Abstract
FBW7 is one of the most well characterized F-box proteins that serve as substrate recognition subunits of SCF (Skp1-Cullin 1-F-box proteins) E3 ubiquitin ligase complexes. SCFFBW7 plays key roles in regulating cell cycle progression, differentiation, and stem cell maintenance largely through targeting a broad range of oncogenic substrates for proteasome-dependent degradation. The identification of an increasing number of FBW7 substrates for ubiquitination, and intensive in vitro and in vivo studies have revealed a network of signaling components controlled by FBW7 that contributes to metabolic regulation as well as its tumor suppressor role. Here we mainly focus on recent findings that highlight a critical role for FBW7 in cancer and metabolism.
Keywords: FBW7, ubiquitination, SCF complex, tumor suppressor, stem cell, metabolism
1. Introduction
1.1 Ubiquitin-proteasome system (UPS)
Protein degradation is often essential for a rapid response to signal transduction and the recycling of amino acids as part of protein turnover. The vast majority of protein degradation is processed by the ubiquitin-proteasome system (UPS) [1]. Ubiquitin is an evolutionally conserved protein of 76 amino acids and covalently linked to target proteins in a multi-step process involving three key enzymes; an ubiquitin-activating enzyme (E1); an ubiquitin-conjugating enzyme (E2) and an ubiquitin ligase (E3). The E1 enzyme activates ubiquitin in an ATP-dependent manner resulting in a thioester bond between the ubiquitin protein and the E1. Sequentially, the C-terminus of ubiquitin binds with the Cys residue in the active site of an E2 enzyme, and then is covalently attached to the ε-amino group of the Lys residue on target molecules by the E3 ubiquitin ligase. Ubiquitin E3 ligases are classified into two major groups; the homologous to the E6AP carboxyl terminus (HECT) domain containing E3s and Really Interesting New Gene (RING) domain containing E3s. In contrast to HECT type E3s that form a thioester bond with ubiquitin, RING-type E3s directly conjugate ubiquitin from E2s to substrates. Although there are only two E1s and thirty-seven E2s, the human genome encodes over six hundred E3s, suggesting that E3 ligases functionally determine the substrate specificity for ubiquitination [2].
Ubiquitin can be added sequentially to form a polyubiquitination chain on the substrate protein. Since ubiquitin has seven Lys residues, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63, various types of polyubiquitination linkages are formed that can result in different physiological outcomes [3]. For instance, Lys48-linked polyubiquitinated proteins are recognized by the 26S proteasome, which is composed of a 20S core sub-complex, the 19S regulatory sub-complex, and the 11S sub-complex. Polyubiquitin chains bind to the 19S core particle and are then cleaved off from substrate proteins. Subsequently, the target protein is unfolded and degraded by peptidase in the 20S core subunit. Recent studies began to reveal that polyubiquitin chains composed of apical chain linkages via the other six Lys residues within ubiquitin are not only involved in protein degradation, but also play important roles in various cellular events including DNA repair response, endocytosis, and signal transduction [4].
1.2 SCF (Skp1-Cullin1-F-box protein) type of E3 ligase complexes
Cullin-RING ubiquitin ligases (CRLs) are one of the RING type E3 ligases, and are composed of a Cullin, RING finger protein, a variable substrate-recognition subunit, and an adaptor subunit. All Cullins contain a conserved domain in their C-terminal regions and binds to either RING-box protein 1 (Rbx1) or Rbx2, which recruits the E2 enzyme to transfer ubiquitin molecules to the target protein. In eukaryotes, eight types of Cullin (Cullin 1, 2, 3, 4A, 4B, 5, 7, and 9) have been identified and each Cullin forms a functionally distinct CRL complex. Since substrate recognition domains specifically recruit target molecules, they determine the substrate selectivity and are the largest contributor to the diversity of cellular functions of CRLs.
CRL1, which is also denoted as the Skp1-Cullin 1-F-box protein (SCF) E3 ligase complex, is the best-characterized member among all CRLs. The SCF complex contains the invariant components S-phase kinase-associated protein 1 (Skp1), Rbx1, and Cullin 1, as well as a variable substrate recognition subunit F-box protein (Figure 1A). So far, 69 F-box proteins have been identified in the human genome, and according to the substrate recognition domains, they are grouped into three major sub-classes; FBXW (WD40 repeat domain), FBXL (leucine-rich repeat domain), and FBXO (other various domains). Importantly, substrate recognition by F-box proteins often involves post-translational modifications of the target proteins such as phosphorylation or glycosylation [5]. SCF complexes often target key molecules involved in cell cycle progression and are thus considered one of the master regulators of the cell cycle machinery [5].
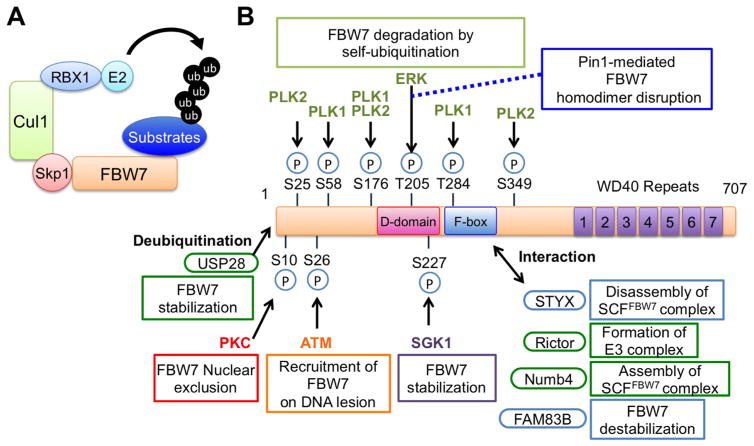
Figure 1.
(A) A schematic illustration of SCFFBW7 E3 ubiquitin ligase complex. (B) Upstream FBW7 signaling that modulates FBW7 function, stability, and subcellular localization. FBW7 E3 ligase activity is coordinately controlled by phosphorylation, ubiquitination, and interaction with key regulatory proteins.
1.3 The F-box protein FBW7
The F-box protein FBW7, also know as FBXW7 and Cdc4, is one of the most well-studied components of the SCF type of E3 ubiquitin ligases (Figure 1A). FBXW7 encodes three splicing variants, FBW7α, β and γ [6]. Each isoform differs in their N-terminal sequence but shares three conserved interaction domains; a D domain for promoting FBW7 dimerization, an F-box domain for recruitment of the SCF complex through Skp1, and a C-terminal WD40 repeat domain for substrate recognition. Thus, all FBW7 isoforms are considered to be functionally identical in principal. However, these isoforms show different subcellular localizations; FBW7α, β and γ localize in the nucleoplasm, cytoplasm and nucleolus, respectively [7]. In addition, tissue distribution also varies among these three isoforms. FBW7α is ubiquitously expressed in mice, whereas FBW7β is exclusively expressed in brain and testis, and FBW7γ is expressed in skeletal muscle and heart [8], which is consistent with the results of human multi-tissue Northern blot analysis [6].
FBW7 substrates typically contain a conserved Cdc4 phosphodegron (CPD) motif (L)-X-pT/pS-P-P-X-pS/pT/E/D (X represents any amino acid) [9, 10]. FBW7 recognizes and ubiquitinates its substrates in response to phosphorylation of this motif. In many cases, GSK3 phosphorylates the CPD motif of FBW7 substrates in concert with priming kinases, which in turn triggers SCFFBW7-directed substrate ubiquitination [11–13].
FBW7 is a well-established tumor suppressor that promotes the degradation of various oncogenic proteins such as cyclin E [14–16], c-Myc [17, 18], c-Jun [11, 19], and MCL1 [20, 21]. FBW7 is located within chromosomal region 4q32 that is frequently lost in cancers [6]. A comprehensive screening of over 1500 human cancers reveal that approximately 6% of all human cancers harbor FBW7 mutations [22]. Notably, mutations were frequently detected in cholangiocarcinomas (35%) and T cell acute lymphoblastic leukemia (T-ALL; 31%), and mutation frequencies in the range 6–9% were found in colon, endometrium, and stomach tumors. In human cancers, the most common missense mutations of FBW7 occurs at R465, R479, and R505 [22, 23], critical residues in the WD40 domain involved in substrate binding (Supplementary Figure 1), which strongly indicates that FBW7 dysfunction leads to tumorigenesis. A mammalian genetic screen for p53-dependent genes involved in tumorigenesis further revealed that Fbw7+/−mice were susceptible to radiation-induced tumorigenesis, and Fbw7+/−Tp53+/− mice have increased susceptibility, suggesting that FBW7 is likely a haploinsufficient tumor suppressor [24].
2. Roles of FBW7 in cancer
2.1 FBW7 downstream substrates
FBW7 targets multiple oncoproteins and oncogenic transcription factors for ubiquitination-mediated proteolysis (Supplementary Table 1). As such, dysregulation of FBW7-dependent proteolysis of these oncogenic proteins contribute to development of various cancers. Given the crucial function of FBW7 as a tumor suppressor, the list of FBW7 substrates still continue to grow (Supplementary Table 1), revealing roles for FBW7 in controlling multiple biological processes such as metastasis, stress responses, and immune functions.
SOX9 is a transcription factor that is involved in cell fate control and is frequently upregulated in various human cancers. In medulloblastoma, missense and nonsense FBW7 mutations are frequent events, and the deficiency of functional FBW7 leads to SOX9 stabilization, which in turn enhances metastatic potential and chemo-resistance [25]. Another independent study indicates that FBW7 is involved in DNA damage-induced SOX9 destabilization, further suggesting that deregulation of FBW7 function leads to cancer therapeutic resistance [26]. Heat shock factor 1 (HSF1) regulates transcription of heat shock proteins [27]. Depletion of FBW7 results in HSF1 stabilization and deregulation of heat shock responses, leading to augmented metastatic potential through altered transcriptional program in melanoma cells [27]. In addition, aberrant HSF1 hyper degradation by FBW7 is reported to contribute to protein misfolding that causes pathological condition of Huntington Disease [28].
The Forkhead transcription factor FOXM1 plays important roles in cell proliferation and cell cycle progression. It was reported that FBW7 negatively regulates Wnt signaling through degrading FOXM1 that promotes Wnt-induced β-catenin transcriptional activity [29]. Furthermore, GATA binding protein 3 (GATA3) is a transcription factor that regulates differentiation of a subtype of T-lymphocytes, and FBW7-mediated GATA3 degradation precisely controls differentiation of T-cell lineages [30]. On the other hand, FBW7 contributes to cilia formation by promoting degradation of NDE1, which is a negative regulator of ciliogenesis [31]. Notably, FBW7 depletion results in shortened cilia length indicating that FBW7 positively regulates cilia formation for proper reception of various extra-cellular stimuli. In addition, although early studies showed that tyrosine-protein phosphatase non-receptor type 11 (PTPN11) mediates the interaction between E3 ligase c-Cbl and its ubiquitin substrate RIG1 to promote its degradation, recent work revealed that FBW7 ubiquitinates PTPN11, which enables RIG1 to escape from c-Cbl-mediated RIG1 ubiquitination [32]. Consequently, myelomonocyte-specific Fbw7 knockout mice exhibit low interferon levels and therefore impaired antiviral immunity. Lastly, SCFFBW7 was reported to catalyze K63 linked poly-ubiquitination of XRCC4 [33]. In this experimental setting, DNA-damage-induced XRCC4 poly-ubiquitination by FBW7 facilitates the interaction of XRCC4 and Ku70/80 to repair DNA lesions.
2.2 Upstream regulation of FBW7 in cancer
FBW7 stability and activity are modulated by FBW7 dimerization, phosphorylation, ubiquitination, or regulatory protein interaction (Figure 1B). Dysfunction of the FBW7 regulatory mechanisms causes oncogenic substrate accumulation, leading to cancer. Disruption of the Cdc4 (Fbw7 yeast homolog) D-domain that is essential for dimer formation results in augmented Cdc4 self-ubiquitination and degradation [34]. Notably, phosphorylation is a key modification for controlling FBW7 protein stability. ERK phosphorylates Thr205 on FBW7 to enhance self-ubiquitination in human pancreatic cancer [35, 36]. Mechanistically, Pin1 binds to the phosphorylated Thr205 site and catalyzes FBW7 isomerization to disrupt FBW7 dimer formation, which in turn promotes FBW7 self-ubiquitination and subsequent degradation [35]. Furthermore, PLK1 catalyzes FBW7 phosphorylation at Ser58/Thr284 to promote FBW7 self-ubiquitination and degradation in human lung cancer [37]. As PLK1 is a direct Myc target gene, PLK1-dependent FBW7 downregulation creates the PLK1-Myc positive feedback loop to promote oncogenic events in lung cancer [37]. Likewise, PLK2-mediated phosphorylation at Ser25/176/349 promotes FBW7 destabilization and subsequent cyclin E accumulation, leading to centriole duplication in U2OS cells [38]. Conversely, SGK1-dependent Ser227 phosphorylation stabilizes FBW7 and enhances FBW7-mediated substrate ubiquitination in HCT116 and HEK293 cells [39, 40]. Besides protein stability control, phosphorylation of FBW7 affects its subcellular localization. To this end, ATM promotes FBW7 phosphorylation at Ser26, recruiting FBW7 to DNA lesions to facilitate the non-homologous end joining (NHEJ) complex formation in HCT116 and MIA PaCa-2 cells [33]. PKC-mediated phosphorylation at Ser10 that lies adjacent to the FBW7α nuclear localization signal (NLS) disrupts the function of the FBW7 NLS, resulting in exclusion of FBW7 from the nucleus in HeLa cells [41].
Moreover, FBW7 E3 ligase activity is controlled via interaction with critical modulators in cells (Figure 1B). To this end, identification of a deubiquitinase (DUB) that antagonizes FBW7 self-ubiquitination is important for understanding FBW7 regulatory mechanisms. Among the almost 100 DUB family members, ubiquitin specific protease 28 (USP28) catalyzes the removal of FBW7 poly-ubiquitin chain[42]. Specifically, USP28 interacts with and deubiquitinates FBW7 to suppress FBW7 self-degradation. At the same time, USP28 deubiquitinates and stabilizes FBW7 substrates in MEF and HeLa cells [43]. This dual regulation induces a transient activation of oncogenic FBW7 substrates and also serves as a safeguard to restrain unnecessarily prolonged oncoprotein stabilization. Thus, deregulated USP28 expression may cause aberrant oncoprotein accumulation, increasing a progression of downstream tumorigenic events [43].
Proper assembly of SCF complexes is a critical determinant of SCFFBW7 catalytic activity. FBW7 is incorporated into the SCF core complex via binding to SKP1 that serves as an adaptor between FBW7 and Cullin1, the scaffolding component of SCF complex. The pseudophosphatase STYX suppresses SCFFBW7 function through competing with SKP1 for binding to the F-box motif within FBW7, resulting in up-regulation of FBW7 substrates including MCL1, c-Myc, and cyclin E in HeLa cells [44]. In support of these biochemical data, STYX and FBW7 expression levels are inversely correlated in breast cancer clinical samples, and high STYX level reflects lower relapse-free survival rate of the patients.
Furthermore, Rictor, a component of mTORC2, was shown to form the E3 ligase complex with FBW7 to direct c-Myc and cyclin E degradation in HCT116 cells, which is independent of mTORC2 formation and function [45]. Moreover, SCFFBW7 is reported to recruit Numb4, a Numb isoform that is critical for cell fate decision, to promote SCF complex assembly thereby enhancing its E3 ligase activity towards Notch degradation in glioblastoma stem-like cells [46]. On the other hand, FAM83B, a chromokinesin interacting protein, interacts with FBW7 to promote FBW7 destabilization and reciprocal mTOR stabilization in breast cancer cell lines [47].
2.3 FBW7 function in somatic and cancer stem cells
Cancer stem cells exhibit similar properties to normal stem cells, such as stem cell pluripotency, self-renewal capacity, and multi-lineage differentiation potential [48, 49]. Multiple studies using in vitro and in vivo models reveal a broad spectrum of the contribution of FBW7 in stem cell maintenance [50], suggesting that FBW7 is involved in cancer stem cell regulation.
In embryonic stem (ES) cells, FBW7 is likely required for pluripotency, but not for self-renewal [51]. FBW7 knockdown affects only c-Myc abundance among the well-documented FBW7 substrates, and FBW7 depletion promotes ES cell differentiation in a c-Myc dependent manner. Consistently, FBW7 depletion enhances induced pluripotent stem cells (iPS cells) generation [51]. These findings highlight an important role of FBW7 in maintaining stem cell pluripotency and reprogramming.
Tissue-specific knockout mouse models have uncovered pivotal roles for Fbw7 in somatic stem cell regulation. To this end, intestine-specific heterozygous Fbw7 knockout mice reveal enhanced stem cell self-renewal, predisposing these mice to colorectal cancer [52, 53]. Fbw7 genetic ablation in the brain induces self-renewal of neuronal stem cells (NSC) due to Notch and c-Jun accumulation, resulting in abnormal brain development [54, 55]. The Notch downstream target Hes5 directly binds the FBW7β promoter to suppress its transcription, suggesting a model that a Notch-Hes5-Fbw7 positive feedback circuit is involved in maintaining proper NSC population [52]. On the other hand, Spermatogonia-specific FBW7 genetic ablation leads to increased self-renewal of spermatogonial stem cells (SSCs) [56] where Fbw7 plays a key role in suppressing self-renewal through c-Myc degradation [56].
Multiple mouse models also reveal that Fbw7 is essential for maintenance of hematopoietic stem cell (HSC). In HSC-specific Fbw7 conditional knockout mice, the HSC population aberrantly decreases due to exit of HSCs from quiescence [57–59]. These results suggest that a considerable portion (~30%) of the Fbw7 ablated mice display a phenotype of leukopenia in part due to aberrant HCS activation and subsequent HCS depletion, and eventually the remaining mice without leukopenia develop T-ALL during the late latency period. Notably, the observed leukopenia in the Fbw7 knockout mice is largely caused by c-Myc accumulation and consequent widespread p53-induced apoptosis [58]. Therefore, this finding indicates that Fbw7 and p53 serve as a fail-safe mechanism to prevent aberrant HSC activation and leukemogenesis. Interestingly, knock-in mice heterozygous for a cancer-derived missense R468C-Fbw7 mutation that is frequently found in the Fbw7 substrate recognition domain display normal bone marrow reconstitution. However, these mice develop T-ALL when crossed with mice carrying an oncogenic Notch1 truncation mutant [60]. These data suggest that the heterozygous hot spot missense mutation of Fbw7 requires additional oncogenic events or mutations to develop leukemia.
2.4 Therapeutic implications of FBW7 signaling
Given the crucial role for FBW7 in regulating tumorigenesis, targeting the FBW7 signaling pathway has attracted significant interest for developing potential effective therapeutics approaches for various types of human cancers.
FBW7 E3 ligase activity is regulated by post-translational modifications, suggesting that aberrant oncogenic upstream signaling might lead to inhibition of FBW7 tumor suppressive function. This also suggests that blockade of the upstream FBW7 negative regulators is a promising therapeutic approach to restore FBW7 tumore suppressive function. For instance, selective PLK1 inhibitors have significant effects on suppressing tumor growth through stabilizing FBW7 thereby downregulating N-Myc [37] (Figure 1B). Alternatively, targeting elevated FBW7 substrates, most of which are oncoproteins, could also be an effective therapeutic strategy in various tumors that associate with FBW7 inactivating mutations. For example, high MCL1 levels are frequently observed in FBW7 mutated tumors, and promote drug resistance [20, 21, 61]. Thus, developing specific MCL1 inhibitors [62] is a vital approach to restore drug sensitivity in FBW7 deficient cancers [62, 63].
3. Roles of FBW7 in metabolism
Downstream substrates of FBW7 such as c-Myc [17, 18] and sterol regulatory element-binding protein (SREBP) [64] play key roles in metabolic pathways, indicating that FBW7 regulates metabolism in both normal and disease conditions. Moreover, a recent study revealed that FBW7 is involved in circadian rhythm that is critical for hepatic metabolic function via targeting the clock protein REV-ERBα for degradation [65].
3.1 FBW7 function in regulating metabolism
3.1.1 c-Myc: the impact of FBW7 on glucose metabolism
The oncoprotein c-Myc is a well-known transcription factor regulating gene expression essential for cell cycle progression. Aside from this, c-Myc contributes to cancer growth by reprogramming cellular metabolism [66]. In particular, c-Myc enhances glucose uptake and glycolysis largely by inducing the expression of glucose transporters (GLUT1, GLUT2 and GLUT4), and a series of glycolytic enzymes such as hexokinase (HK2), phosphofructokinase (PFKM), enolase 1 (ENO1) and pyruvate kinase (PKM2) [67–69]. c-Myc also induces lactate dehydrogenase A (LDH-A) and the lactate transporter MCT1, thereby allowing efflux of glucose-derived carbon as lactate, which is an important process of glycolysis and tumor cell growth [70–72].
A recent study found that pancreatic cancer patients with decreased expression of FBW7 showed increased glucose metabolic activity in the tumor lesion [73]. FBW7 negatively regulates glucose turnover with decreased expression of key enzymes of the glycolysis cascade such as GLUT1, GLUT4, HK2, LDH-A, and LDH-B, in xenograft tumors as well as in pancreatic cancer cell lines. Gene expression profiling data and promoter analysis demonstrate that thioredoxin-binding protein (TXNIP), a suppressor of metabolic transformation, is upregulated by FBW7 ectopic expression and c-Myc degradation [73]. Mechanistically, TXNIP expression is negatively regulated by c-Myc, thereby FBW7 inhibits glucose metabolism by targeting the c-Myc/TXNIP axis in pancreatic cancer.
3.1.2 SREBPs: the impact of FBW7 on lipid metabolism
SREBP family of transcription factors plays a critical role in lipid metabolism by regulating the expression of a range of enzymes required for lipid synthesis. The SREBP family consists of three isoforms: SREBP1a, SREBP1c, and SREBP2 [74]. SREBP1a and 1c are preferentially involved in fatty acid biosynthesis, while SREBP2 primarily regulates genes in the cholesterol biosynthetic pathway [75]. The animal model and pharmacological studies have indicated that upregulation of SREBPs, especially SREBP1c, has a central role in the pathogenesis of the metabolic syndrome [76].
Transcriptionally active nuclear fragments of SREBPs (nSREBPs) are unstable and degraded by the ubiquitin-proteasome pathway [77]. Mechanistically, FBW7 promotes the degradation of nSREBPs in a GSK3 phosphorylation-dependent manner, thereby functioning as a negative regulator of lipid synthesis and metabolism [64, 78, 79]. Although it has been shown that GSK3-mediated phosphorylation of nSREBPs accelerates its turnover by creating a recognition site for FBW7, an additional layer of phosphorylation may antagonize this process during mitosis [80]. Specifically, the mitotic kinases Cdk1 and Plk1 sequentially phosphorylate multiple sites including a residue in close proximity of the CPD motif in SREBP1, which in turn disrupts the interaction of SREBP1 with FBW7 and attenuates FBW7-dependent nSREBP1 degradation during cell division [80]. The expression of SREBP target genes such as fatty acid synthase (FAS) is induced during mitosis, and inactivation of SREBP1 results in mitotic defects, suggesting FBW7-mediated SREBP1 degradation modulates cell division.
3.1.3 Other cell metabolism relevant ubiquitin substrates of FBW7
HIF-1α, a critical regulator of cellular response to hypoxia, exerts two major effects on metabolism. First, HIF-1α enhances glycolytic energy production by transactivating genes involved in extracellular glucose import (e.g. GLUT1) and glycolytic enzymes such as phosphofructokinase 1 (PFK1) and aldolase [81]. Second, HIF-1α downregulates the mitochondrial oxidative phosphorylation by transactivating genes including pyruvate dehydrogenase kinase 1 (PDK1) and MAX interactor 1 (MXI1) [81–84]. These two effects allow tumor cells to adapt hypoxic conditions by reducing O2 demand while still supplying sufficient energy to the cell. Thus, HIF-1α drives major metabolic changes within the tumor that are known as the Warburg effect [81, 85]. Two independent groups have reported that FBW7 can target HIF-1α for proteasomal degradation in a GSK3 phosphorylation-dependent manner, thereby modulating cell growth, migration, and angiogenesis as a negative regulator of HIF-1α [86, 87]. However, further studies are warranted to define the implications of FBW7/HIF-1α pathway in metabolic regulation.
PGC-1α is a key transcriptional coactivator that coordinates energy metabolism. A central feature of PGC-1α is its ability to promote mitochondrial biogenesis and oxidative metabolism [88]. Notably, dysfunction of PGC-1α has been implicated in the pathogenesis of metabolic diseases including diabetes and obesity [89–91]. Indeed, the reduced levels of PGC-1α are observed in pre-diabetic individuals, and considered to result in decreased mitochondrial function and the development of insulin resistance [92, 93]. PGC-1α level is regulated at multiple layers including protein stability. Olson et al. found that PGC-1α contains two CPD motifs, which are phosphorylated by GSK3 and p38 MAPK, respectively, leading to FBW7-dependent ubiquitination and proteasomal degradation of PGC-1α. FBW7 negatively regulates the PGC-1α-dependent transcriptional response to oxidative stress [94]. However, further studies are required to define the physiological relevance of FBW7/PGC-1α axis in metabolism, presumably with engineered mouse models.
mTOR has also been shown to be targeted for proteasomal degradation through a GSK3/FBW7-dependent mechanism, and this pathway contributes to the induction of autophagy [95]. Although a direct role of FBW7-mediated mTOR degradation in metabolism is poorly understood, one might speculate an important role of FBW7 in these metabolic processes given the impact of mTORC1 signaling on various metabolic pathways [96] through regulating FBW7 substrates such as SREBP [64], HIF-1α [86, 87], and PGC-1α [94]. Specifically, mTORC1 signaling can activate SREBP independently through both an S6K1-dependent mechanism [97] and phosphorylation of Lipin1, which inhibits SREBP-dependent transcription [98, 99]. Furthermore, mTORC1 increases the translation of HIF-1α [97], and enhances the transcriptional activity of YY1/PGC-1α complex by directly altering their physical interaction [100]. Thus, FBW7 may negatively regulate a number of metabolic pathways through mTOR degradation.
Furthermore, Ngn3 is a key transcription factor that regulates endocrine cell differentiation. Ngn3 also behaves as a canonical substrate that is degraded by GSK3/FBW7, and loss of FBW7 reprograms adult pancreatic ductal cells into insulin-secreting β cells through Ngn3 accumulation [101]. This evidence suggests that FBW7 can be involved in glycemic control through differentiation program for β cell neogenesis in the pancreas.
3.2 FBW7 function in hepatic metabolism
Liver-specific Fbw7 knockout mice (Fbw7-LKO mice) displayed hepatic steatosis phenotype with increased lipid deposits, suggesting a role for FBW7 in regulating liver metabolism [65, 102]. Mechanistically, hepatic knockout of Fbw7 results in accumulation of nuclear SREBP1, which is accompanied by expression changes of other lipogenic genes such as carbohydrate response element-binding protein (ChREBP), peroxisome proliferator-activated receptor gamma (Pparγ) as well as their downstream targets including fatty acid synthase (Fas), stearoyl CoA desaturase-1 (Scd1), LDL receptor (Ldlr), and HMG-CoA synthase (Hmgcs1), through a negative feedback loop [102]. Another line of in vivo evidence also indicates that FBW7 negatively regulates hepatic lipogenesis by degrading KLF5, an upstream transcription factor of PPARγ2 expression [103]. Given that the KLF5/PPAR axis is a key pathway of adipocyte differentiation and fatty acid oxidation in muscle, FBW7 may have important roles in metabolic pathways in those tissues [104–106].
REV-ERBα is a transcriptional suppressor that forms transcriptional complex integrating circadian rhythm and metabolic pathways [107]. Zhao et al. demonstrated that FBW7α promote the ubiquitination and subsequent degradation of REV-ERBα in a CDK1 phosphorylation-dependent manner, and this pathway contributes to controlling circadian amplitude critical for liver metabolism and whole-body energy homeostasis [65]. They further indicate that Fbw7-LKO mice show lower blood glucose levels and improved glucose tolerance despite a marked increase in hepatic steatosis [65]. This apparent discrepancy is likely attributed to the compromised hepatic gluconeogenesis in Fbw7-LKO mice that display disrupted circadian oscillation of both PEPCK and G6Pase [65]. In addition, FBW7-dependent control of REV-ERBα stability might be another layer of regulation for hepatic metabolism, since hepatic ablation of Fbw7 attenuated the amplitude of diurnal expression of many core clock genes including REV-ERBα targets such as Bmal1 and Clock, and altered expression of a large number of genes controlling liver metabolic pathways [65]. Intriguingly, the amplitude of diurnal expression of Insig2, a key component of the SREBP pathway [108] and also a direct REV-ERBα target gene [107], is reduced in Fbw7-LKO mice [65]. Thus, FBW7 is able to control hepatic SREBP function through regulation of REV-ERBα stability as well as direct control of SREBP protein levels by degradation. This evidence strengthens the physiological role of the FBW7/REV-ERBα pathway in regulating hepatic metabolism associated with whole-body energy homeostasis.
4. Conclusions
Targeting aberrant FBW7 signaling is a promising therapeutic strategy for various types of cancers. Recent studies reveal a broad spectrum of the tissue-specific and context-dependent FBW7 regulatory mechanisms and substrate selectivity regulating tumorigenesis. Thus, comprehensive understanding of FBW7 signaling is required to design effective therapeutic approaches targeting FBW7 deficiency-mediated tumorigenesis. Although a number of studies have focused on identifying targets of FBW7, emerging evidence for upstream regulators of FBW7 may provide new therapeutic options to restore or reactivate FBW7 tumor suppressor function (Figure 1B).
The role of FBW7 in metabolism is receiving increased attention. FBW7 may be capable of governing a broad range of metabolic pathways through targeting many key substrates including c-Myc, HIF-1α, SREBP, PGC-1α, mTOR, and REV-ERBα (Figure 2). Notably, many of these substrates are known to exert potent oncogenic roles through evoking aberrant metabolism. Metabolic alterations are indeed common features of cancer cells and have an important role in the maintenance of malignancies. For example, it has been well documented that the ability of cells to grow during hypoxia is regulated, in part, by metabolic outcomes of crosstalk between c-Myc and HIFs [72]. Thus, loss of FBW7 may contribute to metabolic preferences that benefit tumor growth through deregulation of c-Myc and HIF-1α. It is noteworthy that the discovery of REV-ERBα as a novel downstream substrate of FBW7 revealed a novel role for FBW7 in the regulation of circadian rhythm underlying normal metabolism. Given that changes in metabolism in cancer could be a consequence of a disrupted circadian rhythm, understanding the role of the FBW7/REV-ERBα axis in disease conditions such as cancer as well as metabolic disorders will be of importance for evaluating new therapeutic strategies targeting circadian clocks.
Figure 2.
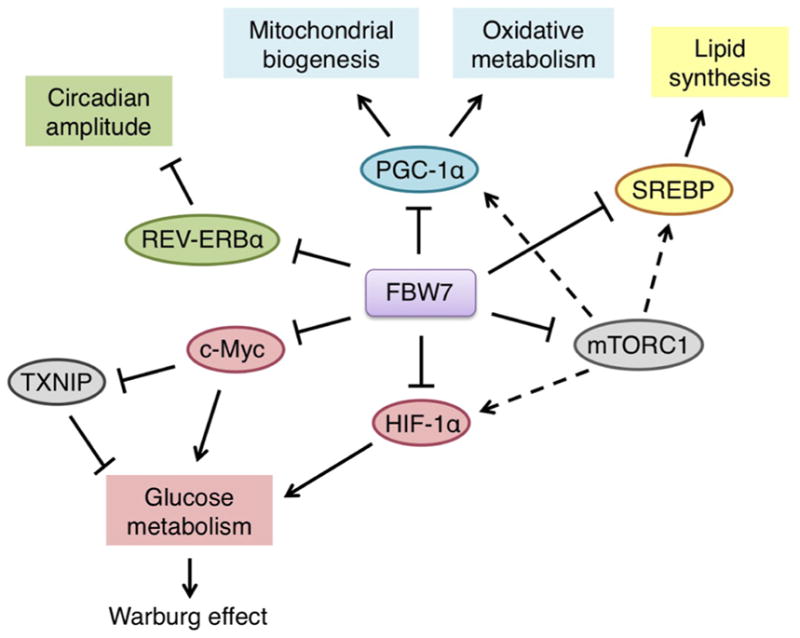
FBW7 ubiquitin substrates and their critical roles in cellular metabolism. FBW7 is capable of governing various metabolic pathways through promoting the degradation of key substrates such as c-Myc, HIF-1α, SREBP, PGC-1α, mTOR, and REV-ERBα, respectively.
In conclusion, recent studies have highlighted the physiological role of FBW7 in cellular maintenance through regulating differentiation, stemness, and metabolism. An ever-growing list of FBW7 substrates and upstream regulators will not only reveal the mechanisms of FBW7-asociated disease, but also offer many novel therapeutic targets to benefit cancer patients in the long run.
Supplementary Material
Highlights.
Physiological role of FBW7 in cellular maintenance through regulating differentiation, stemness, and metabolism.
Targeting aberrant FBW7 signaling is a promising anti-cancer therapeutic strategy
FBW7 is involved in the regulation of circadian rhythm underlying hepatic metabolism associated with whole-body energy homeostasis.
Acknowledgments
The authors apologize to our colleagues whose important work could not cited in this review due to space limitations. The authors thank Dr. Brian J. North for a careful proofreading of manuscript and Dr. Hidefumi Fukushima for critical comments and discussion.
This work was supported by NIH R01 grant (GM094777 and CA177910 to WW), American Cancer Society Research Scholar grants (to WW and HI), Charles H. Hood Foundation Child Health Research Awards Program (to HI). KS was supported by a postdoctoral fellowship from JSPS, The Uehara Memorial Foundation, and The Naito Foundation. NTN was supported by a postdoctoral fellowship from JSPS and The Osamu Hayaishi Memorial Scholarship for Study Abroad.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 3.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nature cell biology. 2016;18(6):579–86. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 4.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nature reviews. Cancer. 2014;14(4):233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spruck CH, Strohmaier H, Sangfelt O, Muller HM, Hubalek M, Muller-Holzner E, Marth C, Widschwendter M, Reed SI. hCDC4 gene mutations in endometrial cancer. Cancer research. 2002;62(16):4535–9. [PubMed] [Google Scholar]
- 7.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Current biology : CB. 2004;14(20):1852–7. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto A, Onoyama I, Nakayama KI. Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochemical and biophysical research communications. 2006;350(1):114–9. doi: 10.1016/j.bbrc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414(6863):514–21. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 10.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112(2):243–56. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 11.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, Clurman BE, Roberts JM. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Molecular cell. 2003;12(2):381–92. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 13.Taira N, Mimoto R, Kurata M, Yamaguchi T, Kitagawa M, Miki Y, Yoshida K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. The Journal of clinical investigation. 2012;122(3):859–72. doi: 10.1172/JCI60818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 15.Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413(6853):311–6. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 16.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413(6853):316–22. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 17.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23(10):2116–25. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303(5662):1374–8. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 20.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–9. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, Belmont LD, Kaminker JS, O'Rourke KM, Pujara K, Kohli PB, Johnson AR, Chiu ML, Lill JR, Jackson PK, Fairbrother WJ, Seshagiri S, Ludlam MJ, Leong KG, Dueber EC, Maecker H, Huang DC, Dixit VM. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 22.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O, Spruck C. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer research. 2007;67(19):9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 23.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer research. 2007;67(12):5611–6. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 24.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432(7018):775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 25.Suryo Rahmanto A, Savov V, Brunner A, Bolin S, Weishaupt H, Malyukova A, Rosen G, Cancer M, Hutter S, Sundstrom A, Kawauchi D, Jones DT, Spruck C, Taylor MD, Cho YJ, Pfister SM, Kool M, Korshunov A, Swartling FJ, Sangfelt O. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. The EMBO journal. 2016;35(20):2192–2212. doi: 10.15252/embj.201693889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong X, Liu W, Song R, Shah JJ, Feng X, Tsang CK, Morgan KM, Bunting SF, Inuzuka H, Zheng XF, Shen Z, Sabaawy HE, Liu L, Pine SR. SOX9 is targeted for proteasomal degradation by the E3 ligase FBW7 in response to DNA damage. Nucleic acids research. 2016;44(18):8855–8869. doi: 10.1093/nar/gkw748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourtis N, Moubarak RS, Aranda-Orgilles B, Lui K, Aydin IT, Trimarchi T, Darvishian F, Salvaggio C, Zhong J, Bhatt K, Chen EI, Celebi JT, Lazaris C, Tsirigos A, Osman I, Hernando E, Aifantis I. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nature cell biology. 2015;17(3):322–32. doi: 10.1038/ncb3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Pastor R, Burchfiel ET, Neef DW, Jaeger AM, Cabiscol E, McKinstry SU, Doss A, Aballay A, Lo DC, Akimov SS, Ross CA, Eroglu C, Thiele DJ. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington's disease. Nature communications. 2017;8:14405. doi: 10.1038/ncomms14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Li Y, Xue J, Gong A, Yu G, Zhou A, Lin K, Zhang S, Zhang N, Gottardi CJ, Huang S. Wnt-induced deubiquitination FoxM1 ensures nucleus beta-catenin transactivation. The EMBO journal. 2016;35(6):668–84. doi: 10.15252/embj.201592810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa K, Shibata K, Matsumoto A, Matsumoto M, Ohhata T, Nakayama KI, Niida H, Kitagawa M. Fbw7 targets GATA3 through cyclin-dependent kinase 2-dependent proteolysis and contributes to regulation of T-cell development. Molecular and cellular biology. 2014;34(14):2732–44. doi: 10.1128/MCB.01549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maskey D, Marlin MC, Kim S, Kim S, Ong EC, Li G, Tsiokas L. Cell cycle-dependent ubiquitylation and destruction of NDE1 by CDK5-FBW7 regulates ciliary length. The EMBO journal. 2015;34(19):2424–40. doi: 10.15252/embj.201490831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, Lai L, Chong Z, He J, Zhang Y, Xue Y, Xie Y, Chen S, Dong P, Chen L, Chen Z, Dai F, Wan X, Xiao P, Cao X, Liu Y, Wang Q. E3 ligase FBXW7 is critical for RIG-I stabilization during antiviral responses. Nature communications. 2017;8:14654. doi: 10.1038/ncomms14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Karnak D, Tan M, Lawrence TS, Morgan MA, Sun Y. FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Molecular cell. 2016;61(3):419–33. doi: 10.1016/j.molcel.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129(6):1165–76. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 35.Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, Shaik S, Lee DY, Finn G, Balastik M, Chen CH, Luo M, Tron AE, Decaprio JA, Zhou XZ, Wei W, Lu KP. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Molecular cell. 2012;46(6):771–83. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji S, Qin Y, Shi S, Liu X, Hu H, Zhou H, Gao J, Zhang B, Xu W, Liu J, Liang D, Liu L, Liu C, Long J, Zhou H, Chiao PJ, Xu J, Ni Q, Gao D, Yu X. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell research. 2015;25(5):561–73. doi: 10.1038/cr.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao D, Yue M, Su H, Ren P, Jiang J, Li F, Hu Y, Du H, Liu H, Qing G. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Molecular cell. 2016;64(3):493–506. doi: 10.1016/j.molcel.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 38.Cizmecioglu O, Krause A, Bahtz R, Ehret L, Malek N, Hoffmann I. Plk2 regulates centriole duplication through phosphorylation-mediated degradation of Fbxw7 (human Cdc4) Journal of cell science. 2012;125(Pt 4):981–92. doi: 10.1242/jcs.095075. [DOI] [PubMed] [Google Scholar]
- 39.Schulein C, Eilers M, Popov N. PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS letters. 2011;585(14):2151–7. doi: 10.1016/j.febslet.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 40.Mo JS, Ann EJ, Yoon JH, Jung J, Choi YH, Kim HY, Ahn JS, Kim SM, Kim MY, Hong JA, Seo MS, Lang F, Choi EJ, Park HS. Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. Journal of cell science. 2011;124(Pt 1):100–12. doi: 10.1242/jcs.073924. [DOI] [PubMed] [Google Scholar]
- 41.Durgan J, Parker PJ. Regulation of the tumour suppressor Fbw7alpha by PKC-dependent phosphorylation and cancer-associated mutations. The Biochemical journal. 2010;432(1):77–87. doi: 10.1042/BJ20100799. [DOI] [PubMed] [Google Scholar]
- 42.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nature cell biology. 2007;9(7):765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 43.Schulein-Volk C, Wolf E, Zhu J, Xu W, Taranets L, Hellmann A, Janicke LA, Diefenbacher ME, Behrens A, Eilers M, Popov N. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell reports. 2014;9(3):1099–109. doi: 10.1016/j.celrep.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 44.Reiterer V, Figueras-Puig C, Le Guerroue F, Confalonieri S, Vecchi M, Jalapothu D, Kanse SM, Deshaies RJ, Di Fiore PP, Behrends C, Farhan H. The pseudophosphatase STYX targets the F-box of FBXW7 and inhibits SCFFBXW7 function. The EMBO journal. 2017;36(3):260–273. doi: 10.15252/embj.201694795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Zhou Y, Evers BM, Wang Q. Rictor regulates FBXW7-dependent c-Myc and cyclin E degradation in colorectal cancer cells. Biochemical and biophysical research communications. 2012;418(2):426–32. doi: 10.1016/j.bbrc.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang X, Xing H, Kim TM, Jung Y, Huang W, Yang HW, Song S, Park PJ, Carroll RS, Johnson MD. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem cells. 2012;30(7):1313–26. doi: 10.1002/stem.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Liu Y, Zhang P, Zhang W, Wang W, Curr K, Wei G, Mao JH. FAM83D promotes cell proliferation and motility by downregulating tumor suppressor gene FBXW7. Oncotarget. 2013;4(12):2476–86. doi: 10.18632/oncotarget.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nature reviews. Cancer. 2008;8(10):755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 49.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–5. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Takeishi S, Nakayama KI. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. British journal of cancer. 2014;111(6):1054–9. doi: 10.1038/bjc.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M, Hochedlinger K, Chen EI, Aifantis I. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell stem cell. 2012;11(6):783–98. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sancho R, Blake SM, Tendeng C, Clurman BE, Lewis J, Behrens A. Fbw7 repression by hes5 creates a feedback loop that modulates Notch-mediated intestinal and neural stem cell fate decisions. PLoS biology. 2013;11(6):e1001586. doi: 10.1371/journal.pbio.1001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, Lewis A, Watson S, Behrens A, Tomlinson I, Nateri AS. FBXW7 influences murine intestinal homeostasis, cancer, targeting Notch, Jun, and DEK for degradation. The Journal of experimental medicine. 2011;208(2):295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoeck JD, Jandke A, Blake SM, Nye E, Spencer-Dene B, Brandner S, Behrens A. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nature neuroscience. 2010;13(11):1365–72. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 55.Matsumoto A, Onoyama I, Sunabori T, Kageyama R, Okano H, Nakayama KI. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. The Journal of biological chemistry. 2011;286(15):13754–64. doi: 10.1074/jbc.M110.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanatsu-Shinohara M, Onoyama I, Nakayama KI, Shinohara T. Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(24):8826–31. doi: 10.1073/pnas.1401837111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reavie L, Della Gatta G, Crusio K, Aranda-Orgilles B, Buckley SM, Thompson B, Lee E, Gao J, Bredemeyer AL, Helmink BA, Zavadil J, Sleckman BP, Palomero T, Ferrando A, Aifantis I. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nature immunology. 2010;11(3):207–15. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, Miyamoto K, Yoshiwara H, Hosokawa K, Nakamura Y, Gomei Y, Iwasaki H, Hayashi Y, Matsuzaki Y, Nakayama K, Ikeda Y, Hata A, Chiba S, Nakayama KI, Suda T. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes & development. 2008;22(8):986–91. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. The Journal of experimental medicine. 2008;205(6):1395–408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, Sandy P, Shen SS, Ferrando A, Aifantis I. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153(7):1552–66. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye M, Zhang Y, Zhang X, Zhang J, Jing P, Cao L, Li N, Li X, Yao L, Zhang J, Zhang J. Targeting FBW7 as a Strategy to Overcome Resistance to Targeted Therapy in Non-Small Cell Lung Cancer. Cancer research. 2017;77(13):3527–3539. doi: 10.1158/0008-5472.CAN-16-3470. [DOI] [PubMed] [Google Scholar]
- 62.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, Chanrion M, Kelly GL, Gong JN, Moujalled DM, Bruno A, Csekei M, Paczal A, Szabo ZB, Sipos S, Radics G, Proszenyak A, Balint B, Ondi L, Blasko G, Robertson A, Surgenor A, Dokurno P, Chen I, Matassova N, Smith J, Pedder C, Graham C, Studeny A, Lysiak-Auvity G, Girard AM, Grave F, Segal D, Riffkin CD, Pomilio G, Galbraith LC, Aubrey BJ, Brennan MS, Herold MJ, Chang C, Guasconi G, Cauquil N, Melchiore F, Guigal-Stephan N, Lockhart B, Colland F, Hickman JA, Roberts AW, Huang DC, Wei AH, Strasser A, Lessene G, Geneste O. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538(7626):477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 63.Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36(6):787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell metabolism. 2005;1(6):379–91. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 65.Zhao X, Hirota T, Han X, Cho H, Chong LW, Lamia K, Liu S, Atkins AR, Banayo E, Liddle C, Yu RT, Yates JR, 3rd, Kay SA, Downes M, Evans RM. Circadian Amplitude Regulation via FBXW7-Targeted REV-ERBalpha Degradation. Cell. 2016;165(7):1644–57. doi: 10.1016/j.cell.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee GY, Chun YS, Shin HW, Park JW. Potential role of the N-MYC downstream-regulated gene family in reprogramming cancer metabolism under hypoxia. Oncotarget. 2016;7(35):57442–57451. doi: 10.18632/oncotarget.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Seminars in cancer biology. 2006;16(4):253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 68.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. The Journal of biological chemistry. 2000;275(29):21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 69.Tarrado-Castellarnau M, de Atauri P, Cascante M. Oncogenic regulation of tumor metabolic reprogramming. Oncotarget. 2016;7(38):62726–62753. doi: 10.18632/oncotarget.10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(13):6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doherty JR, Yang C, Scott KE, Cameron MD, Fallahi M, Li W, Hall MA, Amelio AL, Mishra JK, Li F, Tortosa M, Genau HM, Rounbehler RJ, Lu Y, Dang CV, Kumar KG, Butler AA, Bannister TD, Hooper AT, Unsal-Kacmaz K, Roush WR, Cleveland JL. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer research. 2014;74(3):908–20. doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer cell. 2007;12(2):108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji S, Qin Y, Liang C, Huang R, Shi S, Liu J, Jin K, Liang D, Xu W, Zhang B, Liu L, Liu C, Xu J, Ni Q, Chiao PJ, Li M, Yu X. FBW7 (F-box and WD Repeat Domain- Containing 7) Negatively Regulates Glucose Metabolism by Targeting the c-Myc/TXNIP (Thioredoxin-Binding Protein) Axis in Pancreatic Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(15):3950–60. doi: 10.1158/1078-0432.CCR-15-2380. [DOI] [PubMed] [Google Scholar]
- 74.Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86(11):839–48. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 75.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soyal SM, Nofziger C, Dossena S, Paulmichl M, Patsch W. Targeting SREBPs for treatment of the metabolic syndrome. Trends in pharmacological sciences. 2015;36(6):406–16. doi: 10.1016/j.tips.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Hirano Y, Yoshida M, Shimizu M, Sato R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. The Journal of biological chemistry. 2001;276(39):36431–7. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- 78.Bengoechea-Alonso MT, Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. The Journal of biological chemistry. 2009;284(9):5885–95. doi: 10.1074/jbc.M807906200. [DOI] [PubMed] [Google Scholar]
- 79.Dong Q, Giorgianni F, Beranova-Giorgianni S, Deng X, O'Meally RN, Bridges D, Park EA, Cole RN, Elam MB, Raghow R. Glycogen synthase kinase-3-mediated phosphorylation of serine 73 targets sterol response element binding protein-1c (SREBP-1c) for proteasomal degradation. Bioscience reports. 2015;36(1):e00284. doi: 10.1042/BSR20150234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bengoechea-Alonso MT, Ericsson J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell cycle. 2016;15(20):2753–65. doi: 10.1080/15384101.2016.1220456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nature reviews. Cancer. 2008;8(9):705–13. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 82.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell metabolism. 2006;3(3):177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 83.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell metabolism. 2006;3(3):187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 84.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer cell. 2007;11(5):407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–70. [PubMed] [Google Scholar]
- 86.Cassavaugh JM, Hale SA, Wellman TL, Howe AK, Wong C, Lounsbury KM. Negative regulation of HIF-1alpha by an FBW7-mediated degradation pathway during hypoxia. Journal of cellular biochemistry. 2011;112(12):3882–90. doi: 10.1002/jcb.23321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flugel D, Gorlach A, Kietzmann T. GSK-3beta regulates cell growth, migration and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1alpha. Blood. 2012;119(5):1292–301. doi: 10.1182/blood-2011-08-375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 89.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 90.Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS biology. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. The Journal of clinical investigation. 2006;116(3):615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 93.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes & development. 2008;22(2):252–64. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fu L, Kim YA, Wang X, Wu X, Yue P, Lonial S, Khuri FR, Sun SY. Perifosine inhibits mammalian target of rapamycin signaling through facilitating degradation of major components in the mTOR axis and induces autophagy. Cancer research. 2009;69(23):8967–76. doi: 10.1158/0008-5472.CAN-09-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saxton RA, Sabatini DM. mTOR, Signaling in Growth, Metabolism and Disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39(2):171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shimizu K, Fukushima H, Ogura K, Lien EC, Nihira NT, Zhang J, North BJ, Guo A, Nagashima K, Nakagawa T, Hoshikawa S, Watahiki A, Okabe K, Yamada A, Toker A, Asara JM, Fukumoto S, Nakayama KI, Nakayama K, Inuzuka H, Wei W. The SCFbeta- TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Science signaling. 2017;10(460) doi: 10.1126/scisignal.aah4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450(7170):736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 101.Sancho R, Gruber R, Gu G, Behrens A. Loss of Fbw7 reprograms adult pancreatic ductal cells into alpha, delta and beta cells. Cell stem cell. 2014;15(2):139–53. doi: 10.1016/j.stem.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Onoyama I, Suzuki A, Matsumoto A, Tomita K, Katagiri H, Oike Y, Nakayama K, Nakayama KI. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. The Journal of clinical investigation. 2011;121(1):342–54. doi: 10.1172/JCI40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumadaki S, Karasawa T, Matsuzaka T, Ema M, Nakagawa Y, Nakakuki M, Saito R, Yahagi N, Iwasaki H, Sone H, Takekoshi K, Yatoh S, Kobayashi K, Takahashi A, Suzuki H, Takahashi S, Yamada N, Shimano H. Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7alpha (Fbw7alpha) causes hepatosteatosis through Kruppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor gamma2 (PPARgamma2) pathway but not SREBP-1c protein in mice. The Journal of biological chemistry. 2011;286(47):40835–46. doi: 10.1074/jbc.M111.235283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, Suzuki R, Kitamura T, Akira S, Kadowaki T, Nagai R. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell metabolism. 2005;1(1):27–39. doi: 10.1016/j.cmet.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 105.Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R. SUMOylation of Kruppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-delta. Nature medicine. 2008;14(6):656–66. doi: 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- 106.Drosatos K, Pollak NM, Pol CJ, Ntziachristos P, Willecke F, Valenti MC, Trent CM, Hu Y, Guo S, Aifantis I, Goldberg IJ. Cardiac Myocyte KLF5 Regulates Ppara Expression and Cardiac Function. Circulation research. 2016;118(2):241–53. doi: 10.1161/CIRCRESAHA.115.306383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS biology. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.