Short abstract
Mitophagy is a cellular process by which dysfunctional mitochondria are degraded via autophagy. Increasing empirical evidence proposes that this mitochondrial quality-control mechanism is defective in neurons of patients with various neurodegenerative diseases such as Ataxia Telangiectasia, Alzheimer’s disease, Parkinson’s disease, and Amyotrophic Lateral Sclerosis. Accumulation of defective mitochondria and the production of reactive oxygen species due to defective mitophagy have been identified as causes underlying neurodegenerative disease pathogenesis. However, the reason mitophagy is defective in most neurodegenerative diseases is unclear. Like mitophagy, defects in the ubiquitin/26S proteasome pathway have been linked to neurodegeneration, resulting in the characteristic protein aggregates often seen in neurons of affected patients. Although initiation of mitophagy requires a functional ubiquitin pathway, whether defects in the ubiquitin pathway are causally responsible for defective mitophagy is not known. In this mini-review, we introduce mitophagy and ubiquitin pathways and provide a summary of our current understanding of the regulation of mitophagy by the ubiquitin pathway. We will then briefly review empirical evidence supporting mitophagy defects in neurodegenerative diseases. The review will conclude with a discussion of the constitutively elevated expression of ubiquitin-like protein Interferon-Stimulated Gene 15 (ISG15), an antagonist of the ubiquitin pathway, as a potential cause of defective mitophagy in neurodegenerative diseases.
Impact statement
Neurodegenerative diseases place an enormous burden on patients and caregivers globally. Over six million people in the United States alone suffer from neurodegenerative diseases, all of which are chronic, incurable, and with causes unknown. Identifying a common molecular mechanism underpinning neurodegenerative disease pathology is urgently needed to aid in the design of effective therapies to ease suffering, reduce economic cost, and improve the quality of life for these patients. Although the development of neurodegeneration may vary between neurodegenerative diseases, they have common cellular hallmarks, including defects in the ubiquitin-proteasome system and mitophagy. In this review, we will provide a summary of our current understanding of the regulation of mitophagy by the ubiquitin pathway and discuss the potential of targeting mitophagy and ubiquitin pathways for the treatment of neurodegeneration.
Keywords: Mitochondrial, neurodegeneration, pathology, post-translational, proteasome, proteolysis
Introduction
The major functions of the mitophagy and ubiquitin pathways are to remove dysfunctional/old mitochondria from cells via autophagy,1–6 and to eliminate dysfunctional/misfolded proteins via the 26S proteasome,7–11 respectively. These specific functions enable the mitophagy pathway to regulate mitochondrial quality and the ubiquitin pathway to regulate protein quality in cells. Both healthy mitochondria and proteins are essential for cells to function normally. Therefore, defects in either the mitophagy or ubiquitin pathways are expected to disturb cellular homeostasis and are detrimental to cell survival. Indeed, defects in mitophagy and ubiquitin pathways lead to several human diseases, including cancer12–14 and neurodegenerative diseases.15–20 Although the mitophagy and ubiquitin pathways function to remove abnormal mitochondria and proteins, respectively, there is compelling evidence that the functional ubiquitin pathway is essential for the initiation of mitophagy,1,5,6,21–23 and that both pathways are defective in various neurodegenerative diseases. However, whether mitophagy is defective in neurodegenerative diseases due to defects in the ubiquitin pathway is still unknown. In this mini-review, we summarize major studies regarding the involvement of the ubiquitin pathway in mitophagy initiation and discuss the observed discrepancies and potential reasons for these discrepancies in these studies. Finally, we end this review with a discussion of the constitutively elevated expression of Interferon-Stimulated Gene 15 (ISG15), an antagonist of the ubiquitin pathway,24–26 as a potential cause of defective mitophagy in neurodegenerative diseases.
Mechanisms for mitochondrial quality control
Mitochondria promote cell survival by generating bioenergy in the form of adenosine triphosphate (ATP), which is required for vital cellular functions and can also sense “danger signals” to promote apoptotic cell death.27–29 Mitochondria are present in all human body cell types (except mature red blood cells) to perform these important cell fate-determining functions, and their abundance depends on the role of the cell. For example, mitochondrial content in endothelial cells is modest compared to hepatocytes, cardiac myocytes, and neurons that need constant energy supply for normal functioning. Mitochondria produce ATP in a process called oxidative phosphorylation.30,31 In this process, electrons from NADH and FADH2 formed during fatty acid oxidation, glycolysis, and the TCA cycle (cellular processes that break down proteins, carbohydrates, and fatty acids) are transferred to molecular oxygen (O2) through electron transport carriers (Complexes I, II, III, IV, and V) located in the inner mitochondrial membrane. When electrons reduce molecular O2 to water, a large amount of free energy is released which is then used to synthesize ATP. The transfer of electrons from NADH to O2 through electron transport carriers is not perfect, however, and there is always leakage of electrons at Complexes I and III. This electron leakage results in a partial reduction of O2 to generate a superoxide anion (O2−•), which quickly dismutates to hydrogen peroxide (H2O2) via superoxide dismutase 2 (SOD2) in the mitochondrial matrix and superoxide dismutase 1 (SOD1) in the mitochondrial intermembrane space. H2O2 is then reduced to water or is partially reduced to a hydroxyl radical (OH•). All O2−•, H2O2, and OH• generated in this process are collectively referred to as mitochondrial reactive oxygen species (ROS).32 Low levels of ROS retain physiological functions.33 However, when mitochondria are dysfunctional, levels of ROS increase, which in turn causes oxidative damage to cellular proteins and DNA. Consequently, cellular functions are impaired and cells undergo apoptosis.34–36 Hence, ROS-generating dysfunctional mitochondria are lethal to cells when not adequately removed. Given that neurons have higher energy demands and are post-mitotic, improperly functioning mitochondria can have severe consequences on neuronal survival.
Several mitochondrial quality-control mechanisms that remove dysfunctional mitochondria in cells have been identified.37 First, mitochondria maintain their quality by degrading unfolded, misfolded, or damaged membrane proteins (generated by environmental insults such as oxidative stress, viral infections, etc.) using their proteolytic system.38 Second, during the fusion/fission process, when damage is mild and below a certain threshold, fusion of normal mitochondria and a mitochondrion with mutant/damaged DNA or proteins allows for sharing of components to compensate for these defects.39 Third, a portion of mitochondria can bud off and form mitochondria-derived vesicles under conditions of oxidative stress, which further fuse with lysosomes to degrade oxidized mitochondrial proteins.40 Fourth, damaged mitochondria can also form mitochondrial spheroids that acquire lysosomal markers, become acidic, and undergo limited degradation of mitochondrial proteins.41 Lastly, damaged mitochondria can be degraded via mitophagy in conjunction with the ubiquitin pathway.1,5,21,42–44 In this review, we shall summarize our current understanding of mitophagy regulation via the ubiquitin pathway.
The ubiquitin pathway
Ubiquitin is a highly conserved 8 kDa protein that is ubiquitously expressed in all eukaryotic cells. Ubiquitin and ubiquitin-conjugating enzymes, collectively known as the ubiquitin pathway, regulate many cellular functions including cell cycle progression, development, apoptosis, signal transduction, antigen presentation, among several others, by inducing the signaling or timely degradation of regulatory proteins involved in these processes.7–11 The conjugation of ubiquitin to cellular proteins (ubiquitination) takes place in three steps: In the first step, the C-terminal glycine residues of ubiquitin are activated in an ATP-dependent manner to form a thioester linkage with a cysteine residue of an activating enzyme [E1]. In the second step, the activated ubiquitin is then transferred to one of the several (about a dozen) known carrier enzymes [E2s]. In the third step, ubiquitin is transferred either directly from the E2 carrier enzyme, or indirectly with the help of one of several (roughly 500–1000) known ubiquitin ligases [E3], that in turn, conjugates ubiquitin to target proteins. Although all three conjugating enzymes are necessary for ubiquitin conjugation, the E3 enzyme is key in the ubiquitination process, as this enzyme recognizes and covalently conjugates ubiquitin with high substrate specificity.8,45 The E3 enzyme conjugates either a single ubiquitin (monoubiquitination) or multiple ubiquitins (polyubiquitination) onto its target.11,46,47 In both cases, the first ubiquitin (Ub1) is appended to a specific lysine (Lys) residue on the substrate, and following ubiquitins (Ub2-Ubn) are attached to one of seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, or Lys63) within Ub1. Polyubiquitin chains conjugated through different lysines have distinct functions. For example, Lys48-linked polyubiquitin chains target proteins for degradation via the 26S proteasome.48 Alternatively, Lys63-linked polyubiquitin chains retain a signaling function in various cellular processes such as DNA repair, apoptosis, and autophagy.48 Although other polyubiquitin chains do exist, their functions are not well-defined in cells.49 Details about the structure and function of ubiquitin, ubiquitin-conjugating enzymes, polyubiquitin chains, and the 26S proteasome are extensively reviewed in the literature.48–53 Given the crucial role of the ubiquitin/26S proteasome in normal cell homeostasis, the contribution of defects in this system to the development of several human pathological conditions may come as no surprise. Indeed, the underlying pathology of diseases like cancer and neurodegenerative diseases has been linked to defects in the ubiquitin pathway.
Mitophagy and its regulation by ubiquitin
PTEN-induced putative kinase protein 1 (PINK1) and HECT/RING hybrid ubiquitin E3 ligase Parkin have been recognized as two important players of mitophagy.2,42,54 When mitochondria are healthy (polarized), PINK1 activity is minimized through its degradation by the PARL (presenilin-associated rhomboid-like protein) protease present in the mitochondrial inner membrane space and the 26S proteasome,55,56 and dephosphorylation and autoinhibition minimize Parkin activity. However, when mitochondria are damaged and depolarized, PINK1 is recruited onto the outer mitochondrial membrane and phosphorylates Ser65 of the ubiquitin moiety conjugated to outer mitochondrial membrane proteins. Binding of PINK1-generated phospho-ubiquitin and PINK1-mediated phosphorylation of the Ubl domain at Ser65 of Parkin releases Parkin from its autoinhibited state.57 Fully activated Parkin then appends more cytosolic ubiquitins on several mitochondrial proteins such as Mfn1/2, Miro1/2, VDAC, TOM, Fis1, and mitochondrial hexokinase. PINK1 and Parkin thus modify mitochondrial outer membrane proteins by phospho-ubiquitin polyubiquitin chains on depolarized mitochondria.58–61 These polyubiquitin chains then recruit autophagy receptors NDP52 and optineurin (OPTN) onto outer membrane proteins of depolarized mitochondria, which further recruit autophagy factors Ulk1, Atg14, DFCP1, WIPI-1, and Atg16L1 to Parkin-bound and ubiquitin-labeled mitochondria for their degradation via the autophagy pathway.62 In summary, PINK1 phosphorylates ubiquitin, Parkin amplifies this signal by appending more ubiquitins, these elongated phospho-ubiquitin chains then signal autophagy machinery, and damaged mitochondria are targeted for degradation via autophagy.63
Empirical evidence demonstrates that polyubiquitin chains appended onto multiple outer membrane proteins by Parkin is a prerequisite for mitophagy,21,58–61 as deubiquitination of mitophagy substrates via deubiquitinating enzymes (DUBs) can suppress Parkin-mediated mitophagy.64–67 However, the available information on the nature of polyubiquitin chains and their functions in the initiation and completion of mitophagy is puzzling. For instance, it has been reported that Parkin appends typical Lys48-linked polyubiquitin chains onto outer mitochondrial membrane proteins (e.g. Mfn1 and Mfn2) in response to the treatment of mitochondrial depolarizing agent CCCP.21 Canonically, Lys48 polyubiquitin chains consisting of a minimum of four ubiquitin moieties target proteins for degradation via the proteasome.48 Concurrently, outer mitochondrial proteins are degraded via the proteasome in response to CCCP treatment.1,21 These observations thus led to the conclusion that degradation of mitochondrial outer membrane proteins on depolarizing mitochondria is a prerequisite for mitophagy.1,21 On the other hand, it has also been observed that Mfn2 is not degraded but instead is deubiquitinated (evident by the disappearance of the Mfn2-modified bands on a Western blot) in response to CCCP treatment in conjunction with proteasome inhibitors.22 In another study, Parkin mediates the robust ubiquitination of Mfn1, but not Mfn2, when Parkin is overexpressed in SH-SY5Y cells treated with a proteasome inhibitor.43 Thus, whether outer mitochondrial proteins are indeed degraded via the proteasome before initiation of mitophagy is not clear. It is possible that the observed differences in the Parkin-mediated ubiquitination and degradation of mitochondrial substrates in distinct studies are simply due to variations in drug concentration, treatment time, cell lineages (cancer vs. normal), or the experimental design used in these studies. For example, in some cases, investigators have used a Parkin/PINK1-overexpression approach to investigate the “normal” mechanism of mitophagy in cancer cells. Certainly, utilizing protein overexpression to study cellular functions is feasible, but it has disadvantages. The abnormal expression of a protein can sometimes have a detrimental effect on cellular functions. Overexpression of protein-complex members could cause abnormal activation of a pathway leading to the excessive degradation or inhibition of degradation of intended substrates.68 Notably, endogenous expression of Parkin is moderate (compared to Parkin-overexpressed cells) in primary fibroblasts and induced pluripotent stem-derived neurons.69 It is possible that Parkin does not induce (or minimally induces) the degradation of outer mitochondrial proteins when mitochondria are depolarized. Robust degradation of outer mitochondrial membrane proteins seen in a non-physiological overexpression approach may be due to overactivation of the Parkin/PINK1 pathway in cells. A well-controlled study using proteasome inhibitors with/without depolarizing agents in normal cells may reveal whether Parkin indeed targets mitochondrial proteins on depolarized mitochondria for degradation before mitophagy.
In addition to Lys48 polyubiquitin chains, Parkin also appends atypical polyubiquitin chains that are linked through Lys6321,70 to several outer mitochondrial proteins in cells treated with CCCP in the absence of any mitochondrial stress.43 In a distinct study, Parkin predominantly assembles Lys6 and Lys11 instead of Lys48 and Lys63 polyubiquitin chains in vitro and on depolarized mitochondria.67 More recently, it has been demonstrated that Parkin assembles and USP30 (deubiquitinase) disassembles Lys6-linked polyubiquitin chains, and distally phosphorylated ubiquitin chains impair USP30 activity.71 The overall abundance of Lys6 linkages does not increase with proteasome inhibition suggesting that Lys6 chains on mitochondrial proteins are not for their degradation via the proteasome.72 In general, why Parkin conjugates different types of polyubiquitin chains onto mitochondrial proteins before mitophagy is an intriguing question. Notably, to test the types of Parkin-mediated ubiquitin-modification on outer mitochondrial substrates, some groups have used ubiquitin Lys mutants. However, as pointed out by Kulathu and Komander,49 Lys mutation affects ubiquitin surfaces and influences ubiquitin structure. For example, a mutation on Lys6 prevents assembly of heterotypic Lys11-Lys63 ubiquitin chains by Kaposi Sarcoma virus E3 ligase K5. Together, these results on Parkin-mediated mitophagy have raised some important questions: First, is ubiquitination of mitochondrial proteins indeed necessary for degradation via the proteasome? Second, what are the function of typical (Lys48) and atypical (Lys6, 11, 27, 63) polyubiquitin chains in mitophagy? Third, are polyubiquitin chains appended by Parkin homotypic (same ubiquitin linkages) or heterotypic (different ubiquitin linkages)? Fourth, does Parkin append multiple types of chains simultaneously or sequentially, and onto the same proteins? Finally, what are the cellular determinants that dictate the type of polyubiquitin chain that Parkin conjugates to its substrates? Answers to these questions will help clarify the role of the ubiquitin pathway in regulating normal mitophagy.
Another observation that warrants further exploration in the ubiquitin field is the novel, non-canonical finding that PINK1 phosphorylates ubiquitins on mitochondrial proteins of depolarized mitochondria. One schematic model in the literature shows that PINK1 phosphorylates free ubiquitins.57 Other models show that conjugated ubiquitins are phosphorylated by PINK1.73 Thus, it is not clear whether free ubiquitin, conjugated ubiquitin, or both are substrates of PINK1. Although cellular protein substrates are canonically phosphorylated prior to ubiquitination and degradation, ubiquitins conjugated to these proteins for degradation are not phosphorylated. Moreover, disassembly of ubiquitin chains by DUBs is a prerequisite for the proteasome-mediated degradation of proteins,74 and phospho-ubiquitin chains are resistant to DUBs.75 Therefore, whether these phospho-ubiquitin chains added onto mitochondrial proteins are for degradation, and whether these phospho-ubiquitin chains are disassembled before their degradation via the proteasome, is not clear. Notably, PGAM5, a mitochondrial phosphatase that associates with PINK1,76 may dephosphorylate phospho-ubiquitin before degradation. Evidence that PGAM5 knockout mice exhibit a Parkinson’s disease phenotype supports such a possibility.77 However, PINK1 is stabilized and PGAM5 is degraded when mitochondria are depolarized,78 thus ruling out the possibility of PGAM5 involvement in ubiquitin dephosphorylation on mitochondrial proteins before their degradation. Whether any other unknown specific cellular phosphatase dephosphorylates ubiquitins on substrates before degradation via the 26S proteasome is not known. It is possible that phospho-ubiquitin chains are not degraded, but phosphate groups are added onto ubiquitin chains to protect these substrates from proteasomal degradation, and that the purpose of these degradation-resistant phospho-ubiquitin signals on depolarized mitochondria is to target the damaged mitochondria to undergo autophagy. Indeed, Benischke et al.79 reported that Mfn2 is degraded and that inhibition of autophagy rescues this degradation. Furthermore, Richter et al.80 demonstrated that phospho-ubiquitin chains interact with phosphorylated OPTN to promote selective autophagy of damaged mitochondria.
Finally, although empirical evidence suggests that binding of ubiquitin to Parkin is necessary for its activation, Parkin also interacts with ubiquitin-like proteins (Ubl) SUMO1,81 NEDD8,82 and ISG15.83 SUMOylation increases Parkin’s nuclear localization and auto-ubiquitination,81 and NEDDylation and ISGylation increase Parkin’s E3 ligase activity.82,83 Notably, ISG15 is minimally expressed in normal cells.26 Therefore, whether ISG15 indeed activates Parkin (without its overexpression), and the precise mechanism by which ISG15 and other Ubls (SUMO1 and Nedd8) regulate Parkin activity for mitophagy in normal cells requires further experimentation.
Defects in mitophagy and ubiquitin pathways in neurodegenerative diseases
Defective mitophagy has been linked to several neurodegenerative diseases.84–86 Mutations in PINK1 and Parkin cause familial Parkinson's disease, and the impairment of mitochondrial motility and mitophagy has also been reported in sporadic Parkinson’s disease.87,88 In Huntington’s disease, defects in autophagic cargo recognition lead to accumulation of damaged mitochondria in the cytoplasm.89 Additionally, mutations in Mfn2 cause Charcot-Marie-Tooth type 2A neurodegenerative disease,90 and mutation of ataxin 3 (SCA3/MJD1) impairs Parkin ubiquitination and mitophagy in spinocerebellar ataxia (SCA) type 3 (Machado-Joseph disease).91 Aberrant accumulation of dysfunctional mitochondria due to inadequate mitophagic capacity has also been demonstrated in neurons and brains of patients with Alzheimer’s disease. Notably, accumulation of misfolded protein deposits in affected brain regions has been reported in Alzheimer's disease, Parkinson's disease, Creutzfeldt-Jakob, Huntington’s disease, A-T, and ALS.17 In most cases, proteinaceous deposits were composed of ubiquitin conjugates, suggesting a failure in their degradation via the ubiquitin/26S proteasome. Extensive evidence of mitochondrial dysfunction and defective mitophagy in ALS exists in the literature. For example, in mitochondrial-associated mutant SOD1 (G93A) mice, a mouse model most commonly used to study familial ALS, severe mitochondrial swelling is seen around postnatal day 30 (P30) with motor axon degeneration evidenced around P50, and clinical phenotypes at P60.92,93 Mutant SOD1 mice also exhibited impaired ATP production and an accumulation of damaged mitochondria, suggesting a failure of mitophagy.94 Large vacuoles were also seen via transmission electron microscopy analysis of motor neurons obtained from the spinal cords of mutant SOD1 mice. These vacuoles contained autophagy markers LC3 and beclin 1, further suggesting the presence of defective mitophagy.92 The link between defective mitophagy and ALS is further strengthened by localization of misfolded SOD1 proteins to motor neurons95 and mitochondrial fragmentation96 in vivo. Furthermore, transgenic mice harboring deletions of genes essential for autophagy (e.g. atg5 and atg7) exhibited neurodegenerative phenotypes as well as accumulation of polyubiquitinated proteins.97,98 Interestingly, the same mitochondrial defects were found in skeletal muscle of mutant SOD1 G93A mice,99 and these defects preceded disease onset, suggesting defective mitophagy to be critical in the progression of ALS.92 In ALS patients, mitochondrial damage is evidenced by cristae breakage, matrix dilution, and increases in mitochondrial mass and number.92,100 Importantly, this damage is explicitly seen in severely damaged and dying motor neurons, the cell type principally affected in ALS.92 Accumulation of autophagosomes and autophagolysosomes containing swollen and damaged mitochondria in spinal cords of ALS patients has also been reported,92,100 reflecting defects in mitophagic flux. These autophagosomes and autophagolysosomes have been shown to accumulate throughout the entire neuronal soma, subsequently initiating slow necrosis.100 It is worth noting that, as seen in mutant SOD1 mice, these vesicles also stained positive for autophagic markers, further corroborating the incidence of defective mitophagy in ALS. Two independent groups using human and mouse models have also demonstrated that mitophagy is defective in A-T, a neurodegenerative disease in children similar in pathology to ALS.101,102 Results from our group using human fibroblasts obtained from distinct A-T patients corroborate these results (Desai Lab, unpublished results). Mitochondrial damage manifests in A-T as swollen mitochondria, compromised respiration/ATP synthesis, and increases in mitochondrial mass and number. Interestingly, basal autophagy is increased but mitophagy is decreased in A-T cells.102,103 Together, these results suggest that mitophagy defects are common in neurodegenerative diseases. Unlike Parkinson’s, Charcot-Marie-Tooth type 2A, Machado-Joseph, and Alzheimer’s diseases where causes of defective mitophagy are known (mutations in Parkin/PINK1, Mfn2, ataxin 3, and depletion of Parkin, respectively), the causes underlying defective mitophagy in A-T and ALS are largely unknown. Knowing that the protein and mitochondrial turnover pathways are defective in A-T and ALS and that mitophagy requires a functional ubiquitin pathway, it is plausible that inhibition of the ubiquitin pathway may lead to defective mitophagy in these diseases. Recent studies from our group on ISG15 (Interferon-Stimulated Gene 15), a cellular antagonist of the ubiquitin pathway, support the idea that mitophagy may be defective due to ISG15-mediated impairment of the ubiquitin pathway in A-T and ALS (Desai Lab, unpublished data).
ISG15, a potential modulator of mitophagy in neurodegenerative diseases
ISG15 is an interferon-inducible ubiquitin-like (Ubl) protein. ISG15 is synthesized from the ISG15 gene, and either remains in an intracellular free form, appended to proteins in cells (conjugated form), or secreted from cells (extracellular form) by an unknown mechanism.104–106 ISG15 conjugates to intracellular targets through a multi-enzyme pathway parallel to that of ubiquitin and other Ubl proteins (reviewed in literature104,107–109). ISG15-specific enzymes E1 (UbE1L), E2 (UbcH8), and E3 (HERC5, EFP, and several others) are also IFN-stimulated proteins that conjugate intracellular free ISG15 to cellular proteins, a mechanism referred to as ISGylation.108,110–112 The ISG15 pathway (free ISG15 and its conjugates) is minimally expressed in normal cells. 24,106,113–115 However, the ISG15 pathway is elevated in pathogen-infected cells,116–118 malignancies,25,26 A-T,26 and ALS93 cells. Notably, although ISG15 is a Ubl protein, unlike ubiquitin, which targets proteins for degradation, ISG15 inhibits polyubiquitylation and consequently the ubiquitin-mediated degradation of proteins in cancer25,26 and A-T27 cells. These findings led to our hypothesis that constitutively elevated ISG15 may inhibit ubiquitin-conjugating enzymes (E2 and E3s) and thus polyubiquitylation (a signal for proteasome-mediated degradation) in cells. Literature reports demonstrating decreased polyubiquitylation of cellular proteins in cells expressing ISG1524,26 and ISG15 inhibition of ubiquitin ligase activity119,120 support this hypothesis. Notably, recent studies have demonstrated that ISG15 is conjugated to an artificial proteasome substrate and that endogenous substrates are conjugated to ubiquitin-ISG15 mixed chains in cells.121 Authors have provided empirical evidence that these ubiquitin-ISG15 chains do not serve as a signal for protein degradation. Together, these findings suggest that ISG15 may potentially inhibit the ubiquitin pathway by inhibiting the function of ubiquitin-conjugating enzymes, thus decreasing polyubiquitylation by directly conjugating to cellular substrates and/or by modifying the degradation signal (ubiquitylation) for protein degradation via the 26S proteasome.
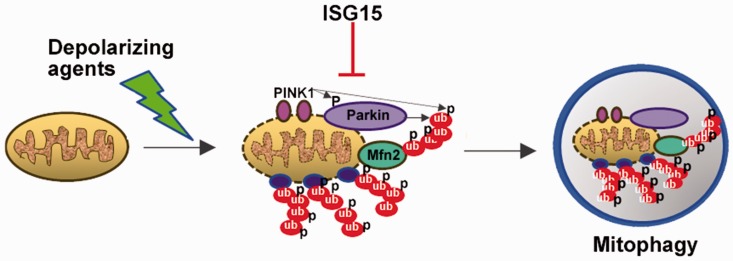
ISG15 is elevated in human A-T cells and mice.26,122 Similarly, interferon signaling is also elevated in mutant SOD1 mice,93,123 and specifically upregulated in their spinal cords approximately 30 days before the presentation of clinical phenotypes. Phosphorylated transcription factors STAT1 and STAT2, downstream targets in Type I Interferon signaling, were also found in the spinal cords of the same transgenic mice,93 further reinforcing the elevation of interferon-stimulated genes in ALS pathology. Wang et al.123 have demonstrated that elevation of ISG15 occurs in inflammatory neuronal injury (e.g. traumatic brain injury). Additionally, ISG15 levels have been shown to be elevated precisely where motor neurons are damaged (in the ventral horn of the spinal cord, where motor neurons reside) suggesting the possibility of ISG15 as a biomarker for neuronal injury and consequently ALS pathology.123 ISG15 was also shown to colocalize with GFAP, an astrocyte marker, suggesting a dialogue between neurons and astrocytes in interferon-stimulated disease processes.93,123 More recent studies from our group have revealed that ISG15 is also constitutively elevated in Parkinson’s and Alzheimer’s cells (Desai Lab, unpublished data). Because the ubiquitin pathway regulates mitophagy, and ISG15 inhibits the ubiquitin pathway, we propose that the constitutively elevated ISG15 pathway inhibits the ubiquitin pathway (polyubiquitination25–27) and consequently ubiquitin-dependent mitophagy, which contributes to neurodegeneration in neurodegenerative diseases (see schematic Figure 1). Recent studies demonstrating the presence of mixed ubiquitin-ISG15 chains on cellular proteins, subsequently inhibiting their degradation via the proteasome, support this model.121 Further studies are needed to confirm this hypothesis, however.
Figure 1.
A proposed model for defective mitophagy in neurodegenerative diseases. Schematic shows that the constitutively elevated ISG15 pathway may inhibit the ubiquitin pathway (polyubiquitination) and consequently ubiquitin-dependent mitophagy in neurodegenerative diseases.
Summary
Neurodegeneration affects millions of people worldwide, especially in the aging population, and the number of affected individuals over the age of 60 is projected to double by 2050. The estimated economic burden in the United States is $1.5 trillion per year, highlighting the necessity for understanding the common mechanisms that underly neurodegeneration. Defects in mitophagy and the ubiquitin pathway have been identified as leading causes of neurodegeneration in several neurodegenerative diseases, rendering pharmacological modulation of the mitophagy and ubiquitin pathways a promising future therapeutic strategy. However, additional understanding of the cellular processes highlighted in this mini-review is required before this becomes a possibility.
Authors’ contributions
SD: Wrote the review.
MJ: Assisted SD to write and proofread the review.
CK: Assisted SD to write the review.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
A-T Children’s Project Foundation, NIH/NINDS R21NS060960, and Consano Crowd Funding grants support this work.
References
- 1.Chan NC, Chan DC. Parkin uses the UPS to ship off dysfunctional mitochondria. Autophagy 2011; 7:771–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci 2012; 125:795–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubli DA, Gustafsson AB. Mitochondria and mitophagy: the yin and yang of cell death control. Circ Res 2012; 111:1208–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxid Redox Signal 2012; 17:794–802 [DOI] [PubMed] [Google Scholar]
- 5.Rub C, Wilkening A, Voos W. Mitochondrial quality control by the Pink1/Parkin system. Cell Tissue Res 2017; 367:111–23 [DOI] [PubMed] [Google Scholar]
- 6.Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomem 2009; 41:499–503 [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A. Early work on the ubiquitin proteasome system, an interview with Aaron Ciechanover. Interview by CDD. Cell Death Differ 2005; 12:1167–77 [DOI] [PubMed] [Google Scholar]
- 8.Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J 1997; 11:1257–68 [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol 1995; 7:215–23 [DOI] [PubMed] [Google Scholar]
- 10.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet 1996; 30:405–39 [DOI] [PubMed] [Google Scholar]
- 11.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70:503–33 [DOI] [PubMed] [Google Scholar]
- 12.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer 2006; 6:776–88 [DOI] [PubMed] [Google Scholar]
- 13.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol 2006; 17:1807–19 [DOI] [PubMed] [Google Scholar]
- 14.Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol 2005; 23:4776–89 [DOI] [PubMed] [Google Scholar]
- 15.Cenini G, Voos W. Role of mitochondrial protein quality control in oxidative stress-induced neurodegenerative diseases. Curr Alzheimer Res 2016; 13:164–73 [DOI] [PubMed] [Google Scholar]
- 16.Dupuis L. Mitochondrial quality control in neurodegenerative diseases. Biochimie 2014; 100:177–83 [DOI] [PubMed] [Google Scholar]
- 17.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis 2010; 15:1292–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther 2012; 342:619–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006; 443:787–95 [DOI] [PubMed] [Google Scholar]
- 20.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med 2004; 10:S10. [DOI] [PubMed] [Google Scholar]
- 21.Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S, Chan DC. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet 2011; 20:1726–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS One 2011; 6:e16746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross JM, Olson L, Coppotelli G. Mitochondrial and ubiquitin proteasome system dysfunction in ageing and disease: two sides of the same coin? Int J Mol Sci 2015; 16:19458–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai SD, Haas AL, Wood LM, Tsai YC, Pestka S, Rubin EH, Saleem A, Nur-E-Kamal A, Liu LF. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res 2006; 66:921–8 [DOI] [PubMed] [Google Scholar]
- 25.Desai SD, Wood LM, Tsai YC, Hsieh TS, Marks JS, Scott GL, Giovanella BC, Liu LF. ISG15 as a novel tumor biomarker for drug sensitivity. Mol Cancer Ther 2008; 7:1430–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood LM, Sankar S, Reed RE, Haas AL, Liu LF. A novel role for ATM in regulating proteasome-mediated protein degradation through suppression of the ISG15 conjugation pathway. PLoS One 2011; 6:e16422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JR, Nunnari J. Mitochondrial form and function. Nature 2014; 505:335–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol 2006; 16:R551–60 [DOI] [PubMed] [Google Scholar]
- 29.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab 2012; 26:711–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraste M. Oxidative phosphorylation at the fin de siecle. Science 1999; 283:1488–93 [DOI] [PubMed] [Google Scholar]
- 31.Fosslien E. Mitochondrial medicine–molecular pathology of defective oxidative phosphorylation. Ann Clin Lab Sci 2001; 31:25–67 [PubMed] [Google Scholar]
- 32.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer 2014; 14:709–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012; 48:158–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao L, Laude K, Cai H. Mitochondrial pathophysiology, reactive oxygen species, and cardiovascular diseases. Vet Clin North Am Small Anim Pract 2008; 38:137–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkinezos IG, Moraes CT. Reactive oxygen species and mitochondrial diseases. Semin Cell Dev Biol 2001; 12:449–57 [DOI] [PubMed] [Google Scholar]
- 36.Rego AC, Oliveira CR. Mitochondrial dysfunction and reactive oxygen species in excitotoxicity and apoptosis: implications for the pathogenesis of neurodegenerative diseases. Neurochem Res 2003; 28:1563–74 [DOI] [PubMed] [Google Scholar]
- 37.Ni HM, Williams JA, Ding WX. Mitochondrial dynamics and mitochondrial quality control. Redox Biol 2015; 4:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker MJ, Tatsuta T, Langer T. Quality control of mitochondrial proteostasis. Cold Spring Harb Perspect Biol 2011; 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 2008; 27:433–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soubannier V, Rippstein P, Kaufman BA, Shoubridge EA, McBride HM. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One 2012; 7:e52830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem 2012; 287:42379–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev 2015; 29:989–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem 2011; 118:636–45 [DOI] [PubMed] [Google Scholar]
- 44.Nguyen TN, Padman BS, Lazarou M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol 2016; 26:733–44 [DOI] [PubMed] [Google Scholar]
- 45.Haas AL. Structural insights into early events in the conjugation of ubiquitin and ubiquitin-like proteins. Mol Cell 2007; 27:174–5 [DOI] [PubMed] [Google Scholar]
- 46.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci 2008; 65:2397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci 2016; 129:875–80 [DOI] [PubMed] [Google Scholar]
- 48.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, Murata S, Hiroaki H, Sato M, Tanaka K, Shirakawa M. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells 2004; 9:865–75 [DOI] [PubMed] [Google Scholar]
- 49.Kulathu Y, Komander D. Atypical ubiquitylation – the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol 2012; 13:508–23 [DOI] [PubMed] [Google Scholar]
- 50.Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev 2001; 21:245–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohtake F, Tsuchiya H. The emerging complexity of ubiquitin architecture. J Biochem 2017; 161:125–33 [DOI] [PubMed] [Google Scholar]
- 52.Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012; 64:136–42 [DOI] [PubMed] [Google Scholar]
- 53.Saeki Y. Ubiquitin recognition by the proteasome. J Biochem 2017; 161:113–24 [DOI] [PubMed] [Google Scholar]
- 54.Tanak A. Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett 2010; 584:1386–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS, Fon EA. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep 2012; 13:378–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 2010; 191:933–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kazlauskaite A, Martínez-Torres RJ, Wilkie S, Kumar A, Peltier J, Gonzalez A, Johnson C, Zhang J, Hope AG, Peggie M, Trost M, van Aalten DM, Alessi DR, Prescott AR, Knebel A, Walden H, Muqit MM. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep 2015; 16:939–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol 2014; 205:143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 2014; 510:162–6 [DOI] [PubMed] [Google Scholar]
- 60.Okatsu K, Koyano F, Kimura M, Kosako H, Saeki Y, Tanaka K, Matsuda N. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J Cell Biol 2015; 209:111–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiba-Fukushima K, Arano T, Matsumoto G, Inoshita T, Yoshida S, Ishihama Y, Ryu KY, Nukina N, Hattori N, Imai Y. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet 2014; 10:e1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015; 524:309–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swatek KN, Komander D. Ubiquitin modifications. Cell Res 2016; 26:399–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014; 510:370–5 [DOI] [PubMed] [Google Scholar]
- 65.Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, D, Strooper B, Verstreken P, Vandenberghe W. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet 2014; 23:5227–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durcan TM, Fon EA. USP8 and PARK2/parkin-mediated mitophagy. Autophagy 2015; 11:428–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durcan TM, Tang MY, Pérusse JR, Dashti EA, Aguileta MA, McLelland GL, Gros P, Shaler TA, Faubert D, Coulombe B, Fon EA. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J 2014; 33:2473–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moriya H. Quantitative nature of overexpression experiments. Mol Biol Cell 2015; 26:3932–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rakovic A, Shurkewitsch K, Seibler P, Grünewald A, Zanon A, Hagenah J, Krainc D, Klein C. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem 2013; 288:2223–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol 2012; 4:pii a011338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gersch M, Gladkova C, Schubert AF, Michel MA, Maslen S, Komander D. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat Struct Mol Biol 2017; 11:920–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011; 44:325–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsuda N, Tanaka K. The PARK2/Parkin receptor on damaged mitochondria revisited-uncovering the role of phosphorylated ubiquitin chains. Autophagy 2015; 11:1700–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics 2011; 10:R110 003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J 2015; 34:307–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, Takeda K, Ichijo H, Lu B, Takahashi R. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet 2010; 6:e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, Zhang Y, Dawson VL, Dawson TM, Lenardo M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson's-like movement disorder. Nat Commun 2014; 5:4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sekine S, Kanamaru Y, Koike M, Nishihara A, Okada M, Kinoshita H, Kamiyama M, Maruyama J, Uchiyama Y, Ishihara N, Takeda K, Ichijo H. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J Biol Chem 2012; 287:34635–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benischke AS, Vasanth S, Miyai T, Katikireddy KR, White T, Chen Y, Halilovic A, Price M, Price F, Jr, Liton PB, Jurkunas UV. Activation of mitophagy leads to decline in Mfn2 and loss of mitochondrial mass in Fuchs endothelial corneal dystrophy. Sci Rep 2017; 7:6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA, Youle RJ, Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 2016; 113:4039–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J Neurosci Res 2006; 84:1543–54 [DOI] [PubMed] [Google Scholar]
- 82.Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J Neurosci Res 2012; 90:1030–42 [DOI] [PubMed] [Google Scholar]
- 83.Im E, Yoo L, Hyun M, Shin WH, Chung KC. Covalent ISG15 conjugation positively regulates the ubiquitin E3 ligase activity of parkin. Open Biol 2016; 6:pii160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez-Vicente M. Neuronal mitophagy in neurodegenerative diseases. Front Mol Neurosci 2017; 10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Palikaras K, Tavernarakis N. Mitophagy in neurodegeneration and aging. Front Genet. 2012; 3:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whitworth AJ, Pallanck LJ. PINK1/Parkin mitophagy and neurodegeneration-what do we really know in vivo? Curr Opin Genet Dev 2017; 44:47–53 [DOI] [PubMed] [Google Scholar]
- 87.Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem Sci 2015; 40:200–10 [DOI] [PubMed] [Google Scholar]
- 88.Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson's disease: the PINK1-parkin link. Biochim Biophys Acta 2011; 1813:623–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci 2010; 13:567–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi BO, Nakhro K, Park HJ, Hyun YS, Lee JH, Kanwal S, Jung SC, Chung KW. A cohort study of MFN2 mutations and phenotypic spectrums in Charcot-Marie-Tooth disease 2A patients. Clin Genet 2015; 87:594–8 [DOI] [PubMed] [Google Scholar]
- 91.Durcan TM, Kontogiannea M, Thorarinsdottir T, Fallon L, Williams AJ, Djarmati A, Fantaneanu T, Paulson HL, Fon EA. The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum Mol Genet 2011; 20:141–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Natale G, Lenzi P, Lazzeri G, Falleni A, Biagioni F, Ryskalin L, Fornai F. Compartment-dependent mitochondrial alterations in experimental ALS, the effects of mitophagy and mitochondriogenesis. Front Cell Neurosci 2015; 9:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang R, Yang B, Zhang D. Activation of interferon signaling pathways in spinal cord astrocytes from an ALS mouse model. Glia 2011; 59:946–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palomo GM, Manfredi G. Exploring new pathways of neurodegeneration in ALS: the role of mitochondria quality control. Brain Res 2015; 1607:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pickles S, Destroismaisons L, Peyrard SL, Cadot S, Rouleau GA, Brown RH, Jr, Julien JP, Arbour N, Vande Velde C. Mitochondrial damage revealed by immunoselection for ALS-linked misfolded SOD1. Hum Mol Genet 2013; 22:3947–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang Z, Wang W, Perry G, Zhu X, Wang X. Mitochondrial dynamic abnormalities in amyotrophic lateral sclerosis. Transl Neurodegener 2015; 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441:880–4 [DOI] [PubMed] [Google Scholar]
- 98.Mendonca DM, Chimelli L, Martinez AM. Expression of ubiquitin and proteasome in motorneurons and astrocytes of spinal cords from patients with amyotrophic lateral sclerosis. Neurosci Lett 2006; 404:315–9 [DOI] [PubMed] [Google Scholar]
- 99.Luo G, Yi J, Ma C, Xiao Y, Yi F, Yu T, Zhou J. Defective mitochondrial dynamics is an early event in skeletal muscle of an amyotrophic lateral sclerosis mouse model. PLoS One 2013; 8:e82112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ruffoli R, Bartalucci A, Frati A, Fornai F. Ultrastructural studies of ALS mitochondria connect altered function and permeability with defects of mitophagy and mitochondriogenesis. Front Cell Neurosci 2015; 9:341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ambrose M, Goldstine JV, Gatti RA. Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum Mol Genet 2007; 16:2154–64 [DOI] [PubMed] [Google Scholar]
- 102.Valentin-Vega YA, Maclean KH, Tait-Mulder J, Milasta S, Steeves M, Dorsey FC, Cleveland JL, Green DR, Kastan MB. Mitochondrial dysfunction in ataxia-telangiectasia. Blood 2012; 119:1490–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Desai SD, Reed RE, Babu S, Lorio EA. ISG15 Deregulates autophagy in genotoxin-treated ataxia telangiectasia cells. J Biol Chem 2013; 288:2388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bogunovic D, Boisson-Dupuis S, Casanova JL. ISG15: leading a double life as a secreted molecule. Exp Mol Med 2013; 45:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.D'Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E, Jr, Borden EC. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J Immunol 1996; 157:4100–8 [PubMed] [Google Scholar]
- 106.Desai SD. ISG15: a double edged sword in cancer. Oncoimmunology 2015; 4:e1052935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dao CT, Zhang DE. ISG15: a ubiquitin-like enigma. Front Biosci 2005; 10:2701–22 [DOI] [PubMed] [Google Scholar]
- 108.Haas AL. ISG15-dependent Regulation In: Mayer RJ, Ciechnover A, Rechsteiner M. (eds) Protein degradation. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co, 2006, pp.103–31 [Google Scholar]
- 109.Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol 2005; 25:6338–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burks J, Reed RE, Desai SD. ISGylation governs the oncogenic function of Ki-Ras in breast cancer. Oncogene 2014; 33:794–803 [DOI] [PubMed] [Google Scholar]
- 111.Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem 2006; 281:4334–8 [DOI] [PubMed] [Google Scholar]
- 112.Narasimhan J, Potter JL, Haas AL. Conjugation of the 15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J Biol Chem 1996; 271:324–30 [DOI] [PubMed] [Google Scholar]
- 113.Bektas N, Noetzel E, Veeck J, Press MF, Kristiansen G, Naami A, Hartmann A, Dimmler A, Beckmann MW, Knüchel R, Fasching PA, Dahl E. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res 2008; 10:R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen RH, Du Y, Han P, Wang HB, Liang FY, Feng GK, Zhou AJ, Cai MY, Zhong Q, Zeng MS, Huang XM. ISG15 predicts poor prognosis and promotes cancer stem cell phenotype in nasopharyngeal carcinoma. Oncotarget 2016; 7:16910–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuo C, Sheng X, Ma M, Xia M, Ouyang L. ISG15 in the tumorigenesis and treatment of cancer: an emerging role in malignancies of the digestive system. Oncotarget 2016; 7:74393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, Mansouri N, Okada S, Bryant VL, Kong XF, Kreins A, Velez MM, Boisson B, Khalilzadeh S, Ozcelik U, Darazam IA, Schoggins JW, Rice CM, Al-Muhsen S, Behr M, Vogt G, Puel A, Bustamante J, Gros P, Huibregtse JM, Abel L, Boisson-Dupuis S, Casanova JL. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 2012; 337:1684–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dai J, Pan W, Wang P. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J 2011; 8:468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lenschow DJ. Antiviral Properties of ISG15. Viruses 2010; 2:2154–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem 2008; 283:8783–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takeuchi T, Yokosawa H. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun 2005; 336:9–13 [DOI] [PubMed] [Google Scholar]
- 121.Fan JB, Arimoto K, Motamedchaboki K, Yan M, Wolf DA, Zhang DE. Identification and characterization of a novel ISG15-ubiquitin mixed chain and its role in regulating protein homeostasis. Sci Rep 2015; 5:12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim CD, Reed RE, Juncker MA, Fang Z, Desai SD. Evidence for the deregulation of protein turnover pathways in Atm-deficient mouse cerebellum: an organotypic study. J Neuropathol Exp Neurol 2017; 76:578–84 [DOI] [PubMed] [Google Scholar]
- 123.Wang RG, Kaul M, Zhang DX. Interferon-stimulated gene 15 as a general marker for acute and chronic neuronal injuries. Sheng Li Xue Bao 2012; 64:577–83 [PMC free article] [PubMed] [Google Scholar]