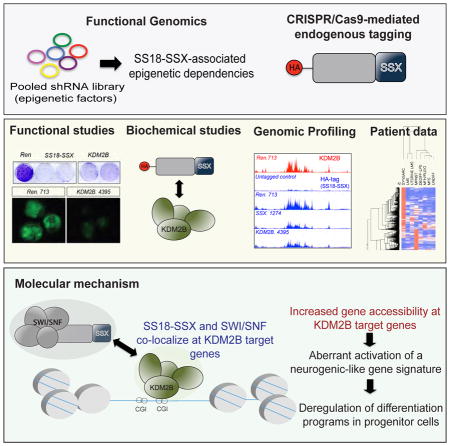
SUMMARY
Synovial sarcoma is an aggressive cancer invariably associated with a chromosomal translocation involving genes encoding the SWI-SNF complex component SS18 and a SSX (SSX1 or SSX2) transcriptional repressor. Using functional genomics, we identify KDM2B – a histone demethylase and component of a non-canonical Polycomb Repressive Complex 1 (PRC1.1) – as selectively required for sustaining synovial sarcoma cell transformation. SS18-SSX1 physically interacts with PRC1.1 and co-associates with SWI/SNF and KDM2B complexes on unmethylated CpG islands. Via KDM2B, SS18-SSX1 binds and aberrantly activates expression of developmentally regulated genes otherwise targets of polycomb-mediated repression, which is restored upon KDM2B depletion leading to irreversible mesenchymal differentiation. Thus, SS18-SSX1 de-regulates developmental programs to drive transformation by hijacking a transcriptional repressive complex to aberrantly activate gene expression.
Graphical Abstract
Banito et al. show that SS18-SSX fusions characteristic of synovial sarcoma associate with KDM2B, a non-canonical polycomb repressive complex 1, to aberrantly activate the expression of developmentally regulated transcription factors that are normally targets of polycomb mediated gene repression.
INTRODUCTION
Soft tissue sarcomas are aggressive cancers afflicting children and young adults that rarely respond to conventional chemotherapy and are often lethal (Helman and Meltzer, 2003; Singer et al., 2000). Many soft tissue sarcomas present with recurrent chromosomal translocations that involve genes encoding proteins thought to drive cancer by perturbing epigenetic regulation of gene expression that, in principle, could be reversed. While the presence of such fusions further underscores the key relationship between cancer genetics and epigenetics during tumorigenesis, the mechanisms by which most chimeric oncoproteins drive oncogenesis remain poorly understood. Consequently, there are no therapeutic strategies to target their activity.
Synovial sarcoma is a paradigm of a gene fusion driven cancer, in which the defining event is the chromosomal translocation t(X,18; p11, q11) that creates an in-frame fusion of SS18 to SSX1, SSX2 or SSX4 (Clark et al., 1994; Ladanyi et al., 2002). SS18-SSX is present in virtually 100% of synovial sarcomas, being the only cytogenetic aberration in most of these tumors characterized by a very low frequency of additional genetic alterations (Nielsen et al., 2015). Accordingly, aberrant expression of the translocated gene product in the myoblast lineage of mice produces tumors that histologically and molecularly resemble the human disease (Haldar et al., 2007).
Unlike oncofusion proteins in other soft tissue sarcomas where a transcription factor is thought to confer target specificity by binding a specific DNA sequence (e.g. PAX3-FOXO1), SS18-SSX lacks a DNA binding domain and is thought to exert its activity by interacting with other chromatin regulators. The SSX1/2 proteins are part of a family of transcriptional repressors and co-localize with polycomb group (PcG) proteins such as RING1B and BMI through unclear mechanisms (dos Santos et al., 2000; Soulez et al., 1999). By contrast, SS18 is a component of mammalian TrxG complexes (such as the SWI/SNF) and, as a consequence, SS18-SSX1/2 interacts with components of the SWI/SNF complex such as hBRM and BRG1 (Kadoch and Crabtree, 2013; Nagai et al., 2001; Thaete et al., 1999)
While PcG proteins lead to chromatin compaction and gene repression, SWI/SNF complexes facilitate transcription by remodeling nucleosomes, thereby promoting gene activation by permitting increased access of transcription factors to their binding sites (Roberts and Orkin, 2004). It remains to be determined precisely how SS18-SSX oncoproteins affect the balance between transcriptional activation via SWI/SNF and PcG-associated gene repression. One study points to the ability of SS18-SSX to repress expression of tumor suppressor genes such as those encoded by the CDKN2A locus, a process depending on SS18-SSX ability to bridge ATF2 targets to TLE1 for recruitment of polycomb repressive complex 2 (PRC2) (Su et al., 2012). However another study suggests that SS18-SSX alters SWI/SNF composition and enhances its ability to oppose the H3K27me3 repressive mark at the SOX2 locus, leading to transcriptional activation (Kadoch and Crabtree, 2013). Efforts to confirm these mechanisms have been thwarted by an inability to identify direct targets of endogenous SS18-SSX. Thus, there is no comprehensive picture of how SS18-SSX binds chromatin, alters transcription, and drives oncogenic transformation. As SS18-SSX is not obviously druggable, such information is necessary for developing rational strategies to disrupt its action in synovial sarcoma.
Efforts to elucidate how cancer-causing mutations sustain the transformed state often reveal potential mechanisms of oncogenesis and rational strategies for therapeutic intervention. As most synovial sarcomas harbor few, if any, genetic alterations besides an SS18-SSX fusion, and because core fusion components are constituents of chromatin regulatory complexes, the goal of this study is to reveal mechanisms mediating global deregulation of epigenetic networks that might be essential for the initiation and maintenance of this disease.
RESULTS
An shRNA screen identifies KDM2B as a specific vulnerability of SS18-SSX driven tumors
To interrogate the epigenetic mechanisms that sustain synovial sarcoma, we performed a pool-based shRNA screen to identify chromatin regulators whose inhibition selectively suppressed the proliferation of synovial sarcoma cells. We transduced a library consisting of ~2400 GFP-coupled shRNAs targeting 400 chromatin regulators into a synovial sarcoma cell line (M5SS1) derived from a murine sarcoma induced by expressing human SS18-SSX2 in the mesenchymal progenitor lineage (Haldar et al., 2007). As a control, we introduced the same library into untransformed C2C12 mouse myoblasts (Figure S1A). Changes in library representation after 16days of continuous passaging in culture were monitored using deep sequencing of shRNA guide strands amplified from genomic DNA. As expected, shRNAs targeting the positive control RPA3 (a replication enzyme) and c-MYC were depleted upon cell passaging in both cell lines, whereas an shRNA targeting p53 was substantially enriched (Figure 1A). Notably, three shRNAs targeting SS18-SSX were strongly depleted in M5SS1 but not in C2C12 cells (Figure 1A), confirming that murine synovial sarcoma cells depend on continuous SS18-SSX expression.
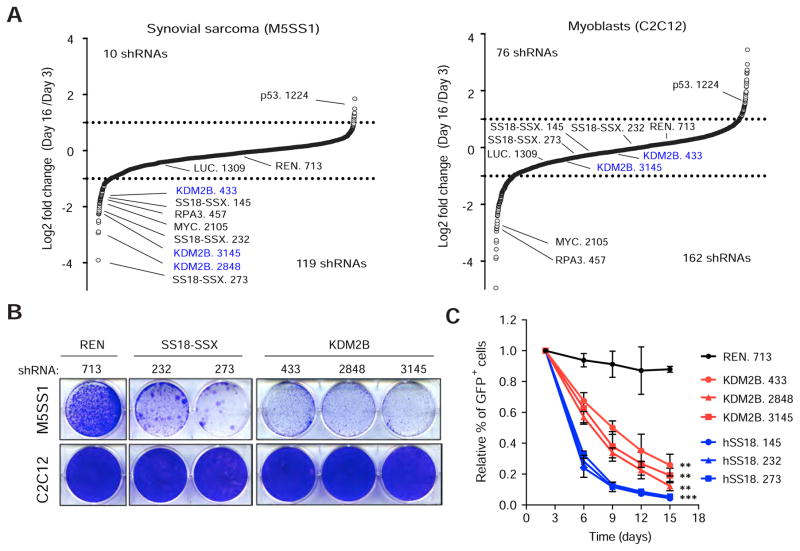
Figure 1. KDM2B is an epigenetic dependency in synovial sarcoma.
(A) Differences in shRNA representation presented as log2 of the ratio between mean average reads at Tf and T0 in synovial sarcoma cells and myoblasts. shRNAs against Renilla and Luciferase (REN. 713 and LUC. 1309) were used as neutral controls. (B) Clonogenic assay of M5SS1 (upper panel) and C2C12 (lower panel) cells transduced with the indicated shRNAs. (C) Validation of the effect of KDM2B shRNAs in mouse synovial sarcoma cells. Data presented as mean ± s.d. (n = 32) ** p value <0.001, ***p value<0.0001, unpaired t-test. See also Figure S1 and Table S1.
To identify additional shRNAs that mimicked those targeting SS18-SSX, we analyzed the sequencing data using specific scoring criteria (see Methods). Some shRNAs were depleted in both M5SS1 and C2C12 cells, including shRNAs targeting the SWI/SNF component BRG1 (SMARC4) (Figure S1B). By contrast, others exhibited mild or no depletion in either cell line, including those targeting PRC2 subunits EZH1/2 and SUZ12. shRNAs that were preferentially depleted in M5SS1 cells included those targeting BRD7 and BRD3 with shRNAs targeting KDM2B being the most potently and consistently depleted (Figures 1A, S1B and Table S1). All three scoring KDM2B shRNAs potently suppressed KDM2B protein expression and selectively impaired the proliferation of synovial sarcoma cells in competition and clonogenic survival assays (Figures 1B,C and Figures S1C,D). Confirming an on-target effect, a non-targetable KDM2B cDNA restored the proliferation of synovial sarcoma cells expressing KDM2B shRNAs (Figure S1E). Thus, KDM2B is required for sustained proliferation of murine synovial sarcoma cells.
KDM2B is required for the maintenance of human synovial sarcoma cells
Next, we examined KDM2B protein expression across different human sarcoma types and tested whether KDM2B was required for the proliferation and tumorigenic potential of human synovial sarcoma cells. Immunohistochemistry of a large panel of human sarcomas revealed that synovial sarcoma cells express high levels of KDM2B (Figure 2A,B). Similarly, KDM2B mRNA levels were higher in synovial sarcoma cell lines than normal fibroblasts or other cancer cell lines (Figure S2A) and, accordingly, analysis of publicly available functional genomics data confirmed that KDM2B is not universally required for cell proliferation (Figure S2B)(Aguirre et al., 2016). RNAi-mediated suppression of KDM2B in a panel of human synovial sarcoma cells triggered proliferative arrest and the acquisition of a fibroblast-like morphology in a manner that was remarkably similar to knockdown of SS18-SSX using either SS18 or SSX1/2 shRNAs (Figures 2C–E and Figure S2C). Accordingly, cells subjected to SS18-SSX or KDM2B inhibition upregulated genes indicative of mesenchymal differentiation, including those encoding certain extracellular matrix proteins and secreted proteins highly expressed in human fibroblasts, such as COL1A1, SERPINE1 (PAI-1), and ACTA2 (α-SMA) and the cell cycle inhibitors CDKN1A and CDKN2B (Figures 2F, G). This cell fate transition was irreversible: cells harboring GFP-coupled, doxycycline (Dox)-inducible shRNAs targeting either SS18-SSX or KDM2B showed proliferative arrest and mesenchymal differentiation upon Dox addition, which was maintained upon shRNA silencing following Dox withdrawal (Figures 2H,I and Figures S2D–E). Apparently, SS18-SSX or KDM2B inhibition disrupts synovial sarcoma maintenance by triggering proliferative arrest and differentiation into a more mesenchymal cell fate.
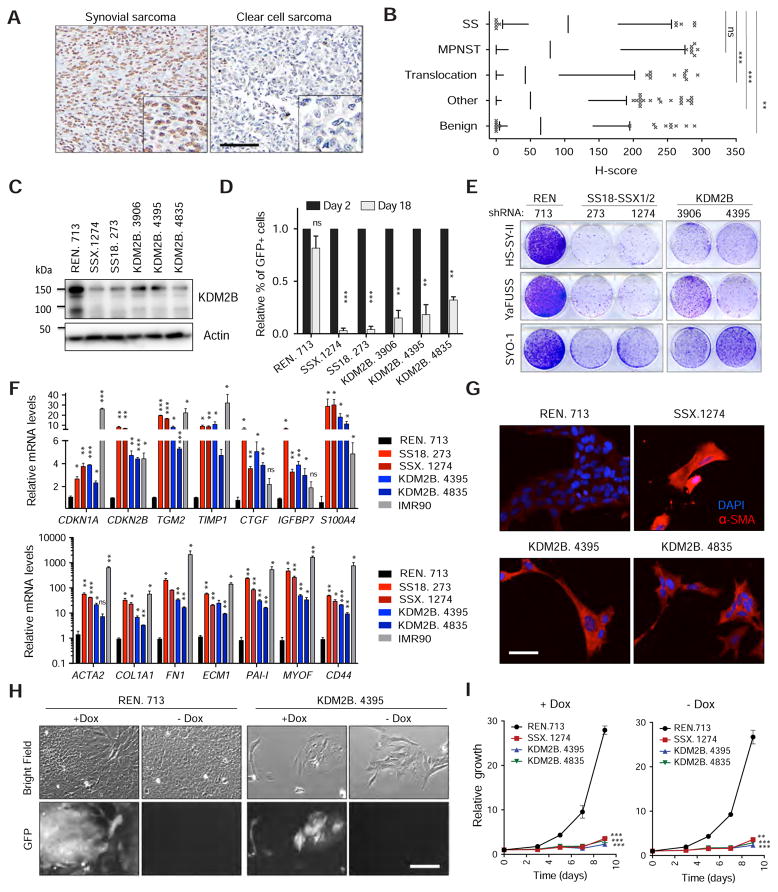
Figure 2. KDM2B inhibition irreversibly triggers mesenchymal differentiation.
(A) Representative image of a KDM2B positive (synovial sarcoma) and of a negative (clear cell sarcoma) sample within the same set of sarcoma tissue microarrays (TMAs) analyzed. Scale bar=100 μm. (B) Quantification of KDM2B IHC in sarcoma TMAs showing H-scores for KDM2B staining in synovial sarcomas (SS, n=58), malignant peripheral nerve sheath tumors (MPNST, n=76), other translocation driven sarcomas (Translocation, n=108), other types of sarcoma (Other, n=209) and benign soft tissue tumors (Benign, n=89). The line in the boxes corresponds to the median. The top and bottom of the boxes correspond to the third and first quartiles, respectively. The lines above and below the box correspond to the 10th and 90th percentile. Data points above or below the limits are considered outliers. (C) Immunoblot for KDM2B in HS-SY-II cells transduced with the indicated shRNAs. (D) Cell competition assay for GFP-linked shRNAs in the HS-SY-II human synovial sarcoma cell line. Relative percentage of GFP+ cells to day 2 following shRNA induction. Data are presented as mean + s.d. (n=2). (E) Clonogenic assay of HS-SY-II, YaFUSS and SYO-1 cells transduced with the indicated shRNAs. (F) Quantitative RT-PCR for the expression of fibroblast-related genes. IMR90 human diploid fibroblasts were used as a positive control. Data presented as mean + s.d. (n=2). (G) Immunofluorescence analysis of alpha smooth muscle actin (α-SMA) in HS-SY-II cells, ten days following transduction with the indicated shRNAs. Scale bar=20 μm. (H) Bright field and GFP images of HS-SY-II cells transduced with TRE-regulated shRNAs cultured with doxycycline (+Dox) or upon doxycycline withdrawal (−Dox), at day 10 after Dox withdrawal. Scale bar=25 μm (I) Growth curves for HS-SY-II cells with (+Dox) or without Dox (-Dox), presented as mean ± s.d. (n = 32). *p value<0.05, **p <0.005 ***p value<0.0005. B. Kruskal-Wallis 1-way ANOVA. D, F, I unpaired t-test. See also Figure S2.
We also examined the impact of SS18-SSX and KDM2B inhibition on tumor growth in vivo. HS-SY-II and SYO-1 synovial sarcoma cell lines expressing the inducible GFP-coupled shRNAs described above were transplanted into immunocompromised mice that were fed a Dox-containing diet to activate shRNA expression (Figure 3A). Tumor xenografts expressing KDM2B shRNAs displayed markedly impaired tumor growth, closely mimicking the effects seen by RNAi-mediated downregulation of SS18-SSX (Figures 3B,C). Importantly, and in contrast to tumors harboring control shRNAs, tumors arising from cells transduced with SS18-SSX or KDM2B shRNAs were composed predominantly of GFP-negative cells that had lost or silenced the shRNA (Figures 3D, E and Figure S2F). Therefore, human synovial sarcoma cells also require KDM2B for tumor maintenance in vivo.
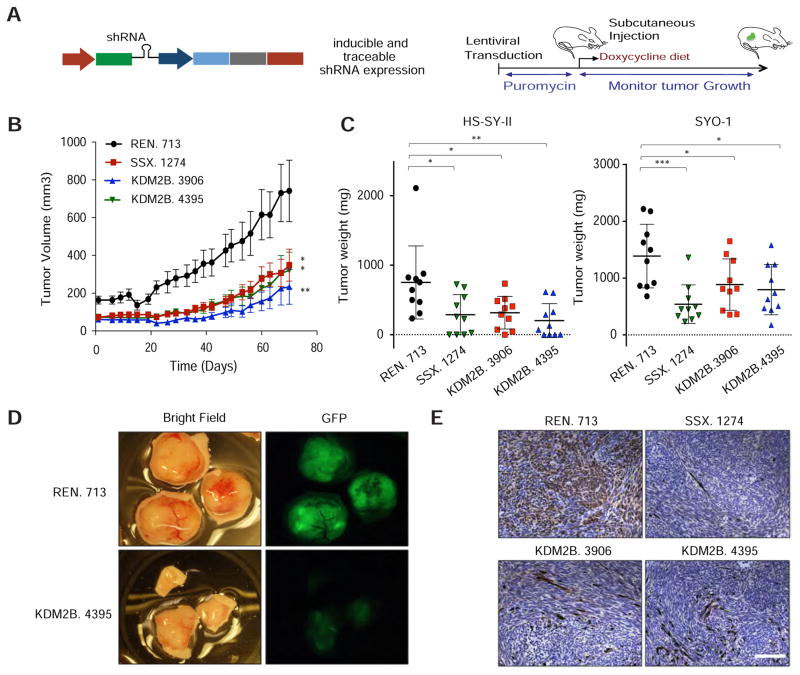
Figure 3. KDM2B is required for synovial sarcoma maintenance in vivo.
(A) Strategy to evaluate the effect of KDM2B knockdown in vivo in HS-SY-II and SYO-1-derived xenografts. See also STAR methods. (B) Tumor volume was measured over time in HS-SY-II xenografts transduced with the indicated shRNAs. Error bars correspond to mean ± s.e.m (n=10). (C) Tumor weight at the final time point. Data represented as mean ± s.d. (n=10). (D) Bright field and GFP images of HS-SY-II-derived tumors. (E) GFP IHC staining of HS-SY-II-derived tumors. Scale bar=150 μm. B, C, and D. Two-tailed t-test *p value<0.05, **p value<0.005, ***p value<0.0005. See also Figure S2.
The DNA-binding domain of KDM2B and PRC1.1 is critical for synovial sarcoma proliferation
KDM2B encodes a histone demethylase that can repress gene expression by demethylating H3K36me2 (He et al., 2008; Tzatsos et al., 2009). In addition, KDM2B is a core component of a poorly understood non-canonical polycomb repressive complex (BCOR complex or PRC1.1) (Gearhart et al., 2006) that, unlike the canonical PRC1, can be recruited to polycomb target sites in a PRC2-independent manner (Blackledge et al., 2014). Whereas the KDM2B demethylase activity requires a JmjC domain, its role in recruiting PRC1.1 involves binding to unmethylated CpG islands (CGIs) via its zinc finger-CxxC (ZF-CxxC) domain (Farcas et al., 2012; He et al., 2013; Wu et al., 2013). To determine whether either or both of these domains are required for synovial sarcoma maintenance, we performed a structure-function analysis that exploits a nuance of CRISPR-Cas9-directed mutagenesis whereby guide RNAs (sgRNAs) targeting essential domains show greater depletion than those targeting dispensable regions in competition assays (Shi et al., 2015) (see STAR Methods). To this end, sgRNAs targeting the 5′ exons of KDM2B and regions encoding the JmjC or the ZF-CxxC domains were introduced into five human synovial sarcoma cell lines expressing Cas9 (Figure 4A).
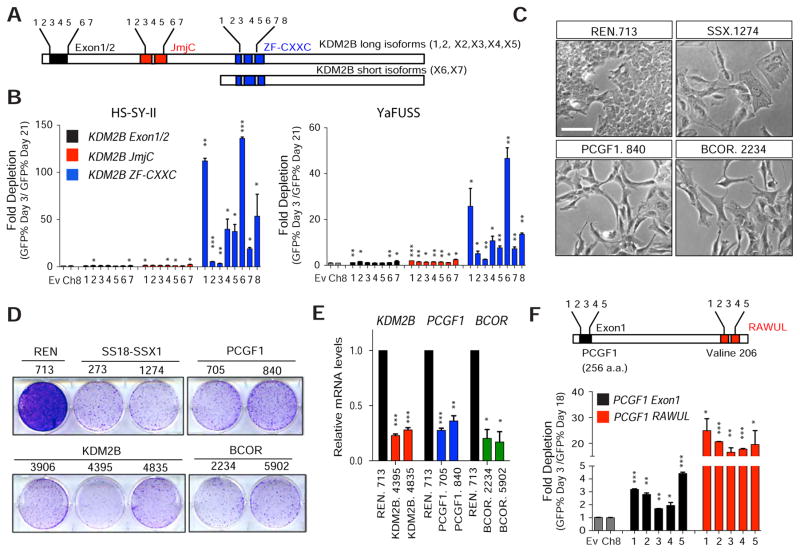
Figure 4. The DNA binding domain of KDM2B and the non-canonical PRC1.1 complex are essential for synovial sarcoma proliferation.
(A) Schematics showing human KDM2B JmjC and ZF-CxxC protein domains in long and short KDM2B isoforms and location of single guide RNAs (sgRNA). (B) Depletion assays based on the % of GFP+ cells at day 3 and at day 21 following sgRNA transduction. Error bars correspond to mean + s.d. of two independent experiments. (C) Bright field images of HS-SY-II cells transduced with the indicated shRNAs 10 days following shRNA induction. Scale bar=25 μm (D) Clonogenic assay of HS-SY-II cells transduced with the indicated shRNAs. (E) Quantitative RT-PCR for KDM2B, PCGF1 and BCOR expression. Error bars correspond to mean +z s.d. (n=2). (F) Schematics of guide RNAs designed against the first exon or against the RAWUL domain of PCGF1 (surrounding Valine 206). Depletion assays based on the % of GFP+ cells at day 3 and day 18 days following sgRNA transduction in HS-SY-II cells. Error bars correspond mean + s.d. (n=2). Unpaired t-test *p value<0.05, **p value<0.005, ***p value<0.0005. See also Figure S3.
Although most sgRNAs tested were potent at producing mutations (Figure S3A), sgRNAs targeting the ZF-CxxC motif showed the most significant depletion over time (Figure 4B and Figure S3B). These observations imply that the DNA binding domain of KDM2B, and not its histone demethylase activity, is crucial for sustaining synovial sarcoma proliferation and are reminiscent of the prevalent role of KDM2B’s ZF-CxxC in promoting gene repression in ESCs cells (He et al., 2013). Accordingly, expression of a JmjC-defective mutant (KDM2BH211A/H222A) or of a JmjC-deficient short KDM2B isoform was as effective as wild-type KDM2B at rescuing the proliferative arrest produced by KDM2B knockdown (Figures S3C–G).
PCGF1, RING1B, and BCOR (and the related protein BCORL1) are additional components of the PRC1.1 complex (Gearhart et al., 2006; Sanchez et al., 2007) that are recruited to unmethylated CGIs by KDM2B. PCGF1 is specific for PRC1.1 and determines the identity of PRC1-like assembly through its ring finger- and WD40-associated ubiquitin-like (RAWUL) domain (Junco et al., 2013). Intriguingly, like KDM2B, BCOR is overexpressed in human synovial sarcoma cell lines (Figure S2A) and patient samples (Kao et al., 2016), and two BCOR shRNAs also depleted in the initial screen or in follow up validation studies (Figure S1B; note that PCGF1 or BCORL1 shRNAs were not present in the library). Furthermore, shRNAs targeting PCGF1 or BCOR induced morphological changes and a proliferative arrest that phenocopied KDM2B knockdown in several human synovial sarcoma cell lines (Figures 4C–E) but did not affect proliferation of normal human fibroblasts (IMR90, Figure S3H). Reinforcing the importance of the PRC1.1 complex, sgRNAs targeting the RAWUL domain of PCGF1, required for PRC1.1 assembly, strongly depleted in CRISPR-Cas9 competition assays in HS-SY-II cells (Figure 4F and Figure S3I–J). We infer that KDM2B sustains synovial sarcoma through its PRC1.1-associated activity.
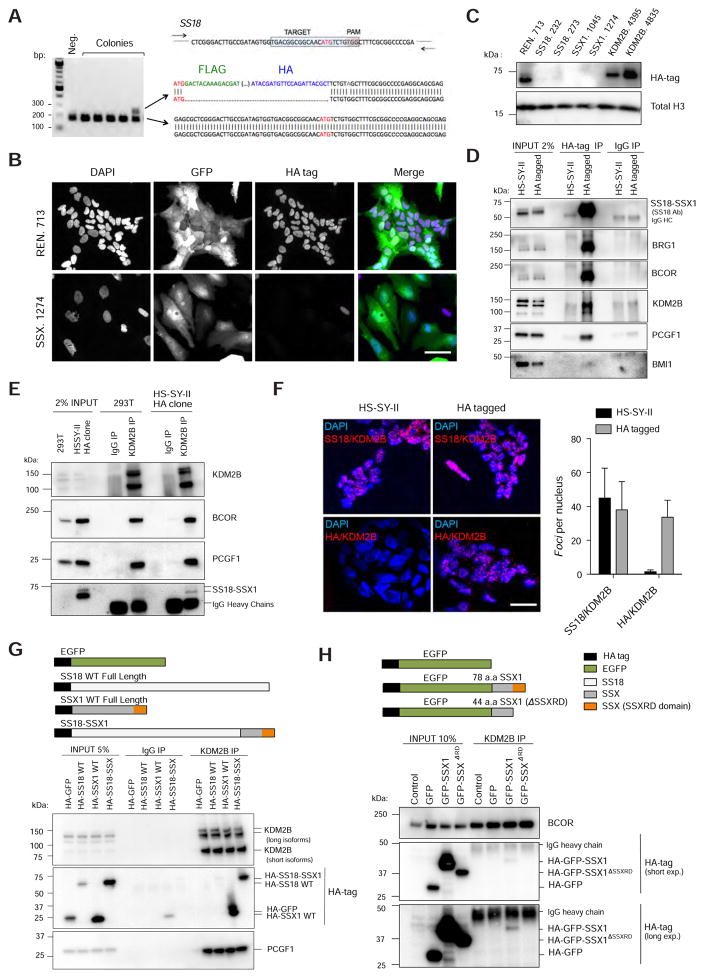
Tagging of endogenous SS18-SSX1 using CRISPR/Cas9 homology directed repair
We next set out to identify the mechanisms whereby SS18-SSX and KDM2B influence transcriptional regulation in synovial sarcoma cells. While studies on candidate target genes indicate that SS18-SSX represses the CDKN2A tumor suppressor locus (Su et al., 2012) and conversely activates SOX2 expression (Kadoch and Crabtree, 2013), the lack of fusion-specific antibodies suitable for chromatin immunoprecipitation (ChIP) has precluded the identification of its targets genome-wide. To overcome this limitation, we applied CRISPR/Cas9 mediated homologous directed repair to knock-in a FLAG-HA tag in the N-terminal coding region of SS18 in HS-SY-II human synovial sarcoma cells (see Star Methods). A positive clone was identified and confirmed to have edited the SS18-SSX1 allele without affecting the wild-type SS18 allele (Figure 5A). Immunofluorescence and immunoblotting confirmed that the HA epitope was depleted by SS18-SSX1 knockdown (Figures 5B and 5C). Importantly, these epitope-tagged cells retained sensitivity to SS18-SSX1 and KDM2B inhibition (Figure S4A).
Figure 5. Endogenous SS18-SSX1 interacts with PRC1.1.
(A) Endogenous SS18-SSX1 in the HS-SY-II human synovial sarcoma cell line was tagged with Flag-HA epitopes using CRISPR/Cas9-mediated homology-directed repair (HDR). An sgRNA targeting the region around the ATG and an ssDNA was used as template. For screening positive colonies PCR primers flanking the ATG site of SS18 were used (represented as arrows). (B) Immunofluorescence analysis showing nuclear staining using an anti HA-tag antibody. Scale bar=25 μm (C) Western blot analysis for HA tag in HA-SS18-SSX1 cells with the indicated shRNAs. (D) Co-IP analysis using an anti-HA tag antibody in HA-SS18-SSX1 clone. HS-SY-II parental cells were used as a negative control. (E) Co-IP using an anti-KDM2B antibody in HA-SS18-SSX clone and 293T cells. (F) Proximity ligation assay images and respective quantification verifying KDM2B and SS18-SSX in situ co-localization using an SS18 specific antibody or an HA tag antibody in untagged (parental) and HA-tagged HS-SY-II cells. Scale bar=25 μm. Error bars correspond to means + s.d. (n=3). (G) Co-IP in 293T expressing the indicated constructs using an anti-KDM2B antibody. (H) Co-IP using an anti-KDM2B antibody in 293T cells expressing GFP fused to the last 78 aminoacids of SS18-SSX1 (SSX1 fragment), and the same fragment lacking the SSXRD domain. See also Figure S4.
SS18-SSX1 interacts with KDM2B-PRC1.1 in human synovial sarcoma cells
We first tested whether SS18-SSX1 and PRC1.1 components interact by performing reciprocal co-immunoprecipitations (Co-IP) with antibodies targeting HA or KDM2B using buffers that contained DNAse to eliminate physical associations mediated by DNA. Like BRG1, KDM2B, BCOR, and PCGF1 were detected in anti-HA IPs of lysates from the HA tagged but not the parental cell line. We did not identify an interaction with BMI1 under the same experimental conditions, suggesting the interaction is specific to PRC1.1. (Figure 5D). SS18-SSX1 was also identified in IPs using a KDM2B antibody, as were PRC1.1 components (Figure 5E). The HA-tagged SS18-SSX1 protein also co-localized with KDM2B in cells as revealed by a proximity ligation assay (PLA) that allows “in situ” detection of two proteins closer than 40 nm (Soderberg et al., 2006) (Figure 5F). This signal was SS18-SSX1/2-specific and dependent: a strong PLA signal was observed in other synovial sarcoma lines using SS18 and KDM2B antibodies but not in MCF7 cells lacking the fusion (Figure S4B), and this signal was abolished upon SS18-SSX1 knockdown using an SSX targeting siRNA (Figure S4C). KDM2B co-immunoprecipitated with epitope-tagged versions of SSX1, but not SS18, in ectopic overexpression assays (Figure 5G), whereas deletion of the C-terminal SSX1 repressive domain (SSXRD)(Lim et al., 1998) in the context of the SS18-SSX1 fusion abolished this interaction (Figure S4D). Furthermore, addition of the SSX1 fragment of the fusion (last 78 amino acids) to GFP enabled it to associate with KDM2B, in an SSXRD-dependent manner (Figure 5H). Thus, SS18-SSX1 directly or indirectly associates with KDM2B via its C-terminal SSX repressive domain.
SS18-SSX1 and KDM2B co-occupy unmethylated CpG islands of developmental transcription factors
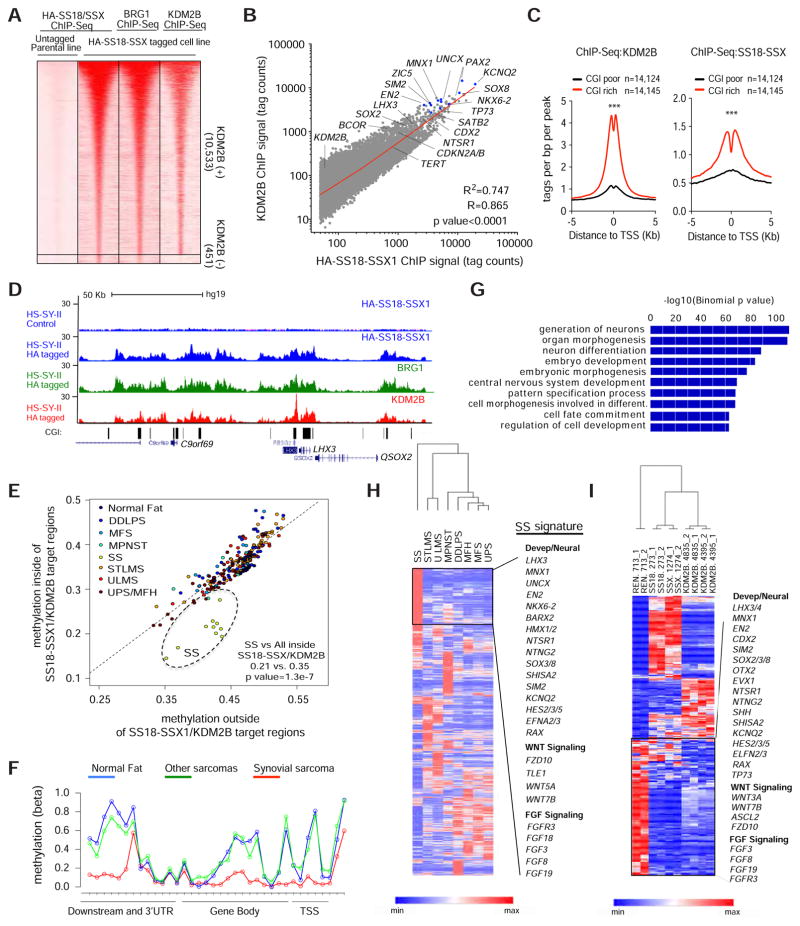
To determine whether SS18-SSX1 and KDM2B occupy similar loci genome-wide, we performed ChIP-Seq using antibodies targeting HA, BRG1 (a SWI/SNF component), and KDM2B in HA-tagged HS-SY-II cells. 10,984 specific SS18-SSX1-occupied regions were identified in HA-tagged cells and, as expected, these loci also contained BRG1 (Figure 6A). Remarkably, there was a quantitative and significant correlation between loci bound by SS18-SSX1 and those bound by KDM2B (Figure 6A and 6B). Likewise, most KDM2B-bound sites in these cells are also bound by SS18-SSX1 and BRG1 (84.4% and 97.0% respectively, Figure S5A).
Figure 6. SS18-SSX1 and KDM2B co-occupy and regulate genes that define a synovial sarcoma signature.
(A) Heat maps showing HA-SS18-SSX1, BRG1 and KDM2B ChIP-Seq signals over the 10,984 HA-enriched regions identified in HA-SS18-SSX tagged cells. Rows correspond to ±5 Kb regions across the midpoint of each HA-enriched region, ranked by increasing HA-SS18-SSX1 signal in the tagged clone. Color shading corresponds to the HA-SS18-SSX1, BRG1 and KDM2B ChIP-Seq read counts in each region. (B) Scatterplot of absolute HA-SS18-SSX1 and KDM2B signals (tag counts) at 10,533 SS18-SSX1/KDM2B co-occupied regions. (C) KDM2B and SS18-SSX1 binding profiles centered on TSS for CpG-rich promoters and CpG-poor promoters. (D) Gene tracks for HA-SS18-SSX1, BRG1 and KDM2B at CGI rich regions at the LHX3 locus. (E) Average methylation (beta) values for regions inside (y-axis) or outside (x-axis) SS18-SSX1/KDM2B occupied regions. Each data point corresponds to an individual patient sarcoma sample. Different sarcoma sub-types are indicated and color-coded. Synovial sarcomas (SS), undifferentiated pleiomorphic sarcomas/malignant fibrous histiocytoma (UPS/MFH), myxofibrosarcomas (MFS), malignant peripheral nerve sheath tumors (MPNST), uterine leiomyosarcomas (ULMS), soft tissue leiomyosarcomas (STLMS), dedifferentiated liposarcomas (DDLPS). (F) Methylation (beta) values across the LHX3 locus in SS compared with normal tissue (Fat) and all other sarcoma subtypes described in (E). (G) Gene ontology analysis of genes associated with all 10,533 SS18-SSX1/KDM2B occupied regions. (H) Unsupervised clustering based on mean RNA-Seq values of sarcoma sub-types analyzed by The Cancer Genome Atlas (TCGA). Synovial Sarcoma (SS, n=10), STLMS (n=72), ULMS (n=32), MPNST (n=10), DDLPS (n=58), MFS (n=25), Malignant fibrous histiocytoma (MFH n=30) and UPS (n=22). (I) Supervised clustering based on the top 200 genes differentially upregulated or downregulated upon KDM2B knockdown. C. Unpaired t-test, ***p value<0.0001. See also Figure S5–S6 and Tables S2–5.
Consistent with the ability of KDM2B to bind to and recruit proteins to CpG islands, SS18-SSX1 and KDM2B were bound predominantly to CpG-rich promoters (Figure 6C) and overlapped with annotated CGIs (Figure 6D, and Figure S5B) that are more hypomethylated in synovial sarcoma compared to normal fat or other sarcoma types (Figure 6E–F and Figure S5C). Furthermore, SS18-SSX1/KDM2B occupancy inversely correlated with DNA methylation levels as genes with the highest SS18-SSX1/KDM2B enrichment showed the lowest overall methylation levels (Figure S5D). Therefore, SS18-SSX1, SWI/SNF, and KDM2B broadly co-occupy genes linked to unmethylated CGIs, suggesting that KDM2B-mediated recognition of these regions is required for SS18-SSX activity.
High SS18-SSX1/KDM2B occupancy is associated with gene activation
Although the previously identified SS18-SSX1/2 targets CDKN2A/B (Su et al., 2012) and SOX2 (Kadoch and Crabtree, 2013) displayed SS18-SSX1 and KDM2B occupancy in synovial sarcoma cells, they were not among the most prominently bound loci identified in our analysis (Figure 6B). Rather, SS18-SSX1 and KDM2B co-occupancy was higher at genes encoding a series of homeobox transcription factors linked to neurogenesis and other developmental processes (EN2, LHX3, UCNX, PAX2, MNX1, ZIC5, KCNQ2, and SIM2) (Figure 6B, Figures S5B and Table S2)(Lee and Pfaff, 2001) and, accordingly, systematic gene ontology analysis using all co-occupied loci identified “neuron differentiation”, “embryo development” and “homeobox” among the most significant categories (Figure 6G and Table S3). Key genes present in these categories were also present in the set of differentially expressed genes that distinguished synovial sarcoma from other sarcoma types (Figure 6H, Figure S5E and Table S4, and the fact that many of these factors are linked to nervous system development likely explains the observation that neurogenesis-related genes are paradoxically upregulated in this disease (Baird et al., 2005; Nagayama et al., 2002). Interestingly, both BCOR and KDM2B are also SS18-SSX1/KDM2B targets, suggesting an auto-regulatory mechanism that could produce the high levels of PRC1.1 components observed in synovial sarcoma cells (Figure 6B and Table S2).
Transcriptional profiling of synovial sarcoma cells expressing either SS18-SSX1 or KDM2B shRNAs revealed a strong correlation between genes that were highly bound by both SS18-SSX1 and KDM2B and whose expression was significantly repressed upon SS18-SSX1 knockdown (Figure S5F). Although the magnitude of the expression changes was lower for KDM2B depletion, the impact of each perturbation on gene expression was remarkably similar (Figure S5G). Co-downregulated genes included the same developmental and neural factors described above, as well as genes involved in FGF and WNT signaling that have been implicated in synovial sarcoma (Barham et al., 2013; Ishibe et al., 2005; Trautmann et al., 2014) (Figure 6I and Figures S5H and Figure S6A–B and Table S5). In contrast, most co-upregulated genes were not SS18-SSX1 direct targets (Figure S6C) and included genes with lower SS18-SSX1/KDM2B-bound levels when compared to down-regulated genes (Figure S6D). Consistent with the results described in Figure 2F and 2G, up-regulated genes included those indicative of mesenchymal differentiation, including genes encoding certain extracellular matrix proteins, secreted factors and cytoskeleton-related proteins (Figures S6E and S6F and Table S5). Apparently, the SS18-SSX1/KDM2B collaboration produces the neural-like gene expression signature that is a hallmark of synovial sarcoma (Nagayama et al., 2002), and its disruption restores a more mesenchymal cell fate.
SS18-SSX1 recruitment to targets genes depends on KDM2B
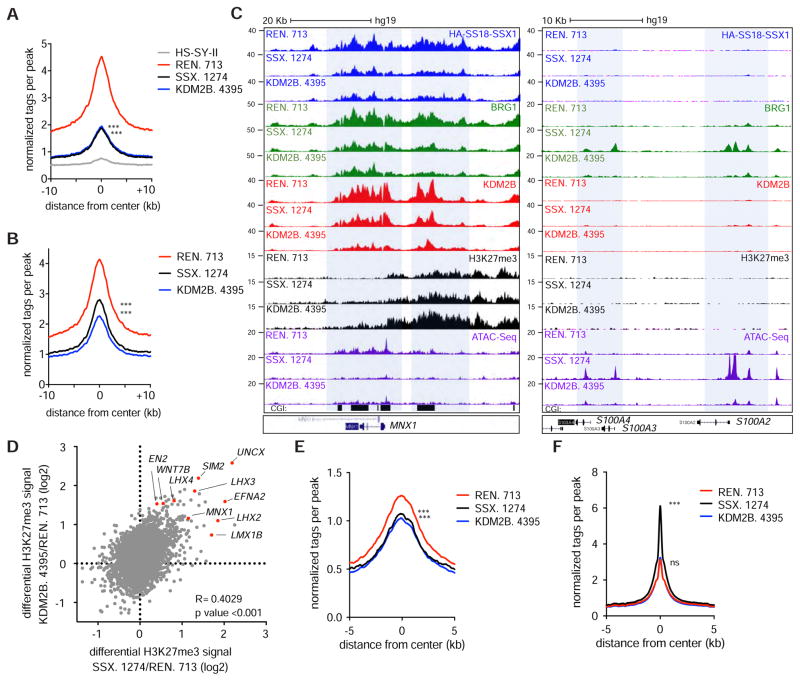
Our results raise the possibility that KDM2B-PRC1.1, via binding to CGIs, mediates SS18-SSX1 recruitment to chromatin thereby dictating target gene identity. To test this hypothesis, we profiled SS18-SSX1 and BRG1 occupancy following either SS18-SSX1 or KDM2B knockdown. As expected, SS18-SSX knockdown reduced the HA-SS18-SSX1 and BRG1 (i.e. SWI/SNF) signal at a large fraction of SS18-SSX1 target loci (Figures 7A–C and Figure S7A). More importantly, KDM2B depletion also reduced SS18-SSX1 and BRG1 binding to loci co-occupied by SS18-SSX1/KDM2B but not loci bound by SS18-SSX1 alone (Figure 7A–C and Figure S7B–D). Conversely, knockdown of either SS18-SSX1 or KDM2B triggered accumulation of the H3K27me3 repressive mark at genomic loci otherwise co-occupied by both proteins, including the same developmentally regulated transcription factors that were targets of the fusion oncoprotein (Figure 7D).
Figure 7. KDM2B recruits SS18-SSX to activate developmentally regulated genes otherwise subjected to polycomb-mediated gene repression.
(A, B) HA-SS18-SSX1 (A) and BRG1 (B) ChIP-Seq enrichment meta-profiles in REN. 713 (control shRNA), SSX.1274 and KDM2B. 4395 conditions representing the average read counts per 20 bp bin across a 20 Kb window centered on 4,567 SSX. 1274 sensitive regions. (C) Gene track for KDM2B, HA-SS18-SSX1 and H3K27me3 ChIP-Seq and ATAC-Seq at the MNX1 and S100A2/4 loci. (D) Scatterplot for differential H3K27me3 levels upon knockdown of SS18-SSX1 or KDM2B at SSX.1274 sensitive regions. Genes with highest gains in H3K27me3 are highlighted. (E, F) ATAC-Seq enrichment meta-profiles representing the average read counts per 20 bp bin across a 10 Kb window centered on 10,984 SS18-SSX1 occupied regions (E) and at 117,459 non-SS18-SSX1 occupied ATAC-Seq peaks (F). Unpaired t-test ***p value<0.0001. See also Figure S7.
That SS18-SSX1-containing SWI/SNF complexes localize to KDM2B-bound developmental genes expressed in synovial sarcoma cells implies that they promote their activation by increasing chromatin accessibility. To directly address this question, we performed Assay for Transposase-Accessible Chromatin sequencing (ATAC-Seq)(Buenrostro et al., 2013) in synovial sarcoma cells in the presence and absence of SS18-SSX1 or KDM2B. As predicted both SS18-SSX1 and KDM2B knockdown resulted in reduced ATAC-Seq signals at regions that were previously occupied by SS18-SSX1 (Figure 7E). These data support a model in which recruitment of SS18-SSX1-containing SWI/SNF complexes via KDM2B promote gene accessibility by opposing H3K27me3-mediated repression.
Surprisingly, SS18-SSX1 and, to a lesser extent, KDM2B knockdown also produced increases in BRG1 signals at a series of new loci associated with genes involved in skeleton and muscle development, including several of the mesenchymal genes we previously noted as upregulated by SS18-SSX1 or KDM2B knockdown (e.g. S100A4; Figure S7C–D). These regions overlapped with regions that gained gene accessibility (Figure 7C) and, accordingly, ATAC-Seq signals were generally increased at non-SS18-SSX1 occupied regions, particularly upon SS18-SSX1 depletion (Figure 7F and Figure S7E). Hence, both SS18-SSX1 and KDM2B inhibition in synovial sarcoma cells triggered a broad reduction of chromatin accessibility at SS18-SSX1/KDM2B targets and a redistribution of SWI-SNF complexes to new loci. Collectively, these data imply that SS18-SSX1 sustains synovial sarcoma by targeting SWI-SNF complexes to polycomb repressive sites via KDM2B. Consequently, KDM2B inhibition triggers cell cycle arrest and terminal differentiation by releasing SS18-SSX1 gene activation complexes and allowing the formation of a repressive chromatin environment at target loci.
DISCUSSION
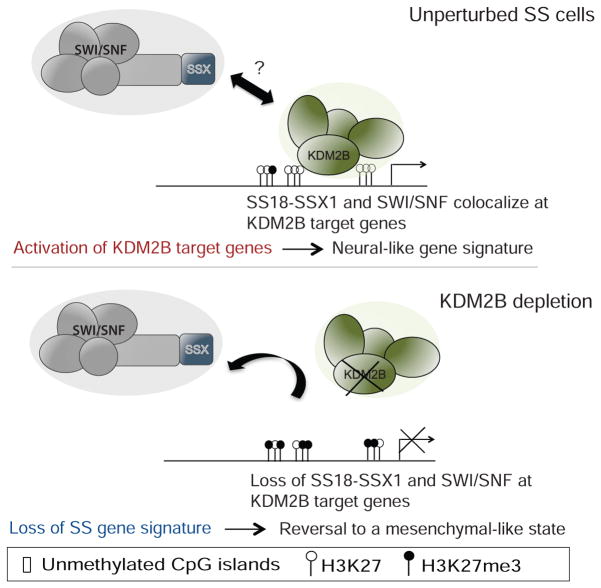
Through functional genomics and mechanistic studies, we identify KDM2B as an epigenetic dependency in synovial sarcoma and reveal how it mediates the oncogenic activity of SS18-SSX1. We propose that the KDM2B-containing PRC1.1 complex promotes recruitment of SS18-SSX1-containing SWI/SNF complexes to unmethylated CpG islands normally subject to polycomb-mediated repression. This process, in turn, enhances gene accessibility leading to aberrant activation of developmentally regulated genes that drive malignancy and underlies the unique transcriptional landscape observed in synovial sarcoma. KDM2B inhibition reverses this program by releasing SS18-SSX1 from chromatin, thereby enabling target gene silencing, re-acquisition of a mesenchymal-like expression program, and irreversible proliferative arrest (Figure 8). While the biochemical details of how SS18-SSX1/2 associates with PRC1.1 remains to be determined, it depends on the SSX fragment and its C-terminal SSXRD domain.
Figure 8. Proposed model for KDM2B-dependent SS18-SSX1 activity.
In synovial sarcoma KDM2B binds unmethylated CpG islands of developmental genes. KDM2B-PRC1.1 facilitates recruitment of SS18-SSX1 containing SWI/SNF complexes by direct or indirect interaction with SS18-SSX1 leading to increased gene accessibility and consequent aberrant activation of developmental genes that would otherwise be repressed. Upon KDM2B inhibition SS18-SSX1 binding is reduced, allowing H3K27me3 gains at a sub-set of SS18-SSX1 targets, reduced gene accessibility and consequent down-regulation of expression of developmental proteins and TFs, possibly allowing re-establishment of normal differentiation programs.
Previous work suggests that SS18-SSX can repress gene expression by enhancing PRC2 activity (Su et al., 2012) or, alternatively activate SOX2 expression by virtue of evicting PRC complexes and repressive marks (Kadoch and Crabtree, 2013). Importantly, these studies focused on single candidate loci and/or relied on ectopic overexpression of SS18-SSX1/2. Furthermore, until now, it has been unclear how SS18-SSX targets SWI/SNF to polycomb-repressed genes. By epitope tagging the endogenous SS18-SSX1 in human sarcoma cells, we were able to evaluate SS18-SSX1 occupancy and its impact on gene expression genome wide. We observe that the most prominent mechanism by which SS18-SSX1 alters target gene expression is by redirecting SWI/SNF to polycomb targets genome wide via KDM2B. Interestingly, KDM2B promotes gene silencing of developmental genes in embryonic stem cells and in some tumorigenic contexts (Andricovich et al., 2016; Farcas et al., 2012; He et al., 2013; Wu et al., 2013). However, in synovial sarcoma, SS18-SSX1 connects SWI/SNF to PRC1.1, turning a non-canonical repressive complex into a potent activator that sustains transformation.
In other cell types, SWI/SNF can oppose polycomb repression by binding and evicting RYBP-containing PRC1 complexes from chromatin (Stanton et al., 2017). We show that, in synovial sarcoma cells, SS18-SSX1 achieves the same end via an entirely different mechanism – by targeting KDM2B PRC1.1 complexes, the SS18-SSX1 fusion facilitates mislocalization of SWI/SNF complexes to polycomb target genes where, as previously described (Kadoch et al., 2017; Stanton et al., 2017), it opposes polycomb-mediated repressive activity. This mechanism leads to the aberrant activation of multiple neurogenic and other transcription factors, which likely explains the curious transcriptional profile associated with synovial sarcoma (Baird et al., 2005; Nagayama et al., 2002).
Our results do not rule out the possibility that altered SWI/SNF composition or gene repression mechanisms contribute to the oncogenic activity of SS18-SSX fusions. Biochemical studies strongly suggest that SS18-SSX1/2 containing SWI/SNF complexes have potentiated activity (Kadoch et al., 2017) and it seems likely that this can potentiate the activity of SS18-SSX1 containing SWI/SNF complexes that are recruited to PRC1.1 targets. In addition, while most repressed genes in SS18-SSX1-driven sarcoma cells are repressed via indirect mechanisms, our analysis identifies a subset of genes that are direct SS18-SSX1 targets. Most of those genes were also occupied by BRG1 and KDM2B, though the magnitude of SS18-SSX1 binding was substantially reduced compared to targets that are directly activated by SS18-SSX1. Perhaps other factors such as transcription factor availability, alterations in the balance of PcG to SWI/SNF activity at particular loci, or increased accessibility to transcriptional repressors ultimately dictate the expression output at SS18-SSX1/2 target genes. Nevertheless, and although SS18-SSX1/KDM2B can be associated with both gene activation and repression, our results suggest that the dominant activity of SS18-SSX1 is to promote gene activation to produce the synovial sarcoma-gene signature that defines and most probably drives the disease.
By virtue of its ZF-CxxC domain, KDM2B has the ability to specifically recognize non-methylated DNA and to recruit chromatin-modifying activities to CGI elements. Accordingly, in synovial sarcoma, SS18-SSX1/KDM2B bind CGI rich genes that are undermethylated in synovial sarcoma patient samples when compared with other sarcoma sub-types. Our model predicts that the impact of SS18-SSX fusions on gene expression will be dictated by the DNA methylation state of the target cell and is intriguing given previous work demonstrating that ectopic SS18-SSX1/2 expression can result in distinct biological outputs and gene expression profiles depending on the cell type (Tamaki et al., 2015). Our results may explain why SS18-SSX1/2 expression does not faithfully recapitulate its activity in synovial sarcoma cells when expressed in certain cell types, and underscore the importance of studying gene fusions in the pathological context in which they occur. By extension, the combination of functional genomics and CRISPR/Cas9-mediated epitope tagging used herein might be effective at elucidating the mechanism of action of other oncogenic fusions.
Since KDM2B protects CGIs from hypermethylation during embryonic development (Boulard et al., 2015) and is required for SS18-SSX recruitment to hypomethylated CGIs, it is possible that particular methylation states present in the cell of origin create a permissive state for SS18-SSX-driven transformation. Such a model parallels recent findings in Ewing sarcoma, in which DNA methylation patterns in patient samples potentially reflect the differentiation state of the cell-of-origin from which the tumor was originally derived (Sheffield et al., 2017). Consistent with this view, studies directing SS18-SSX2 transgenes to various cell types in the skeletal muscle lineage demonstrate that only a subset of cell types are sensitive to oncogenic transformation (Haldar et al., 2007). Regardless, the potential role of intrinsic DNA methylation states in SS18-SSX driven transformation may explain why synovial sarcoma occurs at a particular stage of human development.
Beyond KDM2B, our study reveals the broader importance of the PRC1.1 complex in synovial sarcoma. Indeed, like KDM2B, other PRC1.1-specifc complex proteins BCOR and PCGF1 co-immunoprecipitate with SS18-SSX1, and their disruption phenocopies KDM2B loss on synovial sarcoma cell proliferation and cell fate. Beyond synovial sarcoma, BCOR is translocated to different genes in undifferentiated pediatric sarcomas (BCOR-CCNB3, BCOR-MAML3, ZC3H7B-BCOR)(Peters, 2015; Pierron, 2012; Specht et al., 2016). Additionally, in-frame internal tandem duplications (ITDs) in the PUFD domain of BCOR that interacts with PCGF1 have recently been found in up to 85% of pediatric clear cell sarcoma of the kidney (Roy et al., 2015; Ueno-Yokohata et al., 2015) and in a class of primitive neuroectodermal tumors (CNS-PNET)(Sturm et al., 2016). The repercussions of these mutations on PRC1.1 complex activity are currently unknown, but their presence suggests that alterations in developmental programs controlled by this non-canonical complex broadly contribute to cancer and in particular, pediatric malignancies.
Due to the limited understanding of synovial sarcoma pathogenesis, clinical management consists primarily of surgery and radiotherapy and no effective targeted therapies are currently available. While targeting EZH2 activity are promising strategies for the treatment of SWI/SNF-deficient tumors owing to an increased dependence on PRC2 (Kim et al., 2015), our data suggest that, in synovial sarcoma, direct transcriptional repression is a less prominent feature of SS18-SSX action than its ability to promote gene activation via PRC1.1. As a consequence, alternative strategies to target KDM2B or the broader PRC1.1 complex would more broadly disrupt the genome-wide transcriptional impact of the fusion oncoprotein. While our results suggest that enzymatic inhibition of the histone demethylase function of KDM2B might not prove efficacious, targeted degradation of KDM2B protein, disruption of KDM2B DNA binding, and targeted inhibition of PRC1.1 complex assembly (e.g. by targeting PCGF1 RAWUL domain) (Junco et al., 2013) warrant further investigation. While challenging, our results imply that such approaches would have potent anti-tumor effects by disrupting aberrant differentiation programs and driving synovial sarcoma cells into a non-malignant cell fate.
STAR METHODS
Contact for reagent and resource sharing
Requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Scott Lowe (lowes@mskcc.org).
Experimental model and subject details
Cell lines
M5SS1 synovial sarcoma cells used for shRNA screen were derived from a murine synovial sarcoma and provided by KB Jones and MR Capecchi (Haldar et al., 2007). Human synovial sarcoma cell lines: HS-SY-II (Sonobe et al., 1992), YaFUSS (Ishibe et al., 2005), SYO-1 (Kawai et al., 2004), FUJI (Nojima et al., 1990) and Yamato-SS (Naka et al., 2010) cells were provided by M. Ladanyi and T. Nielsen. Cells were authenticated by quantitative PCR detection of SS18-SSX1/2, which is specific to synovial sarcoma (Figure S2A). Murine myoblasts (C2C12) and human diploid fibroblasts (IMR90, passage 11) were purchased from the American Type Culture Collection (ATCC). Cells were maintained in a humidified incubator at 373°C with 5% CO2, grown in DMEM supplemented with 10% FBS and 1003IU/ml penicillin-streptomycin.
Patient tumor Samples
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded tissue microarrays (TMAs) from the University of British Columbia: TMA 01-003 (synovial sarcoma and differential diagnoses, 82 cases in duplicate) (Nielsen et al., 2003); TMA 03-008 (chondroid tumors, 121 cases in duplicate) (Ng et al., 2005); TMA 06-007 (myxoid liposarcomas, 69 cases in triplicate) (Cheng et al., 2009); TMA 09-006 (epithelioid sarcoma and differential diagnoses, 53 cases in duplicate)(Pacheco and Nielsen, 2012); TMA 10-009 (8 alveolar soft part sarcomas, 2 alveolar rhabdomyosarcomas, 2 desmoplastic small round cell tumors, in triplicate) (Pacheco and Nielsen, 2012); TMA 12-004 (BCL2-positive tumors, 35 cases in triplicate) (Endo et al., 2014); TMA 12-006 (translocation-associated sarcomas, 10 cases in duplicate) (Endo et al., 2014); TMA 12-010 (5 dedifferentiated liposarcomas and 5 undifferentiated pleomorphic sarcomas, in duplicate)(Endo et al., 2014); TMA 14-006 (4 myxoid liposarcomas, 3 myxofibrosarcomas, 3 chondrosarcomas, 1 synovial sarcoma, 1 malignant peripheral nerve sheath tumor, in duplicate) (Endo et al., 2015); TMA 14-007 (dedifferentiated liposarcomas with well-differentiated areas, both components for 57 cases in duplicate); and TMA MPNST (malignant peripheral nerve sheath tumor and differential diagnoses, 176 cases in duplicate) (Terry et al., 2007). Cores measuring 1.0 mm (TMA 14-007) or 0.6mm (all other TMAs) in diameter were derived from representative viable tumor tissue, as identified by a bone and soft tissue subspecialty pathologist (TO Nielsen). TMAs were cut to 4-μm-thick sections, mounted to Fisherbrand™ Superfrost™ Plus charged glass slides (Thermo Fisher Scientific Inc, Waltham, MA), and incubated for 1 h at 60°C. (see methods details for details on Immunohistochemical staining and analysis). Accession of archival patient materials was approved by the Clinical Research Ethics Board of the British Columbia Cancer Agency (protocol H06-00013); due to the historical nature of tissue blocks, the requirement for informed consent was considered infeasible for these retrospective immunohistochemistry studies, and a waiver of consent was approved in this case.
DNA methylation and gene expression analysis of human sarcomas
RNA-Seq and DNA methylation data were from the TCGA Sarcoma cohort (Cancer Genome Atlas Research Network. Electronic address and Cancer Genome Atlas Research, 2017) spanned multiple major sarcoma types. RNA-Seq data comprised 259 samples: synovial sarcoma (SS, n=10), soft tissue leiomyosarcoma (STLMS, n=72) uterine leiomyosarcoma (ULMS, n=32) malignant peripheral nerve sheath tumors (MPNST, 10), dedifferentiated liposarcoma (DDLPS, 58), myxofibrosarcoma (MFS, n=25) malignant fibrous histiocytoma (MFH, n=30) and undifferentiated pleiomorphic sarcomas (UPS, n=22). DNA methylation data comprised 206 samples in the TCGA Sarcoma cohort (Cancer Genome Atlas Research Network, 2017) including dedifferentiated liposarcoma (DDLPS, n=50), uterine leiomyosarcoma (ULMS, n=27), soft-tissue leiomyosarcoma (STLMS, n=53), undifferentiated pleomorphic sarcoma, including malignant fibrous histiocytoma (UPS/MFH, n=44), myxofibrosarcoma (MFS, n=17), malignant peripheral nerve sheath tumor (MPNST, n=5) and synovial sarcoma (SS, n=10). We used Level 3 data that had been pre-processed using standard TCGA protocols. DNA methylation from the Illumina Human Methylation 450K platform for 15 samples of non-cancerous, subcutaneous adipose tissue (pre gastric-bypass) from 15 patients made publicly available (Benton et al., 2015) through the EBI ArrayExpress database (E-MTAB-3052) was used for comparison purposes.
Animal studies
All mouse experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee. Female 5- to 7-week-old athymic Nude-Foxn1nu (Envigo) mice were used for animal experiments with HS-SY-II and SYO-1 human cell lines. To generate xenografts, HS-SY-II and SYO-1 cells were transduced with LT3GEPIR inducible shRNA vectors and selected with puromycin as described in the method details section. Cells (10×106) were harvested on the day of use and injected in growth-factor-reduced Matrigel/PBS (50% final concentration). Each mouse flank was injected subcutaneously. Following inoculation, mice were fed a doxycycline diet (Harlan Laboratories) and monitored daily. Caliper measurements started when tumors became visible. Tumor volumes were calculated using the following formula: tumor volume = (D × d2)/2, in which D and d refer to the long and short tumor diameter, respectively. For immunohistochemistry analysis of HS-SY-II and SYO-1-derived xenografts, tumors were harvested at the final time point of measure. Tissues were fixed overnight in 4% PFA, embedded in paraffin, and cut into 5μm sections. Sections were subjected to haematoxylin and eosin staining, and immunohistochemical staining following standard protocols using an anti-GFP antibody (Cell Signaling, 2956, 1:500).
Method Details
Pooled RNA interference screen
An improved version of a previously described custom shRNA library (Zuber et al., 2011b) targeting 400 (six shRNAs per gene) mouse epigenetic regulators was cloned into the LMN (LTR-miR30-PGK-NeoR-IRES-GFP) vector and transduced in triplicates into M5SS1 mouse synovial sarcoma cell line (Haldar et al., 2007) and C2C12 mouse myoblasts (ATCC). T0 samples (n = 3) were collected three days following transduction. Following sixteen days of culture (Tf), about 30 million shRNA-expressing cells per replicate were collected. Genomic DNA from T0 and Tf samples was isolated and deep-sequencing template libraries were generated by PCR amplification of shRNA guide strands as previously described (Zuber et al., 2011a). Underrepresented shRNAs (<100 normalized reads) at the T0 were discarded resulting in a total of 2307 shRNAs for further analysis (see Supplementary information - Table 1 for a list of all shRNAs and corresponding reads). Using election criteria that required an shRNA depletion averaging greater than twofold at Tf, at least two shRNAs targeting the same gene and for scoring to be specific to M5SS1 cells, and not in C2C12, 14 shRNAs were identified (hSS18, KDM2B (FBXL10), BRD3, BRD7 and PADI4. Please see Table S1 for a list of all shRNAs and corresponding reads in the screen.
Plasmids and viral transduction
All vectors were derived from the Murine Stem Cell Virus (MSCV, Clontech) retroviral vector backbone. miRE-based shRNAs were designed and cloned as previously described (Zuber et al., 2011a) into the LT3GEPIR (TRE3G-GFP-miRE-PGK-PuroR-IRES-rtTA3) vector (Fellmann et al., 2013) (Table S6). The mouse KDM2B cDNA was subcloned from pVXL-Tomato-KDM2B vector (provided by A. Tzatsos) into MSCV-Neo using an EcoRI site. The JmjC and ZF-CxxC were generated from the wild-type KDM2B vector by site directed mutagenesis (Q5 site-directed mutagenesis kit, New England Biolabs). The short isoform of KDM2B was amplified by PRC from M5SS1 cDNA and cloned into MSCV-hygro. All constructs were verified by sequencing. For CRISPR editing constructs see CRISPR/Cas9 genome editing section. Lentiviruses were produced by co-transfection of 293T cells with 10 ug LT3GEPIR construct and helper vectors (6.5ug psPAX2 and 2.5ugVSV-G). For retroviral infection 293T-gag-pol cells were transduced with 20 ug of MSCV vectors and 2.5ug of VSV-G. Transfection of packaging cells was performed using Polyethylenimine (PEI) (Polysciences, 23966-2) by mixing with DNA in a 3:1 ratio. Viral supernatants were collected 48 after transfection, filtered thorough a 0.45 um filter (Millipore) and supplemented with 4 ug/ml of polybrene (Sigma) before adding to target cells.
Competitive proliferation assays and clonogenic assays
For shRNA experiments, human or mouse cells were modified by retroviral or lentiviral transduction followed by drug selection (2μg/ml Puromycin or 100 ug/ml Hygromycin B). LT3GEPIR-Puro-shRNA transduced cells were treated with 1μg/ml doxycycline to induce shRNA expression. For competitive proliferation assays, shRNA-transduced cells were mixed with non-transduced cells (in about a 8:2 ratio) and cultured with doxycycline. The relative percentage of GFP+ cells was determined at day 2 after doxycycline (T0) and after 15–18 days in culture (Tf) (results are relative to T0). The quantification of fluorescent cells was monitored on a Guava EasyCyte (Millipore). Experiments were performed independently two times. For clonogenic assays, cells were seeded in duplicate (10 × 103 to 30 × 103 cells per well of a 6-well plate) and cultured in the presence of doxycycline in complete media for 10–14 days. Cells were fixed with glutaraldehyde (0.5%), stained with 0.5% crystal violet, and photographed using a digital scanner. All experiments were performed at least three times. Representative experiments are shown.
sgRNA design and CRISPR/Cas9 competition assays
To evaluate the role of KDM2B protein domains we exploited the fact that in-frame deletions produced by CRISPR-Cas9-directed mutagenesis (which occur in ~1/3 cases) are more deleterious to protein function when they disrupt critical domains (Shi et al., 2015); consequently, in competition assays, single guide RNAs (sgRNAs) targeting domains crucial for cell proliferation or survival are more depleted than those targeting dispensable regions. sgRNAs targeting the 5′ exons of KDM2B, the regions encoding the JmjC domain, as well as the ZF-CxxC domain (Figure. 4A) were evaluated in 5 human synovial sarcoma cell lines. sgRNAs were cloned by annealing two DNA oligos and ligating into a BsmB1-digested U6-sgRNA-EFS-GFP vector (Addgene #57822). sgRNAs in were designed using http://crispr.mit.edu/andBenchling (https://benchling.com). The majority of sgRNAs used in this study had a quality score above 70 to minimize off-target effects. sgRNAs were designed to target 5′ coding exons of each target gene or functional domains of each protein based on the NCBI database annotation (Table S6). Synovial sarcoma cell lines were transduced with lentiCas9-Blast (Sanjana et al., 2014)(Addgene #52962) and selected using 5ug/ml of blasticidin to generate stable Cas9-expressing cell lines. Cells were consequently transduced with pLKO5.sgRNA.EFS.GFP (Heckl et al., 2014)(Addgene plasmid # 57822) to about 80% transduction efficiency. Quantification of GFP fluorescent cells was monitored on a Guava Easycyte (Millipore) from day 3 following transduction. Fold depletion was calculated as previously described (Shi et al., 2015). T7 assays were performed to evaluate CRISPR/Cas9-mediated gene editing (Table S6). Experiments were performed in two independent duplicates for all cell lines.
Tagging of Endogenous SS18-SSX1 Using CRISPR/Cas9-Mediated Homologous Directed Repair (HDR)
HS-SY-II cells were transfected using lipofectamine 3000 (Thermo Fisher) with pX458-sgSS18 (encoding Cas9, GFP and a sgRNA targeting the N-terminal region of the SS18 gene) (Table S6) and a single stranded DNA template (ssDNA) containing the ATG-FLAG-HA sequence flanked by homology arms to the N-terminal region of SS18:
Sequence for the ssDNA Template Containing Tags and Homology Arms with Mutated PAM Sequence (Silent Mutation)
ggccggcgtctctcccccagtttgccgttcacccggagcgctcgggacttgccgatagtggtgacggcggcaacATG GAC TAC AAA GAC GAT GAC GAC AAG GCC GCA TAC CCA TAC GAT GTT CCA GAT TAC GCT TCT GTA GCT TTC GCG GCC CCG AGG CAG CGA GGC AAG GGG GAG ATC ACT CCC GCT GCG ATT CAG AAG
Three days following transfection cells were single-cell sorted into 96-well plates, for further analysis of single cell clones. Clones were analyzed by immunofluorescence against HA-tag and PCR detection of targeted genomic regions using primers surrounding the ATG region of the SS18 gene (see Table S6). Positive clones were further evaluated by sequencing of PCR amplified genomic regions surrounding the SS18 N-terminal region.
Co-Immunoprecipitation
For co-immunoprecipitation a Nuclear complex Co-IP kit (Active Motif) was used. Cells were collected in cold PBS with phosphatase inhibitors and lysed in hypotonic buffer for 10 min. Isolated nuclei were further incubated in digestion buffer containing DNase for 90 min at 4C. Nuclear lysates were cleared by centrifugation and quantified using DC Protein assay (BioRad); 250–500 μg of protein was incubated with 3μg of antibody (KDM2B Millipore 09-864: HA–tag Cell Signaling 3956, normal rabbit IgG: Santa-Cruz Biotechnologies, sc-2027) in low stringency IP buffer containing 150mM NaCl, 1% detergent and protease inhibitors; and incubated overnight at 4°C with rotation. Next day Protein A/G magnetic beads were washed in low stringency IP buffer and incubated with the immunoprecipitation for 2 hours at 4°C under rotation. Following incubation, beads were washed 3 times in low stringency IP buffer containing BSA and 3 times in low stringency IP buffer without BSA, and boiled in loading dye for 5 minutes, before western blot analysis. Antibodies against PCGF1 (Santa Cruz, 515371), SS18 (Santa Cruz, 390266), BCOR (Santa Cruz, sc-514576) KDM2B (Millipore, 09-864), HA-tag (Cell Signaling, 3724), BRG1 (Santa Cruz, H-88) and BMI1 (Millipore, 05-637) were used. For experiments using transient expression in 293T cells, gene fragments encoding HA tagged versions of SS18, SSX1 wild type versions, SS18-SSX1 C-terminal deletions mutants and GFP fusions, were cloned into EcoRI and XhoI sites in MSCV-puro or pLVX-puro plasmids. 10ug of plasmid was transfected into 293T cells using PEI (as described above), and cells were collected for co-IP analysis 48hours after transfection.
Proximity ligation assay (PLA)
Indicated synovial sarcoma cell lines were seeded at 3 × 104 cells/well in culture treated 8-well chamber slides and treated as previously described (Laporte et al., 2016). Primary antibodies for PLA were used at 1/1000 dilution: SS18 (Santa-Cruz Biotechnologies, sc-28698), KDM2B (Abnova, H00084678-M09), HA (Santa-Cruz Biotechnologies, sc-805). Proximity ligation was performed utilizing the Duolink® In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich, DUO92101-1KT) according to the manufacturer’s protocol. Fluorescence was detected using a Zeiss Axioplan2 microscope at 40x. Images were quantified in triplicate using ImageJ software (NIH) as foci per nucleus, defined as the number of interaction points counted per nucleus. For PLA analysis upon SS18-SSX knockdown, duplex oligo (sense, CAAGAAGCCAGCAGAGGAATT; antisense, UUCCUCUGCUGGCUUCUUGTT) SS18-SSX siRNAs were designed to target the SSX portion of SS18-SSX using the Integrated DNA Technologies RNA interference (RNAi) design tool, and synthesized by Integrated DNA Technologies (IDT). HS-SY-II cells were seeded in 6-well plates. At 60% confluence, cells were transfected with 50pmol siSS18-SSX and 9 μL Lipofectamine RNAiMAX transfection reagent (Invitrogen) in Opti-MEM serum free media (Life Technologies). Protein was harvested 48-hours post transfection, and knockdown confirmed by western blot with an SS18 antibody (Santa Cruz Biotechnologies, sc-28698).
Immunofluorescence and Immunoblotting
Cells were seeded in 96-well imaging plates and fixed the following day with 4% paraformaldehyde (PFA) for 15 minutes. Permeabilization was performed using TritonX (0.1%in PBS) for 10 min followed by incubation with blocking solution (1% BSA, 0.1% Gelatin Fish in PBS) for 30 min. Incubation with Anti-HA-tag antibody (Cell Signaling, 3724, 1:1000) or α-SMA (Sigma clone 1A4, 1:2000) was performed in blocking buffer for 2 hours at RT. Cells were washed, incubated with secondary anti-rabbit Alexa-594 (Invitrogen) and counter stained with DAPI. Image acquisition was performed using IN Cell analyzer 6000 (GE Healthcare Life Sciences). For protein lysates cells were incubated with RIPA buffer supplemented with protease inhibitors (Protease inhibitor tablets, Roche) for 30 min and cleared by centrifugation (15 min 14.000 rpms 4C). Protein was quantified using the DC protein assay (BioRad). The following antibodies were used for immunoblotting: β-ACTIN (ac-15, Sigma), KDM2B (Millipore, 09-864) HA-tag (Cell Signaling, 3724) and Myc-tag (Cell Signaling, 2276).
RNA expression analysis
For quantitative RT-PCR, total RNA was isolated using the RNeasy Mini Kit (Qiagen), and cDNA was obtained using the TaqMan reverse transcription reagents (Applied Biosystems). Real-time PCR was performed in triplicate in two independent experiments using SYBR Green PCR Master Mix (Applied Biosystems) on the ViiA 7 Real-Time PCR System (Invitrogen). β-actin served as an endogenous normalization control. Gene-specific primer sets for mouse and human sequences were designed using NCBI’s qPrimerDepot (http://primerdepot.nci.nih.gov) or described elsewhere (Table S6). For high throughput RNA sequencing, total RNA from two independent experiments (and two shRNAs per gene) was extracted using an RNeasy minikit (Qiagen). Cells transduced with the indicated shRNAs were collected 12 days post-infection. RNA-Seq library construction and sequencing were performed at the integrated genomics operation (IGO) Core at MSKCC according to standard protocols. Poly-A selection was performed. For sequencing approximately 10 million 50 bp paired-end reads were acquired per replicate condition. Resulting RNA-Seq data was analyzed by removing adaptor sequences using Trimmomatic (Bolger et al., 2014). RNA-Seq reads were then aligned to GRCh37.75 (hg19) with STAR (Dobin et al., 2013) and genome-wide transcript counting was performed by HTSeq (Anders et al., 2015) to generate a matrix of fragments per kilobase of exon per million fragments mapped (RPKM). Gene expressions of RNA-Seq data were clustered using hierarchical clustering based on one minus Pearson correlation test using Morpheus (https://software.broadinstitute.org/morpheus/). Gene ontology of shSS18-SSX and shKDM2B co-regulated genes was performed using David functional annotation tool (https://david.ncifcrf.gov/); using a cut off of 2- fold difference in both conditions. For pathway enrichment analysis, the weighted GSEA Pre-ranked mode was used (http://www.broadinstitute.org/gsea).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as previously described (Hatzi et al., 2013). Briefly, HS-SY-II cells were fixed with 1% formaldehyde for 15min and the cross-linking reaction was stopped by adding 125mM glycine. Cells were washed twice with cold PBS and lysed in swelling buffer (150mM NaCl, 1%v/v Nonidet P-40, 0.5% w/v deoxycholate, 0.1% w/v SDS, 50mM Tris pH8, 5mM EDTA) supplemented with protease inhibitors. Cell lysates were sonicated using Covaris E220 Sonicator to generate fragments less than 400 bp. Sonicated lysates were centrifuged, and incubated overnight at 4°C with specific antibodies (BRG1 Abcam 110641; KDM2B Millipore 17-10264, HA-tag Abcam 9110; H3K27me3 Millipore 07-449). Immunocomplexes were recovered by incubation with 30ul protein A/G magnetic beads (Thermofisher) for 2h at 4°C. Beads were sequentially washed twice with RIPA buffer, increasing stringency ChIP wash buffers (150mM NaCl, 250mM NaCl, 250mM LiCl) and finally TE buffer. Immunocomplexes were eluted using elution buffer (1% SDS, 100mM NaHCO3) and cross-linking was reverted by addition of 300mM NaCl and incubation at 65°C overnight. DNA was purified using PCR purification kit (Qiagen). For HA-tag, BRG1 and KDM2B ChIP the same protocol was used with small modifications: cells were pre-fixed for 20 minutes with 1.5mM ethylene glycol bis(succinimidyl succinate) (EGS) (Thermo Scientific) and the washing step containing 250mM LiCl was omitted to increase yield without compromising specificity (as shown by absence of HA ChIP signal in HS-SY-II parental untagged cells). For ChIP-qPCRs a fraction of the ChIP product was used as template in 15 ul real time PCR reactions using SYBR Green PCR Master Mix (Applied Biosystems) on the ViiA 7 Real-Time PCR System (Invitrogen). Input chromatin was used for estimation of relative enrichment. Primer sequences for ChIP-qPCRs are summarized in Table S6.
Assay for Transposase-Accessible Chromatin sequencing (ATAC-Seq)
ATAC-Seq was performed as previously described (Buenrostro et al., 2013). Fifty thousand GFP positive cells were sorted by fluorescence-activated cell sorting (FACS). Cells were lysed in lysis buffer (10mM Tris, pH 7.4; 10mM NaCl; 3mM MgCl2; 0.1% (v/v) IGEPAL) and centrifuged for 10 min (500 x g) to isolate the nuclear fraction. The transposition reaction was performed for 30 minutes at 37°C using the Tn5 Transposase kit from Nextera accordingly to the manufacture’s instructions. Transposed DNA fragments were amplified by PCR using barcoded primers (Buenrostro et al., 2013) and the NEBNext High Fidelity 2X master mix (12 PCR cycles). Amplified libraries were purified using Qiagen MinElute, analyzed using Bioanalyser and combined for Illumina High-throughput sequencing.
ChIP-Seq Library Preparation, Illumina Data Analysis and Peak Detection
ChIP-Seq libraries were prepared at the Center for Epigenetic Research (MSKCC) using the NEBNext® ChIP-Seq Library Prep Master Mix Set for Illumina® (New England BioLabs) following the manufacture’s instructions. Raw reads were trimmed for quality and Illumina adapter sequences using ‘trim_galore’, then aligned to human genome assembly hg19 using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2)(Langmead and Salzberg, 2012) with the default parameters. To avoid clonal artifacts introduced in the library amplification steps, duplicated mapped reads were further discarded using the Picard tool MarkDuplicates (http://broadinstitute.github.io/picard/).
For further analyses, HOMER suit of tools was used (Heinz et al., 2010). Aligned bam files were subjected to peak calling using findPeaks tools with the default setting, except -style histone was implemented to find for broad regions of H3K27me3 peaks. Once ChIP-Seq peaks were identified from HA-SS18/SSX and H3K27me3 ChIP-Seq experiments with individual shRNA expression, all peaks were combined and merged if the maximum distance between peak centers were smaller than 100 bp. With these combined peaks, tag counts were re-calculated and normalized to 10 million reads. The peaks with lower than 50 tag counts of HA-SS18/SSX or H3K27me3 were excluded from further analyses, which finally yielded union peaks of HA-SS18-SSX (n=10,984) and H3K27me3 (n=13,226) ChIP-Seq experiments.
To identify HA-SS18/SSX and KDM2B co-occupancy, tag counts of KDM2B ChIP-Seq experiments were calculated from 10,984 peaks of HA-SS18/SSX ChIP-Seq. If the normalized tag counts of KDM2B were lower than 20, they were considered as HA-SS18/SSX(+) KDM2B(−) regions (n=451), and, if not, considered as HA-SSX/SSX (+) KDM2B (+) regions (n=10,533). To visualize ChIP-Seq tracks, normalized bigWig files were generated with makeBigWig tool. To create metagene plots in Figure 6A, +/− 10 kb from peak center was aligned and binned with 25 bp with annotate Peaks tool, then visualized with Java Tree view. To generate custom gene sets from ChIP-Seq data, genes with the closest TSS from each ChIP-Seq peak were assigned as peak-associated genes or using the GREAT tool (McLean et al., 2010) as explained bellow.
Gene Ontology (GO) analysis of ChIP-Seq regions
GREAT tools were used to predict GO terms of ChIP-Seq enriched regions (http://bejerano.stanford.edu/great/public/html/index.php). 10,533 SS18-SSX and KDM2B co-occupied regions were tested to define associated genes suing GREAT’s default settings (basal plus extension; proximal 5,000-bp upstream and 1,000-bp downstream) but using a more stringent distal extension of 100 kb instead of 1000 kb to include distal regulatory regions but minimize chances of assigning incorrect genes to each regulatory region. This analysis revealed a total of 3883 genes associated with SS18-SSX/KDM2B-bound regions. GO terms from the GO consortium (http://geneontology.org) the molecular signature database (http://software.broadinstitute.org/gsea/msigdb) were plotted as bar graphs with binomial −log10 p values (Figure 6G) or are presented in Table S3. Only the top twenty terms for each category are shown. The total number of genes assigned to 10,984 SS18-SSX-bound regions using GREAT (3928 genes) is presented in Table S2.
Immunohistochemistry of sarcoma tissue microarrays (TMAs)
Immunohistochemical staining was performed using the Ventana DISCOVERY® ULTRA semi-automated staining system (Ventana Medical Systems Inc, Tucson, AZ). Briefly, heat-induced antigen retrieval was performed using the standard Cell Conditioning 1 (CC1, Ventana) protocol. Sections were incubated with goat anti-KDM2B polyclonal antibody (S-15, SantaCruz Biotechnology Inc, Santa Cruz, CA) at 1:25 dilution in DISCOVERY antibody diluent (Ventana) for 2h at room temperature, followed by incubation with AffiniPure rabbit anti-goat IgG (H+L) unconjugated secondary antibody (Jackson ImmunoResearch Laboratories Inc, West Grove, PA) for 32 min at 37°C. Chromogen visualization was performed using the ChromoMap DAB Kit UltraMap anti-rabbit tertiary antibody (Ventana). Slides were counterstained with hematoxylin and mounted. Digital images of immunostained tissue microarrays were acquired using a BLISS imaging microscope (Bacus Laboratories, Lombard, IL, USA). For quantification and statistical analysis, a pathologist experienced in scoring biomarkers in bone and soft tissue tumors scored each core separately, and the average score from all cores of the same tumor was calculated. KDM2B immunopositivity was scored in a semi-quantitative manner for the intensity (0=negative, 1=weak positive, 2=moderate positive, 3=strong positive) and for the percentage of positive sarcoma cell nuclei. These were multiplied to generate an H-score. A Kruskal-Wallis 1-way ANOVA test was used to assess the differences between histological sub-types. Statistically significant difference was defined as p<0.05. Data analysis was performed using IBM® SPSS® statistics software (version 26).
Analysis of DNA methylation and RNA-Seq data from TCGA
RNA-Seq and DNA methylation data were from the TCGA Sarcoma cohort (Cancer Genome Atlas Research Network. Electronic address and Cancer Genome Atlas Research, 2017). RNA-Seq data was downloaded TCGA portal (https://portal.gdc.cancer.gov/projects/TCGA-SARC) (259 samples). Normalized gene counts means values for each sarcoma subtype were used to define an SS gene signature (Table S4). DNA methylation data from the Illumina Human Methylation 450K platform was downloaded from the UCSC Cancer Genome Browser for 206 samples in the TCGA Sarcoma cohort, as described in Experimental model and subject details. We discarded all the probes that were masked as NA (‘Not Available’) for more than 90% of the TCGA samples. A probe is masked as NA at level three of the TCGA database if (a) the detection p value is greater than 0.05 (which means that the measured signal is not significantly different from background), (b) the probe contains known SNPs after comparison with the dbSNP database or (c) the probe contains DNA sequences of known repetitive elements in more than 10 bp of each 50 bp probe sequence. A total of 6,412 regions of the 10,533 regions co-occupied by both SS18-SSX and KDM2B overlapped with at least one of the Illumina probes that remained in the array. For the results shown in Figure 6E, we calculated average DNA methylation levels per sample in co-occupied regions by first computing the average beta value over the 61,577 probes that overlap any of the 6,412 co-occupied regions. DNA methylation levels per sample outside of co-occupied regions were estimated as the average beta value over all the 309,688 probes that did not overlap any of the 6,412 co-occupied regions. For the results shown in Figure S5D, we computed average levels of DNA methylation per region across the 10 synovial samples in the TCGA cohort for each one of the 6,412 regions that overlapped at least one probe in the array. We then compared average levels on DNA methylation in the 500 co-occupied regions with the highest minimal co-occupancy score vs. average DNA methylation levels in the 500 co-occupied regions with the lowest minimal co-occupancy score. For each region, the minimal co-occupancy score was defined as the minimum value in the pair of SS18-SSX and KDM2B ChIP occupancy scores.
Quantification and statistical analysis
Data are expressed as mean ± s.d. or ± s.e.m. as indicated in the figure legends. Group size was determined on the basis of the results of preliminary experiments and no statistical method was used to predetermine sample size. The indicated sample size (n) represents biological replicates. Group allocation and outcome assessment were not performed in a blinded manner. All samples that met proper experimental conditions were included in the analysis. Statistical significance was determined by two-tailed Student’s t-test, and Pearson’s correlation using Prism 6 software (GraphPad Software). Significance was set at P < 0.05.
Data and software availability
RNA-Seq, ChIP-Seq, and ATAC-Seq data where deposited at GEO (GSE108929).
Supplementary Material
shRNA representation for epigenetic screen in myoblasts and synovial sarcoma cells. Related to Figure 1.
List of SS18-SSX target regions and associated genes. Related to Figure 6.
Gene Ontology, Pathway, and Regulatory motif analysis of SS18-SSX1/KDM2B target regions using GREAT. Related to Figure 6.
Genes upregulated in synovial sarcoma - TCGA sarcoma data set. Related to Figure 6.
RNA-Seq normalized gene counts for HS-SY-II cells transduced with SS18-SSX or KDM2B shRNAs. Related to Figure 6.
List of DNA oligos used in this study. Related to STAR Methods.
SIGNIFICANCE.
Somatic gene fusions are hallmarks of soft tissue sarcomas but little is known about how they exert their oncogenic effects. Using unbiased genetic screening and extensive genomic analyses in synovial sarcoma cells expressing the endogenous SS18-SSX1 oncoprotein, we identify KDM2B as a key mediator of SS18-SSX1 recruitment and reveal how this interaction hijacks the functions of the KDM2B-PRC1.1 repressive complex to aberrantly activate gene expression. Accordingly, KDM2B inhibition recapitulates the consequences of SS18-SSX1 inhibition resulting in loss of the synovial sarcoma-specific gene signature and irreversible acquisition of mesenchymal-like features. These studies reveal a mechanism by which an oncogenic fusion exploits antagonism between SWI-SNF and polycomb complexes to sustain transformation and identify the KDM2B-PRC1.1 as a candidate therapeutic target.
Highlights.
An RNAi screen identifies KDM2B as an epigenetic dependency in synovial sarcoma
KDM2B depletion abolishes neurogenic programs inducing mesenchymal differentiation
KDM2B-PRC1.1 recruits SS18-SSX and SWI/SNF to unmethylated CpG islands
SS18-SSX hijacks a repressive complex to aberrantly activate gene expression
Acknowledgments
The authors are grateful to Charles Sherr, Neal Poulin and members of the Lowe laboratory for comments and discussions. Amaia Lujambio for help with the analysis of shRNA screening data, Tatsuo Ito and Shinji Kohsaka for human synovial sarcoma cell lines, Alexandros Tzatsos for sharing KDM2B-expressing vectors, and Yilong Zou for help designing the strategy for endogenous HA-tagging. AB was supported by an EMBO long-term fellowship. This work was supported by an NIH P01 (4P01CA013106-45), the Comprehensive Cancer Support Grant (P30 CA00848-49), and a project grant from the Liddy Shriver Sarcoma Initiative (PI, Nielsen). A.N.L, A.R.D and T.O.N were supported by the Canadian Cancer Society Research Institute (Grant # 701582) and the Terry Fox Research Institute (TFF 105265 New Frontiers in Cancer). C.R.V and J.S.R are supported by the The Christina Renna Foundation, The Clark Gillies Foundation, The Friends of T.J. Foundation, and the The Michelle Paternoster Foundation. S.W.L. is the Geoffrey Beene chair for Cancer Biology and an Investigator in the Howard Hughes Medical Institute. S.W.L. would like to dedicate this work to Enrique (Henry) Cepero.
Footnotes
AUTHOR CONTRIBUITIONS
AB conceived the study, designed and performed experiments and wrote the manuscript. X.L., K.H., C.H., D.F.T., N.T., M.L., N.T., R.C., C.R.V. assisted in experiments, produced reagents or edited the manuscript. A.L. performed proximity ligation assays, assisted in experiments and provided reagents. J.R., C.C. and F.S.V analyzed ChIP-Seq, RNA-Seq and sarcoma methylation DNA data, respectively. A.R.D. performed IHC for sarcoma tumors. S.W.L conceived and supervised the study and wrote the manuscript. All authors read the manuscript.
DECLARATION OF INTEREST
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre AJ, Meyers RM, Weir BA, Vazquez F, Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andricovich J, Kai Y, Peng W, Foudi A, Tzatsos A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J Clin Invest. 2016;126:905–920. doi: 10.1172/JCI84014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- Barham W, Frump AL, Sherrill TP, Garcia CB, Saito-Diaz K, VanSaun MN, Fingleton B, Gleaves L, Orton D, Capecchi MR, et al. Targeting the Wnt pathway in synovial sarcoma models. Cancer Discov. 2013;3:1286–1301. doi: 10.1158/2159-8290.CD-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, Griffiths L, Hoffman EP, Stubbs RS, Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 2015;16:8. doi: 10.1186/s13059-014-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet. 2015;47:479–485. doi: 10.1038/ng.3272. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Electronic address e.d.s.c, Cancer Genome Atlas Research N. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171:950–965. e928. doi: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dodge J, Mehl E, Liu S, Poulin N, van de Rijn M, Nielsen TO. Validation of immature adipogenic status and identification of prognostic biomarkers in myxoid liposarcoma using tissue microarrays. Hum Pathol. 2009;40:1244–1251. doi: 10.1016/j.humpath.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos NR, de Bruijn DR, Kater-Baats E, Otte AP, van Kessel AG. Delineation of the protein domains responsible for SYT, SSX, and SYT-SSX nuclear localization. Exp Cell Res. 2000;256:192–202. doi: 10.1006/excr.2000.4813. [DOI] [PubMed] [Google Scholar]
- Endo M, de Graaff MA, Ingram DR, Lim S, Lev DC, Briaire-de Bruijn IH, Somaiah N, Bovee JV, Lazar AJ, Nielsen TO. NY-ESO-1 (CTAG1B) expression in mesenchymal tumors. Mod Pathol. 2015;28:587–595. doi: 10.1038/modpathol.2014.155. [DOI] [PubMed] [Google Scholar]
- Endo M, Su L, Nielsen TO. Activating transcription factor 2 in mesenchymal tumors. Hum Pathol. 2014;45:276–284. doi: 10.1016/j.humpath.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- Ishibe T, Nakayama T, Okamoto T, Aoyama T, Nishijo K, Shibata KR, Shima Y, Nagayama S, Katagiri T, Nakamura Y, et al. Disruption of fibroblast growth factor signal pathway inhibits the growth of synovial sarcomas: potential application of signal inhibitors to molecular target therapy. Clin Cancer Res. 2005;11:2702–2712. doi: 10.1158/1078-0432.CCR-04-2057. [DOI] [PubMed] [Google Scholar]
- Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21:665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Williams RT, Calarco JP, Miller EL, Weber CM, Braun SM, Pulice JL, Chory EJ, Crabtree GR. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Sung YS, Zhang L, Jungbluth AA, Huang SC, Argani P, Agaram NP, Zin A, Alaggio R, Antonescu CR. BCOR Overexpression Is a Highly Sensitive Marker in Round Cell Sarcomas With BCOR Genetic Abnormalities. Am J Surg Pathol. 2016;40:1670–1678. doi: 10.1097/PAS.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai A, Naito N, Yoshida A, Morimoto Y, Ouchida M, Shimizu K, Beppu Y. Establishment and characterization of a biphasic synovial sarcoma cell line, SYO-1. Cancer Lett. 2004;204:105–113. doi: 10.1016/j.canlet.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, Wang W, Haswell JR, Walensky LD, Hahn WC, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, Healey JH, Brennan MF, Bridge JA, Neff JR, Barr FG, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte AN, Ji JX, Ma L, Nielsen TO, Brodin BA. Identification of cytotoxic agents disrupting synovial sarcoma oncoprotein interactions by proximity ligation assay. Oncotarget. 2016;7:34384–34394. doi: 10.18632/oncotarget.8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lim FL, Soulez M, Koczan D, Thiesen HJ, Knight JC. A KRAB-related domain and a novel transcription repression domain in proteins encoded by SSX genes that are disrupted in human sarcomas. Oncogene. 1998;17:2013–2018. doi: 10.1038/sj.onc.1202122. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, Minami A, Hiraga H, Nishihara H, Sawa H, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama S, Katagiri T, Tsunoda T, Hosaka T, Nakashima Y, Araki N, Kusuzaki K, Nakayama T, Tsuboyama T, Nakamura T, et al. Genome-wide analysis of gene expression in synovial sarcomas using a cDNA microarray. Cancer Res. 2002;62:5859–5866. [PubMed] [Google Scholar]
- Naka N, Takenaka S, Araki N, Miwa T, Hashimoto N, Yoshioka K, Joyama S, Hamada K, Tsukamoto Y, Tomita Y, et al. Synovial sarcoma is a stem cell malignancy. Stem Cells. 2010;28:1119–1131. doi: 10.1002/stem.452. [DOI] [PubMed] [Google Scholar]
- Ng TL, Gown AM, Barry TS, Cheang MC, Chan AK, Turbin DA, Hsu FD, West RB, Nielsen TO. Nuclear beta-catenin in mesenchymal tumors. Mod Pathol. 2005;18:68–74. doi: 10.1038/modpathol.3800272. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, O’Connell JX, Gilks CB, Sorensen PH, Linn S, West RB, Liu CL, Botstein D, Brown PO, et al. Tissue microarray validation of epidermal growth factor receptor and SALL2 in synovial sarcoma with comparison to tumors of similar histology. Am J Pathol. 2003;163:1449–1456. doi: 10.1016/S0002-9440(10)63502-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, Poulin NM, Ladanyi M. Synovial sarcoma: recent discoveries as a roadmap to new avenues for therapy. Cancer Discov. 2015;5:124–134. doi: 10.1158/2159-8290.CD-14-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Wang YS, Abe S, Matsuno T, Yamawaki S, Nagashima K. Morphological and cytogenetic studies of a human synovial sarcoma xenotransplanted into nude mice. Acta Pathol Jpn. 1990;40:486–493. doi: 10.1111/j.1440-1827.1990.tb01590.x. [DOI] [PubMed] [Google Scholar]
- Pacheco M, Nielsen TO. Histone deacetylase 1 and 2 in mesenchymal tumors. Mod Pathol. 2012;25:222–230. doi: 10.1038/modpathol.2011.157. [DOI] [PubMed] [Google Scholar]
- Peters TL. BCOR-CCNB3 fusions are frequent in undifferentiated sarcomas of male children. Mod Pathol. 2015;28:575–586. doi: 10.1038/modpathol.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron G. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex--chromatin and cancer. Nat Rev Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Roy A, Kumar V, Zorman B, Fang E, Haines KM, Doddapaneni H, Hampton OA, White S, Bavle AA, Patel NR, et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat Commun. 2015;6:8891. doi: 10.1038/ncomms9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Pierron G, Klughammer J, Datlinger P, Schonegger A, Schuster M, Hadler J, Surdez D, Guillemot D, Lapouble E, et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat Med. 2017;23:386–395. doi: 10.1038/nm.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S, Demetri GD, Baldini EH, Fletcher CD. Management of soft-tissue sarcomas: an overview and update. Lancet Oncol. 2000;1:75–85. doi: 10.1016/s1470-2045(00)00016-4. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sonobe H, Manabe Y, Furihata M, Iwata J, Oka T, Ohtsuki Y, Mizobuchi H, Yamamoto H, Kumano O, Abe S. Establishment and characterization of a new human synovial sarcoma cell line, HS-SY-II. Lab Invest. 1992;67:498–505. [PubMed] [Google Scholar]
- Soulez M, Saurin AJ, Freemont PS, Knight JC. SSX and the synovial-sarcoma-specific chimaeric protein SYT-SSX co-localize with the human Polycomb group complex. Oncogene. 1999;18:2739–2746. doi: 10.1038/sj.onc.1202613. [DOI] [PubMed] [Google Scholar]
- Specht K, Zhang L, Sung YS, Nucci M, Dry S, Vaiyapuri S, Richter GH, Fletcher CD, Antonescu CR. Novel BCOR-MAML3 and ZC3H7B-BCOR Gene Fusions in Undifferentiated Small Blue Round Cell Sarcomas. Am J Surg Pathol. 2016;40:433–442. doi: 10.1097/PAS.0000000000000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, Hodges C, Calarco JP, Braun SM, Ku WL, Kadoch C, Zhao K, Crabtree GR. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DT, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Sampaio AV, Jones KB, Pacheco M, Goytain A, Lin S, Poulin N, Yi L, Rossi FM, Kast J, et al. Deconstruction of the SS18-SSX fusion oncoprotein complex: insights into disease etiology and therapeutics. Cancer Cell. 2012;21:333–347. doi: 10.1016/j.ccr.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Fukuta M, Sekiguchi K, Jin Y, Nagata S, Hayakawa K, Hineno S, Okamoto T, Watanabe M, Woltjen K, et al. SS18-SSX, the Oncogenic Fusion Protein in Synovial Sarcoma, Is a Cellular Context-Dependent Epigenetic Modifier. PLoS One. 2015;10:e0142991. doi: 10.1371/journal.pone.0142991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry J, Saito T, Subramanian S, Ruttan C, Antonescu CR, Goldblum JR, Downs-Kelly E, Corless CL, Rubin BP, van de Rijn M, et al. TLE1 as a diagnostic immunohistochemical marker for synovial sarcoma emerging from gene expression profiling studies. Am J Surg Pathol. 2007;31:240–246. doi: 10.1097/01.pas.0000213330.71745.39. [DOI] [PubMed] [Google Scholar]
- Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet. 1999;8:585–591. doi: 10.1093/hmg/8.4.585. [DOI] [PubMed] [Google Scholar]
- Trautmann M, Sievers E, Aretz S, Kindler D, Michels S, Friedrichs N, Renner M, Kirfel J, Steiner S, Huss S, et al. SS18-SSX fusion protein-induced Wnt/beta-catenin signaling is a therapeutic target in synovial sarcoma. Oncogene. 2014;33:5006–5016. doi: 10.1038/onc.2013.443. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci U S A. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno-Yokohata H, Okita H, Nakasato K, Akimoto S, Hata J, Koshinaga T, Fukuzawa M, Kiyokawa N. Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat Genet. 2015;47:861–863. doi: 10.1038/ng.3338. [DOI] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011a;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011b;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shRNA representation for epigenetic screen in myoblasts and synovial sarcoma cells. Related to Figure 1.
List of SS18-SSX target regions and associated genes. Related to Figure 6.
Gene Ontology, Pathway, and Regulatory motif analysis of SS18-SSX1/KDM2B target regions using GREAT. Related to Figure 6.
Genes upregulated in synovial sarcoma - TCGA sarcoma data set. Related to Figure 6.
RNA-Seq normalized gene counts for HS-SY-II cells transduced with SS18-SSX or KDM2B shRNAs. Related to Figure 6.
List of DNA oligos used in this study. Related to STAR Methods.