Abstract
Liquid biopsy provides minimally invasive and readily obtainable access to tumor-associated biological material in blood or other body fluids. These samples provide important insights into cancer biology, such as primary tumor heterogeneity, real-time tumor evolution, response to therapy, including immunotherapy, and mechanisms of cancer metastasis. Initial biological materials studied were circulating tumor cells and circulating nucleic acids, including circulating tumor DNA and microRNAs; more recently, studies have expanded to investigate extracellular vesicles, such as exosomes, microvesicles, and large oncosomes; tumor-derived circulating endothelial cells; and tumor-educated platelets. Even with an ongoing ambitious investment effort to develop liquid biopsy as an early cancer detection test in asymptomatic individuals, current challenges remain regarding how to access and analyze rare cells and tumor-derived nucleic acids in cancer patients. Technologies and associated bioinformatic tools are continuously evolving to capture these rare materials in an unbiased manner and to analyze them with high confidence. After first presenting recent applications of liquid biopsy, this review will discuss aspects affecting the field, including tumor heterogeneity, single cell analyses, and associated computational tools that will shape the future of liquid biopsy, with resultant opportunities and challenges.
Keywords: Liquid biopsy, circulating tumor DNA, circulating tumor cells, tumor heterogeneity, single cell analysis
The term Liquid Biopsy originally referred to the clinical use of blood samples for the capture and study of circulating tumor cells (CTCs) to monitor cancer patient treatment response and for marker detection associated with sensitivity or resistance to specific therapies.1 More recently, liquid biopsy encompasses myriad assays that measure tumor-derived cells or products in blood and other body fluids that, in general, may be obtained serially and easily with minimal patient risk. Most commonly studied are CTCs and circulating nucleic acids, such circulating tumor DNA (ctDNA) and microRNAs (miRNAs).2 However, the liquid biopsy field also comprises analyses of extracellular vesicles (EVs), including exosomes, microvesicles, large oncosomes (LO), and apoptotic bodies.3–10 In addition to distinct populations of immune-derived cells11–13, other cells in circulation garnering particular interest are tumor-derived circulating endothelial cells (CECs)14,15; and tumor-educated platelets (TEPs).16–18
Liquid Biopsy: Multifaceted Applications
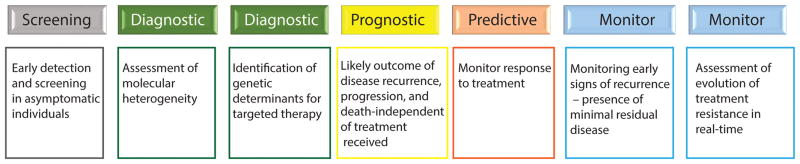
Tissue biopsy provides a snapshot of genetic alterations at the time of sampling. The information gleaned from such an approach is of limited use over time, as tumors constantly evolve to become highly heterogeneous. The tissue sample itself may represent a single geographic location within a heterogeneous tumor, and shed cells may form secondary or tertiary metastases which themselves become heterogeneous due to differences in microenvironmental cues and/or drug selection pressures and then seed further metastases.19,20 Longitudinal tracking of genetic and epigenetic alterations relevant to cancer through liquid biopsy is highly desirable for multiple applications, which include assessment of molecular heterogeneity of primary and secondary tumors, identification of genetic determinants for targeted therapy, cancer surveillance and monitoring for relapse, monitoring treatment response (chemo-, radiation-, and immunotherapy), assessment of resistant tumor evolution in real-time due to treatment pressure, and an aspiring application to screen asymptomatic individuals for early detection of cancer (Figure 1).
FIGURE 1.
Multiple applications for liquid biopsy as a tool for cancer screening, surveillance, disease monitoring, and identification of new therapeutic targets during disease evolution.
Blood is the most widely used body fluid for liquid biopsy. However, other body fluids such as urine, tears, saliva, pleural effusions, and cerebrospinal fluid harbor tumor-derived nucleic-acid, cells, and/or other materials.2,21 Sampling of blood or body fluids that directly drain tumor sites may yield higher quantities of CTCs or DNA of tumor origin than peripheral blood or plasma.22,23 For example, in patients with multiple types of primary and metastatic brain tumors, cerebrospinal fluid (CSF) showed higher concentration of ctDNA than plasma for disease restricted to the brain and meninges (in contrast to high tumor burden from visceral disease, which was then reflected in plasma ctDNA). CSF ctDNA was more representative of genomic alterations in the brain, a site difficult to biopsy, than plasma, and showed potentially actionable mutations not present in plasma.23 Recently, another interesting body fluid was added to the list. Researchers at Children’s Hospital Los Angeles reported detection of ctDNA in aqueous humor, the clear fluid between the cornea and the lens of the eye, in pediatric patients with retinoblastoma undergoing enucleation or intravitreous drug injections.24 Using whole genome sequencing of ctDNA, the authors were able to identify copy number variations (CNV) that were representative of the tumors. The authors noted that they were not successful in finding CTCs in aqueous humor and blood, nor ctDNA in blood.
Currently, there is an ambitious program underway sponsored by GRAIL, Inc., called the Circulating Cell-free Genome Atlas Study (CCGA, ClinicalTrials.gov Identifier: NCT02889978) that proposes to collect de-identified biospecimens and clinical data from 10,500 subjects with multiple types of malignancies and from 4500 representative subjects without cancer to develop and evaluate models that distinguish cancer and non-cancer as well as tissue of origin.25 It is hoped that deep sequencing of circulating cell-free nucleic acids (cfNAs) will provide useful information for future development of assays geared toward early detection of cancer. In another trial, the STRIVE study (ClinicalTrials.gov Identifier: NCT03085888), GRAIL is hoping to prospectively enroll 120,000 participants who are undergoing screening mammography and will perform high-intensity sequencing to characterize cfNAs that classify cancer from non-cancer, again with the aim of developing a blood test for the early detection of breast cancer.26 Other groups have demonstrated the capability of circulating microRNA (miRNA) as markers for diagnosing early-stage ovarian27 and gastrointestinal28 cancer.
Single Cell Analysis to Decipher Tumor Heterogeneity
Cancer is a complex disease that is characterized by high diversity among different cancers (intertumor heterogeneity) and within a single tumor (intratumor heterogeneity). Intertumor heterogeneity is the basis for the molecular classification of cancer.29–31 Single cell analysis shines limited light on intertumor heterogeneity, and hence it is not well suited to answer many questions related to cancer classification. However, single cell analysis plays a crucial role in deciphering intratumor heterogeneity, as clearly demonstrated by multiregion sequencing of primary tumor and comparison to metastatic foci.32–33 In addition to tissue biopsy, CTCs and ctDNA are used to understand the intratumor heterogeneity of primary tumor and metastatic disease progression.34 However, it is worth noting that although liquid biopsy alone cannot completely dissect interlesion heterogeneity,20 single cell profiling of CTCs shed from primary and/or metastatic tumors display heterogeneity among individual CTCs even within a single blood draw and, in cases of advanced disease, this may reflect the diversity of tumor cells remaining which would require further treatment.35
Although CTCs and ctDNA are considered complementary approaches to liquid biopsy, and use of ctDNA is fast gaining in popularity because of the relative ease of sample prep compared to technical challenges in capturing rare cancer cells, there are still some potential advantages for analyzing CTCs. First, while CTCs and ctDNA are both present in blood containing high background of non-specific biological materials such as white blood cells and normal or non-tumor-derived genomic DNA that comprises the majority of circulating cell free DNA (cfDNA), enriching for CTCs from blood will increase the signal to noise ratio. Second, functional assays are possible on enriched viable CTCs, including propagation in vitro and in vivo. In contrast, there still exist numerous technical challenges due to the presence of huge background DNA, in the determination of ctDNA alterations. In one large study using a targeted gene panel, over 37% of patients with multiple types of cancer had no detectable ctDNA test alterations and 24% had variants of unknown significance.36 Other studies have revealed cases of metastatic cancer in which one ctDNA marker shows evidence of treatment response while another may remain abnormally high, suggesting metastatic heterogeneity and that current treatment was only ablating a portion of the cancer cells.37 Future drug testing of any residual live cells would be a valuable tool for selecting follow-on therapeutic regimen. Typically, targeted sequencing of ctDNA for known cancer-related genes is performed, although this approach might restrict detection of novel mutations.38
Viable CTCs correlate with existing live tumor burden, while ctDNA concentration is thought to be generally associated with tumor death: it is posited that ctDNA is released from dying cancer cells either by apoptosis or necrosis;39,40 although others have argued that viable cancer cells may also release ctDNA.41 As the half-life of ctDNA is just under 2 hours,40,42 timing of blood collection relative to chemotherapy dosing may impact its measurement. Finally, thresholds for ctDNA response to a therapy remain to be standardized. Despite these limitations, it has been demonstrated that, when measureable, ctDNA nicely reflects tumor burden.43
About 10–20% of patients with non-small cell lung cancer (NSCLC) have EGFR mutations and may be candidates for EGFR tyrosine kinase inhibitor therapy. Thus, the cobas EGFR Mutation Test v2 has recently been FDA-approved for identifying patients with NSCLC who may harbor EGFR mutations, including the T790M mutation, that would make them candidates for different EGFR tyrosine kinase inhibitor therapies (note, the test is approved for patients with tumors not amenable to biopsy because of the 40% false-negative rate associated with this test).44–46 However, most lung cancers and other tumors do not contain actionable mutations that would specify a particular therapy. Although mutations may exist at the single cell level, ctDNA reflects an amalgam of DNA released from mutationally heterogeneous populations of cancer cells. Thus the concept of ctDNA alteration thresholds will become important. Finally, a critical concept for clinical utility of either liquid biopsy component will be in determining which drugs can be used to target specific tumor biological features or pathways in residual cancer cells. Live residual CTCs can potentially reveal this heterogeneous biology, which might then affect choice of specific and combinational treatments.
Multiple authors have demonstrated clear biological heterogeneity within tumor tissue using single-cell RNA sequencing (RNA-seq). Patel et al. analyzed 430 single cells obtained directly from five primary glioblastomas and reported extensive intratumoral heterogeneity in regulatory programs key to glioblastoma biology.47 Similarly, Tirosh et al. employed RNA-seq to analyze 4645 single cells isolated from 19 patients, and profiled malignant, immune, stromal, and endothelial cells.48 The authors noted that malignant cells within the same tumor displayed transcriptional heterogeneity associated with the cell cycle, spatial context, and a drug-resistance program. Tumor-infiltrating T cells showed exhaustion and variability across patients. The biological heterogeneity identifiable through transcriptional and proteomic profiling of individual CTCs shed from residual tumor still in the body may elucidate combinations of therapies that should be tried. We also expect that in the future, live CTCs themselves might be individually assayed and/or propagated for drug testing or drug discovery.
CTC enrichment for single cell analysis
For CTCs, there are numerous enrichment technologies (some commercial) with different pros and cons.49,50 These technologies can be broadly binned into two categories: affinity-based and label-free. Label-free approaches are desirable as they less biased in enrichment with respect to surface marker selection, but instead depend on size- or density-selection of CTCs or their intrinsic dielectric properties. Toner’s group reported an inertial focusing-enhanced microfluidic CTC capture platform, also known as the “CTC-iChip”, that can operate in a tumor epitope independent manner.51 In this platform, the nucleated white blood cells (WBCs; tagged with magnetic beads) and CTCs are separated from red blood cells (RBCs) by using deterministic lateral displacement. Subsequently, the nucleated cells are directed to a microfluidic channel using inertial focusing, and finally WBCs are depleted by applying a magnetic field. Extensive validation of this platform has been demonstrated for breast,52 pancreas,53 and prostate cancer.54 Recently, Nagrath’s group reported a label-free microfluidic platform, known as Labyrinth, that employs enhanced Dean flow fractionation (DFF) to enrich CTCs from breast cancer blood samples.55 The authors analyzed the gene expression of these enriched CTCs using a targeted TaqMan-based stem cell gene signature panel on C1™ Single-Cell Auto Prep system and BioMark System (Fluidigm).
Commercial label-free microfluidic systems for CTC enrichment include ClearCell® FX1 System (Clearbridge BioMedics), ApoStream® technology (ApoCell), VTX-1 platform (Vortex Biosciences), the C-Prep Genesis (Celsee Diagnostics), the Liquid Biopsy Platform (Cynvenio), and the Parsortix System (ANGLE), as well as multiple filter-based systems such as ISET (Rarecells Diagnostics), ScreenCell (ScreenCell), CellSieve (Creatv Microtech), and faCTChecker (Circulogix Inc.). Ramalingam et al. integrated ClearCell® FX System with the Polaris™ system (Fluidigm), and analyzed the full-length mRNA expression profile of individual CTCs from triple-negative (TNBC) and ER+/PR+/HER2− breast cancer patients.56 O’Shannessy et al. used the dielectrophoresis (DEP)-based ApoStream system to isolate CTCs from multiple epithelial cancers and confirmed the expression of folate receptor alpha (FRα) in CTCs from NSCLC lung, breast, and ovarian cancer patients.57 Powell et al. transcriptionally profiled single CTCs from the blood of women with aggressive primary and metastatic breast cancer using 87 cancer-associated and reference genes, finding a subset of 31 highly expressed genes that distinguished different subpopulations of CTCs; these included genes associated with metastasis (NPTN, S100A4, S100A9) and epithelial-mesenchymal transition (VIM, TGFβ1, ZEB2, FOXC1, CXCR4).35 Moreover, the expression profiles of single CTCs were markedly distinct from that of single cancer cells from cell lines or primary breast cancer cultures. In contrast, Pixberg et al. analyzed DNA methylation patterns in individual CTCs from patients with metastatic breast and castration-resistant prostate cancer. They reported epigenetic heterogeneity among CTCs, but surprisingly noted a relative lack of methylation at the promoter regions of three EMT-suppressing genes when inactivation by promoter hypermethylation was expected.58 This suggests that more work needs to be done on global epigenetic analyses to understand tumor cell shedding and metastasis at the single cell level.
Recently, Liu et al. demonstrated a targeted sequencing workflow, optimizing DNA extraction methods and evaluating whole genome amplification methods for use with targeted next-generation sequencing (NGS) panels on rare pooled CTCs from the blood of patients with metastatic colorectal cancer, captured label-free using laminar microvortices in a microfluidic device.59 This same label-free platform captured fresh CTCs for individual cell analysis using a single-cell multiplex protein assay.60
Sensitivity issues
Technologies for ctDNA detection are either based on polymerase chain reaction (PCR) or next-generation sequencing (NGS). The sensitivity of PCR (~ 1%) and its variant (droplet digital PCR; sensitivity ≤ 0.01%) is better compared to NGS-based methods (~2%).40 Oxnard’s group implemented Bio-Rad Laboratories’ droplet digital PCR into clinical practice for detection of KRAS and EGFR mutations in blood for non-small cell lung cancer (NSCLC) patients, and suggested that an EGFR T790M mutation that may be missed by tissue genotyping of a heterogeneous tumor may be detected in plasma.61 To improve the sensitivity of NGS-based methods, researchers added an additional PCR-based preamplification step prior to library preparation. This pre-amplification step improved the mutation detection down to 0.6% allele frequency from 1.6 ng DNA input for a previously published microfluidic multiplex PCR-based (MMP) target enrichment method.62 New methods to determine the tissue of origin of ctDNA are emerging based on DNA methylation patterns63 or nucleosome positioning.39
Bioinformatics Tools for Liquid Biopsy
Single cell/CTC variant callers have many pitfalls, as these analysis tools were originally developed for bulk sequencing data analyses. Current variant callers such as VarScan 2,64 Genome Analysis Tool Kit (GATK),65 and the Bayesian classifier method for very low allele fraction mutation detection, MuTect66 do not account for allelic dropout (ADO), coverage non-uniformity, allelic imbalance, and false-positive sequencing errors, which are characteristics of single cell DNA sequencing data. Monovar is a unique variant caller specifically developed to analyze single cell data to detect and genotype single-nucleotide variants.67 Recently, researchers reported SiNVICT (single nucleotide variant and indel caller for circulating tumor DNA), a computational method designed for sensitive and specific analysis of tumor-derived ctDNA, including ctDNA originating from multiple tumor subclones within a background of non-tumor-related circulating cfDNA. It detects single nucleotide variants (SNVs) and indels at very low variant allele percentages (as low as 0.5%) in situations of very high read depth and very low dilutions by accounting for false positives from mapping errors and other artifacts, including strand bias and low base quality at read ends.68
Filtering data by applying suitable criteria such as read coverage, sequencing quality score (e.g., Q20 indicates a 1 in 100 incorrect base call or an inferred base call accuracy of 99%), percentage reads mapped to genome, is important for single cell analysis. These criteria are subjective among research groups, and there is no standard for analysis filters. In addition, current methods for whole-genome amplification (WGA) are not perfect, and most existing methods introduce some amplification bias and chimeric DNA rearrangement. Hence, it is essential to optimize workflow to reduce bias and improve the read coverage.59
For single cell gene expression profiling, typical pitfalls include low mRNA capture efficiency, dropouts, incorrect global normalization methods. In the last few years, there has been a huge surge in single cell analysis platforms. Broadly, the chemistry for single cell mRNA-seq can be categorized as either full-length mRNA-seq or end-counting methods (3′ and 5′ end-counting). The mRNA capture efficiency for CEL-seq (cell expression by linear amplification and sequencing) protocol is ~ 3%,69 and ~ 7% for inDrop (indexing droplets),70 suggesting that these methods may only detect medium to highly abundant transcripts. The best reported capture efficiency is for Smart-seq2 full-length mRNA-seq protocol (~20%),71 also confirmed to be most sensitive but somewhat more costly in a systematic comparison of multiple single-cell RNA sequencing technologies.72 These data indicate that a majority of expressed transcripts in single cells may not be detected by current methods. Other high throughput droplet-based methods may be used to generate transcriptomic data for thousands of single cells, as in the study of immune cell subpopulations from blood or bone marrow.73
Computational methods are also heavily used to understand the concordance in mutation profile between cfDNA and tumor tissue. Towards this, Adalsteinsson et. al. reported ichorCNA, a software that quantifies the fraction of tumor-derived DNA in cfDNA without prior knowledge of tumor mutations by utilizing ultra-low-pass whole genome sequencing (ULP-WGS) to analyze somatic copy number alterations. In 1439 blood samples from 520 patients with metastatic breast or prostate cancer, the author noted that 34% of patients had sufficient (≥ 10%) tumor-derived DNA to perform standard depth whole-exome sequencing (WES), allowing an untargeted approach to ctDNA analyses and showing high concordance between WES of ctDNA and metastasis tissue.74 In a prior work, the same group performed whole-exome sequencing on individual CTCs from patients with metastatic prostate cancer using a census-based sequencing approach, where a sample was sequenced by preparing multiple independent individual CTC libraries, each exhibiting non-uniform coverage, with variants called only if present in multiple libraries. This approach works only for patients with at least five CTCs and emphasizes early metastatic trunk mutations over private mutations as the latter, which may be present in single CTCs are difficult to differentiate from sequencing errors associated with low input DNA.75
Conclusions – Opportunities and Challenges
Liquid biopsy is an active and advancing field, academically and commercially. There are many new platforms still being developed and tested for isolation and analysis of populations of circulating cells, nucleic acids, extracellular vesicles, and especially pertinent, investigations of ways in which these cells or tumor products interact with normal tissue, cancerous tissue, and importantly, immune cells in tissue, bone marrow, and bloodstream, and how this interaction may be leveraged for optimizing cancer care.
However, despite excellent demonstration of potential clinical applicability and promise, studies showing evidence of clinical utility of liquid biopsy to change or reject a therapy are still ongoing. Liquid biopsy in its current format remains an investigational tool. Part of the reason lies in the dearth of therapies for end-stage and generally chemoresistant cancers: earlier diagnosis of dismal disease does not impact clinical outcome when patients are switched from one ineffective therapy to another ineffective, and potentially more toxic, therapy. There is, however, hope that newer immunotherapies and drugs that affect the tumor microenvironment will changes the therapeutic landscape and increase response to therapy.
That said, liquid biopsy will most certainly play an important and evolving role in the future diagnosis and care of cancer patients.
Acknowledgments
This review was supported in part by NCI IMAT Grant R21CA177447 (SSJ), Susan G. Komen Foundation SAB1500003 (SSJ), and the John and Marva Warnock Research Fund (SSJ).
References
- 1.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 3.Pink RC, Elmusrati AA, Lambert D, et al. Royal Society Scientific Meeting: Extracellular vesicles in the tumour microenvironment. Phil Trans R Soc B. 2017;373:20170066. doi: 10.1098/rstb.2017.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menck K, Scharf C, Bleckmann A, et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J Mol Cell Biol. 2015;7:143–153. doi: 10.1093/jmcb/mju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menck K, Bleckmann A, Wachter A, et al. Characterisation of tumour-derived microvesicles in cancer patients’ blood and correlation with clinical outcome. J Extracell Vesicles. 2017;6:1340745. doi: 10.1080/20013078.2017.1340745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minciacchi VR, You S, Spinelli C, et al. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 2015;6:11327–11341. doi: 10.18632/oncotarget.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minciacchi VR, Spinelli C, Reis-Sobreiro M, et al. MYC mediates large oncosome-induced fibroblast reprogramming in prostate cancer. Cancer Res. 2017;77:2306–2317. doi: 10.1158/0008-5472.CAN-16-2942. [DOI] [PubMed] [Google Scholar]
- 10.Zweemer AJM, French CB, Mesfin J, Gordonov S, Meyer AS, Lauffenburger DA. Apoptotic bodies elicit Gas6-mediated migration of AXL-expressing tumor cells. Mol Cancer Res. 2017;15:1656–1666. doi: 10.1158/1541-7786.MCR-17-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akyüz N, Brandt A, Stein A, et al. T-cell diversification reflects antigen selection in the blood of patients on immune checkpoint inhibition and may be exploited as liquid biopsy biomarker. Int J Cancer. 2017;140:2535–2544. doi: 10.1002/ijc.30549. [DOI] [PubMed] [Google Scholar]
- 12.Beausang JF, Wheeler AJ, Chan NH, et al. T cell receptor sequencing of early-stage breast cancer tumors identifies altered clonal structure of the T cell repertoire. Proc Natl Acad Sci U S A. 2017;114:E10409–E10417. doi: 10.1073/pnas.1713863114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams DL, Martin SS, Alpaugh RK, et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc Natl Acad Sci U S A. 2014;111:3514–3519. doi: 10.1073/pnas.1320198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cima I, Kong SL, Sengupta D, et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci Transl Med. 2016;8:345ra89. doi: 10.1126/scitranslmed.aad7369. [DOI] [PubMed] [Google Scholar]
- 15.Lin PP, Gires O, Wang DD, et al. Comprehensive in situ co-detection of aneuploid circulating endothelial and tumor cells. Sci Rep. 2017;7:9789. doi: 10.1038/s41598-017-10763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. 2017;36:263–272. doi: 10.1007/s10555-017-9674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Best MG, Sol N, In ‘t Veld SGJG, et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell. 2017;32:238–252. e9. doi: 10.1016/j.ccell.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang G, Yuan M, Chen M, et al. Integrating multiple fitting regression and Bayes decision for cancer diagnosis with transcriptomic data from tumor-educated blood platelets. Analyst. 2017;142:3588–3597. doi: 10.1039/c7an00944e. [DOI] [PubMed] [Google Scholar]
- 19.Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor evolution as a therapeutic target. Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-17-0343. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Cheng X, Chen Z, et al. Circulating tumor cells in peripheral and pulmonary venous blood predict poor long-term survival in resected non-small cell lung cancer patients. Sci Rep. 2017;7:4971. doi: 10.1038/s41598-017-05154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nature Communications. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry JL, Xu L, Murphree A, et al. Potential of aqueous humor as a surrogate tumor biopsy for retinoblastoma. JAMA Ophthalmology. 2017;135:1221–1230. doi: 10.1001/jamaophthalmol.2017.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.https://clinicaltrials.gov/ct2/show/NCT02889978
- 26.https://clinicaltrials.gov/ct2/show/NCT03085888
- 27.Elias KM, Fendler W, Stawiski K, et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. eLife. 2017;6:e28932. doi: 10.7554/eLife.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging role of microRNAs as liquid biopsy biomarkers in gastrointestinal cancers. Clinical Cancer Research. 2017;23:2391. doi: 10.1158/1078-0432.CCR-16-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perou CM, Jeffrey SS, van de Rijn M, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava A, Kulkarni C, Mallick P, et al. Building trans-omics evidence: using imaging and ‘omics’ to characterize cancer profiles. Pac Symp Biocomput. 2018;23:377–387. [PubMed] [Google Scholar]
- 32.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupont Jensen J, Laenkholm A-V, Knoop A, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 34.Kidess-Sigal E, Liu HE, Triboulet MM, et al. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: Comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget. 2016;7:85349–85364. doi: 10.18632/oncotarget.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell AA, Talasaz AH, Zhang H, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwaederle M, Chattopadhyay R, Kato S, et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer Research. 2017;77:5419. doi: 10.1158/0008-5472.CAN-17-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 38.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. 2007;53:2215. doi: 10.1373/clinchem.2007.092734. [DOI] [PubMed] [Google Scholar]
- 42.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol. 2015;26:1883–1889. doi: 10.1093/annonc/mdv270. [DOI] [PubMed] [Google Scholar]
- 46.Odogwu L, Mathieu L, Goldberg KB, et al. FDA benefit-risk assessment of osimertinib for the treatment of metastatic non-small cell lung cancer harboring epidermal growth factor receptor T790M mutation. Oncologist. 2017 Dec 14; doi: 10.1634/theoncologist.2017-0425. pii: theoncologist.2017–0425. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murlidhar V, Rivera-Báez L, Nagrath S. Affinity versus label-free isolation of circulating tumor cells: who wins? Small. 2016;12:4450–4463. doi: 10.1002/smll.201601394. [DOI] [PubMed] [Google Scholar]
- 50.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10:374–394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Translat Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, Bardia A, Aceto N, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ting DT, Wittner BS, Ligorio M, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin E, Rivera-Báez L, Fouladdel S, et al. High-throughput microfluidic labyrinth for the label-free isolation of circulating tumor cells. Cell Syst. 2017;5:295–304. e4. doi: 10.1016/j.cels.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Ramalingam N, Lee YF, Szpankowski L, et al. Abstract 2923: Label-free enrichment and integrated full-length mRNA transcriptome analysis of single live circulating tumor cells from breast cancer patients. Cancer Research. 2017;77:2923. [Google Scholar]
- 57.O’Shannessy DJ, Davis DW, Anderes K, Somers EB. Isolation of circulating tumor cells from multiple epithelial cancers with ApoStream(®) for detecting (or monitoring) the expression of folate receptor alpha. Biomark Insights. 2016;11:7–18. doi: 10.4137/BMI.S35075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pixberg CF, Raba K, Müller F, et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene. 2017;36:3223. doi: 10.1038/onc.2016.480. [DOI] [PubMed] [Google Scholar]
- 59.Liu HE, Triboulet M, Zia A, et al. Workflow optimization of whole genome amplification and targeted panel sequencing for CTC mutation detection. NPJ Genom Med. 2017;2:34. doi: 10.1038/s41525-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sinkala E, Sollier-Christen E, Renier C, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:14622. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping as a sensitive and specific tool for guiding lung cancer care. JAMA oncology. 2016;2:1014–22. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bourgon R, Lu S, Yan Y, et al. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next-generation sequencing. Clin Cancer Res. 2014;20:2080–2091. doi: 10.1158/1078-0432.CCR-13-3114. [DOI] [PubMed] [Google Scholar]
- 63.Guo S, Diep D, Plongthongkum N, et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nature Genetics. 2017;49:635. doi: 10.1038/ng.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zafar H, Wang Y, Nakhleh L, Navin N, Chen K. Monovar: single-nucleotide variant detection in single cells. Nat Methods. 2016;13:505–507. doi: 10.1038/nmeth.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kockan C, Hach F, Sarrafi I, et al. SiNVICT: ultra-sensitive detection of single nucleotide variants and indels in circulating tumour DNA. Bioinformatics. 2017;33:26–34. doi: 10.1093/bioinformatics/btw536. [DOI] [PubMed] [Google Scholar]
- 69.Grun D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- 70.Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 72.Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65:631–643. e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 73.Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lohr JG, Adalsteinsson VA, Cibulskis K, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]