ABSTRACT
Hot-air hand dryers in multiple men's and women's bathrooms in three basic science research areas in an academic health center were screened for their deposition on plates of (i) total bacteria, some of which were identified, and (ii) a kanamycin-resistant Bacillus subtilis strain, PS533, spores of which are produced in large amounts in one basic science research laboratory. Plates exposed to hand dryer air for 30 s averaged 18 to 60 colonies/plate; but interior hand dryer nozzle surfaces had minimal bacterial levels, plates exposed to bathroom air for 2 min with hand dryers off averaged ≤1 colony, and plates exposed to bathroom air moved by a small fan for 20 min had averages of 15 and 12 colonies/plate in two buildings tested. Retrofitting hand dryers with HEPA filters reduced bacterial deposition by hand dryers ∼4-fold, and potential human pathogens were recovered from plates exposed to hand dryer air whether or not a HEPA filter was present and from bathroom air moved by a small fan. Spore-forming colonies, identified as B. subtilis PS533, averaged ∼2.5 to 5% of bacteria deposited by hand dryers throughout the basic research areas examined regardless of distance from the spore-forming laboratory, and these were almost certainly deposited as spores. Comparable results were obtained when bathroom air was sampled for spores. These results indicate that many kinds of bacteria, including potential pathogens and spores, can be deposited on hands exposed to bathroom hand dryers and that spores could be dispersed throughout buildings and deposited on hands by hand dryers.
IMPORTANCE While there is evidence that bathroom hand dryers can disperse bacteria from hands or deposit bacteria on surfaces, including recently washed hands, there is less information on (i) the organisms dispersed by hand dryers, (ii) whether hand dryers provide a reservoir of bacteria or simply blow large amounts of bacterially contaminated air, and (iii) whether bacterial spores are deposited on surfaces by hand dryers. Consequently, this study has implications for the control of opportunistic bacterial pathogens and spores in public environments including health care settings. Within a large building, potentially pathogenic bacteria, including bacterial spores, may travel between rooms, and subsequent bacterial/spore deposition by hand dryers is a possible mechanism for spread of infectious bacteria, including spores of potential pathogens if present.
KEYWORDS: spores, Bacillus, hand dryers, infection control, Bacillus subtilis, pathogens
INTRODUCTION
Hands contaminated with pathogenic microbes play a major role in the transmission of bacteria in health care institutions, the food industry, and community and domestic settings (1, 2). The microbial population of the skin comprises both resident and transient pathogenic and nonpathogenic floras (3). Transient floras colonize the superficial layers of the skin, are more easily removed by hand washing, and may be transferred by direct contact between human hands and the environment, as well as to patients (3–6). Importantly, transient floras include microorganisms associated with nosocomial infection, such as Staphylococcus aureus, enterococci, Pseudomonas spp., Klebsiella spp., and Acinetobacter spp. (3). Consequently, hand washing is essential in minimizing transmission of such pathogenic bacteria by hands, and many studies have focused on topics such as hand-washing techniques (7), selection and handling of hand-washing agents (8–10), and methods to improve hand hygiene adherence for health care workers (11–14). Hand drying is the important last stage of the hand-washing process and must decrease the risk of cross-contamination since transmission of microorganisms is more effective in wet than dry environments (15, 16).
Methods for hand drying vary considerably and include cloth or paper towels and hot-air or jet-air dryers (7–10, 17–25). These hand-drying methods differ in their abilities to aerosolize bacteria and thus the potential to transmit microorganisms, with hot-air dryers giving more aerosolization than paper towels (10, 17–25). Indeed, several studies have found that hot-air dryers (here, hand dryers) can disperse bacteria from hands and contaminate the surrounding area (26–28). However, it is also possible that forced air from hand dryers deposits environmental bacteria on well-washed hands. Indeed, a recent report (29) suggested that hand dryers could deposit both pathogenic and nonpathogenic bacteria on the hands and bodies of users. This information raises several important questions, including the following. (i) Does the forced air from hand dryers disperse aerosolized, environmental microorganisms on surfaces during drying? (ii) Are the hand dryers a potential bacterial reservoir?
RESULTS
Hand dryers deposit bacteria on agar plates in multiple bathrooms.
Analyses of colonies on rich, double-strength Schaeffer's glucose (2×SG) medium plates left open for 2 min in men's and women's bathrooms (36 in total) with hand dryers off in three buildings in the basic science research areas of the University of Connecticut (UConn) School of Medicine found an average of 0 to 1 colony/plate (Table 1, environmental samples). Extending exposure of duplicate open plates for 18 h in two bathrooms with hand dryers off gave an average of only 6 colonies/plate (Table 1). However, when plates were exposed to air from hand dryers for 30 s in the 36 bathrooms located on different floors of the three buildings tested (Fig. 1), there was an average of 18, 24, and 60 colonies/plate in each building tested (range, 3 to 254 colonies/plate) in two different experiments separated by 3 to 4 weeks (Table 1). Bathrooms in research areas above the academic building (Fig. 1) had significantly greater average numbers of colonies/plate, i.e., 60, than bathrooms in two other basic science research areas (Fig. 1, buildings 1 and 2), which averaged 24 and 18 colonies/plate, respectively (P < 0.001) (Table 1). Notably, in two of the three buildings in which bacteria dispersed by hand dryers were measured, there was no statistically significant difference between the numbers of bacteria dispersed in men's and women's bathrooms (data not shown), and this subject was not studied further.
TABLE 1.
Bacteria and spore recovery from bathroom hand dryer air or environmental aira
| Location | No. and types of bathrooms | Environmentb |
Hand dryersc |
|||
|---|---|---|---|---|---|---|
| Total no. of colonies/plate | No. of KSF/plate | Total no. of colonies/plated | No. of KSF/platee | % KSFf | ||
| Building 1 | 7 M, 7 W | 0.21 ± 0.57 | 0 | 17.7 ± 10.1 | 0.68 ± 0.93 | 3.85 |
| Building 2 | 7 M, 7W | 0 | 0 | 23.8 ± 23.3 | 1.16 ± 1.07 | 4.87 |
| Academic building | 4 M, 4 W | 1 ± 1.46 | 0 | 59.5 ± 60.2 | 1.5 ± 1.4 | 2.52 |
Plates were exposed to the bathroom environment for 2 min or for 30 s under hand dryers in men's (M) or women's (W) bathrooms on various floors in research buildings 1 and 2 and on research floors above the academic building, and total bacteria and levels of Kmr sporeformers (KSF) were quantified, all as described in Materials and Methods. Values for total colony number and number of Kmr sporeformers are averages ± standard deviations.
The range of numbers of colonies per individual open plate exposed to the bathroom environment for 2 min was 0 to 3. Exposure of two open plates to bathroom environmental air for 18 h in two bathrooms yielded 2, 4, 8, and 10 colonies (average, 6 ± 3.65).
The range of total colonies per individual plate exposed to a bathroom hand dryer air for 30 s was 3 to 254.
The plates exposed to bathroom hand dryer air in research areas above the academic building had significantly higher numbers of total colonies than the those in research building 1 (P < 0.001) and research building 2 (P < 0.001). There was no statistical difference between the numbers of colonies recovered from bathrooms in research building 1 and those from building 2 (P = 0.235).
Hand dryer air in research building 2 bathrooms had significantly fewer Kmr sporeformers than either bathrooms in research building 1 (P = 0.025) or bathrooms in the research area above the academic building (P = 0.016). There was no difference in numbers of Kmr sporeformer recovered from bathrooms in building 1 and those for the research areas above the academic building (P = 0.440).
There was also a statistically significant difference in the percentages of Kmr sporeformers recovered from hand dryer air in bathrooms in buildings 1 and 2 (P = 0.044) and the percentages in building 1 and above the academic building (P < 0.001), but not for the percentages for building 2 and above the academic building (P = 0.306).
FIG 1.
Approximate relative locations of bathrooms on various floors in different basic science research areas where bacterial deposition by hand dryer air was measured, including above the academic (A) building, research buildings 1 and 2, and the laboratory on the 2nd floor of building 1 that produces spores. All buildings are connected by hallways, and the rise between floors in all buildings is ∼14 feet.
These bacterial counts were obtained from plates ∼12 in. from the hand dryer air outlet. Since hands below a hand dryer may be closer than 12 in., we also compared bacterial deposition after exposure of plates to hand dryer air 2 in. or 12 in. from the nozzle (see Table S1 in the supplemental material). While more bacteria were recovered from plates closer to the hand dryer nozzle, this difference did not reach statistical significance (P = 0.175).
Source of aerosolized bacteria deposited on surfaces by hand dryers.
If the bacteria found on plates exposed to hand dryer air are coming from room air passing through hand dryers, then HEPA filters in the hand dryers should reduce bacterial surface deposition. Indeed, this is reported to reduce bacteria passing through the hand dryers used in this work by ≥99.9% (30). Therefore, we compared bacterial recovery from five hand dryers before and after retrofitting the dryers with HEPA filters. While bacterial recoveries prior to HEPA filter retrofit were comparable to those seen in previous experiments, bacterial counts at 9 days after HEPA filter retrofitting were reduced ∼4-fold, and this difference was significant (P < 0.001) (Table 2; note that for the hand dryer data in Table 1, air exposure was 30 s whereas it was 1 min for the data in Table 2). Thus, the HEPA filter retrofit reduced, but did not eliminate, microbe deposition by hand dryers.
TABLE 2.
Bacterial deposition by bathroom hand dryers with and without HEPA filters
| Hand dryer location (floor) | Building no. | Bathroom typea | No. of colonies/plateb |
|
|---|---|---|---|---|
| No HEPA filterc | With HEPA filterd | |||
| 2nd | 1 | M | 44, 64 | 16, 13 |
| 2 | M | 77, 41 | 19, 13 | |
| 2 | W | 56, 72 | 11, 21 | |
| 3rd | 1 | M | 50, 89 | 14, 16 |
| 4th | 1 | M | 65, 40 | 11, 14 |
| Overall avg ± SDe | 59.8 ± 16.5 | 14.8 ± 3.3 | ||
M, men's bathroom; W, women's bathroom.
Two plates were exposed to hand dryers for 1 min in men's (M) or women's (W) bathrooms in various locations, and the numbers of bacteria deposited were determined as described in Materials and Methods. Values are for each of two plates.
Measured 2 days before HEPA filter retrofit.
Measured 9 days after HEPA filter retrofit.
The difference in the number of colonies/plate with and without HEPA filters was statistically significant (P < 0.001).
A second possible source of bacteria deposited on plates by hand dryers is from buildup of bacteria in the hand dryers themselves and then dispersal by the forced air. To test this possibility, the inner surface of hand dryer nozzles from eight different bathrooms, four with and four without HEPA filters, were swabbed to recover bacteria as described in Materials and Methods. However, an average of only ∼4 colonies was recovered per bathroom (data not shown). Thus, it seems extremely unlikely that hand dryers carry within the nozzle a significant reservoir of bacteria that are aerosolized and deposited on plates.
The data above indicated that much, and perhaps all, of the bacteria deposited on plates by hand dryer air is coming from the bathroom air by either passing through hand dryers without HEPA filters or by being pulled into the air coming out of hand dryer nozzles by convection. To test this directly, two Luria broth (LB) (see Materials and Methods) and two blood agar plates in each of five bathrooms in research buildings 1 and 2 were exposed to air blown by small sanitized fans (Fig. S1) for 20 min as described in Materials and Methods. These bathroom air exposures gave averages of 15 and 12 colonies/plate in research buildings 1 and 2, respectively (Table 3) (range, 2 to 33 colonies/plate), demonstrating that the small fan is capable of deposition of aerosolized bacteria from bathroom air. These results did not differ significantly from those for bacterial colonies deposited by hand dryers from the same bathrooms when calculations of the colonies deposited by hand dryers and small fans were corrected for the times for air exposure and rates of airflow from these two sources (Table 3).
TABLE 3.
Bacterial recovery from hand dryer air and bathroom air moved by small fans
| Floor | Bacterial counts by source and location (no. of colonies/plate)a |
|||
|---|---|---|---|---|
| Hand dryer air |
Bathroom air |
|||
| Building 1 | Building 2 | Building 1 | Building 2 | |
| 1st | 19, 5, 23, 36 | 8, 6, 16, 17 | 19, 33, 14, 22 | 14, 17, 24, 23 |
| 2nd | 38, 2, 35, 35 | 27, 37, 17, 16 | 16, 21, 25, 30 | 18, 21, 13, 8 |
| 3rd | 6, 0, 18, 31 | 7, 12, 12, 11 | 19, 8, 15, 20 | 7, 5, 9, 11 |
| 4th | 3, 4, 10, 11 | 10, 10, 10, 12 | 6, 5, 3, 7 | 23, 6, 11, 9 |
| 5th | 16, 11, 11, 19 | 26, 22, 12, 16 | 6, 8, 8, 14 | 2, 3, 4, 12 |
| Avg ± SDb | 16.7 ± 12.5 | 15 ± 7.7 | 15 ± 8.6 | 12 ± 7.0 |
| Corrected avg ± SDc | 13.4 ± 10.0 | 2.2 ± 6.2 | 15 ± 8.6 | 12 ± 7.0 |
Bacteria were collected on two occasions separated by ∼1 week in women's bathrooms in research buildings 1 and 2, with either a 30-s exposure to hand dryer air or to air moved for 20 min by small fans as described in Materials and Methods. For each sampling occasion, duplicate plates were collected. Values are for each of four plates.
These differences were not statistically significant (P = 0.235).
The average number of colonies/plate was corrected for the ∼50-fold greater airflow on plates from hand dryers and the 40-fold longer exposure of plates to air from small fans which meant that the average numbers of colonies/plate in 30 s from hand dryer air were divided by 1.25 to correct for the differences in airflow and exposure time using air moved by hand dryers and small fans.
Bacillus subtilis PS533 is deposited on surfaces by hand dryers.
One of the organisms recovered from multiple bathrooms was B. subtilis (see below), the species of strain PS533 used to prepare large amounts of spores in a research laboratory on the 2nd floor of research building 1 (Fig. 1). B. subtilis is nonpathogenic, and its spores are found throughout the environment, including the human gut, and are also present in many probiotic preparations ingested by both animals and humans (31). Since B. subtilis PS533 is kanamycin resistant (Kmr) because it contains the plasmid pUB110 (32), we determined numbers of Kmr and spore-forming colonies deposited on plates by hand dryers in different bathrooms (Table 1). Of note, we recovered Kmr sporeformers from all floors of the three research buildings tested, regardless of distance from the spore laboratory on the 2nd floor of building 1 (Fig. 1). While there was no statistical relationship by floor between the distance of the bathroom from the spore laboratory and the number of Kmr spore-forming bacteria recovered (Table S2 and data not shown), there was a statistically significant increased percentage of Kmr spores recovered from building 1 bathrooms overall compared with that from bathrooms above the academic building and building 2 (Table 1). We next extracted plasmid DNA from 25 Kmr spore formers, and all had a plasmid that comigrated with plasmid pUB110 (Fig. 2 and data not shown). In addition, 12 of these plasmids that were randomly chosen had a BamH1/EcoR1 restriction enzyme digestion pattern consistent with pUB110 (Fig. 2 and data not shown), strongly indicating that these isolates are strain PS533 bacteria. In total, these latter isolates comprised ∼2.5 to 5% of all bacteria deposited on plates by hand dryer air. Kmr spore-forming B. subtilis bacteria, most likely PS533, also comprised 3 to 4% of strains isolated from bathroom air blown on plates by small fans (see below).
FIG 2.
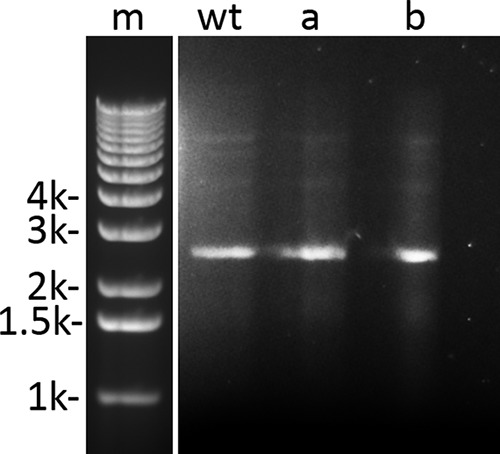
Gel electrophoresis of plasmid DNA from cells of some Kmr spore-forming colonies dispersed by hand dryers in various bathrooms in basic science research areas at the UConn School of Medicine. Plasmid DNA was isolated from cells and subjected to agarose gel electrophoresis, and the gel was stained and photographed as described in Materials and Methods. Migration positions of double-stranded DNA markers (m) of various thousands (k) of bp are shown on the left side of the figure. Lanes wt (wild type), a, and b have plasmid DNA isolated from cells of B. subtilis PS533 (wild type), and two randomly selected isolates that were Kmr, bacillus-like, and spore forming collected after exposure to hand dryer air in a women's bathroom on the 1st floor of building 2 (a) and a men's bathroom on the 6th floor of building 1 (b).
Aerosolized spores are deposited on surfaces by hand dryers.
We next wanted to determine whether B. subtilis PS533 and perhaps other sporeformers were deposited on surfaces as spores or as growing or sporulating cells. To answer this question, microbes from hand dryer air in a bathroom in building 2 were collected in liquid and in control liquid open to bathroom air as described in Materials and Methods. Aliquots of the liquid were heated to kill growing mesophilic bacteria but not spores, and aliquots of heat-treated control and hand dryer air-exposed liquid were plated for colony recovery, again as described in Materials and Methods. No colonies were recovered from the control liquid, while 16 colonies were recovered from the two plates spread with 200 μl of hand dryer air-exposed samples (40 colonies/ml). All but two of these colonies were Kmr, and all but one formed significant amounts of spores on LB plates. Analysis of the bacteria in these 16 colonies by a matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) Biotyper (see below) found that one colony could not be identified, one of the non-Kmr organisms was Bacillus pumilus, and the remainder were B. subtilis colonies. These data confirm that at least some of the B. subtilis bacteria recovered from agar plates are deposited on surfaces as spores by hand dryers. However, since the total number of bacteria in the liquid exposed to hand dryer air was not determined, it was not possible to directly determine the percentage of total bacteria dispersed by hand dryer that were present as spores.
To identify spores present in bathroom air itself, small fans were used to blow air in two bathrooms for 1 h at two LB plates in each bathroom; after incubation for 48 h at 37°C, these four plates had an average of 57 colonies. The air blown in each bathroom was also directed at one sterile nitrocellulose filter on purified agarose plates lacking germinants such that spores that adhered to the filters would not germinate. The filters were heated at 70°C for 45 min to kill cells, but not spores, of mesophilic species and then transferred to LB plates that were incubated at 37°C for 48 h. The 11 colonies that grew on the two heated filters were then picked on LB and LB plus Km (10 μg/ml) plates, as well as 2×SG sporulation medium plates. After incubation for 24 to 48 h at 37°C, 4 colonies were Kmr, and microscopic examination showed that all 11 colonies had formed spores and that the spore and cell morphologies of 10 of the 11, including the 4 Kmr colonies, were indistinguishable from those of B. subtilis. Identification of 7 of these colonies (3 Kmr colonies and 4 colonies sensitive to kanamycin) showed that all were B. subtilis bacteria (data not shown; see below). Kmr sporeformers were ∼3.5% of total colonies deposited from environmental air as, for the two plates examined, there were 4 Kmr spore formers from 114 (2 plates × 57 colonies each) total colonies.
Diverse bacteria are deposited on plates by hand dryers and directly from bathroom air.
A variety of floras were recovered from blood agar plates exposed to hand dryer air, with and without HEPA filters, while only 2 colonies were recovered from MacConkey plates that select for enteric Gram-negative organisms (Table 4) (note that only one of these colonies could be identified). A sample of 70 colonies from hand dryer air in different bathrooms with or without HEPA filters, selected based on differing colony morphologies, was subjected to identification by the MALDI-TOF Biotyper as described in Materials and Methods. Eight colonies were not identified and might be environmental organisms not present in the database used for bacterial identification. The 62 identified bacteria represented 21 species, with a wide variety of environmental bacteria, including Staphylococcus aureus (Table 4). While several other potential opportunistic human pathogens were recovered from multiple bathrooms, we did not identify all bacteria from all bathrooms, so these data almost certainly underrepresent bacterial diversity across bathrooms (see Discussion).
TABLE 4.
Bacteria recovered from agar plates exposed to hand dryers with and without HEPA filters or to bathroom aira
| Bacteriab | Present after dryer air exposure |
Present after bathroom air exposure | |
|---|---|---|---|
| Without HEPA filter (n = 53)c | With HEPA filter (n = 17) | ||
| Acinetobacter baumannii† | √ | ||
| Acinetobacter radioresistens | √ | ||
| Bacillus cereus† | √ | √ | |
| Bacillus infantis | √ | ||
| Bacillus licheniformis† | √ | ||
| Bacillus marisflavi | √ | ||
| Bacillus megaterium† | √ | √ | √ |
| Bacillus pumilus† | √ | √ | |
| Bacillus simplex† | √ | √ | |
| Bacillus subtilis† | √ | √ | |
| Erwinia sp. | √ | √ | |
| Exiguobacterium aurantiacum | √ | ||
| Kocuria rhizophila† | √ | ||
| Micrococcus luteus† | √ | √ | |
| Pantoea septica‡ | √ | ||
| Paracoccus yeei | √ | ||
| Pseudomonas luteola | √ | ||
| Roseomonas mucosa | √ | ||
| Staphylococcus aureus† | √ | √ | |
| Staphylococcus capitis† | √ | √ | |
| Staphylococcus epidermidis† | √ | ||
| Staphylococcus hominis† | √ | √ | √ |
| Staphylococcus pasteuri | √ | ||
| Staphylococcus simulans | √ | ||
Colonies were recovered on blood agar or MacConkey plates exposed to hand dryer air or bathroom air moved by small fans, and organisms were identified as described in Materials and Methods.
Organisms shown in boldface are reported in the literature to cause invasive disease in susceptible hosts. †, recovered from multiple bathrooms; ‡, recovered from MacConkey agar (the only other colony recovered from these plates could not be identified).
n, number of colonies tested.
A sample of 34 colonies deposited on plates by small fans directly from bathroom air, including 7 colonies deposited as spores as described above, was also subjected to identification. The 7 colonies deposited as spores were all B. subtilis bacteria, and 25 of the remaining 27 colonies were identified, including potential human pathogens such as Acinetobacter baumannii (Table 4).
DISCUSSION
The data reported in this communication demonstrate that sporeformers, including a laboratory strain of B. subtilis, were found on plates exposed to hand dryer air or air moved by small fans in bathrooms at multiple locations in basic research areas at the UConn School of Medicine, including areas far from where these spores were produced. Indeed, Kmr sporeformers were found in bathrooms on floors above the academic building at least 440 feet from the laboratory on the 2nd floor of building 1 where the Kmr B. subtilis spores are produced, i.e., 370 lateral feet and ∼70 feet above the spore laboratory given a rise of ∼14 feet/floor (Fig. 1). Perhaps the air-handling system in the basic research areas is dispersing these spores/sporeformers throughout the building. The bathrooms on floors above the academic building had increased numbers of bacterial colonies recovered, and whether this is related to air circulation between research buildings, bathroom utilization, or another explanation remains unclear. One reason hand dryers may disperse so many bacteria is the large amount of air that passes through hand dryers, ∼19,000 linear feet/min at the nozzle (30). The convection generated by high airflow below the hand dryer nozzles could also draw in room air.
Several results from this study indicate that bacteria dispersed by hand dyers are from general bathroom air passing through the hand dryer. These results include the following observations: (i) very low numbers of bacteria were found on hand dryers' internal nozzle surface; (ii) there was an ∼4-fold reduction in bacteria deposited by air from hand dryers retrofitted with HEPA filters; and (iii) when corrected for airflow and exposure time, there were similar levels of bacterial deposition from bathroom air moved by small fans and hand dryers. Heat treatment of samples from hand dryer air or directly from bathroom air and identification of collected heat-resistant forms further indicated that many of the sporeformers were dispersed by hand dryers as spores, in particular, some spores produced at high levels in a basic research laboratory.
A number of the bacteria that were deposited by hand dryers with and without HEPA filters and from bathroom air directly are skin flora and/or environmental organisms that are reported in the literature to potentially cause invasive human disease, including bloodstream, ocular, and peritoneal infections (33–37). In most of these cases, patients acquiring these diseases had a predisposition to infection, such as a permanent catheter, a defect in the immune system, underlying chronic disease (e.g., cancer), or repeated blood-borne exposure to bacteria through intravenous drug use (36, 38). It is certainly clear that hand washing can reduce the risk of infections (39). However, the deposition of potentially pathogenic bacteria on the hands after hand washing to remove transient floras reduces the effectiveness of hand washing and is concerning for these individuals as well as their contacts as bacterial colonization of the hands could potentially lead to transmission of infectious organisms (e.g., S. aureus) to other individuals and surfaces. Importantly, we did not identify all bacteria in the environment and instead subjectively chose unique colony morphologies for identification. Thus, the identified organisms are almost certainly a subset of the population of deposited bacteria, which is likely to vary over time. While HEPA filters reduced the numbers of bacteria deposited on the agar plates, potential pathogens were still recovered (Table 4). Thus, HEPA filters in hand dryers most likely reduce the number of potentially pathogenic bacteria with the potential to colonize hands but do not eliminate the risk entirely.
Another notable finding in this work is that we have tracked levels of a single bacterial strain used in only one research laboratory into bathrooms in the whole basic research facility, including bathrooms far from the laboratory where this strain is used. Presumably, this organism is also in air throughout the basic science research areas, but we have not tested this. This organism was almost certainly dispersed throughout bathrooms in the research areas as spores, which would easily survive desiccation in room air, as well the elevated temperatures in hand dryer air; however, growing or stationary-phase bacteria would not be nearly so hardy as spores. However, the facile dispersion of one bacterial strain throughout a research facility should probably be a concern to risk assessors and risk managers when dispersion of potentially pathogenic bacteria is considered (40–43). In addition, the new work also suggests that hand dryers in bathrooms can contribute to bacterial deposition on users as well as to bacterial dispersion, including that of bacterial spores, as has been suggested by other work (26, 27, 40, 42, 44). Consequently, this work also highlights another factor that should be considered in discussions on use of hand dryers in bathrooms instead of other methods of hand drying such as paper towels. Indeed, paper towel dispensers have recently been added to all 36 bathrooms in basic science research areas in the UConn School of Medicine surveyed in the current study.
Remaining questions include the following. (i) What are the levels of spores of anaerobes in the environmental air in the basic research areas of the UConn School of Medicine, as well as in other academic medical centers, and what percentage is captured on sampling plates below hand dryers with or without HEPA filters? Notably, levels of bacteria reported in indoor air in buildings are generally higher than the values we captured during hand dryer exposure (45–48). (ii) What are levels of growing bacteria and spores dispersed by hand dryers in clinical environments? The data in this report indicate that spores can be dispersed over large distances in buildings, possibly by air-handling systems, and deposited on surfaces by hand dryers. Previous work has shown that lidless toilets can disperse Clostridium difficile spores into bathroom air (49), and spores of this organism can be dispersed in air by patients with C. difficile infection in an inpatient setting (43). Since the current work shows that spores in bathroom air can be deposited on surfaces from the air by hand dryers, this suggests another means of C. difficile transmission and one that may not be interrupted by either hand washing or traditional surface decontamination methods. The role of this potential mode of C. difficile transmission is worthy of future study.
MATERIALS AND METHODS
Measurement of bacterial dispersion on plates in bathrooms.
We surveyed 36 hot-air Xlerator hand dryers (model XL-SB; Excel Dryer, Inc., East Longmeadow, MA, USA) without HEPA filters (but see below) in 18 men's and 18 women's bathrooms in or adjacent to two basic science research areas in the UConn School of Medicine (building 2, with 14 bathrooms on floors 1 to 7 and building 1 with 14 bathrooms on floors 1 to 7) and in areas above the academic building (8 bathrooms; floors 4 to 7). These bathrooms are interconnected through hallways (Fig. 1) but are not within clinical areas. Access to these bathrooms was unrestricted during sampling of hand dryer air or bathroom air itself (see below). Unless otherwise noted, hand dryer airflow was directed onto rich 2×SG medium agar plates (50) for 30 s with covers off and held in gloved hands ∼12 in. from the air outlet of the dryers. However, in a few experiments, hand dryer air exposure was for 1 min, or the plates exposed were 5% tryptic soy broth-sheep's blood agar plates (Hardy Diagnostics, Santa Maria, CA) or were MacConkey's agar (Remel, Lenexa, Kansas) to select for enteric Gram-negative organisms. Plates were then covered and incubated for ∼40 h at 37°C, and colonies were counted. Two 2×SG plates were also incubated open for 2 min in each bathroom, routinely with the hand dryers off. Duplicate 2×SG plates were also left open for 2 min in the laboratory where PS533 spores were prepared but not adjacent to areas where spores were being isolated and purified, and again ≤1 colony/plate was recovered (data not shown).
HEPA filters were retrofitted in hand dryers in five bathrooms according to the manufacturers' specifications. These hand dryers were tested 2 days before and 9 days after this retrofit as described above, but exposure of plates was for 1 min in order to increase the numbers of colonies obtained from the hand dryers that contained HEPA filters.
To test for bacteria in bathroom air, we determined the numbers of bacteria deposited on plates by small fans (Haptime portable mini fan, model YGH-539; Haptimetech, China). The fans were sanitized by swabbing all exposed surfaces with 50% ethanol and were placed ∼3.5 in. above open 2×SG plates on bathroom counters (see Fig. S1 in the supplemental material); airflow at the edge of these fans (diameter, ∼2.5 in) was measured at 400 linear ft/min with a TSI VelociCalc thermo-anemometer (TSI, Inc., Shoreview, MN) but was slightly lower at the center. After exposure to air moved by these fans, for 20 min unless noted otherwise, plates were closed, incubated for ∼40 h at 37°C, and colonies were counted.
To assess the presence of bacteria on the inner surfaces of hand dryer nozzles, cotton swabs were moistened in sterile phosphate-buffered saline ([PBS] 10 mM KPO4 buffer, pH 7.8, 150 mM NaCl) and used to swab the entire inner surface of the hand dryer nozzles in four bathrooms for ∼20 s. The swabs were placed in 300 μl of PBS for 15 min and vortexed for ∼30 s, and then 200 μl was spread on 2×SG medium plates, the plates were incubated for ∼36 h, and the colonies were counted.
Identification of B. subtilis PS533 among bacteria dispersed by hand dryers on plates.
To look for B. subtilis PS533 in bacteria dispersed by hand dryer air, colonies from overnight incubations of plates exposed to hand dryer air were picked onto LB plates (51) (per liter, 5 g of yeast extract, 10 g of tryptone, 15 g of agar, and 150 mmol of NaCl) with Km (10 mg/liter) and incubated overnight at 37°C. The morphology of cells in Kmr colonies was then determined by phase-contrast microscopy. Cells with a bacillus-like morphology were further tested for spore formation by picking cells onto 2×SG plates, incubating the plates for 48 h at 37°C to allow sporulation, and then examining colonies for spores by phase-contrast microscopy. Some sporulating Kmr colonies were also tested for plasmid pUB110 by plasmid isolation and purification from cells grown overnight in LB at 37°C using a QIAprep Spin Miniprep kit for plasmid DNA purification (Qiagen, Valencia, CA). Purified plasmid and plasmid digested with the restriction enzymes BamH1 and EcoR1, as well as purified plasmid pUB110 from growing cells of B. subtilis strain PS533, were subjected to 1% agarose gel electrophoresis with DNA markers. Gels were stained with ethidium bromide and photographed.
Identification of spores dispersed by hand dryer air.
To determine if spore-forming bacteria deposited on plates by hand dryers were deposited as spores or as growing or stationary-phase cells, we took advantage of the fact that spores are much more resistant to wet heat than are germinated spores or growing or stationary-phase cells of the same organism (52–54). Twenty milliliters of sterile PBS was placed in each of three 1-liter sterile lyophilization flasks, and the open flasks were held 10 to 12 in. below the dryer and given 10 min of hand dryer air exposure. As a control, another three flasks with liquid were left open to room air on the counter during the course of the experiment. The total volume per flask of PBS left after hand dryer air exposure was ∼14 ml; 500 μl was removed, heated at 65°C for 30 min to kill germinated spores or growing or stationary-phase cells (53, 54), and cooled to 23°C. Two hundred microliters of each heat-treated PBS sample was then plated in duplicate on blood agar plates that were incubated for ∼20 h at 37°C, and colonies were counted.
To directly identify spores present in bathroom air itself, the small fans described above were used to blow air in two bathrooms for 1 h at 2 LB medium plates in each bathroom to determine total bacteria deposited. The bathroom air in the same two bathrooms was also blown by small fans for 1 h at sterile nitrocellulose filters on purified agarose plates lacking all germinants, such that spores that adhered to the filters would not germinate. The filters were then heated at 70°C for 45 min to kill cells of mesophilic species, but not spores, and the filters were transferred to LB medium plates that were incubated at 37°C for 48 h. Heat treatment of the damp filters was more severe than that of the liquid samples because dry-growing cells are more resistant to heat than cells in liquid (52, 53). The colonies that grew on the heated filters were then picked on LB and LB plus Km (10 μg/ml) plates, as well as 2×SG sporulation medium plates.
Identification of bacteria from bathroom air.
Colonies were prepared for definitive identification of various bacteria as follows: (i) air from hand dryers with and without HEPA filters was directed for 1 min at blood agar plates or MacConkey's agar plates; (ii) colonies from heat-resistant organisms deposited in liquid by hand dryer air were picked to blood agar plates; (iii) bathroom air moved by small fans was directed for 1 h at blood agar plates; or (iv) colonies from spores deposited on filters on purified agarose plates by bathroom air moved by small fans were picked onto blood agar plates. Plates were incubated at 37°C for 24 to 40 h, selected individual colonies of various representative morphologies were picked to fresh blood agar plates, and plates were incubated at 37°C overnight. Bacteria in the resultant colonies were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry using a MALDI Biotyper instrument (Bruker GmbH, Bremen, Germany) in the Hartford Hospital clinical microbiology laboratory (55).
Statistical analysis.
To compare the bacterial colony levels between buildings, floors, distance from hand dryer, and the use of the small fan, a generalized linear model with Poisson distribution, a log link function, and a scaled deviance for overdispersion was used. The comparison of pre-HEPA to post-HEPA filter bacterial levels was performed using generalized estimating equations with Poisson distribution, log link, and an unstructured correlation matrix with robust estimator. Descriptive statistics include means and standard deviations. All analyses were conducted in SPSS, version 24, and statistical significance was set at 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the many individuals who prepared plates, exposed plates to hand dryers, facilitated retrofitting of hand dryers with HEPA filters, and measured airflow generated by the small fans.
L.D.C.H.-E. was supported by a scholarship from the Consejo Nacional de Ciencia y Tecnologia in Mexico.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00044-18.
REFERENCES
- 1.Curtis V, Cairncross S. 2003. Effect of washing hands with soap on diarrhoea risk in the community: a systematic review. Lancet Infect Dis 3:275–281. doi: 10.1016/S1473-3099(03)00606-6. [DOI] [PubMed] [Google Scholar]
- 2.Larson EL, Gomez-Duarte C, Lee LV, Della-Latta P, Kain DJ, Keswick BH. 2003. Microbial flora of the hands of homemakers. Am J Infect Control 31:72–79. doi: 10.1067/mic.2003.33. [DOI] [PubMed] [Google Scholar]
- 3.Noble WC, Somerville DA. 1974. Microbiology of human skin. WB Saunders, Philadelphia, PA. [Google Scholar]
- 4.Pittet D, Dharan S, Touveneau S, Sauvan V, Perneger TV. 1999. Bacterial contamination of the hands of hospital staff during routine patient care. Arch Intern Med 159:821–826. doi: 10.1001/archinte.159.8.821. [DOI] [PubMed] [Google Scholar]
- 5.Lowbury EJL, Lilly HA, Bull JP. 1964. Disinfection of hands: removal of transient organisms. Br Med J 2:230–233. doi: 10.1136/bmj.2.5403.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crabtree TD, Pelletier SJ, Pruett TL. 2001. Surgical antisepsis, p 919–934. In Block SS. (ed), Disinfection, sterilization, and preservation, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 7.Smith S. 2009. A review of hand washing techniques in primary care and community settings. J Clin Nurs 18:786–790. doi: 10.1111/j.1365-2702.2008.02546.x. [DOI] [PubMed] [Google Scholar]
- 8.Edson RS, Bundrick JB, Litin SC. 2011. Clinical pearls in infectious diseases. Mayo Clin Proc 86:245–248. doi: 10.4065/mcp.2010.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sickbert-Bennett E, Weber D, Gergen-Teague M, Sobsey M, Samsa G, Rutala W. 2005. Comparative efficacy of hand hygiene agents in the reduction of bacteria and viruses. Am J Infect Control 33:67–77. doi: 10.1016/j.ajic.2004.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todd E, Greig J, Michaels B, Bartleson C, Smith D, Holah J. 2010. Outbreaks where food workers have been implicated in the spread of foodborne disease, part 11: use of antiseptics and sanitizers in community settings and issues of hand hygiene compliance in health care and food industries. J Food Prot 73:2306–2320. doi: 10.4315/0362-028X-73.12.2306. [DOI] [PubMed] [Google Scholar]
- 11.Erasmus V, Daha TJ, Brug H, Richardus JH, Behrendt MD, Vos MC, van Beeck EF. 2010. Systematic review of studies on compliance with hand hygiene guidelines in hospital care. Infect Control Hosp Epidemiol 31:283–294. doi: 10.1086/650451. [DOI] [PubMed] [Google Scholar]
- 12.Haas J, Larson E. 2007. Measurement of compliance with hand hygiene. J Hosp Infect 66:6–14. doi: 10.1016/j.jhin.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 13.O'Boyle C, Henly S, Larson E. 2001. Understanding adherence to hand hygiene recommendations: the theory of planned behavior. Am J Infect Control 29:352–360. doi: 10.1067/mic.2001.18405. [DOI] [PubMed] [Google Scholar]
- 14.Pittet D, Hugonnet S, Harbarth S, Mourouga P, Sauvan V, Touveneau S, Perneger TV. 2000. Effectiveness of a hospital-wide programme to improve compliance with hand hygiene. Lancet 356:1307–1312. doi: 10.1016/S0140-6736(00)02814-2. [DOI] [PubMed] [Google Scholar]
- 15.Patrick DR, Findon G, Miller TE. 1997. Residual moisture determines the level of touch-contact-associated bacterial transfer following hand washing. Epidemiol Infect 119:319–325. doi: 10.1017/S0950268897008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merry AF, Miller TE, Findo G, Webster CS, Neff SP.. 2001. Touch contamination levels during anaesthetic procedures and their relationship to hand hygiene procedures: a clinical audit. Br J Anaesth 87:291–294. doi: 10.1093/bja/87.2.291. [DOI] [PubMed] [Google Scholar]
- 17.Ansari SA, Springthorpe VS, Sattar SA, Tostowaryk W, Wells GA. 1991. Comparison of cloth, paper, and warm air drying in eliminating viruses and bacteria from washed hands. Am J Infect Control 19:243–249. doi: 10.1016/S0196-6553(05)80256-1. [DOI] [PubMed] [Google Scholar]
- 18.Blackmore MA. 1989. A comparison of hand drying methods. Cater Health 1:189–198. [Google Scholar]
- 19.Harrison WA, Griffith CJ, Ayers T, Michaels B. 2003. Bacterial transfer and cross-contamination potential associated with paper-towel dispensing. Am J Infect Control 31:387–391. doi: 10.1067/mic.2003.81. [DOI] [PubMed] [Google Scholar]
- 20.Huang C, Ma W, Stack S. 2012. The hygienic efficacy of different hand-drying methods: a review of the evidence. Mayo Clin Proc 87:791–798. doi: 10.1016/j.mayocp.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews JA, Newsom SW. 1987. Hot air electric hand driers compared with paper towels for potential spread of airborne bacteria. J Hosp Infect 9:85–88. doi: 10.1016/0195-6701(87)90101-0. [DOI] [PubMed] [Google Scholar]
- 22.Meers PD, Leong KY. 1989. Hot-air hand driers. J Hosp Infect 14:169–171. doi: 10.1016/0195-6701(89)90121-7. [DOI] [PubMed] [Google Scholar]
- 23.Taylor JH, Brown KL, Toivenen J, Holah JT. 2000. A microbiological evaluation of warm air hand driers with respect to hand hygiene and the washroom environment. J Appl Microbiol 89:910–919. doi: 10.1046/j.1365-2672.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto Y, Ugai K, Takahashi Y. 2005. Efficiency of hand drying for removing bacteria from washed hands: comparison of paper towel drying with warm air drying. Infect Control Hosp Epidemiol 26:316–332. doi: 10.1086/502546. [DOI] [PubMed] [Google Scholar]
- 25.Gustafson DR, Vetter EA, Larson DR, Ilstrup DM, Maker MD, Thompson RL, Cockerill FR III. 2000. Effects of 4 hand-drying methods for removing bacteria from washed hands: a randomized trial. Mayo Clin Proc 75:705–708. doi: 10.4065/75.7.705. [DOI] [PubMed] [Google Scholar]
- 26.Best EL, Redway K. 2015. Comparison of different hand-drying methods: the potential for airborne microbe dispersal and contamination. J Hosp Infect 89:215–217. doi: 10.1016/j.jhin.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Best EL, Parnell P, Wilcox MH. 2014. Microbiological comparison of hand-drying methods: the potential for contamination of the environment, user, and bystander. J Hosp Infect 88:199–206. doi: 10.1016/j.jhin.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Redway K, Dawdar S. 2008. A comparative study of three different hand drying methods: paper towel, warm air dryer, jet air dryer. European Tissue Symposium, Brussels, Belgium. [Google Scholar]
- 29.Alharbi SA, Salmen SH, Chinnathambi A, Alharbi NS, Zayed ME, Al-Johny BO, Wainwright M. 2016. Assessment of the bacterial contamination of hand air dryer in washrooms. Saudi J Biol Sci 23:268–271. doi: 10.1016/j.sjbs.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Excel Dryer, Inc. 2017. Xlerator and Xleratoreco hand dryer. Excel Dryer, Inc, East Longmeadow, MA: https://www.exceldryer.com/wp-content/uploads/pdf/Excel-Dryer-Accessories.pdf. [Google Scholar]
- 31.Cutting SM. 2011. Bacillus probiotics. Food Microbiol 28:214–220. doi: 10.1016/j.fm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J Bacteriol 178:3486–3495. doi: 10.1128/jb.178.12.3486-3495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhende M, Karpe A, Arunachalam S, Therese KL, Biswas J. 2017. Endogenous endophthalmitis due to Roseomonas mucosa presenting as a subretinal abscess. J Ophthalmic Inflamm Infect 7:5. doi: 10.1186/s12348-017-0123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Courjaret JC, Drancourt M, Hoffart L. 2014. Paracoccus yeei keratitis in a contact lens wearer. Eye Contact Lens 40:e21–e22. doi: 10.1097/ICL.0b013e31829e8fc7. [DOI] [PubMed] [Google Scholar]
- 35.Moissenet D, Becker K, Mérens A, Ferroni A, Dubern B, Vu-Thien H. 2012. Persistent bloodstream infection with Kocuria rhizophila related to a damaged central catheter. J Clin Microbiol 50:1495–1498. doi: 10.1128/JCM.06038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer G, Campbell W, Jenks J, Beesley C, Katsivas T, Hoffmaster A, Mehta SR, Reed S. 2016. Persistent Bacillus cereus bacteremia in 3 persons who inject drugs, San Diego, California, USA. Emerg Infect Dis 22:1621–1623. doi: 10.3201/eid2209.150647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goel N, Munshi LB, Thyagarajan B. 2016. Intravenous drug abuse by patients inside the hospital: a cause for sustained bacteremia. Case Rep Infect Dis 2016:1738742. doi: 10.1155/2016/1738742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fox C, Wavra T, Drake DA, Mulligan D, Bennett YP, Nelson C, Kirkwood P, Jones L, Bader MK. 2015. Use of a patient hand hygiene protocol to reduce hospital-acquired infections and improve nurses' hand washing. Am J Crit Care 24:216–224. doi: 10.4037/ajcc2015898. [DOI] [PubMed] [Google Scholar]
- 40.Reshetin VP, Regens JL. 2003. Simulation modeling of anthrax spore dispersion in a bioterrorism incident. Risk Anal 23:1135–1145. doi: 10.1111/j.0272-4332.2003.00387.x. [DOI] [PubMed] [Google Scholar]
- 41.Bishop AH, Stapleton HL. 2016. Aerosol and surface deposition characteristics of two surrogates of Bacillus anthracis spores. Appl Environ Microbiol 82:6682–6690. doi: 10.1128/AEM.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts K, Smith CF, Snelling AM, Kerr KG, Banfield KR, Sleigh PA, Beggs CB. 2008. Aerial dissemination of Clostridium difficile spores. BMC Infect Dis 8:7. doi: 10.1186/1471-2334-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best EL, Fawley WN, Parnell P, Wilcox MH. 2010. The potential for airborne dispersal of Clostridium difficile from symptomatic patients. Clin Infect Dis 50:1450–1457. doi: 10.1086/652648. [DOI] [PubMed] [Google Scholar]
- 44.Wilcox MH, Best EL, Parnell P. 2017. Pilot study to determine whether microbial contamination levels in hospital washrooms are associated with hand-drying method. J Hosp Infect 97:201–203. doi: 10.1016/j.jhin.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Ijaz MK, Zargar B, Wright KE, Rubino JR, Sattar SA. 2016. Generic aspects of the airborne spread of human pathogens indoors and emerging air decontaminant technologies. Am J Infect Control 44:S109–S120. doi: 10.1016/j.ajic.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prussin AJ II, Garcia EB, Marr LC. 2015. Total virus and bacteria concentrations in indoor and outdoor air. Environ Sci Technol Lett 2:84–88. doi: 10.1021/acs.estlett.5b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephens B. 2016. What have we learned about the microbiomes of indoor environments. mSystems 1:e00083-16. doi: 10.1128/mSystems.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai FC, Macher JM. 2005. Concentrations of airborne culturable bacteria in 100 large US office buildings from the BASE study. Indoor Air 15(Suppl 9):71–81. doi: 10.1111/j.1600-0668.2005.00346.x. [DOI] [PubMed] [Google Scholar]
- 49.Best EL, Sandoe JAT, Wilcox MH. 2012. Potential for aerosolization of Clostridium difficile after flushing toilets: the risk of toilet lids in reducing environmental contamination risk. J Hosp Infect 80:1–5. doi: 10.1016/j.jhin.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450. In Harwood CR, Cutting SM (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 51.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 52.Setlow P, Johnson EA. 2012. Spores and their significance, p 45–79. In Doyle MP, Buchanan R (ed), Food microbiology: fundamentals and frontiers, 4th ed ASM Press, Washington, DC. [Google Scholar]
- 53.Setlow P. 2016. Spore resistance properties, p 201–215. In Eichenberger P, Driks A (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 54.Paidhungat M, Setlow B, Driks A, Setlow P. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J Bacteriol 182:5505–5512. doi: 10.1128/JB.182.19.5505-5512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croxatto V, Prod'hom G, Greub G. 2012. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.