Abstract
Overexpression of c-Myc is involved in the tumorigenesis of B-lineage acute lymphoblastic leukemia (B-ALL), but the mechanism is not well understood. In the present study, a c-Myc-knockdown model (Raji-KD) was established using Raji cells, and it was indicated that c-Myc regulates the expression of genes associated with cell cycle progression in G2/M-phase, cyclin D kinase (CDK)1 and cyclin B1, by modulating 60 kDa Tat-interactive protein (TIP60)/males absent on the first (MOF)-mediated histone H4 acetylation (AcH4), which was then completely restored by re-introduction of the c-Myc gene into the Raji-KD cells. The expression of CDK1 and cyclin B1 was markedly suppressed in Raji-KD cells, resulting in G2/M arrest. In comparison to Raji cells, the proliferation of Raji-KD cells was significantly reduced, and it was recovered via re-introduction of the c-Myc gene. In the tumorigenesis assays, the loss of c-Myc expression significantly suppressed Raji cell-derived lymphoblastic tumor formation. Although c-Myc also promotes Raji cell apoptosis via the caspase-3-associated pathway, CDK1/cyclin B1-dependent-G2/M cell cycle progression remains the major driving force of c-Myc-controlled tumorigenesis. The present results suggested that c-Myc regulates cyclin B1- and CDK1-dependent G2/M cell cycle progression by TIP60/MOF-mediated AcH4 in Raji cells.
Keywords: c-Myc, B-lineage acute lymphoblastic leukemia, cyclin D kinase 1, cyclin B1, G2/M arrest, histone H4 acetylation
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in pediatric patients, whose malignant white blood cells continuously multiply and lead to an excess of lymphoblasts in peripheral blood and bone marrow (1). During the past decade, several genetic and transcriptional factors have been identified as aberrant in B-ALL, which is a common subtype of ALL. Among these factors, overexpression of c-Myc is the main characteristic of B-ALL (2,3).
The c-Myc protein belongs to a larger family of the helix-loop-helix leucine zipper (HLHzip) transcription factors. The C-terminal basic region HLHzip domain and the N-terminal transcriptional activation domain (TAD) are essential for the activation and repression of target genes (4). c-Myc forms a heterodimer with Max to bind target DNA sequences through the canonical E box DNA binding motif (CACGTG) (5). Transcription/transformation-associated protein (TRRAP), a subunit of different histone acetyltransferases, is a coactivator with the TAD domain of c-Myc (6). Most estimates suggest that c-Myc-regulated genes are generally involved in cell-cycle progression and metabolism, which includes cyclin (CCN) D kinases (CDKs), cyclins and ribosomal RNAs. The target genes of c-Myc affect the multiple cellular processes by direct or indirect pathways in a diversity of cancer cell types. When directly activating the oncogenic pathway, c-Myc upregulates a series of transcriptional programs, which influences physiological processes including metabolic adaptation, cell division and survival (5,7). When acting indirectly, c-Myc alters microRNA and long non-coding RNA expression patterns or histone modifications, which impacts target gene expression in various carcinoma types (8,9).
In contrast to the tightly regulated expression of c-Myc in normal cells, it is frequently dysregulated in human cancers. Abundant evidence has indicated that c-Myc is pathologically activated in numerous types of human malignancy (5,9,10,11). c-Myc mRNA has been detected to be 5- to 40-fold overexpressed in 60–80% of colon carcinomas (12), 29% of prostate cancers (13), 40% of ovarian cancers (14), 23% of lung carcinomas (15), 61% of nodular melanomas and in 30% of metastases (10). In virtually all Burkitt's lymphoma, the c-Myc gene is translocated to one of the immunoglobulin loci enhancers that drive the high expression levels of c-Myc mRNA and protein. A typical translocation of c-Myc into the immunoglobulin heavy chain locus is observed in ~80% of Burkitt's lymphomas. Variant translocations of c-Myc into either the κ or λ light chain locus each occur at a frequency of ~10% (16,17). However, the association between c-Myc overexpression and tumorigenesis has remained to be fully elucidated. In this light, the present study on the function of c-Myc aimed to contribute to the current knowledge on the molecular mechanisms underlying the genesis of B-ALL.
Materials and methods
Mice
Severe combined immunodeficient (SCID) mice were purchase from Beijing Huafu Kang Company (Beijing, China) and maintained in a specific-pathogen-free environment at 24–26°C with a relative humidity of 60–65%. They were provided with water and food ad libitum throughout the experimental period. The procedures for handling animals complied with the Current Laboratory Animal Laws and Regulations, Policies and Administration in China. All experiments were approved by the Animal Ethics Committee of Dalian Medical University (no. AEE17013).
Antibodies
Mouse monoclonal anti-c-Myc (9E10; cat. no. E1809) antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA); mouse monoclonal anti-cyclin E1 (HE12; cat. no. 4129S), anti-cyclin B1 (V152; cat. no. 4135S), anti-caspase-3 (3G2; cat. no. 9668), anti-caspase-8 (1C12; cat. no. 9746), anti-caspase-9 (C9; cat. no. 9508) antibodies, and rabbit polyclonal anti-cyclin D kinase (CDK)1 (cat. no. 9112) and anti-phospho-(p)CDK1 (Tyr15) (cat. no. 9111S) antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Rabbit polyclonal anti-acetyl-histone H4 (cat. no. 06-866) was purchased from EMD Millipore (Billerica, MA, USA). Mouse monoclonal anti-c-Myc (9E10 cat. no. ab32)-chromatin immunoprecipitation (ChIP) grade antibody and rabbit polyclonal anti-GAPDH antibody were purchased Abcam (Cambridge, UK). Fluorescein isothiocyanate (FITC)-labeled immunoglobulin (Ig)G (cat. no. 11-4011-85) was obtained from e-Bioscience (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Horseradish peroxidase (HRP)-conjugated rabbit (cat. no. A0208) and mouse (cat. no. A0216) IgG antibodies were from Beyotime Institute of Biotechnology (Haimen, China).
Patient samples
The clinical manifestations and examination results of B-ALL patients newly diagnosed and treated at Dalian Municipal Central Hospital (Dalian, China) from May 2015 to January 2016 were retrospectively analyzed. Bone marrow aspiration and biopsy were obtained from all B-ALL patients (n=12; 7 women and 5 men; mean age, 23 years; range, 18–35 years) and individuals with anemia (n=4; 2 women and 2 men; mean age, 22 years; range, 18–30 years). All B-ALL patients newly diagnosed and have not received any medication prior to sample collection. All investigations were performed either for diagnostic purposes or with residual material obtained through diagnostic procedures. All experiments were approved by the Ethics Committee of Dalian Municipal Central Hospital (approval no. YN2016-019-01).
Cells and culture conditions
Raji cells were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 tissue culture medium supplemented with 2 mM glutamine (both from Thermo Fisher Scientific, Inc.), 50 mM 2-mercaptoethanol (Fluka, Buchs, Switzerland), 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin.
Transient transfection of c-Myc small interfering (si)RNA
Cells (5×104/well) were seeded into six-well plates and allowed to grow to 90% confluence. Transient transfections of c-Myc siRNAs were performed for 6 h with TransIT-TKO transfection reagent (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's instructions. The siRNAs were designed to form 19-bp double-stranded RNA with 2 thymine overhangs at each 3′ end of RNA. The following 3 targeting sequences of c-Myc siRNA were used: siRNA 1 sense, 5′-GCUUCACCAACAGGAACUAUU-3′ and antisense, AACGAAGUGGUUGUCCUUGAU (region: 586–605 bp); siRNA 2 sense, 5′-GGCGAACACACAACGUCUUUU-3′ and antisense, 5′-AACCGCUUGUGUGUUGCUGUU (region: 1,636–1,655 bp); and siRNA 3 sense, 5′-GGAAACGACGAGAACAGUUUU-3′, and antisense, 5′-AACCUUUGCUGCUCUUGUCAA-3′ (region: 1,831–1,850 bp). Alexa 488-conjugated siRNA duplex (Qiagen, Hilden, Germany) was used to determine the transfection efficiency.
Establishment of Raji cells with c-Myc knockdown (Raji-KD cells) and those with c-Myc knockdown and subsequent restoration of c-Myc expression (Raji-KD-Re cells)
A retroviral vector carrying siRNA targeting c-Myc was constructed as follows. A 21-nucleotide sequence (siRNA 3 region: 1,831–1,850) of the c-Myc complementary (c)DNA was inserted in the sense and antisense directions into the pSINsi-mU6 cassette vector (recombinant retroviral vector; Takara Bio, Inc.), containing the mouse U6 promoter. The recombinant retroviruses were generated by co-transfection of the vector mixture into 293 cells (Genomeditech, Shanghai, China), including recombinant retroviral vector, pE-eco vector (ecotropic env) and pGP vector (gag-pol; cat. no. 6161; Takara Bio, Inc.). Recombinant retroviral particles containing the target sequence or a mock control were transfected into the parental Raji cells, and the geneticin (G418)-resistant clones were selected as transfected cells. The Raji-derived cells, transfected with the plasmid expressing siRNA that targeted c-Myc, are referred to hereafter as 'Raji-KD'. Next, pseudotyped retroviral vectors (cat. no. 6161; Takara Bio, Inc.) were generated for transfection of the designated mock cells.
For c-Myc reintroduction, the open reading frame (ORF) of c-Myc was cloned into the ClaI site of the pLHCXsi-mU6-c-Myc expression vector, which is resistant to the siRNA expressed in the c-Myc-knockdown cells. For this purpose, c-Myc shRNA containing multiple mutations were introduced into pLHCX vector (cat. no. 631511; Clontech Laboratories, Inc., Mountainview, CA, USA; five point mutations in the 1,831–1,850 region) that did not alter the original amino acid residues. Raji-KD cells with c-Myc restoration, referred to as 'Raji-KD-Re cells', were established by the transfection of pLHCXsi-mU6-c-Myc into Raji-KD cells.
Western blot analysis
Cell lysates were prepared with ice-cold buffer (50 mM Tris-HCl, 150 mM of NaCl, 1% Triton X-100, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, supplemented with 0.1 mM phenylmethylsulfonylflouride, 1 mM dithiothreitol and a mixture of protease and phosphatase inhibitors (Pierce; Thermo Fisher Scientific, Inc.). The cell lysate was cleared by centrifugation at 12,000 × g for 10 min at 4°C. The proteins were determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). A total of 10 µg protein per lane in 4X Laemmli loading buffer was resolved by 10% SDS-PAGE gel and immunostained with. The protein samples were subjected to 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Immobilon-P; 0.45 µm; EMD Millipore) at 240 mA for 60 min. Blots were blocked at 37°C for 2 h with 5% skimmed milk in 10 mM Tris-HCl (pH 7.5) with 150 mM NaCl and 0.1% Tween-20 (TBST) for caspase antibodies or with 5% bovine serum albumin (BSA; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in TBS-T for other antibodies. Following incubation with the appropriate primary antibodies overnight at 4°C (dilution, 1:4,000 dilution or 1:3,000 for caspase antibodies), the slides were washed. After washing, the blots were incubated at 37°C for 1 h with the HRP-conjugated anti-rabbit IgG (1:8,000 dilution) or anti-mouse IgG (1:8,000 dilution) secondary antibodies. Finally, specific proteins were visualized using an enhanced chemiluminescence system (GE Healthcare, Little Chalfont, UK).
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) and RT-quantitative (q)PCR
RNA was isolated from each cell population using TRIzol reagent (Thermo Fisher Scientific, Inc.). First-strand cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) and the oligo(dT) 18 primer. The mixture without SuperScript II and RNasin was heated to 70°C for 5 min to unfold the secondary structures of the RNA. After adding SuperScript II and RNasin, reactions were performed for 70 min at 42°C and for 15 min at 70°C in a 20-µl reaction volume. Prepared cDNAs were stored at −20°C.
For PCR, GAPDH served as an internal control gene. The thermocycling conditions were as follows: 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. The PCR products were separated by electrophoresis on 2% agarose gels and images were captured under ultraviolet light.
Real-time qPCR was performed in triplicate in a reaction volume of 20 µl (20 µmol/l of primers and 10 µl of Master Mix (Takara Bio, Inc.), which was adapted from the standard protocol provided by SYBR-Green PCR Master Mix. The PCR procedure was performed on an Applied Biosystems Prism 7000 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min, with a subsequent standard dissociation run to obtain melting curve profiles of the amplicons. Using the 2−∆∆Cq (18) method, relative internal mRNA expression of target genes was normalized to GAPDH.
Primer sequences used in the PCR assays were as follows (5′-3′): CDK1 (NG_029877.1) forward, GAAATTGAGCGGAGAGCGAC and reverse, CCGTTCCTCAATACTCGCCC; CDKN1A (NG_009364.1) forward, GCGGAGTGGAGTAAGTTCGT and reverse, TGGCGTAAAGGACCTGAACC; CDKN1B (NG_016341.1) forward, CGCTCGCCAGTCCATTTG and reverse, AAAGACACAGACCCCGACG; CCNG1 (XM_011534685) forward, GATCAGGGCCGAGTTGTCTC and reverse, GAGGAGAGGGGACTCGTAGG; retinoblastoma 1 (RB1; NG_009009.1) forward, CCCTGTTTCAATTTATCAGGC and reverse, TCACCCCAGATTAGTTTAGGC; tumor protein (TP)53 (NG_017013.2) forward, CTCAGACACTGGCATGGTGT and reverse, GTGGGGATCCAGCATGAGAC; CCNG2 (NM_004354.2) forward, CCTCCTGTGCCATTCAACCA and reverse, CCAAGTCAACGGGGGTAAGG; CCNB1 (NM_031966.3) forward, TGAGAGCCATCCTAATTGACT and reverse, CAATTATTCTGCATGAACCGAT; c-Myc (NM_002467.4) forward, AATGTCAAGAGGCGAACACAC and reverse, ATTGTTTTCCAACTCCGGGAT; histone acetyl transferase (HAT) [K(Lysine) acetyltransferase 5 (kat5); NM_001206833.1] forward, CATTGCCTGTCCTCTACCTG and reverse, ACTCTTGTTCTTACGTCCATC; HAT (kat8; NM_032188.2) forward, GCAAGATCACTCGCAACCAA and reverse, ATCAATTTCGTAGTTCCCGAT; GAPDH (NM_001289746.1) forward, AGATCATCAGCAATGCCTCCTG and reverse, ATGGCATGGACTGTGGTCATG.
Proliferation assay
An MTT assay was performed to assess viability of Raji, Raji-KD and Raji-KD-Re cells. The tetrazo-lium salt MTT is taken up by viable cells and reduced to a formazan residue by functional mitochondria of living cells. For the MTT assay, cells were seeded into a 96-well flat bottom microtiter plate. A total of 10 µl per well of a 5 mg/ml solution of MTT (Sigma-Aldrich; Merck KGaA) in PBS was added for the last 4 h of incubation. Subsequently, the plate was centrifuged 2,500 × g, at 4°C for 10 min, the media were removed and dimethylsulfoxide was added to each well to dissolve the precipitate. The absorbance of each well was read at 490 nm with a Benchmark microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
Bromodeoxyuridine (BrdU) assay
BrdU labelling was performed using a BrdU assay kit (CycLex Co., Ltd., Nagano, Japan) according to the manufacturer's protocol. Briefly, cells were seeded in 96-well plates and exposed to 10 µm of BrdU for 8 h at 37°C. The cells were fixed with denaturing solution for 30 min at room temperature and incubated with 50 µl anti-BrdU monoclonal antibody for 1 h at room temperature. Following washing, the plates were incubated with 50 µl HRP conjugated anti-mouse IgG at 37°C for 1 h. The plates were visualized by incubation with 50 µl substrate reagent for 15 min. The absorbance was measured at a wavelength of 450 nm.
Cell-cycle and apoptosis analysis
The cells were harvested, fixed with ice-cold 70% ethanol for 2 h, and single-cell suspensions were prepared. After extensive washing, the cells were resuspended in PBS containing propidium iodide (PI) and RNase A (both from Sigma-Aldrich; Merck KGaA), incubated for 1 h at room temperature and analyzed using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The cell-cycle-phase distribution was analyzed with CellQuest software 3.3 (BD Biosciences).
For analysis of apoptosis, 5×105 cells were harvested. Subsequently, PI and Annexin V-FITC double staining were performed at room temperature for 15 min. Finally, cells were washed three times with PBS and analyzed by flow cytometry (BD Biosciences).
PCR array
Total RNA was extracted from cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A Cell Cycle PCR Array (PAHS-020) was used to simultaneously examine the mRNA levels of 96 genes, including 6 'housekeeping genes' for noralization, in 96-well plates, according to the protocol of the manufacturer (cat. no. OHS-020; Zhejiang Kangchen Biotech Co., Ltd., Wuhan, China). The Cq value was calculated for each sample as the difference in gene expression between Raji and Raji-KD cells.
ChIP assay
During the culturing process of Raji cells and their derivatives, formaldehyde was added to the medium at a final concentration of 1%. Cross-linking was allowed to proceed for 10 min at room temperature and stopped by addition of glycine to a final concentration of 125 mM, followed by an additional incubation for 5 min at 25°C. Fixed cells were washed twice with PBS and harvested in SDS buffer [50 mM Tris-HCl (pH=8.0), 0.5% SDS, 100 mM NaCl, 5 mM EDTA and aforementioned protease inhibitors]. Cells were pelleted by 1,500 × g centrifugation for 5 min at 4°C and suspended in 4 ml IP buffer [100 mM Tris (pH 8.6), 0.3% SDS, 1.7% Triton X-100 and 5 mM EDTA]. The cells were then disrupted by sonication for 10 sec in a Branson 250 sonicator (Branson, Danbury, CT, USA) at a power setting of 3 and duty cycle of 100%, yielding genomic DNA fragments with a bulk size of 100–400 bp. The lysate was then diluted with IP buffer to a final volume of 1 ml. For each immunoprecipitation, 1 ml diluted lysate was precleared by adding 30 µl blocked protein G beads [50% slurry protein A-Sepharose (GE Healthcare); 0.5 mg/ml fatty acid-free BSA and 0.2 mg/ml salmon sperm DNA (both from Sigma-Aldrich; Merck KGaA) in Tris-EDTA (TE) buffer]. Samples were immunoprecipitated overnight at 4°C with 1:500 anti-c-Myc antibody or 1:200 anti-AcH4 antibody). Immune complexes were recovered by adding 30 µl blocked protein G beads and incubated for 6 h at 4°C. Beads were washed and eluted, and cross links were reversed. This included successive washes in 1 ml Mixed Micelle Buffer [20 mM Tris (pH 8.1), 150 mM NaCl, 5 mM EDTA, 5% w/v sucrose, 1% Triton X-100 and 0.2% SDS], buffer 500 [50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid at pH 7.5, 0.1% w/v deoxycholic acid, 1% Triton X-100, 500 mM NaCl, and 1 mM EDTA], LiCl Detergent Wash Buffer [10 mM Tris-HCl (pH 8.0), 0.5% deoxycholic acid, 0.5% Nonidet P-40, 250 mM LiCl and 1 mM EDTA] and TE (pH 7.5). The eluted material was phenol/chloroform-extracted and ethanol-precipitated. DNA was re-suspended in 100 µl water (19). PCR was performed as described above using 1 µl DNA and 800 nM aforementioned primers diluted to a final volume of 20 µl in SYBR-Green Reaction Mix (Takara Bio Inc.). Using the 2−∆∆Cq (18) method, relative internal mRNA expression of target genes was normalized to GAPDH.
Immunostaining and immunofluorescence analysis
Cells were grown on glass coverslips for 24 h. For cell immunostaining, cells were fixed in 4% neutral-buffered paraformaldehyde at 4°C for 20 min. Subsequently, the cells were blocked with the 5% FBS in PBS for 1 h at room temperature. Anti-acetyl-histone H4 antibody (1:250) staining was performed overnight at 4°C, whereas the FITC-labeled IgG (1:250) staining was performed for 1 h at room temperature. Finally, the slides were visualized with 3,3′-diaminobenzidine. All the slides were observed under a fluorescence microscope (Olympus, Tokyo, Japan) and images were captured.
In vivo tumor formation assays (Xenograft model)
Male SCID mice (age, 6 weeks, average weight 25 g; n=11 mice/group) were allowed to acclimatize to the laboratory environment for 1 week. To establish the xenograft tumor formation model, 1×107 Raji cells and Raji-KD cells were injected into the right and left sides axilla of a site in each mouse, respectively. Over 30 days, tumor formation at the site of injection and at distant tissue sites was monitored. Tumor size was measured using a caliper and tumor volume was estimated using the following equation: V=π/6xaxbxc (V, volume of the tumor; a, the longest radial line; b, the shortest radial line; c, thickness).
To assess the efficiency of the retroviral vector-mediated c-Myc-siRNA in suppressing lymphoblastic tumor growth in vivo, 2×106 of Raji cells were inoculated into the bilateral hind axilla of 7-week-old SCID mice (n=3 mice/group). A total of 50 µl retroviral vector (0.5 µg/µl) was injected once after 24 h inoculation. Mice were injected with c-Myc shRNA retrovirus on the right axilla or with the pseudotyped retrovirus on the left. Over 45 days, the tumor weight and the date at which a palpable tumor first arose was recorded.
Immunohistochemistry
For immunohistochemical detection of protein in the xenograft tumors, 5-mm sections taken from formalin-fixed paraffin blocks were mounted on glass slides and incubated at 56°C overnight. The slides were then de-paraffinized with xylene and dehydrated with alcohol. After washing with tap water, the slides were incubated in 0.3% hydrogen peroxide to block endogenous peroxidase activity. After washing with PBS, the slides were blocked with 5% skimmed milk in PBS, followed by incubation with the primary antibodies (c-Myc, 1:250; cyclin B1, 1:250; p-CDK1, 1:250; CDK1, 1:250; cyclin E1, 1:250) for 1 h at room temperature. Next, the slides were washed in PBS and incubated with HRP-conjugated rabbit or mouse IgG antibodies (1:500) for 30 min at room temperature. Finally, the antibody binding was visualized using a diaminobenzidine system and counterstained with Giemsa staining at 37°C for 5 mins. Bone marrow from B-ALL patients was smeared and stained as described above.
Semi-quantitative analysis of the immunohistochemical staining in tissues
The intensity of positive staining in tissue sections was analyzed via the integrated optical density (IOD) using the Image-Pro Plus 5.1 software (Media Cybernetics, Inc., Rockville, MD, USA). In brief, 4 images for each of 4 individual tumor samples were analyzed in a blinded manner. All of the images were captured using the same microscope and camera settings. Image-Pro Plus 5.1 software was used to calculate the average IOD per stained area (IOD/µm2) to quantify the staining intensity.
Statistical analysis
Each experiment was performed at least three times. Values are expressed as the mean ± standard error mean. Statistical analyses were performed with the statistical software package GraphPad Prism 5 (GraphPad Inc., La Jolla, CA, USA). The statistical significance was assessed by one-way analysis of variance with Student-Newman-Keuls post-hoc test. The level of differences between two groups was analyzed by Student's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Loss of the c-Myc gene significantly reduces the growth rate of Raji cells
To explore the function of c-Myc, specific c-Myc-siRNA fragments corresponding to nucleotides 586–605, 1,636–1,655 and 1,831–1,850 of a human c-Myc ORF (NM_002467) were designed. In the RT-PCR analysis, the siRNA fragment (1,831–1,850) exhibited a stronger inhibitory effect than other siRNA fragments (Fig. 1A). However, this effect was short-lived (about 3 days by transient transfection; data not shown). It is important to assess the role of c-Myc under experimental conditions where the effect of endogenous c-Myc is completely eliminated. To stably silence c-Myc gene expression, c-Myc knockdown cells were established, which were referred to as 'Raji-KD cells', by transfection with a replication-defective retrovirus encoding a c-Myc small hairpin (sh)RNA. For c-Myc reintroduction, an ORF of the c-Myc gene was cloned into the ClaI site of a pLHCXsi-mU6-c-Myc expression vector that is resistant to the siRNA expressed in Raji-KD cells, and cells transfected with the two were referred to as 'Raji-KD-Re cells'. As presented in Fig. 1B, the mRNA expression of the c-Myc gene was significantly downregulated in the Raji-KD cells after c-Myc shRNA transduction. The loss of mRNA expression was then restored in the Raji-KD-Re cells via the introduction of the c-Myc gene. Again, the c-Myc protein expression was significantly down-regulated from the Raji-KD cells as evidenced by western blot analysis, and partly restored in the Raji-KD-Re cells (Fig. 1C).
Figure 1.
Knockdown of c-Myc significantly reduces the proliferation rate of Raji cells. (A) Three different c-Myc siRNAs were transiently transfected into Raji cells. mRNA expression of c-Myc was determined by RT-PCR. GAPDH was used as an internal loading control. (B) mRNA expression of c-Myc was determined by RT-qPCR with normalization to GAPDH expression. (C) Western blot analysis was used to determine c-Myc expression in Raji, Raji-KD and Raji-KD-Re cells. GAPDH served as a loading control. (D) MTT assays for Raji, Raji-KD and Raji-KD-Re cells. OD(490) values were measured using a microplate reader. (E) Effect of c-Myc-knockdown on cell proliferation was measured using BrdU assays. Values are expressed as the mean ± standard error of the mean. **P<0.01, ***P<0.001. RT-PCR, semi-quantitative reverse transcription PCR; RT-qPCR, RT-quantitative PCR; PCR, polymerase chain reaction; siRNA, small interfering RNA; OD(490), optical density at 490 nm; Raji-KD, Raji cells with c-Myc knockdown; Raji-KD-Re, Raji-KD with re-expression of c-Myc.
Previous studies have demonstrated that c-Myc has a critical role in regulating cell proliferation and growth (20). Removal of c-Myc results in a marked prolongation of the doubling time. In the case of Rat1 fibroblasts, the doubling time increases from ~16 to ~50 h without c-Myc, indicating that the loss of c-Myc expression has an impact the proliferation of cells. From the MTT assay, it was indicated that in contrast to the mock-transfected cells, the growth rates of the Raji-KD cells were significantly reduced (>50% inhibition) after 72 h, and were restored by c-Myc re-expression in the Raji-KD-Re cells (Fig. 1D). Furthermore, the populations of Raji, Raji-KD and Raji-KD-Re cells were detected immunochemically using a bromodeoxyuridine cell proliferation assay, which mirrored the results of the MTT assay, suggesting that c-Myc affected the cell proliferation by inhibiting DNA synthesis (Fig. 1E).
Loss of c-Myc decreases the expression of cell cycle-associated genes and induces G2/M-phase arrest
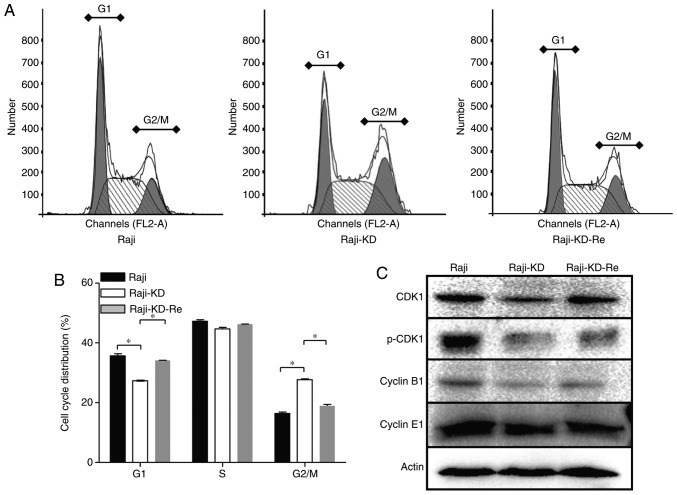
Previous studies suggest that the regulation of cell proliferation by c-Myc involves several key components of cell-cycle regulatory molecules (21,22). To determine whether c-Myc influences cell proliferation via inducing cell-cycle arrest, the three types of cells were processed for cell-cycle analysis by flow cytometry. The cell-cycle profiles of Raji-KD cells were obviously different from those of Raji and Raji-KD-Re cells (Fig. 2A and B). Loss of c-Myc expression resulted in a significant G2/M-phase arrest, while the G1-phase population was decreased (Fig. 2A and B).
Figure 2.
Inhibition of the c-Myc gene induces G2/M arrest and downregulates the expression of CDKs in Raji cells. PI staining and flow cytometric analysis were performed to measure the DNA content of Raji, Raji-KD and Raji-KD-Re cells. (A) Cell cycle analysis of Raji, Raji-KD and Raji-KD-Re cells. (B) Statistical analysis of flow cytometry results. The percentages of cells in different cell cycle phases (G1, S or G2/M) are displayed. Values are expressed as the mean ± standard error of the mean. *P<0.05. (C) Western blot analysis of CDK1, p-CDK1, cyclin B1 and cyclin E1 protein expression. GAPDH severed as a loading control. PI, propidium iodide; Raji-KD, Raji cells with c-Myc knockdown; Raji-KD-Re, Raji-KD with re-expression of c-Myc; p-CDK, phosphorylated cyclin D kinase.
Activation of CDK1 and cyclin B1 is a major checkpoint of transition from the G2-phase to mitosis (G2/M), which governs most of the processes involved in mitotic initiation and progression (23,24). To assess why knockdown of the c-Myc gene induced G2/M arrest, the expression of CDK1 and cyclin B1 in the Raji-derived cells was examined. The loss of c-Myc caused a reduction in the levels of CDK1, p-CDK1 and cyclin B1 in Raji-KD cells, while no significant difference was identified in cyclin E1 expression. Of note, the c-Myc shRNA-associated reduction in the expression of CDK1, p-CDK1 and cyclin B1 was attenuated by re-expression of c-Myc (Fig. 2C), suggesting that the cell-cycle arrest in G2/M-phase may be attributed to the downregulation of CDK1, p-CDK1 and cyclin B1 (Fig. 2A and B). To further elucidate the underlying mechanisms of the cell-cycle arrest caused by the suppression of c-Myc, cell cycle-associated gene expression was examined between Raji cells and Raji-KD cells via a PCR array. A total of 28 genes with a 2-fold difference in their expression levels were identified. Among them, 24 genes [CCNG1, CCNG2, HUS1 checkpoint clamp component, RB1, RB-like 2, BRCA2 and -1 DNA repair associated, CDK1, S-phase kinase-associated protein 2, CDKN1B, CDK6, caspase-3, growth arrest and DNA-damage-inducible α, meiotic recombination 11 homolog A (S. cerevisiae), MRE11 homolog double strand break repair nuclease, MDM2 protooncogene, CCNF, CDK8, WEE1 G2 checkpoint kinase, cell division cycle 6, hypoxanthine phosphoribosyl transferase 1, checkpoint kinase 1, transcription factor Dp-2 and TP53], which are involved in cell cycle regulation, were downregulated in Raji-KD cells, whereas 4 genes (CDKN1A, CCND2, CCND1 and baculoviral IAP repeat containing 5), which function in cell differentiation, were upregulated (Table I). CDKN1A (also known as p21) serves as an inhibitor of cellular proliferation in response to DNA damage (25). CCND1 and CCND2 are components of the cyclinD/CDK4/CDK6 complexes, and the latter phosphorylates and inhibits members of the retinoblastoma protein family and regulates the cell cycle during G1/S transition (26). These results demonstrated that downregulation of c-Myc induces G2/M arrest by regulating the expression of CDK1 and cyclin B1, which are capable of accelerating cell-cycle progression.
Table I.
Up- and downregulated genes in Raji-KD cells.
| Gene name | Definition | GenBank ID | Fold change (Raji-KD/Raji) |
|---|---|---|---|
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | NM_000389 | 6.10 |
| CCND2 | Cyclin D2 | NM_001759 | 3.31 |
| CCND1 | Cyclin D1 | NM_053056 | 3.03 |
| BIRC5 | Baculoviral IAP repeat containing 5 | NM_001012270 | 2.12 |
| CCNG1 | Cyclin G1 | NM_004060 | −6.90 |
| CCNG2 | Cyclin G2 | NM_004354 | −5.22 |
| HUS1 | HUS1 checkpoint clamp component | NM_004507 | −3.75 |
| RB1 | RB transcriptional corepressor 1 | NM_000321 | −3.69 |
| RBL2 | RB transcriptional corepressor like 2 | NM_005611 | −3.44 |
| BRCA2 | BRCA2, DNA repair associated | NM_000059 | −3.40 |
| CDK1 | Cyclin dependent kinase 1 | NM_001170406 | −3.34 |
| SKP2 | S-phase kinase-associated protein 2 | NM_001243120 | −2.90 |
| CDKN1B | Cyclin dependent kinase inhibitor 1B | NM_004064 | −2.79 |
| CDK6 | Cyclin dependent kinase 6 | NM_001145306 | −2.78 |
| CASP3 | Caspase-3 | NM_004346 | −2.66 |
| GADD45A | Growth arrest and DNA damage inducible α | NM_001199741 | −2.60 |
| MRE11A | MRE11 homolog, double strand break repair nuclease | NM_005590 | −2.54 |
| BRCA1 | BRCA1, DNA repair associated | NM_007294 | −2.40 |
| MAD2L1 | Mitotic arrest deficient 2 like 1 | NM_002358 | −2.24 |
| MDM2 | MDM2 proto-oncogene | NM_001145337 | −2.13 |
| CCNF | Cyclin F | NM_001761 | −2.03 |
| CDK8 | Cyclin dependent kinase 8 | NM_001260 | −2.00 |
| WEE1 | WEE1 G2 checkpoint kinase | NM_001143976 | −2.00 |
| CDC6 | Cell division cycle 6 | NM_001254 | −2.00 |
| HPRT1 | Hypoxanthine phosphoribosyl transferase 1 | NM_000194 | −2.00 |
| CHEK1 | Checkpoint kinase 1 | NM_001114121 | −2.00 |
| TFDP2 | Transcription factor Dp-2 | NM_001178138 | −2.00 |
| TP53 | Tumor protein p53 | NM_000546 | −2.00 |
Raji-KD, Raji cells with c-Myc knockdown.
c-Myc induces cell cycle-associated gene expression by transcriptionally regulating HAT
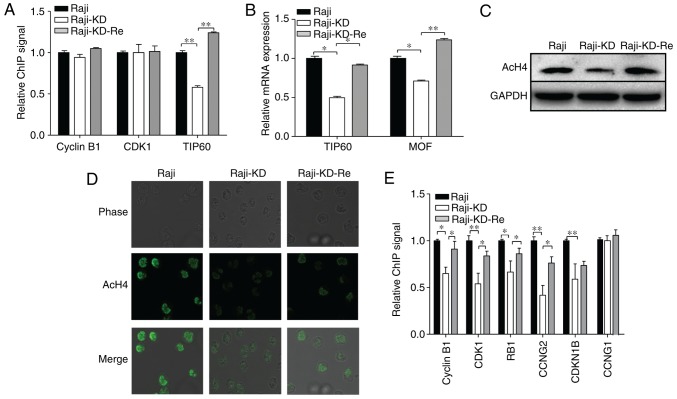
As a transcription factor, c-Myc usually binds to specific sites in the target gene promoters, acts as a general amplifier of transcription, and is known to alter histone modifications, thereby having a role in remodeling target chromatin (19,27). To assess whether c-Myc directly impacts gene transcription in Raji cells, a ChIP assay was used to evaluate the c-Myc-dependent regulation of cell cycle-associated genes. Based on a PCR array and western blot analysis of cell-cycle-associated genes (Fig. 2C and Table I), cyclin B1 and CDK1 were selected as the target genes. The real-time PCR-amplified cyclin B1 and CDK1 signals from the c-Myc-ChIP assay exhibited no significant differences between Raji, Raji-KD and Raji-KD-Re cells; however, the PCR-amplified gene signals of TIP60 immunoprecipitated with c-Myc antibodies were reduced in the Raji-KD cells compared with those in Raji cells, and were significantly increased in Raji-KD-Re cells (Fig. 3A). In addition, the mRNA levels of TIP60 and MOF were assessed in the three groups of cells, indicating that knockdown of the c-Myc gene resulted in decreased expression of TIP60 and MOF (Fig. 3B). Of note, the level of AcH4 was markedly decreased in the Raji-KD cells, as evidenced by western blot analysis (Fig. 3C) and immunostaining assays (Fig. 3D). These results suggest that c-Myc regulates the transcription of cell cycle-associated genes through an indirect AcH4-associated mechanism in Raji cells. To further evaluate AcH4-dependent changes in cell cycle-associated gene expression, ChIP assays using AcH4 antibodies and PCR analysis were performed. As presented in Fig. 3E, the gene signals of cyclin B1, CDK1, RB1, CCNG2 and cyclin dependent kinase inhibitor 1B (CDKN1B) from the amplification of AcH4-ChIPed sequences were reduced in the Raji-KD cells compared with Raji cells and restored in Raji-KD-Re cells, while the gene signals for CCNG1 were not affected. It is conceivable that c-Myc regulates the expression of cell cycle-associated genes, including cyclin B1, CDK1, RB1 and CCNG2, by controlling the HAT-mediated acetylation of histone H4 in target chromatin.
Figure 3.
Loss of c-Myc suppresses the expression of CDK by transcriptionally regulating HAT that targets histone H4. (A) c-Myc-ChIP assay with Raji, Raji-KD and Raji-KD-Re cells. Following formaldehyde cross-linking and DNA fragmentation, samples were immunoprecipitated with antibody specific for c-Myc. The c-Myc binding signal for TIP60 was reduced in Raji-KD cells, while that for cyclin B1 and CDK1 did not change. (B) mRNA expression of TIP60 and MOF was determined by reverse transcription-quantitative polymerase chain reaction. GAPDH expression was used for normalization. (C) Western blot analysis of AcH4 expression in Raji, Raji-KD and Raji-KD-Re cells. GAPDH served as a loading control. (D) Confocal microscopy analysis of Raji, Raji-KD and Raji-KD-Re cells after staining with anti-AcH4 antibody (magnification, ×400). (E) AcH4-ChIP assay of Raji, Raji-KD and Raji-KD-Re cells. The AcH4 binding signals of cell cycle-associated genes were reduced by the knockdown of c-Myc gene. Values are expressed as the mean ± standard error of the mean. *P<0.05, **P<0.01. Raji-KD, Raji cells with c-Myc knockdown; Raji-KD-Re, Raji-KD with re-expression of c-Myc; CDK, cyclin D kinase; ChIP, chromatin immunoprecipitation; AcH4, acetylated histone H4; TIP60, 60 kDa Tat-interactive protein; MOF, males absent on the first; RB, retinoblastoma; CCN, cyclin. CDKN1B, cyclin dependent kinase inhibitor 1B.
Knockdown of c-Myc suppresses apoptosis by regulating caspase-3 expression
In addition to the function of c-Myc in cell growth and proliferation, numerous studies have reported that c-Myc induces cell apoptosis (4,5,28). To investigate the effect of c-Myc on cell apoptosis, the three cell groups of the present study were analyzed by flow cytometry after Annexin V and PI staining. As presented in Fig. 4A and B, the percentage of early apoptotic cells (PI− Annexin V+ cells) in Raji, Raji-KD and Raji-KD-Re cells was 2.56, 2.39 and 3.7%, respectively, and the percentage of late apoptotic cells (PI+ Annexin V+ cells) was 6.91, 2.42 and 3.16%, respectively (Fig. 4B). The apoptotic rate in the Raji-KD cells was significantly lower than that in Raji cells and was partially restored in the Raji-KD-Re cells, which indicated that c-Myc is involved in cell apoptosis. To explore the underlying mechanism, the expression of caspase-3, -8 and -9 was examined. In contrast to the Raji cells, knockdown of the c-Myc gene led to a significant decrease in pro-caspase-3 expression in Raji-KD cells (Fig. 4C), while the expression of caspase-8 or 9 exhibited no difference among the Raji, Raji-KD and Raji-KD-Re cells. These results indicate that c-Myc promotes the apoptosis of Raji cells by regulating the expression of activated caspase-3, which has a key role in the execution of multiple apoptotic pathways.
Figure 4.
Knockdown of c-Myc suppresses the apoptosis of Raji-KD cells by regulating caspase-3 expression. (A) Flow cytometric analysis was performed to measure the apoptotic rate of Raji, Raji-KD and Raji-KD-Re cells after PI and Annexin V staining. (B) Quantitative analysis of flow cytometry results. Values are expressed as the mean ± standard error of the mean. *P<0.05, **P<0.01. (C) Western blot analysis of caspase-3, caspase-8 and caspase-9 expression in Raji, Raji-KD and Raji-KD-Re cells. Caspase-3 showed two forms of pro caspase and cleaved caspase. GAPDH served as a loading control. PI, propidium iodide; Raji-KD, Raji cells with c-Myc knockdown; Raji-KD-Re, Raji-KD with re-expression of c-Myc.
c-Myc expression is essential for the Raji cells-derived tumor formation
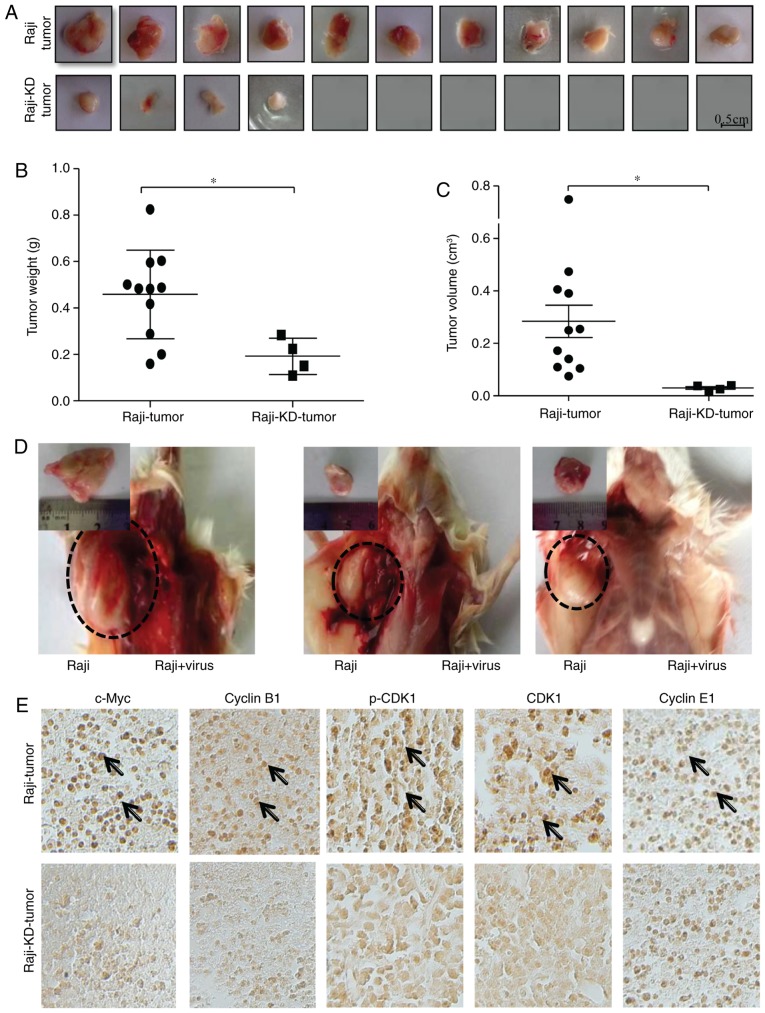
It is well known that c-Myc has a significant role in promoting tumor formation in vivo. To examine the influence of c-Myc knockdown in Raji cells on xenograft tumor formation, a xenograft model of B-ALL was established in SCID mice. As presented in Fig. 5A, Raji cells rapidly formed tumors with no notable rejection following subcutaneous injection into the left axilla of mice, whereas the incidence of tumor formation was significantly suppressed in mice that were injected with Raji-KD cells over a period of 30 days. Only 4 small tumors (4 tumors/11 mice) were observed in the mice that were transplanted with Raji-KD cells (Fig. 5A). No adverse events were noted in mice injected with Raji cells expressing c-Myc shRNA. After 30 days, the volume and weight of the tumors were measured. It was apparent that the tumors of the Raji-KD cell group were smaller and lighter than those of the Raji cells group (Fig. 5B and C), suggesting that the tumor-forming ability of Raji-KD was severely reduced by knockdown of c-Myc. Since the abovementioned results indicated that c-Myc promotes tumor growth in vitro by modulating the cell cycle, the efficiency of injection of the c-Myc shRNA retrovirus in inhibiting tumor formation was then assessed in vivo. During the first two days after inoculation of Raji cells into the bilateral hind axilla, mice were injected with c-Myc shRNA retrovirus on the right axilla or with a pseudotyped retrovirus on the left over. As presented in Fig. 5D, injection of the c-Myc shRNA retrovirus markedly inhibited the tumor formation in vivo. The weights and volumes of the tumors were 1.27±0.85 g and 1.23±1.14 (cm3) in mice injected with Raji cells, while the tumor was not generated following injection with c-Myc shRNA retroviruses (Table II). Based on the immunohistochemical analysis, the expressions of cyclin B1, CDK1 and p-CDK1 were suppressed in Raji cell-derived tumors following the retroviral vector-mediated c-Myc-siRNA injection, while no significant difference was identified in cyclin E1 expression (Fig. 5E). These results are consistent with the observation that the expression of these proteins was significantly suppressed in Raji cells afterc-Myc-siRNA injection vs. Raji cells (Fig. 2C). The present results suggested that loss of c-Myc suppressed tumorigenesis, probably and at least in part via downregulation of the expression of G2/M checkpoint-associated genes, including CDK1 and cyclin B1.
Figure 5.
Knockdown of c-Myc suppresses the tumorigenesis of Raji cells in a SCID mouse xenograft model. (A-C) At 30 days after injection of 1×107 Raji or Raji-KD cells into the axilla, tumors were harvested. Tumor formation at the site of injection was monitored. (A) Representative images of tumors formed in the mice. (B) Tumor weight in the two groups. The weight of Raji-KD tumors was lower than that of Raji tumors. (C) Tumor volume in the two groups. The volumes of Raji-KD tumors were smaller than those of Raji tumors. Values are expressed as the mean ± standard error of the mean. *P<0.05. (D) Representative images of tumors grown in the animals at 45 days after injection of 2×106 Raji cells into the bilateral axillary sites of the hind legs (circles with broken lines). On the first two days after inoculation of Raji cells, mice were injected in to the axilla with c-Myc shRNA retrovirus on the right or with the pseudotyped retrovirus on the left. Magnified images display the respective tumors after dissection. The tumor formation of Raji cells was inhibited by injection of c-Myc shRNA retrovirus. (E) Immunohistochemical staining of Raji (upper panels) and Raji-KD (lower panels) xenograft tumor tissues [from (A)] for c-Myc, CDK1, p-CDK1, cyclin B1 and cyclin E1 (magnification, ×200). Immunopositivity was indicated by a brown stain (arrows). shRNA, small hairpin RNA; p-CDK1, phosphorylated cyclin D kinase 1; Raji-KD, Raji cells with c-Myc knockdown.
Table II.
Injection of c-Myc shRNA retrovirus completely inhibits tumor formation in vivo.
| Cells | Tumor formation time (days) | Tumor weight (g) | Tumor volume (cm3) |
|---|---|---|---|
| Raji (2×106) | 45 | 1.27±0.85 | 1.23±1.14 |
| Raji (2×106) + shRNA | 45 | 0.00 | 0.00 |
shRNA, small hairpin RNA.
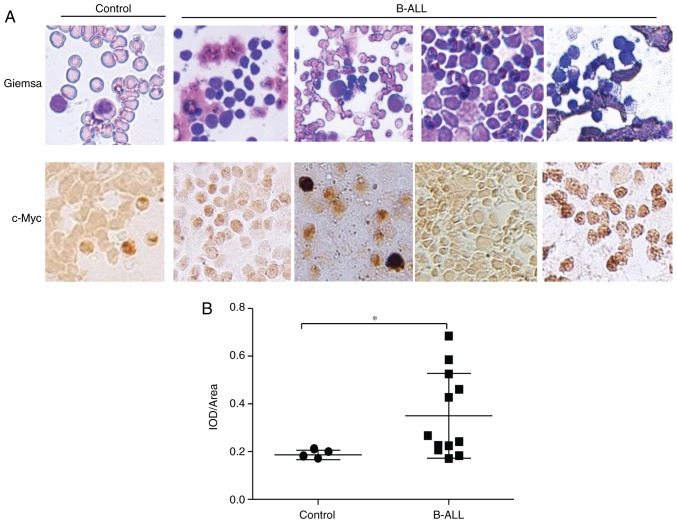
As reported previously, overexpression of c-Myc is the major characteristic of B-ALL (2). To further confirm the tumor promoter effect of c-Myc in B cell lymphoma tumorigenesis, c-Myc expression in the bone marrow aspirations and biopsies of B-ALL patients was examined. The immunohistochemistry assays using anti-c-Myc antibody indicated that c-Myc was overexpressed in the B-ALL samples, compared with that in the samples from individuals with anemia (Fig. 6A). Semi-quantitative IOD analysis also confirmed significantly enhanced anti-c-Myc antibody staining in the patient group (Fig. 6B). These results indicated that the expression of c-Myc is constitutively activated in the process of B-ALL tumorigenesis, and that c-Myc RNA interference may serve as a potential therapeutic strategy for B-ALL.
Figure 6.
Immunohistochemical analysis of bone marrow cells from patients with B-ALL and individuals with anemia for c-Myc. (A) Cytogenetic analysis of biopsy samples by classical Giemsa staining (upper panels) and immunostaining with anti-c-Myc antibody (lower panels). Representative images are displayed (magnification, ×200). (B) Comparison of average IOD/area of c-Myc antibody staining in biopsy samples between controls and B-ALL patients. *P<0.05. IOD, integrated optical density; B-ALL, B-lineage acute lymphoblastic leukemia.
Discussion
The c-Myc protein has multiple functions in tumorigenesis and progression, and c-Myc transgenic mice, carrying c-Myc linked to the intron enhancer of the immunoglobulin heavy chain gene, developed clonal B-cell malignancies (29). Furthermore, when lymphoblastoid cells were transfected with a constitutively expressed c-Myc gene, the cells became tumorigenic in nude mice (30). Understanding c-Myc tumorigenic activity requires experimentally tractable models. To date, the study of c-Myc target genes has not been standardized as to the cell types used or the expression systems employed. With this regard, the present study established a novel B-ALL cell model (Raji-KD) with knockdown of the c-Myc gene, and demonstrated that c-Myc regulates the CDK1/cyclin B1-dependent G2/M cell cycle progression via controlling histone H4 acetylation.
A deletion of c-Myc in mice was lethal in homozygotes between 9.5 and 10.5 days of gestation, and the embryos were generally smaller and retarded in their development compared with their littermates (31), indicating that c-Myc is involved in the regulation of the cell cycle and cell proliferation. Prior study into the mechanisms underlying the effects of c-Myc have revealed that c-Myc-null rat fibroblasts display a prolonged G2-phase (32). Song et al (33) reported that knockdown of c-Myc led to a growth inhibition and cell cycle arrest at G2/M-phase in Jijoye cells, but the role of c-Myc in the G2/M-phase has remained elusive. The present study indicated that loss of c-Myc controlled the expression of genes associated with the G2/M-phase transition, including cyclin B1 and CDK1, by modulating TIP60-mediated histone H4 acetylation in specific chromosomal regions, which resulted in a marked G2/M-phase arrest in Raji-KD cells. Other studies have reported that loss of c-Myc function impedes G1-phase progression (28,34). Felsher et al (35) has reported that overexpression of c-Myc causes TP53-dependent G2-phase arrest in normal fibroblasts. These observations suggest that c-Myc has different functions in diverse tumor cell types. Furthermore, the CDK inhibitors p27 and p21 appear to be critical targets of c-Myc. c-Myc cooperates with Zn-finger transcription factor Miz-1 to repress the transcription of cell-cycle inhibitors, including p15, p21 and p27 (5,36). The present study reported that suppression of c-Myc induced the upregulation of p21 and the downregulation of p27 in Raji-KD cells. However, further study is required to determine whether G2/M arrest induced by the loss of c-Myc is associated with changes in p27/p21 expression.
Distinct HATs, including TIP60, MOF, Kat2B and Kat2A, associate with c-Myc TAD as the TRRAP, and form the different HAT complexes that regulate histone H3 and H4 acetylation (6,27,37). Recruitment of TIP60 to c-Myc is significant for gene regulation by c-Myc (27,37). In the present study, a c-Myc-ChIP assay indicated that c-Myc directly regulates TIP60 expression. Reduction of MOF and AcH4 are known to correlate with the reduced transcription of certain genes and with G2/M-phase arrest (38). However, the universal mechanism underlying c-Myc-mediated G2/M-phase arrest remains to be elucidated. In the present study, the AcH4-ChIP assay indicated that the transcription of cyclin B1, CDK1, RB1, CCNG2 and CDKN1B was controlled by TIP60 and/or MOF-mediated histone H4 acetylation, which was regulated by c-Myc. The unique means by which c-Myc was demonstrated to regulate cell cycle-associated factors is indicative of previously unsuspected links between c-Myc, HATs, AcH4 and G2/M-phase arrest.
Cell-cycle arrest in G2/M-phase usually reflects a requirement for repairing cell damage; if not repaired, apoptotic mechanisms are often activated (39). The present study indicated that downregulation of c-Myc in Raji-KD cells induced decreases in cell apoptosis. c-Myc induces apoptosis through various mechanisms; for instance, it activates upstream events in the apoptotic cascade, including death receptor Fas and Fas ligand (40). Furthermore, numerous target genes of c-Myc that regulate mitochondrial function have been implicated in c-Myc-induced apoptosis (41). In addition, various target genes of c-Myc that regulate the cell cycles, including CDC25A, have been implicated in cell apoptosis (42). In the present study, TP53 expression was also downregulated in the Raji-KD cells. Furthermore, knockdown of c-Myc decreased the expression of pro-caspase-3, but did not affect the expression of caspase-8 and 9 in Raji cells. Although the detailed molecular mechanisms of how c-Myc causes a decrease in caspase-3 levels remains to be determined, elucidation of the links between c-Myc and the caspase family may clarify the mechanisms underlying c-Myc-induced apoptosis.
B-ALL is an aggressive form of Non-Hodgkin lymphoma and is the most common type of childhood cancer (43). The survival rates of B-ALL patients require to be improved via development of novel, innovative therapies. To elucidated the biological functions of c-Myc in B-ALL, an experimentally tractable model is required. In this regard, to determine the role of c-Myc in modulating lymphoma cell behavior, the present study used Raji cells to generate Raji-KD and Raji-KD-Re cells. It was revealed that c-Myc recruited HAT that induced histone H4 acetylation in specific chromatin regions, thereby opening chromatin structures and amplifying the expression of target genes. Knockdown of the c-Myc gene led to the downregulation of the expression of CDK1 and cyclin B1 and resulted in G2/M-phase arrest by histone H4 acetylation. Of note, loss of c-Myc function by anti-sense suppression significantly inhibited the tumorigenesis of Raji cells in vivo. Indeed, c-Myc was overexpressed in the B-ALL samples, compared to the samples from individuals with anemia (Fig. 6). It is consistent with the reports that c-Myc contributes to the lymphoma tumorgenesis (44). The therapeutic strategies of c-Myc knockdown may be valuable for establishing novel methods for B-ALL therapy.
Acknowledgments
The authors gratefully acknowledge Professor Jianzhong Qin (Department of Biological Sciences, Dalian University, Dalian, China) for supportive comments regarding the manuscript.
Footnotes
Funding
This study is supported by National Nature Science Foundation of China (grant nos. 31570797, 31270864, 30972675 and 31101717), the Science and Technology Planning Project of Dalian City (grant no. 2010J21DW011) and the Natural Science Foundation of Liaoning Province (grant no. 2015020253).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author upon reasonable request.
Authors' contributions
WL was desponsible for conception and designed of the present study. YY, KX, ZL, WD, WZ, ZS, SS and TM performed experiments; WL wrote the manuscript, which was reviewed by all authors.
Ethics approval and consent to participate
The procedures for handling animals complied with the Current Laboratory Animal Las and Regulations, Policies and Administration in China. All experiments were approved by the Animal Ethics Committee of Dalian Medical University (no. AEE17013).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Villarreal-Martinez L, Jaime-Pérez JC, Rodríguez-Martínez M, González-Llano O, Gómez-Almaguer D. Acute lymphoblastic leukemia of childhood presenting as aplastic anemia: Report of two cases. Rev Bras Hematol Hemoter. 2012;34:165–167. doi: 10.5581/1516-8484.20120037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liew M, Rowe L, Clement PW, Miles RR, Salama ME. Validation of break-apart and fusion MYC probes using a digital fluorescence in situ hybridization capture and imaging system. J Pathol Inform. 2016;7:20. doi: 10.4103/2153-3539.181764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Teriete P, Hu A, Raveendra-Panickar D, Pendelton K, Lazo JS, Eiseman J, Holien T, Misund K, Oliynyk G, et al. Direct inhibition of c-Myc-Max heterodimers by celastrol and celastrol-inspired triterpenoids. Oncotarget. 2015;6:32380–32395. doi: 10.18632/oncotarget.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta. 2014;1839:395–405. doi: 10.1016/j.bbagrm.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013;3:a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Bennett RL, May WS. c-Myc and caspase-2 are involved in activating Bax during cytotoxic drug-induced apoptosis. J Biol Chem. 2008;283:14490–14496. doi: 10.1074/jbc.M801107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C, Wang J, Tan Q, Cheng Y, Xia E, et al. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am J Transl Res. 2017;9:533–545. [PMC free article] [PubMed] [Google Scholar]
- 10.Treszl A, Adány R, Rákosy Z, Kardos L, Bégány A, Gilde K, Balázs M. Extra copies of c-myc are more pronounced in nodular melanomas than in superficial spreading melanomas as revealed by fluorescence in situ hybridisation. Cytometry B Clin Cytom. 2004;60:37–46. doi: 10.1002/cyto.b.20016. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Pérez MV, Henley AB, Arsenian-Henriksson M. The MYCN protein in health and disease. Genes (Basel) 2017;8:E113. doi: 10.3390/genes8040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Yoo HM, Choi I, Pyun KH, Byun SM, Ha H. Interleukin 4-induced proliferation in normal human keratinocytes is associated with c-myc gene expression and inhibited by genistein. J Invest Dermatol. 1996;107:367–372. doi: 10.1111/1523-1747.ep12363346. [DOI] [PubMed] [Google Scholar]
- 13.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol. 1998;153:141–148. doi: 10.1016/S0002-9440(10)65554-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nupponen NN, Hyytinen ER, Kallioniemi AH, Visakorpi T. Genetic alterations in prostate cancer cell lines detected by comparative genomic hybridization. Cancer Genet Cytogenet. 1998;101:53–57. doi: 10.1016/S0165-4608(97)00060-5. [DOI] [PubMed] [Google Scholar]
- 15.Gugger M, Burckhardt E, Kappeler A, Hirsiger H, Laissue JA, Mazzucchelli L. Quantitative expansion of structural genomic alterations in the spectrum of neuroendocrine lung carcinomas. J Pathol. 2002;196:408–415. doi: 10.1002/path.1065. [DOI] [PubMed] [Google Scholar]
- 16.Hecht JL, Aster JC. Molecular biology of Burkitt's lymphoma. J Clin Oncol. 2000;18:3707–3721. doi: 10.1200/JCO.2000.18.21.3707. [DOI] [PubMed] [Google Scholar]
- 17.Neri A, Barriga F, Knowles DM, Magrath IT, Dalla-Favera R. Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt lymphoma. Proc Natl Acad Sci USA. 1988;85:2748–2752. doi: 10.1073/pnas.85.8.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 19.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer D, Chung YM, Hu MC. Effects of low-dose bisphenol A on DNA damage and proliferation of breast cells: The role of c-Myc. Environ Health Perspect. 2015;123:1271–1279. doi: 10.1289/ehp.1409199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap CS, Peterson AL, Castellani G, Sedivy JM, Neretti N. Kinetic profiling of the c-Myc transcriptome and bioinformatic analysis of repressed gene promoters. Cell Cycle. 2011;10:2184–2196. doi: 10.4161/cc.10.13.16249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo HR, Kim J, Bae S, Soh JW, Lee YS. Cdk5-mediated phosphorylation of c-Myc on Ser-62 is essential in transcriptional activation of cyclin B1 by cyclin G1. J Biol Chem. 2008;283:15601–15610. doi: 10.1074/jbc.M800987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu YX, Manley JL. New insights into mitotic chromosome condensation: A role for the prolyl isomerase Pin1. Cell Cycle. 2007;6:2896–2901. doi: 10.4161/cc.6.23.4977. [DOI] [PubMed] [Google Scholar]
- 25.Ehedego H, Boekschoten MV, Hu W, Doler C, Haybaeck J, Gaβler N, Müller M, Liedtke C, Trautwein C. p21 ablation in liver enhances DNA damage, cholestasis, and carcinogenesis. Cancer Res. 2015;75:1144–1155. doi: 10.1158/0008-5472.CAN-14-1356. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Di Liberto M, Jayabalan D, Liang J, Ely S, Bretz J, Shaffer AL, III, Louie T, Chen I, Randolph S, et al. Prolonged early G(1) arrest by selective CDK4/CDK6 inhibition sensitizes myeloma cells to cytotoxic killing through cell cycle-coupled loss of IRF4. Blood. 2012;120:1095–1106. doi: 10.1182/blood-2012-03-415984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravens S, Yu C, Ye T, Stierle M, Tora L. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenetics Chromatin. 2015;8:45. doi: 10.1186/s13072-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Nussenzweig MC, Schmidt EV, Shaw AC, Sinn E, Campos-Torres J, Mathey-Prevot B, Pattengale PK, Leder P. A human immunoglobulin gene reduces the incidence of lymphomas in c-Myc-bearing transgenic mice. Nature. 1988;336:446–450. doi: 10.1038/336446a0. [DOI] [PubMed] [Google Scholar]
- 30.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 32.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/MCB.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song A, Ye J, Zhang K, Sun L, Zhao Y, Yu H. Lentiviral vector-mediated siRNA knockdown of c-MYC: Cell growth inhibition and cell cycle arrest at G2/M phase in Jijoye cells. Biochem Genet. 2013;51:603–617. doi: 10.1007/s10528-013-9590-0. [DOI] [PubMed] [Google Scholar]
- 34.Schorl C, Sedivy JM. Loss of protooncogene c-Myc function impedes G1 phase progression both before and after the restriction point. Mol Biol Cell. 2003;14:823–835. doi: 10.1091/mbc.E02-10-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM. Overexpression of MYC causes 53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci USA. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vo BT, Wolf E, Kawauchi D, Gebhardt A, Rehg JE, Finkelstein D, Walz S, Murphy BL, Youn YH, Han YG, et al. The interaction of Myc with Miz1 defines medulloblastoma subgroup identity. Cancer Cell. 2016;29:5–16. doi: 10.1016/j.ccell.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf E, Lin CY, Eilers M, Levens DL. Taming of the beast: Shaping Myc-dependent amplification. Trends Cell Biol. 2015;25:241–248. doi: 10.1016/j.tcb.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith ER, Cayrou C, Huang R, Lane WS, Côté J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozenblat S, Grossman S, Bergman M, Gottlieb H, Cohen Y, Dovrat S. Induction of G2/M arrest and apoptosis by sesquiterpene lactones in human melanoma cell lines. Biochem Pharmacol. 2008;75:369–382. doi: 10.1016/j.bcp.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Wiese KE, Haikala HM, von Eyss B, Wolf E, Esnault C, Rosenwald A, Treisman R, Klefström J, Eilers M. Repression of SRF target genes is critical for Myc-dependent apoptosis of epithelial cells. EMBO J. 2015;34:1554–1571. doi: 10.15252/embj.201490467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 42.Shen T, Huang S. The role of Cdc25A in the regulation of cell proliferation and apoptosis. Anticancer Agents Med Chem. 2012;12:631–639. doi: 10.2174/187152012800617678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228–233. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]