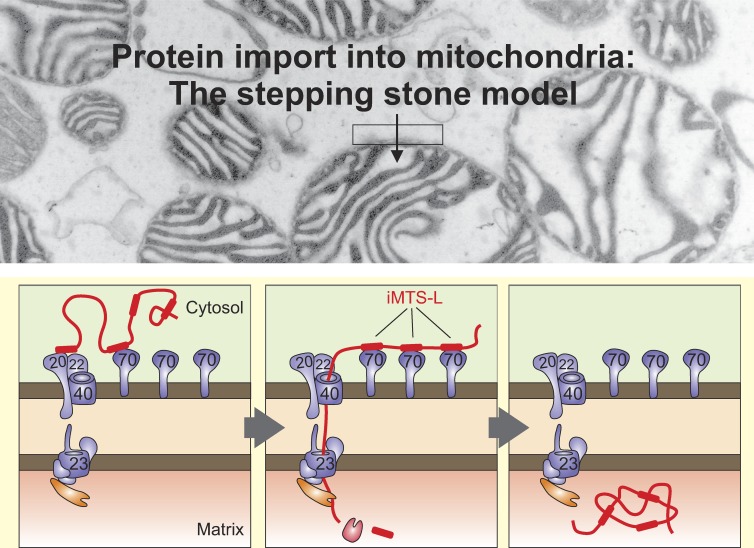
N-terminal matrix-targeting signals (MTSs) are critical for mitochondrial protein import. Backes et al. identified additional internal MTS-like sequences scattered along the sequences of mitochondrial proteins. By binding to Tom70 on the mitochondrial surface, these sequences support the import process.
Abstract
The biogenesis of mitochondria depends on the import of hundreds of preproteins. N-terminal matrix-targeting signals (MTSs) direct preproteins to the surface receptors Tom20, Tom22, and Tom70. In this study, we show that many preproteins contain additional internal MTS-like signals (iMTS-Ls) in their mature region that share the characteristic properties of presequences. These features allow the in silico prediction of iMTS-Ls. Using Atp1 as model substrate, we show that iMTS-Ls mediate the binding to Tom70 and have the potential to target the protein to mitochondria if they are presented at its N terminus. The import of preproteins with high iMTS-L content is significantly impaired in the absence of Tom70, whereas preproteins with low iMTS-L scores are less dependent on Tom70. We propose a stepping stone model according to which the Tom70-mediated interaction with internal binding sites improves the import competence of preproteins and increases the efficiency of their translocation into the mitochondrial matrix.
Graphical Abstract
Introduction
The intracellular sorting of newly synthesized proteins relies on targeting information encoded in their amino acid sequences. Proteins that are translocated as unfolded polypeptides typically use N-terminal targeting sequences that can be decoded during their synthesis on the ribosome. Well-studied examples for such N-terminal targeting sequences are the signal sequences of proteins of the ER (Blobel and Dobberstein, 1975; Schibich et al., 2016), leader peptides of proteins of the bacterial periplasm (Wickner et al., 1978), transit peptides of chloroplast proteins (Lubben et al., 1988), and presequences or matrix-targeting sequences (MTSs), which direct proteins to mitochondria (Hartl et al., 1986; Hurt et al., 1986; von Heijne, 1986a). Receptors on the surface of the target compartment recognize these signals and pass them on to protein-conducting channels through which the precursor proteins are threaded. Finally, processing peptidases remove the targeting sequences, and the mature proteins are folded with assistance of chaperones.
Proteins that are transported in a folded conformation (such as in the case of nuclear proteins) often use more complex internal signals, which are displayed on the 3D protein surface (De Robertis et al., 1978; Lee et al., 2006). These signals are part of the mature protein structure and are normally not removed by proteases.
Mitochondria are comprised of 800–1,500 different proteins (Mootha et al., 2003; Sickmann et al., 2003; Rhee et al., 2013; Morgenstern et al., 2017). About two thirds of these proteins are synthesized as precursors with N-terminal presequences that are both necessary and sufficient for their import (Wiedemann and Pfanner, 2017). These signals form amphipathic helices with one positively charged and one hydrophobic surface (von Heijne, 1986a). They are of variable length, typically between 8 and 70 amino acids, cleaved by the matrix processing peptidase (MPP), and degraded by the presequence peptidase PreP (Vögtle et al., 2009; Alikhani et al., 2011; Mossmann et al., 2014). Presequences are recognized by Tom20 and Tom22, receptors of the translocase of the outer membrane of mitochondria (TOM) complex, and are directed into the translocation pore formed by the β-barrel protein Tom40 (Rimmer et al., 2011; Shiota et al., 2011). The inner membrane translocase or TIM23 complex together with the import motor completes protein translocation into the matrix (Malhotra et al., 2013; Banerjee et al., 2015; Ramesh et al., 2016; Backes and Herrmann, 2017; Schendzielorz et al., 2017). The inner membrane harbors a second, independent translocase, the TIM22 complex, that inserts hydrophobic carrier proteins into the inner membrane (Rehling et al., 2003; Hasson et al., 2010; Wrobel et al., 2013). TIM22 substrates lack N-terminal presequences but carry internal targeting signals that are recognized on the surface of the mitochondria by a dedicated TOM receptor called Tom70 (Sirrenberg et al., 1996). Tom70, such as its paralog Tom71, has a tetratricopeptide structure and can, together with cytosolic Hsp70 and Hsp90 chaperones, recruit and stabilize its substrate proteins on the mitochondrial surface (Young et al., 2003; Fan et al., 2011; Hoseini et al., 2016; Zanphorlin et al., 2016; Xue et al., 2017). Tom70 and Tom20/Tom22 partially overlap in their substrate spectrum so that Tom70 is not essential as long as Tom20/Tom22 receptors are present (Ramage et al., 1993).
Some recent studies suggest that the mature parts of mitochondrial precursor proteins play a critical role in the efficiency of the translocation reaction that cannot simply be explained by the absence or presence of tightly folded translocation-resisting regions (Yamamoto et al., 2009; Schendzielorz et al., 2017). In this study, we show that sequences with MTS-like features are not confined to the N termini of mitochondrial proteins but are also frequently present in the mature parts of a subset of precursor proteins. These internal MTS-like signals (iMTS-Ls) show affinity for the Tom70 receptor and increase the efficiency of protein translocation. Our study points to a novel as yet unknown category of mitochondrial import signals that mimic the sequence properties of classical mitochondrial targeting sequences and which are present in the majority of matrix proteins.
Results
The matrix protein Atp25 has an internal presequence-like segment that improves its import efficiency
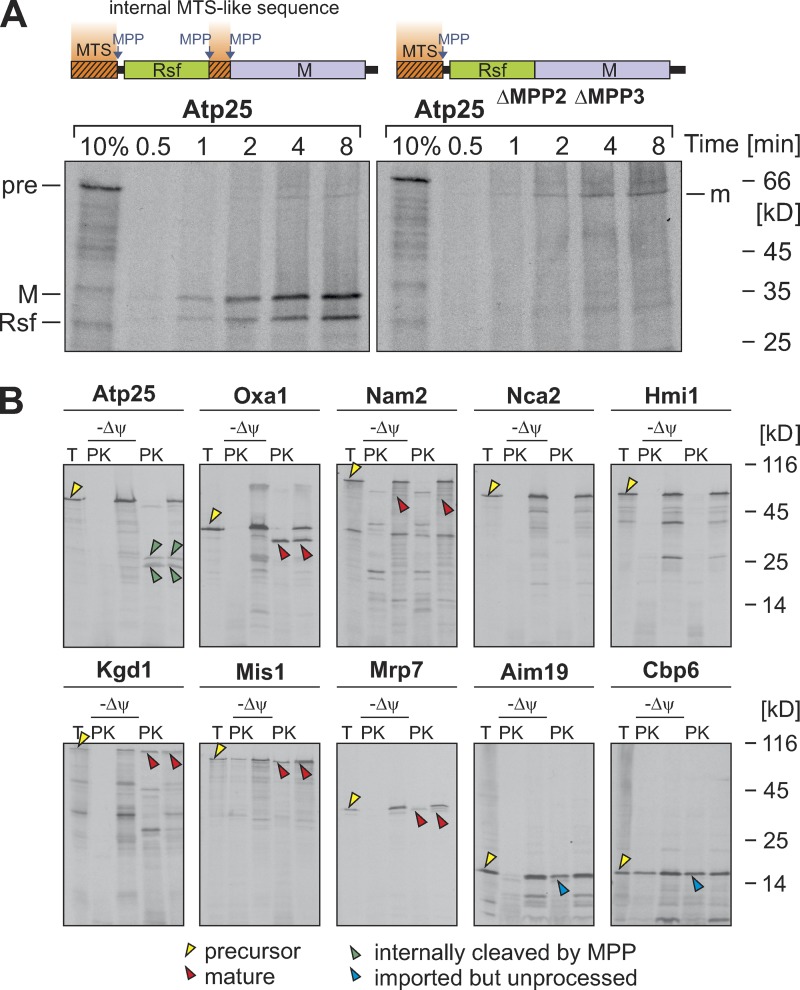
Atp25 is a composite matrix protein that employs three MPP-processing sites to generate two mature matrix-localized fragments referred to as the Rsf and M domains (Woellhaf et al., 2016). Deletion of the internal region in Atp25 that contains the two MPP cleavage sites led to the generation of one matured fusion protein (Fig. 1 A, m). To our knowledge, this was a very exceptional organization of a precursor protein because only very few tandem proteins were identified thus far: established examples are Arg6-Arg7 of Saccharomyces cerevisiae (Boonchird et al., 1991) and Rsm22-Cox11, Cox15-Yah1, YKR070w-Crd1, and Aco2-Mrpl49 of Schizosaccharomyces pombe (Khalimonchuk et al., 2006). To identify further tandem proteins, we screened the mitochondrial proteome of yeast for proteins that contain internal sequences that adhere to the MPP consensus R(AFLR)(FLY)↓(AKLS)(HQST) (Vögtle et al., 2009). We subcloned the coding sequences of these proteins (Nam2, Nca2, Hmi1, Kgd1, Mis1, Mrp7, Aim19, and Cbp6) as well as those of Atp25 (a tandem protein) and Oxa1 (a nontandem protein), generated radiolabeled proteins by in vitro transcription/translation reactions, and incubated them with isolated mitochondria (Fig. 1 B). Most of these precursors were imported into mitochondria, but for none of these cases did we observe internal cleavage by MPP. Thus, the tandem organization of Atp25 and Arg6-Arg7 are presumably rare exceptions. Obviously, nontandem organization of mitochondrial precursor proteins is strongly favored unless, such as in the case of Atp25, there is a good reason (Woellhaf et al., 2016) for such a composite structure.
Figure 1.
Atp25 is a rare example for a tandem precursor protein in yeast. (A) Atp25 and Atp25ΔMPP2ΔMPP3 (lacking amino acids 279–293; Woellhaf et al., 2016) were synthesized in the presence of [35S]methionine and incubated with isolated mitochondria for the times indicated. Nonimported material was removed by degradation with proteinase K (PK) before samples were analyzed by SDS-PAGE and autoradiography. Precursor (pre) and the M and Rsf domains as well as the N-terminally cleaved mature (m) species are indicated. 10% of the radiolabeled precursor protein used per time point was loaded for control. (B) The indicated proteins were radiolabeled and incubated with isolated mitochondria in the presence or absence of membrane potential (Δψ). The samples were split, and one fraction was treated with proteinase K. A 20% total (T) of the precursor protein used per import reaction was loaded for control. Yellow arrowheads depict precursor proteins, red arrowheads indicate N-terminally matured proteins, and green arrowheads indicate internally matured proteins. The presequences of Aim19 and Cbp6 were not cleaved by MPP (Vögtle et al., 2009); the imported species of these proteins are indicated by blue arrowheads. No protease-protected forms of Nca2 and Hmi1 were observed, indicating that these proteins were not imported in our in vitro import system.
To our surprise, we realized that the internal presequence-like structure in Atp25 was not only important for its internal processing but also for its overall import into mitochondria. As obvious from Fig. 1 A, Atp25 was efficiently imported, resulting in two strong signals of the Rsf and the M domain, respectively. In contrast, the matured species of Atp25ΔMPP2ΔMPP3 that acquired a protease-inaccessible location was very faint, indicating that this protein was barely imported. Low levels were not caused by rapid degradation as the imported Atp25ΔMPP2ΔMPP3 protein was stable under the import conditions used (Fig. S1 A). Obviously, the internal region in Atp25 that contains the two internal MPP cleavage sites contributes to the import competence of the protein. We showed in a previous study that this internal region has the ability to serve as an N-terminal MTS (Woellhaf et al., 2016). Thus, the presence of the internal mitochondrial targeting-like sequence in Atp25 considerably improved the import efficiency of this protein, at least in the in vitro assay used in this study.
iMTS-Ls can be predicted by TargetP
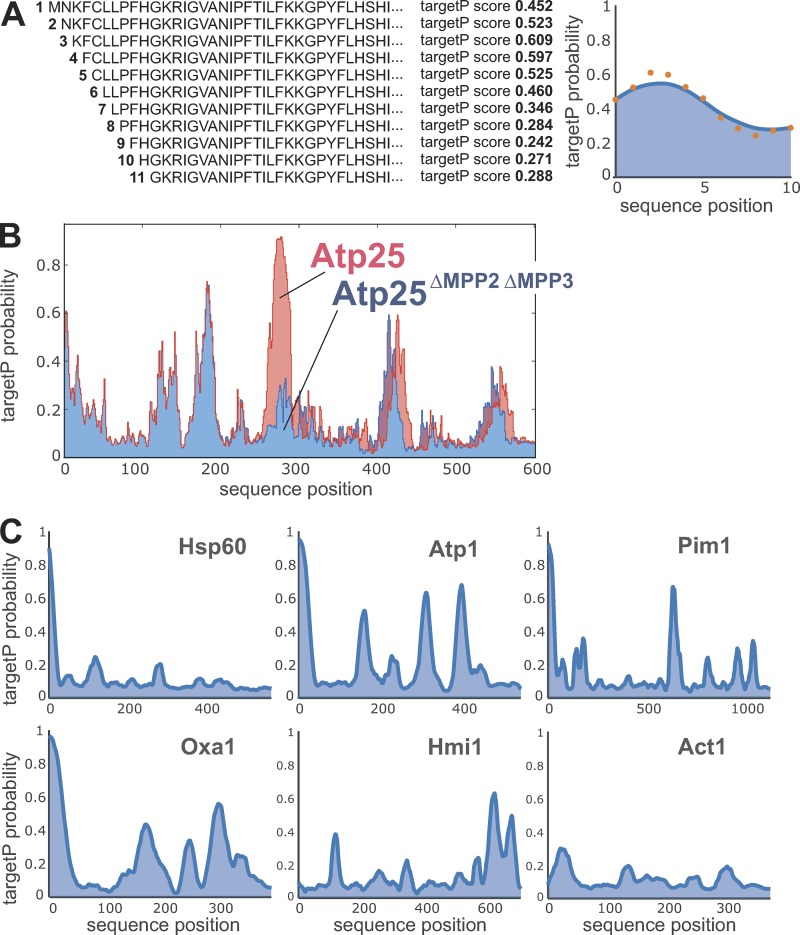
Several established algorithms exist to predict the presence of N-terminal MTSs on precursor proteins. Initially, algorithms were developed that searched for specific features of targeting signals such as helicity, positive charge, amphipathy, or the presence or absence of specific amino acid residues (McGeoch, 1985; von Heijne, 1986b). More recently, neural networks such as the TargetP algorithm were trained on sets of proteins of known cellular localization to predict the presence or absence of MTSs with high confidence (Nakai and Kanehisa, 1992; Emanuelsson et al., 2000, 2007; Habib et al., 2007). To identify potential internal targeting information in proteins, we used the TargetP predictive score consecutively for each residue in a given protein (Fig. 2 A), leading to a profile reflecting MTS-like properties. This approach was able to recognize the iMTS-Ls in Atp25 and showed that this stretch of high iMTS-L profile score was absent in Atp25ΔMPP2ΔMPP3 (Fig. 2 B).
Figure 2.
The TargetP algorithm can be used to predict iMTS-Ls throughout protein sequences. (A) The TargetP algorithm is designed to calculate prediction scores for N-terminal sequences. To calculate scores for internal regions, we consecutively N-terminally truncated the sequences and calculated the corresponding TargetP scores for each position (orange dots), leading to a characteristic profile that shows internal regions in proteins with presequence-like properties. Profiles are smoothed using a Savitzky-Golay filter (blue line). (B) Raw TargetP profiles in Atp25 predict with accuracy the iMTS of the protein, which contains the two internal MPP cleavage sites (Woellhaf et al., 2016). In the Atp25ΔMPP2ΔMPP3 variant, the internal presequence-like region was deleted, and hence, the internal region with the very high TargetP scores is missing. (C) Smoothed TargetP profiles of mitochondrial preproteins without (Hsp60) and with (Atp1, Pim1, Oxa1, and Hmi1) iMTS-Ls. The TargetP profile of the cytosolic protein actin (Act1) is shown for comparison.
We wondered whether similar iMTS-Ls are also present in other matrix proteins. In some preproteins such as Hsp60, the N-terminal MTS is the only segment of the protein with a TargetP score >0.3 (Fig. 2 C). However, most matrix proteins contained iMTS-Ls in addition to their N-terminal MTS (Table S1). As examples, profiles for the mitochondrial proteins Atp1, Pim1, Oxa1, and Hmi1 are shown. Hmi1 is an unconventional precursor that carries its targeting signal at its C terminus (Lee et al., 1999). The profile of the cytosolic protein actin (Fig. 2 C, Act1) is shown for comparison. It should be noted that internal sequences of high iMTS-L score were also found in several cytosolic proteins and thus, these sequences, unlike N-terminal MTSs, were no reliable indication of mitochondrial localization.
Many matrix proteins contain iMTS-Ls in their mature part
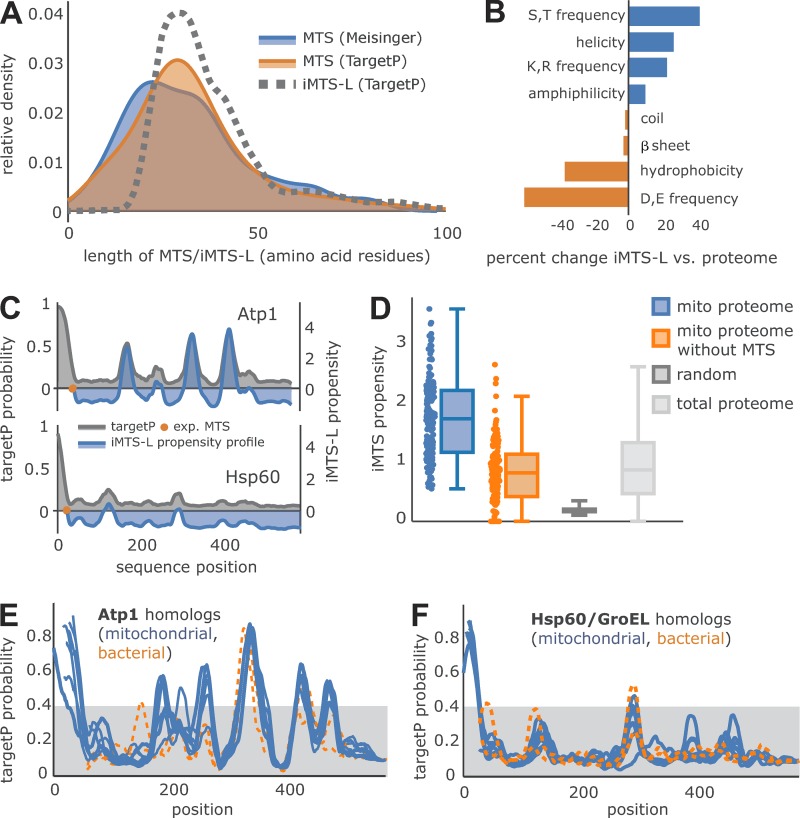
Next, we analyzed the structural features of the iMTS-Ls in more detail. We found that the iMTS-Ls are spread over the length of mature regions of mitochondrial proteins (Fig. S1 B and Table S1). We defined the width of the iMTS-Ls as the number of amino acids under a peak of scores higher than a random TargetP score. It is a priori not obvious from the nature of the TargetP algorithm that this setting captured the correct beginning and end of the relevant signals: Commonly, a high TargetP score is assigned to the first amino acid of an N-terminal sequence patch indicating the presence of an MTS. However, our current approach can predict the length of MTS as well as iMTS-Ls under the TargetP scoring peak. The width showed some variation, but most ranged between 20 and 30 residues, and thus, the lengths of iMTS-Ls resembled that of N-terminal MTSs analyzed by using the same parameters (Fig. 3 A) and the experimentally verified length determined by the published mitochondrial N proteome (Vögtle et al., 2009). To analyze the specific characteristics of iMTS-Ls, we compared different features of these sequences with those of the total yeast proteome (Fig. 3 B). We found that iMTS-Ls had low frequencies of aspartate and glutamate but high frequencies of arginine, lysine, and hydroxylated amino acids. They are predominantly helical, of amphipathic nature, and show low hydrophobicity scores. Thus, they clearly mimic N-terminal presequences, with the only obvious difference being that they are not N-terminal.
Figure 3.
Many precursor proteins contain iMTS-Ls. (A) Kernel density estimation of the length distribution shows the agreement of experimentally verified MTS length (blue; Vögtle et al., 2009), TargetP-predicted MTS (orange), and the length of iMTS-Ls predicted by our approach (dotted gray line). (B) Sequence property differences between iMTS-Ls and the total yeast proteome are shown in percent change. (C) The iMTS-L propensity profile (blue) for two selected proteins, Atp1 and Hsp60, is plotted against the smoothed TargetP profile (gray). The experimentally verified end of the MTS (Vögtle et al., 2009) is marked (orange dot) and matches the x axis intersection of the corresponding iMTS-L propensity profile. (D) The iMTS-L propensity calculated for experimentally verified mitochondrial matrix proteins (blue; Vögtle et al., 2009) shows a clear difference from random iMTS-L propensity calculated based on a sequence biased by yeast proteome amino acid frequency. However, the iMTS-L propensity of the total yeast proteome is similar to that of mature regions of matrix proteins. (E and F) Atp1 and Hsp60/GroEL sequences of S. cerevisiae (NP_009453 and NP_013360), Kluyveromyces lactis (XP_454248 and XP_455510), Candida glabrata (XP_449761 and XP_448482), S. pombe (CAB11207 and NP_592894), Drosophila (NP_726243 and NP_511115), Homo sapiens (AAH11384 and P10809), Arabidopsis thaliana (P92549 and AEE76842), Chlamydomonas reinhardtii (EDP07337 and XP_001691353), E. coli (WP_021537164 and ABF67773), and Rickettsia prowazekii (WP_004599658 and AMS12509) were aligned. iMTS-L propensity scores of mitochondrial and bacterial homologues were calculated and are shown as blue and orange traces, respectively.
Next, we generated 10,000 random protein sequences reflecting the mitochondrial proteome in length and amino acid frequency and calculated their iMTS-L scores. The comparison of the iMTS-L scores/profiles of mitochondrial proteins with these values allowed us to calculate a measure for each protein combining length and scores of the iMTS-Ls in their sequences, which we called iMTS-L propensity (Fig. 3 C). These propensities allowed it to compare different proteins by a single parameter. A list with mitochondrial proteins of particularly high or low iMTS-L propensities is shown in Table 1. For instance, Atp1 (the F1α subunit of the FoF1 ATPase) had an iMTS-L propensity of 1.57, whereas Hsp60 had 0.18 (Table S2). There was no obvious correlation between the iMTS-L propensities of yeast proteins with their lengths (Fig. S1 C). Most proteins contain considerably higher iMTS-L scores than randomly generated sequences of the same length and amino acid content. iMTS-L sequences were not only enriched in mitochondrial proteins but also in the total yeast proteome (Fig. 3 D), suggesting that iMTS-Ls are a structural sequence feature of more general distribution. This is also supported by the observation that the distribution and positions of iMTS-Ls were conserved not only among mitochondrial homologues of Atp1 and Hsp60/GroEL but even in their bacterial counterparts (Fig. 3, E and F, orange lines).
Table 1. Proteins with the highest or lowest iMTS-L propensity among all 135 analyzed mitochondrial proteins.
| Standard name | iMTS-L propensity |
|---|---|
| Mss18 | 2.48 |
| Qcr2 | 2.35 |
| Idh2 | 2.33 |
| Mba1 | 2.32 |
| Ndi1 | 1.86 |
| Arg5,6 | 1.73 |
| Mrs2 | 1.69 |
| Pkp1 | 1.67 |
| Mss51 | 1.65 |
| Cox15 | 1.57 |
| Mgm101 | 1.57 |
| Atp1 | 1.57 |
| Ism1 | 1.54 |
| Yme2 | 1.50 |
| Psd1 | 1.47 |
| Rmd9 | 1.44 |
| Coq2 | 1.43 |
| Dss1 | 1.41 |
| Msy1 | 1.39 |
| Kgd2 | 1.39 |
| Fmp16 | 0.00 |
| Sdh4 | 0.00 |
| Atp5 | 0.00 |
| Cox4 | 0.00 |
| Pdx1 | 0.00 |
| Atp12 | 0.00 |
| Atp14 | 0.00 |
| Cox8 | 0.00 |
| Cpr3 | 0.00 |
| Cox5a | 0.00 |
| Grx5 | 0.00 |
| Isu1 | 0.00 |
| Atp15 | 0.00 |
| Stf1 | 0.00 |
| Trx3 | 0.00 |
| Sod2 | 0.00 |
| Nfu1 | 0.00 |
| Atp16 | 0.00 |
| Inh1 | 0.08 |
| Ppa2 | 0.15 |
iMTS-Ls have the potential to function as mitochondrial targeting sequences
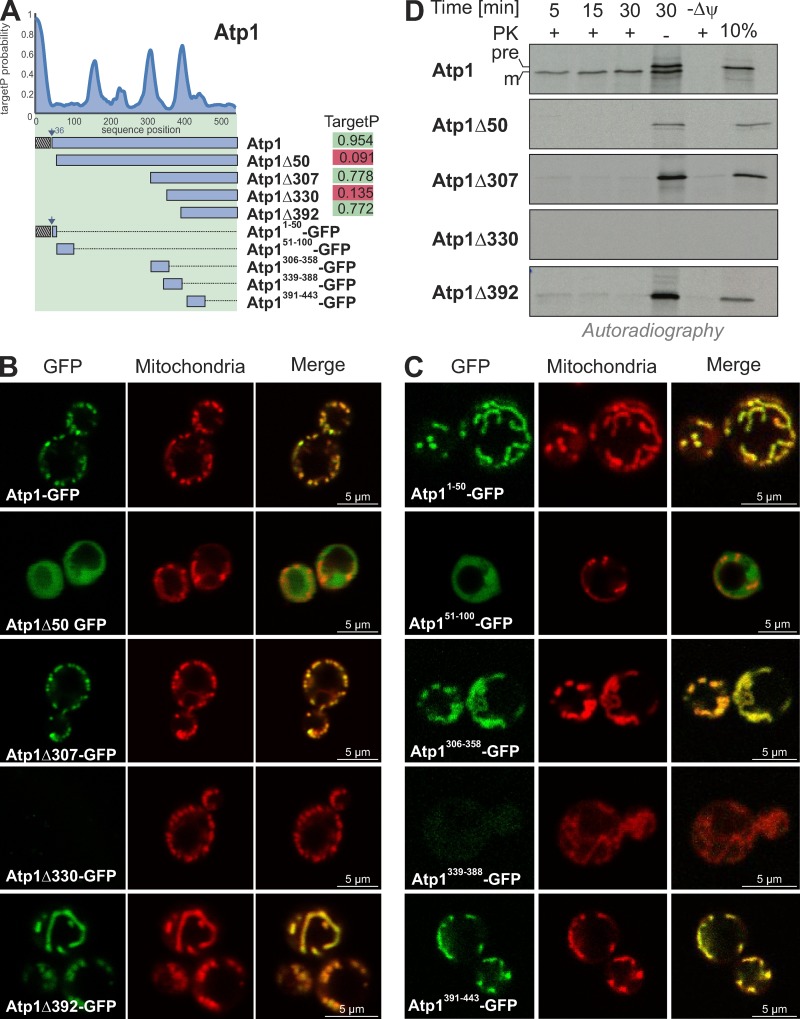
The similarity of the properties of iMTS-Ls to canonical mitochondrial presequences inspired us to test whether iMTS-Ls can target proteins to mitochondria if placed at the N terminus of a protein. To this end, we used Atp1 as a model protein. This protein contains several iMTS-Ls, two very prominent ones starting with residues 308 and 393 (Fig. 4 A). We generated constructs for the cytosolic expression of N-terminally truncated versions of Atp1, which either started with an iMTS-L sequence (Atp1Δ307 and Atp1Δ392) or with a sequence of low TargetP score (Atp1Δ50 and Atp1Δ330) fused to GFP. As shown in Fig. 4 B, Atp1-GFP, Atp1Δ307-GFP, and Atp1Δ392-GFP colocalized with mitochondria, whereas Atp1Δ50-GFP remained in the cytosol. For Atp1Δ330-GFP, no expression was detected. The iMTS-Ls were obviously sufficient for mitochondrial targeting because we observed that fusion proteins consisting of 50-residue segments of Atp1 and GFP confirmed the localization that was found with the N-terminally truncated Atp1 versions (Fig. 4 C). Thus, the iMTS-Ls in Atp1 can efficiently target GFP to mitochondria in vivo.
Figure 4.
iMTS-Ls have mitochondrial targeting potential if present at the N terminus. (A) TargetP profile of Atp1. Overview and TargetP scores of N-terminally truncated Atp1 variants. (B and C) Fusion proteins of N-terminally truncated Atp1 variants and GFP were expressed in yeast cells. Representative fluorescence microscopy images of yeast cells show the intracellular distribution of GFP (green) and mitochondria stained with rhodamine B hexylester (red). Please note that Atp1-GFP, Atp1Δ307-GFP, and Atp1Δ392-GFP as well as Atp11–50-GFP, Atp1306–358-GFP, and Atp1391–443-GFP colocalize with mitochondria, whereas Atp1Δ50-GFP and Atp11–50-GFP are present in the cytosol. Atp1Δ330-GFP and Atp1339–388-GFP failed to be expressed. (D) N-terminally truncated variants of Atp1 were synthesized in reticulocyte lysate in the presence of [35S]methionine and incubated for different times with isolated WT mitochondria. For the sample labeled with –ψ, the mitochondrial membrane potential was dissipated by the addition of valinomycin. Nonimported protein was removed by treatment with proteinase K (PK) before the samples were visualized by SDS-PAGE and autoradiography. 10% of the radiolabeled protein used per import lane was shown for control. Atp1 was efficiently imported into mitochondria. Only very minor amounts of Atp1Δ307-GFP and Atp1Δ392-GFP were taken up, and Atp1Δ50-GFP was not imported. Atp1Δ330-GFP failed to be synthesized.
Next, we tested whether these sequences can serve as MTSs in vitro. We synthesized Atp1 and its N-terminally truncated variants in the presence of [35S]methionine and incubated these variants with yeast mitochondria. Atp1 was efficiently imported into the mitochondria, where its presequence was cleaved off by MPP, resulting in a mature form (Fig. 4 D, m). The N-terminally truncated variants of Atp1 were either not imported (Atp1Δ50 and Atp1Δ330) or imported only with very low efficiency (Atp1Δ307 and Atp1Δ392). Even after an incubation of 30 min, most of these proteins remained accessible to added protease. This shows that iMTS-Ls, when placed to the N terminus, can efficiently mediate the targeting of proteins to mitochondria. However, they apparently are not able to drive the complete import of proteins into mitochondria, suggesting that they differ in certain critical properties. For example, iMTS-Ls might be unable to open the protein-conducting channel of the TIM23 complex or to activate the import motor, processes which still are poorly understood (Truscott et al., 2001; Okamoto et al., 2002; Chacinska et al., 2005; Meinecke et al., 2006; Longen et al., 2014; Ramesh et al., 2016; Schendzielorz et al., 2017; Ting et al., 2017).
iMTS-Ls can serve as binding regions for TOM receptors
A previous study by Yamamoto et al. (2009) reported that the outer membrane receptor Tom70 interacts with the mature parts of some mitochondrial precursor proteins. In particular, they observed that the import of Atp1 into Δtom70 mitochondria occurred only with reduced efficiency and that the cytosolic receptor domain of Tom70 helps to prevent the aggregation of the Atp1 precursor in vitro. We therefore wondered whether the iMTS-Ls in Atp1 might represent binding sites for Tom70.
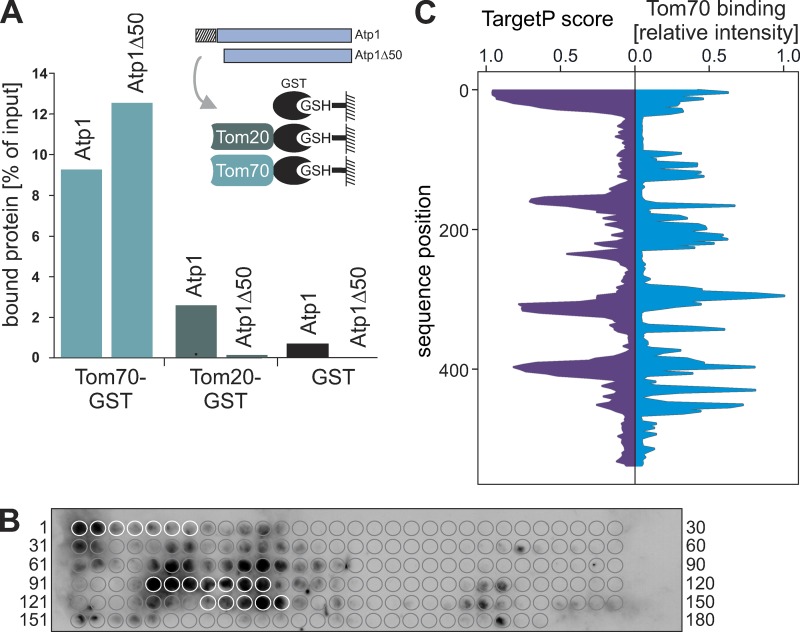
To test a potential binding of the Atp1 protein to Tom70, we incubated radiolabeled Atp1 and Atp1Δ50 with either purified recombinant GST, Tom70-GST, or Tom20-GST coupled with glutathione (GSH) beads (Hoseini et al., 2016). Atp1Δ50 lacks the N-terminal 50 residues of Atp1, including the 35 residues of the presequence (Vögtle et al., 2009). Tom20 showed a strong preference for the presequence-containing Atp1 precursor (Fig. 5 A), consistent with the well-documented function of Tom20 in presequence recognition (Brix et al., 1999; Yamamoto et al., 2011; Shiota et al., 2015). Interestingly, Tom70 efficiently bound Atp1 as well as Atp1Δ50, suggesting that the presequence of Atp1 is not essential for Tom70 binding. Neither Atp1 nor Atp1Δ50 were recovered with GST control beads.
Figure 5.
Tom70 binds to specific regions in the mature part of Atp1. (A) The cytosolic receptor domains of Tom70 and Tom20 were purified as GST fusion proteins and bound to GSH beads. Radiolabeled Atp1 or Atp1Δ50 was incubated with the indicated beads for 10 min at room temperature. Beads were then pelleted by centrifugation, the supernatant fractions were discarded, and the pellet fractions were analyzed by SDS-PAGE and autoradiography. The results were quantified using ImageJ. (B) 20-mer peptides shifted by three amino acids along the entire 545 residues of the Atp1 sequence were covalently bound to a membrane and incubated with purified Tom70-GST as used in A. After extensive washing, the bound Tom70 was detected by Western blotting using a specific GST antibody. Circles indicate the location of each single peptide on the membrane. White circles represent the peptides of the N-terminal presequence of Atp1 (peptides 1–7) and the iMTS-L sequences presented in Fig. 4 A (peptides 95–101 and 128–132). (C) Binding of Tom70-GST was quantified, and the relative intensity of each peptide spot was plotted (blue trace). For comparison, the profile of Atp1 TargetP scores is shown (purple trace).
To identify the regions in Atp1 that are recognized by Tom70, we used a peptide scan approach that had been successfully used in the past to determine Tom70 binding sites in mitochondrial carrier proteins (Brix et al., 1999). Peptides of 20-amino-acid residues each were covalently bound to a membrane. In total, 176 peptides were synthesized, each shifted by three amino acids along the entire 545 residues of the Atp1 sequence. The membrane was incubated with purified Tom70-GST. After extensive washing, the bound Tom70 was detected by Western blotting (Fig. 5 B). We observed that several regions in Atp1 were efficiently bound by Tom70, which very well reflected the patterns of the MTS/iMTS-L sequences (Fig. 5 C). This suggests that the iMTS-Ls in Atp1, and presumably also in other mitochondrial precursor proteins, represent Tom70 binding regions in these proteins.
The presence or absence of iMTS-L sequences in Atp1 and Hsp60 correlates with their Tom70 dependence
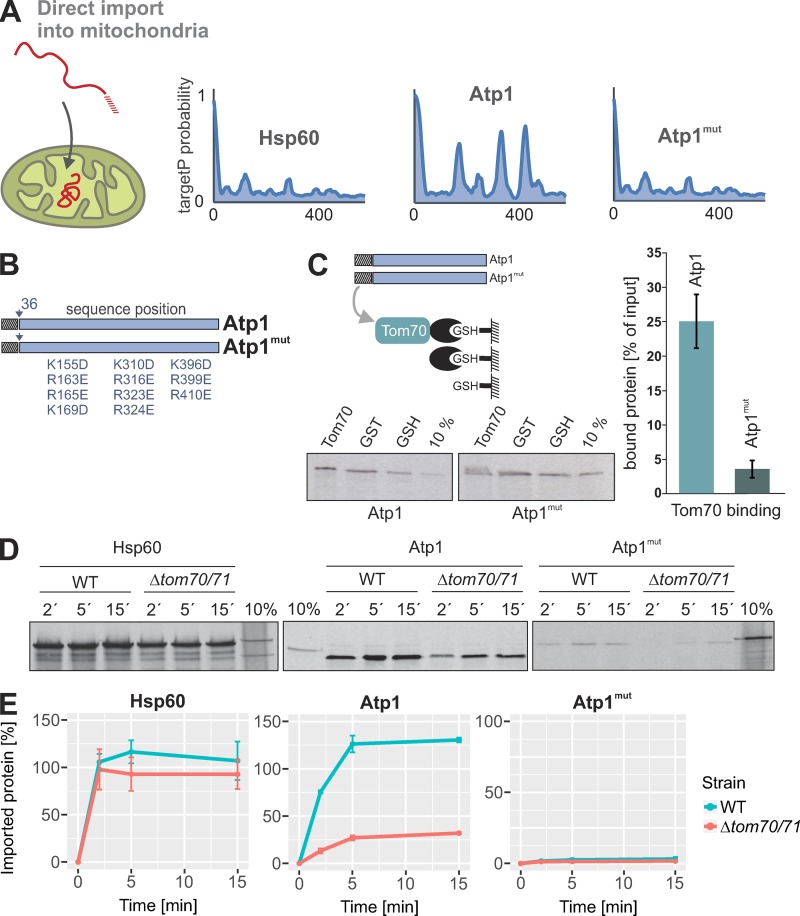
Next, we tested the relevance of iMTS-Ls for protein import reactions in vitro. We produced radiolabeled Atp1 (as a protein with iMTS-Ls) and Hsp60 (as a protein without such sequences) as well as a mutated version of Atp1 in which its three iMTS-Ls were mutated (Atp1mut; Fig. 6, A and B). At least in vitro, this Atp1mut version hardly bound to Tom70, whereas Atp1 did (Fig. 6 C), supporting the idea that the iMTS-Ls support the association of precursor proteins with the Tom70 receptor.
Figure 6.
The presence of MTS-like sequences in the mature part of mitochondrial proteins influences the import behavior. (A and B) TargetP probabilities of Hsp60, Atp1, and an Atp1mut version of Atp1 in which the iMTS-Ls were mutated by replacing positively charged residues by negative ones. (C) Radiolabeled Atp1 or Atp1mut precursor was incubated with empty GSH beads or beads coupled with GST or GST-Tom70. Beads were pelleted by centrifugation, extensively washed, and analyzed by SDS-PAGE and autoradiography. Lanes labeled “10%” show 10% of the radiolabeled precursor proteins used per reaction. The levels of proteins bound to Tom70 relative to those bound to empty beads were quantified from three independent experiments. Shown are means and SD. (D) The indicated model proteins were radiolabeled in reticulocyte lysate and incubated for the indicated times with WT and Δtom70/Δtom71 mitochondria at 25°C. Mitochondria were incubated with 100 µg/ml proteinase K for 30 min on ice to remove nonimported material and analyzed by SDS-PAGE and autoradiography. Lanes labeled “10%” show 10% of the radiolabeled precursor proteins used per time point. (E) The experiments shown in C were repeated three times and quantified. Intensities were normalized to the 10% control of the corresponding precursor protein. Means and SEM are shown.
To assess the Tom70 dependence of these proteins, Hsp60, Atp1, and Atp1mut were incubated with isolated WT and Δtom70/Δtom71 mitochondria for different times before nonimported protein was degraded (Fig. 6, D and E). Hsp60 was efficiently imported into WT and Δtom70/Δtom71 mitochondria. In contrast, efficient import of Atp1 required the presence of Tom70/Tom71, consistent with an earlier study that showed that Tom70 can prevent the aggregation of Atp1 precursor (Yamamoto et al., 2009). Interestingly, when the iMTS-Ls in Atp1 were mutated, the import competence of the precursor protein was almost completely abolished (Fig. 6 E; compare 10% of the total to the protease-inaccessible protein after 15 min of import). From this, we conclude that Atp1 requires the outer membrane receptor Tom70/Tom71 as well as its iMTS-Ls to be efficiently imported into mitochondria.
iMTS-Ls maintain nonimported Atp1 precursor in an import-competent conformation
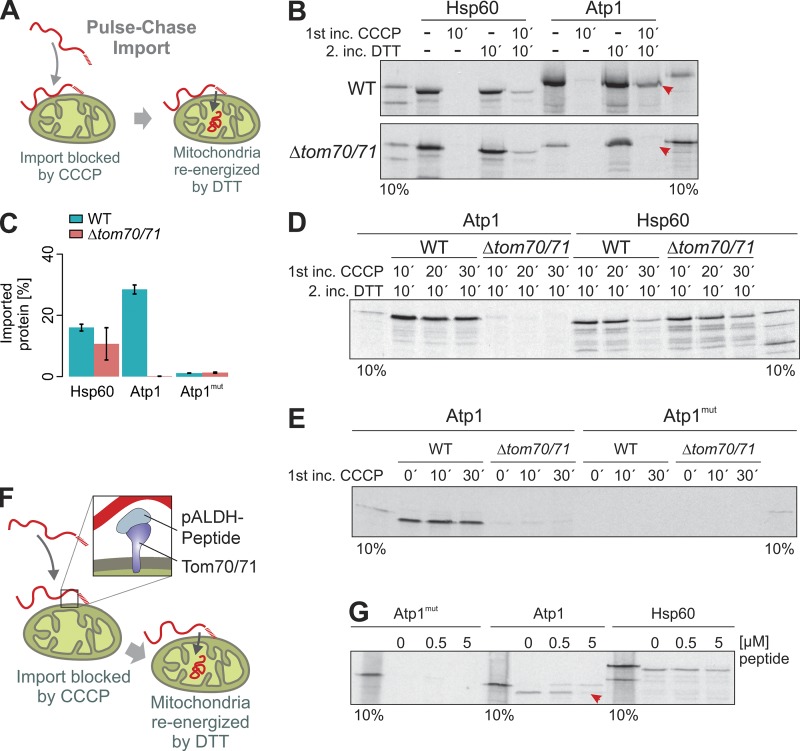
The reduced efficiency by which Atp1 is imported into Δtom70/Δtom71 mitochondria suggests a direct role of Tom70 that is mediated via interaction with its iMTS-Ls. We used a pulse-chase assay that had been developed to study the relevance of Tom70 binding to precursors bound to the mitochondrial surface (Hines and Schatz, 1993). To this end, the mitochondrial membrane potential was dissipated by carbonyl cyanide chlorophenyl hydrazine (CCCP) before radiolabeled Hsp60 or Atp1 were bound to WT or Δtom70/Δtom71 mitochondria. Mitochondria were reisolated and reenergized by treatment with DTT, NADH, and ATP (Fig. 7, A–C). Atp1 was efficiently chased into mitochondria only in the presence of Tom70/Tom71 (Fig. 7 B, arrowheads), which obviously were crucial to maintain Atp1 import competence on the mitochondrial surface. In contrast, Hsp60 remained largely import competent also in Δtom70/Δtom71 mitochondria unless it was incubated with nonenergized mitochondria for prolonged time (Fig. 7 D).
Figure 7.
Tom70 is essential to maintain the Atp1 precursor in an import-competent conformation. (A) Model of the pulse-chase assay used in the following experiments. The membrane potential of isolated mitochondria was dissipated by addition of CCCP. Radiolabeled precursor proteins were added. After incubation of the mitochondria for 10–30 min, CCCP was quenched by DTT, and the mitochondria were reenergized. (B–E) WT or Δtom70/71 mitochondria were preincubated with CCCP for 10–30 min in the presence of radiolabeled Hsp60, Atp1, and Atp1mut (pulse). Mitochondria were reisolated and reenergized by treatment with DTT. After incubation (chase) for 10 min at 25°C, nonimported protein was removed by protease treatment, and samples were analyzed by SDS-PAGE and autoradiography. C shows means and SEM of three replicates. Arrowheads indicate the Atp1 protein that only was chased into mitochondria if Tom70/Tom71 were present. (F and G) A peptide from the rat pALDH presequence that was shown to work as a competitive inhibitor of the Tom70 receptor (Melin et al., 2015) was added to isolated mitochondria to the pulse-chase reaction with Atp1mut, Atp1, and Hsp60 precursors. The presence of 5 µM pALDH peptide prevented the import of Atp1 (red arrowheads).
The Atp1 version in which the iMTS-Ls were mutated was not imported in the pulse-chase experiment. Obviously, this variant lost its minor import competence during the preincubation completely, irrespective of the presence of Tom70/Tom71 (Fig. 7, C and E).
To exclude that the pronounced defect of the pulse-chase import into Δtom70/Δtom71 mitochondria is caused by pleiotropic problems of this mutant, we generated a peptide aldehyde dehydrogenase (pALDH) from which it was previously shown that it binds to the substrate-binding groove of Tom70 (Melin et al., 2015) and which hence competes for Tom70 binding with precursor proteins. Addition of excess amounts of this peptide to WT mitochondria during preincubation with precursor proteins compromised the import of Atp1 but not that of Hsp60 (Fig. 7, F and G). This nicely confirmed that Tom70 binding is critical to maintaining Atp1 in an import-competent state, for which iMTS-Ls are of critical relevance.
Tom70 supports the import of proteins with high iMTS-L propensities
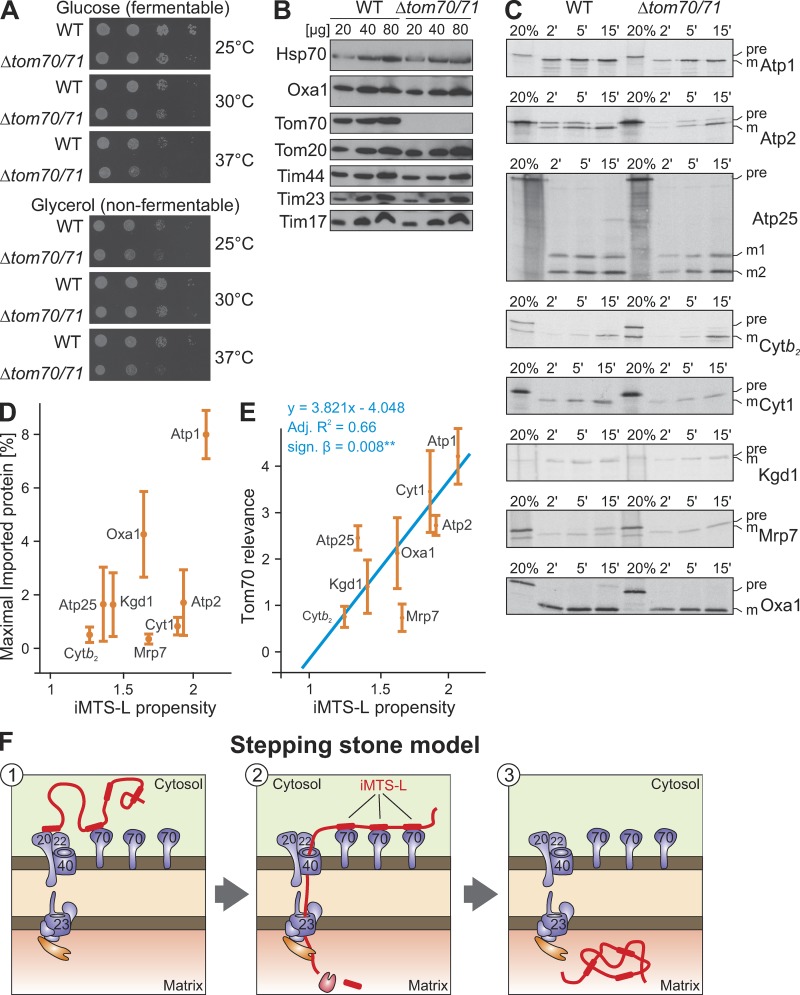
Tom70 and its paralog Tom71 are not essential proteins, and in most genetic backgrounds, Δtom70/Δtom71 double mutants grow efficiently even under nonfermentative conditions (Fig. 8 A). To analyze the relevance of Tom70 in more detail, we purified WT and Δtom70/Δtom71 mitochondria, which contained similar levels of Tom20 as well as of the TIM23 subunits Tim44, Tim23, and Tim17 (Fig. 8 B). These mitochondria were incubated with radiolabeled precursor forms of Atp1, Atp2, Atp25, cytochrome b2, cytochrome c1, Kgd1, Mrp7, and Oxa1 for 2, 5, and 15 min, and the amounts of fully imported proteins were visualized after proteolytic digestion of nonimported proteins (Fig. 8 C). The signals of three replicates were quantified (see Fig. S2 for examples), and the amounts of maximally imported proteins were calculated. These amounts differed considerably between the different proteins, and there was no significant correlation with their respective iMTS-L propensities (Fig. 8 D). However, when we compared the ratios of the maximal imported protein of WT to that of Δtom70/Δtom71 mitochondria (Fig. 8 E), there was a convincing correlation between Tom70 dependence and the iMTS-L propensity. The larger the iMTS-L propensities of the precursor proteins tested, the more their import was improved by the presence of Tom70/Tom71. This is consistent with a role of Tom70 in promoting the import of mature parts of mitochondrial precursor proteins, presumably via its interaction with iMTS-L sequences (Fig. 8 F).
Figure 8.
The presence of Tom70 supports the import of precursor proteins with high iMTS-L propensity scores. (A) WT and Δtom70/Δtom71 were grown to log phase in galactose medium before tenfold serial dilutions were spotted onto plates containing glucose or glycerol medium. Plates were incubated at the indicated temperatures for 2 d. Mutants lacking Tom70 and its paralog Tom71 show growth defects at 37°C. (B) Mitochondria were isolated from WT and Δtom70/Δtom71 cells. 20, 40, and 80 µg of mitochondrial proteins were resolved by SDS-PAGE and analyzed by Western blotting. (C) The indicated proteins were radiolabeled in reticulocyte lysate and incubated for the indicated times with WT and Δtom70/Δtom71 mitochondria at 25°C. Mitochondria were reisolated, incubated with 100 µg/ml proteinase K for 30 min on ice to remove nonimported material, and analyzed by SDS-PAGE and autoradiography. Lanes labeled “20%” show 20% of the radiolabeled precursor proteins used per time point. (D) The import experiments shown in C were repeated three times. The signals were quantified and fitted with the Michaelis-Menten equation y = (x + imax)/(x + i50%) from which maximal imported amounts imax were deduced (Fig. S2). Absolute amounts of imported proteins and iMTS-L propensities show no significant relatedness (y = 5.051x − 5.399; adjusted R2 = 0.23; signal β = 0.125). (E) In contrast, regression analysis revealed a significant relationship between the iMTS-L propensity of a protein and its Tom70 relevance (y = 3.821x − 4.048; adjusted R2 = 0.66; signal β = 0.008**), which we define as the protein maximally imported into WT mitochondria divided by that of Δtom70/Δtom71 mitochondria. Error bars denote SD. (F) Hypothetical model for the role of iMTS-Ls as stepping stones for Tom70 interaction during protein import. Binding of Tom70 to the iMTS-Ls prevents premature folding or aggregation of the precursors and maintains their import competence.
Discussion
Protein targeting relies on the specific recognition of precursor proteins by receptors of the destination membrane. Proteins of the mitochondrial matrix are characterized by N-terminal MTSs, which bind to the Tom20/Tom22 receptor on the mitochondrial surface. These N-terminal signals are both necessary and sufficient for mitochondrial protein targeting.
In this study, we describe additional as yet unidentified iMTS-L sequences that are scattered over the mature part of mitochondrial precursor proteins. Previous studies already reported the presence of latent targeting signals in cytosolic proteins (Hurt and Schatz, 1987) as well as in Escherichia coli DNA-derived sequences (Baker and Schatz, 1987). In this study, we show that iMTS-Ls mimic the structure of presequences in respect to their length, amphipathicity, positive charge distribution, and high content of hydroxylated amino acid residues. Despite their similarity to MTSs, the iMTS-Ls of Atp1 failed to target proteins to the matrix in in vitro import experiments even if they were presented N-terminally, and thus apparently are not necessarily fully functional presequences. Nevertheless, these sequences were able to target GFP fusion proteins to mitochondria in vivo, indicating that they interact with proteins on the mitochondrial surface.
We identified Tom70 as an efficient and specific iMTS-L binding partner that, potentially in cooperation with the other TOM receptors, interacts with these regions during preprotein import into mitochondria (Fig. 8 F). The interaction of Tom70 with the iMTS-Ls is obviously not essential for protein translocation into mitochondria because mutants lacking Tom70 still import matrix proteins (Ramage et al., 1993; Gärtner et al., 1995; Yamamoto et al., 2009) and because a small number of matrix proteins lack iMTS-Ls completely (Tables 1 and S2). It appears conceivable that the binding of iMTS-Ls can delay the passage of precursors across the outer membrane and hence might be counterproductive for the import of proteins that are not prone to adopt import-incompetent conformations. Our understanding of the interactions of mitochondrial surface receptors with their substrates is still incomplete. However, substrate release from TOM receptors was reported to be triggered by an interaction of the cytosolic domains of Tom70 and Tom20 (Fan et al., 2011).
It was shown that Tom70 improves the import of only certain precursor proteins (Yamamoto et al., 2009; Horvath et al., 2012). The import of some matrix proteins such as Atp1 into Tom70-deficient mitochondria was reported to be strongly reduced, consistent with what we observed in this study. Interestingly, the mature part but not the presequences was found to determine the Tom70 dependence of these preproteins, which were proposed to aggregate if Tom70 is absent (Yamamoto et al., 2009). Hsp90 and Hsp70 chaperones, which directly interact with Tom70, support Tom70 in this process (Young et al., 2003; Bhangoo et al., 2007; Li et al., 2009; Fan et al., 2011; Hoseini et al., 2016). Tom70 consists primarily of tetratricopeptide repeat motifs (Chan et al., 2006; Wu and Sha, 2006), a structure characteristic for many interactors of Hsp70 and Hsp90 proteins. Moreover, Δtom70 deletion mutants show strong synthetic negative growth defects with mutants lacking cytosolic chaperones such as the cytosolic J protein Djp1 (Papić et al., 2013). Tom70 might constitute a platform on the mitochondrial surface that interacts with specific precursor proteins and with different cytosolic chaperones. It appears likely that the iMTS-Ls identified in this study serve as Tom70 binding sites distributed like stepping stones in the sequence of precursor proteins, which keep precursors unfolded and import competent during their translocation into mitochondria (Fig. 8 F). According to this hypothesis, the predominant function of iMTS-Ls is not that of mitochondrial targeting signals but rather that of unfolding signatures in precursor proteins, which facilitate the import of potentially aggregation-prone or multidomain precursors. Our analysis suggests that iMTS-Ls are not a characteristic feature of mitochondrial proteins but rather are distributed generally in proteins. It appears likely that Tom70 had evolved to exploit this pervasive feature in proteins to prevent premature folding before import into mitochondria. Cytosolic chaperones and folding factors such as the recently described ubiquilins (Itakura et al., 2016) might support Tom70 in this function.
Because presequences are both necessary and sufficient for mitochondrial targeting, the relevance of the mature part of proteins was not carefully analyzed thus far. It is well established that tightly folded protein domains such as those of the methotrexate-bound dihydrofolate reductase domain or titin can prevent or slow down protein translocation into mitochondria (Wienhues et al., 1991; Gaume et al., 1998; Sato et al., 2005; Yagawa et al., 2010). Moreover, it was shown that the recognition of mature stretches of precursors by the mitochondrial chaperone machinery influences their import efficiency (Okamoto et al., 2002; Schendzielorz et al., 2017). The identification of iMTS-Ls in this study is the first approach to systematically identify specific biogenesis signals in the mature parts of mitochondrial proteins. Interestingly, internal targeting sequences were recently also identified in the mature parts of secretory proteins, where they might serve a similar purpose (Chatzi et al., 2017). It will be exciting to study their specific relevance for the mitochondrial protein import in more detail in the future.
Materials and methods
Yeast strains and plasmids
All yeast strains used in this study were based on the WT strain W303-1A (Sherman, 1963). The Δtom70/Δtom71 strain, which was a gift from D. Rapaport (University of Tübingen, Tübingen, Germany), was described previously (Kondo-Okamoto et al., 2008). All strains were grown on YP (1% yeast extract and 2% peptone) medium containing 2% galactose (Altmann et al., 2007). DNA sequences corresponding with regions coding for Atp1 or fragments thereof were amplified by PCR and cloned into the EcoRI and BamHI sites of a pYX122 vector.
For confocal microscopy, 3 ml yeast culture (OD600 0.8) was incubated for 5 min with 30 µl of 10 µM rhodamine B hexylester dissolved in DMSO.
The Atp25-coding region or fragment was amplified by PCR and cloned into pGEM4 (Promega) using the EcoRI and HindIII restriction sites. For construction of the Atp25ΔMPP2 ΔMPP3 version, the sequence encoding for amino acid residues 279–293 were deleted and replaced by the restriction sites of XbaI and SalI (Woellhaf et al., 2016).
Import of radiolabeled proteins into isolated mitochondria
Import reactions were essentially performed as described previously (Weckbecker et al., 2012) in the following import buffer: 500 mM sorbitol, 50 mM Hepes, pH 7.4, 80 mM KCl, 10 mM magnesium acetate, and 2 mM KH2PO4. Mitochondria were energized by addition of 2 mM ATP and 2 mM NADH before radiolabeled precursor proteins were added. To dissipate the membrane potential, a mixture of 1 µg/ml valinomycin, 8.8 µg/ml antimycin, and 17 µg/ml oligomycin was added to the mitochondria. Precursor proteins were incubated with mitochondria for different times at 25°C before nonimported protein was degraded by addition of 100 µg/ml proteinase K.
CCCP chase experiment
Isolated mitochondria were preincubated in import buffer containing 50 µM CCCP for 5 min at 25°C. Radiolabeled precursor proteins were added. After incubation for 10–30 min at 25°C, mitochondria were reisolated by centrifugation for 10 min at 30,000 g. Pellets were resuspended in import buffer containing 6% fatty acid–free BSA and 5 mM DTT. 2 mM ATP and 2 mM NADH were added to energize the mitochondria. After incubation for 10 min at 25°C, mitochondria were treated with proteinase K to remove nonimported material.
Protein purification of Tom70-GST, Tom20-GST, and GST
E. coli cells harboring the pGEX4T1 GST-Tom20 cytosolic domain, pGEX4T1 GST-Tom70 cytosolic domain, or pGEX4T1-GST plasmid (Hoseini et al., 2016) were grown to an OD600 of 0.8 at 37°C, and 0.5 mM IPTG was added. After incubation for 16 h, 50 ml of the culture was harvested. The cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.1% NP-40, and 1 mM β-mercaptoethanol) and incubated with 1 mg/ml lysozyme for 30 min at room temperature. After one step of freeze thawing and 15 cycles of sonification for 1 s at 60% duty level, the lysate was cleared at 25,000 g for 5 min at 4°C. The supernatant was incubated with GSH Sepharose 4B beads (GE Healthcare) and washed 3× with PBS. Bound GST fusion proteins were eluted by incubation with elution buffer (50 mM Tris-HCl, pH 8.0, 25 mM GSH, and 1 mM DTT) for 30 min at 4°C. Purification efficiency was analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Binding assay
N-terminally GST-tagged cytosolic domains of Tom20 or Tom70 were expressed in E. coli cells and purified with GSH beads. 15 µl of the GSH Sepharose with bound GST-tagged proteins or GST alone were washed two times with 200 µl import buffer. 200 µl import buffer containing 0.5 µl radiolabeled Atp1, Atp1Δ50, or Atp1mut was subsequently added to the GSH Sepharose and incubated 10 min on an end-over-end rotator at room temperature. The Sepharose was pelleted by centrifugation, washed with import buffer, and boiled in 25 µl sample buffer containing 50 mM DTT.
Dot blot assay
Peptides of Atp1 with a length of 20 amino acids each were synthesized on a cellulose membrane as described previously (Weckbecker et al., 2012). The amino acid frame was shifted by three amino acids from one spot to the next. The peptide spots covered the whole amino acid sequence of Atp1. The membrane was incubated with methanol for 2 min at room temperature, subsequently washed 2 min with H2O, and equilibrated in binding buffer (20 mM Tris-HCl, pH 7.0, and 200 mM NaCl) for 20 min. After this, the membrane was incubated for 3 h with 0.5 µM of recombinantly purified GST-Tom70 dissolved in binding buffer. The membrane was washed twice for 10 min with binding buffer and twice with TBS (10 mM Tris-HCl, pH 7.4, and 150 mM NaCl) and then was subjected to immunoblotting against GST.
iMTS-L profile generation
Suffix sequences of the given protein were subjected to TargetP prediction (TargetP 1.1 standalone software package; DTU Bioinformatics) in standard FastA format. The resulting mitochondrial targeting peptide probability of the suffix sequence was used as positional information and concatenated to generate the raw iMTS-L scoring sequence. A Savitzky–Golay filtering step with the successive subsets of adjacent data points was applied. A window size of the expected value of the length distribution of known MTSs and a quadratic polynomial was used to smooth the raw profile into the iMTS-L probability profile , where i is the position in the amino acid sequence.
iMTS-L propensity calculation
The score was normalized to allow the comparison between amino acid sequences of different lengths. The normalization uses expected value µiMTS-L and SD σiMTS-L were calculated over random sequences (with n = 5,000) using the amino acid frequencies of the whole yeast proteome as follows:
From the score, it was possible to define an overall iMTS-L propensity by summing over all the amino acids of a sequence that had an iMTS-L score higher than those of random sequences (i.e., those positions i where ), excluding the MTS at sequence start, by using the equation
Analysis and sequence property calculation
The iMTS-Ls of a given sequence were detected by searching for regions of positive that were flanked by zero or negative regions. Afterward, the sequence properties were independently deduced from two sets of amino acid sequences containing all identified iMTSs and all yeast proteins. A composition vector for each set was generated and multiplied by the helicity index (Koehl and Levitt, 1999), amphiphilicity index (Cornette et al., 1987), coil index (Ptitsyn and Finkelstein, 1983), β sheet propensity (Crawford et al., 1973), and hydrophibicity index (Fasman, 1989). Analyses and calculations were performed using Microsoft F# functional programming language with the bioinformatics library BioFSharp (available on GitHub at https://github.com/CSBiology/BioFSharp) and the graphical library FSharp.Plotly (available on GitHub at https://github.com/muehlhaus/FSharp.Plotly).
Online supplemental material
The supplemental material provides additional information on the distribution of iMTS-L scores in proteins (Fig. S1) and the import kinetics corresponding with Fig. 8 (Fig. S2) as well as lists of iMTS-L propensities for mitochondrial proteins in S. cerevisiae (Tables S1 and S2).
Supplementary Material
Acknowledgments
We thank Doron Rapaport for the Δtom70/Δtom71 deletion strain and for E. coli strains to express Tom70-GST, Tom20-GST, and GST. We are grateful to Sabine Knaus for technical assistance and to Bruce Morgan for discussion.
The work was supported by grants from the Deutsche Forschungsgemeinschaft (He2803/9-1 and IRTG 1830 to J.M. Herrmann) and the Landesschwerpunkt BioComp.
The authors declare no competing financial interests.
Author contributions: S. Hess generated mutants, recombinant proteins, cell fractions, and antibodies. F. Boos analyzed the data and designed experiments. S. Backes purified mitochondria, synthesized radiolabeled proteins, and performed in vitro import experiments. M.W. Woellhaf, S. Gödel, and T. Mühlhaus generated the iMTS-L prediction algorithm and analyzed the targeting signals in Atp25. S. Gödel characterized the biochemical properties of iMTS-Ls and assessed the relation between iMTS-L propensity and import efficiency. M. Jung generated the peptide scans for Atp1. T. Mülhaus and J.M. Herrmann analyzed the data, designed the experiments, and wrote the draft of the manuscript to which all other authors contributed.
References
- Alikhani, N., Berglund A.K., Engmann T., Spånning E., Vögtle F.N., Pavlov P., Meisinger C., Langer T., and Glaser E.. 2011. Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J. Mol. Biol. 410:400–410. 10.1016/j.jmb.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Altmann, K., Dürr M., and Westermann B.. 2007. Saccharomyces cerevisiae as a model organism to study mitochondrial biology. In Mitochondria. Practical Protocols. Vol. 372. Leister D., and Herrmann J.M., editors. Humana Press, Totowa, New Jersey. 81–90. 10.1007/978-1-59745-365-3_6 [DOI] [PubMed] [Google Scholar]
- Backes, S., and Herrmann J.M.. 2017. Protein Translocation into the Intermembrane Space and Matrix of Mitochondria: Mechanisms and Driving Forces. Front. Mol. Biosci. 4:83. 10.3389/fmolb.2017.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A., and Schatz G.. 1987. Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc. Natl. Acad. Sci. USA. 84:3117–3121. 10.1073/pnas.84.10.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, R., Gladkova C., Mapa K., Witte G., and Mokranjac D.. 2015. Protein translocation channel of mitochondrial inner membrane and matrix-exposed import motor communicate via two-domain coupling protein. eLife. 4:e11897. 10.7554/eLife.11897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo, M.K., Tzankov S., Fan A.C., Dejgaard K., Thomas D.Y., and Young J.C.. 2007. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell. 18:3414–3428. 10.1091/mbc.E07-01-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel, G., and Dobberstein B.. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67:835–851. 10.1083/jcb.67.3.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonchird, C., Messenguy F., and Dubois E.. 1991. Determination of amino acid sequences involved in the processing of the ARG5/ARG6 precursor in Saccharomyces cerevisiae. Eur. J. Biochem. 199:325–335. 10.1111/j.1432-1033.1991.tb16128.x [DOI] [PubMed] [Google Scholar]
- Brix, J., Rüdiger S., Bukau B., Schneider-Mergener J., and Pfanner N.. 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274:16522–16530. 10.1074/jbc.274.23.16522 [DOI] [PubMed] [Google Scholar]
- Chacinska, A., Lind M., Frazier A.E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H.E., Truscott K.N., Guiard B., et al. . 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 120:817–829. 10.1016/j.cell.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Chan, N.C., Likić V.A., Waller R.F., Mulhern T.D., and Lithgow T.. 2006. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J. Mol. Biol. 358:1010–1022. 10.1016/j.jmb.2006.02.062 [DOI] [PubMed] [Google Scholar]
- Chatzi, K.E., Sardis M.F., Tsirigotaki A., Koukaki M., Šoštarić N., Konijnenberg A., Sobott F., Kalodimos C.G., Karamanou S., and Economou A.. 2017. Preprotein mature domains contain translocase targeting signals that are essential for secretion. J. Cell Biol. 216:1357–1369. 10.1083/jcb.201609022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornette, J.L., Cease K.B., Margalit H., Spouge J.L., Berzofsky J.A., and DeLisi C.. 1987. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J. Mol. Biol. 195:659–685. 10.1016/0022-2836(87)90189-6 [DOI] [PubMed] [Google Scholar]
- Crawford, J.L., Lipscomb W.N., and Schellman C.G.. 1973. The reverse turn as a polypeptide conformation in globular proteins. Proc. Natl. Acad. Sci. USA. 70:538–542. 10.1073/pnas.70.2.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis, E.M., Longthorne R.F., and Gurdon J.B.. 1978. Intracellular migration of nuclear proteins in Xenopus oocytes. Nature. 272:254–256. 10.1038/272254a0 [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen H., Brunak S., and von Heijne G.. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005–1016. 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Brunak S., von Heijne G., and Nielsen H.. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2:953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Fan, A.C., Kozlov G., Hoegl A., Marcellus R.C., Wong M.J., Gehring K., and Young J.C.. 2011. Interaction between the human mitochondrial import receptors Tom20 and Tom70 in vitro suggests a chaperone displacement mechanism. J. Biol. Chem. 286:32208–32219. 10.1074/jbc.M111.280446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasman, G.D., editor. 1989. Prediction of Protein Structure and the Principles of Protein Conformation. Springer US, Boston, MA. 10.1007/978-1-4613-1571-1 [DOI] [Google Scholar]
- Gärtner, F., Voos W., Querol A., Miller B.R., Craig E.A., Cumsky M.G., and Pfanner N.. 1995. Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants: Independence from receptors, but requirement for matrix hsp70 translocase function. J. Biol. Chem. 270:3788–3795. 10.1074/jbc.270.8.3788 [DOI] [PubMed] [Google Scholar]
- Gaume, B., Klaus C., Ungermann C., Guiard B., Neupert W., and Brunner M.. 1998. Unfolding of preproteins upon import into mitochondria. EMBO J. 17:6497–6507. 10.1093/emboj/17.22.6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S.J., Neupert W., and Rapaport D.. 2007. Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol. 80:761–781. 10.1016/S0091-679X(06)80035-X [DOI] [PubMed] [Google Scholar]
- Hartl, F.U., Schmidt B., Wachter E., Weiss H., and Neupert W.. 1986. Transport into mitochondria and intramitochondrial sorting of the Fe/S protein of ubiquinol-cytochrome c reductase. Cell. 47:939–951. 10.1016/0092-8674(86)90809-3 [DOI] [PubMed] [Google Scholar]
- Hasson, S.A., Damoiseaux R., Glavin J.D., Dabir D.V., Walker S.S., and Koehler C.M.. 2010. Substrate specificity of the TIM22 mitochondrial import pathway revealed with small molecule inhibitor of protein translocation. Proc. Natl. Acad. Sci. USA. 107:9578–9583. 10.1073/pnas.0914387107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines, V., and Schatz G.. 1993. Precursor binding to yeast mitochondria. A general role for the outer membrane protein Mas70p. J. Biol. Chem. 268:449–454. [PubMed] [Google Scholar]
- Horvath, S.E., Böttinger L., Vögtle F.N., Wiedemann N., Meisinger C., Becker T., and Daum G.. 2012. Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 287:36744–36755. 10.1074/jbc.M112.398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseini, H., Pandey S., Jores T., Schmitt A., Franz-Wachtel M., Macek B., Buchner J., Dimmer K.S., and Rapaport D.. 2016. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 283:3338–3352. 10.1111/febs.13813 [DOI] [PubMed] [Google Scholar]
- Hurt, E.C., and Schatz G.. 1987. A cytosolic protein contains a cryptic mitochondrial targeting signal. Nature. 325:499–503. 10.1038/325499a0 [DOI] [PubMed] [Google Scholar]
- Hurt, E.C., Soltanifar N., Goldschmidt-Clermont M., Rochaix J.-D., and Schatz G.. 1986. The cleavable pre-sequence of an imported chloroplast protein directs attached polypeptides into yeast mitochondria. EMBO J. 5:1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura, E., Zavodszky E., Shao S., Wohlever M.L., Keenan R.J., and Hegde R.S.. 2016. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol. Cell. 63:21–33. 10.1016/j.molcel.2016.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk, O., Ott M., Funes S., Ostermann K., Rödel G., and Herrmann J.M.. 2006. Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe. Eukaryot. Cell. 5:997–1006. 10.1128/EC.00092-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehl, P., and Levitt M.. 1999. Structure-based conformational preferences of amino acids. Proc. Natl. Acad. Sci. USA. 96:12524–12529. 10.1073/pnas.96.22.12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto, N., Shaw J.M., and Okamoto K.. 2008. Tetratricopeptide repeat proteins Tom70 and Tom71 mediate yeast mitochondrial morphogenesis. EMBO Rep. 9:63–69. 10.1038/sj.embor.7401113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., and Chook Y.M.. 2006. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 126:543–558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.M., Sedman J., Neupert W., and Stuart R.A.. 1999. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem. 274:20937–20942. 10.1074/jbc.274.30.20937 [DOI] [PubMed] [Google Scholar]
- Li, J., Qian X., Hu J., and Sha B.. 2009. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284:23852–23859. 10.1074/jbc.M109.023986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longen, S., Woellhaf M.W., Petrungaro C., Riemer J., and Herrmann J.M.. 2014. The disulfide relay of the intermembrane space oxidizes the ribosomal subunit mrp10 on its transit into the mitochondrial matrix. Dev. Cell. 28:30–42. 10.1016/j.devcel.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Lubben, T.H., Theg S.M., and Keegstra K.. 1988. Transport of proteins into chloroplasts. Photosynth. Res. 17:173–194. 10.1007/BF00047688 [DOI] [PubMed] [Google Scholar]
- Malhotra, K., Sathappa M., Landin J.S., Johnson A.E., and Alder N.N.. 2013. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat. Struct. Mol. Biol. 20:965–972. 10.1038/nsmb.2613 [DOI] [PubMed] [Google Scholar]
- McGeoch, D.J. 1985. On the predictive recognition of signal peptide sequences. Virus Res. 3:271–286. 10.1016/0168-1702(85)90051-6 [DOI] [PubMed] [Google Scholar]
- Meinecke, M., Wagner R., Kovermann P., Guiard B., Mick D.U., Hutu D.P., Voos W., Truscott K.N., Chacinska A., Pfanner N., and Rehling P.. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science. 312:1523–1526. 10.1126/science.1127628 [DOI] [PubMed] [Google Scholar]
- Melin, J., Kilisch M., Neumann P., Lytovchenko O., Gomkale R., Schendzielorz A., Schmidt B., Liepold T., Ficner R., Jahn O., et al. . 2015. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim. Biophys. Acta. 1853:1850–1859. 10.1016/j.bbamcr.2015.04.021 [DOI] [PubMed] [Google Scholar]
- Mootha, V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M., et al. . 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 115:629–640. 10.1016/S0092-8674(03)00926-7 [DOI] [PubMed] [Google Scholar]
- Morgenstern, M., Stiller S.B., Lübbert P., Peikert C.D., Dannenmaier S., Drepper F., Weill U., Höß P., Feuerstein R., Gebert M., et al. . 2017. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Reports. 19:2836–2852. 10.1016/j.celrep.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossmann, D., Vögtle F.N., Taskin A.A., Teixeira P.F., Ring J., Burkhart J.M., Burger N., Pinho C.M., Tadic J., Loreth D., et al. . 2014. Amyloid-β peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 20:662–669. 10.1016/j.cmet.2014.07.024 [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa M.. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 14:897–911. 10.1016/S0888-7543(05)80111-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., Brinker A., Paschen S.A., Moarefi I., Hayer-Hartl M., Neupert W., and Brunner M.. 2002. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 21:3659–3671. 10.1093/emboj/cdf358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papić, D., Elbaz-Alon Y., Koerdt S.N., Leopold K., Worm D., Jung M., Schuldiner M., and Rapaport D.. 2013. The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol. Cell. Biol. 33:4083–4094. 10.1128/MCB.00227-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn, O.B., and Finkelstein A.V.. 1983. Theory of protein secondary structure and algorithm of its prediction. Biopolymers. 22:15–25. 10.1002/bip.360220105 [DOI] [PubMed] [Google Scholar]
- Ramage, L., Junne T., Hahne K., Lithgow T., and Schatz G.. 1993. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 12:4115–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh, A., Peleh V., Martinez-Caballero S., Wollweber F., Sommer F., van der Laan M., Schroda M., Alexander R.T., Campo M.L., and Herrmann J.M.. 2016. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J. Cell Biol. 214:417–431. 10.1083/jcb.201602074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling, P., Model K., Brandner K., Kovermann P., Sickmann A., Meyer H.E., Kühlbrandt W., Wagner R., Truscott K.N., and Pfanner N.. 2003. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science. 299:1747–1751. 10.1126/science.1080945 [DOI] [PubMed] [Google Scholar]
- Rhee, H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., and Ting A.Y.. 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 339:1328–1331. 10.1126/science.1230593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer, K.A., Foo J.H., Ng A., Petrie E.J., Shilling P.J., Perry A.J., Mertens H.D., Lithgow T., Mulhern T.D., and Gooley P.R.. 2011. Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J. Mol. Biol. 405:804–818. 10.1016/j.jmb.2010.11.017 [DOI] [PubMed] [Google Scholar]
- Sato, T., Esaki M., Fernandez J.M., and Endo T.. 2005. Comparison of the protein-unfolding pathways between mitochondrial protein import and atomic-force microscopy measurements. Proc. Natl. Acad. Sci. USA. 102:17999–18004. 10.1073/pnas.0504495102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendzielorz, A.B., Schulz C., Lytovchenko O., Clancy A., Guiard B., Ieva R., van der Laan M., and Rehling P.. 2017. Two distinct membrane potential-dependent steps drive mitochondrial matrix protein translocation. J. Cell Biol. 216:83–92. 10.1083/jcb.201607066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibich, D., Gloge F., Pöhner I., Björkholm P., Wade R.C., von Heijne G., Bukau B., and Kramer G.. 2016. Global profiling of SRP interaction with nascent polypeptides. Nature. 536:219–223. 10.1038/nature19070 [DOI] [PubMed] [Google Scholar]
- Sherman, F. 1963. Respiration-deficient mutants of yeast. I. Genetics. Genetics. 48:375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota, T., Mabuchi H., Tanaka-Yamano S., Yamano K., and Endo T.. 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. USA. 108:15179–15183. 10.1073/pnas.1105921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota, T., Imai K., Qiu J., Hewitt V.L., Tan K., Shen H.H., Sakiyama N., Fukasawa Y., Hayat S., Kamiya M., et al. . 2015. Molecular architecture of the active mitochondrial protein gate. Science. 349:1544–1548. 10.1126/science.aac6428 [DOI] [PubMed] [Google Scholar]
- Sickmann, A., Reinders J., Wagner Y., Joppich C., Zahedi R., Meyer H.E., Schönfisch B., Perschil I., Chacinska A., Guiard B., et al. . 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 100:13207–13212. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg, C., Bauer M.F., Guiard B., Neupert W., and Brunner M.. 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 384:582–585. 10.1038/384582a0 [DOI] [PubMed] [Google Scholar]
- Ting, S.Y., Yan N.L., Schilke B.A., and Craig E.A.. 2017. Dual interaction of scaffold protein Tim44 of mitochondrial import motor with channel-forming translocase subunit Tim23. eLife. 6:e23609. 10.7554/eLife.23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott, K.N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A.J., Rassow J., Pfanner N., and Wagner R.. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8:1074–1082. 10.1038/nsb726 [DOI] [PubMed] [Google Scholar]
- Vögtle, F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., and Meisinger C.. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 139:428–439. 10.1016/j.cell.2009.07.045 [DOI] [PubMed] [Google Scholar]
- von Heijne, G. 1986a. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. 1986b. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683–4690. 10.1093/nar/14.11.4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckbecker, D., Longen S., Riemer J., and Herrmann J.M.. 2012. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 31:4348–4358. 10.1038/emboj.2012.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, W., Mandel G., Zwizinski C., Bates M., and Killick T.. 1978. Synthesis of phage M13 coat protein and its assembly into membranes in vitro. Proc. Natl. Acad. Sci. USA. 75:1754–1758. 10.1073/pnas.75.4.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, N., and Pfanner N.. 2017. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 86:685–714. 10.1146/annurev-biochem-060815-014352 [DOI] [PubMed] [Google Scholar]
- Wienhues, U., Becker K., Schleyer M., Guiard B., Tropschug M., Horwich A.L., Pfanner N., and Neupert W.. 1991. Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J. Cell Biol. 115:1601–1609. 10.1083/jcb.115.6.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellhaf, M.W., Sommer F., Schroda M., and Herrmann J.M.. 2016. Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein. Mol. Biol. Cell. 27:3031–3039. 10.1091/mbc.E16-07-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel, L., Trojanowska A., Sztolsztener M.E., and Chacinska A.. 2013. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol. Biol. Cell. 24:543–554. 10.1091/mbc.E12-09-0649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., and Sha B.. 2006. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13:589–593. 10.1038/nsmb1106 [DOI] [PubMed] [Google Scholar]
- Xue, Q., Pei H., Liu Q., Zhao M., Sun J., Gao E., Ma X., and Tao L.. 2017. MICU1 protects against myocardial ischemia/reperfusion injury and its control by the importer receptor Tom70. Cell Death Dis. 8:e2923. 10.1038/cddis.2017.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagawa, K., Yamano K., Oguro T., Maeda M., Sato T., Momose T., Kawano S., and Endo T.. 2010. Structural basis for unfolding pathway-dependent stability of proteins: vectorial unfolding versus global unfolding. Protein Sci. 19:693–702. 10.1002/pro.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., Uchida M., Esaki M., Nishikawa S., Yoshihisa T., et al. . 2009. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284:31635–31646. 10.1074/jbc.M109.041756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H., Itoh N., Kawano S., Yatsukawa Y., Momose T., Makio T., Matsunaga M., Yokota M., Esaki M., Shodai T., et al. . 2011. Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria. Proc. Natl. Acad. Sci. USA. 108:91–96. 10.1073/pnas.1014918108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., Hoogenraad N.J., and Hartl F.U.. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 112:41–50. 10.1016/S0092-8674(02)01250-3 [DOI] [PubMed] [Google Scholar]
- Zanphorlin, L.M., Lima T.B., Wong M.J., Balbuena T.S., Minetti C.A., Remeta D.P., Young J.C., Barbosa L.R., Gozzo F.C., and Ramos C.H.. 2016. Heat shock protein 90 kDa Hsp90 has a second functional interaction site with the mitochondrial import receptor Tom70. J. Biol. Chem. 291:18620–18631. 10.1074/jbc.M115.710137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.