Abstract
Bromodomain and extra-terminal (BET) family proteins are key regulators of gene expression in cancer. Herein, we utilize BRD4 profiling to identify critical pathways involved in pathogenesis of chronic lymphocytic leukemia (CLL). BRD4 is over-expressed in CLL and is enriched proximal to genes up-regulated or de novo expressed in CLL with known function in disease pathogenesis and progression. These genes, including key members of the BCR signaling pathway, provide rationale for this therapeutic approach to identify new targets in alternative types of cancer. Additionally, we describe PLX51107, a structurally distinct BET inhibitor with novel in vitro and in vivo pharmacologic properties that emulates or exceeds the efficacy of BCR signaling agents in pre-clinical models of CLL. Herein, the discovery of the involvement of BRD4 in the core CLL transcriptional program provides a compelling rationale for clinical investigation of PLX51107 as epigenetic therapy in CLL and application of BRD4 profiling in other cancers.
Keywords: Chronic lymphocytic leukemia (CLL), Bromo and extra terminal domain family proteins (BRD and BET), BET inhibitors (e.g. PLX51107), Epigenetic modulation, Oncogenic Drivers
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is characterized by increased B-cell receptor (BCR) signaling, defective apoptosis, and disrupted immune effector function (1–3). Ibrutinib, a BCR pathway inhibitor that targets the Bruton’s tyrosine kinase (BTK), interferes with BCR signaling to impair microenvironment-derived survival signals and reverse disease-mediated immune suppression (4,5). In CLL patients, ibrutinib demonstrates dramatic and protracted efficacy with a tolerable toxicity profile (5–8) and thus has transformed the therapeutic landscape of this disease. The limitations of ibrutinib include a low frequency of complete response and frequent relapse in patients with high risk disease including those with del(17)(p13.1) (9). Relapse often results from BTK mutations that preclude ibrutinib binding and allow re-activation of BCR signaling. This underscores the importance of BCR signaling and attractiveness of novel therapies that can simultaneously target both BCR signaling and multiple other genes involved in CLL pathogenesis. Additionally, 5 to 10 percent of patients with CLL transform to a rapidly growing aggressive lymphoma, a syndrome known as Richter’s Transformation (RT), which is resistant to aggressive immuno-chemotherapy and associated with a short overall survival (~6 months). RT is the most common type of progression now observed in CLL patients responding to targeted therapies such as ibrutinib and venetoclax (10,11).
To date the molecular mechanisms driving the establishment and progression of CLL remain to be fully elucidated. Aberrant expression of anti-apoptotic (e.g. BCL2, TRAIL, and MCL1) (12–14), pro-survival (e.g. IL4R, IL21R) (15,16), and cell-trafficking proteins (e.g. CCR7, and CXCR4) (17,18) has been associated with aggressive disease behavior. Additionally, gene expression profiling and pathway analysis has established BCR signaling as a central driver of CLL (19). Furthermore, next-generation sequencing efforts have identified a relatively small number of recurrently mutated genes of potential pathogenetic relevance that affect cellular functions including DNA damage and cell cycle control (e.g. TP53 and ATM), Notch signaling, mRNA processing (e.g. SF3B1 and XPO1), B-cell transcription (e.g. IKZF3), and chromatin remodeling (e.g. ARID1A and CHD2) (20). Despite the relatively low mutational load compared to other tumors, CLL presents with substantial clinical and biological heterogeneity. In addition, some patients develop a more aggressive disease course despite the absence of high-risk genetic alterations, suggesting that factors other than genetic lesions could play a crucial role in CLL pathogenesis and outcome. For example, epigenetic mechanisms such as aberrant promoter methylation that lead to abnormal expression of TCL1A (21), ZAP70 (22), microRNAs (miR155 and miR21) (23), BCL2 (13) and long noncoding RNAs are implicated in CLL pathogenesis. Therefore, a deeper understanding of the epigenetic mechanisms controlling the aberrant expression of these pathogenic factors could provide a novel axis for targeted therapies to reverse the transcriptional abnormalities seen in CLL. Recent DNA methylation and ATAC-seq studies in CLL revealed that methylation profiles and genome-wide chromatin accessibility can more precisely differentiate CLL subtypes and predict clinical course compared to gene expression profiles (24). Hence, the biological differences between the major subtypes of CLL are imprinted in the epigenome.
The BET subfamily of proteins (BRD2, BRD3, BRD4 and BRDT) are transcriptional regulators that bind to acetylated histones and recruit the positive transcription elongation factor complex (P-TEFb) and RNA Pol II (25,26) to control gene expression at those regions (27,28). BRD4 associates with all active promoters and a significant fraction of active enhancers in both normal and transformed cell types (29–31). Despite its role as a general transcriptional regulator, inhibition of BRD4 has shown selective anti-cancer activity in a variety of pre-clinical models. This therapeutic window versus normal tissues has been attributed to the high BRD4 load at super-enhancer (SE) regions regulating cancer-specific genes (i.e. MYC, and BCL6), making the expression of those genes exquisitely sensitive to pharmacological targeting of BET proteins. While the pro-tumor role of BET proteins has been shown in many highly proliferative cancers, predominately using cell lines, no studies have examined the involvement of these epigenetic modifiers in a slowly proliferating disease like CLL. CLL therefore represents an important disease to interrogate the potential role of BET proteins in tumorigenesis and the relevance of therapeutic targeting. Furthermore, if pathways such as BCR signaling, which are validated as therapeutically relevant in CLL, were shown to be highly transcriptionally enhanced by BRD4, it would provide rationale for utilizing BRD4 profiling to explore for relevant therapeutic targets in other cancers.
A number of first generation small-molecule BET inhibitors sharing the same benzodiazepine scaffold (i.e. JQ1, iBET762, and OTX015) have demonstrated anti-tumor activity in different preclinical models of solid and hematologic malignancies by dissociating BET proteins from SE regions with consequent suppression of cMYC and NFκB activity (32–37). The most mature data is derived with OTX015 in acute leukemia (38) or in lymphoma and multiple myeloma (39) (ClinicalTrials.gov, NCT01713582). In the lymphoma study, hematologic dose-limiting toxicities (DLTs) at pharmacologically effective doses necessitated intermittent dosing. Even with intermittent dosing, there were on-target DLTs in addition to non-DLT gastrointestinal toxicities that significantly hampered study adherence. The few clinical responses were observed only at dose levels or schedules in which DLTs occurred. These results suggest that while OTX015 has clinical activity in lymphoid malignancies, its therapeutic index is narrow. Following on the exceptional therapeutic index of ibrutinib in B-cell malignancies, there is a need to develop BET inhibitors with a broader therapeutic index to improve safety while maintaining efficacy.
Given the promise of BET targeting in rapidly proliferative tumors, we applied an integrative functional epigenomic approach to examine the genome-wide chromatin distribution of BRD4 in slowly-proliferating primary CLL tumor cells. We demonstrate over-expression of BRD4 and characterize its asymmetric binding to a small set of enhancer regions (SEs) that control nearby genes implicated in CLL pathogenesis and disease progression (i.e. BCR signaling-associated genes, MIR155, MIR21, and IKZF1). To develop BET inhibitors with an improved therapeutic index and pharmaceutical profile over current clinical compounds, we applied scaffold-based and crystallography-guided drug design (40). This effort yielded PLX51107, a potent and selective small molecule BET inhibitor with a different chemotype from the previously published benzodiazepine-based BET inhibitors. Notably, the BRD4-overloaded SE regions were subsequently shown to have exquisite sensitivity to PLX51107 compared to other enhancers, explaining the selective effect of BET inhibitors on CLL specific oncogenic pathways. Furthermore, PLX51107 demonstrated potent antitumor activity in pre-clinical models of CLL and RT with a short terminal half-life that differentiates it from other agents currently under clinical development.
RESULTS
BRD4 binds genome-wide to transcriptionally active genes in CLL
Studies exploring the role of BET proteins as transcriptional co-activators in slowly-proliferative cancers such as CLL have yet to be reported. Their contribution to relevant pathogenic driving pathways could provide an avenue to identify new therapies. BRD4 and other bromodomain-containing proteins such as BRD2, BRD3, p300 and CREBBP binding protein (CPB) are ubiquitously expressed in primary CLL cells (Fig. 1A and Supplementary Fig. S1A). Notably, BRD4 is significantly over-expressed in CLL cells when compared to normal B-cells (Supplementary Fig. S1B and S1C) while no statistically significant difference in BRD4 expression was observed between naïve and memory B cells (Supplementary Fig. S1D).
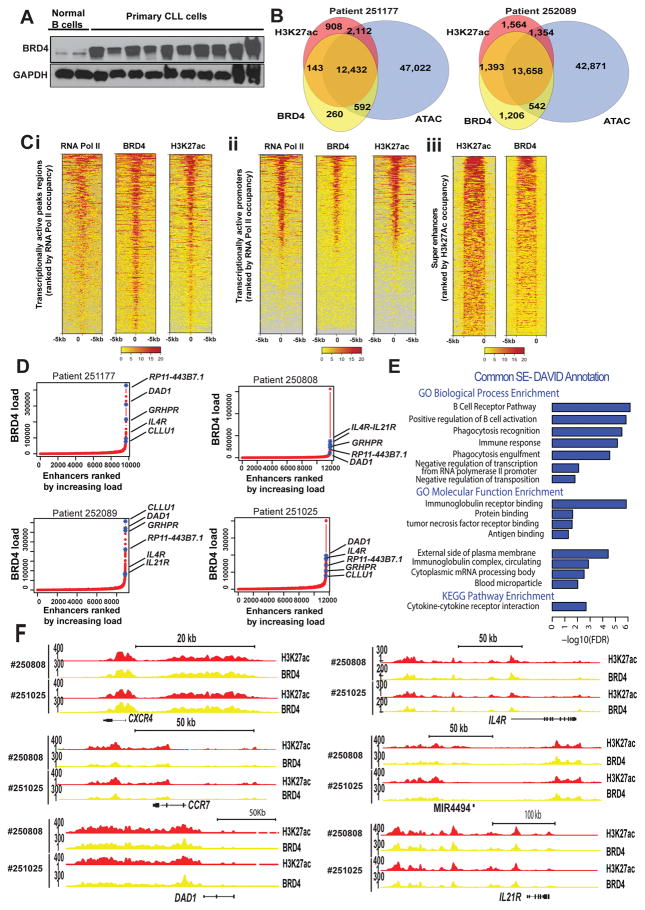
Figure 1. BRD4 expression and genomic distribution in B- CLL.
A. BRD4 protein expression level in B-cells isolated from normal donors or CLL patients; B. Representative Venn diagram of BRD4 and H3K27ac binding (ChIP-seq), and chromatin accessibility (ATAC-seq) for two CLL patient samples. Approximately 94% of BRD4 binding occurs within H3K27 acetylation regions, 81% of H3K27 acetylation regions contain BRD4, and 93% of H3K27 acetylation occurs within open chromatin regions; Ci–iii. Representative heatmap of ChIP-seq read densities for RNA Pol II (transcriptionally active), BRD4 and H3K27ac in patient-derived CLL cells ranked by decreasing combined occupancy signals centered ± 5 kb relative to transcription starting site (TSS) of transcriptionally active peak regions (Ci) and promoter regions (Cii); Heatmap showing ChIP-seq read densities (anchored 10 kb center region with 5 kb flanking on either side) across H3K27ac peaks located in SEs in CLL patient cells (Ciii); D. Hockey stick plot for BRD4 loading across enhancers of four CLL patient cells. SEs are defined as enhancers surpassing the inflection point. On average 3% of enhancers were identified as SEs and contain ~30% of all enhancer-bound BRD4 in CLL patient cells (a total of 3.2, 4, 1.7, and 3.2% of enhancers contain 29.2, 37.7, 23.2, and 28.1% of all enhancer-bound BRD4 in the CLL patient samples #251177, #252089, #250808, #251025 respectively. There are ~397 common annotated regions (coding and noncoding genes) contained within SE regions across all samples. Top BRD4 high loaded enhancers are indicated; E. DAVID database was used for the Gene Ontology (GO) functional annotation cluster analysis. The Fig. depicts the major biological and molecular pathways associated with BRD4 inhibition; F. ChIP-seq binding density for H3K27ac (red) and BRD4 (blue) at the enhancer of CXCR4, CCR7, DAD1, IL4R, MIR4494, and IL21R. The x-axis of the tracks shows genomic position, and the y-axis shows ChIP-seq signal (rpm/bp).
Cancer cell lines differ drastically in genomic and transcriptional profile from their originating primary tissue (41,42) making their relevance to understand the tumor epigenome limited. We used primary CLL B-cells derived from four patients (patient details summarized in Supplementary Table S1) to assess chromatin accessibility genome-wide, using ATAC-seq, and to identify regions in the epigenome that are accessible to transcription factor binding. In the same samples we probed for BRD4 expression at mRNA and protein level (Supplementary Fig. S1B and S1C) and the genome-wide localization of BRD4 and its abundance in enhancer areas using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) and examining both the histone H3K27ac chromatin mark and RNA Pol II as measures of downstream transcription. The genome-wide correlation between BRD4 occupancy and H3K27ac is strong, with the majority of H3K27ac occurring within open chromatin regions (Fig. 1B). In contrast, genes not expressed in CLL (e.g. EGFR, Myoglobin, etc.) are not identified in this analysis. Rank ordering of enhancer regions by H3K27ac enrichment reveals that about 94% of all chromatin-bound BRD4 resides in regions marked by H3K27ac, indicating the predominant recruitment of BRD4 at active enhancers in human CLL. Accordingly, BRD4 co-localizes with RNA Pol II at transcriptionally active peak regions, promoters and SEs across the CLL genome (Fig. 1C). For each patient sample, on average, 300 out of 10,500 enhancer regions (~3%) show a distinctly larger size and higher BRD4 load compared to typical enhancer and are henceforth defined as SEs based on previously published definition of these regions (30) These SEs account for ~30% of all enhancer bound BRD4 in CLL cells. Next using published data of BRD4 ChIP-seq in a non-tumor B cell line(43) we have analyzed the genome wide BRD4 load across enhancers in normal B cells and compared this to CLL cells under basal and CpG stimulated conditions. Overall in normal B cells BRD4 enhancer regions are only modestly loaded with BRD4 compared to CLL (Supplementary Fig. S1E, p=0 in all samples). As expected, a number of common SEs between CLL and normal B cells are observed, possibly defining typical B-cell lineage regions (Supplementary Fig. S1F). However, there are numerous of SEs that are unique to CLL (Supplementary Fig. S1G).
The topmost BRD4-enriched SEs in CLL, common among all patient samples, include IL4R, IL21R, CLLU1, GRHPR, and RP11-443B7.1, a novel noncoding gene located in chromosome 1 (Fig. 1D). In addition, common highly ranked BRD4-dense enhancers are near genes with established roles in CLL biology and tumor microenvironment interactions and reported aberrant expression in CLL (e.g. CXCR4, CCR7, IKZF3, IL4R, IL2R, and IL21R) or de-novo regulated in CLL (e.g. MIR21).
BRD4 binding profile of these regions confirms a preferential overload in CLL compared to normal B cells. Functional enrichment analysis (Fig. 1E and Supplementary Table S2) of genes nearby SEs common among all patient samples revealed significant enrichment in these regions for genes involved in the BCR signaling pathway (e.g. PTPN6, BLK, BCL2) and immune response (e.g. IL2RA, IL19, IL10, IL4R, IL24, TNF superfamily, CCR7, CXCR5, and B2M). As expected, large BRD4-enriched SEs in CLL and normal B cells were also found adjacent to the IGH loci similar to what has been described in multiple myeloma (35). Examples of select tracks for highly ranked BRD4-dense enhancers are illustrated in Fig. 1F and Supplementary Fig. S1I.
PLX51107 is as a novel BET inhibitor with attractive pharmaceutical properties
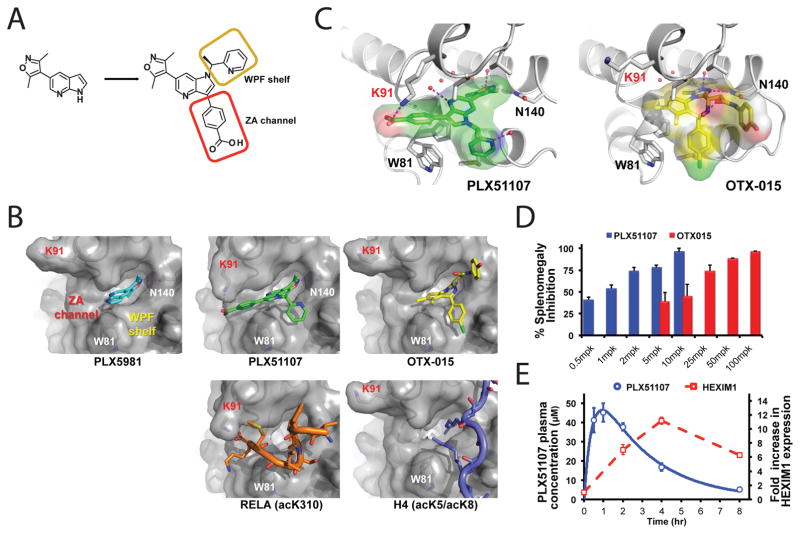
Scaffold screening using low-affinity binding assays followed by co-crystallography identified a 213 Da micromolar hit (PLX5981) consisting of 3,5-dimethylisoxazole coupled to 7-azaindole (Fig. 2A). The isoxazole moiety mimics the disposition of an N-ε-acetyl-lysine while the 7-azaindole presents a facile framework to access both the Trp81-Pro82-Phe83 (WPF) shelf and the ZA channel (a specificity loop defined by αZ and αA helices) in BRD4 (32). Structure-guided chemistry exploration, including the replacement of 7-azaindole by 4-azaindole, yielded PLX51107, a potent BET inhibitor (Fig. 2A) with optimized pharmacokinetic properties. PLX51107 interacts with the WPF-shelf and the ZA-channel with its pyridylethyl (R1) and benzoic acid (R3) side chains, respectively (Fig. 2B). In particular, the benzoic acid traverses the ZA-channel and forms a salt bridge with Lys91 revealing a level of structural plasticity in BRD4 that has not been observed with other BET inhibitors (Fig. 2B, 2C, and Supplementary Fig. S2). While the binding of known BET inhibitors (e.g. OTX015) is confined to the space occupied by a typical extended diacetylated histone peptide, PLX51107 represents a new class of inhibitors that engage additional binding elements in the ZA channel (Fig. 2B). The PLX51107-bound structure of BRD4 resembles the conformation induced by the binding of acetylated Lys310 peptide from RelA protein (Fig. 2B and Supplementary Table S3) (44).
Figure 2. Scaffold-based discovery and cellular profile of PLX51107 as a non-benzodiazepine BET inhibitor targeting the inducible ZA channel.
A. The chemical structures of PLX5981 (scaffold) and PLX51107; B. Co-structures of BRD4-BD1 with different inhibitors and substrate peptides. The 7-azaindole core of PLX5981 presents a facile framework to access both the WPF shelf and the ZA channel. PLX51107 was developed from the modified 4-azaindole scaffold. The benzoic acid of PLX51107 traverses the ZA channel and forms a salt bridge with Lys91. Similar induced-conformational change was observed in the co-structure of BRD4-BD1 with the acetylated RelA acK310 peptide (PDB: 4KV1). Known BET inhibitors including OTX015 bind in the same space as extended H4 histone substrate (PDB: 3UVW); C. Detailed interactions of BRD4-BD1 with PLX51107 and OTX015; D. In Ba/F3-induced mouse splenomegaly model, PLX51107 is 10 fold more potent than OTX015; E. Transcriptional changes (represented by HEXIM1) in MV4-11 tumor xenograft substantially out-live the plasma drug levels of PLX51107. A single dose of PLX51107 was administered. AUC0–24h = 90,100 ng•h/mL (or 205 μM•hr), half-life T1/2 = 2.8 h.
PLX51107 exhibits low nanomolar potency in blocking interactions mediated by the four BET family members. Binding affinity (Kd) measurements using isolated bromodomains (Supplementary Table S4) show that PLX51107 has a modest preference for bromodomain-1 (BD1) versus bromodomain-2 (BD2) within each BET protein (Kd = 1.6, 2.1, 1.7, and 5 nM for BD1 and 5.9, 6.2, 6.1 and 120 nM for BD2 of BRD2, BRD3, BRD4, and BRDT, respectively). Among non-BET proteins, PLX51107 shows significant interactions only with the bromodomains of CBP and EP300 (p300) (Kd in the 100 nM range). PLX51107 has a single chiral center and bears the (S)-enantiomer. The (R)-enantiomer is 5–10 fold less potent than PLX51107 across all BET bromodomains. Profiling PLX51107 in a Leukemia and Lymphoma panel reveals broad activity in various malignant cell lines (Supplementary Table S5).
Preclinical in vivo studies in the Ba/F3 (murine interleukin-3 dependent pro-B cell line) splenomegaly mouse model (45) show that PLX51107 is well tolerated and is active at 10-fold lower doses than OTX015 (Fig. 2D). Approximately 75% inhibition of splenomegaly was achieved with the 2 mg/kg PLX51107 dose, which was associated with an AUC0–24 of 17,700 ng•h/mL (or 40 μM•h). This value defines the threshold exposure required for PLX51107 to be effective in vivo. The same level of efficacy was achieved with 25 mg/kg OTX015 and the associated steady state AUC0–24 was 11,900 ng•h/mL (or 24 μM•h).
BET proteins have important roles in normal tissues; strong and prolonged suppression of their functions have often-untoward effects in multiple tissues (46). Importantly, the relatively short half-life of PLX51107 (<3 h in rodents and dogs) may underlie an improved therapeutic index, as epigenetic deviations tend to be long-lived. To support this notion, MV4-11 cells xenografted in SCID mice treated with PLX51107 were monitored for transcriptional changes over time. As shown in Fig. 2E transcriptional changes of HEXIM1, a strong pharmacodynamic marker for monitoring target engagement of BRD4 inhibitors in tumors (47), substantially out-live the plasma drug levels.
A set of cell culture experiments was performed to investigate the effect of treatment duration on the pharmacological activity of PLX51107. Short-term (4h) treatment of PLX51107 resulted in robust change in PD markers (c-MYC was used as a representative, Supplementary Fig. S2C) but did not induce an immediate apoptotic response (Supplementary Fig. S2D). Induction of apoptosis occurred at after prolonged treatment (16 hours or more of continuous exposure). A delay between the exposure to a pro-death stimulus and the onset of effector activation is inherent to the apoptotic program. To take this into consideration, flow cytometry analysis of tumor cells treated with PLX51107 for 4 or 8 hours followed by a washout period of 20 or 16 hours respectively was performed. Four hours treatment did not induce apoptosis but longer (8 h) treatment did (Supplementary Fig. S3E). Note that PLX51107 and OTX-015 have similar residence time (Supplementary Fig. S3F). These results suggest that short-term target engagement may generate desired pharmacological effect without inducing non-specific killing. A BET inhibitor with a short half-life is expected to have reduced risk of toxic effects associated with long-term inhibition of BET family proteins(38,39).
BET inhibition exerts in vitro anti-proliferative effects in CLL
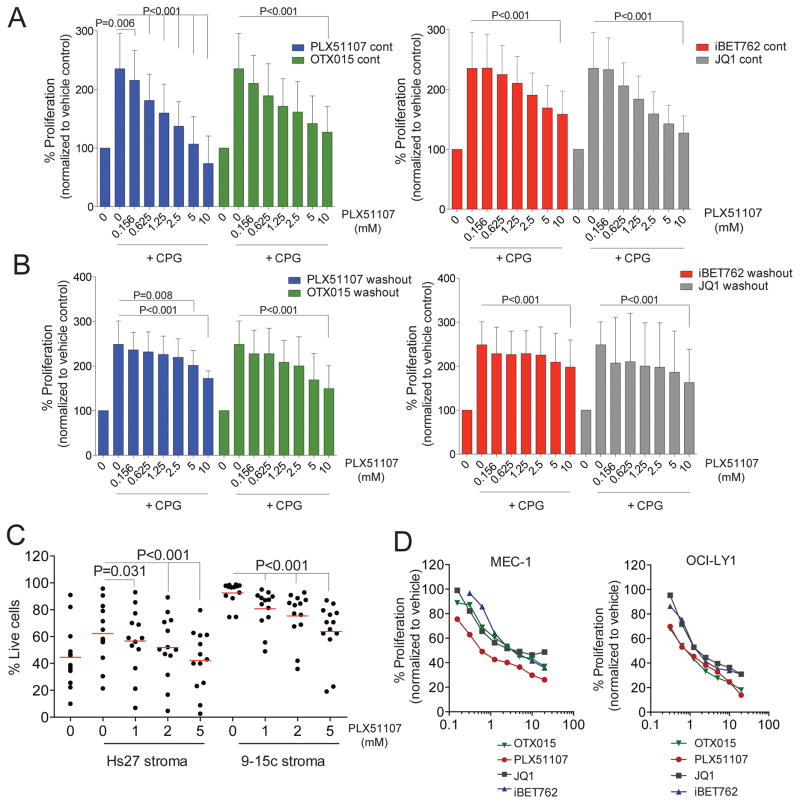
CLL has been considered an accumulative disease that derives from an inherent defect in apoptosis rather than increased proliferation rate, based on the observation that the majority of the circulating CLL cells are arrested in the G0/G1 phase. Nevertheless, recent studies have shown that CLL disease development is a dynamic process, wherein microenvironmental cell-cell interactions and accessory signals within pseudofollicular proliferation centers are essential for the expansion and survival of CLL cells (48,49). These signals have also been shown to promote acquisition of chemoresistance. We set out to determine whether BRD4 inhibition could attenuate CLL cell survival and proliferation when stimulated with CpG oligonucleotides, a well-characterized TLR9 agonist that promotes in vitro B-CLL cell proliferation (50) and mimics the effect of pseudo-proliferation centers where CLL expands (6,51,52). PLX51107 prevents CpG-induced cell proliferation (Fig. 3A and 3B) in primary CLL cells in a dose dependent fashion under both continuous and washout conditions. Similar trends were observed with other commercially available BRD4 inhibitors. However at higher concentrations the decrease was significantly greater with PLX compared to the other three drugs (p<0.001 for all comparisons under both continuous and washout conditions). Inhibition of proliferation or toxicity was not observed in CLL cells under basal condition. PLX51107 induces accumulation of p21 (cyclin-dependent kinase inhibitor) and IκBα, reduced levels of cMYC, and modulation of pro- and anti-apoptotic proteins (Supplementary Fig. S3B, S3C, and S3D).
Figure 3. Cytotoxic effect of BET inhibition in malignant B-cell lines and CLL patient-derived B-cells is independent of survival signals.
A–B. Dose-dependent decrease of proliferation of CpG stimulated primary CLL cells upon BRD4 inhibition with increasing doses of PLX51107, OTX015, JQ1, or iBET762 for 72h continuously (A) or for 4h followed by washout (B); Symbols represents individual patient data, black lines represent averages; C. CD19 positive cells from CLL patients (n=13) were co-culture with human Hs27 or murine 9–15c bone marrow-derived stroma and incubated with PLX51107 (1, 2 or 5μM) for 72 h after which CLL cell viability was determined by Annexin-V/PI staining. Red lines represent averages; D. Dose dependent decrease of proliferation of MEC-1, OCI-LY1 treated with increasing doses of PLX51107, OTX015, JQ1, or iBET762 for 72h (n=3 independent experiments per cell line). p<0.001 for all inhibitor and each cell line.
Inhibition of proliferation with PLX51107 was also observed in CpG stimulated normal B cells under continuous but was not washout condition (Supplementary Figure S3A).
Immune dysfunction is typically observed in CLL and leads to the increased number of infections and second malignancies seen in this disease. In addition, this decreased T-cell immune surveillance permits the survival of leukemia cells. Treatment agents capable of preserving T cells are therefore desired. As shown in Figure S3E and S3F PLX51107 had no anti-proliferative effect in healthy T-cells stimulated with CD3/CD28 agonist antibodies under washout conditions. Furthermore, cytokine production by T-cells was also not disrupted by PLX51107. The differential result of short-term (washout) versus long-term (continuous) inhibition of BRD4 by PLX51107 on normal B and T cells suggest that short-term BRD4 engagement may generate desired pharmacological effect in CLL without inducing non-specific killing and provides support to improved therapeutic index of BET inhibition of agents having a short terminal half-life. We next investigated the effect of PLX51107 on CLL cells in the presence of stromal protection. While primary CLL cells co-cultured on stromal cells (9–15c and Hs27) are protected from spontaneous apoptosis, treatment with PLX51107 induces significant cytotoxicity on CLL cells (Fig. 3C) with no effect on stromal cell viability (Supplementary Fig. S3G). This suggests that BET inhibition can overcome stromal protection that contributes to in vivo CLL cell survival and confers resistance to therapeutics used in this disease. Dose-dependent inhibition of cell growth (p<0.001 for all comparisons except at 0.15uM) with accumulation of cells in G0/G1 phase and increased apoptosis (Fig. 3D, and Supplementary Figure S3H, and S3I) was also observed in malignant B-cell cell lines (MEC-1 and OCI-LY1) with lower IC50 for PLX51107 compared to iBET762, JQ1, or OTX015. Collectively, these data demonstrate the sensitivity of proliferating CLL cells to BET inhibition and also the ability of BET inhibition to overcome microenvironment-derived resistance to apoptosis. Similar findings were observed with the highly active BCR targeting CLL therapeutic ibrutinib pre-clinically (6).
BET inhibition selectively modulates CLL driver genes
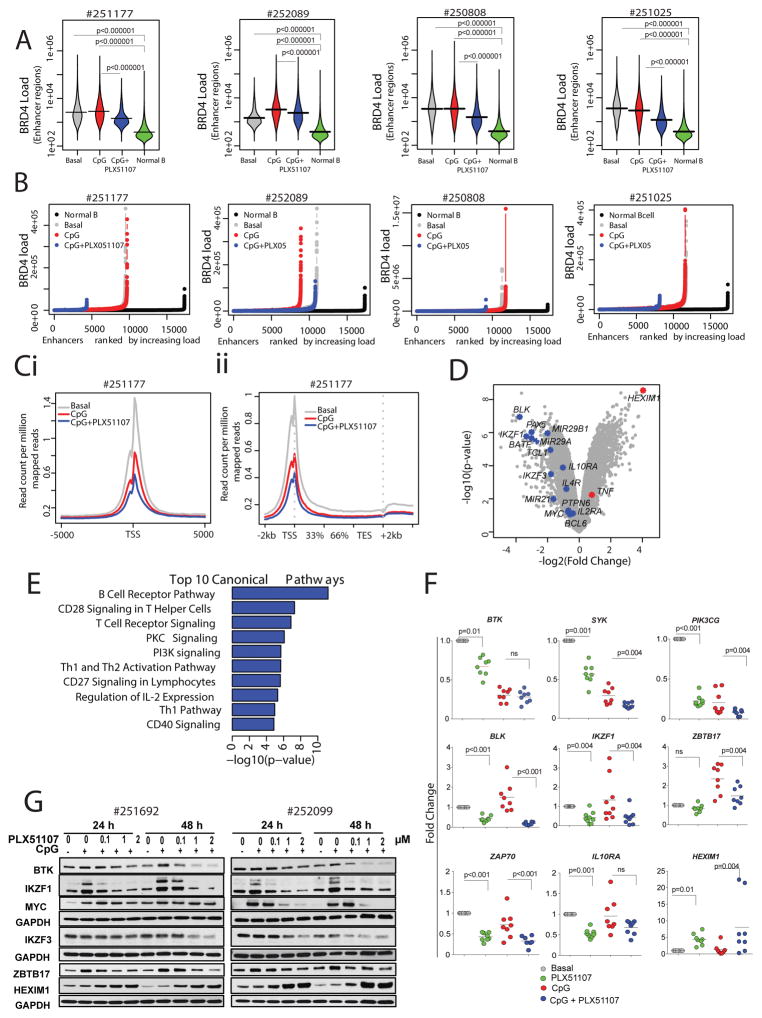
To gain mechanistic insight to the consequences of BET inhibition on genome-wide BRD4 binding, we augmented ChIP-seq studies to include CpG-stimulated primary CLL cells with or without PLX51107 treatment. BRD4 occupancy was globally decreased (p<10−6) by PLX51107 in three out of four samples (Supplementary Fig. S4A). More importantly, regardless of the global picture, BRD4 load was decreased in enhancer regions of all samples (Figure 4A) with significant loss of the SE regions following PLX51107 treatment (Fig. 4A and 4B). When compared to normal B cell enhancer regions of CLL samples were also significantly overloaded (p<10−6) (Fig. 4A and 4B).
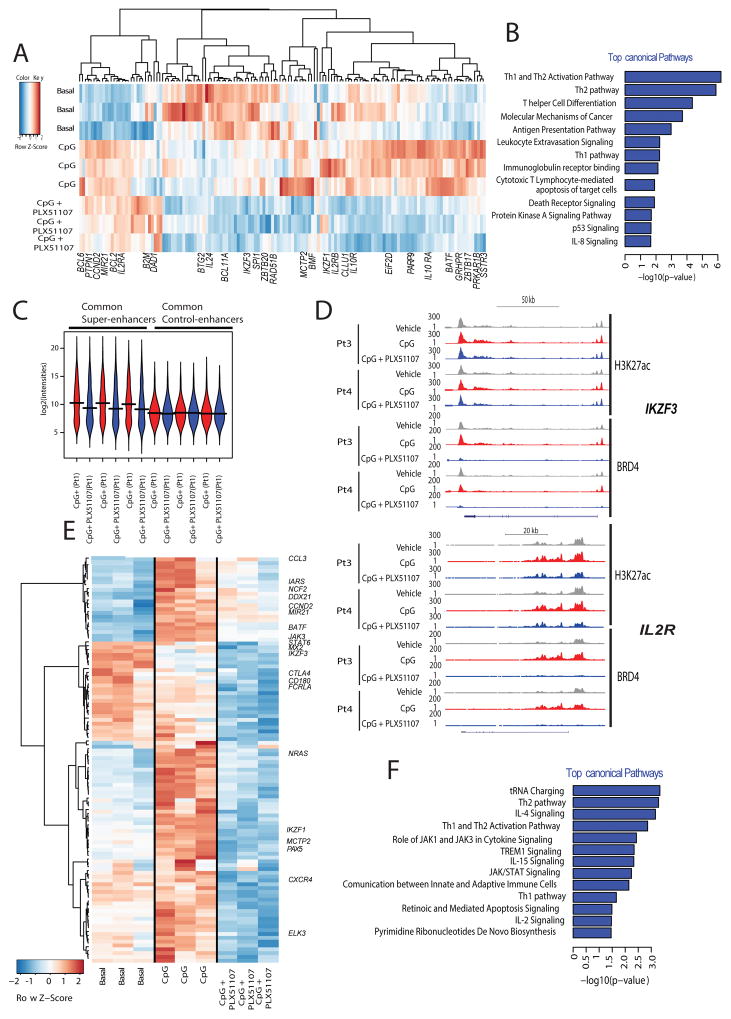
Figure 4. Epigenetic characterization of BET-inhibition in primary CLL.
A. Bean plots showing distribution of BRD4 peak scores at enhancers regions of patient derived CLL samples following 4h treatment (vehicle = basal, CpG = 3.2 μM CpG stimulation or CpG+ PLX51107) or a normal B cell lines. Horizontal bars mark the mean scores); B. Hockey stick plot for BRD4 loading across enhancers of four CLL patient samples following treatment; Ci. Metagene representation of RNA Pol II occupancy following the indicated treatment conditions created from normalized genome-wide average of reads at gene promoters centered on a ±5 kb window around the TSS. The y-axis shows average signal (rpm/bp); Cii. Metagene representation of RNA Pol II occupancy at gene bodies following stimulation and the indicated treatment conditions in primary CLL cells. The x-axis shows gene bodies region flanked by ± 2 kb of adjacent sequence and the y-axis shows average signal (rpm/bp); D. Microarray analysis using GeneChip Human Transcriptome Array HTA 2.0 (Affymetrix®) was performed for patient derived CLL cells stimulated with 3.2 μM CpG and treated with either DMSO vehicle (VEH) or 1 μM PLX51107 for 4h. Volcano plot representation of microarray data for CLL cells treated with or without PLX51107. Gene expression profiles were plotted according to the log2 fold change (x-axis) and log10-unadjusted p-value (y-axis). Few CLL relevant genes are highlighted. Experiments were performed as triplicates, each representing a pool of 2–3 CLL patient samples; E. The 10 top-ranked canonical pathways based on IPA p-values; F. Quantitative real time-PCR analysis of the indicated genes in PLX51107 treated CpG-stimulated CLL patient-derived B-cells (n=8–10). Red lines represent averages. G. Representative immunoblot analysis of protein levels of BTK, IKZF1, cMYC, IKZF3, ZBTB17, and HEXIM1 following PLX51107 treatment (0.1, 1, or 2 μM) in CpG-stimulated CLL cells (24 and 48 h, n=5).
A reduced RNA Pol II occupancy following PLX51107 treatment was observed at gene promoters centered ± 5 kb relative to the transcriptional start site (TSS) (Fig. 4Ci) and across gene bodies (Fig. 4Cii) indicating a reduced transcription rate at the nearby genes. We next performed microarrays analysis on CpG-stimulated primary CLL cells with or without PLX51107 treatment to gain functional insight on the transcriptional effects of BET inhibition. Microarray analysis revealed that more than 1300 genes were significantly differentially expressed (>2-fold change, P < 0.01; 745 up- and 616 down-regulated genes) (Fig. 4D and Supplementary Fig. S4B). Comprehensive Ingenuity Pathway Analysis (IPA) of all the modulated genes (up and down) revealed that BRD4 inhibition affects multiple cellular pathways including BCR, PKC, PI3K/AKT, CD40, IL4, IL2, P53, and death receptor signaling (Fig. 4E and Supplementary Table S6A). Inhibition of BRD4, IFNG, TNF, IL12, CD40LG, and FLT3LG, as well as activation of EGR2, FOXO3, CD38, IL1RN, and IL5 pathways were among the most significant effects predicted by differentially expressed genes (Supplementary Table S6B).
A variety of genes were modulated by BRD4 including CXCR5, CXCR4, IKZF1, IKZF3, TCL1, CD180, CCL3, PI3KCG, CTLA4, STAT6, and BLNK. Validation of microarray data was performed for select genes by quantitative real-time PCR at 4h (Fig. 4F and Supplementary Fig. S4C) and/or protein expression analysis at 24h and 48h in primary CLL cells (Fig. 4G and Supplementary Fig. S4D) as well as malignant B-cell lines (Supplementary Fig. S4E). Of note overexpression of de-novo upregulation of many of these genes has been well-described in CLL compared to normal B-cells and is also supported by our own data (Supplementary Fig S5A and S5B)(53–55). Many of these were also shown to be essential for CLL proliferation and associated with a more aggressive disease phenotype and poor response to therapy (56–58). These support the hypothesis that that BRD4 controls the expression of many CLL tumor specific genes and that BRD4 inhibition targets molecular defects in CLL in addition to B-cell lineage pathways.
An integrative approach was then applied to assess the link between BRD4 genomic load and gene expression. First, overlap between changes in BRD4 SEs and gene expression data was assessed. SEs whose ranking decreased upon BET inhibition in at least three out of the four patients samples were selected, then the expression profiles of 166 genes within 5 kb of these SEs were analyzed. Heat-maps of unsupervised clustering of these genes with the leading edge genes annotated and the top canonical pathways identified by IPA analysis are shown respectively in Fig. 5A, 5B and Supplementary Table S7A. The top signaling pathways identified by IPA to be differentially modulated by PLX51107 include activation of STAT3, BCL6, and CBFB as well as inhibition of IL1, IL4, and IL21 (Supplementary Tables S7B and S7C). Lymphopoiesis is the top biologic function predicted to be decreased upon BRD4 inhibition as supported by changes in expression of ARID5B, B2M, BATF, BCL11A, BCL2, BCL6, CCND2, CD44, EIF2AK4, ETV6, and HLA (Supplementary Table S7C). PLX51107 selectively decreased the transcript abundance of 397 genes regulated by BRD4-loaded SEs, in contrast to 205 control genes contained within the least BRD4-loaded enhancers (lower quartile of enhancer regions) in all samples (Fig. 5C). Examples of select gene tracks are shown in Fig. 5D.
Figure 5. BRD4 inhibition modulates key CLL pathways.
A. Gene expression heatmap of the common 142 genes contained within BRD4 SEs that are ranked lower upon PLX51107 treatment; B. The top-ranked canonical pathways based on IPA p-values are illustrated; C. Mean transcript abundance of the genes associated with the most and the least BRD4-loaded enhancers in all primary CLL cells treated with vehicle or PLX51107 (4h); D. ChIP-seq binding density for H3K27ac and BRD4 following treatment with PLX51107at the enhancer of IKZF3, and IL2R. The x-axis of the tracks shows genomic position, and the y-axis shows ChIP-seq signal (rpm/bp); E. Microarray expression profiles of 106 genes that are directly regulated by BRD4 (2 fold decrease and p<0.05 ) out of 537 common genes with decreased BRD4 load across three out of four analyzed CLL samples; F. The top-ranked canonical pathways regulated by BRD4 based on IPA p-values are illustrated.
Next, overlap between changes in individual BRD4 binding sites and gene expression was evaluated for annotated genes. First, BRD4 peaks (a) within 5 kb of a gene’s TSS, (b) within 5kb of an H3K27ac mark, and (c) with at least two fold decrease in peak score upon PLX51107 treatment were selected, revealing 537 common genes with decreased BRD4 peaks across the four patient samples. Microarray data was then utilized to identify genes with decreased expression (two fold decrease and p<0.05) upon treatment. As a result, 106 genes that are directly regulated by BRD4 were identified (Fig. 5E). Among the top canonical pathways identified by IPA are IL4 /IL15/IL2 signaling, and JAK/STAT signaling (Fig. 5F and Supplementary Table S8A). IPA upstream regulator analysis revealed that BET inhibition also resulted in inhibition of the BCR-complex, STAT1, VEGF-A, IFNL1, IL21, IFN-gamma, MYC, and activation of IL1RN (Supplementary Table S8B). Additionally, IPA biological function analysis recognized decrease in cell survival, cell viability, cell migration, leukopoiesis, and lymphocyte migration upon BET inhibition (Supplementary Table S8C). Taken together, these data indicate that BRD4 loading at select CLL enhancers underlies the pathway-specific transcriptional consequences of BET inhibition. Moreover, these data raise the possibility that BET inhibition may have broad activity in in vivo models of CLL similar to what we observed with BTK inhibitors such as ibrutinib pre-clinically (6).
PLX51107 demonstrates in vivo antitumor effects in preclinical models of CLL and aggressive lymphoma
We next evaluated the translational potential of BET inhibition in a mouse model of CLL that has been used extensively to evaluate experimental therapeutics for this disease (59–61). Eμ-TCL1 transgenic mice spontaneously develop a CD5/CD19 positive CLL-like leukemia that express BRD4 (Supplementary Fig. S6A) and respond to therapies proven effective in CLL (e.g. ibrutinib) (61,62). PLX51107 induce a dose-dependent cell growth inhibition of B-cells derived from Eμ-TCL1 mice with active disease (Supplementary Fig. S6B), with modulation of BRD4 targets such as Myc, Hexim1 while Eμ-TCL1 transgene expression remains (Supplementary Fig. S6C). To evaluate BRD4 in vivo target modulation mice with de novo leukemia, defined as > 60 WBC count, > 60% CD19/CD5/CD45 positive peripheral blood lymphocytes (PBL) and splenomegaly, were treated with PLX51107 via oral gavage (20 mg/kg daily) for eight days. Compared to vehicle-treated mice, PLX51107 significantly reduced leukemic disease burden in peripheral blood and spleen (Fig. 6A and B and Supplementary Fig. S6D) with modulation of BRD4 targets (Fig. 6C). PLX51107 (20 mg/kg, qd) was also more effective than OTX015 (50 mg/kg, qd) in reducing tumor burden (Supplementary Fig. S6E, and S6F) and in inducing in vivo CLL target modulation in a thirty day study (Supplementary Fig. S6G).
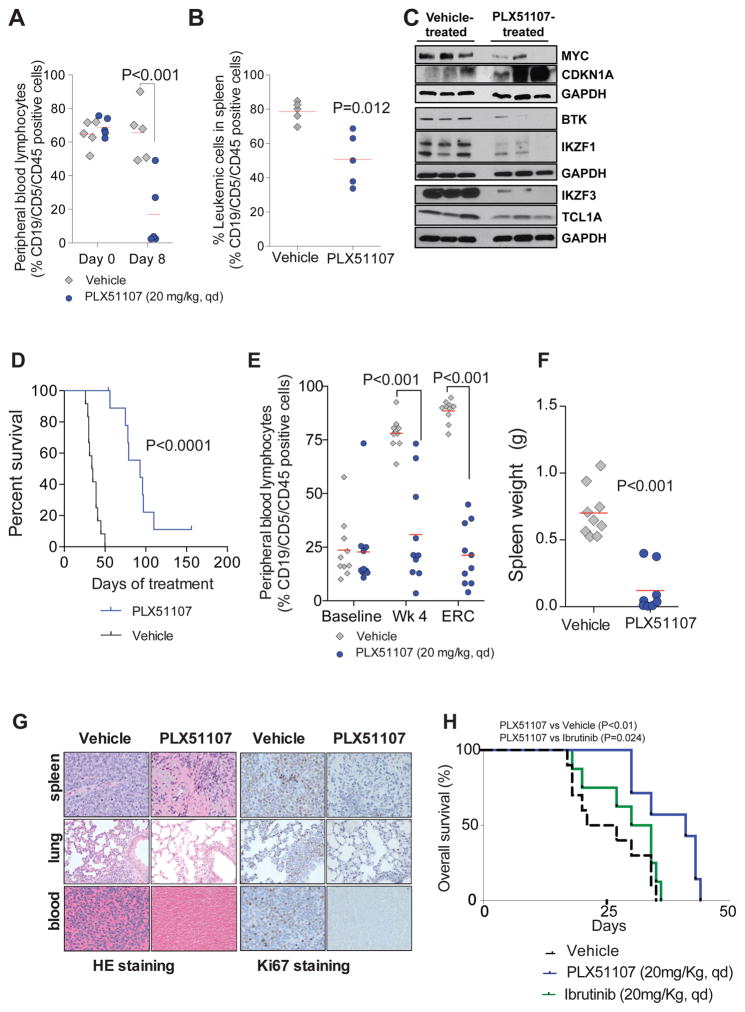
Figure 6. Targeting BRD4 with PLX51107 proves potent anti-leukemic effects in disease models of aggressive CLL and Richter’s Transformation.
A–C. Pharmacodynamic evaluation of antitumor effects of PLX51107 in Eμ-TCL1 with advanced leukemia. Mice were stratified according to leukemic peripheral blood lymphocytes (PBLs) and spleen palpation score to receive either vehicle or PLX51107 (20 mg/kg, qd, oral gavage) for 8 d: PLX51107 reduced leukemic cells in systemic circulation (A) and locally in spleen (B). Red line represents average; C. Representative immunoblot analysis of relative protein levels of cMYC, P21, BTK, IKZF1, IKZF3 and TCL1A protein at end of 8-d study; D–G. Using an adaptive transfer model of Eμ-TCL1, recipient wild type mice were randomized to receive vehicle (n=12) or PLX51107 (20 mg/kg, qd, oral gavage, n=10) at leukemia onset and disease progression was measured by flow cytometry as % CD19/CD5/CD45 positive PBL. Treatment was ended at 150 days. Kaplan-Meier curve showing overall survival (OS) (p<0.0001); Median OS: 93 days (PLX51107), 34 days (vehicle); (D), decreased % of circulating leukemic PBL (E), and reduced spleen mass (F). G. HE and Ki67 staining of spleen, lung and blood from the PLX51107-treated mice are depleted of lymphocytes, and Ki67 staining is mostly absent; H. Kaplan-Meier curve showing overall survival for C57BL/6 mice engrafted with Eμ-TCL1 leukemic splenocytes treated with Ibrutinib or PLX51107 (20 mg/kg, qd, oral gavage) at leukemia onset. Median OS: 41 days (PLX51107, n=7), 32 days (Ibrutinib, n=8) and 21 days (vehicle, n=7). PLX51107 significantly increased survival compared with vehicle (p=0.024) and Ibrutinib (p=0.049). Survival comparisons for (D), and (H) were made with the log-rank test. P-values have been adjusted for multiple comparisons.
Next we evaluated the long term in vivo therapeutic effects of PLX51107 utilizing an adoptive transfer model (61,63) in which leukemic cells from a Eμ-TCL1 mouse are engrafted into healthy wild type recipients of the same background. This produces a homogeneous population with same pathological findings to the Eμ-TCL1 transgenic mice and a more rapid disease acquisition (weeks instead of months), although time to disease and disease progression can vary depending on the donor used for the engraftment mimicking the heterogeneous natural history of CLL. PLX51107 significantly prolonged survival compared to vehicle control (Fig. 6D) with marked decrease in leukemia cells in blood (Fig. 6E) and reduction of spleen size (Fig. 6F). In addition, pathologic exam of spleens, bronchus-associated lymphoid tissue, alveolar septa of the lungs and blood from the cardiac chambers of PLX51107-treated mice revealed depletion of infiltrating leukemic cells identifiable by HE staining and generally negative for Ki67, a classic proliferation marker (Fig. 6G). We next tested the effect of PLX51107 or Ibrutinib in a cohort of mice with engrafted leukemic cells from a different Eμ-TCL1 donor. PLX51107 significantly prolonged survival compared to Ibrutinib (Fig. 6H) with significant reduction of spleen weight and blood leukemia cells (Supplementary Fig. S6H and S6I).
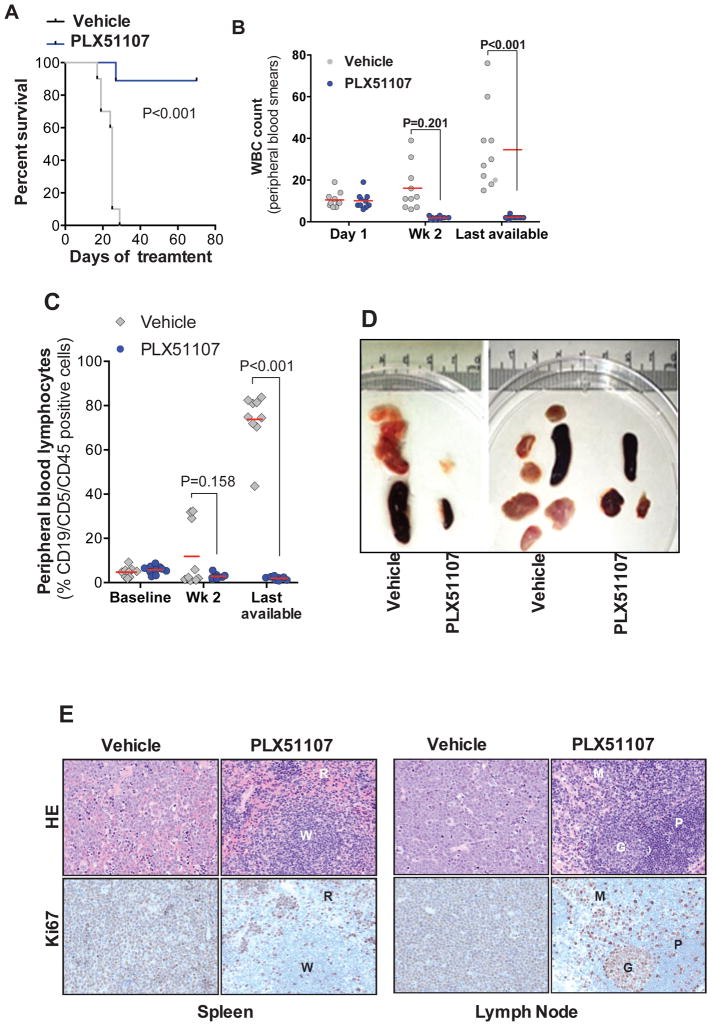
To assess the effects of BET inhibition in a highly penetrant, malignant leukemia/lymphoma phenotype analogous to high-grade lymphoma (37) we utilized the Eμ-Myc/TCL1 adoptive transfer mouse model which we recently introduced as a potential model for RT (59). Ex vivo treatment of Eμ-Myc/TCL1 derived splenocytes with PLX51107 (8h) suggests that both Eμ-Myc and Eμ-TCL1 transgenes are not affected by BET inhibition (Supplementary Fig. 7), confirming previous findings with JQ1 jn a Eμ-Myc model of lymphoma (64). PLX51107 treatment (70 days) significantly prolonged survival (Fig. 7A) and decreased tumor burden (Fig. 7B and 7C) with marked reduction in spleen size and lymph node masses (Fig. 7D). Histologically, atypical neoplastic lymphocytes were absent in spleens (Fig. 7E), wherein tissue architecture was preserved. Ki67 staining revealed scattered proliferating cells predominantly in the red pulp corresponding mostly to foci of extramedullary hematopoiesis. Similarly, lymph nodes of PLX51107 treated mice were devoid of neoplastic cell infiltration, and Ki67 expression was largely restricted to germinal centers with few positive cells in the medulla and paracortical zones. Daily administration of PLX51107 was well tolerated and resulted in physiologically relevant plasma levels (AUC0–24h = 96,700 h•ng/mL or 220 μM•h) in wild type B6 mice. Collectively, these data provide support for application of PLX51107 in clinical trials directed at both CLL and RT.
Figure 7. PLX51107 demonstrates in vivo anti-tumor effects in preclinical model of aggressive leukemia/lymphoma.
A–D Using the Eμ-Myc/TCL1 adoptive transfer model of high grade lymphoma analogous to RT, recipient wild type mice were randomized to receive vehicle (n=10) or PLX51107 (20 mg/kg, qd, oral gavage, n=10) at disease onset (≥ 5% CD19/CD5/CD45 positive PBL and/or increased WBC count), and disease progression was measured by flow cytometry as % CD19/CD5/CD45 positive PBL. PLX51107 prolonged survival (A), decreased peripheral WBC counts (B), reduced % CD19/CD5/CD45 positive PBL (C), and reduced spleen and lymph node masses (D); E. HE and Ki67 staining of spleen and lymph node from the PLX51107-treated mice are depleted of atypical neoplastic lymphocytes with preserved tissue architecture. Spleens from vehicle-treated mice are effaced with large, neoplastic lymphocytes, which are diffusely positive for Ki67. Conversely, atypical neoplastic lymphocytes are absent in spleens of PLX51107-treated mice. Splenic architecture is preserved with clearly demarcated red pulp (R) and white pulp (W) areas and scattered Ki67 positive cells are predominantly in the red pulp, and the majority of which correspond to foci of extramedullary hematopoiesis. Lymph nodes from vehicle-treated mice are distorted by neoplastic lymphocytes, which are diffusely positive for Ki67. By comparison, these neoplastic cells are absent in the lymph nodes of PLX51107-treated mice, wherein Ki67 expression is largely restricted to germinal centers (G) with few positive cells in the medulla (M) and paracortical zones (P).
DISCUSSION
A central role for BET bromodomains in governing the transcriptional landscape of many rapidly proliferating hematopoietic cancers has recently been recognized (65). Here, we utilize CLL as a model of slowly proliferating tumor cells to study the relevance of BET bromodomains in driving the expression of genes of the CLL transcriptional program. BRD4 over-expression has been shown to potentially act as an early driving event in lymphomagenesis and relapse-associated events (66) suggesting a role for BRD4 in leukemia disease progression and resistance. Here, we provide mechanistic evidence of BET bromodomains as transcriptional co-activators at SEs that regulate both important pathways (BCR signaling) and genes (TCL1, MCL1, MIR155, IKZF3, etc.) that are known to contribute to the development and progression of this disease. We hypothesize that for other slowly-proliferating neoplasms where distinct driving pathogenic pathways have not been identified, study of BET SE biology and pharmacologic interrogation (BET profiling) may allow identification of new therapeutic targets.
BET inhibition caused proliferation arrest in primary CLL cells even in the presence of microenvironment protection, and also significantly improved survival in two independent in vivo models of aggressive CLL.
Our comprehensive functional epigenetic and biochemical studies reveal insight into BET inhibition of oncogenic pathways and identify previously unrecognized targets of BRD4 in CLL. Similar to what was previously reported in other cancers, we observed the presence of a small subset of genes with a disproportionate BRD4 load at their proximal enhancers that are particularly sensitive to BET inhibition.
As these SEs are found adjacent to genes aberrantly expressed, de-novo regulated or with known oncogenic roles in CLL we hypothesized that SE analysis in CLL could identify previously unrecognized CLL tumor dependencies. Accordingly, we observed direct interaction of BRD4 at regulatory regions of the MIR21, IL4R, IL21R, IKZF3, IKZF1, mir155, and TCL1A genes. The de-novo expression of MIR155 and MIR21 in CLL has been reported and associated with a more aggressive disease phenotype and poor response to therapy (57,58). CLL cells express significantly higher levels of IL4R compared with normal B cells (67), resulting in an increased STAT6-dependent signaling pathway. TCL1A overexpression is also associated with rapid disease progression and poor response to therapy in CLL (68–70). Furthermore gain of function mutations in TCL1A are found in a small subset of CLL patients (71); TCL1A remains an un-druggable target to date. CCND2 is upregulated in CLL (72) and is essential for BCR-induced proliferation and CD5-positive B-cell development (56). Although dysregulation of all the above genes has been documented in CLL (73), their mechanisms of regulation as well as strategies to modulate their expression remain mostly unknown. Here we demonstrate that MIR21, IL4R, IL21R, MIR155, and TCL1A are all regulated by SEs controlled by BRD4 and can be targeted by PLX51107 in vitro and in vivo in CLL models. Mutations in IKZF3 have recently been proposed as putative CLL drivers (20,74) and IKZF3 upregulation has been associated with adverse outcome (75). The observed transcriptional modulation of the B-cell specific transcription factors IKZF1 and IKZF3 by BRD4 suggests a therapeutic benefit of BET inhibitors alone or in combination with immunomodulatory drugs (IMiDs) in CLL. To this extent, synergistic cytotoxicity of BRD4 inhibitors and the IMiD lenalidomide has recently been demonstrated in preclinical models of primary effusion lymphoma, an aggressive type of non-Hodgkin lymphoma (76).
Our group has defined mutations of either BTK or PLCG2 as mechanisms of resistance to ibrutinib in CLL(77). The transcriptional profiling of PLX51107-treated CLL cells revealed downregulation of multiple core pathways in CLL including B-cell development, BCR and TLR/MYD88 signaling, and cytokine/chemokine interactions. Given the central role of BCR activation in CLL cell maintenance and expansion, our observation that BRD4 regulates multiple components of the BCR pathway (e.g. PLCG2, ZAP70, and PI3K) not only emphasizes an important role of BRD4 in CLL pathology but supports the clinical investigation of BET inhibitors alone or in combination with BCR-targeting agents (e.g. idelalisib, and ibrutinib) to overcome resistance and achieve deeper and more durable responses in relapsed/refractory CLL and other B-cell malignancies. This is of critical importance given the increasing number of patients relapsing on targeted therapies including the BTK inhibitor ibrutinib (7,78). Moreover, the proximal BCR pathway kinases SYK and LYN were reported to be critical for the activation of mutant PLCG2 protein(79), suggesting that intervention with PLX51107 has the potential to overcome acquired ibrutinib resistance that frequently results from reactivation of BCR signaling. This approach was recently shown to synergistically induce apoptosis in ibrutinib-resistant mantle cell lymphoma (80) and diffuse large B–cell lymphoma (81). Further studies to confirm the therapeutic benefit of this combination in CLL are warranted.
The tool compound JQ1 has been recently reported to downregulate PD-L1 expression in the Eμ-Myc B-cell lymphoma model (82). Here, we show similar effects of the clinical agent PLX51107 on PD-L1 in primary CLL cells. Furthermore, we observed this effect on additional checkpoint molecules (CD200, LAG3, CTLA4). The regulation of these factors by BRD4 on CLL cells has not been reported to date and points to the potential role of this agent in reversing tumor immune suppression. Comprehensive analysis for the potential immunomodulatory effects of BRD4 inhibition and their functional relevance in CLL are needed and our data and supports the clinical application of PLX51107 to target tumor immune evasion alone or in combination with immune-modulatory agents in CLL.
Collectively, our findings demonstrate that BRD4 is enriched at hallmark genes of CLL biology, and that BET inhibition is capable of downregulating multiple survival pathways in malignant CLL B-cells. This mechanism of action provides a strong rationale for human clinical investigation of BET inhibitors to target the neoplastic B-cells and their supportive microenvironment in relapsed/refractory CLL and other aggressive B-cell malignancies including RT.
METHODS
Patient sample processing, cell lines, and cell culture
Blood was obtained from healthy volunteers or CLL patients following written informed consent under a protocol approved by the Institutional Review Board (IRB) of The Ohio State University (OSU; Columbus, OH) in accordance with the Declaration of Helsinki. All patients examined had CLL as defined by the 2008 IWCLL criteria (51), and cells were isolated and cultured as previously described (83). The CLL leukemic cell line MEC-1 was obtained from DSMZ (Braunschweig, Germany) in 2015. MEC-1 cell identity was confirmed in May 2015 via fluorescence in situ hybridization (FISH) using CLL probe panel, i.e. CEP12/13q14/13q34, ATM/p53, BCL6, MYC, IGH/CCND1 (Abbott Molecular, Des Plaines, IL), and 6q21 (Kreatech, Buffalo Grove, IL) (results for 200 analyzed cells were reported). A large number of vials were cryopreserved. For experiments cells were resuscitated, cultured in RPMI (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), and used within 3–4 weeks from thawing. DLBCL cell line OCI-LY1 was obtained from Ontario Cancer Institute (Ontario, Canada) in 2016. OCI-LY1 cell identity was confirmed in May 2016 by exome-sequencing and mutation status was compared to publically available exome sequencing data sets (84,85) and a large number of vials was banked. For experiment cells were cultured in IMDM (Invitrogen) supplemented with 20% FBS and used within 3–4 weeks from thawing. The human bone marrow stromal Hs27-EGFP cell line was authenticated and provided by Dr. Torok-Storb (Fred Hutchinson Cancer Research Center) (86) in 2010. The murine marrow-derived mesenchymal 9–15c cell line (82) was obtained from RIKEN cell bank (Ibaraki, Japan) in 2012 (87). A large stock was cryopreserved, For experiments cells were resuscitated, maintained in RPMI supplemented with 10% FBS as previously described (88) and used within 3–4 weeks from thawing. All cell lines were confirmed to be mycoplasma negative using the MycoAlert™ Mycoplasma Detection Kit from Lonza (Rockland, ME) according to manufacturer instructions and used within 3–4 weeks from thawing. See Supplementary information for details on sample processing.
Mouse B-cell isolation
Primary B-cells were freshly isolated from mouse (WT B6 or TCL1-Tg) spleens using the EasySep™ mouse pan B-cell isolation kit (STEMCELL Technologies) according to the manufacturer’s instructions. B-cell purity was checked using anti-CD19-PE (BD Pharmingen) staining by flow cytometry and isolated B-cells were used for immunoblot analysis of BRD4 levels.
Reagents and antibodies
The BRD4 inhibitor PLX51107 was provided by Plexxikon Inc. (Berkeley, CA). JQ1 and iBET762 were purchased from Selleck Chemicals (Houston, TX). OTX015 was purchased from Advanced ChemBlocks Inc. (Burlingame, CA). See Supplementary information for a detailed list of reagents and antibodies.
Immunoblot analysis
Proteins extracted from whole-cell lysates were resolved by SDS-PAGE and transferred on nitrocellulose membrane as previously described (89).
Proliferation assay and flow cytometric studies
MTS and Annexin V-FITC flow based assays was performed to determine cell proliferation and death as previously described (90). See Supplementary information for details.
Animal studies
Experiments were approved by The Ohio State University Institutional Animal Care and Use Committee (IACUC). For engraftment studies, C57BL/6 wild type mice (Jackson Laboratory, Bar Harbor, ME) were engrafted with 1E7 cells by tail vein injection of splenocytes derived from Eμ-TCL1 or Eμ-Myc/TCL1 mice with active disease. At the onset of leukemia (Eμ-TCL1: ≥10% CD19/CD5/CD45 positive circulating cells; Eμ-Myc/TCL1: WBC count ≥ 8 and/or ≥ 5% CD19/CD5/CD45 positive circulating cells) mice were randomized to receive treatments as indicated. Vehicle = 10% N-Methyl-2-pyrrolidone (NMP) plus diluent (40% PEG400, 5% TPGS, 5% Poloxamer 407 and 50% water). Mice were sacrificed when meeting early removal criteria (ERC: >20% weight loss, impaired motility, splenomegaly and evident tumor masses), and tissues were collected for further analysis.
RNA extraction and microarray analysis
Total RNA enriched in small RNA (>200 bp) was isolated from primary CLL cells following drug treatment using miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the supplier’s protocol. Total RNA concentration was measured by using Nano Drop UV/Vis spectrophotometer (2000c, Thermo Scientific). RNA quality was assessed by capillary electrophoresis with a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Afterwards, the RNA was pooled (2 μg/ patient sample), creating 3 pools per treatment group. Prior to hybridization, RNA integrity and comparability were tested by BioAnalyzer and only samples with RNA integrity numbers (RIN) > 8.0 were used in microarray experiments. cDNA synthesis, labelling and the basic microarray analysis was performed by the Genomics Shared Resource at The Ohio State University Comprehensive Cancer Center. Generation of double-stranded cDNA, preparation and labelling of cRNA, hybridization to GeneChip® Human Transcriptome Array 2.0 (HTA 2.0) (Affymetrix, Santa Clara, CA, USA) and washing was performed according to the standard Affymetrix protocol. The arrays were scanned using a GeneChip® Scanner 3000 (Affymetrix). Signal intensities were analyzed by Affymetrix Expression Console software.
Quantitative Real-Time PCR
Real-time PCR was performed using cDNA prepared as described (83) using TaqMan gene expression assays. See Supplementary information for a detailed list of primers.
ChIP-Seq and Data Processing
Primary CLL cells (1E7 cells per condition) were treated with vehicle (DMSO), or 1μM PLX51107 with or without CpG oligonucleotides (3.2 μM) for 4 h. Cells were fixed with 1% formaldehyde for 15 min and quenched with 0.125 M glycine. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. Lysates were sonicated and the DNA sheared to an average length of 300–500 bp. See supplementary information for detailed cross-link procedure. Genomic DNA regions of interest were isolated using 4μg antibody against BRD4, H3K27ac and RNA Pol II. Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinks were reversed by incubation overnight at 65°C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation.
Illumina sequencing libraries were prepared from the ChIP and input DNAs by the standard consecutive enzymatic steps of end-polishing, dA-addition, and adaptor ligation. After a final PCR amplification step, the resulting DNA libraries were quantified and sequenced on Illumina’s NextSeq 500. Sequence reads were aligned to the reference genome, peak locations were identified and annotated for further analysis. See Supplementary information for additional details.
ATAC-seq and data processing
Accessible chromatin mapping was performed using the ATAC-seq method as previously described (24,91). See Supplementary information for details.
Data integration and visualization
In-house shell and R scripts (https://www.r-project.org) were used for data integration. To describe genome-wide correlation between BRD4 occupancy, H3K27 acetylation and chromatin accessibility, number of peaks/hotspots within 5 kb of one another were counted and presented as a Venn diagram. To study functional consequences of BRD4 inhibition with PLX51007 treatment, data integration process was always centered around annotated genes within 5 kb of BRD4 peaks, H3K27ac mark and expressed in microarray data. IGV (http://www.broadinstitute.org/igv/) was used for visualization. Annotation files were downloaded from UCSC.
Functional annotation
DAVID functional annotation tool was used to identify enriched gene ontology (GO) terms in a given gene list (92). Ingenuity Pathway Analysis (IPA, (Qiagen Bioinformatics) software was used for functional annotation of differentially expressed genes regulated by BRD4 inhibition. Fold change information from microarray gene expression data was also provided as an input to IPA. Significantly overrepresented canonical pathways were reported. Upstream transcriptional regulators that could explain the observed changes in gene expression were identified by IPA upstream regulator analysis with a z-score cutoff of +2 for activation and −2 for inhibition. Overrepresented diseases and biological functions with a predicted activation state were identified by IPA diseases/biofunctions analysis with a z-score cutoff of 2 for increase and −2 for decrease.
Statistical analysis
All analyses were performed using SAS/STAT software, version 9.2 (SAS Institute, Inc., Cary, NC). See Supplementary materials and methods for details.
Published data
All raw sequence data is available in GEO via accession number GSE109411.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
To date, functional studies of BRD4 in CLL are lacking. Through integrated genomic, functional and pharmacological analyses we uncover the existence of BRD4 regulated core CLL transcriptional programs and present preclinical proof-of-concept studies validating BET inhibition as an epigenetic approach to target BCR signaling in CLL.
Acknowledgments
We are grateful to the patients and healthy volunteers who provided blood for the above studies and to the OSU Comprehensive Cancer Center Leukemia Tissue Bank (supported by NCI P30 CA016058) for sample procurement. X-ray diffraction data were collected at beamline 8.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory), beamline 08ID-1 at Canadian Light Source and beamline 7.1 at the Stanford Synchrotron Radiation Lightsource (a directorate of the SLAC National Accelerator Laboratory). Authors would also like to acknowledge Alan Flechtner, HTL and The Ohio State University Veterinary Histology Laboratory for their assistance in all immunohistochemical studies, also supported by NCI P30 CA016058. This work was supported by the National Cancer Institute (K99/R00 CA208017, R01 CA177292, R01 CA206658, R01 CA214046, and R35 CA198183), a Research Scholar grant (RSG) 129863-RSG-16-158-01-CDD from the American Cancer Society, NCI P30 (CA 016058), The OSU Comprehensive Cancer Center using Pelotonia funds, and further research support to the Byrd Laboratory from the Four Winds Foundation, the D. Warren Brown Foundation, the Connie Brown CLL Research fund, and the Sullivan CLL Research Foundation. This study makes use of RNA-seq data generated by the Blueprint Consortium. A full list of the investigators who contributed to the generation of the data is available from www.blueprint-epigenome.eu. Funding for the project was provided to the Blueprint Consortium by the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no 282510 BLUEPRINT.
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
Ben Powell, Jiazhong Zhang, Yan Ma, Ying Zhang, Wayne Spevak, Songyuan Shi, Paul Severson, Rafe Shellooe, Heidi Carias, Garson Tsang, Ken Dong, Todd Ewing, Adhirai Marimuthu, Christina Tantoy, Jason Walters, Laura Sanftner, Hamid Rezaei, Marika Nespi, Bernice Matusow, Gaston Habets, Prabha Ibrahim, Chao Zhang, and Gideon Bollag are employees of Plexxikon Inc.
References
- 1.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nature reviews Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 2.Riches JC, Gribben JG. Understanding the immunodeficiency in chronic lymphocytic leukemia: potential clinical implications. Hematology/oncology clinics of North America. 2013;27:207–35. doi: 10.1016/j.hoc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Chiorazzi N, Rai KR, Ferrarini M. Chronic Lymphocytic Leukemia. New England Journal of Medicine. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 4.Dubovsky JA, Flynn R, Du J, Harrington BK, Zhong Y, Kaffenberger B, et al. Ibrutinib treatment ameliorates murine chronic graft-versus-host disease. The Journal of clinical investigation. 2014;124:4867–76. doi: 10.1172/JCI75328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubovsky JA, Beckwith KA, Natarajan G, Woyach JA, Jaglowski S, Zhong Y, et al. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122:2539–49. doi: 10.1182/blood-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herman SE, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–96. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. The New England journal of medicine. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraietta JA, Beckwith KA, Patel PR, Ruella M, Zheng Z, Barrett DM, et al. Ibrutinib enhances chimeric antigen receptor T-cell engraftment and efficacy in leukemia. Blood. 2016;127:1117–27. doi: 10.1182/blood-2015-11-679134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woyach JA, Johnson AJ. Targeted therapies in CLL: mechanisms of resistance and strategies for management. Blood. 2015 doi: 10.1182/blood-2015-03-585075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maddocks KJ, Ruppert AS, Lozanski G, Heerema NA, Zhao W, Abruzzo L, et al. Etiology of Ibrutinib Therapy Discontinuation and Outcomes in Patients With Chronic Lymphocytic Leukemia. JAMA oncology. 2015;1:80–7. doi: 10.1001/jamaoncol.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stilgenbauer S, Eichhorst B, Schetelig J, Coutre S, Seymour JF, Munir T, et al. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. The Lancet Oncology. 2016;17:768–78. doi: 10.1016/S1470-2045(16)30019-5. [DOI] [PubMed] [Google Scholar]
- 12.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. The Journal of clinical investigation. 2007;117:112–21. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–8. [PubMed] [Google Scholar]
- 14.Secchiero P, Tiribelli M, Barbarotto E, Celeghini C, Michelutti A, Masolini P, et al. Aberrant expression of TRAIL in B chronic lymphocytic leukemia (B-CLL) cells. Journal of cellular physiology. 2005;205:246–52. doi: 10.1002/jcp.20392. [DOI] [PubMed] [Google Scholar]
- 15.Browning RL, Byrd WH, Gupta N, Jones J, Mo X, Hertlein E, et al. Lenalidomide Induces Interleukin-21 Production by T Cells and Enhances IL21-Mediated Cytotoxicity in Chronic Lymphocytic Leukemia B Cells. Cancer immunology research. 2016;4:698–707. doi: 10.1158/2326-6066.CIR-15-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrer G, Bosch R, Hodgson K, Tejero R, Roue G, Colomer D, et al. B cell activation through CD40 and IL4R ligation modulates the response of chronic lymphocytic leukaemia cells to BAFF and APRIL. British journal of haematology. 2014;164:570–8. doi: 10.1111/bjh.12645. [DOI] [PubMed] [Google Scholar]
- 17.Ghobrial IM, Bone ND, Stenson MJ, Novak A, Hedin KE, Kay NE, et al. Expression of the chemokine receptors CXCR4 and CCR7 and disease progression in B-cell chronic lymphocytic leukemia/ small lymphocytic lymphoma. Mayo Clinic proceedings. 2004;79:318–25. doi: 10.4065/79.3.318. [DOI] [PubMed] [Google Scholar]
- 18.Till KJ, Lin K, Zuzel M, Cawley JC. The chemokine receptor CCR7 and alpha4 integrin are important for migration of chronic lymphocytic leukemia cells into lymph nodes. Blood. 2002;99:2977–84. doi: 10.1182/blood.v99.8.2977. [DOI] [PubMed] [Google Scholar]
- 19.Herishanu Y, Perez-Galan P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–74. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuille MR, Condie A, Stone EM, Wilsher J, Bradshaw PS, Brooks L, et al. TCL1 is activated by chromosomal rearrangement or by hypomethylation. Genes, chromosomes & cancer. 2001;30:336–41. doi: 10.1002/gcc.1099. [DOI] [PubMed] [Google Scholar]
- 22.Claus R, Lucas DM, Ruppert AS, Williams KE, Weng D, Patterson K, et al. Validation of ZAP-70 methylation and its relative significance in predicting outcome in chronic lymphocytic leukemia. Blood. 2014;124:42–8. doi: 10.1182/blood-2014-02-555722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guinn D, Ruppert AS, Maddocks K, Jaglowski S, Gordon A, Lin TS, et al. miR-155 expression is associated with chemoimmunotherapy outcome and is modulated by Bruton’s tyrosine kinase inhibition with Ibrutinib. Leukemia. 2015;29:1210–3. doi: 10.1038/leu.2014.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rendeiro AF, Schmidl C, Strefford JC, Walewska R, Davis Z, Farlik M, et al. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nature communications. 2016;7:11938. doi: 10.1038/ncomms11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8758–63. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert reviews in molecular medicine. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Molecular and cellular biology. 2008;28:967–76. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–82. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. The Journal of biological chemistry. 2012;287:43137–55. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Molecular cell. 2014;54:728–36. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. The Lancet Haematology. 2016;3:e186–95. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 39.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. The Lancet Haematology. 2016;3:e196–204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 40.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, et al. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene. 2014;33:2395–404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercher T, Wernig G, Moore SA, Levine RL, Gu TL, Frohling S, et al. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood. 2006;108:2770–9. doi: 10.1182/blood-2006-04-014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, et al. Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell reports. 2014;8:1919–29. doi: 10.1016/j.celrep.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin X, Huang X, Uziel T, Hessler P, Albert DH, Roberts-Rapp LA, et al. HEXIM1 as a Robust Pharmacodynamic Marker for Monitoring Target Engagement of BET Family Bromodomain Inhibitors in Tumors and Surrogate Tissues. Molecular cancer therapeutics. 2017;16:388–96. doi: 10.1158/1535-7163.MCT-16-0475. [DOI] [PubMed] [Google Scholar]
- 48.Burger JA, Gribben JG. The microenvironment in chronic lymphocytic leukemia (CLL) and other B cell malignancies: insight into disease biology and new targeted therapies. Seminars in cancer biology. 2014;24:71–81. doi: 10.1016/j.semcancer.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 49.Ramsay AD, Rodriguez-Justo M. Chronic lymphocytic leukaemia--the role of the microenvironment pathogenesis and therapy. British journal of haematology. 2013;162:15–24. doi: 10.1111/bjh.12344. [DOI] [PubMed] [Google Scholar]
- 50.Decker T, Schneller F, Sparwasser T, Tretter T, Lipford GB, Wagner H, et al. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood. 2000;95:999–1006. [PubMed] [Google Scholar]
- 51.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swerdlow SH, Murray LJ, Habeshaw JA, Stansfeld AG. Lymphocytic lymphoma/B-chronic lymphocytic leukaemia--an immunohistopathological study of peripheral B lymphocyte neoplasia. British journal of cancer. 1984;50:587–99. doi: 10.1038/bjc.1984.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohle R, Failenschmid C, Bautz F, Kanz L. Overexpression of the chemokine receptor CXCR4 in B cell chronic lymphocytic leukemia is associated with increased functional response to stromal cell-derived factor-1 (SDF-1) Leukemia. 1999;13:1954–9. doi: 10.1038/sj.leu.2401602. [DOI] [PubMed] [Google Scholar]
- 54.Burkle A, Niedermeier M, Schmitt-Graff A, Wierda WG, Keating MJ, Burger JA. Overexpression of the CXCR5 chemokine receptor, and its ligand, CXCL13 in B-cell chronic lymphocytic leukemia. Blood. 2007;110:3316–25. doi: 10.1182/blood-2007-05-089409. [DOI] [PubMed] [Google Scholar]
- 55.Fayad L, Keating MJ, Reuben JM, O’Brien S, Lee BN, Lerner S, et al. Interleukin-6 and interleukin-10 levels in chronic lymphocytic leukemia: correlation with phenotypic characteristics and outcome. Blood. 2001;97:256–63. doi: 10.1182/blood.v97.1.256. [DOI] [PubMed] [Google Scholar]
- 56.Solvason N, Wu WW, Parry D, Mahony D, Lam EW, Glassford J, et al. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. International immunology. 2000;12:631–8. doi: 10.1093/intimm/12.5.631. [DOI] [PubMed] [Google Scholar]
- 57.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. The New England journal of medicine. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 58.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–9. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers KA, El-Gamal D, Bonnie HK, Zachary HA, Virginia GM, Rose M, et al. The Eμ-Myc/TCL1 Transgenic Mouse As a New Aggressive B-Cell Malignancy Model Suitable for Preclinical Therapeutics Testing. Blood. 2015;126:2752. [Google Scholar]
- 60.Zhong Y, El-Gamal D, Dubovsky JA, Beckwith KA, Harrington BK, Williams KE, et al. Selinexor suppresses downstream effectors of B-cell activation, proliferation and migration in chronic lymphocytic leukemia cells. Leukemia. 2014 doi: 10.1038/leu.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6955–60. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson AJ, Lucas DM, Muthusamy N, Smith LL, Edwards RB, De Lay MD, et al. Characterization of the TCL-1 transgenic mouse as a preclinical drug development tool for human chronic lymphocytic leukemia. Blood. 2006;108:1334–8. doi: 10.1182/blood-2005-12-011213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Molecular and cellular biology. 2011;31:2641–52. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hogg SJ, Newbold A, Vervoort SJ, Cluse LA, Martin BP, Gregory GP, et al. BET Inhibition Induces Apoptosis in Aggressive B-Cell Lymphoma via Epigenetic Regulation of BCL-2 Family Members. Molecular cancer therapeutics. 2016;15:2030–41. doi: 10.1158/1535-7163.MCT-15-0924. [DOI] [PubMed] [Google Scholar]
- 65.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. The New England journal of medicine. 2012;367:647–57. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 66.Jiang Y, Redmond D, Nie K, Eng KW, Clozel T, Martin P, et al. Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas. Genome biology. 2014;15:432. doi: 10.1186/s13059-014-0432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aguilar-Hernandez MM, Blunt MD, Dobson R, Yeomans A, Thirdborough S, Larrayoz M, et al. IL-4 enhances expression and function of surface IgM in CLL cells. Blood. 2016;127:3015–25. doi: 10.1182/blood-2015-11-682906. [DOI] [PubMed] [Google Scholar]
- 68.Browning RL, Geyer SM, Johnson AJ, Jelinek DF, Tschumper RC, Call TG, et al. Expression of TCL-1 as a potential prognostic factor for treatment outcome in B-cell chronic lymphocytic leukemia. Leukemia research. 2007;31:1737–40. doi: 10.1016/j.leukres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herling M, Patel KA, Khalili J, Schlette E, Kobayashi R, Medeiros LJ, et al. TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia. 2006;20:280–5. doi: 10.1038/sj.leu.2404017. [DOI] [PubMed] [Google Scholar]
- 70.Herling M, Patel KA, Weit N, Lilienthal N, Hallek M, Keating MJ, et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood. 2009;114:4675–86. doi: 10.1182/blood-2009-03-208256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pekarsky Y, Palamarchuk A, Maximov V, Efanov A, Nazaryan N, Santanam U, et al. Tcl1 functions as a transcriptional regulator and is directly involved in the pathogenesis of CLL. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19643–8. doi: 10.1073/pnas.0810965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delmer A, Ajchenbaum-Cymbalista F, Tang R, Ramond S, Faussat AM, Marie JP, et al. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood. 1995;85:2870–6. [PubMed] [Google Scholar]
- 73.Teitell MA. The TCL1 family of oncoproteins: co-activators of transformation. Nature reviews Cancer. 2005;5:640–8. doi: 10.1038/nrc1672. [DOI] [PubMed] [Google Scholar]
- 74.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 75.Nuckel H, Frey UH, Sellmann L, Collins CH, Duhrsen U, Siffert W. The IKZF3 (Aiolos) transcription factor is highly upregulated and inversely correlated with clinical progression in chronic lymphocytic leukaemia. British journal of haematology. 2009;144:268–70. doi: 10.1111/j.1365-2141.2008.07442.x. [DOI] [PubMed] [Google Scholar]
- 76.Gopalakrishnan R, Matta H, Tolani B, Triche T, Jr, Chaudhary PM. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene. 2016;35:1797–810. doi: 10.1038/onc.2015.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. The New England journal of medicine. 2014;370:2286–94. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Byrd JC, Brown JR, O’Brien S, Barrientos JC, Kay NE, Reddy NM, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. The New England journal of medicine. 2014;371:213–23. doi: 10.1056/NEJMoa1400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shan C, Lin J, Hou JQ, Liu HY, Chen SB, Chen AC, et al. Chemical intervention of the NM23-H2 transcriptional programme on c-MYC via a novel small molecule. Nucleic acids research. 2015 doi: 10.1093/nar/gkv641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun B, Shah B, Fiskus W, Qi J, Rajapakshe K, Coarfa C, et al. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood. 2015 doi: 10.1182/blood-2015-04-639542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11365–70. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell reports. 2017;18:2162–74. doi: 10.1016/j.celrep.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120:4621–34. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3879–84. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature genetics. 2011;43:830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Graf L, Iwata M, Torok-Storb B. Gene expression profiling of the functionally distinct human bone marrow stromal cell lines HS-5 and HS-27a. Blood. 2002;100:1509–11. doi: 10.1182/blood-2002-03-0844. [DOI] [PubMed] [Google Scholar]
- 87.Yamada Y, Sakurada K, Takeda Y, Gojo S, Umezawa A. Single-cell-derived mesenchymal stem cells overexpressing Csx/Nkx2. 5 and GATA4 undergo the stochastic cardiomyogenic fate and behave like transient amplifying cells. Experimental cell research. 2007;313:698–706. doi: 10.1016/j.yexcr.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 88.El-Gamal D, LaFollette TD, Lai H, Chenglong L, Sampath D, Lehman A, et al. Expression of PRMT5 in B-Cell Chronic Lymphocytic Leukemia and Its Significance in Disease Progression and Richter’s Transformation. Blood. 2014;124:2197. [Google Scholar]
- 89.Lapalombella R, Yu B, Triantafillou G, Liu Q, Butchar JP, Lozanski G, et al. Lenalidomide down-regulates the CD20 antigen and antagonizes direct and antibody-dependent cellular cytotoxicity of rituximab on primary chronic lymphocytic leukemia cells. Blood. 2008;112:5180–9. doi: 10.1182/blood-2008-01-133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herman SE, Gordon AL, Wagner AJ, Heerema NA, Zhao W, Flynn JM, et al. Phosphatidylinositol 3-kinase-delta inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116:2078–88. doi: 10.1182/blood-2010-02-271171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.