Abstract
Rationale and Objectives
The purpose of this study is to improve accuracy of near-term breast cancer risk prediction by applying a new mammographic image conversion method combining with a two-stage artificial neural network (ANN) based classification scheme.
Materials and Methods
The dataset included 168 negative mammography screening cases. In developing and testing our new risk model, we first converted the original grayscale value (GV) based mammographic images into optical density (OD) based images. For each case, our computer-aided scheme then computed two types of image features representing bilateral asymmetry and the maximum of the image features computed from GV and OD images, respectively. A two-stage classification scheme consisting of three ANNs was developed. The first stage includes two ANNs trained using features computed separately from GV and OD images of 138 cases. The second stage includes another ANN to fuse the prediction scores produced by two ANNs in the first stage. The risk prediction performance was tested using the rest 30 cases.
Results
with the two-stage classification scheme, the computed area under the receiver operating characteristic curve was AUC=0.816±0.071, which was significantly higher than the AUC values of 0.669±0.099 and 0.646±0.099 achieved using two ANNs trained using GV features and OD features, respectively (p < 0.05).
Conclusion
This study demonstrated that applying an optical density image conversion method can acquire new complimentary information to those acquired from the original images. As a result, fusion image features computed from these two types of images enabled to yield significantly higher performance in near-term breast cancer risk prediction.
Keywords: Breast cancer, Computer-aided detection (CAD), Risk stratification, Image conversion, Feature analysis
I. INTRODUCTION
Due to the low cancer detection yield (i.e., < 0.5%) and high false-positive recall rates (i.e., ≥ 10%), efficacy of current population-based mammography screening paradigm remains quite controversial1–4. In order to solve this clinical dilemma, developing a new optimal personalized breast cancer screening paradigm to detect early cancers has attracted extensive research interest recently5,6. The goal of developing this new screening paradigm is to more effectively identify a small fraction of women who have significantly higher risk of developing breast cancer in the near-term than the average population. As a result, each woman should have an adaptive screening interval. Only a small fraction of “high-risk” women should be screened more frequently, whereas the vast majority of “low-risk” women should be screened at longer intervals until their near-term cancer risk significantly increases in new assessment4, 8.
The success of establishing an optimal personalized breast cancer screening paradigm depends on a reliable risk prediction model. Although many epidemiology study-based breast cancer risk models, such as Gail, Claus, and Tyrer-Cuzick model1,7 have been developed, none of them has a discriminatory power to determine which women should receive screening mammography in the near-term10. Thus, establishing an optimal personalized breast cancer screening paradigm requires to develop and test new risk prediction models that have significantly higher discriminatory power in predicting the risk of individual women developing breast cancer in the near-term8. Based on the computed quantitative image features, several research groups have developed and tested a number of new risk stratification models to predict breast cancer risk9–13.
In our studies, we hypothesized that bilateral mammographic density or tissue based image feature asymmetry between the left and right breasts is an important radiographic image phenotype related to abnormal biologic processes that may lead to cancer development. Thus, increase in bilateral mammographic density asymmetry could be an important indicator of developing breast abnormality or cancer 4. To test this hypothesis, we have developed several computer-aided image processing and feature analysis schemes to detect bilateral mammographic density related image feature asymmetry between the left and right breasts, and preliminarily investigated the association between bilateral mammographic image feature asymmetry scores and the risk of a woman developing breast cancer in a near-term (e.g., ≤ 3 years) after a negative screening examination14–16. Another type of features are maximum features, which also can be used to detect structural and textural changes within breast regions due to abnormalities that are starting to develop. The greater one of the two values of the same feature was chosen to represent the region-based maximum feature value of the two bilateral breasts. It is observed that abnormalities that start to develop in one breast can be detected sensitively by extracting the maximum features of both breasts8. However, how to optimally detect bilateral mammographic asymmetry features and maximum features and thus improve accuracy in predicting near-term breast cancer risk remains a difficult and unsolved issue.
In this study, we focused on investigating a new approach aiming to more accurately detect bilateral mammographic image features and testing whether it can help further improve the performance of applying quantitative image feature analysis methods to predict near-term breast cancer risk. In previous studies, the features used for predicting breast cancer risk were only computed on the original mammographic images. In fact, the original mammographic images could be converted or mapped into other images, which may be used to compute features with higher discriminatory power4,15,17. Thus, we proposed to convert the original grayscale values (GV) mammograms into optical density (OD) images. We computed asymmetry features and maximum features on these two different types of GV and OD images, and then trained and tested two artificial neural networks (ANNs) using these two groups of image features computed from the OD images and the GV images, respectively. Therefore, we investigated whether applying this image conversion approach yield higher performance in predicting near-term breast cancer risk. Moreover, we developed and tested a unique two-stage classification scheme. The first stage includes the two ANNs trained using the GV features and the OD features separately. The outcomes of the two ANNs were then fused into the third ANN in the second stage to generate the final risk prediction score for each testing case. As a result, we also investigated whether applying this two-stage classification scheme further improve risk prediction performance.
II. MATERIALS AND METHODS
II.A. A dataset
Our risk model was built based only on the bilateral image features and their asymmetry computed from the negative screening mammograms. We retrospectively assembled a testing image dataset for this study, which includes two sequential mammographic screening examinations acquired from 168 women. Each examination includes four full-field digital mammography (FFDM) images representing the craniocaudal (CC) and mediolateral oblique (MLO) view of the left and right breasts. All the images were acquired using Hologic Selenia FFDM systems (Hologic Inc., Bedford, MA, USA) with 12-bit resolution. From the results of the latest FFDM examinations on record (or namely, the “current” images), 83 were positive cases (with cancer detected and verified) and 85 were negative cases (cancer-free). TABLE I summarizes the distribution of women’s age, breast density (BIRADS rating), and lesion type that were detected on the “current” examinations.
TABLE I.
Distribution of women’s age, breast density (BIRADS rating), and lesion type that were detected on the “current” examinations.
| Risk factor | Positive cases | Negative cases |
|---|---|---|
| Woman’s age (years old) | ||
| A≤40 | 4 (4.8%) | 8 (9.4%) |
| 40<A≤50 | 25 (30.1%) | 38 (44.7%) |
| 50<A≤60 | 22 (26.5%) | 21 (24.7%) |
| 60<A≤70 | 19 (22.9%) | 10 (11.8%) |
| 70<A≤80 | 11 (13.3%) | 7 (8.2%) |
| A>80 | 2 (2.4%) | 1 (1.2%) |
| Density BIRADS rating | ||
| Almost all fatty tissue (1) | 3 (3.6%) | 1 (1.2%) |
| Scattered fibroglandular densities (2) | 22 (26.5%) | 20 (23.5%) |
| Heterogeneously dense (3) | 56 (67.5%) | 57 (67.1%) |
| Extremely dense (4) | 2 (2.4%) | 7 (8.2%) |
| Type of Lesion | ||
| No lesion (0) | 2 (2.4%) | 71 (83.5%) |
| Mass/Assymetry (1) | 30 (36.1%) | 7 (8.2%) |
| Calcifications (2) | 26 (31.3%) | 5 (5.9%) |
| Architectural Distortion (3) | 1 (1.2%) | 0 (%) |
| Calcifications + mass/assym (4) | 24 (28.9) | 2 (2.4%) |
For each case, the most recent (or first “prior”) FFDM screening images were also retrieved. The intervals between the “prior” and “current” examinations are 410.0±51.7 days and 400.1±45.0 days for positive cases and negative cases, respectively. All “prior” FFDM examinations were interpreted and rated by radiologists as “negative” or “definitely benign” (i.e., screening BIRADS 1 or 2). Thus, all these “prior” examinations were not recalled. Thus, only these “prior” negative screening images were used for computing mammographic image features and building our new near-term breast cancer risk prediction model. Although all these “prior” images were negative, they were divided into two different classes based on their status changes in the next sequential FFDM examinations (“current” images). The cases were divided into subgroups of “positive” (cancer detected in “current” images) and “negative” (i.e., remained cancer-free) cases.
Although a FFDM examination typically includes four images of CC and MLO view of left and right breasts, applying a computerized scheme to detect mammographic density or tissue related image features using CC view images is often more accurate and reliable than using MLO view images as demonstrated in the previous studies15. Hence, due to the use of a limited dataset, we only selected and processed a pair of bilateral CC view FFDM images of left and right breasts from each of the “prior” examinations in this study.
II.B. Methods
The new near-term breast cancer risk prediction model was proposed to detect the cases with high possibility of having highly suspicious breast abnormalities that are detectable in the next sequential screening examination. Specifically, our new risk model was developed using four steps including (1) segmenting the whole and dense breast regions from the GV images, (2) creating OD images on the segmented breast regions, (3) computing two groups of features on the GV images and the OD images, respectively, and (4) optimizing a two-stage ANN based classification scheme.
II.B.1 Breast region segmentation
For each case, a pair of bilateral CC view FFDM images of the left and right breasts were analyzed. We first applied an automatic segmentation scheme on each image to extract the whole breast region as described in Ref 18,19. In brief, a gray level histogram of the image was plotted and an iterative searching method was used to detect the smoothest curvature between the breast tissue and background or air region. The pixels in the background were discarded and the skin region was removed by a morphological erosion operation.
Second, we segmented the dense breast region from each image. The dense breast region was defined as the region of the breast that encompasses the pixel values above the mean value of the whole breast region. Unlike the previous studies in which the thresholds used for segmenting dense regions of the left and right breast mammographic images were separately selected from each image, which generally have different values, we in this study defined a new method to segment dense regions of left and right breast mammographic images using a single “mutual” threshold instead of two different individual thresholds. The mean grayscale value of all pixels in the two whole breast regions of the left and right mammographic images was defined as the “mutual” threshold.
II.B.2 Optical density image generation
We generated an OD image for each GV image. There are a number of published methods that have been used to convert grayscale values into optical densities. The first one is a relative optical density (ROD). All the original FFDM images were acquired with 12-bit resolution. The grayness is expressed as a gray value (GV) on the linear scale between 0 and 4095. The GV was transformed into the ROD with the following equation, which is a nonlinear transformation of GV values to OD values without the use of a standard20–23.
| (1) |
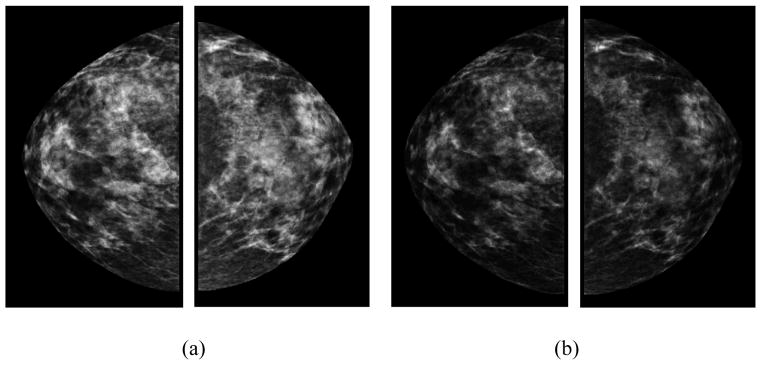
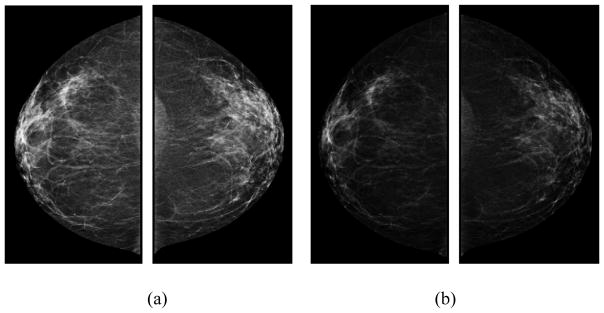
The ROD units display the same log-reciprocal relationship to light transmittance as the true OD and are reported to be closely correlated with the true OD values20. In this study, we used (1) to convert GV images into OD images. The purpose of these conversions is to account for the nonlinear relationship between the darkness of a digital image and the number of particles striking the photodetector from radioactive decay. Because these transformations are nonlinear, they have the effect of making high values in the data disproportionately higher, which may provide useful and complementary information to the original mammographic images. For example, FIG. 1 presents a positive case showing a set of bilateral original grayscale value images and a set of bilateral optical density images. FIG. 2 shows a negative case. Two examples show visual difference between GV and OD images, which will generate different feature values.
FIG. 1.
An example of a positive case showing (a) bilateral original grayscale value images and (b) optical density images in CC view.
FIG. 2.
An example of a negative case showing (a) bilateral original grayscale value images and (b) optical density images in CC view.
II.B.3 Image feature computation
In the literature, various studies have been conducted to compute and assess many mammographic density-related image features24–27. In this study, we computed some mammographic image features that have been proposed in the literature, as well as redefined some existing features and explored some new features that have never been examined and used before for the task of breast cancer risk prediction. The image features were computed from both GV images and OD images. Specifically, our computer-aided scheme computed the same 148 features from each of the 4 pairs of different ROIs namely, (1) the segmented whole breast regions and (2) the segmented dense breast regions in the bilateral GV images, which are named as WBR-GV and DBR-GV images, respectively (In the following, these features will be name as , (i = 1, 2, …, 148×2)), (3) the segmented whole breast regions and (4) the segmented dense breast regions in the bilateral OD images, which are named as WBR-OD and DBR-OD images, respectively (In the following, these features will be name as , (i = 1, 2, …, 148×2)).
Each group of 148 features were divided into two distinct types namely: (1) 76 bilateral mammographic tissue asymmetry features, which were represented by the absolute subtraction of two matched bilateral feature values computed from the left and right images; (2) 72 maximum features, which were represented by the greater feature value of two matched bilateral feature values. In the following, the two types of image features will be mentioned as asymmetry features and maximum features, respectively.
Type 1: Asymmetry features
The asymmetry features can be divided into a number of subgroups. The first subgroup include13 image statistics based features ( and , i = 1, 2, …, 13)15. The second subgroup includes 4 fractal dimension related breast tissue composition features ( and , i = 14, 15, 16, 17)17. The third subgroup has 39 texture related image features ( and , i = 18, 19, …, 56)28, which include 8 Gray-level co-occurrence matrix based features, 13 Gray-level run-length matrix based features, 13 Gray-level size zone matrix based features, and 5 Neighborhood gray-tone difference matrix based features. The fourth subgroup includes 16 features ( or , i = 57, 58, …, 72), which we modified to compute a variety of percentage of mammographic density (PMD) for mimicking BIRADS used in clinical practice. The last subgroup includes 4 newly explored features. Specific definition of the features could be seen in Appendix A.
From each pair of matched ROIs of left and right breasts, the first 72 asymmetry features ( to (or to )) were independently computed for the left and right breasts. Each bilateral mammographic density asymmetric feature was computed by the absolute difference (subtraction) of two feature values computed from the two matched bilateral images. The last 4 asymmetry features were computed directly from each pair of matched bilateral images.
Type 2: Maximum features
This type of features includes another set of 72 features ( to (or to )), which were similar to the features in type 1 ( to (or to )). From each of two matched ROIs of left and right breasts, each of the 72 features was also independently computed. Then, the greater one of the two values of the same feature was chosen to represent the region-based maximum feature value of the two bilateral breasts. As reported in Ref 8, it is observed that abnormalities that start to develop in one breast can be detected more sensitively by extracting the maximum features of both breasts. The maximum features can be used to detect structural and textural changes within breast regions due to abnormalities that are starting to develop, but have not matured or developed fully yet.
II.B.4 Risk prediction and performance assessment
All above computed image features of 168 cases were normalized based on their mean values and standard deviations to the range of 0 to 1. The normalized image features were then divided into two groups: features of 138 training cases and features of 30 testing cases (15 positive cases and 15 negative cases). Because there may be some redundant features that are of low discriminatory power for breast cancer risk prediction, we selected two small sets of “optimal” image features with higher discriminatory power from the complete sets of GV features and OD features of the 138 training cases, respectively. The feature selection were performed using a publicly available WEKA data mining and machine learning software package30. In the WEKA Explorer window, we chose “CfsSubsetEval” as attribute evaluator and “BestFirst” as search method. “CfsSubsetEval” means evaluating the worth of a subset of attributes by considering the individual predictive ability of each feature along with the degree of redundancy between them). “BestFirst” means searching the space of attribute subsets by greedy hillclimbing augmented with a backtracking facility.
In order to investigate predictive power of the selected features, we tested 3 simple feature fusion methods (taking the average, maximum, and minimum value of the selected features) 29 on the same-label(number) GV and OD features of the 30 testing cases to generate 3 sets of new classification scores. These scores were used as potential image based markers for predicting near-term breast cancer risk. The prediction performance was evaluated using AUC. The AUCs were computed applying a publically available ROC curve fitting program (ROCKIT, http://www-radiology.uchicago.edu/krl/, University of Chicago).
Many computer aided schemes have been developed for disease diagnosis and prediction using different classifiers including linear discriminant analysis (LDA), support vector machines (SVMs), artificial neural networks (ANNs), Bayesian belief networks, and rule-based classifiers. In the literature, ANNs are the most popular classifiers that have been widely explored for classification 8, 16, 28, 35. In this study, the “optimal” GV and OD features (selected from the 138 training cases) were also used to train a “two-stage” classification scheme including 3 ANNs to predict near-term breast cancer risk. As shown in FIG. 3. The first stage has 2 ANNs that were independently trained using the GV features and the OD features, respectively. Specifically, the first ANN was trained using the selected “optimal” GV features; while the second ANN was trained using the selected “optimal” OD features. In the second stage, we trained the third ANN using the 2 risk prediction scores produced by the 2 ANNs trained in the first stage as input. In order to investigate performance of the “two-stage” classification scheme, we tested the trained ANNs on the selected small sets of GV and OD features of the 30 testing cases that have the same labels(numbers) with the features selected from the 138 training cases to generate 1 set of new classification scores. The scores were used as potential image based markers for predicting near-term breast cancer risk.
FIG. 3.
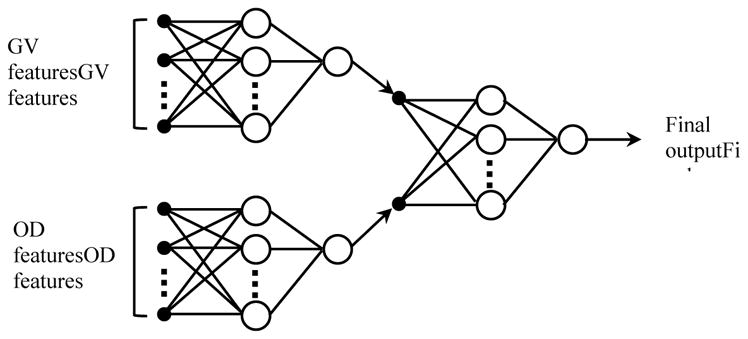
A two-stage “scoring fusion” RBFN-based ANN classification scheme, whereby the final classification score is derived by optimally fusing the prediction scores produced by 2 ANNs trained using GV features and OD features, respectively.
All ANN based training and testing experiments were performed using WEKA. In the WEKA Explorer window, we chose Gaussian Radial Basis Function Network (RBFN) as the classifier. RBFN is a forward-feed 3-layer ANN which uses radial basis functions as activation functions32, which has been used in numerous applications for the pattern identification tasks. It has drawn much attention due to its good generalization ability and simple network structure33. Thus, an optimally trained RBFN enables to accelerate learning process and minimize the risk of trapping inside the local minimum. Past research of universal approximation theorems on RBFN has shown that any nonlinear function over a compact set with arbitrary accuracy can be approximated by RBFN34, which makes it a powerful tool for hazard risk assessment35.
AUC of the ROC curve was also used to assess the discriminatory power or classification performance of the prediction scores generated by the two-stage scheme, as well as that generated by the one-stage scheme based on GV features, OD features, and both GV and OD features. In addition, since in breast cancer screening practice, the healthcare professional or the women who participate in mammography screening need to make a binary decision (“Yes” or “No”) on whether an individual women need to take another annual mammography screening following a recent negative screening result in question, we also assessed and compared the overall prediction accuracies on all 30 testing cases, as well as on 15 “positive” cases and 15 “negative” cases by applying an operation threshold of 0.5 on the prediction scheme or model-generated risk prediction scores, which range from 0 to 1, to divide the 30 testing cases into two prediction case groups (with and without image detectable cancer in the next sequential annual screening). The computed accuracy results are tabulated and compared.
III. RESULTS
TABLE II lists two small sets of individual OD and GV features that were selected based on the 138 training cases. One set (on the right) of 9 features were selected from the pool of GV features. Among the 9 features, 5 ones are asymmetry features and 4 ones are maximum features; 4 were computed from the dense breast regions and 5 were computed from the whole breast regions. Another set (on the left of TABLE II) of 10 features were selected from the pool of OD features. Among the 10 features, 7 ones are asymmetry features and 3 ones are maximum features; 5 were computed from the dense breast regions and 5 were computed from the whole breast regions.
TABLE II.
Mammographic image features computed from GV images and OD images that were selected.
| OD features | GV features | ||||
|---|---|---|---|---|---|
| Feature number | ROI | Feature subgroup | Feature number | ROI | Feature subgroup |
| 52 | WBR | asymmetry | 59 | WBR | asymmetry |
| 59 | WBR | asymmetry | 60 | WBR | asymmetry |
| 60 | WBR | asymmetry | 100 | WBR | maximum |
| 85 | WBR | maximum | 128 | WBR | maximum |
| 86 | WBR | maximum | 136 | WBR | maximum |
| 157 | DBR | asymmetry | 157 | DBR | asymmetry |
| 175 | DBR | asymmetry | 158 | DBR | asymmetry |
| 218 | DBR | asymmetry | 220 | DBR | asymmetry |
| 219 | DBR | asymmetry | 253 | DBR | maximum |
| 230 | DBR | maximum | |||
TABLE III compares AUC values of different schemes. In the middle column and the left column, AUC values of 4 different schemes are presented. Based on the GV features (middle column), AUC values ranged from 0.550±0.105 to 0.669±0.099. Based on the OD features (left column), AUC values ranged from 0.605±0.107 to 0.646±0.099. For each of the two sets of AUCs, the one yielded by the ANN based one-stage classification scheme is higher than those yielded by the simple fusion methods, but the differences are not statistically significant. As a whole, there is not a significant difference between the two sets of AUC values. The one-stage classification scheme based on both GV and OD features yielded an AUC value of 0.697±0.098 (presented in the right column). The two-stage classification scheme yielded an AUC value of 0.816±0.071 (presented in the right column), which is significantly higher than the AUC values yielded by either of two one-stage classification schemes using GV features and OD features each (P<0.05). The AUC value of the two-stage classification scheme is also much higher than that of the GV-and-OD-feature based one-stage classification scheme, but the difference is not statistically significant (P>0.05).
TABLE III.
Comparison of AUC values and corresponding standard deviations of 5 different prediction schemes by using GV features and OD features.
| Feature fusion method | Based on OD features | Based on GV features | Based on GV and OD features |
|---|---|---|---|
| Scheme 1 (Taking maximum value) | \ | 0.550±0.105 | \ |
| Scheme 2 (Taking minimum value) | \ | 0.660±0.104 | \ |
| Scheme 3 (Taking mean value) | 0.605±0.107 | 0.621±0.105 | \ |
| Scheme 4 (One-stage classification scheme) | 0.646±0.099 | 0.669±0.099 | 0.697±0.098 |
| Scheme 5 (Two-stage classification scheme) | \ | \ | 0.816±0.071 |
Note: “\” means no corresponding AUC values or AUC≤0.5
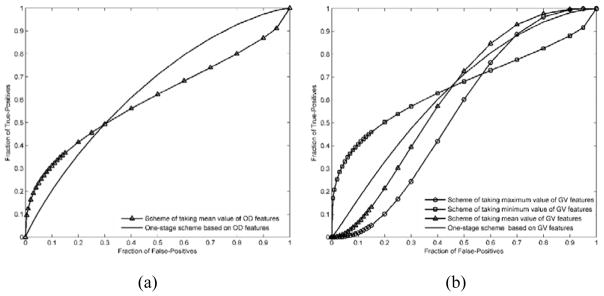
FIG. 4 shows and compares 2 sets of ROC curves generated using the classification scores yielded by 4 classification schemes corresponding to the AUC data shown in TABLE III. FIG. 5 shows and compares four ROC curves. One of the ROC curves was generated using the classification scores yielded by the two-stage classification scheme, while other three were generated using the classification scores yielded by the one-stage classifiers trained using GV features, OD features, both GV and OD features, respectively.
FIG. 4.
Comparison of receiver operating characteristic (ROC) curves generated by 4 breast cancer risk prediction schemes in which (a) and (b) corresponding to the ROC curves generated using OD features and GV features, respectively.
FIG. 5.
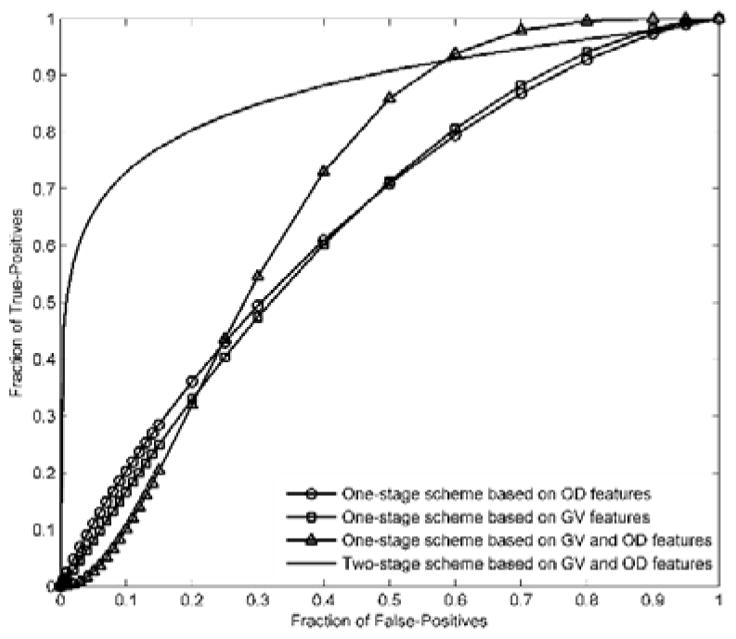
Comparison of ROC curves generated by the proposed two-stage classification scheme and the one-stage classification scheme using GV features an OD features, respectively.
TABLE IV summarizes the overall prediction accuracy on the complete 30 testing cases, as well as the prediction accuracy for the 15 positive cases and 15 negative cases, which were yielded by the GV feature based one-stage classifier, the OD feature based one-stage classifier, the OD and GV feature based one-stage classifier, and the two-stage classification scheme, respectively. The results in TABLE IV show that the group of prediction accuracy values yielded by the OD feature based one-stage classifier are the same with those yielded by the GV feature based one-stage classifier. For both of the two one-stage classifiers, the prediction accuracy for the positive cases are relatively low (lower than 60%). By using the two-stage classification scheme, the prediction accuracy was significantly increased (p < 0.01). The results in TABLE IV also show that, by using the two-stage classification scheme, the overall prediction accuracy, the prediction accuracy for the positive cases, and the prediction accuracy for the negative cases are quite comparable (80%, 80%, and 80%).
TABLE IV.
Comparison of the overall prediction accuracy, prediction accuracy for the case groups with and without breast cancer of 4 different prediction schemes.
| Feature fusion method | Overall prediction accuracy | prediction accuracy for cases with cancer | prediction accuracy for cases without cancer |
|---|---|---|---|
| One-stage classifier trained using OD features | 63.3% | 53.3% | 73.3% |
| One-stage classifier trained using GV features | 63.3% | 53.3% | 73.3% |
| One-stage classifier trained using GV and OD features | 56.7% | 53.3% | 60% |
| Two-stage classification scheme | 80% | 80% | 80% |
FIG. 6 shows and compares four groups of prediction accuracy values generated using the classification scores yielded by the OD feature based one-stage classifier, the GV feature based one-stage classifier, the OD and GV feature based one-stage classifier, and the two-stage classification scheme, respectively.
FIG. 6.
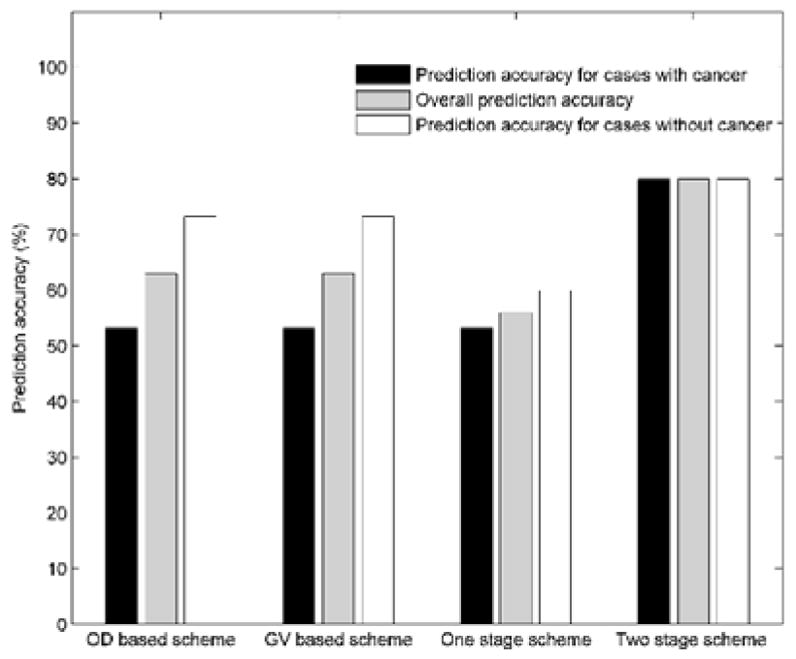
Comparison of the overall prediction accuracy, prediction accuracy for the case groups with and without breast cancer of 4 different prediction schemes.
IV. DISCUSSION
This study is a part of our continuing effort to develop and optimize new quantitative image analysis methods to more accurately predict near-term breast cancer risk, which is an important prerequisite to help develop and establish a new optimal personalized breast cancer screening paradigm. Comparing to the previous studies, the unique contribution of this study is that we successfully tested and demonstrated that converting the original grayscale value mammographic images into optical density images and analyzing image features computed from the optical density images could play a surprisingly important role to significantly improve performance of near-term breast cancer risk prediction. To the best our knowledge, no similar approach or study has been previously reported in the literature.
The motivation of testing this new approach came from our previous observation and experience in developing mammographic image feature analysis based breast cancer risk prediction methods. We observed that in the some studies related to image-based breast cancer risk prediction in the literature4,15,17, the original mammographic images were converted into other types of images and the image features computed from the converted images provided importantly supplementary information to yield higher discriminatory power to those computed from the original mammographic images. However, how to optimally convert the original mammographic images is important, which determines the discriminatory power of the features computed from the converted images. In this study, we tested a new method to convert the original grayscale value mammographic images into optical density based images, compute features from the optical density images, and then fuse these features with other features computed from the original mammographic images using a two-stage ANN based classification scheme for breast cancer risk prediction. The purpose of doing these is to account for the nonlinear relationship between the darkness of the digital images and the number of particles striking the photodetector from radioactive decay. Because the transformation is nonlinear, it enables to generate low correlated image features computed from the original and converted images, which may have potential to acquire useful and complementary information computed from the original mammographic images.
From our study results, we can make the following new and interesting observations. First, we observed that the classifier trained using the GV features generated comparable discriminatory power scores with that trained using the OD features. However, since two groups of image features contain different but complementary information, comparing to train and optimize a one-stage ANN based classifier using only GV features or OD features, developing a two-stage ANN based classifier that fuses the prediction outcomes of 2 ANNs, which were separately trained with GV features and OD features, enabled to yield significantly higher cancer risk prediction performance.
Second, we also observed that by using the proposed two-stage classification scheme, the prediction accuracy for the “negative” cases could be increased to some extent (73.3% to 80%). This indicates that by using the proposed scheme, the prediction result is more accurate by a narrow margin in identifying the women with low risk of having mammography detectable breast cancers in the next sequential FFDM screening. This is a potentially advantage of applying this risk prediction model in the real screening environment due to the low cancer prevalence rate (i.e., < 0.5%) in the screening environment.
Despite the promising results and new observations, this is a laboratory based retrospective data analysis study with a number of limitations. First, the dataset used in this study is small and could not adequately cover the diversity of cases in screening environment. In our future study, we will continue to cooperate with doctors and collect new cases to expand the size of our dataset. Particularly, we will increase the number of negative cases to better represent the cancer prevalence ratio in a mammographic screening environment. Second, the FFDM images used were all acquired with Hologic Selenia FFDM systems. In order to use the proposed schemes to images acquired with other FFDM units, we need to study preprocessing methods to process different images. Third, our model was only applied to predict the risk of having mammography-detectable cancer in the next sequential mammography screening following a negative screening of interest. Whether the similar model can be developed and applied to predict risk in a relatively longer time period (i.e., 2–5 years) has not been tested.
V. CONCLUSIONS
In summary, in this study we tested and demonstrated a new image conversion and feature analysis approach to compute and search for relevant image features with higher discriminatory power from two different types of images, and to improve performance in predicting near-term breast cancer risk. The initial testing results are promising. However, the robustness of this new approach and risk prediction model (or classifier) needs to be further evaluated before it can be clinically acceptable to help establish a new optimal and personalized breast cancer screening paradigm in the future.
Acknowledgments
This work is supported in part by Grants R01 CA160205 and R01 CA197150 from the National Cancer Institute, National Institutes of Health, USA.
APPENDIX A
In this study, we computed some mammographic image features that have been proposed in the literature, as well as redefined some existing features and explored some new features that have never been examined and used before for the task of breast cancer risk prediction. The image features were computed from both GV images and OD images. Specifically, our computer-aided scheme computed the same 148 features from each of the 4 pairs of different ROIs namely, (1) the segmented whole breast regions and (2) the segmented dense breast regions in the bilateral GV images, which are named as WBR-GV and DBR-GV images, respectively (In the following, these features will be name as , (i = 1, 2, …, 148×2)), (3) the segmented whole breast regions and (4) the segmented dense breast regions in the bilateral OD images, which are named as WBR-OD and DBR-OD images, respectively (In the following, these features will be name as , (i = 1, 2, …, 148×2)).
Each group of 148 features were divided into two distinct types namely: (1) 76 bilateral mammographic tissue asymmetry features, which were represented by the absolute subtraction of two matched bilateral feature values computed from the left and right images; (2) 72 maximum features, which were represented by the greater feature value of two matched bilateral feature values. In the following, the two types of image features will be mentioned as asymmetry features and maximum features, respectively.
Type 1: Asymmetry features
The asymmetry features can be divided into a number of subgroups. The first subgroup include13 image statistics based features ( and , i = 1, 2, …, 13) 15, which are average local value fluctuation of gray level histogram, mean of gray level histogram values, standard deviation of gray level histogram values, statistics based features computed from the pixel value distributions of the ROI (region of interest) including mean value, variance, standard deviation, skewness, kurtosis, energy, and entropy, statistics based features computed from the local pixel value fluctuation map of the ROI including mean value, standard deviation, and skewness. The second subgroup includes 4 fractal dimension related breast tissue composition features ( and , i = 14, 15, 16, 17)17, which are estimated with variation method, mathematical morphology, two slopes of fitting lines using textural edgeness and Gaussian subtraction.
The third subgroup has 39 texture related image features ( and , i = 18, 19, …, 56) 28, which include 8 Gray-level co-occurrence matrix based features namely, Energy, Contrast, Correlation, Homogeneity, Variance, Sum Average, Entropy, Dissimilarity, 13 Gray-level run-length matrix based features namely, Short Run Emphasis (SRE), Long Run Emphasis (LRE), Gray-Level Non-uniformity (GLN), Run-Length Non-uniformity (RLN), Run Percentage (RP), Low Gray-Level Run Emphasis (LGRE), High Gray-Level Run Emphasis (HGRE), Short Run Low Gray-Level Emphasis (SRLGE), Short Run High Gray-Level Emphasis (SRHGE), Long Run Low Gray-Level Emphasis (LRLGE), Long Run High Gray-Level Emphasis (LRHGE), Gray-Level Variance (GLV), and Run-Length Variance (RLV), 13 Gray-level size zone matrix based features namely, Small Zone Emphasis (SZE), Large Zone Emphasis (LZE), Gray-Level Non-uniformity (GLN), Zone-Size Non-uniformity (ZSN), Zone Percentage (ZP), Low Gray-Level Zone Emphasis (LGZE), High Gray-Level Zone Emphasis (HGZE), Small Zone Low Gray-Level Emphasis (SZLGE), Small Zone High Gray-Level Emphasis (SZHGE), Large Zone Low Gray-Level Emphasis (LZLGE), Large Zone High Gray-Level Emphasis (LZHGE), Gray-Level Variance (GLV), and Zone-Size Variance (ZSV), as well as, 5 Neighborhood gray-tone difference matrix based features namely, Coarseness, Contrast, Busyness, Complexity, and Strength.
The fourth subgroup includes 16 features ( or , i = 57, 58, …, 72), which we modified to compute a variety of percentage of mammographic density (PMD) for mimicking BIRADS used in clinical practice. These features are defined and computed as following:
| (1) |
| (2) |
| (3) |
| (4) |
Where, NU is the total number of pixels in the ROI of left or right breast; NB is the total number of pixels in the ROIs of both left and right breasts; NHA is the number of pixels in the ROI of left or right breast with gray value larger than the average value of all pixels in the ROIs of both left and right breasts; NHM is the number of pixels in the ROI of left or right breast with gray value larger than the median value of all pixels in the ROIs of both left and right breasts. These 4 features are redefined from a feature used in Ref 15.
Next, to mimic BIRADS of mammographic density, we computed 3 features.
| (5) |
Specifically, IS is the average gray value of all breast tissue pixels in one image and IK are the average gray values of the pixels whose gray values are under the threshold of K = 25%, 50%, 75% of the maximum breast tissue pixel value in the image, respectively. In addition, we also computed other 3 features.
| (6) |
Where NS is the total number of breast tissue pixels in a single image and NK are numbers of pixels whose gray values are under the threshold of K = 25%, 50%, 75% of the maximum pixel value in the image, respectively. Similarly, we computed another set of 6 features combining all pixels in two bilateral images.
| (7) |
| (8) |
Where IB is the average pixel value of total breast tissue pixels (NB) of both left and right mammograms.
The last subgroup includes 4 newly explored features, which are defined and computed as following:
| (9) |
| (10) |
Where and are the maximum breast tissue pixel values in the left and right images, respectively; GVh is the number of gray levels in the ROI of left or right breast, in which the maximum pixel value is higher. Specifically, if , GVh refers to the number of gray levels in the ROI of the left breast; otherwise GVh refers to the number of gray levels in the ROI of the right breast.
| (11) |
| (12) |
Where, Ng is the number of pixels in the ROI of higher maximum pixel value with gray values higher than the maximum pixel value of another ROI; Nh is the total number of pixels in the ROI of higher maximum pixel value. Specifically, if , Ng refers to the number of pixels in the ROI of the left breast with gray values higher than and Nh refers to the number of pixels in the ROI of the left breast; otherwise Ng refers to the number of pixels in the ROI of the right breast with gray values higher than and Nh refers to the number of pixels in the ROI of the right breast.
From each pair of matched ROIs of left and right breasts, the first 72 asymmetry features ( to (or to )) were independently computed for the left and right breasts. Each bilateral mammographic density asymmetric feature was computed by the absolute difference (subtraction) of two feature values computed from the two matched bilateral images. The last 4 asymmetry features were computed directly from each pair of matched bilateral images. The 76 asymmetry features which will be computed from the segmented whole breast regions (WBR-GV) are listed in table I.
| Feature group | Feature number | Description |
|---|---|---|
| Image statistics based features | 1–13 | Average local value fluctuation of gray level histogram, Mean of gray level histogram values, Standard deviation of gray level histogram values; Statistics based features computed from the pixel value distributions of the ROI (region of interest) including Mean value, Variance, Standard deviation, Skewness, Kurtosis, Energy, Entropy; Statistics based features computed from the local pixel value fluctuation map of the ROI including mean value, Standard deviation, Skewness |
| Breast tissue composition features | 14–17 | Fractal dimension estimated with variation method, Fractal dimension estimated with mathematical morphology, Slope estimated by fitting lines using textural edgeness, and Slope estimated by fitting lines using Gaussian subtraction |
| Texture related image features | 18–56 | |
| Gray-level co-occurence matrix based features | 18–25 | Energy, Contrast, Correlation, Homogeneity, Variance, Sum Average, Entropy, and Dissimilarity |
| Gray-level run-length matrix based features | 26–38 | Short Run Emphasis (SRE), Long Run Emphasis (LRE), Gray-Level Non-uniformity (GLN), Run-Length Non-uniformity (RLN), Run Percentage (RP), Low Gray-Level Run Emphasis (LGRE), High Gray-Level Run Emphasis (HGRE), Short Run Low Gray-Level Emphasis (SRLGE), Short Run High Gray-Level Emphasis (SRHGE), Long Run Low Gray-Level Emphasis (LRLGE), Long Run High Gray-Level Emphasis (LRHGE), Gray-Level Variance (GLV), and Run-Length Variance (RLV) |
| Gray-level size zone matrix based features | 39–51 | Small Zone Emphasis (SZE), Large Zone Emphasis (LZE), Gray-Level Non-uniformity (GLN), Zone-Size Non-uniformity (ZSN), Zone Percentage (ZP), Low Gray-Level Zone Emphasis (LGZE), High Gray-Level Zone Emphasis (HGZE), Small Zone Low Gray-Level Emphasis (SZLGE), Small Zone High Gray-Level Emphasis (SZHGE), Large Zone Low Gray-Level Emphasis (LZLGE), Large Zone High Gray-Level Emphasis (LZHGE), Gray-Level Variance (GLV), and Zone-Size Variance (ZSV) |
| Neighbourhood gray-tone difference matrix based features | 52–56 | Coarseness, Contrast, Busyness, Complexity, and Strength |
| Features for mimicking BIRADS | 57–72 | Ratio between the number of pixels in the ROI with gray value larger than the average value of all pixels and the total number of pixels in the ROI, Ratio between the number of pixels in the ROI with gray value larger than the average value of all pixels and the total number of pixels in the ROIs of both left and right breasts, Ratio between the number of pixels in the ROI with gray value larger than the median value of all pixels and the total number of pixels in the ROI, Ratio between the number of pixels in the ROI with gray value larger than the median value of all pixels and the total number of pixels in the ROIs of both left and right breasts; Ratio between the average value of pixels under threshold of 25% of the maximum pixel value and the average pixel value of the whole ROI, Ratio between the average value of pixels under threshold of 50% of the maximum pixel value and the average pixel value of the whole ROI, Ratio between the average value of pixels under threshold of 75% of the maximum pixel value and the average pixel value of the whole ROI, Ratio between the number of pixels under 25% threshold and the total number of pixels of the whole ROI, Ratio between the number of pixels under 50% threshold and the total number of pixels of the whole ROI, Ratio between the number of pixels under 75% threshold, and the total number of pixels of the whole ROI; Ratio between the average gray value of the pixels in the ROI whose gray value is under the threshold of 25% of the maximum pixel value and the average gray value of all pixels in the ROIs of both left and right breasts, Ratio between the average gray value of the pixels in the ROI whose gray value is under the threshold of 50% of the maximum pixel value and the average gray value of all pixels in the ROIs of both left and right breasts, Ratio between the average gray value of the pixels in the ROI whose gray value is under the threshold of 75% of the maximum pixel value and the average gray value of all pixels in the ROIs of both left and right breasts, Ratio between the number of the pixels in the ROI with gray values under the threshold of 25% of the maximum pixel value and the total number of pixels in the ROIs of both left and right breasts, Ratio between the number of the pixels in the ROI with gray values under the threshold of 50% of the maximum pixel value and the total number of pixels in the ROIs of both left and right breasts, Ratio between the number of the pixels in the ROI with gray values under the threshold of 75% of the maximum pixel value and the total number of pixels in the ROIs of both left and right breasts |
| Newly defined features | 73–76 | Absolute difference of the maximum pixel values in the ROI of the left breast and that of the right breast, Ratio between the absolute difference of the maximum pixel values in the ROI of the left breast and that of the right breast and the number of gray levels in the ROI of left or right breast whose maximum pixel value is higher, The number of the pixels in the ROI of higher maximum pixel value with gray values higher than the maximum pixel value of another ROI, Ratio between the number of the pixels in the ROI of higher maximum pixel value with gray values higher than the maximum pixel value of another ROI and the total number of pixels in the ROI of higher maximum pixel value. |
Type 2: Maximum features
This type of features includes another set of 72 features ( to (or to )), which were similar to the features in type 1 ( to (or to )). From each of two matched ROIs of left and right breasts, each of the 72 features was also independently computed. Then, the greater one of the two values of the same feature was chosen to represent the region-based maximum feature value of the two bilateral breasts.
References
- 1.Amir E, Freedman OC, Seruga B, Evans DG. Assessing women at high risk of breast cancer: a review of risk assessment models. J Natl Cancer Inst. 2010;102:680–691. doi: 10.1093/jnci/djq088. [DOI] [PubMed] [Google Scholar]
- 2.Nelson HD, Tyne K, Naik A, et al. U.S. Preventive Services Task Force. Screening for breast cancer: an update for the U.S. Preventive Services Task Force. Ann Intern Med. 2009;151:727–737. doi: 10.1059/0003-4819-151-10-200911170-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlin L, Hall FM. More mammography muddle: emotions, politics, science, costs and polarization. Radiology. 2010;255:311–316. doi: 10.1148/radiol.10100056. [DOI] [PubMed] [Google Scholar]
- 4.Zheng B, Tan M, Ramalingam P, Gur D. Association between Computed Tissue Density Asymmetry in Bilateral Mammograms and Near-term Breast Cancer Risk. The Breast Journal. 2014;20:249–257. doi: 10.1111/tbj.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brawley OW. Risk-based mammography screening: an effort to maximize the benefits and minimize the harms. Ann Intern Med. 2012;156:662–663. doi: 10.7326/0003-4819-156-9-201205010-00012. [DOI] [PubMed] [Google Scholar]
- 6.Gail MH. Personalized estimates of breast cancer risk in clinical practice and public health. Stat Med. 2011;30:1090–1104. doi: 10.1002/sim.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boughey JC, Hartmann LC, Anderson SS, et al. Evaluation of the Tyrer-Cuzick (International Breast Cancer Intervention Study) model for breast cancer risk prediction in women with atypical hyperplasia. J Clin Oncol. 2010;28:3591–3596. doi: 10.1200/JCO.2010.28.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M, Pu J, Cheng S, et al. Assessment of a Four-View Mammographic Image Feature Based Fusion Model to Predict Near-Term Breast Cancer Risk. Annals of Biomedical Engineering. 2015;40:2416–2428. doi: 10.1007/s10439-015-1316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen M, Karemore G, Loog M, et al. A novel and automatic mammographic texture resemblance marker is an independent risk factor for breast cancer. Cancer Epidemiol. 2011;35:381–387. doi: 10.1016/j.canep.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Haberle L, Wagner F, Fasching PA, et al. Characterizing mammographic images by using generic texture features. Breast Cancer Res. 2012;14:R59. doi: 10.1186/bcr3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun W, Qian W, Zhang J, et al. Using multi-scale texture and density features for near-term breast cancer risk analysis. Med Phys. 2015;42:2853–2862. doi: 10.1118/1.4919772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan M, Qian W, Pu J, et al. A new approach to develop computer-aided detection schemes of digital mammograms. Phys Med Biol. 2015;60:4413–4427. doi: 10.1088/0031-9155/60/11/4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian W, Sun W, Zheng B. Improving the efficacy of mammography screening: the potential and challenge of developing new computer-aided detection approaches. Expert Rev Med Devices. 2015;12:497–499. doi: 10.1586/17434440.2015.1068115. [DOI] [PubMed] [Google Scholar]
- 14.Tan M, Zheng B, Leader JK, Gur D. Association between changes in mammographic image features and risk for near-term breast cancer development. IEEE Transactions on Medical Imaging. 2016;35:1719–1728. doi: 10.1109/TMI.2016.2527619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Lederman D, Tan J, Wang X, Zheng B. Computerized prediction of risk for developing breast cancer based on bilateral mammographic breast tissue asymmetry. Medical Engineering & Physics. 2011;33:934–942. doi: 10.1016/j.medengphy.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng B, Sumkin JH, Zuley ML, Wang X, et al. Bilateral mammographic density asymmetry and breast cancer risk: a preliminary assessment. European Journal of Radiology. 2012;81:3222–3228. doi: 10.1016/j.ejrad.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y, Wang X, Hardesty LA, et al. Computerized assessment of tissue composition on digitized mammograms. Acad Radiol. 2002;9:898–905. doi: 10.1016/s1076-6332(03)80459-2. [DOI] [PubMed] [Google Scholar]
- 18.Tan M, Pu J, Zheng B. Reduction of false-positive recalls using a computerized mammographic image feature analysis scheme. Phys Med Biol. 2014;59:4357–4373. doi: 10.1088/0031-9155/59/15/4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng B, Sumkin JH, Zuley ML, et al. Computer-aided detection of breast masses depicted on full-field digital mammograms: a performance assessment. Br J Radiol. 2012;85:e153–e161. doi: 10.1259/bjr/51461617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BASKIN DG, STAHL WL. Fundamentals of quantitative autoradiography by computer densitometry for in situ hybridization, with emphasis on 33P. J Histochem Cytochem. 1993;41(12):1767–1776. doi: 10.1177/41.12.8245425. [DOI] [PubMed] [Google Scholar]
- 21.Vizi S, Palfi A, Hatvani L, Gulya K. Methods for quantification of in situ hybridization signals obtained by film autoradiography and phosphorimaging applied for estimation of regional levels of calmodulin mRNA classes in the rat brain. Brain Research Protocols. 2001;8:32–44. doi: 10.1016/s1385-299x(01)00082-4. [DOI] [PubMed] [Google Scholar]
- 22.Stanley EL. Statistical evaluation of methods for quantifying gene expression by autoradiography in histological sections. BMC Neuroscience. 2009;10:5. doi: 10.1186/1471-2202-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arpad P, Hatvani L, Gulya K. A New Quantitative Film Autoradiographic Method of uantifying mRNA Transcripts for In Situ Hybridization. The Journal of Histochemistry & Cytochemistry. 1998;46(10):1141–1149. doi: 10.1177/002215549804601006. [DOI] [PubMed] [Google Scholar]
- 24.Gierach GL, Li H, Loud JT, et al. Relationships between computer-extracted mammographic texture pattern features and BRCA1/2 mutation status: a cross-sectional study. Breast Cancer Res. 2014;16:1–16. doi: 10.1186/s13058-014-0424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertrand KA, Tamimi RM, Scott CG, et al. Mammographic density and risk of breast cancer by age and tumor characteristics. Breast Cancer Res. 2013;15:R104–R104. doi: 10.1186/bcr3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heine JJ, Scott CG, Sellers TA, et al. A novel automated mammographic density measure and breast cancer risk. J Natl Cancer Inst. 2012;104:1028–1037. doi: 10.1093/jnci/djs254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Szekely L, Eriksson L, et al. High-throughput mammographic-density measurement: a tool for risk prediction of breast cancer. Breast Cancer Res. 2012;14:1–12. doi: 10.1186/bcr3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallières M, Freeman CR, Skamene SR, EI Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Physics in Medicine & Biology. 2015;60:5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 29.Lederman D, Zheng B, Wang X, Wang XH, Gur D. Improving breast cancer risk stratification using resonance-frequency electrical impedance spectroscopy through fusion of multiple classifiers. Ann Biomed Eng. 2011;39:931–945. doi: 10.1007/s10439-010-0210-4. [DOI] [PubMed] [Google Scholar]
- 30.Witten I, Frank E, Hall MA. Data Mining: Practical Machine Learning Tools and Techniques. 3. Elsevier; New York, NY: http://www.cs.waikato.ac.nz/ml/weka/ [Google Scholar]
- 31.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence. 1997;97:273–324. [Google Scholar]
- 32.Fang Y, Fei J, Ma K. Model reference adaptive sliding mode control using RBF neural network for active power filter. Electrical Power and Energy Systems. 2015;73:249–258. [Google Scholar]
- 33.Liu J. Radial Basis Function (RBF) Neural Network Control for Mechanical Systems Design, Analysis and Matlab Simulation. Springer; Heidelberg New York Dordrecht London: [Google Scholar]
- 34.Hartman EJ, Keeler JD, Kowalski JM. Layered neural networks with Gaussian hidden units as universal approximations. Neural Comput. 1990;2:210–215. [Google Scholar]
- 35.Liu D, Wang D, Wu J, Wang Y. A risk assessment method based on RBF artificial neural network-cloud model for urban water hazard. Journal of Intelligent & Fuzzy Systems. 2014;27:2409–2416. [Google Scholar]