perturbs telomere homeostasis by a stochastic increase in deletional recombination. Both abiotic and biotic stressors, including genome instability, are associated with elevated HR in plants (Bennetzen 2000; Vicient 2010). While increased HR is beneficial to enhance the capacity of individuals to adapt to adverse conditions, it may also stimulate telomere deletion. There is mounting evidence that telomeres are responsive to environmental stress through recombinational mechanisms. For example, both telomere shortening and increased telomere sister chromatid exchange are triggered by ionizing radiation and oxidative stress in human cells (Berardinelli et al. 2010; Coluzzi et al. 2017). In addition, telomeres in plants deficient in the TEN1 component of the CST telomere end-binding complex undergo acute shortening upon heat shock; telomere truncation coincides with an increased level of ECTCs (Lee et al. 2016).
It is possible that the developmental defects associated with protracted DDM1 deficiency may not only reflect misregulation of development-specific genes, but also the consequences of a metastable genome (Ronemus et al. 1996; Zhang et al. 2016). The RAM is extraordinarily sensitive to DNA damage, and undergoes PCD in response to genotoxic stress (Fulcher and Sablowski 2009). We found that RAMs of G6 ddm1-2 mutants are more susceptible to DNA damage than wild-type or earlier generation of ddm1-2 mutants. These results argue that the genome of G6 ddm1 mutants, while not grossly compromised, is intrinsically unstable and prone to PCD. Truncated telomeres have the potential to further jeopardize metastable stem cell genomes in G6 ddm1-2 mutants by accelerating PCD (Lendvay et al. 1996; Hemann et al. 2001), potentially contributing to the developmental defects observed in ddm1-2. From an evolutionary perspective, elimination of severely compromised stem cells would be advantageous by preventing the accumulation of mutations (Fulcher and Sablowski 2009). Consequently, the seemingly damaging outcome of telomere truncation in late generation ddm1-2 mutants or other backgrounds with genome instability may stimulate culling of damaged stem cells and thereby help to sustain the integrity of the remaining stem cell reservoir.
Experimental procedures
Plant materials
Ddm1-2 seeds (G2 and G6) were a gift from Dr. Keith Slotkin (Ohio State University). G2 seeds were propagated for successive generations. An additional independent ddm1-2 line was also grown. Both lines were propagated until the terminal generation (G6 or G7). Plants were grown in soil under long-day conditions (16 h light/8 h dark) at 23 °C. For experiments using seedlings, seeds were sterilized using 50% bleach with 0.1% Triton-X 100 and plated on Murashige and Skoog (MS) medium with 0.7% agar (Caisson Labs). The stn1-1 and ku70 mutant alleles were previously described (Song et al. 2008; Zellinger et al. 2007).
Telomere analysis
DNA from whole plants was extracted using 2 × CTAB (100 mM Tris–HCl, 1.4 M NaCl, and 20 mM EDTA). Terminal restriction fragment (TRF) analysis was performed using 50 μg genomic DNA (gDNA) digested with Tru1l, resolved on an 0.8% agarose gel and hybridized with [32P] 5′ end-labeled (T3AG3)4 probe (Fitzgerald et al. 1999). The TeloTool was used for TRF quantification (Gohring et al. 2014). The mean TRF, standard deviation, the range, the max signal intensity were calculated using the TRF profile of individual plants. Average telomere length was calculated using the mean TRF of at least five individual plants of the same genetic background. Primer extension telomere repeat amplification (PETRA) was performed with 2 μg of gDNA and subtelomere specific primers as described (Heacock et al. 2004). Telomere fusion PCR (TF-PCR) was performed with subtelomere specific primers (Heacock et al. 2004).
G-overhang status was monitored using an in-gel hybridization assay (Heacock et al. 2007). EtBr staining was performed prior to the in-gel hybridization to obtain loading controls. Single-stranded G-overhang signals were quantified using Quantity One (Bio-Rad) and then normalized to the corresponding EtBr signals. The G-overhang signal obtained from wild-type plants was set to 1 and mutant samples were normalized to this value. Telomeric Circle Amplification (TCA) was performed as previously described (Zellinger et al. 2007). Telomeric circle signals were normalized to linear telomere signals in the absence of phi29 polymerase.
Quantitative telomeric repeat amplification protocol (qTRAP)
To measure telomerase activity, total protein was extracted from flowers of individual plants (Fitzgerald et al. 1996). qTRAP was carried out as previously described (Kannan et al. 2008), using a Dynamo HS SYBR Green qPCR kit (Thermo Fisher).
RT‑PCR and TERRA detection
Total RNA was extracted from plant tissues using a Directzol RNA kit (Zymo Research). Reverse transcription was performed with 1 μg total RNA with the qScript cDNA SuperMix (Quanta Biosciences). mRNA levels were assessed by quantitative PCR with primers described previously to detect telomere-related transcripts (Cifuentes-Rojas et al. 2011; Leehy et al. 2013) or DDR transcripts (Boltz et al. 2012; Cifuentes-Rojas et al. 2012; Hashimura and Ueguchi 2011), using SsoAdvanced Universal Supermix (Bio-Rad). RNA from at least three individual plants was used for each genetic background and at least two technical replicates were run for each reaction. Expression levels were averaged and normalized to GAPDH. Wild-type was set to 1 and mutant samples were compared to this value. To detect transcription of retrotransposons, primers recognizing AtMu1 and ATGP3 were used as described previously (Hirochika et al. 2000; Tsukahara et al. 2009).
TERRA was monitored by northern blot using 10 μg of total RNA. RNA was resolved on a 1% agarose gel, transferred to a nylon membrane and hybridized with a 32P 5′ labeled (CCC TAA A)5 probe as previously described (Zellinger et al. 2007).
Propidium iodide staining and cytogenetics
Sterilized seeds were grown in liquid MS culture for 4–5 days. For zeocin treatment, seedlings were transferred to fresh liquid MS culture either with or without 20 μM zeocin (Invitrogen) and treated for 4 h. After zeocin treatment, seedlings were immersed in 10 μg/ml propidium iodide solution at room temperature in the dark for 2 min, and then rinsed twice with H2O. Individual roots were separated and transferred to a slide in a drop of H2O. Images were obtained using a fluorescence microscope with a Zeiss filter set. ImageJ was used to adjust the brightness and contrast of images.
Supplementary Material
Key message.
Prolonged hypomethylation of DNA leads to telomere truncation correlated with increased telomere recombination, transposon mobilization and stem cell death.
Acknowledgments
We thank Keith Slotkin for providing mutant seeds, Lanying Zeng for sharing her fluorescence microscope, members of the Shippen lab for insightful comments and Jeff Kapler and Eugene Shakirov for critically reading the manuscript. This work was supported by the National Institutes of Health (GM065383 to D.E.S.).
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00299-017-2245-6) contains supplementary material, which is available to authorized users.
Accession numbers Sequence data from this article can be found in the Arabidopsis Genome Initiative or Gen-Bank/EMBL databases under the following accession numbers: DDM1 (AT5G66750), CTC1 (AT4G09680), STN1 (AT1G07130) and TEN1 (AT1G56260), POT1a (AT2G05210), TER1 (HQ401284), TER2 (AT5G16850) and Ku70 (AT1G16970).
Author contribution statement XX and DES conceived and designed the study. XX performed the experiments and analyzed data. XX and DES wrote the manuscript.
References
- Arnoult N, Van Beneden A, Decottignies A. Telomere length regulates TERRA levels through increased trimethylation of telo-meric H3K9 and HP1alpha. Nat Struct Mol Biol. 2012;19:948–956. doi: 10.1038/nsmb.2364. [DOI] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun. 2014;5:5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Finke A, Scheid MO, Pecinka A. Meristem-specific expression of epigenetic regulators safeguards transposon silencing in Arabidopsis. EMBO Rep. 2014;15:446–452. doi: 10.1002/embr.201337915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- Berardinelli F, Antoccia A, Cherubini R, De Nadal V, Gerardi S, Cirrone GA, Tanzarella C, Sgura A. Transient activation of the ALT pathway in human primary fibroblasts exposed to high-LET radiation. Radiat Res. 2010;174:539–549. doi: 10.1667/RR2127.1. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Boltz KA, Leehy K, Song X, Nelson AD, Shippen DE. ATR cooperates with CTC1 and STN1 to maintain telomeres and genome integrity in Arabidopsis. Mol Biol Cell. 2012;23:1558–1568. doi: 10.1091/mbc.E11-12-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc Natl Acad Sci USA. 2011;108:73–78. doi: 10.1073/pnas.1013021107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cifuentes-Rojas C, Nelson AD, Boltz KA, Kannan K, She X, Shippen DE. An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 2012;26:2512–2523. doi: 10.1101/gad.202960.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi E, Buonsante R, Leone S, Asmar AJ, Miller KL, Cimini D, Sgura A. Transient ALT activation protects human primary cells from chromosome instability induced by low chronic oxidative stress. Sci Rep. 2017;7:43309. doi: 10.1038/srep43309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- Fairlie J, Harrington L. Enforced telomere elongation increases the sensitivity of human tumour cells to ionizing radiation. DNA Repair. 2015;25:54–59. doi: 10.1016/j.dnarep.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA. 1996;93:8449–8454. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, McKnight TD, Shippen DE. Characterization and developmental patterns of telomerase expression in plants. Proc Natl Acad Sci USA. 1996;93:14422–14427. doi: 10.1073/pnas.93.25.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA. 2009;106:20984–20988. doi: 10.1073/pnas.0909218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Gohring J, Fulcher N, Jacak J, Riha K. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 2014;42:e21. doi: 10.1093/nar/gkt1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Hashimura Y, Ueguchi C. The Arabidopsis MERISTEM DISORGANIZATION 1 gene is required for the maintenance of stem cells through the reduction of DNA damage. Plant J. 2011;68:657–669. doi: 10.1111/j.1365-313X.2011.04718.x. [DOI] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock ML, Idol RA, Friesner JD, Britt AB, Shippen DE. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 2007;35:6490–6500. doi: 10.1093/nar/gkm472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Okamoto H, Kakutani T. Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP. Telomeric strategies: means to an end. Annu Rev Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Flowers SK, Munakata K, Richards EJ. Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc Natl Acad Sci USA. 1996;93:12406–12411. doi: 10.1073/pnas.93.22.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Nelson AD, Shippen DE. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Mol Cell Biol. 2008;28:2332–2341. doi: 10.1128/MCB.01490-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazda A, Zellinger B, Rossler M, Derboven E, Kusenda B, Riha K. Chromosome end protection by blunt-ended telomeres. Genes Dev. 2012;26:1703–1713. doi: 10.1101/gad.194944.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini-Denchi E, Sfeir A. Stop pulling my strings—what telomeres taught us about the DNA damage response. Nat Rev Mol Cell Biol. 2016;17:364–378. doi: 10.1038/nrm.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Xie X, Yang K, Zhang J, Lee SY, Shippen DE. Dynamic interactions of Arabidopsis TEN1: stabilizing telomeres in response to heat stress. Plant Cell. 2016 doi: 10.1105/tpc.16.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leehy KA, Lee JR, Song X, Renfrew KB, Shippen DE. MERISTEM DISORGANIZATION1 encodes TEN1, an essential telomere protein that modulates telomerase processivity in Arabidopsis. Plant Cell. 2013;25:1343–1354. doi: 10.1105/tpc.112.107425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2ΔB-mediated telomere shortening. Mol Cell Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Longhese MP. DNA damage response at functional and dysfunctional telomeres. Genes Dev. 2008;22:125–140. doi: 10.1101/gad.1626908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev Genet. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci. 2015;40:504–515. doi: 10.1016/j.tibs.2015.06.003. [DOI] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- Nelson AD, Shippen DE. Surprises from the chromosome front: lessons from Arabidopsis on telomeres and telomerase. Cold Spring Harb Symp Quant Biol. 2012;77:7–15. doi: 10.1101/sqb.2013.77.017053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrocka A, Polanska P, Majerova E, Janeba Z, Fajkus J, Fojtova M. Compromised telomere maintenance in hypomethylated Arabidopsis thaliana plants. Nucleic Acids Res. 2014;42:2919–2931. doi: 10.1093/nar/gkt1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90:93–107. doi: 10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 2012;8:e1002747. doi: 10.1371/journal.pgen.1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fransz PF, Paszkowski J, Scheid MO. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- Questa JI, Fina JP, Casati P. DDM1 and ROS1 have a role in UV-B induced- and oxidative DNA damage in A. thaliana. Front Plant Sci. 2013;4:420. doi: 10.3389/fpls.2013.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew KB, Song X, Lee JR, Arora A, Shippen DE. POT1a and components of CST engage telomerase and regulate its activity in Arabidopsis. PLoS Genet. 2014;10:e1004738. doi: 10.1371/journal.pgen.1004738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:611–615. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Shippen DE. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagboom PE, Droog S, Boomsma DI. Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–426. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- Vaquero-Sedas MI, Vega-Palas MA. Determination of Arabidopsis thaliana telomere length by PCR. Sci Rep. 2014;4:5540. doi: 10.1038/srep05540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Sedas MI, Gamez-Arjona FM, Vega-Palas MA. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011;39:2007–2017. doi: 10.1093/nar/gkq1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Sedas MI, Luo C, Vega-Palas MA. Analysis of the epigenetic status of telomeres by using ChIP-seq data. Nucleic Acids Res. 2012;40:e163. doi: 10.1093/nar/gks730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Vaquero A, Bonora G, Morselli M, Vaquero-Sedas MI, Rubbi L, Pellegrini M, Vega-Palas MA. Novel features of telomere biology revealed by the absence of telomeric DNA methylation. Genome Res. 2016;26:1047–1056. doi: 10.1101/gr.202465.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM. Transcriptional activity of transposable elements in maize. BMC Genom. 2010;11:601. doi: 10.1186/1471-2164-11-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Watson JM, Riha K. Comparative biology of telomeres: where plants stand. FEBS Lett. 2010;584:3752–3759. doi: 10.1016/j.febslet.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Bulankova P, Riha K, Shippen DE, Vyskot B. Telomerase-independent cell survival in Arabidopsis thaliana. Plant J. 2005;43:662–674. doi: 10.1111/j.1365-313X.2005.02479.x. [DOI] [PubMed] [Google Scholar]
- Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jacobsen SE. Genetic analyses of DNA methyltransferases in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol. 2006;71:439–447. doi: 10.1101/sqb.2006.71.047. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li Y, Xu T, Srivastava AK, Wang D, Zeng L, Yang L, He L, Zhang H, Zheng Z, et al. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov. 2016;2:16027. doi: 10.1038/celldisc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


