Abstract
Sheep flystrike is caused by parasitic flies laying eggs on soiled wool or open wounds, after which the hatched maggots feed on the sheep flesh and often cause large lesions. It is a significant economic problem for the livestock industry as infestations are difficult to control due to ongoing cycles of larval development into flies followed by further egg laying. We therefore screened venom fractions from the Australian theraphosid spider Coremiocnemis tropix to identify toxins active against the sheep blowfly Lucilia cuprina, which is the primary cause of flystrike in Australia. This screen led to isolation of two insecticidal peptides, Ct1a and Ct1b, that are lethal to blowflies within 24 h of injection. The primary structure of these peptides was determined using a combination of Edman degradation and sequencing of a C. tropix venom-gland transcriptome. Ct1a and Ct1b contain 39 and 38 amino acid residues, respectively, including six cysteine residues that form three disulfide bonds. Recombinant production in bacteria (Escherichia coli) resulted in low yields of Ct1a whereas solid-phase peptide synthesis using native chemical ligation produced sufficient quantities of Ct1a for functional analyses. Synthetic Ct1a had no effect on voltage-gated sodium channels from the American cockroach Periplanata americana or the German cockroach Blattella germanica, but it was lethal to sheep blowflies with an LD50 of 1687 pmol/g.
Keywords: Spider venom, Insecticidal peptide, Bioinsecticide, Coremiocnemis tropix, Lucilia cuprina, Flystrike
1. Introduction
Management of arthropod pests such as flies, lice and ticks constitutes a significant cost to livestock farmers. In Australia, the sheep blowfly Lucilia cuprina is the major cause of flystrike, which results in annual economic losses of more than USD $200 million (Sackett et al., 2006). Mulesing, in which strips of loose, wool-bearing skin are removed from around the breech, in combination with docking tails at the correct length, has been effective in controlling breech strike (Sandeman et al., 2014; Tyrell et al., 2014). However, mulesing is a highly controversial practice (Sneddon and Rollin, 2009) and attempts are being made to phase out the practice in both Australia and New Zealand (Sandeman et al., 2014; Tyrell et al., 2014). This will inevitably increase reliance on chemical insecticides and insect growth regulators for blowfly control, even though the effectiveness of many of these compounds is being diminished by the development of resistance (Sandeman et al., 2014). Hence, there is significant interest in the development of novel approaches to protect sheep against flystrike.
Spiders have proven to be an excellent source of insecticidal compounds (King and Hardy, 2013). Spiders are the most speciose venomous animal and, along with predatory beetles, the most successful insect predators on the planet (Windley et al., 2012). Their venoms are extremely complex chemical arsenals comprised of salts, small organic molecules, peptides and proteins (Escoubas and Rash, 2004; Schroeder et al., 2008; Kuhn-Nentwig et al., 2011; King and Hardy, 2013). However, the major components of most spider venoms are small, disulfide-rich peptides (King and Hardy, 2013), with most venoms containing many hundreds to more than 1000 peptides (Escoubas et al., 2006). As a whole, spider venoms have been predicted to contain as many as 10 million unique peptides (King, 2011), most of which are expected to be insecticidal, but less than <0.1% of this chemical diversity has been explored to date. Nevertheless, numerous insecticidal peptides have been isolated from spider venom with a broad range of phyletic selectivity (King and Hardy, 2013; Bende et al., 2014; Herzig et al., 2016). Recently, an insecticidal spider-venom peptide was approved for control of crop pests by the United States Environmental Protection Agency (Herzig et al., 2014).
Here we show that venom from the Australian theraphosid spider Coremiocnemis tropix (Araneae, Theraphosidae), which was first described in 2005 (Raven, 2005) and subsequently reported to be insecticidal to crickets (order Orthoptera) and mealworms (order Coleoptera) (Gentz et al., 2009; Herzig and Hodgson, 2009), contains compounds that are active against sheep blowflies (order Diptera). We isolated two disulfide-rich peptides, Ct1a and Ct1b, that are lethal when injected into adult L. cuprina. Sequencing of these peptides revealed that they are paralogs comprised of 38–39 residues. Each peptide contains six cysteine residues that form three disulfide bonds, but the cysteine pattern does not conform to the canonical inhibitor cystine knot framework (Pallaghy et al., 1994; King et al., 2002). We show that Ct1a does not target insect voltage-gated sodium channels, which are the primary target of many spider-venom peptides as well as commonly employed dipteran-active insecticides such as pyrethroids and oxadiazines.
2. Materials and methods
2.1. Spider collection, identification and venom preparation
Female C. tropix spiders were collected from Cairns, Queensland, Australia and identified by Dr Robert Raven (Queensland Museum, Brisbane, Australia). Venom was extracted as previously described by applying mild electrical stimulation to the chelicerae (Herzig and Hodgson, 2009). The expressed venom was then lyophilized and stored at −20 °C until required.
2.2. Chemicals
All chemicals were purchased from Sigma-Aldrich Australia (Castle Hill, NSW, Australia), Sigma-Aldrich USA (St Louis, MO, USA) or Merck Chemicals (Kilsyth, Victoria, Australia) with the exception of isopropyl β-D-1-thiogalactopyranoside (IPTG) and streptomycin (Life Technologies, Mulgrave, Victoria, Australia), tetrodotoxin (Alomone Labs, Israel), and high-performance liquid chromatography (HPLC)-grade acetonitrile (RCI Labscan, Bangkok, Thailand). Recombinant His6-TEV protease (EC 3.4.22.44) was produced in-house.
2.3. Isolation of native Ct1a and Ct1b
Pooled, lyophilized C. tropix venom (5 mg) was fractionated using a Phenomenex Jupiter C18 reverse phase (RP) HPLC semipreparative column (250 × 10 mm, 5 μm particle size; Phenomenex, Sydney, NSW, Australia) connected to a Shimadzu 10A series HPLC system. A linear gradient of solvent B (90% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA)) in solvent A (0.1% TFA in water) was used such that the percentage of acetonitrile was increased from 5 to 20% over the first 10 min, then from 20 to 35% over the subsequent 50 min. Peptide elution was monitored at 214 nm and the flow rate was 2 mL/min.
A RP-HPLC fraction that was lethal to blowflies within 24 h of injection was further fractionated using cation-exchange chromatography (CEX) using a polysulfoethyl column (4.6 × 100 mm, 3 μm particle size, 300 Å pore size) with the following gradient of CEX solvent B (1 M KCl, 20 mM KH2PO4, 20% ACN, pH 2.7) in CEX solvent A (20 mM KH2PO4, 20% ACN, pH 2.7): 0% B for 3 min, then 0–50% B over 50 min. Peptide elution was monitored at 214 nm and the flow rate was 1 mL/min.
2.4. MALDI-TOF mass spectrometry
The masses and purity of native venom peptides were determined via matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) using a Model 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA, USA). Samples were spotted with α-cyano-4-hydroxy-cinnamic acid (CHCA) matrix (6 mg/mL in 50/50 ACN/H2O with 5% formic acid), and spectra were collected in positive reflector mode. Reported MALDI-TOF masses are for the monoisotopic [M+H]+ ions.
The mass of synthetic peptides and fragments were determined using electrospray ionization (ESI) MS using an API2000 spectrometer (Applied Biosystems) in positive ion mode (m/z 500–1800, declustering potential 10–20 V). Samples in ACN/H2O containing 0.1% TFA were directly injected into the ion source.
2.5. Edman sequencing of Ct1a and Ct1b
N-terminal Edman sequencing, including prior reduction and alkylation of peptides with tris(2-carboxyethyl)phosphine (TCEP)/iodoethanol, was performed by the Australian Proteome Analysis Facility (Sydney, NSW, Australia). Peptide samples (30 μL) were loaded onto precycled, Biobrene-treated discs and subjected to 40 cycles of automated N-terminal sequencing on an Applied Biosystems 494 Procise Protein Sequencing System.
2.6. Preparation and sequencing of venom-gland cDNA library
Three days after exhaustive milking, nine female C. tropix spiders were anesthetized, then the venom glands were removed and total RNA extracted using TRIzol® (Life Technologies). An Oligotex direct mRNA mini kit (Qiagen Australia) was used to perform mRNA enrichment from total RNA. RNA quality and concentration was determined using a Bioanalyzer 2100 pico chip (Agilent Technologies, Santa Clara, CA, USA).
Approximately 100 ng of mRNA was used to construct a cDNA library using standard Roche protocols for cDNA library preparation via emulsion PCR. Sequencing was performed at the Brisbane node of the Australian Genome Research Facility using a Roche GS-FLX sequencer. Low quality sequences were filtered out using the Fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit) with a minimum Phred score cut-off of 25; sequences with less than 75 bp were also discarded. De novo assembly was performed using MIRA v4.0.2 (Chevreux et al., 2004) using the following parameters: project = –RL5Ctropix –job = est, denovo, accurate –parameters = –GE:not = 4 \ 454_SETTINGS –AS:mrpc = 1 AL:mrs = 99, egp = 1 –readgroup = Data_454 –data = GFKSPG_CtropixRL5_trimmed4.fastq –technology = 454. The assembled data set was visualized using Tablet (Milne et al., 2009) or Geneious v5.4 (Kearse et al., 2012) software. Consensus sequences were submitted to Blast2GO (www.blast2go.com) to acquire BLAST information and functional annotations (Conesa et al., 2005). Signal and propeptide sequences were determined using the SpiderP algorithm (Wong et al., 2013). The entire C. tropix venom-gland transcriptomic data set has been submitted to the European Nucleotide Archive under accession number PRJEB15661.
2.7. Production of recombinant Ct1a
A synthetic gene encoding Ct1a, with codons optimized for expression in E. coli, was cloned into the pLIC-MBP expression vector by GeneArt (Invitrogen, Regensburg, Germany). This vector contains a MalE signal sequence for periplasmic export, a His6 tag for affinity purification, a maltose-binding protein (MBP) to enhance stability and solubility, and a tobacco etch virus (TEV) protease recognition sequence directly preceding the Ct1a gene. The plasmid was transformed into E. coli strain BL21 (λDE3) for recombinant toxin production. Cultures were grown at 37 °C with shaking in Luria-Bertani medium supplemented with 100 μg/mL ampicillin. Expression of the His6-MBP-Ct1a fusion protein was induced with 0.5 mM IPTG at an OD600 ~1, then the culture was grown at 20 °C overnight. Cells were harvested the following day by centrifugation at 5000 g for 15 min.
The cell pellet was resuspended in equilibration buffer (20 mM Tris, 250 mM NaCl, pH 8), then the cells were lysed by mechanical disruption at 27 kpsi using a TS Series Cell Disrupter (Constant Systems Ltd, Daventry, UK). The fusion protein was captured by passing the lysate over Ni-NTA Superflow resin (Qiagen, Chadstone, Australia), followed by washing with 5 mM imidazole to remove non-specifically bound proteins. The fusion protein was then eluted with 250 mM imidazole, concentrated to 10 mL using a 30 kDa cutoff spin filter, and the buffer exchanged to remove imidazole. Reduced and oxidized gluthathione were then added to 3.0 and 0.3 mM respectively, to facilitate TEV protease activity and folding of the protein. Approximately 200 μg of TEV protease was added for each litre of bacteria culture, then the cleavage reaction was allowed to proceed for >12 h at room temperature. The liberated recombinant peptide was purified via RP-HPLC using a Phenomenex C4 column (250 × 4.6 mm, 10 μm) at a flow rate of 5 mL/min with a gradient of 15–45% solvent B (0.043% TFA in 90% ACN) in solvent A (0.05% TFA in water) over 30 min.
2.8. Production of synthetic Ct1a
Ct1a was chemically synthesised via solid-phase peptide synthesis (SPPS) by native chemical ligation of two unprotected peptide segments: Ct1a(1–24)-α-thioester and Ct1a(25–39). Ct1a(1–24)-α-thioester (LFECSFSCDIKKNGKPCKGSGEKK-[COS]-Tyr) was assembled using manual Boc solid-phase peptide synthesis (SPPS) on HO-CH2-Pam-polystyrene resin (Johnson and Kent, 2006). The following side-chain protection groups were used: Asp(OcHex), Asn(Xan), Cys(4-MeBzl), Glu(OcHxl), Lys(2Cl-Z), Ser(Bzl), Tyr(Br-Z). After chain assembly, side-chains were deprotected and the peptide was simultaneously cleaved from the resin by treatment with anhydrous HF containing 10% (v/v) p-cresol for 1 h at 0 °C. HF was evaporated under reduced pressure. The crude product was precipitated, washed with chilled diethylether, dissolved in 50% (v/v) aqueous ACN containing 0.1% TFA (v/v), and then lyophilized.
Ct1a(25–39) (CSGGWRCKMNFCVKV) was synthesised using automated Fmoc SPPS using standard protocols. The following side-chain protecting groups were used: Arg(Pbf), Asn(Trt), Cys(Trt), Lys(Boc), Ser(tBu), and Trp(Boc). After chain assembly, peptides were cleaved from the resin using a cocktail of TFA/triisopropylsilane/water (95:2.5:2.5) for 1.5 h at 22 °C. TFA was removed under vacuum and the peptide was precipitated by adding chilled diethylether. After filtration, the crude peptide was dissolved in 50% (v/v) aqueous ACN containing 0.1% TFA (v/v), filtered, and then lyophilized.
Crude peptides were purified by preparative RP-HPLC on a Shimadzu Prominence system using a Zorbax C18 column (22.1 × 250 mm), and characterized by analytical RP-HPLC and electrospray ionization MS (ESI-MS). Fractions containing the desired peptide were pooled, lyophilized and stored at −20 °C. The masses of the purified synthetic peptides determined using ESI-MS agreed with the predicted values: Ct1a(1–24)-α-thioester (measured 2885.1 ± 0.3 Da; calculated 2885.3); Ct1a(25–39) (measured 1717.8 ± 0.3 Da; calculated 1718.1 Da).
Native chemical ligation of Ct1a(1–24)-α-thioester and Ct1a(25–39) was performed using a previously described protocol (Johnson and Kent, 2006). Briefly, 8.6 μmol (25 mg) of Ct1a(1–24)-α-thioester and 8.6 μmol (14.8 mg) of Ct1a(25–39) were combined and dissolved in 5.0 mL of ligation buffer consisting of 6 M guanidine hydrochloride, 200 mM Na2HPO4, 30 mM TCEP and 25 mM 4-mercaptophenylacetic acid (MPAA), pH 7.1. The solution was stirred under argon and the reaction was monitored by HPLC. Ligation was found to be quantitative after 6 h. The solution was then acidified by adding TFA, syringe filtered, and the full-length Ct1a peptide purified via RP-HPLC using an Agilent Zorbax 300SB C3 column (9.4 × 250 mm, 5 μm). Fractions containing the desired linear peptide were lyophilized and stored at −20 °C. The final yield of Ct1a was 48% (4.0 μmol/17.5 mg). The mass of synthetic Ct1a determined using ESI-MS agreed with the predicted value (measured 4334.3 ± 0.5 Da; calculated 4334.2 Da).
2.9. Folding of synthetic Ct1a
2.3 μMol (9.8 mg) of reduced Ct1a was dissolved in 8 mL of 6 M guanidine hydrochloride (pH ~5) to give a 1.2 mg/mL peptide solution. The peptide solution was then added to 160 mL of oxidation buffer comprised of 50 mM Na2HPO4, 8 mM reduced L-glutathione (GSH), and 1 mM oxidised L-glutathione (GSSG), pH 7.5. The solution was degassed by helium sparging for 30 min. Peptide oxidation was performed at 22 °C and monitored using RP-HPLC. The reaction reached equilibrium after 24 h, yielding a dominant product with an average mass of 4328.2 Da, indicative of the formation of three disulfide bonds. After addition of 1.5 mL of TFA, the mixture was purified by RP-HPLC using an Agilent Zorbax 300SB C3 column (9.4 × 250 mm, 5 μm). Fractions were lyophilized, then the most potent fraction identified via injection into houseflies (Musca domestica). MALDI-TOF MS was used to confirm that this fraction contained the expected molecular mass for oxidised Ct1a
2.10. Activity of Ct1a against sheep blowflies
Synthetic Ct1a was dissolved in insect saline (Eitan et al., 1990) and injected into the ventro-lateral thoracic region of adult sheep blowflies (L. cuprina; mass 24.8–30.1 mg) according to the method described previously (Bende et al., 2013). A maximum of 2 μL was injected per fly using a 1.0 mL Terumo Insulin syringe with a fixed 29-gauge needle fitted to an Arnold hand micro-applicator (Burkard Manufacturing, Rickmansworth, England). After injection, flies were individually housed in 2 mL tubes and paralytic effects and lethality were determined after 24 h. A total of three tests were carried out and for each test nine doses of Ct1a (n = 10 flies per dose) and the appropriate control (insect saline; n = 20 flies) were used. The dose that paralysed or killed 50% of flies (PD50 and LD50, respectively) was calculated as previously described (Herzig and Hodgon, 2008).
2.11. Effect of Ct1a on sodium channel currents in Periplanata americana neurons
Dorsal unpaired median (DUM) neurons were isolated from unsexed adult American cockroaches (Periplaneta americana) as described previously (Bende et al., 2013). Briefly, terminal abdominal ganglia were removed and placed in normal insect saline (NIS) containing (in mM): NaCl 180, KCl 3.1, N-hydroxyethylpiperazine-N-ethanesulfonic acid (HEPES) 10 and D-glucose 20. Ganglia were then incubated in 1 mg/mL collagenase (type IA) for 40 min at 29 °C. Following enzymatic treatment, ganglia were washed twice in NIS and resuspended in NIS supplemented with 4 mM MgCl2, 5 mM CaCl2, 5% foetal bovine serum and 1% penicillin/streptomycin (NIS+) and triturated through a fire-polished Pasteur pipette. The resultant cell suspension was then distributed onto 12-mm diameter glass coverslips pre-coated with 2 mg/mL concanavalin A (type IV). DUM neurons were maintained in NIS + at 29 °C and 100% humidity.
Ionic currents were recorded in voltage-clamp mode using the whole-cell patch-clamp technique employing version 10.2 of the pCLAMP data acquisition system (Molecular Devices, Sunnyvale, CA). Data were filtered at 10 kHz with a low-pass Bessel filter with leakage and capacitive currents subtracted using P–P/4 procedures. Digital sampling rates were set between 15 and 25 kHz depending on the length of the protocol. Single-use 0.8–1.5 MΩ electrodes were pulled from borosilicate glass and fire-polished prior to current recordings. Liquid junction potentials were calculated using JPCALC, and all data were compensated for these values. Cells were bathed in external solution through a continuous pressurized perfusion system at 1 mL/min, while toxin solutions were introduced via a wide bore gravity fed perfusion needle at ~80 μL/min (Automate Scientific, San Francisco, CA). Control data were not acquired until at least 10 min after whole-cell configuration was achieved to eliminate the influence of fast time-dependent shifts in steady-state inactivation resulting in rundown of sodium currents (INa) from voltage-gated sodium (NaV) channels. All experiments were performed at ambient room temperature (20–23 °C). To record INa, the external bath solution contained (in mM): NaCl 80, CsCl 5, CaCl2 1.8, tetraethylammonium chloride (TEA-Cl) 50, 4-aminopyridine (4-AP) 5, HEPES 10, NiCl2 0.1, and CdCl2 1, adjusted to pH 7.4 with 1 M NaOH. The pipette solution contained (in mM): NaCl 34, CsF 135, MgCl2 1, HEPES 10, ethylene glycol-bis (2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) 5, and ATP-Na2 3, adjusted to pH 7.4 with 1 M CsOH. To eliminate any influence of differences in osmotic pressure, all internal and external solutions were adjusted to 400 ± 5 mOsmol/L with sucrose. Experiments were rejected if leak currents exceeded 1 nA or currents showed signs of poor space clamping.
Peak current amplitude was analysed offline using AxoGraph X v1.5.3 (Molecular Devices). All curve-fitting was performed using PRISM 6 for Windows (GraphPad Software Inc., San Diego, CA). Nonlinear least-squares regression was used to fit I/V curves whilst linear least-squares regression was used to fit voltage-dependent block data. Comparisons of two sample means were made using a paired Student’s t-test. A test was considered to be significant when p < 0.05. All data represent the mean ± SEM of n independent experiments.
2.12. Activity of Ct1a on cloned Blattella germanica sodium channels
The DNA sequence of the B. germanica NaV channel (BgNaV1) (Dong, 1997) was confirmed by automated DNA sequencing and cRNA was synthesised using T7 polymerase (mMessage mMachine kit, Life Technologies) after linearizing DNA with NotI. BgNaV1 was expressed in Xenopus oocytes together with the TipE subunit (Feng et al., 1995) (1:5 molar ratio). After cRNA injection, oocytes were incubated at 17 °C (in 96 mM NaCl, 2 mM KCl, 5 mM HEPES, 1 mM MgCl2 and 1.8 mM CaCl2, 50 μg/mL gentamycin, pH 7.6 with NaOH) for 1–2 days, then currents were measured using two-electrode voltage-clamp recording techniques (OC-725C, Warner Instruments, Hamden, CT, USA) with a 150-μL recording chamber. Data were filtered at 4 kHz and digitized at 20 kHz using pCLAMP software (Molecular Devices). Microelectrode resistances were 0.5–1 MΩ when filled with 3 M KCl. The external recording solution contained (in mM): 96 NaCl, 2 KCl, 5 HEPES, 1 MgCl2 and 1.8 CaCl2, pH 7.6 with NaOH. All experiments were performed at room temperature (~22 °C). Leak and background conductances, identified by blocking the channel with tetrodotoxin (TTX), were subtracted for currents shown.
Voltage–activation relationships were obtained by measuring steady-state currents and calculating conductance. In brief, oocytes were depolarized in 5 mV increments from −90 mV to 5 mV for 50 ms from a holding potential of −90 mV, followed by a depolarizing pulse to −15 mV for 50 ms. Peak current from these steps was converted to conductance and normalized to generate the G-V relationship, while peak current from the −15 mV depolarization step was normalized to yield the steady-state inactivation (SSI) relationship. Protocols for other measurements are described in the legend to Fig. 5. After addition of Ct1a to the recording chamber, equilibration between toxin and the channel was monitored using weak depolarizations elicited at 5-s intervals. For all channels, voltage–activation relationships were recorded in the absence and presence of toxin. Off-line data analysis was performed using Clampfit 10 (Molecular Devices) and Origin 8.0 (Originlab, Northampton, MA, USA).
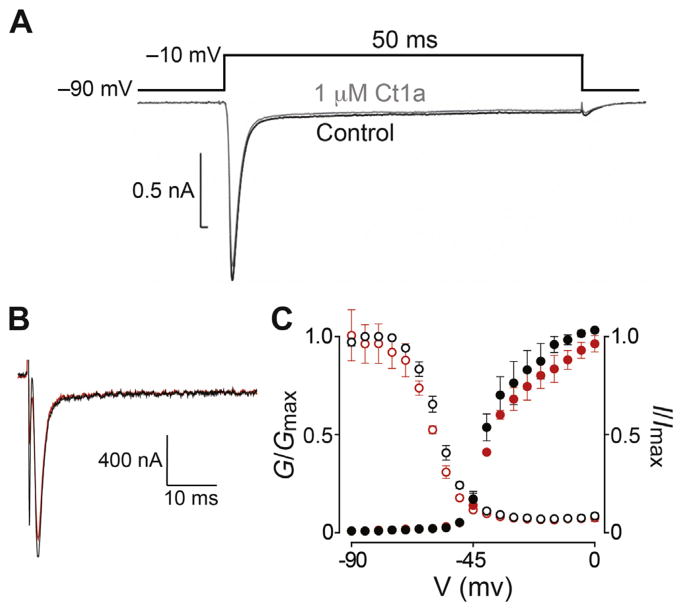
Fig. 5. Effect of Ct1a on insect NaV channels.
(A) Representative trace showing the lack of effect of Ct1a on NaV channel currents in P. americana DUM neurons recording using the whole-cell patch clamp technique. A standard test pulse to −10 mV from a holding potential of −90 mV (protocol shown above the traces) was used to elicit an inward INa represented by the superimposed traces before (black) and following a 5 min exposure (grey) to 1 μM Ct1a. The experiment was performed on three independent cells (n = 3). (B) Two-electrode voltage-clamp electrophysiology was used to examine the effect of Ct1a on BgNaV1 expressed in Xenopus oocytes. Shown are representative traces from a single oocyte, obtained from the −15 mV step of a conductance-voltage (G–V) step series (panel C), with the current before and after application of 1 μM Ct1a shown in black and red, respectively. (C) Normalized G–V relationship (G/Gmax, closed circles) and steady-state inactivation (SSI) relationship (I/Imax, open circles) before (black) and after (red) addition of 1 μM Ct1a. In both cases the toxin effect was normalized to control. Data are from two-electrode voltage-clamp of Xenopus oocytes expressing BgNaV1. Oocytes were depolarized in 5-mV increments from −90 mV to 5 mV for 50 ms from a holding potential of −90 mV, followed by a depolarizing pulse to −15 mV for 50 ms. Peak current from these step series was converted to conductance and normalized to generate the G–V relationship, while peak current from the −15 mV depolarization step was normalized to yield the SSI relationship. Data points are mean ± SEM (n = 3). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Results
3.1. Toxin isolation and sequencing
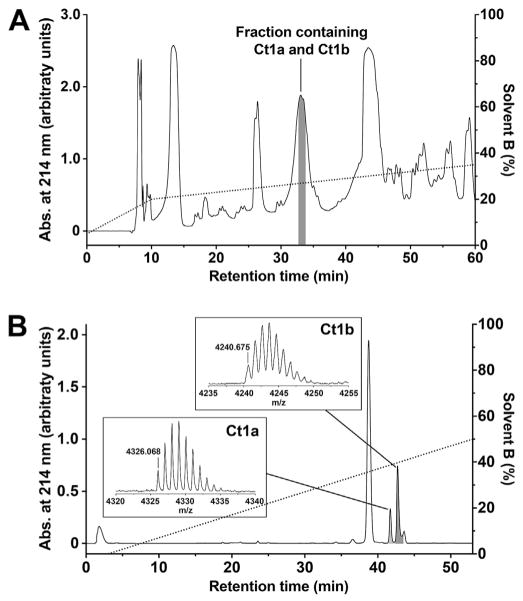
We screened 54 fractions resulting from RP-HPLC separation of C. tropix venom for insecticidal activity by injection into sheep blowflies (n = 3). Of these, seven fractions induced reversible paralysis while two caused paralysis leading to less than 35% mortality at 24 h post-injection. Only one fraction eluting at ~33 min (Fig. 1A) induced rapid paralysis of flies followed by death within 24 h. This fraction was further separated using cation exchange chromatography, which led to isolation of two active peptides tentatively named Ct1a and Ct1b (Fig. 1B).
Fig. 1. Isolation of Ct1a and Ct1b from C. tropix venom.
(A) Chromatogram showing fractionation of C. tropix venom using C18 RP-HPLC. The dotted line shows the gradient of solvent B (right ordinate axis). A single peak with retention time of ~33 min (highlighted in grey) was active against L. cuprina. (B) Chromatogram resulting from fractionation of the insect-active RP-HPLC peak using cation-exchange chromatography, which yielded two fractions that were active against L. cuprina (highlighted in grey). The dotted line shows the gradient of solvent B (right ordinate axis). The insets show MALDI-TOF MS spectra of the active venom peptides Ct1a and Ct1b. All masses are for monoisotopic M+H+ ions.
MALDI-TOF MS analysis of Ct1a yielded a monoisotopic mass of 4325.068 Da (inset, Fig. 1B). Edman sequencing of reduced and alkylated Ct1a returned the N-terminal sequence LFECSFSCDIKKNGKPCKGSGEKKCSGGWRCKMNFCVK, which gives a calculated monoisotopic mass of 4225.964 Da. This calculated mass is 99.104 Da less than the monoisotopic mass of Ct1a. We therefore conclude that the Edman degradation-derived sequence is lacking a C-terminal valine residue and that the complete sequence of Ct1a is LFECSFSCDIKKNGKPCKGSGEKKCSGGWRCKMNFCVKV; the calculated monoisotopic mass for this sequence is 4325.032 Da, which is only 0.036 Da less than the mass of native Ct1a. The presence of a C-terminal valine residue in Ct1a was later confirmed by analysis of a C. tropix venom-gland transcriptome (see Section 3.2 below).
MALDI-TOF MS analysis of Ct1b yielded a monoisotopic mass of 4239.675 Da (inset, Fig. 1B). Edman sequencing of reduced and alkylated Ct1b returned the N-terminal sequence FECSLSCDIKKNGKPCKGSGEKKCSGGWRCKMNFCLK with a calculated monoisotopic mass of 4092.991 Da, which is 146.684 Da less than the monoisotopic mass of Ct1b. We therefore conclude that the Edman degradation-derived sequence of Ct1b lacks a C-terminal phenylalanine and the complete sequence for this toxin is FECSLSCDIKKNGKPCKGSGEKKCSGGWRCKMNFCLKF; the calculated monoisotopic mass for this peptide 4239.979 Da, which is only 0.304 Da higher than the mass of native Ct1b. The presence of a C-terminal phenylalanine in Ct1b was later confirmed by analysis of a C. tropix venom-gland transcriptome (see Section 3.2 below). Ct1a and Ct1b are clearly paralogs with an overall amino acid sequence identity of 90% (Fig. 2A).
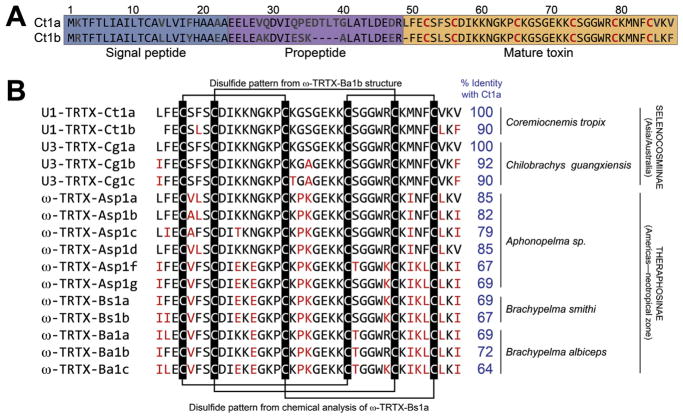
Fig. 2. Sequences of Ct1a, Ct1b, and orthologs.
(A) Amino acid sequences of the prepropeptide precursors encoding Ct1a and Ct1b. Numbering above the alignment refers to Ct1a. (B) Alignment of the sequences of the mature Ct1a and Ct1b toxins with orthologs from other theraphosid spiders. Residues that differ from those in the corresponding positions in Ct1a are highlighted in red. The disulfide-bond pattern from the NMR-derived structure of ω-TRTX-Ba1b (Corzo et al., 2009) and from chemical analysis of ω-TRTX-Bs1a (Kaiser et al., 1994) are shown above and below the sequence alignment, respectively. Percent identity with Ct1a is shown in blue to the right of the alignment. Taxonomic information (species and subfamily) and geographic distribution are provided at far right. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Sequencing of a C. tropix venom-gland transcriptome
ESTs obtained from the venom glands of nine individuals were sequenced and assembled using MIRA software (v4.0.2) (Chevreux et al., 2004), which produced a total of 10924 clusters and 4193 singlets with an average cluster length of 545 bp. Functional characterization and annotation of the dataset was performed using Blast2GO (Conesa et al., 2005). Clusters and singlets were grouped into three categories: (1) proteins and other enzymes; (2) toxins and toxin-like peptides; (3) sequences with no hits to databases (data not shown). At the same time, we used in-house scripts to generate a list of toxin and toxin-like sequences. Ct1a and Ct1b sequences were specifically searched against this list, and from this file, a total of 22 (clusters + singlets) consensus sequences were found to encode Ct1a, whereas 89 (clusters + singlets) were found to encode Ct1b. No additional paralogs were identified.
The full-length venom-gland transcripts indicate that Ct1a and Ct1b are initially produced as larger prepropeptides that are post-translationally processed to yield the mature toxins, which is typical for spider-venom peptides (Sollod et al., 2005). The signal peptide and propeptide regions were predicted using the SpiderP algorithm (Wong et al., 2013), which yielded a mature toxin sequence which matched that determined via Edman degradation (Fig. 2A).
A search of the ArachnoServer spider-toxin database (Wood et al., 2009; Herzig et al., 2011) yielded several near-identical orthologs of Ct1a and Ct1b (Fig. 2B). Most surprisingly, Ct1a is 100% identical to a mature toxin (U3-TRTX-Cg1a) of unknown function predicted from a venom-gland transcriptome of the Chinese tarantula Chilobrachys guangxiensis, and 90–92% identical with predicted paralogs of this toxin (U3-TRTX-Cg1b and U3-TRTX-Cg1c) (Chen et al., 2008). Ct1a and Ct1b are also have high levels of sequence identity (≥64%) with toxins isolated from the Mexican red kneed tarantula Brachypelma smithi (Kaiser et al., 1994), the Mexican golden redrump tarantula Brachypelma albiceps (Corzo et al., 2009), and a tarantula of unclear taxonomic status (Aphonopelma sp.) (Savel-Niemann, 1989; Nason et al., 1994) (Fig. 2B). All other previously isolated spider toxins have less than 60% sequence identity with Ct1a and Ct1b. Thus, Ct1a and Ct1b appear to be members of a family of spider toxins that are taxonomically restricted to tarantulas (family Theraphosidae). Notably, this family of toxins has a conserved pattern of six cysteine residues (Fig. 2B) that does not conform to the inhibitor cystine knot motif (Pallaghy et al., 1994) that is commonly found in spider toxins (King et al., 2002; King and Hardy, 2013).
3.3. Production of recombinant Ct1a
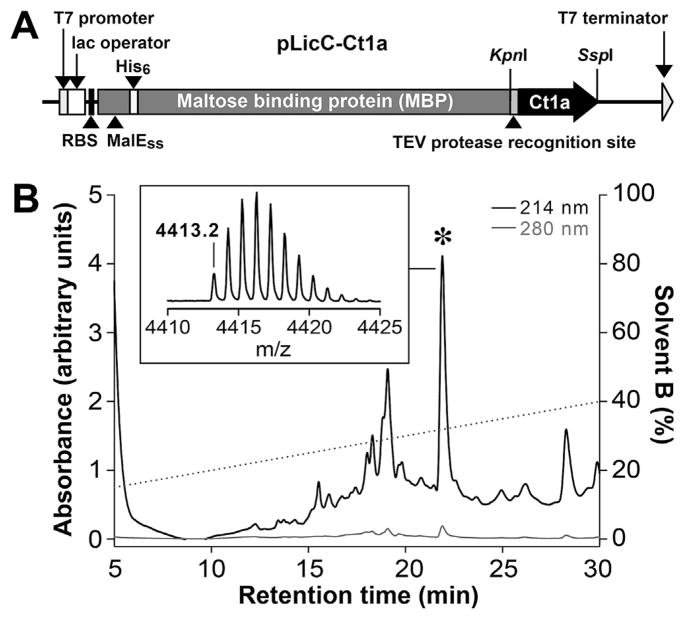
Recombinant Ct1a was produced via expression of a His6-MBP-Ct1a fusion protein in the periplasm of E. coli using a protocol described for production of disulfide-rich venom peptides (Klint et al., 2013). This system employs an IPTG-inducible construct containing a MalE signal sequence (Fig. 3A) that allows export of the fusion protein from the cytoplasm to the periplasm, where the protein machinery for disulfide-bond formation is located. The peptide was liberated from the fusion protein by addition of TEV protease and purified using RP-HPLC (Fig. 3B). The recombinant Ct1a was active in preliminary tests on sheep blowflies (data not shown). However, due to low yields of recombinant peptide (~100 μg/L of culture), Ct1a was subsequently produced using SPPS and the synthetic peptide was used for all future experiments.
Fig. 3. Production of recombinant Ct1a.
(A) Schematic representation of the pLicC–Ct1a vector used for periplasmic expression of recombinant Ct1a. The coding region includes a MalE signal sequence (MalESS) for periplasmic export, a His6 affinity tag, an MBP fusion tag and a codon-optimized gene encoding Ct1a, with a TEV protease recognition site inserted between the MBP and toxin-coding regions. The locations of key elements of the vector are shown, including the ribosome-binding site (RBS), T7 promoter and lac operator. (B) RP-HPLC chromatogram showing purification of Ct1a following removal of the His6-MBP tag by TEV protease. The dotted line shows the gradient of solvent B (right ordinate axis). Asterisk denotes the peak corresponding to correctly folded Ct1a. Inset is a MALDI-TOF MS spectrum showing the [M+H]+ ion for the purified recombinant toxin (observed = 4413.2 Da; calculated = 4413.0 Da). Note that the recombinant Ct1a contains a non-native N-terminal serine residue and hence has a slightly higher mass than native Ct1a.
3.4. Chemical synthesis of Ct1a
Chemical synthesis of Ct1a was achieved by native chemical ligation of two unprotected polypeptide segments: Ct1a(1–24)-α-thioester and Ct1a(25–39). Synthesis of the individual peptide segments and their subsequent ligation proceeded well and afforded the fully reduced full-length polypeptide in good yield (48%, based on starting peptide segments). Folding of the polypeptide chain and formation of the three disulfide bonds were carried out in a redox buffer as outlined in Section 2.9. The reaction reached equilibrium after 24 h and resulted in a mixture of disulfide isomers. Toxicity assays in houseflies (data not shown) were used to identify the correctly folded isomer, which was isolated in 10% yield.
3.5. Toxicity of synthetic Ct1a to sheep blowflies
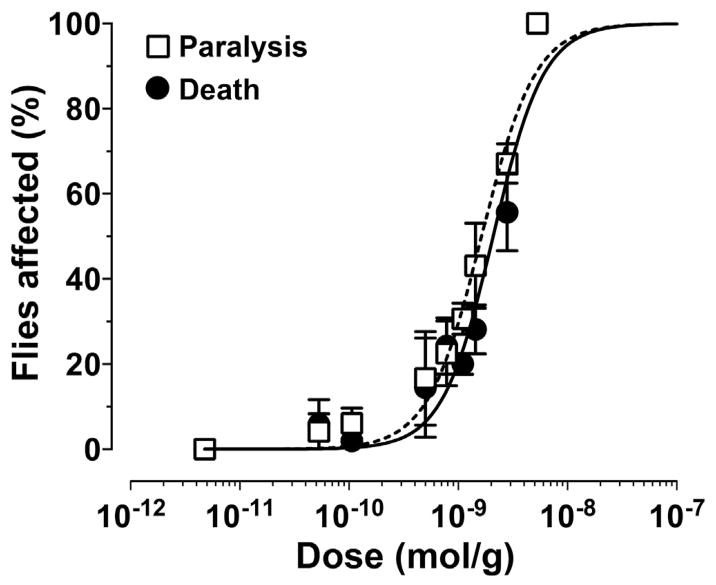
Ct1a induced contractile paralysis in adult L. cuprina blowflies within minutes after injection. The observed paralysis was irreversible and eventually led to death. The calculated LD50 at 24 h (1688 ± 64 pmol/g, n = 3) was only slightly higher than the respective PD50 (1335 ± 132 pmol/g, n = 3) (Fig. 4), indicating that the great majority of flies die within 24 h after injection.
Fig. 4. Insecticidal activity of Ct1a.
Synthetic Ct1a was intra-thoracically injected into adult L. cuprina blowflies then paralysis (□) and lethality (●) were measured at 24 h post-injection. Data points are mean ± SEM. Fitting of dose-response curves yielded PD50 and LD50 values of 1335 ± 132 pmol/g and 1688 ± 64 pmol/g, respectively.
3.6. Effect of synthetic Ct1a on insect NaV channels
In contrast to vertebrates, insects express only a single NaV channel subtype and consequently they are extremely sensitive to modulation of its activity. It is therefore not surprising that NaV channel modulators are disproportionately represented in the venoms of arthropod predators (Billen et al., 2008; King et al., 2008a) such as spiders (Klint et al., 2012) and scorpions (Gurevitz et al., 2007). Thus, we used two approaches to examine the activity of Ct1a against insect NaV channels.
We initially used patch-clamp electrophysiology to examine the effect of Ct1a on NaV channel currents in DUM neurons isolated from the American cockroach P. americana. Application of 1 μM Ct1a had minimal effect on NaV channel currents in P. americana DUM neurons, reducing peak current by less than 10% (Fig. 5A) and shifting V0.5 by only 1.3 mV in the hyperpolarizing direction (not shown).
It has been shown that some spider toxins that target insect NaV channels are ineffective against P. americana and triatomine bugs due to rare sequence variations in the domain II voltage sensor of their NaV channels (Billen et al., 2008; Herzig et al., 2016). For example, the spider toxins μ-DGTX-Dc1a (Bende et al., 2014) and μ-TRTX-Ae1a (Herzig et al., 2016) are potent inhibitors of the BgNaV1 channel from the German cockroach Blattella germanica whereas P. americana is resistant to these toxins. Thus, we next used two-electrode voltage-clamp electrophysiology to examine the effect of Ct1a on the cloned BgNaV1 channel heterologously expressed in Xenopus oocytes. At concentrations up to 1 μM, synthetic Ct1a had no significant effect (<10% inhibition) on sodium currents mediated by BgNaV1 (Fig. 5B) and it did not affect conductance or steady-state inactivation (SSI) of the channel (Fig. 5C). Fitting of a Boltzmann equation to the G–V relationships yielded values for half-maximal activation (V1/2) of −39 ± 1 mV (slope 5.3 ± 0.8) and −38 ± 1 mV (slope 5.8 ± 0.7) in the absence and presence of Ct1a, respectively. Fitting of a Boltzmann equation to the SSI curves yielded V1/2 values for inactivation of −58 ± 1 mV (slope 4.9 ± 0.2) and −60 ± 1 mV (slope 5.0 ± 0.2) in the absence and presence of Ct1a, respectively. Thus, we conclude that insect NaV channels are not the molecular target of Ct1a.
4. Discussion
4.1. Ct1a and Ct1b are dipteran-active insecticidal peptides
Flystrike caused by sheep blowflies (L. cuprina) is a huge economic burden for the Australian livestock industry, resulting in significant reduction of wool quality and quantity as well as decreased ewe fertility and even death of livestock (Kongsuwan et al., 2005). In order to find novel molecules with the potential to treat flystrike, we established a toxicity assay using adult sheep blowflies for identifying insecticidal venom peptides (Bende et al., 2013). In this study, we used this assay to identify two paralogous insecticidal toxins, Ct1a and Ct1b, from venom of the Australian tarantula C. tropix. Both of these peptide toxins induced irreversible contractile paralysis in sheep blowflies which led to death within 24 h of injection.
Synthetic Ct1a was insecticidal to L. cuprina with an LD50 of 1687 pmol/g. Only three spider toxins have been previously shown to be lethal to L. cuprina, with LD50 values of 198 pmol/g for U1-agatoxin-Ta1a (Ta1a) (Undheim et al., 2015), 231 pmol/g for μ-diguetoxin-Dc1a (Bende et al., 2014), and 278 pmol/g for κ-hexatoxin-Hv1c (de Araujo et al., 2013). The spider venom peptides μ-cyrtautoxin-As1a and μ-segestritoxin-Sf1a both paralyse L. cuprina, without causing lethality, with PD50 values of 700 pmol/g (Bende et al., 2013) and 2229 pmol/g (Bende et al., 2015), respectively. Thus, we consider Ct1a to be a moderately potent insecticidal peptide with respect to its activity on blowflies. However, its potential for development as a bioinsecticide is enhanced by the fact that the orthologous toxin ω-TRTX-Ba1b causes no adverse effects when injected either intracranially or intraperitoneally into mice (Corzo et al., 2009), suggesting that Ct1a may also be devoid of vertebrate toxicity.
Ct1a is identical to U3-TRTX-Cg1a, a toxin with unknown molecular target and function predicted from a venom-gland transcriptome of the Chinese tarantula Chilobrachys guangxiensis. Interestingly, the genera Coremiocnemis and Chilobrachys belong to the same taxonomic subfamily (Selenocosmiinae), which is restricted to Asia and Australia. All other orthologs of Ct1a/1b are found in members of Theraphosinae, a theraphosid subfamily that is largely restricted to neotropical regions of the Americas (Fig. 2B). We propose that these toxins have a general function linked to prey capture in these subfamilies of theraphosid spiders as several of these peptides have been shown to be insecticidal. For example, ω-TRTX-Ba1b (72% identity with Ct1a) is insecticidal to crickets (Acheta domesticus) with a LD50 of 2.1 nmol/g (Corzo et al., 2009), while ω-TRTX-Asp1g (69% identity with Ct1a) is lethal to the American cockroach P. americana (Herzig et al., 2011). However, the current work represents the first time that any member of this family of toxins has been shown to be lethal to an agriculturally important dipteran pest.
4.2. Molecular target of Ct1a/1b
The molecular target of Ct1a/1b remains to be determined, but we ruled out NaV channels as a possible target. Our data are consistent with the report that ω-TRTX-Ba1a and -Ba1b, which are 69–72% identical to Ct1a (see Fig. 2B), have no effect on the Drosophila NaV channel (Para/TipE) or mammalian NaV1.2 and NaV1.5 channels (Corzo et al., 2009). The high level of homology between Ct1a and ω-TRTX-Asp1b (82% identity; see Fig. 2B) suggests that voltage-gated calcium (CaV) channels are the most likely target for Ct1a. Thus, the next logical step is to test Ct1a against insect CaV channels, which we were unable to do in the current study due to insufficient amounts of recombinant or synthetic Ct1a. Investigation is warranted into better techniques to obtain sufficient Ct1a to facilitate elucidation of its structure, molecular target, and mechanism of action. In the absence of a defined molecular target, the rational names for Ct1a and Ct1b based on the nomenclature recommended for spider toxins (King et al., 2008b) are U1-theraphotoxin-Ct1a (U1-TRTX-Ct1a) and U1-theraphotoxin-Ct1b (U1-TRTX-Ct1b), respectively.
4.3. Structure of Ct1a/1b
Toxins in the Ct1a family contain a conserved pattern of six cysteine residues (CX3CX8C7CX5CX4CX3; see Fig. 2B) that does not conform to the inhibitor cystine knot (ICK) motif commonly found in spider toxins (Pallaghy et al., 1994; Saez et al., 2010; King and Hardy, 2013), as evident from the lack of a cysteine doublet at positions 3 and 4. A three-dimensional structure has been determined for only one member of this toxin family, namely ω-TRTX-Ba1b, for which the NMR data revealed a non-ICK fold with a disulfide connectivity of C1–C3, C2–C5, C4–C6 (Corzo et al., 2009). However, this pattern differs from the disulfide connectivity of C1–C4, C2–C5, C3–C6 that was chemically determined for the closely related ortholog ω-TRTX-Bs1a (Kaiser et al., 1994), which is 92% identical. It is possible, but unlikely, that these toxins have different disulfide connectivities (and therefore different 3D folds), so the reason for the different conclusions relating to the disulfide connectivity of this toxin family remains to be determined. It is often difficult to accurately determine disulfide connectivities using NMR-based dipolar couplings (Mobli and King, 2010) while chemical methods for determination of disulfide bonds are susceptible to disulfide scrambling unless performed at low pH (Gray, 1993). Thus, determination of the structure of additional members of this toxin family and/or careful chemical analysis will be required to resolve the conundrum of whether members of this family have a conserved or variable disulfide-bond pattern.
Acknowledgments
We thank the Australian Grains Research & Development Corporation (GRDC) for financial support, Dr Robert Raven (Queensland Museum, Brisbane, Australia) for identification of C. tropix specimens, Dr Geoff Brown (Department of Agriculture, Fisheries and Forestry, Brisbane, Australia) for supply of blowflies, and Prof. Ke Dong (Michigan State University) for sharing BgNaV1/TipE clones. This research was facilitated by access to the Australian Proteome Analysis Facility, which is supported under the Australian Government’s National Collaborative Research Infrastructure Strategy.
Footnotes
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.toxicon.2016.10.013.
References
- Bende N, Dziemborowicz S, Herzig V, Ramanujam V, Brown GW, Bosmans F, Nicholson GM, King GF, Mobli M. The insecticidal spider toxin SFI1 is a knottin peptide that blocks the pore of insect voltage-gated sodium channels. FEBS J. 2015;282:904–920. doi: 10.1111/febs.13189. [DOI] [PubMed] [Google Scholar]
- Bende NS, Kang E, Herzig V, Bosmans F, Nicholson GM, Mobli M, King GF. The insecticidal neurotoxin Aps III is an atypical knottin peptide that potently blocks insect voltage-gated sodium channels. Biochem Pharmacol. 2013;85:1542–1554. doi: 10.1016/j.bcp.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bende NS, Dziemborowicz S, Mobli M, Herzig V, Gilchrist J, Wagner J, Nicholson GM, King GF, Bosmans F. A distinct sodium channel voltage-sensor locus determines the insect selectivity of the spider toxin Dc1a. Nat Commun. 2014;5:4350. doi: 10.1038/ncomms5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billen B, Bosmans F, Tytgat J. Animal peptides targeting voltage-activated sodium channels. Curr Pharm Des. 2008;14:2492–2502. doi: 10.2174/138161208785777423. [DOI] [PubMed] [Google Scholar]
- Chen J, Deng M, He Q, Meng E, Jiang L, Liao Z, Rong M, Liang S. Molecular diversity and evolution of cystine knot toxins of the tarantula Chilobrachys jingzhao. Cell Mol Life Sci. 2008;65:2431–2444. doi: 10.1007/s00018-008-8135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Corzo G, Bernard C, Clement H, Villegas E, Bosmans F, Tytgat J, Possani LD, Darbon H, Alagón A. Insecticidal peptides from the theraposid spider Brachypelma albiceps: an NMR-based model of Ba2. Biochim Biophys Acta. 2009;1794:1190–1196. doi: 10.1016/j.bbapap.2009.04.004. [DOI] [PubMed] [Google Scholar]
- de Araujo AD, Herzig V, Windley MJ, Dziemborowicz S, Mobli M, Nicholson GM, Alewood PF, King GF. Do vicinal disulfide bridges mediate functionally important redox transformations in proteins? Antioxid Redox Signal. 2013;19:1976–1980. doi: 10.1089/ars.2013.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong K. A single amino acid change in the para sodium channel protein is associated with knockdown-resistance (kdr) to pyrethroid insecticides in German cockroach. Insect Biochem Mol Biol. 1997;27:93–100. doi: 10.1016/s0965-1748(96)00082-3. [DOI] [PubMed] [Google Scholar]
- Eitan M, Fowler E, Herrmann R, Duval A, Pelhate M, Zlotkin E. A scorpion venom neurotoxin paralytic to insects that affects sodium current inactivation: purification, primary structure, and mode of action. Biochemistry. 1990;29:5941–5947. doi: 10.1021/bi00477a009. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Rash L. Tarantulas: eight-legged pharmacists and combinational chemists. Toxicon. 2004;43:555–574. doi: 10.1016/j.toxicon.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Sollod BL, King GF. Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Feng G, Deak P, Chopra M, Hall LM. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- Gentz MC, Jones A, Clement H, King GF. Comparison of the peptidome and insecticidal activity of venom from a taxonomically diverse group of theraphosid spiders. Toxicon. 2009;53:496–502. doi: 10.1016/j.toxicon.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Gray WR. Disulphide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–1748. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevitz M, Karbat I, Cohen L, Ilan N, Kahn R, Turkov M, Stankiewicz M, Stuhmer W, Dong K, Gordon D. The insecticidal potential of scorpion β-toxins. Toxicon. 2007;49:473–489. doi: 10.1016/j.toxicon.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Herzig V, Hodgon WC. Neurotoxic and insecticidal properties of venom from the Australian theraphosid spider Selenotholus foelschei. Neurotoxicology. 2008;29:471–475. doi: 10.1016/j.neuro.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Herzig V, Hodgson WC. Intersexual variations in the pharmacological properties of Coremiocnemis tropix (Araneae, Theraphosidae) spider venom. Toxicon. 2009;53:196–205. doi: 10.1016/j.toxicon.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Herzig V, Wood DL, Newell F, Chaumeil PA, Kaas Q, Binford GJ, Nicholson GM, Gorse D, King GF. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2011;39:D653–D657. doi: 10.1093/nar/gkq1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig V, Bende NS, Alam MS, Tedford HW, Kennedy RM, King GF. Methods for deployment of spider venom peptides as bioinsecticides. In: Dhadialla TS, Gill SS, editors. Advances in Insect Physiology: Insect Midgut and Insecticidal Proteins. Vol. 47. Elsevier; London: 2014. pp. 389–411. [Google Scholar]
- Herzig V, Ikonomopoulou M, Smith JS, Dziemborowicz S, Gilchrist J, Kukhn-Nentwig L, Rezende FO, Moreira LA, Nicholson GM, Bosmans F, King GF. Isolation of an insecticidal sodium channel toxin with remarkable phyletic specificity from the African theraphosid spider Augacephalus ezendami. Sci Rep. 2016;6:29538. doi: 10.1038/srep29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Kent SBH. Insights into the mechanism and catalysis of the native chemical ligation reaction. J Am Chem Soc. 2006;128:6640–6646. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- Kaiser II, Griffin PR, Aird SD, Hudiburg S, Shabanowitz J, Francis B, John TR, Hunt DF, Odell GV. Primary structures of two proteins from the venom of the Mexican red knee tarantula (Brachypelma smithii) Toxicon. 1994;32:1083–1093. doi: 10.1016/0041-0101(94)90392-1. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GF, Tedford HW, Maggio F. Structure and function of insecticidal neurotoxins from Australian funnel-web spiders. J Toxicol-Toxin Rev. 2002;21:359–389. [Google Scholar]
- King GF, Escoubas P, Nicholson GM. Peptide toxins that selectively target insect NaV and CaV channels. Channels. 2008a;2:100–116. doi: 10.4161/chan.2.2.6022. [DOI] [PubMed] [Google Scholar]
- King GF, Gentz MC, Escoubas P, Nicholson GM. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon. 2008b;52:264–276. doi: 10.1016/j.toxicon.2008.05.020. [DOI] [PubMed] [Google Scholar]
- King GF. Venoms as a platform for human drugs: translating toxins into therapeutics. Expert Opin Biol Ther. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- King GF, Hardy MC. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu Rev Entomol. 2013;58:475–496. doi: 10.1146/annurev-ento-120811-153650. [DOI] [PubMed] [Google Scholar]
- Klint JK, Senff S, Rupasinghe DB, Er SY, Herzig V, Nicholson GM, King GF. Spider-venom peptides that target voltage-gated sodium channels: pharmacological tools and potential therapeutic leads. Toxicon. 2012;60:478–491. doi: 10.1016/j.toxicon.2012.04.337. [DOI] [PubMed] [Google Scholar]
- Klint JK, Senff S, Saenz NJ, Seshadri R, Lau HY, Bende NS, Undheim EA, Rash LD, Mobli M, King GF. Production of recombinant disulfide-rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli. PLos One. 2013;8:e63865. doi: 10.1371/journal.pone.0063865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuwan K, Gough J, Kemp D, McDevitt A, Akhurst R. Characterization of a new Bacillus thuringiensis endotoxin, Cry47Aa, from strains that are toxic to the Australian sheep blowfly, Lucilia cuprina. FEMS Microbiol Lett. 2005;252:127–136. doi: 10.1016/j.femsle.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Kuhn-Nentwig L, Stocklin R, Nentwig W. Venom composition and strategies in spiders: is everything possible. In: Casas J, editor. Spider Physiology and Behaviour—physiology. Vol. 40. Elsevier; 2011. pp. 1–86. [Google Scholar]
- Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet—next generation sequence assembly visualization. Bioinformatics. 2009;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobli M, King GF. NMR methods for determining disulfide-bond connectivities. Toxicon. 2010;56:849–854. doi: 10.1016/j.toxicon.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Nason DM, Phillips D, Saccomano NA, Volkmann RA. Calcium channel blocking polypeptides from Theraphosidae Aphonopelma. WO1994010196 A1 Patent. 1994
- Pallaghy PK, Nielsen KJ, Craik DJ, Norton RS. A common structural motif incorporating a cystine knot and a triple-stranded β-sheet in toxic and inhibitory polypeptides. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven RJ. A new tarantula species from northern Australia (Araneae, Theraphosidae) Zootaxa. 2005;1004:15–28. [Google Scholar]
- Sackett D, Holmes P, Abbott K, Jephcott S, Barber M. Assessing the Economic Cost of Endemic Disease on the Profitability of Australian Beef Cattle and Sheep Producers. Meat & Livestock Australia; Sydney: 2006. [Google Scholar]
- Saez NJ, Senff S, Jensen JE, Er SY, Herzig V, Rash LD, King GF. Spider-venom peptides as therapeutics. Toxins. 2010;2:2851–2871. doi: 10.3390/toxins2122851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeman RM, Levot GW, Heath AC, James PJ, Greeff JC, Scott MJ, Batterham P, Bowles VM. Control of the sheep blowfly in Australia and New Zealand—are we there yet? Int J Parasitol. 2014;44:879–891. doi: 10.1016/j.ijpara.2014.08.009. [DOI] [PubMed] [Google Scholar]
- Savel-Niemann A. Tarantula (Eurypelma californicum) venom, a multicomponent system. Biol Chem Hoppe-Seyler. 1989;370:485–498. doi: 10.1515/bchm3.1989.370.1.485. [DOI] [PubMed] [Google Scholar]
- Schroeder FC, Taggi AE, Gronquist M, Malik RU, Grant JB, Eisner T, Meinwald J. NMR-spectroscopic screening of spider venom reveals sulfated nucleosides as major components for the brown recluse and related species. Proc Natl Acad Sci U S A. 2008;105:14283–14287. doi: 10.1073/pnas.0806840105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon J, Rollin B. Mulesing and animal ethics. J Ag Environ Ethics. 2009;23:371–386. [Google Scholar]
- Sollod BL, Wilson D, Zhaxybayeva O, Gogarten JP, Drinkwater R, King GF. Were arachnids the first to use combinatorial peptide libraries? Peptides. 2005;26:131–139. doi: 10.1016/j.peptides.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Tyrell LD, Larsen JW, Anderson N. Breech-strike on mulesed, clipped and unmulesed Merino ewes and hoggets in south-eastern Australia. Aust Vet J. 2014;92:348–356. doi: 10.1111/avj.12228. [DOI] [PubMed] [Google Scholar]
- Undheim EA, Grimm LL, Low CF, Morgenstern D, Herzig V, Zobel-Thropp P, Pineda SS, Habib R, Dziemborowicz S, Fry BG, Nicholson GM, Binford GJ, Mobli M, King GF. Weaponization of a hormone: convergent recruitment of hyperglycemic hormone into the venom of arthropod predators. Structure. 2015;23:1283–1292. doi: 10.1016/j.str.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Windley MJ, Herzig V, Dziemborowicz SA, Hardy MC, King GF, Nicholson GM. Spider-venom peptides as bioinsecticides. Toxins. 2012;4:191–227. doi: 10.3390/toxins4030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Hardy MC, Wood D, Bailey T, King GF. SVM-based prediction of propeptide cleavage sites in spider toxins identifies toxin innovation in an Australian tarantula. PLoS One. 2013;8:e66279. doi: 10.1371/journal.pone.0066279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DL, Miljenovic T, Cai S, Raven RJ, Kaas Q, Escoubas P, Herzig V, Wilson D, King GF. ArachnoServer: a database of protein toxins from spiders. BMC Genomics. 2009;10:375. doi: 10.1186/1471-2164-10-375. [DOI] [PMC free article] [PubMed] [Google Scholar]