Abstract
N-linked glycosylation is an enzymatic reaction in which an oligosaccharide is transferred en bloc onto an asparagine residue of an acceptor polypeptide, catalyzed by oligosaccharyltransferase (OST). Despite the available crystal structures, the role of the external loop EL5, which is critical for the catalytic cycle, is enigmatic as EL5 in the crystal structures is partially absent or blocks a pathway of lipid-linked oligosaccharide to the active site. Here we report the molecular origin of EL5 conformational changes through a series of molecular dynamics simulations of a bacterial OST, Campylobacter lari PglB. The simulations reveal that the isoprenoid moiety of lipid-linked oligosaccharide favorably binds to a hydrophobic groove of the PglB transmembrane domain. This binding triggers the conformational changes of the transmembrane domain and subsequently impairs the structural stability of EL5, leading to disordered EL5 with open conformations that are required for correct placement of the oligosaccharide in the active site.
Keywords: external loop 5, lipid-linked oligosaccharide, molecular dynamics simulation, N-linked glycosylation, oligosaccharyltransferase
Introduction
The covalent attachment of an oligosaccharide onto an asparagine residue in a protein, N-glycosylation, is a fundamental posttranslational modification that affects cellular functions (Weerapana and Imperiali 2006; Moremen et al. 2012) and occurs across all domains of life in a highly conserved manner (Schwarz and Aebi 2011; Aebi 2013). Asparagine residues, as part of a consensus sequon, are chemically modified with carbohydrates from lipid-linked oligosaccharides (LLOs) (Figure 1A). The N-glycosylation sequon is generally a consecutive sequence of Asn-X-Ser/Thr (X ≠ Pro), but other nonstandard motifs (such as Asn-X-Cys, Asn-Gly and Asn-X-Val) occur with very low frequency (Moremen et al. 2012). An LLO is the assembly of an oligosaccharide with a lipid-phospho carrier, which is typically dolichol pyrophosphate in eukaryotes and archaea, and undecaprenyl pyrophosphate (UndPP) in eubacteria (Supplementary data, Figure S1), although there are exceptions (e.g., dolichol monophosphate in methanogenic archea (Larkin et al. 2013)).
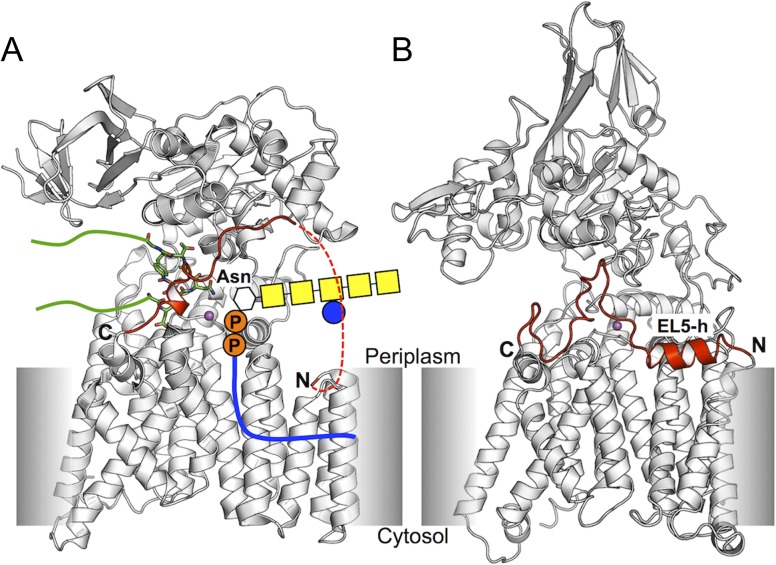
Fig. 1.
Crystal structures of oligosaccharyltransferase. (A) C. lari PglB (ClPglB). The red dotted line indicates missing residues in the crystal structure. Binding of a donor LLO is schematically represented in the structure. A stick-represented short peptide linked with green lines indicates a nascent acceptor polypeptide. (B) A. fulgidus AglB. (AfAglB). EL5-h is an α-helix in the N-terminal half of EL5. EL5s are colored red in both structures. The N- and C-termini of EL5 are marked by N and C, respectively. Divalent cations, which form the catalytic site with proximate residues, are shown in magenta sphere. This figure is available in black and white in print and in color at Glycobiology online.
Oligosaccharyltransferase (OST), a membrane protein, catalyzes the en bloc transfer of oligosaccharides in LLOs onto the side-chain amide groups of asparagine residues in nascent polypeptides. The eukaryotic OST is a heptameric protein complex containing STT3 (staurosporine and temperature sensitivity 3), which is the largest and most conserved catalytic subunit of OST (Burda and Aebi 1999; Yan and Lennarz 2002). The archaeal and bacterial OSTs are single subunit enzymes homologous to the STT3 subunit of the eukaryotic OST (Kelleher and Gilmore 2006), which are referred to as “AglB” (archaeal glycosylation B) and “PglB” (protein glycosylation B), respectively. All OST catalytic enzymes share a common topology signature of an N-terminal transmembrane (TM) part and a soluble C-terminal domain (Figure 1). The similarity in sequence and topology indicates that catalytic OSTs from distinct species share a common reaction mechanism.
Lizak et al. reported the X-ray structure of a full-length bacterial OST, the PglB protein of Campylobacter lari (ClPglB). The structure is a complex of ClPglB with the hexapeptide DQNATF, a sequon-containing sequence (PDB id: 3rce) (Lizak et al. 2011) (Figure 1A). In this structure, the TM domain contains 13 helices (TM1 to TM13). The fifth external loop (EL5, residues 282−323) between TM9 and TM10 is partially disordered, and the coordinates of its N-terminal 24 residues (283–306) are missing in the structure file. Based on the crystal structure, Lizak et al. proposed a catalytic cycle for the glycosylation mechanism. Step1: In the absence of bound acceptor peptide (i.e., nascent protein), the full-length EL5 is flexible and not structured. The C-terminal half of EL5 is ordered upon peptide binding, resulting in the formation of the catalytic site where a divalent metal ion is coordinated with the surrounding acidic residues (Supplementary data, Figure S2A). Step 2: An LLO binds to OST, forming a ternary complex (Supplementary data, Figure S2B). An N-glycosidic linkage between the amide nitrogen of the asparagine side chain of the acceptor peptide and the C1 carbon of the first saccharide moiety of the LLO is formed at the active site (Supplementary data, Figure S2C). Step 3: The glycosylated peptide and the cleaved lipid-linked pyrophosphate are dissociated from ClPglB.
Matsumoto et al. reported two crystal structures of a full-length AglB from Archaeoglobus fulgidus (AfAglB) under different crystallization conditions (Matsumoto et al. 2013). One crystal form (PDB id: 3waj) is an AfAglB complex with a sulfate ion next to the divalent metal ion. In this structure, the entire EL5 is disordered. The other crystal structure (PDB id: 3wak) corresponds to the apo form in which no ligands (e.g., acceptor peptide and sulfate ion) are bound. In this structure, the entire EL5 is well structured with a parallel, partially membrane-buried α-helix in the N-terminal half (EL5-h) (Figure 1B). Based on the previous ClPglB and their AfAglB structures, the authors proposed a refined glycosylation mechanism. Step 1: EL5 is structured in the absence of substrates (apo state). Step 2: The binding of sequon in an acceptor peptide makes the N-terminal half of EL5 flexible and disordered (Supplementary data, Figure S2A: peptide-bound state). Step 3: An LLO binds to the binary complex (Supplementary data, Figure S2B: ternary complex state). Step 4: The acceptor protein is glycosylated (Supplementary data, Figure S2C) and the dynamic motions of Ser/Thr-binding pocket facilitate the release of the glycosylated products (Nyirenda et al. 2013) (transition state). Step 5: The glycosylated protein is detached from OST, and the lipid phosphate is a leaving product, interacting with an adjacent positively charged residue (Arg426 in AfAglB and Arg375 in ClPglB) (Supplementary data, Figure S2D). EL5 is entirely disordered in this step (lipid-phosphate bound state). The authors also mentioned that the binding of LLO to ClPglB or AfAglB might precede that of the acceptor peptide in both proposed glycosylation mechanisms.
These available structures and catalytic cycle models have shed light on the complex mechanism of OST action. Arguably, however, nearly the entire mechanism is unknown at the molecular level. Kern et al. performed a computational study on LLO in membrane bilayers of three different lipid types, showing its oligosaccharide orientations parallel to the bilayer surface (Kern et al. 2014). They also suggested the potential role of the isoprenyl moiety as a tentacle to search for a binding site in the TM domain of OST. Pedebos et al. simulated ClPglB with its peptide acceptor or glycopeptide bound forms, suggesting motions likely associated with the glycopeptide detachment and identifying transient states and an octahedral metal coordination at the catalytic site (Pedebos et al. 2015). However, their systems do not include polyisoprenyl pyrophosphate and thus the study has limitation on exploring its biological role.
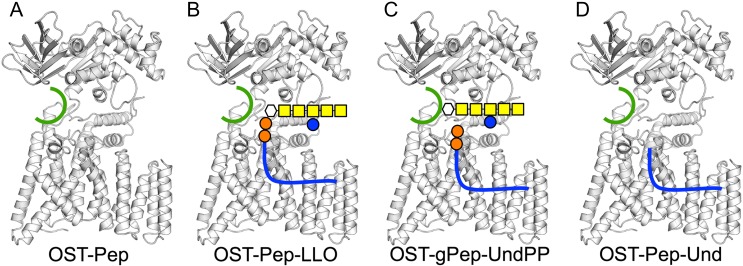
In particular, the least understood feature is the role of the external loop EL5. The crystal structures (ordered EL5 in apo AfAglB and entirely or partially disordered EL5 in other crystal structures) indicate that EL5 must undergo a large conformational change during the reaction cycle to properly position the oligosaccharide moiety of LLO at the catalytic site. This is a critical question in deciphering the N-glycosylation mechanism, but the EL5 conformational change mechanism is enigmatic. This study aims to explore the function of EL5 by performing a series of molecular dynamics (MD) simulations for ClPglB in complex with: (i) only a short sequon-containing peptide (Pep: DQNATF); (ii) Pep with a bacterial LLO (Supplementary data, Figure S1); and (iii) a glycopeptide (gPep) with an undecaprenyl pyrophosphate (UndPP) moiety (Figure 2A−C). In addition, we perform MD simulations of an artificial ternary complex of ClPglB with Pep and an undecaprenyl (Und) moiety (Figure 2D) to specifically investigate the role of Und upon its binding to ClPglB. Collectively, our key finding is that binding of the Und moiety to a hydrophobic TM groove in ClPglB induces the conformational changes of the TM domain, resulting in unstructured EL5 in the N-terminal half.
Fig. 2.
Schematic representations of simulation systems in this study. (A) OST-Pep, a binary complex of ClPglB with peptide (green). (B) OST-Pep-LLO, a ternary complex of ClPglB with peptide and LLO. (C) OST-gPep-UndPP, a ternary complex of ClPglB with glycopeptide and undecaprenyl pyrophosphate. (D) OST-Pep-Und, a ternary complex of ClPglB with peptide and undecaprenyl moiety. This figure is available in black and white in print and in color at Glycobiology online.
Results
LLO binding makes EL5 unstructured and open conformations
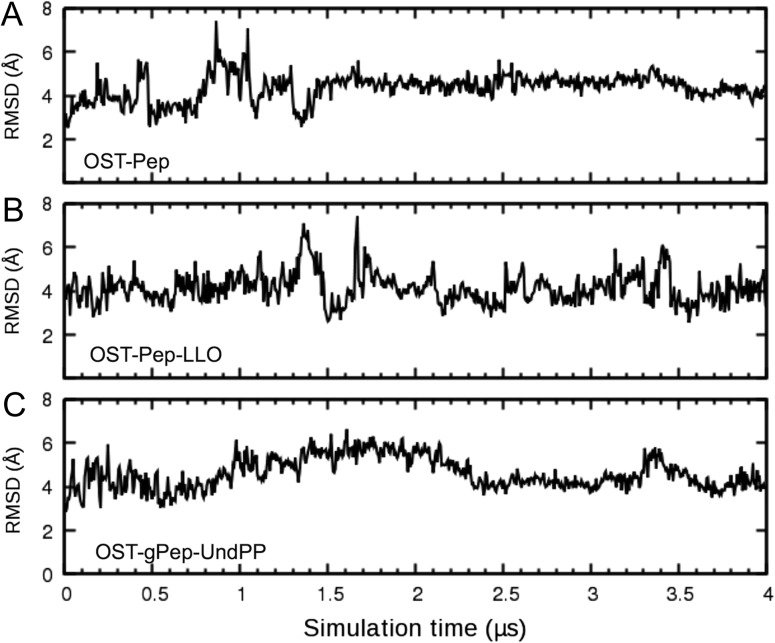
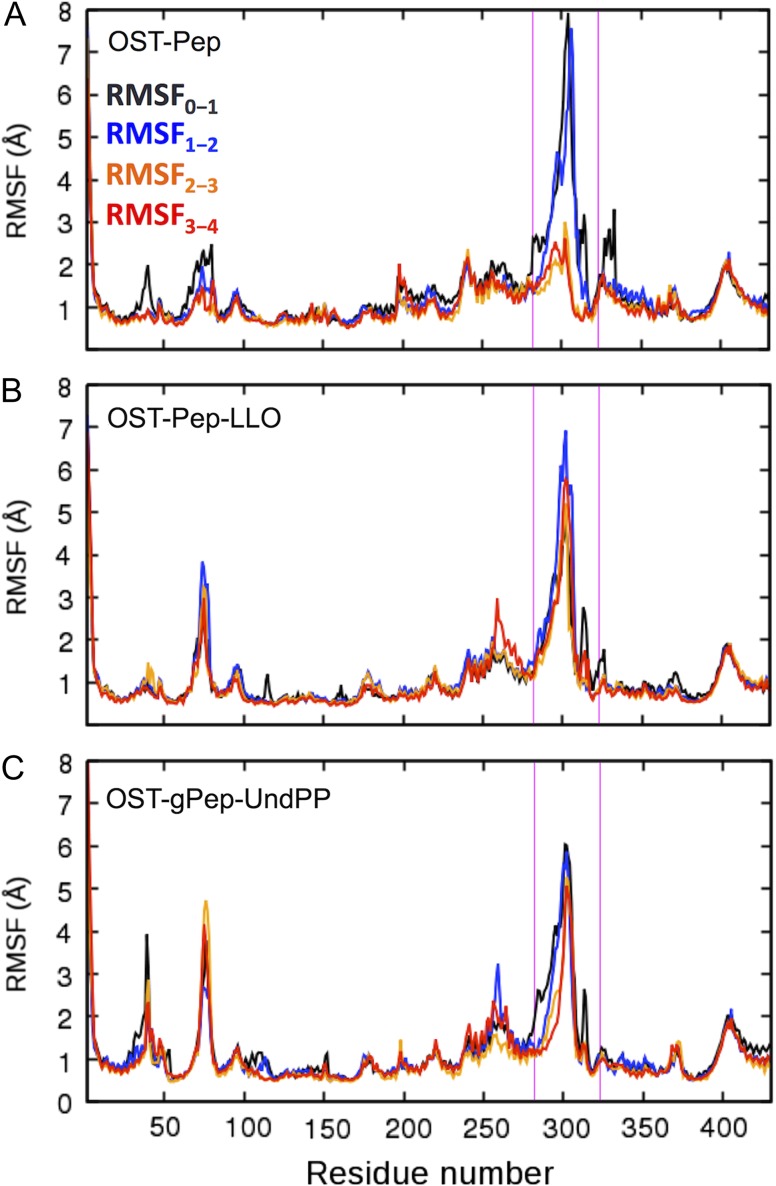
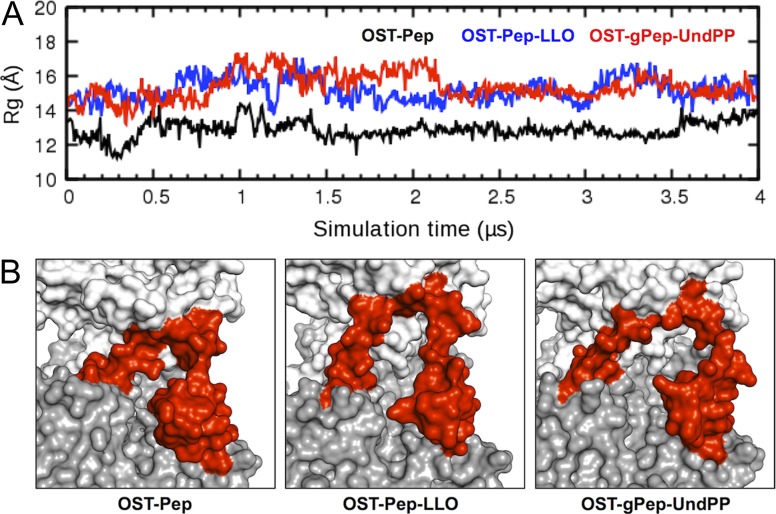
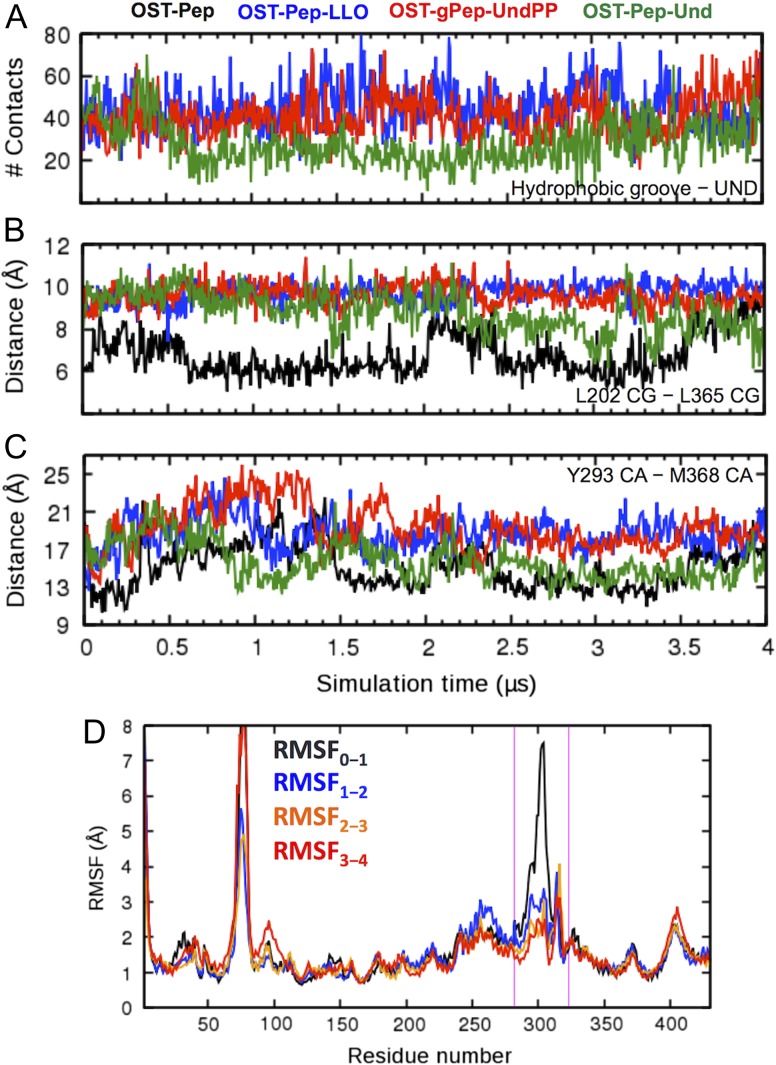
Figure 3 shows the time-series of root-mean-square deviation (RMSD) of EL5 from the initial structures in three ClPglB complex systems (OST-Pep, OST-Pep-LLO, and OST-gPep-UndPP). The RMSD of OST-Pep becomes stable after 1.5 μs, while OST-Pep-LLO and OST-gPep-UndPP show large RMSD fluctuations. This difference in EL5 dynamics is also confirmed in Figure 4 by the root-mean-square fluctuation (RMSF) of Cα atoms in different simulation periods. In OST-Pep, the RMSF profiles in the EL5 region significantly decrease after 2 μs, indicating ordering of EL5. On the contrary, OST-Pep-LLO and OST-gPep-UndPP do not show any decrease in RMSF for this region, indicating that LLO or glycosylated products (N-linked oligosaccharide moiety and undecaprenyl pyrophosphate) in ClPglB make EL5 more dynamic in the presence of Pep. EL5 shows the largest RMSFs during the entire simulations of all three systems. When we focus on the EL5 region, its N-terminal missing residues in the crystal structure show distinctly larger fluctuations than the rest of EL5 (Supplementary data, Figure S3) in OST-Pep-LLO and OST-gPep-UndPP. The initial simulation structures and snapshots at 2 μs and 4 μs are shown in Supplementary data, Figure S4 to illustrate the EL5 structural differences in the three systems. Based on these data, the existence of LLO or the glycosylated products appears to make the N-terminal half of EL5 more unstructured. Figure 5A shows the time-series of the radius of gyration (Rg) of EL5. The ternary complexes show comparable Rg values (15.15 ± 0.60 Å for OST-Pep-LLO and 15.29 ± 0.51 Å for OST-gPep-UndPP), but larger ones than the binary complex (12.95 ± 0.39 Å). The structure figures in Figure 5B clearly show that EL5 of the ternary structures has more open conformations.
Fig. 3.
RMSD time-series of EL5. (A) OST-Pep. (B) OST-Pep-LLO. (C) OST-gPep-UndPP. The RMSD was calculated using the Cα atoms of EL5 with respect to the initial structure. The RMSD standard deviations for the last 2 μs are 0.34 Å for OST-Pep, 0.62 Å for OST-Pep-LLO, and 0.53 Å for OST-gPep-UndPP.
Fig. 4.
RMSF plots in different simulation periods (RMSF0-1: 0−1 μs, RMSF1-2: 1−2 μs, RMSF2-3: 2−3 μs, and RMSF3-4: 3−4 μs). (A) OST-Pep. (B) OST-Pep-LLO. (C) OST-gPep-UndPP. The magenta lines in each plot indicate the EL5 region (residues 282−323). This figure is available in black and white in print and in color at Glycobiology online.
Fig. 5.
Conformational changes of EL5. (A) Radius of gyration (Rg) time-series of EL5 in OST-Pep (black), OST-Pep-LLO (blue), and OST-gPep-UndPP (red). The Rg was calculated using the Cα atoms of EL5. (B) The last snapshot of each system is shown in surface representation. The transmembrane domain (residues 2−432), periplasmic domain (433−712), and EL5 (282−323) are colored gray, white, and red, respectively. This figure is available in black and white in print and in color at Glycobiology online.
LLO binding triggers TM domain movements, inducing EL5 conformational changes
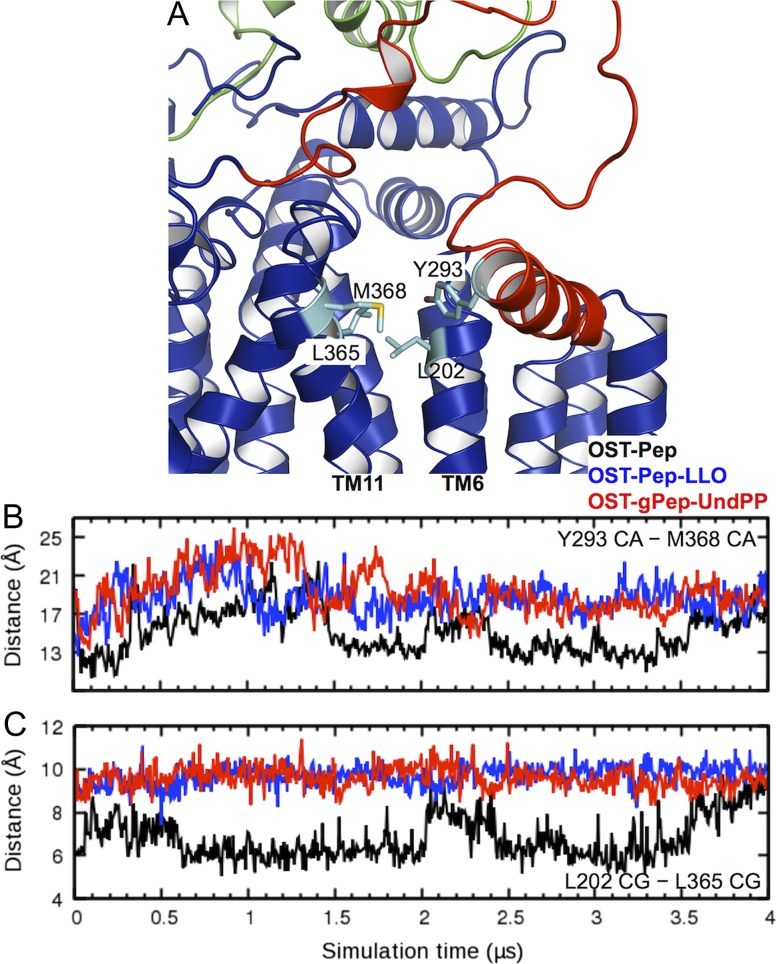
To understand why the EL5 dynamics in OST-Pep differs from those in OST-Pep-LLO and OST-gPep-UndPP, heavy atom contacts between EL5 and the rest of ClPglB were analyzed to identify residues with distinct interaction patterns in between the binary and ternary complexes (Supplementary data, Figure S5). This analysis reveals a notable feature of Y293 in the N-terminal half of EL5, where the residue has frequent contacts with the rest of ClPglB in OST-Pep, but not in OST-Pep-LLO and OST-gPep-UndPP. During our simulations, EL5 has a stable helix (EL5-h; residues 285−294) in its N-terminal half (Supplementary data, Figure S6). Y293 is located at the C-terminus of EL5-h and turns out to dominantly interact with S201 and M368 (contact population = 63.6% and 34.1%, respectively). M368 is a residue comprising the hydrophobic groove of ClPglB (L202, A205, P361, L365, M368 and F376) (Lizak et al. 2011) and S201 is a consecutive residue to a hydrophobic groove residue L202 (Figure 6A). Therefore, the displacement of EL5-h from the hydrophobic groove was measured by the Cα distance between Y293 and M368 in Figure 6B, showing that large deviations of EL5-h from the hydrophobic groove are associated with the open conformations of EL5 in the ternary complexes.
Fig. 6.
Deviations of EL5-h from the hydrophobic groove and its conformational changes. (A) A representative structure to show the key residue positions. (B) Time-series of the Cα distance between Y293 and M368 in OST-Pep (black), OST-Pep-LLO (blue), and OST-gPep-UndPP (red). (C) Time-series of the Cγ distance between L202 and L365. This figure is available in black and white in print and in color at Glycobiology online.
Figure 6C is the time-series of the Cγ distance between L202 and L365. Both residues are part of the hydrophobic groove, but each belongs to different TM helices (L202 in TM6 and L365 in TM11). In the ternary complexes, the distances are less variable and higher than those in the binary complex during the simulations. Moreover, the L202−L365 Cγ distance profiles show clear correlations with the Y293−M368 Cα distances, i.e., the deviations of EL5-h from the hydrophobic groove (Figure 6B), after 1.5 μs (which is an approximate simulation time needed for sufficient equilibration of EL5). The L202−L365 Cγ distances appear to be stabilized before the Y293−L368 Cα distances, suggesting the stability of the TM helices before EL5 stability. This also suggests that the conformational changes in the TM domain induce those in EL5, once EL5 is stabilized. Interestingly, the TM domain in OST-Pep shows a breathing motion with the concerted conformational changes of EL5 even after 1.5 μs, indicating that EL5 is ordered but not completely rigid. Despite such breathing motions, the extent of the deviations from the hydrophobic groove is smaller than those in the ternary complexes.
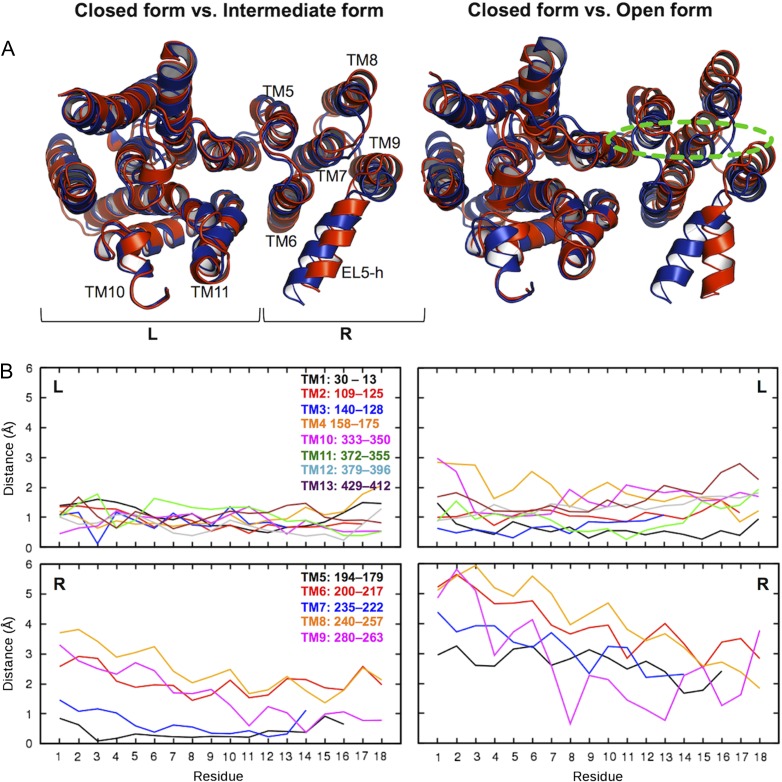
Based on the L202−L365 Cγ distance, the EL5 conformations can be loosely classified into “closed,” “intermediate,” and “open” forms (Figure 7A) to characterize if the conformational changes in the TM domain are associated only with M6 and TM11 or all TM helices. Figure 7B shows the Cα-atom displacements of the closed form from the intermediate or open form for each TM helix. Clearly, increased L202−L365 Cγ distances, i.e., opening of the hydrophobic glove, involves movements of the entire TM domain, not just TM6 and TM11. In particular, the TM movements are mainly driven by the right TM bundle (TM5−TM9). The extents of the movements are more dominant on the periplasmic side than the cytoplasmic one.
Fig. 7.
Conformational changes of TM domain. (A) Superposed structure pairs. The closed form is colored blue. The intermediate (left) and open (right) forms are colored red. Dotted green line indicates an approximate position of the carbohydrate moiety of LLO. These representative structures were chosen based on the L202−L365 Cγ distance in Figure 6C: the closed form (2.58 μs in OST-Pep, showing the minimum distance within 2.5−3.0 μs), the intermediate form (2.40 μs in OST-Pep, showing the maximum distance within 2.0−2.5 μs), and the open form (3.81 μs in OST-Pep-LLO, showing the maximum distance within 3.0−4.0 μs). “R” and “L” in the plots indicate right and left TM helix bundle, respectively. (B) Plots of Cα-atom displacements of the closed form from the intermediate or open form for each TM helix. The residue numbers in the plots are not the residue sequences of ClPglB, but counted from the periplasmic side for each TM helix. This figure is available in black and white in print and in color at Glycobiology online.
Undecaprenyl binding to hydrophobic groove causes TM domain movements
The larger movements of the TM domain between the open and closed forms are shown in Figure 7, and the right TM bundle in OST-Pep-LLO and OST-gPep-UndPP is more separated from the left TM bundle than that in OST-Pep. EL5 is tethered to TM9 and TM10, each of which belongs to the right and left TM bundle, respectively. The N-terminus of EL5-h also has contacts with TM6 in the right TM bundle both in the open and closed forms (Figure 7A: Supplementary data, Figure S5). The large outward movements of the right TM bundle in the ternary complexes appear to impair structured EL5, leading to more disordered, open conformations. Based on these observations, it is clear that the existence of LLO or the glycosylated products triggers this large conformational change in the TM domain.
However, it is still unclear which LLO moiety (oligosaccharide, pyrophosphate or undecaprenoid) in the ternary complexes is the main cause of the TM movements. To address this, we performed additional 4-μs MD simulations of an artificial system, OST-Pep-Und, in which oligosaccharide and pyrophosphate are removed from LLO and only the undecaprenyl moiety is bound to the binary complex (Figure 2D). In OST-Pep-LLO and OST-gPep-UndPP, the undecaprenyl moiety stably binds to the hydrophobic groove of ClPglB with contacts persistent during the entire simulation (Figure 8A). In particular, R375 near to a divalent cation forms strong electrostatic interactions with the pyrophosphate group, which contributes to maintaining the stability of the undecaprenyl moiety in the hydrophobic groove (Supplementary data, Figure S7). In contrast, the undecaprenyl moiety in OST-Pep-Und, which does not have the pyrophosphate group, does not show such a consistent level of contacts, losing them after 0.5 μs and partially regaining after 3.0 μs. Figure 8B and C shows that the decrease in the contact numbers between the undecaprenyl moiety and hydrophobic groove in OST-Pep-Und results in the decrease in the L202−L365 Cγ and Y293−M368 Cα distances, indicating transition from the open form to the closed or intermediate form. The RMSF plots of OST-Pep-Und (Figure 8D) show much less fluctuations in EL5 after 1 μs, as in OST-Pep (Figure 4A), suggesting that the loss of interactions by the undecaprenyl moiety with the hydrophobic groove triggers inward movements between the right and left TM bundle, making EL5 more ordered. Consequently, these data indicate that the binding of the undecaprenyl moiety of LLO to the hydrophobic groove is the cause of the outward conformational changes of the TM domain, inducing disordered EL5 with open conformations.
Fig. 8.
Comparison of OST-Pep-Und (green) with OST-Pep (black), OST-Pep-LLO (blue), and OST-gPep-UndPP (red). (A) Time-series of contact numbers between undecaprenyl moiety and hydrophobic groove. (B) Time-series of the Cγ distance between L202 and L365. (C) Time-series of the Cα distance between Y293 and M368. (D) RMSF plots of OST-Pep-Und during different simulation periods. The magenta lines indicate the EL5 region. This figure is available in black and white in print and in color at Glycobiology online.
Undecaprenyl binds to hydrophobic groove more favorably than phospholipids
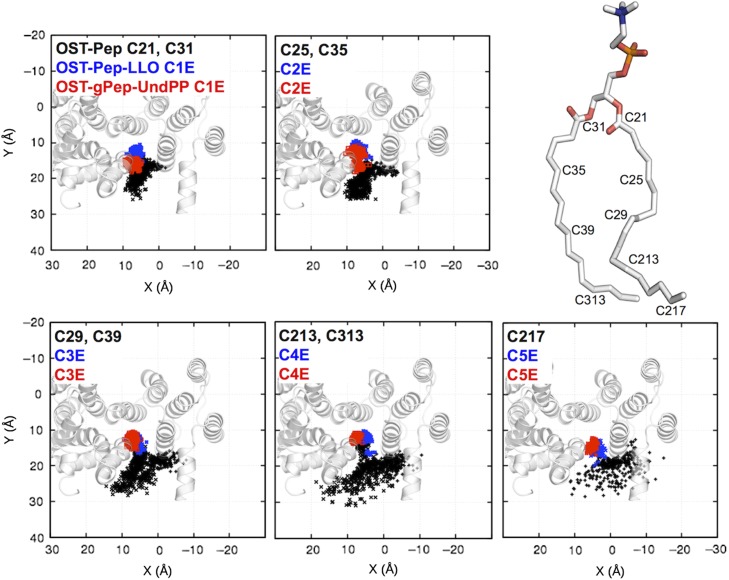
One may think that phospholipids in the membrane could also bind to the hydrophobic groove of OST-Pep and then induce a disordered EL5 through TM domain movements. Indeed, a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) molecule moved into and stayed bound to the hydrophobic groove during the OST-Pep simulation (Supplementary data, Figure S8). Figure 9 shows the distributions of the XY locations of a few selected carbon atoms in a POPC (OST-Pep) and in undecaprenyl moieties (OST-Pep-LLO and OST-gPep-UndPP; see Supplementary data, Figure S1 for the carbon atom name for the undecaprenyl moiety). Clearly, the undecaprenyl moieties bind deeper inside the hydrophobic groove than the POPC molecule. In addition, the motions of the undecaprenyl tails are more restricted within the hydrophobic groove, suggesting its tighter binding. The polyisoprenyl pyrophosphate in an LLO, which has a more negatively charged pyrophosphate group and whose tail is more hydrophobic, likely replaces a membrane phospholipid in the hydrophobic groove to form a ternary complex. The tighter binding of the bulkier polyisoprenyl chain expands the hydrophobic groove and triggers outward movements of TM helices adjacent to the hydrophobic groove, leading to TM domain-scale outward movements and subsequently disordered EL5 with open conformations.
Fig. 9.
Distributions of the XY locations of a few selected carbon atoms in a POPC in OST-Pep (black) and in undecaprenyl moieties in OST-Pep-LLO (blue) and OST-gPep-UndPP (red). Each figure contains carbon atoms that are in equivalent Z-positions in both POPC and undecaprenyl tails. The TM domain in cartoon representation is incorporated into each figure with the same orientation as in Figure 7A. See Supplementary data, Figure S9 for the XY locations of isoprenoid units 6–11. This figure is available in black and white in print and in color at Glycobiology online.
Discussion and conclusions
Our previous simulations showed that an LLO oligosaccharide is preferentially oriented to be parallel to the bilayer surface (Kern et al. 2014). Using the LLO conformations, we generated the ternary complexes of ClPglB in complex with an acceptor peptide and a bacterial LLO or the glycosylated products to investigate the dynamics and structures of EL5 in this study. The results show that the undecaprenyl chain favorably binds to a hydrophobic groove in ClPglB. This tight binding expands the hydrophobic groove, forcing the right TM bundle to move outwards from the left TM bundle. Such TM movements impair the stability of structured EL5, which is tethered by two TM helices each from the right and left TM bundle, consequently inducing disordered EL5 with open conformations.
Matsumoto et al. recently reported the crystal structure of a binary complex of AfAglB cross-linked with an acceptor peptide (Matsumoto et al. 2017). In this structure, the N-terminal half of EL5 was disordered, whereas the C-terminal half was ordered, which is the same partially structured EL5 as in the crystal structure of ClPglB with an acceptor peptide, indicating an intrinsic property of EL5 upon acceptor peptide binding. Based on our simulation results and the available crystal structures, we propose an updated N-glycosylation mechanism by bacterial OST, focusing on EL5 conformations. In its apo form, OST has fully ordered EL5 (based on the crystal of apo AfAglB). Upon a nascent polypeptide chain binding to OST, the N-terminal half of EL5 is still structured, but becomes more dynamic (Supplementary data, Figure S2A). The isoprenyl moiety shows high flexibility inside the bilayer hydrophobic core (Kern et al. 2014) and can likely recognize OST by replacing a bound phospholipid in the hydrophobic groove. This isoprenyl moiety binding triggers outward movements between the right and left TM bundle. Electrostatic interactions of the LLO pyrophosphate group with an adjacent positively charged residue (R375 in ClPglB) maintain the isoprenoid moiety in the hydrophobic groove more stably, facilitating the TM domain movements. As a result, EL5 tethered between the right and left TM bundle becomes disordered with open conformations. The open conformations likely allow the oligosaccharide moiety of LLO to move in and properly position at the catalytic site for glycosylation (Supplementary data, Figure S2B). The isoprenyl moiety is still embedded in the hydrophobic groove with the pyrophosphate group interacting with the positively charged residue. The N-terminal half of EL5 remains disordered in the presence of the isoprenyl pyrophosphate moiety. Oligosaccharide is covalently attached to the polypeptide. Meanwhile, the cleaved isoprenyl pyrophosphate remains on OST with EL5 whose N-terminal half is still disordered (Supplementary data, Figure S2C). The N-glycosylated polypeptide and cleaved isoprenyl pyrophosphate leave OST, and the EL5 goes back to the fully structured conformation.
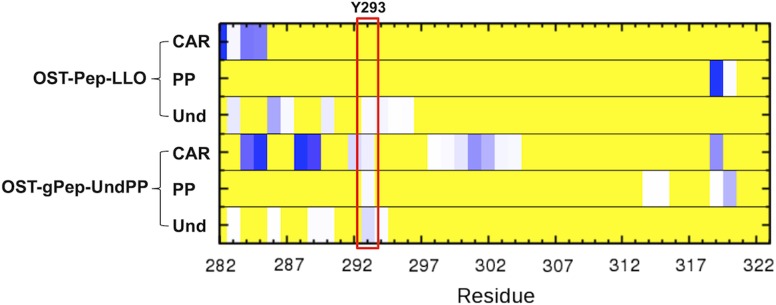
Lizak et al. reported that a conserved Y293 in EL5 is a functionally essential residue for N-glycosylation. It was shown that Y293 could be replaced by other aromatic residues, suggesting that the residue favorably interacts with the sugar rings of an oligosaccharide moiety in the LLO substrate (Lizak et al. 2014) through electrostatic and electronic complementarity between carbohydrates and aromatics residues (Hudson et al. 2015). It is interesting that our analysis also characterizes Y293 as a specific residue that has distinct contact patterns with the rest of ClPglB between OST-Pep and OST-Pep-LLO/OST-gPep-UndPP. The characterization of the heavy atom contact frequencies between the carbohydrate moiety and EL5 (Figure 10) indicates that the contacts between the carbohydrate moiety and Y293 are not observed in OST-Pep-LLO and not significant in OST-gPep-UndPP. This suggests that a major role of Y293 is to facilitate the penetration of the carbohydrate moiety through disordered EL5 to form a ternary complex, rather than to increase catalytic efficiency in the ternary complex.
Fig. 10.
Frequencies of heavy atom contacts of EL5 with carbohydrate moiety (CAR), pyrophosphate (PP), and undecaprenyl moiety (Und). The frequencies were averaged during the last 2 μs simulations. The existence of heavy atom contacts (distance cutoff = 4.5 Å) for each snapshot was characterized and then the fraction of the contacts was calculated over the last 2 μs. If there is no contact during the last 2 μs, the fraction is 0 (yellow). If the fraction is larger than 0, it is colored white to dark blue. This figure is available in black and white in print and in color at Glycobiology online.
Lizak et al. also defined a new biologically important motif named the Tyr-plug, which has four aromatic residues (Y288, F292, Y293, and F295) in EL5 and is highly conserved in bacterial homologs of PglB. Based on the experimental results, they suggested that the Tyr-plug has a defined conformation during catalysis (Lizak et al. 2014). Supporting this, our simulation results show that EL5 has a stable helix (EL5-h) in its N-terminal half (Supplementary data, Figure S6), which contains all four Tyr-plug residues. The presence of EL5-h is also consistent with the crystal structure of apo AfAglB (Matsumoto et al. 2013).
POPC is a common zwitterionic lipid used in MD simulations. However, the main phospholipid components of the inner membrane of Gram-negative bacteria are negatively charged phosphatidylglycerol (PG) and zwitterionic phosphatidylethanolamine (PE). In this context, our simulation systems were not built based on more biologically plausible phospholipid composition for Gram-negative bacterial protein ClPglB. However, we believe that our simulation results explain biological phenomena in that lipid phosphatidyl group conserved in these different phospholipids is the only moiety associated with binding to hydrophobic groove and electrostatic interactions with R375 in ClPglB. It might be interesting to see if the current simulation results depend on the lipid types. Petrou et al. recently reported the crystal structures of aminoarabinose transferase ArnT from Cupriavidus metallidurans (CmArnT), showing a structural rearrangement of periplasmic loop 4 upon undecaprenyl phosphate binding (Petrou et al. 2016). Interestingly, CmArnT has a similar fold to bacterial OSTs as well as an undecaprenyl moiety-involved conformational change of a periplasmic loop, although both enzymes have largely different functions.
Although it is not the main goal of this study, Pedebos et al. observed that the divalent cation at the catalytic site is firmly stable in their simulations of ClPglB bound to its peptide acceptor (Pedebos et al., 2015). The catalytic site in ClPglB has four negatively charged residues (Asp56, Asp154, Asp156 and Glu319); Asp56, Asp154 and Glu319 coordinate the divalent cation (Mg2+ in this study) in the crystal structure. These residues are not directly associated with the large conformational changes in EL5 and TM domain because Glu319 is a residue in the stable C-terminal region of EL5 and Asp56, Asp154, and Asp156 are all located in the left TM bundle. The distances between the residues and divalent cation show that Mg2+ is stably coordinated with the proximate residues during the simulations and the overall coordinate patterns are similar between different systems (Supplementary data, Figure S10). It appears that the distances from our MD simulations are consistently shorter than the crystal ones. It is not clear whether this difference results from the low resolution of the crystal structure (3.4 Å) or inaccuracy in treating metal ion-related interactions in the MD simulations. Interestingly, Asp156 Oδ1 coordinates Mg2+, which is not observed in the crystal structure, and the interaction patterns by Glu319 differ between the binary complex (OST-Pep) and ternary complexes (OST-Pep-LLO and OST-gPep-UndPP). It will be computationally challenging, but interesting to explore the allosteric structural changes of the active site accompanied with the large conformational changes of EL5 using quantum mechanics calculations.
In conclusion, our simulation results indicate that the conformational changes of the TM domain triggered by undecaprenyl moiety binding induce disordered EL5 in ClPglB. Although the data provide new insight into the N-glycosylation mechanism, there are still open questions to be addressed. For example, the binding of LLO to ClPglB might precede that of a nascent polypeptide, but it is unclear whether the well-ordered EL5 in apo form affords to have open conformations by the isoprenoid moiety binding. Additional crystal structures combined with sophisticated computer simulations could clarify the remaining questions.
Materials and methods
Preparation of initial structures, equilibration and production
We used an X-ray structure (PDB id: 3rce) of a binary complex of ClPglB (missing 24 N-terminus residues (283–306) of EL5) and a short acceptor peptide DQNATF{4-NO2} (containing a chemically modified phenylalanine). The modified phenylalanine in the peptide was replaced with phenylalanine by removing the nitrogen dioxide group. We removed the N- and C-terminal Gly residues of the peptide and capped them with acetyl (ACE) and N-methyl (CT3) groups, respectively, to keep the termini neutral. To model the missing 24 residues of EL5, we generated a three-dimensional (3D) structure of ClPglB from the amino acid sequence using I-TASSER (Roy et al. 2010) and then copied the coordinates of residues 283–306 into the crystal structure. Mg2+ was used for a divalent metal ion in the catalytic site.
To generate a ternary complex of ClPglB with the peptide and an LLO, we performed molecular docking using G1Gn5B1-PP-Und (Weerapana et al. 2005) (Supplementary data, Figure S1); see our previous study (Kern et al. 2014) or Supplementary data for the details of the docking procedure. Briefly, a set of bacterial LLO conformations was obtained from MD simulations of the LLO in lipid bilayers (Kern et al. 2014). A best docking pose was chosen based on the lowest number of bad contacts between ClPglB and G1Gn5B1-PP. Bad contacts between ClPglB and the undecaprenyl moiety were unavoidable due to the conformational flexibility of the lipid chain in the absence of ClPglB. Therefore, we manually adjusted the dihedral angles in the undecaprenyl moiety to minimize the bad contacts. This ternary structure was embedded into an explicit POPC bilayer with explicit water molecules (Supplementary data, Figure S11) using the Membrane Builder module (Jo et al. 2007, 2009) in CHARMM-GUI (Jo et al. 2008) and equilibrated for 370 ns using NAMD (Phillips et al. 2005; Lee et al. 2016). This equilibrated ternary structure is named OST-Pep-LLO (Pep: peptide) (Figure 2B). Starting from the equilibrated ternary structure, we prepared additional two initial structures by removing LLO (OST-Pep, Figure 2A) or covalently attaching an oligosaccharide to the peptide (OST-gPep-UndPP, gPep: glycopeptide, UndPP: undecaprenyl pyrophosphate, Figure 2C). We also prepared OST-Pep-Und by removing G1Gn5B1-PP (Und: undecaprenyl moiety, Figure 2D). The simulation system information is shown in Supplementary data, Table S1. Each simulation system was further equilibrated for 100 ns using NAMD and simulated up to 4 μs on Anton (Shaw et al. 2008) for the data presented in this paper.
MD simulation setup
Each system was shortly relaxed for 300 ps by applying various restraints to the protein backbone, side-chain, water, ions, and lipid tail/head atoms at 303.15 K with default options in the standard 6-step Membrane Builder equilibration inputs using CHARMM (Brooks et al. 2009) before NAMD simulations. The hydrogen geometry was kept fixed using SHAKE (Ryckaert et al. 1977). This short relaxation was performed in constant particle number, volume, and temperature (NVT) conditions. The TIP3P model (Jorgensen et al. 1983) was used for explicit water molecules. The system size was determined to have at least 15 Å between the protein and its periodic image in each XYZ direction (measured system size is 100 × 100 × 126 Å3), and 150 mM KCl was added for physiological salt concentration. The CHARMM36 force field (MacKerell et al. 1998, 2004; Guvench et al. 2008, 2009; Best et al. 2012) was used for the proteins, carbohydrates and lipids. The particle mesh Ewald algorithm (Essmann et al. 1995) was applied to calculate electrostatic forces, and the van der Waals interactions were smoothly switched of at 10–12 Å by a force-switching function (Steinbach and Brooks 1994). NAMD (Phillips et al. 2005) was used for the constant particle number, pressure and temperature (NPT) equilibrations of the initial ternary system (370 ns) and each system (100 ns) derived from the initial ternary system. The Langevin coupling coefficient was set to 1 ps–1 and a Nosé-Hoover Langevin-piston (Martyna et al. 1994; Feller et al. 1995) was used to maintain constant pressure (1 bar) with a piston period of 50 fs and a piston decay of 25 fs.
For Anton simulations, the NVT ensemble was used with the temperature maintained at 300 K using the Nosé-Hoover method. The time step was 2 fs and trajectories were saved every 240 ps. The short-range forces and long-range electrostatics were evaluated every 2 fs and 6 fs, respectively. The short-range nonbonded and electrostatic interactions were calculated with a cutoff of 9.52 Å. The long-range electrostatic interactions were calculated using the k-Gaussian Split Ewald method (Shan et al. 2005). SHAKE was used to constrain all bonds involving hydrogen atoms.
Supplementary Material
Acknowledgments
The Anton machine at PSC was generously made available by D.E. Shaw Research. We thank Seonghoon Kim for preforming Anton simulations.
Supplementary data
Funding
This work was supported in part by the National Science Foundation (DBI-1707207), the National Institutes of Health (GM087519), and XSEDE (MCB070009). Anton computer time was provided by the National Center for Multiscale Modeling of Biological Systems (MMBioS) through the National Institutes of Health (P41GM103712-S1) from the National Institutes of Health and the Pittsburgh Supercomputing Center (PSC).
Conflict of interest statement
None declared.
Abbreviation
LLOs, lipid-linked oligosaccharides; MD, molecular dynamics; NPT, number, pressure and temperature; NVT, number, volume, and temperature; OST, oligosaccharyltransferase; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; RMSD, root-mean-square deviation; RMSF, root-mean-square fluctuation; TM, transmembrane; UndPP, undecaprenyl pyrophosphate.
References
- Aebi M. 2013. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 1833:2430–2437. [DOI] [PubMed] [Google Scholar]
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD Jr. 2012. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 8:3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL 3rd, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S et al. . 2009. CHARMM: The biomolecular simulation program. J Comput Chem. 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Aebi M. 1999. The dolichol pathway of N-linked glycosylation. Biochim Biophys Acta. 1426:239–257. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J Chem Phys. 103:8577–8593. [Google Scholar]
- Feller SE, Zhang YH, Pastor RW, Brooks BR. 1995. Constant-pressure molecular-dynamics simulation – the Langevin piston method. J Chem Phys. 103:4613–4621. [Google Scholar]
- Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD Jr. 2008. Additive empirical force field for hexopyranose monosaccharides. J Comput Chem. 29:2543–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvench O, Hatcher ER, Venable RM, Pastor RW, Mackerell AD. 2009. CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J Chem Theory Comput. 5:2353–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson KL, Bartlett GJ, Diehl RC, Agirre J, Gallagher T, Kiessling LL, Woolfson DN. 2015. Carbohydrate-aromatic interactions in proteins. J Am Chem Soc. 137:15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W. 2007. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE. 2:e880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. 2008. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 29:1859–1865. [DOI] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W. 2009. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys J. 97:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 79:926–935. [Google Scholar]
- Kelleher DJ, Gilmore R. 2006. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 16:47R–62R. [DOI] [PubMed] [Google Scholar]
- Kern NR, Lee HS, Wu EL, Park S, Vanommeslaeghe K, MacKerell AD Jr, Klauda JB, Jo S, Im W. 2014. Lipid-linked oligosaccharides in membranes sample conformations that facilitate binding to oligosaccharyltransferase. Biophys J. 107:1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Chang MM, Whitworth GE, Imperiali B. 2013. Biochemical evidence for an alternate pathway in N-linked glycoprotein biosynthesis. Nat Chem Biol. 9:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y et al. . 2016. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 12:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP. 2011. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 474:350–355. [DOI] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Zinne D, Michaud G, Schubert M, Chen F, Bucher M, Darbre T, Zenobi R, Reymond J-L et al. . 2014. A catalytically essential motif in external loop 5 of the bacterial oligosaccharyltransferase PglB. J Biol Chem. 289:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S et al. . 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- MacKerell AD Jr, Feig M, Brooks CL 3rd. 2004. Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc. 126:698–699. [DOI] [PubMed] [Google Scholar]
- Martyna G, Tobias DJ, Klein ML. 1994. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 101:4177–4189. [Google Scholar]
- Matsumoto S, Shimada A, Nyirenda J, Igura M, Kawano Y, Kohda D. 2013. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci USA. 110:17868–17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Taguchi Y, Shimada A, Igura M, Kohda D. 2017. Tethering an N-glycosylation sequon-containing peptide creates a catalytically competent oligosaccharyltransferase complex. Biochemistry. 56:602–611. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, Nairn AV. 2012. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat Rev Mol Cell Biol. 13:448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyirenda J, Matsumoto S, Saitoh T, Maita N, Noda NN, Inagaki F, Kohda D. 2013. Crystallographic and NMR evidence for flexibility in oligosaccharyltransferases and its catalytic significance. Structure. 21:32–41. [DOI] [PubMed] [Google Scholar]
- Pedebos C, Arantes PR, Giesel GM, Verli H. 2015. In silico investigation of the PglB active site reveals transient catalytic states and octahedral metal ion coordination. Glycobiology. 25:1183–1195. [DOI] [PubMed] [Google Scholar]
- Petrou VI, Herrera CM, Schultz KM, Clarke OB, Vendome J, Tomasek D, Banerjee S, Rajashankar KR, Dufrisne MB, Kloss B et al. . 2016. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 351:608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert JP, Ciccotti G, Berendsen HJC. 1977. Numerical-integration of Cartesian equations of motion of a system with constraints – molecular-dynamics of N-alkanes. J Comput Phys. 23:327–341. [Google Scholar]
- Schwarz F, Aebi M. 2011. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 21:576–582. [DOI] [PubMed] [Google Scholar]
- Shan YB, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. 2005. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J Chem Phys. 122:54101–54113. [DOI] [PubMed] [Google Scholar]
- Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC et al. . 2008. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 51:91–97. [Google Scholar]
- Steinbach PJ, Brooks BR. 1994. New spherical-cutoff methods for long-range forces in macromolecular simulation. J Comput Chem. 15:667–683. [Google Scholar]
- Weerapana E, Glover KJ, Chen MM, Imperiali B. 2005. Investigating bacterial N-linked glycosylation: Synthesis and glycosyl acceptor activity of the undecaprenyl pyrophosphate-linked bacillosamine. J Am Chem Soc. 127:13766–13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. 2006. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology. 16:91R–101R. [DOI] [PubMed] [Google Scholar]
- Yan G, Lennarz WJ. 2002. Studies on the function of oligosaccharyl transferase subunits – Stt3p is directly involved in the glycosylation process. J Biol Chem. 277:47692–47700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.